In this study, Chen and colleagues investigated the biological function of ferredoxin reductase (FDXR), a target of p53. They generated a Fdxr-deficient mouse model and found that the signal from FDXR to iron homeostasis and the p53 pathway was transduced by ferredoxin 2, a substrate of FDXR, and that p53 played a role in iron homeostasis and was required for FDXR-mediated iron metabolism, suggesting that the FDXR–p53 loop is critical for tumor suppression via iron homeostasis.
Keywords: FDXR, p53, FDX1, FDX2, IRP2, iron homeostasis, mRNA translation
Abstract
Ferredoxin reductase (FDXR), a target of p53, modulates p53-dependent apoptosis and is necessary for steroidogenesis and biogenesis of iron–sulfur clusters. To determine the biological function of FDXR, we generated a Fdxr-deficient mouse model and found that loss of Fdxr led to embryonic lethality potentially due to iron overload in developing embryos. Interestingly, mice heterozygous in Fdxr had a short life span and were prone to spontaneous tumors and liver abnormalities, including steatosis, hepatitis, and hepatocellular carcinoma. We also found that FDXR was necessary for mitochondrial iron homeostasis and proper expression of several master regulators of iron metabolism, including iron regulatory protein 2 (IRP2). Surprisingly, we found that p53 mRNA translation was suppressed by FDXR deficiency via IRP2. Moreover, we found that the signal from FDXR to iron homeostasis and the p53 pathway was transduced by ferredoxin 2, a substrate of FDXR. Finally, we found that p53 played a role in iron homeostasis and was required for FDXR-mediated iron metabolism. Together, we conclude that FDXR and p53 are mutually regulated and that the FDXR–p53 loop is critical for tumor suppression via iron homeostasis.
The p53 tumor suppressor is a transcription factor and can be activated in response to an array of stresses, such as DNA damage (Nelson and Kastan 1994), oncogene activation (Lowe and Ruley 1993), and hypoxia (Graeber et al. 1996). p53 transcriptional activity is also modulated by cellular redox state and the abundance of ADP and iron in response to altered energy and iron metabolism (Vogelstein et al. 2000; Abeysinghe et al. 2001; Liang and Richardson 2003; Whitnall et al. 2006; Yang et al. 2006; Kawauchi et al. 2008; Dongiovanni et al. 2010; Puzio-Kuter 2011; Shen et al. 2014). Upon activation, p53 induces a plethora of genes for prosurvival (such as p21), proapoptotic (such as Puma), and many other biological responses (Riley et al. 2008; Vousden and Prives 2009). p53 also regulates several aspects of cellular metabolism critical for development and various disease processes, especially cancer (Vogelstein et al. 2000; McLure et al. 2004; Yang et al. 2006; Kawauchi et al. 2008; Puzio-Kuter 2011; Shen et al. 2014; Funauchi et al. 2015).
Iron is essential for a variety of cellular and biochemical activities, including energy production and biogenesis of iron–sulfur (Fe-S) clusters (Hentze et al. 2010; Wang and Pantopoulos 2011; Lawen and Lane 2013). On the other hand, an excess amount of iron is known to cause an array of abnormalities (Hentze et al. 2010; Wang and Pantopoulos 2011; Lawen and Lane 2013). The role of iron overload in cancer development is evidenced by the fact that patients suffering from hereditary hemochromatosis, a genetic disease characterized by morbid iron accumulation, are prone to hepatocellular carcinoma (HCC) and colorectal carcinoma (Shaheen et al. 2003; Beutler 2006; Simcox and McClain 2013). Additionally, high intake of dietary iron is associated with an increased risk of cancer (Nishina et al. 2008; Pietrangelo 2009; Sorrentino et al. 2009). Thus, proper control of iron homeostasis is critical for suppressing tumorigenesis. Systemic iron homeostasis is controlled primarily by hepcidin, a peptide hormone secreted from the liver (Nemeth et al. 2004). However, cellular iron homeostasis is controlled primarily by iron regulatory protein 1 (IRP1) and IRP2 (Hentze et al. 1989; Rouault et al. 1990). As an RNA-binding protein, IRP1/2 regulates gene expression through mRNA stability and/or translation, including transferrin receptor 1 (TfR1) and ferritin heavy chain 1 (FTH1) (Butt et al. 1996; Henderson et al. 1996; Rouault 2006). Interestingly, iron homeostasis is also modulated by the p53 pathway potentially via post-transcriptional regulation of TfR1 and FTH1 expression (Zhang et al. 2008).
Ferredoxin reductase (FDXR), a mitochondrial flavoprotein, transfers an electron from NADPH to ferredoxin 1 (FDX1) and FDX2 and then to cytochrome P450 for steroidogenesis and biogenesis of Fe-S clusters and heme A (Brandt and Vickery 1992; Lange et al. 2000; Muller et al. 2001; Sheftel et al. 2010; Shi et al. 2012). We and others found that FDXR is a p53 target and sensitizes tumor cells to 5-fluorouracil- and H2O2-induced apoptosis (Hwang et al. 2001; Liu and Chen 2002). Through interaction with the Fhit tumor suppressor, FDXR modulates apoptosis induced by Fhit (Trapasso et al. 2008). Through interaction with MPZL, a mitochondrial protein and reactive oxygen species (ROS) inducer, FDXR regulates epidermal cell differentiation via mitochondria (Bhaduri et al. 2015). FDXR is also found to be the most consistent internal biodosimetry marker in the peripheral blood following radiation therapy (Abend et al. 2016; Edmondson et al. 2016). Moreover, FDXR is a potential marker for efficacy of chemotherapy (Yu et al. 2003; Okumura et al. 2015). These studies indicate that FDXR has multiple cellular and biochemical activities, but its biological function has not been explored in vivo. Using both FDXR-deficient cell lines and mouse models, we made novel observations that FDXR and p53 are mutually regulated and that the FDXR–p53 loop is critical for tumor suppression via iron homeostasis.
Results
Fdxr is essential for embryonic development
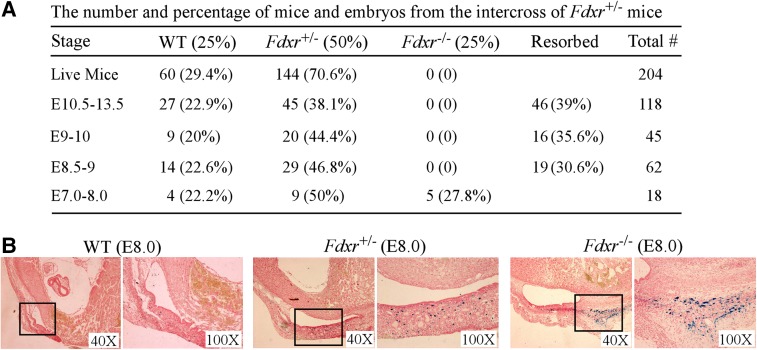
To explore the biological function of FDXR, we generated Fdxr-deficient mice in which the Fdxr gene was deleted through homologous recombination (Supplemental Fig. S1A,B). We found that Fdxr+/− mice were healthy and fertile and did not exhibit obvious abnormality within the first 6 mo. However, no single Fdxr−/− mouse was found among 204 newborn animals from intercrosses of Fdxr+/− mice (Fig. 1A), suggesting that complete loss of Fdxr induces embryonic lethality. Thus, we isolated embryos at various developmental stages and showed that Fdxr−/− embryos were alive at embryonic days 7.0–8.0 (E7.0–E8.0) but resorbed at E8.5–E13.5 (Fig. 1A; Supplemental Fig. S1C). We also found that E8.0 Fdxr−/− embryos exhibited severe developmental defects, whereas E7.5 Fdxr−/− embryos failed to develop normal layers of embryonic and extraembryonic tissues, including visceral endoderm, mesoderm, embryonic ectoderm, extraembryonic ectoderm, and extraembryonic visceral endoderm (Supplemental Fig. S1D).
Figure 1.
Loss of Fdxr leads to embryonic lethality potentially due to iron accumulation. (A) The number and percentage of embryos and live offspring from the intercrosses of Fdxr+/− mice. (B) Fdxr deficiency leads to iron overload in E8.0 embryos. Wild-type, Fdxr+/−, and Fdxr−/− embryos were stained with Prussian blue. The right panels represent higher magnification of the boxed regions in the left panels of each set. Blue staining represents iron accumulation.
Since FDXR is necessary for biogenesis of Fe-S clusters and is a critical modulator of cellular iron homeostasis (Lange et al. 2000; Sheftel et al. 2010; Shi et al. 2012), we examined whether embryonic lethality is associated with aberrant iron metabolism in Fdxr−/− embryos. Thus, Pearl's Prussian blue staining was performed and showed that iron accumulation was extensive in Fdxr−/− embryos, moderate in Fdxr+/− embryos, and sparse in wild-type embryos (Fig. 1B). Thus, aberrant iron accumulation in developing Fdxr−/− embryos would generate reactive radicals and oxidative stress, leading to embryonic lethality at E8.5 (Fig. 1A), which is similar to what was observed in FBXL5−/− embryos that died at E8.5 (Moroishi et al. 2011; Ruiz et al. 2013).
Mice deficient in Fdxr are prone to spontaneous tumors and liver abnormalities
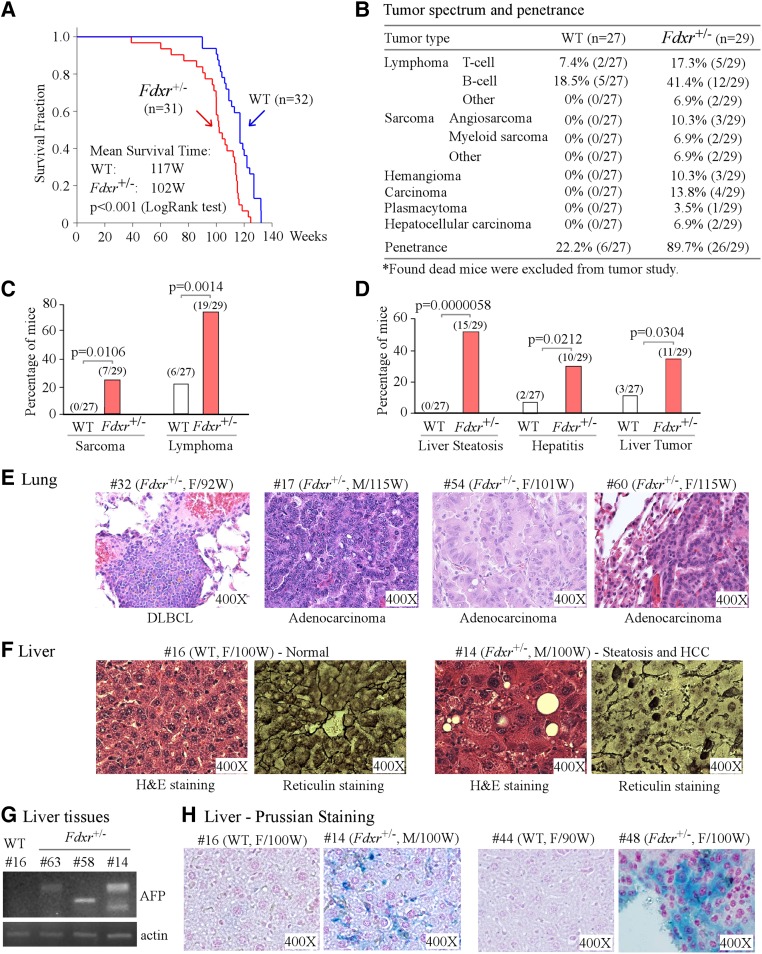
A cohort of wild-type (n = 32) and Fdxr+/− (n = 31) mice was generated to examine median survival, tumor incidence, and other abnormalities (Supplemental Tables S1, S2). We found that the median survival for Fdxr+/− mice (102 wk) was significantly shorter than that for wild-type mice (117 wk; P < 0.001 by log rank test) (Fig. 2A). We also found that Fdxr+/− mice were prone to a broad spectrum of spontaneous tumors, especially high-grade malignant sarcoma (seven out of 29 mice), lung carcinoma (four out of 29 mice), liver tumors (11 out of 29 mice; two HCCs out of 11 liver tumors), and high-grade lymphoma (19 out of 29 mice) (Fig. 2B–F; Supplemental Fig. S2A–I; Supplemental Tables S1, S2).
Figure 2.
Mice deficient in Fdxr are prone to spontaneous tumors. (A) Kaplan-Meier survival curve for wild-type and Fdxr+/− mice. (B) Mice deficient in Fdxr are prone to spontaneous tumors. Tumor spectrum and penetrance in a cohort of wild-type and Fdxr+/− mice are shown. (C) The numbers and percentages of wild-type and Fdxr+/− mice with sarcoma and lymphoma. (D) The numbers and percentages of wild-type and Fdxr+/− mice with liver steatosis, hepatitis, and liver tumors. (E) Representative images of hematoxylin and eosin (H&E)-stained lung tumors in Fdxr+/− mice. (F) Representative images of a normal liver from a wild-type mouse and a liver tumor from a Fdxr+/− mouse that were stained with H&E (left panel of each set) or reticulin (right panel of each set). (G) The level of α-fetoprotein (AFP) transcript in liver tissues is increased by Fdxr deficiency. The levels of AFP and actin transcripts were measured in wild-type and Fdxr+/− mouse livers. (H) Fdxr deficiency leads to iron overload in liver tissues. Livers from wild-type and Fdxr+/− mice were stained with Prussian blue. Blue staining represents iron accumulation in Fdxr+/− hepatocytes.
Among 29 Fdxr+/− mice whose organs were available for histological analysis, 15 had steatosis, 10 had chronic hepatitis, and 11 had liver tumors (Fig. 2D; Supplemental Table S2). Fisher's exact test showed that the difference between wild-type and Fdxr+/− mice is significant in liver steatosis (P = 0.0014), hepatitis (P = 0.0212), and liver tumors (P = 0.0304). Since reticulin staining is used to differentiate HCC from normal livers (Bergman et al. 1997), both wild-type and Fdxr+/− livers were stained for reticulin. We found that wild-type livers showed a well-preserved reticulin network (Fig. 2F; Supplemental Fig. S2C). In contrast, Fdxr+/− livers had decreased reticulin staining along with widened trabeculae, encompassing three or more cell layers (Fig. 2F; Supplemental Fig. S2C). Additionally, we found that the transcript for α-fetoprotein (AFP), an HCC biomarker (Farinati et al. 2006), was highly induced in Fdxr+/− livers compared with that in wild-type livers (Fig. 2G). Since aberrant iron metabolism leads to liver abnormalities (Andrews 1999; Fleming and Ponka 2012; Simcox and McClain 2013), Pearl's Prussian blue staining was performed and showed that iron accumulation was extensive in Fdxr+/− hepatocytes compared with that in wild-type hepatocytes (Fig. 2H; Supplemental Fig. S2D). Since mutant p53 is known to promote HCC development, RT–PCR was performed to determine the p53 status in these 11 Fdxr+/− liver tumor tissues. No mutation was detected in the p53 DNA-binding domain, where the vast majority of mutations occurs, suggesting that p53 mutation is likely rare in Fdxr-deficient cells and may not play a role in tumorigenesis induced by Fdxr deficiency.
Iron metabolism and the p53 pathway are regulated by FDXR deficiency
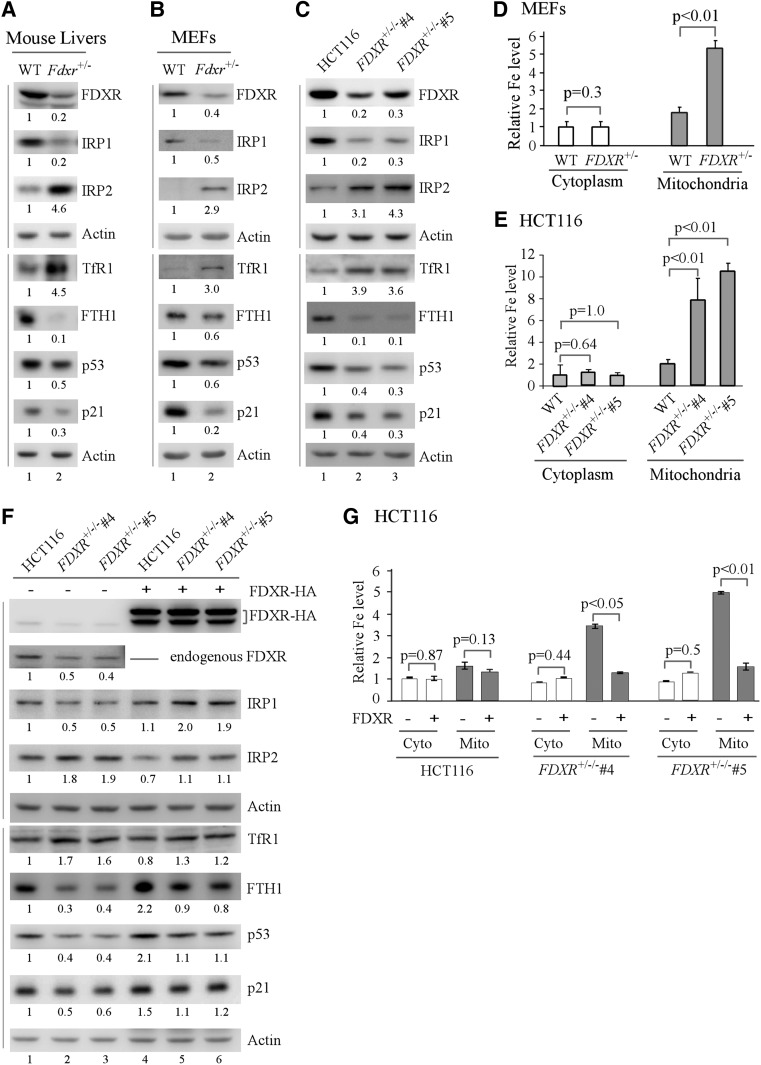
To determine what pathway is responsible for aberrant iron accumulation in developing embryos, several master regulators for iron metabolism were examined in wild-type and Fdxr+/− littermate liver tissues. We found that Fdxr deficiency led to increased expression of IRP2 and TfR1 but decreased expression of IRP1 and FTH1 (Fig. 3A; Supplemental Fig. S3A). We also found that knockdown of FDXR in HepG2 and HCT116 cells led to similar regulation for IRP1/2, TfR1, and FTH1 (Supplemental Fig. S3B,C). Next, the effect of Fdxr deficiency on iron metabolism was examined in wild-type and Fdxr-deficient mouse embryonic fibroblasts (MEFs). Due to the requirement of FDXR for embryonic development (Fig. 1), only Fdxr+/− MEFs were generated (Fig. 3B). As in Fdxr-deficient liver tissues, IRP2 and TfR1 were increased, whereas IRP1 and FTH1 were decreased by Fdxr deficiency in MEFs (Fig. 3B).
Figure 3.
Iron metabolism and the p53 pathway are regulated by FDXR. (A–C) Western blots were prepared using extracts from wild-type and Fdxr+/− mouse livers (A) and MEFs (B) and wild-type and FDXR+/−/− HCT116 cells (C). The blots were probed with antibodies against FDXR, IRP1, IRP2, TfR1, FTH1, p53, p21, and actin, respectively. (D,E) FDXR deficiency leads to mitochondrial iron accumulation. The level of cytosolic and mitochondrial iron (Fe2+) was measured by QuantiChrom iron assay in a pair of wild-type and Fdxr+/− littermate MEFs (D) and in isogenic control and FDXR+/−/− HCT116 cells (E). The level of cytosolic iron in wild-type MEFs and isogenic control HCT116 cells (the first left column) was set at 1.0. Data are mean ± SD from three independent experiments. (F) Ectopic expression of FDXR restores near-normal expression of iron regulatory proteins and p53. Isogenic control and FDXR+/−/− HCT116 cells were transfected with 1 µg of control pcDNA3 (−) or a vector expressing FDXR with an HA tag at its C terminus (FDXR-HA) (+) for 24 h followed by Western blot analysis with various antibodies as indicated. (G) Ectopic expression of FDXR abrogates mitochondrial iron overload in FDXR-deficient cells. The experiment was performed as in F. The level of cytosolic and mitochondrial iron (Fe2+) was measured in isogenic control and FDXR+/−/− HCT116 cells transfected with control pcDNA3 (−) or a vector expressing FDXR-HA (+). (First left column) The level of cytosolic iron in isogenic control HCT116 cells transfected with control pcDNA3 was set at 1.0. Data are mean ± SD from three independent experiments.
To explore how FDXR deficiency modulates iron metabolism, CRISPR–cas9 was used to generate multiple FDXR-deficient HCT116 cell lines. Interestingly, the HCT116 cell line carries three copies of the FDXR gene (Masramon et al. 2000; Hwang et al. 2001). We were able to generate multiple clones in which two of the three FDXR alleles were deleted. However, we were unable to generate a single clone in which all three alleles were knocked out. These observations are consistent with a previous report that FDXR is necessary for cell survival (Hwang et al. 2001) as well as with the above observation that Fdxr is necessary for embryonic development (Fig. 1). Two FDXR+/−/− HCT116 clones (clones #4 and #5) were selected for further studies. Sequence analysis showed that allele #1 had no deletion or mutation, allele #2 had a 20-nucleotide (nt) out-of-frame deletion in exon 9, and allele #3 had a 16-nt out-of-frame deletion in exon 9. Using these genetically defined HCT116 cells, we found that FDXR deficiency led to increased expression of IRP2 and TfR1 but decreased expression of IRP1 and FTH1 (Fig. 3C). To confirm the observation that Fdxr deficiency leads to iron accumulation in developing embryos (Fig. 1), QuantiChrom iron assay was performed and showed that FDXR deficiency markedly increased the level of iron in mitochondria but not in cytoplasm in MEFs and HCT116 cells (Fig. 3D,E).
p53 expression and activity are often regulated by its own targets (Jin and Levine 2001; Brooks and Gu 2011; Zhang et al. 2011) and thus possibly by FDXR. To test this, we examined the levels of p53 and p21 proteins and found that p53 expression was inhibited by FDXR deficiency along with decreased expression of p21 in FDXR-deficient liver tissues and cells (Fig. 3A–C; Supplemental Fig. S3A–C). To confirm the role of FDXR in mitochondrial iron homeostasis and the p53 pathway, we examined whether ectopic expression of FDXR would restore normal iron metabolism and p53 expression. Indeed, we found that, in FDXR+/−/− cells, the levels of IRP2, TfR1, FTH1, p53, and p21 were restored to near normal by ectopic expression of FDXR (Fig. 3F, cf. lanes 2,3 and 5,6, respectively). Interestingly, IRP1 was induced by FDXR (Fig. 3F), potentially due to increased association of IRP1 with Fe-S clusters, which enhances IRP1 stability (Hentze et al. 2010). However, in isogenic control cells, ectopic expression of FDXR slightly suppressed IRP2 and TfR1 expression but had little if any effect on IRP1 (Fig. 3F, cf. lanes 1 and 4). Additionally, p53, p21, and FTH1 were induced by FDXR (Fig. 3F, cf. lanes 1 and 4). Since FTH1 is a putative p53 target (Funauchi et al. 2015), it is likely that the induction of p21 and FTH1 is due to increased expression of p53 (Fig. 3F, cf. lanes 1 and 4). Moreover, we found that mitochondrial iron overload in FDXR+/−/− cells (both clones #4 and #5) was eliminated by ectopic expression of FDXR (Fig. 3G). In contrast, in isogenic control cells, FDXR had no effect on iron homeostasis (Fig. 3G).
As a major regulator of iron homeostasis and a mediator of ROS-mediated apoptosis (Hwang et al. 2001; Liu and Chen 2002), FDXR may play a role in p53-dependent ferroptosis (Jiang et al. 2015), which is characterized by lipid peroxidation, generation of lipid-based ROS, and its dependency on iron (Yang and Stockwell 2016). To test this, we examined whether FDXR modulates the extent of ferroptosis induced by RSL3 and Erastin. RSL3 is a glutathione peroxidase 4 inhibitor, whereas Erastin is an inhibitor of the cysteine/glutamate antiporter, both of which are known inducers of ferroptosis (Jiang et al. 2015). Briefly, we found that RSL3- and erastin-induced ferroptosis was inhibited by FDXR deficiency in both MEFs and HCT116 cells (Supplemental Fig. S4A–H). Surprisingly, we found that RSL- and Erastin-induced ferroptosis was also inhibited by ectopic expression of FDXR in HCT116 cells (Supplemental Fig. S4I–L). Consistent with these observations, cell proliferation and colony formation were increased by FDXR deficiency in MEFs and HCT116 cells (Supplemental Fig. S4M–O). These data suggest that FDXR may play a role in p53-dependent ferroptosis.
The signal from FDXR to iron homeostasis and the p53 pathway is transduced by FDX2
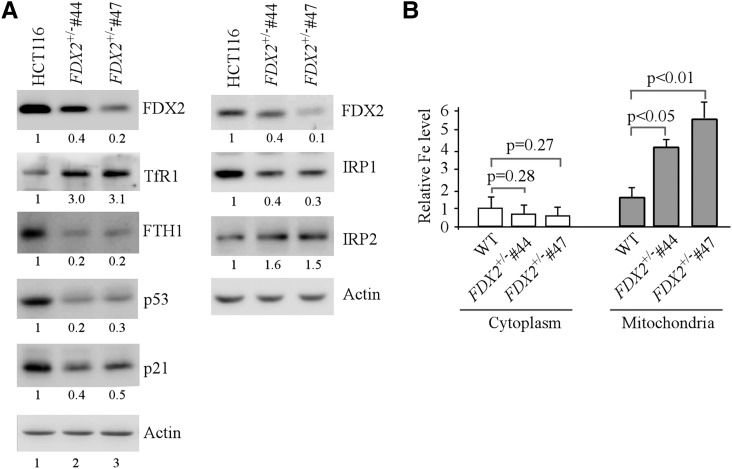
Both FDX1 and FDX2 are bona fide FDXR substrates (Lange et al. 2000; Sheftel et al. 2010). To determine which substrate is involved in iron homeostasis and the p53 pathway, multiple FDX1- and FDX2-deficient HCT116 cell lines were generated. FDX1−/− clones #18 and #19 had a 32-nt out-of-frame deletion in exon 1 and were chosen for further studies. However, we were able to generate only FDX2+/− but not FDX2−/− clones, suggesting that, like FDXR, FDX2 may be required for cell survival. FDX2+/− clones #44 and #47 had a 19-nt out-of-frame deletion in exon 1 and were chosen for further studies. We found that, like FDXR deficiency, FDX2 deficiency led to increased expression of IRP2 and TfR1 but decreased expression of IRP1 and FTH1 (Fig. 4A). Similar results were detected in HCT116 cells in which FDX2 was knocked down by two separate siRNAs (Supplemental Fig. S5). In addition, FDX2 deficiency led to decreased expression of p53 and p21 (Fig. 4A) as well as mitochondrial iron overload (Fig. 4B). In contrast, knockout or transient knockdown of FDX1 had no effect on the level of IRP1, IRP2, p53, and p21 and slightly increased the level of TfR1 but significantly decreased the level of FTH1 (Supplemental Fig. S6A,B). Moreover, FDX1 knockout had no effect on iron homeostasis in HCT116 cells (Supplemental Fig. S6C).
Figure 4.
FDX2 transduces FDXR signals to regulate iron homeostasis and the p53 pathway. (A) FDX2 regulates iron regulatory proteins and the p53 pathway. Cell lysates were collected from isogenic control HCT116 cells and FDX2+/− #44/#47 HCT116 cells and subjected to Western blot analysis with various antibodies as indicated. (B) FDX2 deficiency leads to mitochondrial iron overload. The level of cytosolic and mitochondrial iron was measured in isogenic control and FDX2+/− #44/#47 cells. (First left column) The level of cytosolic iron in isogenic control cells was set at 1.0. Data are mean ± SD from three independent experiments.
FDXR regulates iron homeostasis and p53 expression through IRP2
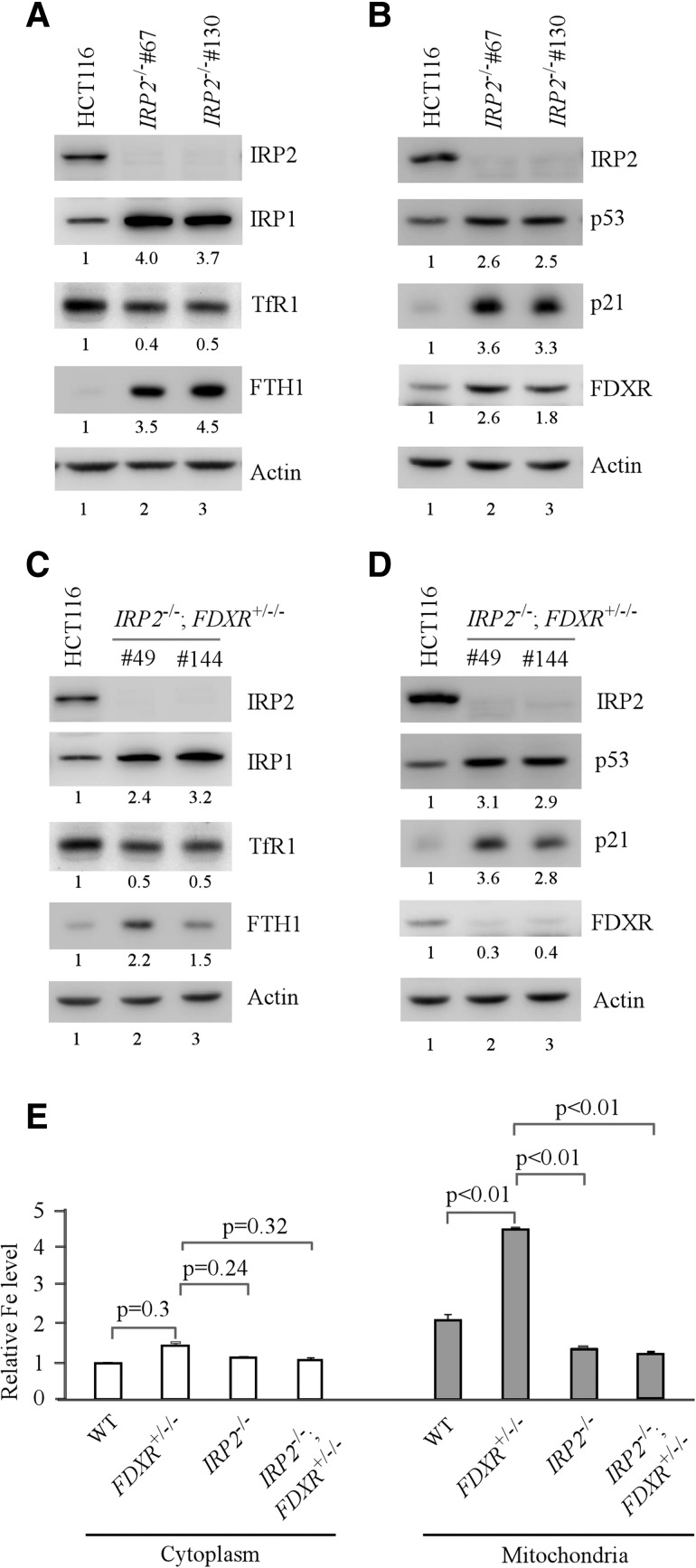
IRP2 is a master regulator of key genes in cellular iron metabolism, including TfR1 and FTH1 (Butt et al. 1996; Henderson et al. 1996). Since FDXR deficiency induces IRP2 expression and mitochondrial iron overload (Fig. 3), we postulate that IRP2 may mediate FDXR-dependent iron homeostasis. To test this, multiple IRP2 knockout HCT116 cell lines were generated. Clones #67 and #130, which had a 25-nt and 44-nt out-of-frame deletion in exon 3, respectively, were chosen for further studies. We found that TfR1 was decreased, whereas FTH1 was increased by IRP2 knockout (Fig. 5A), consistent with previous studies (LaVaute et al. 2001; Cooperman et al. 2005; Galy et al. 2005). We also found that IRP1 was accumulated in IRP2 knockout cells (Fig. 5A), suggesting that the level of Fe-S clusters is abundant for IRP1 protein stability. Most importantly, we found that IRP2 knockout markedly induced p53 expression along with its targets, p21 and FDXR (Fig. 5B).
Figure 5.
FDXR regulates iron homeostasis and the p53 pathway through IRP2. (A–D) Iron regulatory proteins and the p53 pathway are regulated by IRP2. Western blots were prepared with lysates from isogenic control HCT116 cells (A–D), IRP2−/− cells (A,B), and IRP2−/−;FDXR+/−/− cells (C,D) and then probed with antibodies against IRP1, IRP2, FDXR, TfR1, FTH1, p53, p21, and actin, respectively. (E) IRP2 knockout abolishes mitochondrial iron overload in FDXR-deficient cells. The levels of cytosolic and mitochondrial iron were measured in isogenic control, IRP2−/−, FDXR+/−/−, and IRP2−/−;FDXR+/−/− HCT116 cells. (First left column) The level of cytosolic iron in isogenic control cells was set at 1.0. Data are mean ± SD from three independent experiments.
To confirm the above observation, we generated multiple IRP2−/−;FDXR+/−/− HCT116 cell lines in which both alleles of the IRP2 gene were deleted (44-nt deletion in exon 3), whereas two of the three alleles of the FDXR gene were deleted (20-nt deletion in one allele and 16-nt deletion in the other allele). Thus, clones #49 and #144 were selected for further studies. We showed that in IRP2−/−;FDXR+/−/− cells, IRP2 knockout was still capable of increasing expression of IRP1, FTH1, p53, and p21 but decreasing expression of TfR1 (Fig. 5C,D). We also showed that IRP2 knockout had no effect on intracellular iron content, consistent with a recent study (Cloonan et al. 2016). However, IRP2 knockout reversed mitochondrial iron overload by FDXR deficiency to near normal in IRP2−/−;FDXR+/−/− cells, suggesting that IRP2 is an important mediator acting downstream from FDXR (Fig. 5E).
Next, we determined whether increased expression of p53 is responsible for induction of p21 and FDXR in IRP2 knockout cells. We found that, upon knockdown of p53, p21 and FDXR were not induced in IRP2−/− cells (Supplemental Fig. S7A). In contrast, IRP2 knockout was still capable of inducing IRP1 and FTH1 and repressing TfR1 expression (Supplemental Fig. S7B). We note that as a putative target of p53 (Funauchi et al. 2015), FTH1 was reduced by p53 knockdown but still induced by IRP2 knockout independent of p53 (Supplemental Fig. S7B). As a control, we found that knockdown of p53 led to decreased expression of p21 and FDXR in control HCT116 cells (Supplemental Fig. S7A).
p53 expression is regulated by IRP2 via mRNA translation
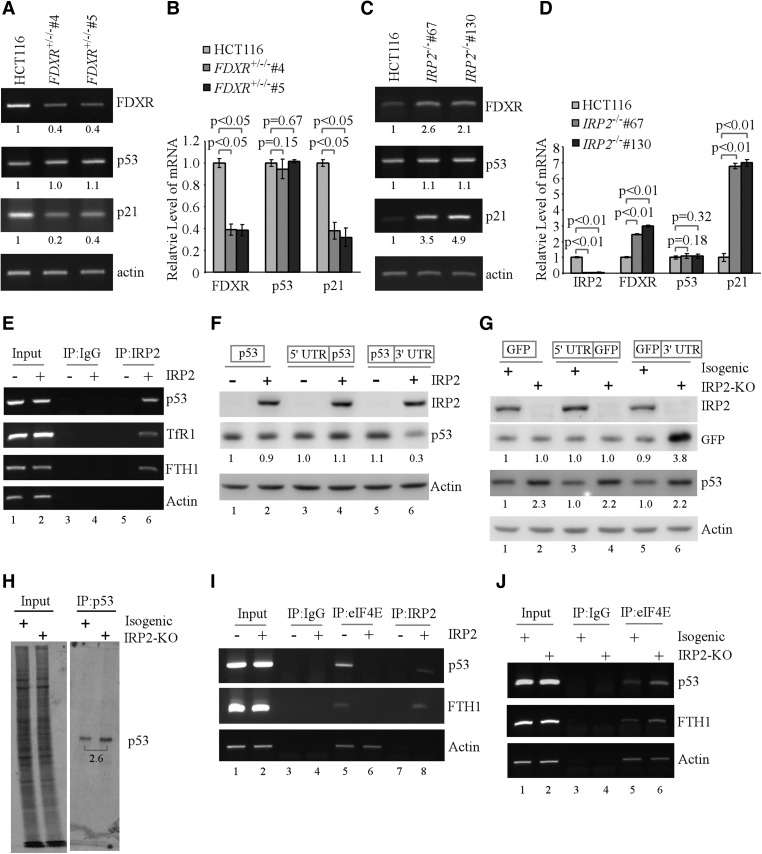
p53 expression is subject to transcriptional and post-transcriptional regulations (Zhang and Chen 2008; Vousden and Prives 2009). Thus, semiquantitative and quantitative RT–PCR were performed and showed that the level of p53 transcript was not changed in FDXR+/−/− HCT116 cells (Fig. 6A,B), FDXR knockdown HCT116 cells (Supplemental Fig. S8A), or FDXR+/− MEFs (Supplemental Fig. S8B). Similarly, the level of p53 transcript was not altered in IRP2−/− HCT116 cells (Fig. 6C,D). As a control, the levels of FDXR and p21 transcripts were found to be decreased in FDXR-deficient cells (Fig. 6A,B; Supplemental Fig. S8A,B) but increased in IRP2−/− cells (Fig. 6C,D). These results indicate that IRP2 may regulate p53 expression through a translational mechanism.
Figure 6.
p53 is regulated by IRP2 via mRNA translation. (A,B) FDXR deficiency has no effect on the level of p53 transcript. Semiquantitative (A) and quantitative (B) RT–PCR were performed to measure the levels of FDXR, p53, p21, and actin transcripts in isogenic control and FDXR+/−/− #4/#5 cells. (C,D) IRP2 deficiency has no effect on the level of p53 transcript. Semiquantitative (C) and quantitative (D) RT–PCR were performed to measure the levels of IRP2, FDXR, p53, p21, and actin transcripts in isogenic control and IRP2−/− #67/#130 cells. (E) RNA immunoprecipitation (RNA-IP) was performed with DNAase I-treated extracts from HCT116 cells that were transfected with control pcDNA3 (−) or a vector expressing HA-tagged IRP2 (+) for 24 h. Total RNAs were purified from IRP2 immunocomplexes and subjected to RT–PCR to measure the levels of p53, TfR1, FTH1, and actin mRNAs. (F) p53-null H1299 cells were transfected with control pcDNA3 (−) or a vector expressing HA-IRP2 (+) along with a reporter that contains the p53-coding region alone or together with the p53 5′ or 3′ untranslated region (UTR). Twenty-four hours after transfection, cell lysates were collected and subjected to Western blot analysis to detect HA-IRP2, p53, and actin. (Lane 1) The level of p53 protein in H1299 cells transfected with control pcDNA3 and a reporter that contains the p53-coding region alone was set at 1.0. (G) Isogenic control (−) and IRP2 knockout (+) HCT116 cells were transfected with a reporter that contains the GFP-coding region alone or together with the p53 5′ or 3′ UTR. Twenty-four hours after transfection, cell lysates were collected and subjected to Western blot analysis to detect IRP2, GFP, p53, and actin. (Lane 1) The level of GFP protein in isogenic control cells transfected with a reporter that contains GFP coding alone was set at 1.0. (H) Isogenic control and IRP2−/− HCT116 cells were labeled with 35S-methionine for 10 min. Cell lysates were isolated and used for immunoprecipitation with 1.0 µg of anti-p53. The immunoprecipitates were separated on 8% SDS-PAGE. p53 was visualized by autoradiography. The relative level of p53 was measured by densitometry. (I) HCT116 cells were transfected with control pcDNA3 (−) or a vector expressing HA-IRP2 (+) for 24 h. Using a control IgG, anti-HA that recognizes IRP2, or anti-eIF4E, RNA-IP was performed with DNAaseI-treated extracts from HCT116 cells transfected with pcDNA3 or HA-IRP2-expressing pcDNA3. Total RNAs were purified from immunocomplexes and subjected to RT–PCR to measure the levels of p53, FTH1, and actin mRNAs. (J) RNA-IP was performed with DNAase I-treated extracts from isogenic control (−) and IRP2−/− (+) HCT116 cells with control IgG or anti-eIF4E. Total RNAs were purified from immunocomplexes and subjected to RT–PCR to measure the levels of p53, FTH1, and actin mRNAs.
As an RNA-binding protein, IRP2 may bind to p53 mRNA and then regulate p53 expression. To test this, IRP2 immunoprecipitation (IRP2-IP) was performed and showed that the p53 transcript was highly enriched in IRP2 immunoprecipitates (Fig. 6E). As a control, TfR1 and FTH1 mRNAs, both of which are known to be recognized by IRP2 (Butt et al. 1996; Henderson et al. 1996), were also enriched in IRP2 immunoprecipitates (Fig. 6E). To identify a region in p53 mRNA that is responsive to IRP2, multiple reporters were generated (shown in Supplemental Fig. S8C). We found that in p53-null H1299 cells, ectopic expression of IRP2 inhibited p53 expression from a vector that carries a p53-coding region plus its 3′ untranslated region (UTR) (Fig. 6F, cf. lanes 5 and 6). However, IRP2 had no effect on p53 expression from a reporter that carries a p53-coding region alone or together with the p53 5′ UTR in p53-null H1299 cells (Fig. 6F, lanes 1–4). Conversely, we found that in HCT116 cells, IRP2 knockout increased GFP expression from a reporter that carries a GFP-coding region plus the p53 3′ UTR (Fig. 6G, lanes 5,6). However, IRP2 knockout had no effect on GFP expression from a reporter that carries a GFP-coding region alone or together with the p53 5′ UTR in HCT116 cells (Fig. 6G, lanes 1–4). Additionally, we showed that in HCT116 cells, endogenous p53 expression was induced by IRP2 knockout regardless of the transfection with a GFP reporter (Fig. 6G). Since the p53 3′ UTR is responsive to IRP2 (Fig. 6F,G), we searched for a consensus iron-responsive element (IRE) (Butt et al. 1996; Henderson et al. 1996). Indeed, we found such a conserved IRE (CAGUGU) located at nucleotide 2344 in the p53 3′ UTR (Supplemental Fig. S8C). Thus, we generated a GFP reporter that carries the p53 3′ UTR but without such a conserved IRE (Supplemental Fig. S8C). Since a poly(U) region is recognized by Rbm38 to regulate p53 mRNA translation (Zhang et al. 2011), we generated a GFP reporter that carries the p53 3′ UTR but without the poly(U) (Supplemental Fig. S8C). We showed that in IRP2−/− HCT116 cells, ectopic expression of IRP2 inhibited GFP expression from a reporter that carries an intact or poly(U)-deleted p53 3′ UTR (Supplemental Fig. S8D, cf. lanes 3,7 and 4,8, respectively). In contrast, ectopic expression of IRP2 had a weakened effect on GFP expression from a reporter that contains an IRE-deleted p53 3′ UTR in IRP2−/− HCT116 cells (Supplemental Fig. S8D, cf. lanes 5 and 6). To further confirm the regulation of p53 mRNA translation by IRP2, we measured the newly synthesized 35S-labeled p53 protein in control and IRP2 knockout HCT116 cells. We found that the level of de novo synthesized p53 protein was highly increased by IRP2 knockout (Fig. 6H).
Since the binding of eIF4E to the mRNA 5′ cap is a rate-limiting step for mRNA translation, we examined whether the binding of eIF4E to the p53 5′ cap is modulated by IRP2. Indeed, we found that in HCT116 cells, the binding of eIF4E to p53 mRNA was suppressed by ectopic expression of IRP2 (Fig. 6I, cf. lanes 5 and 6) but increased by IRP2 knockout (Fig. 6J, cf. lanes 5 and 6). Since FTH1 mRNA translation is regulated by IRP2 (Gray and Hentze 1994), the level of FTH1 mRNA associated with eIF4E was examined as a control and found to be decreased by ectopic expression of IRP2 but increased by IRP2 knockout (Fig. 6I,J, cf. lanes 5 and 6).
p53 plays a role in iron homeostasis and is required for FDXR-mediated iron metabolism
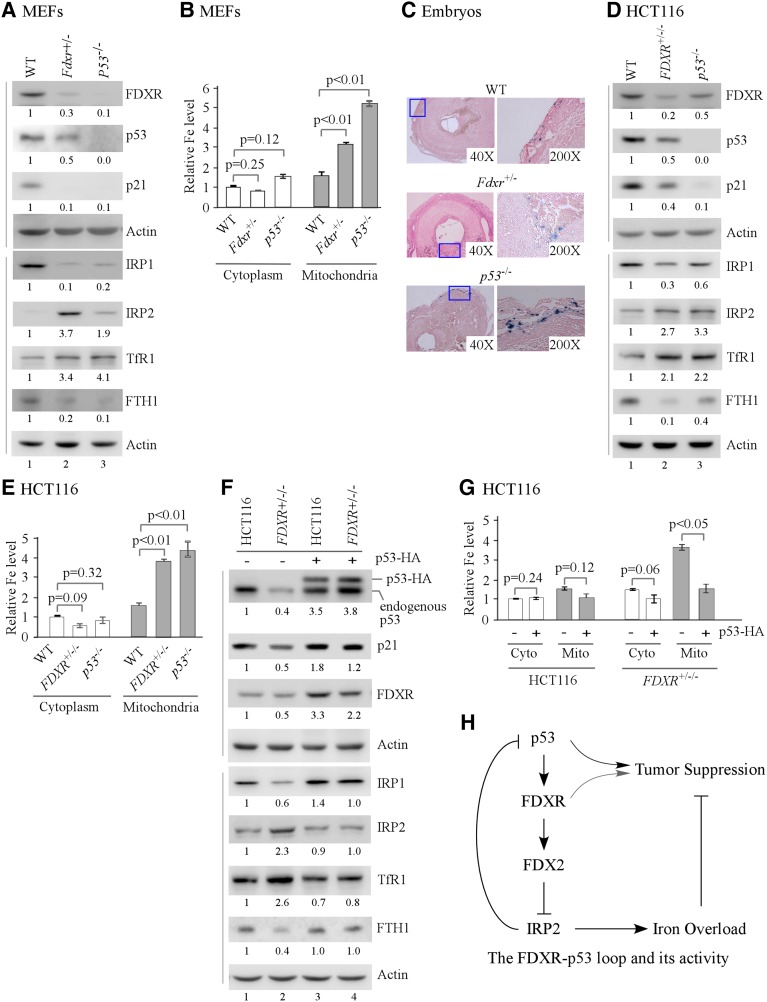
Since FDXR deficiency leads to decreased p53 expression along with mitochondrial iron overload (Fig. 3), we hypothesized that p53 plays a role in iron homeostasis. Indeed, we found that in MEFs, p53 deficiency led to increased expression of IRP2 and TfR1 but decreased expression of IRP1 and FTH1 (Fig. 7A). As a control, the levels of FDXR and p21 were decreased in p53-deficient MEFs, as expected (Fig. 7A). We also found that, like FDXR deficiency, p53 deficiency led to mitochondrial iron overload in MEFs (Fig. 7B) and iron overload in E8.0 p53−/− embryos (Fig. 7C). Additionally, iron overload was detected in Fdxr+/−, p53+/−, Fdxr+/−;p53+/−, and Fdxr+/−;p53−/− embryos (Supplemental Fig. S9A). We note that, like Fdxr+/− embryos, p53−/− embryos can tolerate a moderate increase of iron, since p53−/− embryos are known to develop normally (Donehower et al. 1992).
Figure 7.
Mitochondrial iron homeostasis is regulated by p53, and FDXR-mediated iron metabolism is p53-dependent. (A) Iron regulatory proteins are regulated by p53 in MEFs. Western blots were prepared using extracts from wild-type, Fdxr+/−, and p53−/− MEFs. The blots were probed with antibodies against FDXR, p53, p21, IRP1, IRP2, TfR1, FTH1, and actin, respectively. (B) Loss of p53 leads to mitochondrial iron overload in MEFs. The levels of cytosolic and mitochondrial iron were measured in wild-type, Fdxr+/−, and p53−/− MEFs. (First left column) The level of cytosolic iron in wild-type MEFs was set at 1.0. Data are mean ± SD from three independent experiments. (C) Loss of p53 leads to a mild iron overload in E8.0 embryos. Prussian blue staining was performed with wild-type, Fdxr+/−, and p53−/− embryos. The right panels represent the boxed regions in the left panels of each set at higher magnification. Blue staining represents iron accumulation. (D) Iron regulatory proteins are regulated by p53 in HCT116 cells. Western blots were prepared using extracts from isogenic control, FDXR+/−/−, and p53−/− HCT116 cells and probed with various antibodies. (E) Loss of p53 leads to mitochondrial iron overload in HCT116 cells. The levels of cytosolic and mitochondrial iron were measured in isogenic control, FDXR+/−/−, and p53−/− HCT116 cells. (First left column) The level of cytosolic iron in isogenic control cells was set at 1.0. Data are mean ± SD from three independent experiments. (F) Ectopic expression of p53 restores near-normal expression of iron regulatory proteins in FDXR+/−/− HCT116 cells. Isogenic control and FDXR+/−/− HCT116 cells were transfected with control pcDNA3 (−) or a vector expressing p53 (+). Cell lysates were collected and subjected to Western blot analysis with various antibodies as indicated. (G) Ectopic expression of p53 restores near-normal iron metabolism in FDXR+/−/− HCT116 cells. The levels of cytosolic and mitochondrial iron were measured in isogenic control and FDXR+/−/− HCT116 cells transfected with control pcDNA3 (−) or a vector expressing HA-tagged p53 (+). (First left column) The level of cytosolic iron in isogenic control HCT116 cells transfected with control pcDNA3 was set at 1.0. Data are mean ± SD from three independent experiments. (H) A model of the FDXR–IRP2–p53 loop in iron homeostasis and tumor suppression.
Next, the role of p53 in iron homeostasis was examined in HCT116 cells in which the p53 gene was deleted by CRISPR–cas9 (Fig. 7D) or knocked down by p53 siRNA (Supplemental Fig. S9B). We found that p53 deficiency led to increased expression of IRP2 and TfR1 but decreased expression of IRP1 and FTH1 in HCT116 cells (Fig. 7D; Supplemental Fig. S9B), consistent with the above observation in p53−/− MEFs. As a control, we found that p53 deficiency led to decreased expression of p21 and FDXR (Fig. 7D; Supplemental Fig. S9B). Additionally, we found that p53 deficiency led to mitochondrial iron overload in HCT116 cells (Fig. 7E; Supplemental Fig. S9C), consistent with the observation in p53−/− MEFs.
The above observations prompted us to hypothesize that loss of p53 plays a role in mitochondrial iron overload in FDXR-deficient cells. To test this, p53 was ectopically expressed in wild-type and FDXR+/−/− HCT116 cells. As expected, p21 and FDXR were induced by ectopic expression of p53 in both wild-type and FDXR+/−/− cells (Fig. 7F). Interestingly, we found that the levels of IRP1 and FTH1, both of which were markedly decreased by FDXR deficiency (Fig. 7F, cf. lanes 1 and 2), were increased to near normal by ectopic expression of p53 (Fig. 7F, cf. lanes 2 and 4). Similarly, the levels of IRP2 and TfR1, both of which were increased by FDXR deficiency (Fig. 7F, cf. lanes 1 and 2), were reduced to near normal by ectopic expression of p53 (Fig. 7F, cf. lanes 2 and 4). Furthermore, we found that mitochondrial iron overload by FDXR deficiency was restored to near normal by ectopic expression of p53 (Fig. 7G, last two columns). Together, our results suggest that p53 plays a role in iron homeostasis and mediates FDXR-dependent iron metabolism.
Discussion
This study showed that FDXR and p53 are mutually regulated and that the FDXR–p53 loop is critical for tumor suppression via iron homeostasis based on the following evidence: (1) Loss of Fdxr leads to embryonic lethality and cell death. (2) Mice heterozygous in Fdxr have a short life span and are prone to spontaneous tumors and liver abnormalities. (3) FDXR deficiency leads to aberrant iron metabolism and increased expression of IRP2 that represses p53 mRNA translation. (4) p53 plays a role in iron homeostasis and mediates FDXR-dependent iron metabolism. The hypothesis is summarized in Figure 7H.
The FDXR–p53 loop and mitochondrial iron overload
Loss of FDXR leads to mitochondrial iron overload, which is likely responsible for embryonic lethality (Fig. 1B). Our observation is consistent with previous studies that disruption of genes related to iron metabolism leads to embryonic lethality at E8.5–E12.5 due to oxidative stress (Donovan et al. 2005; Mao et al. 2010; Moroishi et al. 2011; Ruiz et al. 2013). We also found that p53 plays a role in iron homeostasis and is required for FDXR-mediated iron metabolism (Fig. 7). Thus, our findings strengthen the idea that p53 is a critical modulator of mitochondrial integrity and energy and iron metabolism (Zhou et al. 2003; Matoba et al. 2006; Funauchi et al. 2015).
Mitochondrial iron overload is observed in a number of diseases, including sideroblastic anemia and cancer. Mitochondrial iron overload can be induced by increased import, decreased export, or both, but it remains unclear what drives mitochondrial iron accumulation (Huang et al. 2011; Rouault 2016). Import of iron to mitochondria is controlled primarily by inner mitochondrial matrix proteins: mitoferrin 1 and mitoferrin 2. Indeed, mitoferrin expression is found to be altered in FDX1/2 knockdown cells (Shi et al. 2012). While mitoferrin is regulated primarily by transcription, it is not known what controls mitoferrin transcription (Rouault 2016). Iron export is known to be controlled by ABCB7, an ABC transporter in the mitochondrial matrix and a potential Fe-S exporter. Interestingly, IRP2 knockout reverses mitochondrial iron overload by FDXR deficiency to near normal (Fig. 5). Additionally, loss of FDXR leads to increased expression of IRP2, which suppresses p53 expression (Figs. 3–5). Thus, our data suggest that IRP2 has a novel activity as a mitochondrial iron regulator. Moreover, mitochondrial membrane potential, which is known to be regulated by FDXR and p53 via ROS production (Li et al. 1999; Charlot et al. 2004), may play a role in mitochondrial iron import and export. Together, we speculated that IRP2 and p53 coordinately regulate these critical iron importers and exporters through transcription, RNA stability, and/or translation.
FDXR, a potential mediator of p53-dependent ferroptosis
Previously, we and others showed that FDXR is a mediator of p53-dependent ROS-induced apoptosis (Hwang et al. 2001; Liu and Chen 2002) and modulates Fhit-mediated apoptosis (Trapasso et al. 2008). Additionally, through interaction with MPZL, FDXR plays a role in ROS-induced epidermal cell differentiation via mitochondria (Bhaduri et al. 2015). Here, we found that, by modulating RSL3- and Erastin-induced ferroptosis, FDXR plays a role in p53-dependent ferroptosis (Supplemental Fig. S4). Interestingly, RSL3- and Erastin-induced ferroptosis are suppressed by FDXR deficiency as well as FDXR overexpression. As a common target of the p53 family (Liu and Chen 2002) and a mediator of p53-dependent apoptosis and ferroptosis, further studies are warranted to determine the mechanism by which FDXR modulates ferroptosis, whether p63 and p73 are capable of inducing ferroptosis via FDXR, and how FDXR coordinately regulates p53-dependent ROS-mediated apoptosis and ferroptosis.
IRP2 and p53 mRNA translation
We found that ectopic expression of IRP2 inhibits, whereas knockout of IRP2 increases, p53 expression in the presence and absence of FDXR (Fig. 5; Supplemental Fig. S8). Considering that IRP2 is markedly induced along with decreased p53 expression by FDXR deficiency, we conclude that p53 expression is regulated primarily by IRP2 in FDXR-deficient cells. Additionally, we found that ectopic expression of IRP2 inhibits, whereas loss of IRP2 increases, the association of eIF4E with the p53 5′ cap (Fig. 6I,J), indicating that IRP2 represses p53 mRNA translation in a cap-dependent manner. These results are consistent with a previous report that the binding of IRP2 with an IRE on its target mRNA prevents the assembly of the 43S translation preinitiation complex (Gray and Hentze 1994). Generally, IRP2 recognizes an IRE located in the 5′ UTR for mRNA translation and an IRE in the 3′ UTR for mRNA stability (Butt et al. 1996; Henderson et al. 1996). Interestingly, we found that a putative IRE in the p53 3′ UTR is responsive to IRP2 (Fig. 5; Supplemental Fig. S8). We note that the p53 3′ UTR is recognized by and responsive to RBM38 and RPL26 for translational control (Takagi et al. 2005; Zhang et al. 2011). Thus, we hypothesize that p53 mRNA translation is uniquely regulated through its 3′ UTR by multiple RNA-binding proteins. Nevertheless, it remains possible that IRP1, which is likely to be regulated by FDXR, may modulate p53 translation through binding to the p53 5′ UTR.
FDXR deficiency and tumorigenesis
Mice deficient in Fdxr are prone to spontaneous tumors and liver abnormalities (Fig. 2; Supplemental Fig. S2; Supplemental Table S2). Since p53 expression is suppressed by FDXR deficiency, we postulate that decreased expression of p53 is a major contributing factor. However, compared with p53-deficient mice (Donehower and Lozano 2009), we found that mice deficient in Fdxr are prone to a broader spectrum of tumors, including lung adenocarcinoma and HCC (Fig. 2; Supplemental Fig. S2; Supplemental Tables S1, S2), suggesting that other pathways altered by Fdxr deficiency may play a role in tumorigenesis. First, we hypothesize that mitochondrial iron overload leads to increased liver inflammation (hepatitis), which would synergize with loss of p53 to promote cell transformation. Second, FDXR is a key regulator of steroidogenesis in mitochondria (Muller et al. 2001). Thus, Fdxr deficiency would alter lipid metabolism, leading to liver steatosis as observed in Fdxr-deficient mice (Fig. 2; Supplemental Fig. S2; Supplemental Table S2). Since liver steatosis predisposes livers to hyperplasia and eventually HCC (Meynard et al. 2014), we hypothesize that liver steatosis would synergize with loss of p53 to promote cell (hepatocyte) transformation. Third, FDXR is known to interact with Fhit and may play a role in Fhit-mediated tumor suppression (Trapasso et al. 2008). Thus, we hypothesize that loss of p53 in FDXR-deficient cells may synergize with loss of Fhit-mediated tumor suppression to promote tumorigenesis. Fourth, increased expression of IRP2 in FDXR-deficient cells can promote c-Myc expression (Wu et al. 1999; Maffettone et al. 2010), which would cooperate with loss of p53 to promote tumorigenesis.
FDX2—the primary transducer of FDXR for iron homeostasis and tumor suppression
Both FDX1 and FDX2 can carry out biogenesis of Fe-S clusters (Shi et al. 2012), but FDX2 is necessary and sufficient for such an activity (Sheftel et al. 2010). Here, we showed that FDX1 is not required for cell survival and has no effect on iron homeostasis and p53 expression (Supplemental Fig. S6). Since FDX1 is necessary and sufficient for steroidogenesis in mitochondria (Sheftel et al. 2010), we hypothesize that loss of the signal from FDXR to FDX1 is likely responsible for liver steatosis in Fdxr-deficient mice (Fig. 2; Supplemental Fig. S2; Supplemental Table S2). In contrast, FDX2 is required for cell survival, and FDX2 deficiency leads to mitochondrial iron overload and decreased p53 expression. Additionally, FDX2 deficiency leads to increased IRP2 expression (Fig. 4A; Supplemental Fig. S5). These data indicate that FDX2 is the primary substrate to transduce the FDXR signal via IRP2 to iron homeostasis, p53 expression, and tumor suppression.
Conclusion and future direction
Our data together with published studies indicate that the FDXR–p53 loop plays a critical role in iron homeostasis for tumor suppression. As a target of the p53 family, we speculate that FDXR may engage other p53 family members (that is, p63 and p73) to regulate iron homeostasis. Importantly, we speculate that FDXR would modulate mutant p53 gain of function through iron metabolism. Indeed, a recent study showed that FDXR knockdown suppresses T47D mammary tumor cell growth and xenograft (Zhang et al. 2015), potentially due to decreased expression of mutant p53 (L194F) in T47D cells. Thus, further studies are warranted to determine whether iron homeostasis is regulated by other p53 family pathways via FDXR and whether the FDXR–p53 loop can be explored for managing tumors that carry wild-type p53 as well as tumors addicted to mutant p53.
Materials and methods
Reagents
Proteinase inhibitor cocktail, RNase A, and protein A/G beads were purchased from Sigma. Potassium ferrocyanide, nuclear Fast Red, and hematoxylin and eosin (H&E) were purchased from Fisher Scientific.
Fdxr- and p53-deficienct mouse models
Fdxr+/− mice were generated by the Mouse Biology Program at University of California at Davis. The targeting vector carries the entire Fdxr locus in which exons 1–12 were replaced by LacZ-poly(A) and hUbDpro-neo-poly(A) cassettes. Mutant embryonic stem cells were microinjected into C57BL/6Ntac blastocytes to generate chimeras. The resulting chimeras were crossed with C57BL/6 mice for germline transmission, and the offspring were backcrossed with C57BL/6 mice for more than six generations. The p53+/− mice (on a C57BL/6 background) were purchased from the Jackson Laboratory. All animal procedures were approved by the University of California at Davis Institutional Animal Care and Use Committee.
Cell culture
HCT116, HepG2, H1299, and their derivatives were cultured in DMEM (Dulbecco's modified Eagle's medium, Invitrogen) supplemented with 10% fetal bovine serum (Hyclone). Wild-type and Fdxr+/− MEFs were generated as described previously (Zhang et al. 2011) and cultured in DMEM supplemented with 10% fetal bovine serum, 55 µM β-mercaptoethanol, and MEM nonessential amino acid solution (Cellgro).
Plasmid construction and cell line generation
FDXR, FDX1, FDX2, IRP2, and p53 guide RNAs (gRNAs) were designed using CRISPR design tool (http://crispr.mit.edu) and are listed in Supplemental Table S3. To generate a vector expressing a single gRNA (sgRNA), two 25-nt oligos were annealed and then cloned into the pSpCas9(BB) (Ran et al. 2013) sgRNA expression vector. To generate a knockout cell line, HCT116 cells were transfected with a gRNA-expressing vector and selected with puromycin. Cells deficient in FDXR, FDX1, FDX2, IRP2, or p53 were confirmed by genotyping, sequencing, and Western blot analysis. The primers used for sequencing FDXR, FDX1, FDX2, IRP2, and p53 are listed in Supplemental Table S4.
pcDNA3 vectors carrying a p53- or GFP-coding region alone or together with the p53 5′ or 3′ UTR (Supplemental Fig. S8C) were used as described previously (Zhang et al. 2013). GFP reporters that carry a p53 3′ UTR with deletion of the putative IRE or poly(U) region were generated by two-step PCR reactions (Supplemental Fig. S8C). The pcDNA4 vector expressing HA-tagged FDXR was generated by subcloning the HA-FDXR from pUHD 10-3-HA-FDXR (Liu and Chen 2002) to the pcDNA4 vector via EcoRI. The primers are listed in Supplemental Table S5.
Western blot analysis
Western blot was performed as described (Zhang et al. 2014). Antibodies against p53, p21, IRP2, and TfR1 were purchased from Santa Cruz Biotechnology. Anti-mouse p53 (1C12) was purchased from Cell Signaling Technology. Antibodies against FDXR, FDX1, FDX2, IRP1, and FTH1 were purchased from Abcam. Anti-HA was purchased from Covance. Anti-actin and HRP-conjugated secondary antibodies against rabbit or mouse IgG were purchased from Bio-Rad. The immunoreactive bands were visualized by enhanced chemoluminescence (Thermo Fisher Scientific, Inc.) and quantified by densitometry with the BioSpectrum 810 imaging system (UVP LLC).
RNA isolation and RT–PCR analysis
Total RNAs were extracted from cells using TRIzol (Invitrogen Life Technologies) according to the manufacturer's instructions. cDNA was synthesized using M-MLV reverse transcriptase kit (Promega Corporation) according to the manufacturer's protocol. The levels of p53, p21, FDXR, and actin transcripts were measured by semiquantitative and/or quantitative PCR with primers listed in Supplemental Tables S6 and S7.
RNAi
Scrambled siRNA and siRNAs against FDXR (siFDXR#1 and siFDXR#2), FDX2 (siFDX2#1 and siFDX2#2), and p53 (sip53) were purchased from Dharmacon and are listed in Supplemental Table S8. For siRNA transfection, siLentFectTM lipid reagent (Bio-Rad) was used according to the user's manual.
Histological analysis, iron histochemistry, and reticulin staining
Mouse tissues or embryos were fixed in 10% (w/v) neutral buffered formalin, processed, and embedded in paraffin blocks. Embedded tissues were sectioned (5 µm) and stained with H&E. For iron histochemistry, the level of iron was detected by Prussian blue staining using freshly prepared 5% potassium ferrocyanide and 5% hydrochloric acid. Thirty minutes after staining, tissue sections were rinsed with water and counterstained with nuclear Fast Red, dehydrated, and covered. Wild-type, Fdxr+/−, and/or p53−/−, p53+/−, Fdxr+/−;p53+/−, and Fdxr+/−;p53−/− mouse embryos and tissues were stained simultaneously for comparison. For reticulin staining, embedded tissues were oxidized, sensitized with ferric ammonium sulfate, and stained with silver. The silver was then reduced with formalin to its visible metallic state. Tissue sections were rinsed with water and counterstained with nuclear Fast Red.
Iron measurement
The level of iron in mitochondria and the cytoplasm was detected by QuantiChrom iron assay kit (Bioassay) following the manufacturer's instructions. Fe3+ in the sample was reduced to Fe2+, which was then recognized by a chromogen to form a blue-colored complex. Briefly, mitochondria and cytoplasm fractions were isolated from cells or liver tissues as described previously (Frezza et al. 2007; Wieckowski et al. 2009). Iron levels in mitochondria and cytosolic fractions were determined by measuring the value of absorption at 590 nm using a microplate reader (Bio-Rad).
RNA immunoprecipitation (RNA-IP) assay
RNA-IP was carried out as described previously (Peritz et al. 2006). Briefly, cell extracts were prepared with immunoprecipitation buffer (10 mm HEPES at pH 7.0, 100 mm KCl, 5 mm MgCl2, 0.5% NP-40, 1 mm DTT) and then incubated with 2 µg of anti-HA (IRP2), anti-eIF4E, or an isotype control IgG overnight at 4°C. The RNA–protein immunocomplexes were brought down by protein G beads. RT–PCR analysis was carried out to determine RNA transcripts associated with IRP2 or eIF4E.
Statistical analysis
For iron concentration, data are presented as mean ± SD. Statistical significance was determined by Student's t-test. For Kaplan-Meyer survival analysis, log-rank test was performed. Values of P < 0.05 were considered significant. Fisher's exact test was used for comparison of tumors from different genotypes.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants CA076069, CA195828, and CA093373. Y.Z., Y.Q., J.Z., W.Y., Y.-S.J., G.L., M.C., and E.H. performed the experiments. Y.Z., Y.Q., J.Z., W.Y., Y.-S.J., J.W., M.C., E.H., K.L., and X.C. designed the experiments and analyzed the data. Y.Z. and X.C. wrote the manuscript. J.Z., Y.Q., J.W., G.L., M.C., and E.H. proofread and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.299388.117.
References
- Abend M, Badie C, Quintens R, Kriehuber R, Manning G, Macaeva E, Njima M, Oskamp D, Strunz S, Moertl S, et al. 2016. Examining radiation-induced in vivo and in vitro gene expression changes of the peripheral blood in different laboratories for biodosimetry purposes: first RENEB gene expression study. Radiat Res 185: 109–123. [DOI] [PubMed] [Google Scholar]
- Abeysinghe RD, Greene BT, Haynes R, Willingham MC, Turner J, Planalp RP, Brechbiel MW, Torti FM, Torti SV. 2001. p53-independent apoptosis mediated by tachpyridine, an anti-cancer iron chelator. Carcinogenesis 22: 1607–1614. [DOI] [PubMed] [Google Scholar]
- Andrews NC. 1999. Disorders of iron metabolism. N Engl J Med 341: 1986–1995. [DOI] [PubMed] [Google Scholar]
- Bergman S, Graeme-Cook F, Pitman MB. 1997. The usefulness of the reticulin stain in the differential diagnosis of liver nodules on fine-needle aspiration biopsy cell block preparations. Mod Pathol 10: 1258–1264. [PubMed] [Google Scholar]
- Beutler E. 2006. Hemochromatosis: genetics and pathophysiology. Annu Rev Med 57: 331–347. [DOI] [PubMed] [Google Scholar]
- Bhaduri A, Ungewickell A, Boxer LD, Lopez-Pajares V, Zarnegar BJ, Khavari PA. 2015. Network analysis identifies mitochondrial regulation of epidermal differentiation by MPZL3 and FDXR. Dev Cell 35: 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Vickery LE. 1992. Expression and characterization of human mitochondrial ferredoxin reductase in Escherichia coli. Arch Biochem Biophys 294: 735–740. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. 2011. p53 regulation by ubiquitin. FEBS Lett 585: 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt J, Kim HY, Basilion JP, Cohen S, Iwai K, Philpott CC, Altschul S, Klausner RD, Rouault TA. 1996. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc Natl Acad Sci 93: 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlot JF, Pretet JL, Haughey C, Mougin C. 2004. Mitochondrial translocation of p53 and mitochondrial membrane potential (ΔΨm) dissipation are early events in staurosporine-induced apoptosis of wild type and mutated p53 epithelial cells. Apoptosis 9: 333–343. [DOI] [PubMed] [Google Scholar]
- Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabon MA, Konrad C, Polverino F, Siempos II, Perez E, et al. 2016. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med 22: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. 2005. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 106: 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Lozano G. 2009. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer 9: 831–841. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- Dongiovanni P, Fracanzani AL, Cairo G, Megazzini CP, Gatti S, Rametta R, Fargion S, Valenti L. 2010. Iron-dependent regulation of MDM2 influences p53 activity and hepatic carcinogenesis. Am J Pathol 176: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. 2005. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab 1: 191–200. [DOI] [PubMed] [Google Scholar]
- Edmondson DA, Karski EE, Kohlgruber A, Koneru H, Matthay KK, Allen S, Hartmann CL, Peterson LE, DuBois SG, Coleman MA. 2016. Transcript analysis for internal biodosimetry using peripheral blood from neuroblastoma patients treated with (131)I-mIBG, a targeted radionuclide. Radiat Res 186: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinati F, Marino D, De Giorgio M, Baldan A, Cantarini M, Cursaro C, Rapaccini G, Del Poggio P, Di Nolfo MA, Benvegnu L, et al. 2006. Diagnostic and prognostic role of α-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol 101: 524–532. [DOI] [PubMed] [Google Scholar]
- Fleming RE, Ponka P. 2012. Iron overload in human disease. N Engl J Med 366: 348–359. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. 2007. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2: 287–295. [DOI] [PubMed] [Google Scholar]
- Funauchi Y, Tanikawa C, Yi Lo PH, Mori J, Daigo Y, Takano A, Miyagi Y, Okawa A, Nakamura Y, Matsuda K. 2015. Regulation of iron homeostasis by the p53–ISCU pathway. Sci Rep 5: 16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy B, Ferring D, Minana B, Bell O, Janser HG, Muckenthaler M, Schumann K, Hentze MW. 2005. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 106: 2580–2589. [DOI] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ. 1996. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379: 88–91. [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. 1994. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J 13: 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Menotti E, Kuhn LC. 1996. Iron regulatory proteins 1 and 2 bind distinct sets of RNA target sequences. J Biol Chem 271: 4900–4908. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Seuanez HN, O'Brien SJ, Harford JB, Klausner RD. 1989. Chromosomal localization of nucleic acid-binding proteins by affinity mapping: assignment of the IRE-binding protein gene to human chromosome 9. Nucleic Acids Res 17: 6103–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C. 2010. Two to tango: regulation of mammalian iron metabolism. Cell 142: 24–38. [DOI] [PubMed] [Google Scholar]
- Huang ML, Lane DJ, Richardson DR. 2011. Mitochondrial mayhem: the mitochondrion as a modulator of iron metabolism and its role in disease. Antioxid Redox Signal 15: 3003–3019. [DOI] [PubMed] [Google Scholar]
- Hwang PM, Bunz F, Yu J, Rago C, Chan TA, Murphy MP, Kelso GF, Smith RA, Kinzler KW, Vogelstein B. 2001. Ferredoxin reductase affects p53-dependent, 5-fluorouracil-induced apoptosis in colorectal cancer cells. Nat Med 7: 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. 2015. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Levine AJ. 2001. The p53 functional circuit. J Cell Sci 114: 4139–4140. [DOI] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. 2008. p53 regulates glucose metabolism through an IKK–NF-κB pathway and inhibits cell transformation. Nat Cell Biol 10: 611–618. [DOI] [PubMed] [Google Scholar]
- Lange H, Kaut A, Kispal G, Lill R. 2000. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci 97: 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVaute T, Smith S, Cooperman S, Iwai K, Land W, Meyron-Holtz E, Drake SK, Miller G, Abu-Asab M, Tsokos M, et al. 2001. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet 27: 209–214. [DOI] [PubMed] [Google Scholar]
- Lawen A, Lane DJ. 2013. Mammalian iron homeostasis in health and disease: uptake, storage, transport, and molecular mechanisms of action. Antioxid Redox Signal 18: 2473–2507. [DOI] [PubMed] [Google Scholar]
- Li PF, Dietz R, von Harsdorf R. 1999. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J 18: 6027–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SX, Richardson DR. 2003. The effect of potent iron chelators on the regulation of p53: examination of the expression, localization and DNA-binding activity of p53 and the transactivation of WAF1. Carcinogenesis 24: 1601–1614. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X. 2002. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 21: 7195–7204. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev 7: 535–545. [DOI] [PubMed] [Google Scholar]
- Maffettone C, Chen G, Drozdov I, Ouzounis C, Pantopoulos K. 2010. Tumorigenic properties of iron regulatory protein 2 (IRP2) mediated by its specific 73-amino acids insert. PLoS One 5: e10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, McKean DM, Warrier S, Corbin JG, Niswander L, Zohn IE. 2010. The iron exporter ferroportin 1 is essential for development of the mouse embryo, forebrain patterning and neural tube closure. Development 137: 3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masramon L, Ribas M, Cifuentes P, Arribas R, Garcia F, Egozcue J, Peinado MA, Miro R. 2000. Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet Cytogenet 121: 17–21. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. 2006. p53 regulates mitochondrial respiration. Science 312: 1650–1653. [DOI] [PubMed] [Google Scholar]
- McLure KG, Takagi M, Kastan MB. 2004. NAD+ modulates p53 DNA binding specificity and function. Mol Cell Biol 24: 9958–9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynard D, Babitt JL, Lin HY. 2014. The liver: conductor of systemic iron balance. Blood 123: 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. 2011. The FBXL5–IRP2 axis is integral to control of iron metabolism in vivo. Cell Metab 14: 339–351. [DOI] [PubMed] [Google Scholar]
- Muller JJ, Lapko A, Bourenkov G, Ruckpaul K, Heinemann U. 2001. Adrenodoxin reductase–adrenodoxin complex structure suggests electron transfer path in steroid biosynthesis. J Biol Chem 276: 2786–2789. [DOI] [PubMed] [Google Scholar]
- Nelson WG, Kastan MB. 1994. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol 14: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. 2004. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306: 2090–2093. [DOI] [PubMed] [Google Scholar]
- Nishina S, Hino K, Korenaga M, Vecchi C, Pietrangelo A, Mizukami Y, Furutani T, Sakai A, Okuda M, Hidaka I, et al. 2008. Hepatitis C virus-induced reactive oxygen species raise hepatic iron level in mice by reducing hepcidin transcription. Gastroenterology 134: 226–238. [DOI] [PubMed] [Google Scholar]
- Okumura H, Uchikado Y, Omoto I, Motomura M, Kita Y, Sasaki K, Noda M, Arigami T, Uenosono Y, Baba K, et al. 2015. Ferredoxin reductase is useful for predicting the effect of chemoradiation therapy on esophageal squamous cell carcinoma. Anticancer Res 35: 6471–6474. [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. 2006. Immunoprecipitation of mRNA-protein complexes. Nat Protoc 1: 577–580. [DOI] [PubMed] [Google Scholar]
- Pietrangelo A. 2009. Iron in NASH, chronic liver diseases and HCC: how much iron is too much? J Hepatol 50: 249–251. [DOI] [PubMed] [Google Scholar]
- Puzio-Kuter AM. 2011. The role of p53 in metabolic regulation. Genes Cancer 2: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR–Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412. [DOI] [PubMed] [Google Scholar]
- Rouault TA. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2: 406–414. [DOI] [PubMed] [Google Scholar]
- Rouault TA. 2016. Mitochondrial iron overload: causes and consequences. Curr Opin Genet Dev 38: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Tang CK, Kaptain S, Burgess WH, Haile DJ, Samaniego F, McBride OW, Harford JB, Klausner RD. 1990. Cloning of the cDNA encoding an RNA regulatory protein—the human iron-responsive element-binding protein. Proc Natl Acad Sci 87: 7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JC, Walker SD, Anderson SA, Eisenstein RS, Bruick RK. 2013. F-box and leucine-rich repeat protein 5 (FBXL5) is required for maintenance of cellular and systemic iron homeostasis. J Biol Chem 288: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen NJ, Silverman LM, Keku T, Lawrence LB, Rohlfs EM, Martin CF, Galanko J, Sandler RS. 2003. Association between hemochromatosis (HFE) gene mutation carrier status and the risk of colon cancer. J Natl Cancer Inst 95: 154–159. [DOI] [PubMed] [Google Scholar]
- Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R. 2010. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci 107: 11775–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Sheng X, Chang Z, Wu Q, Wang S, Xuan Z, Li D, Wu Y, Shang Y, Kong X, et al. 2014. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep 7: 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. 2012. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta 1823: 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox JA, McClain DA. 2013. Iron and diabetes risk. Cell Metab 17: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino P, D'Angelo S, Ferbo U, Micheli P, Bracigliano A, Vecchione R. 2009. Liver iron excess in patients with hepatocellular carcinoma developed on non-alcoholic steato-hepatitis. J Hepatol 50: 351–357. [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63. [DOI] [PubMed] [Google Scholar]
- Trapasso F, Pichiorri F, Gaspari M, Palumbo T, Aqeilan RI, Gaudio E, Okumura H, Iuliano R, Di Leva G, Fabbri M, et al. 2008. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J Biol Chem 283: 13736–13744. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K. 2011. Regulation of cellular iron metabolism. Biochem J 434: 365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitnall M, Howard J, Ponka P, Richardson DR. 2006. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc Natl Acad Sci 103: 14901–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. 2009. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4: 1582–1590. [DOI] [PubMed] [Google Scholar]
- Wu KJ, Polack A, Dalla-Favera R. 1999. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 283: 676–679. [DOI] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. 2016. Ferroptosis: death by lipid peroxidation. Trends Cell Biol 26: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. 2006. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol 8: 1074–1083. [DOI] [PubMed] [Google Scholar]
- Yu J, Marsh S, Ahluwalia R, McLeod HL. 2003. Ferredoxin reductase: pharmacogenomic assessment in colorectal cancer. Cancer Res 63: 6170–6173. [PubMed] [Google Scholar]
- Zhang J, Chen X. 2008. Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr Mol Med 8: 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang W, Tsuji Y, Torti SV, Torti FM. 2008. Post-transcriptional modulation of iron homeostasis during p53-dependent growth arrest. J Biol Chem 283: 33911–33918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, Skorupski K, Chen H, Chen X. 2011. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev 25: 1528–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang J, Chen X, Cho SJ, Chen X. 2013. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1. Genes Dev 27: 2246–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan W, Chen X. 2014. P63 regulates tubular formation via epithelial-to-mesenchymal transition. Oncogene 33: 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang C, Chen X, Takada M, Fan C, Zheng X, Wen H, Liu Y, Wang C, Pestell RG, et al. 2015. EglN2 associates with the NRF1–PGC1α complex and controls mitochondrial function in breast cancer. EMBO J 34: 2953–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Kachhap S, Singh KK. 2003. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis 18: 287–292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.