Ogami et al. highlight a critical role for Mtr4/ZFC3H1 in nuclear surveillance of naturally unstable lncRNAs to prevent their accumulation, transport to the cytoplasm, and resultant disruption of protein synthesis.
Keywords: Mtr4, ZFC3H1, exosome, lncRNA, polyadenylation
Abstract
Many long noncoding RNAs (lncRNAs) are unstable and rapidly degraded in the nucleus by the nuclear exosome. An exosome adaptor complex called NEXT (nuclear exosome targeting) functions to facilitate turnover of some of these lncRNAs. Here we show that knockdown of one NEXT subunit, Mtr4, but neither of the other two subunits, resulted in accumulation of two types of lncRNAs: prematurely terminated RNAs (ptRNAs) and upstream antisense RNAs (uaRNAs). This suggested a NEXT-independent Mtr4 function, and, consistent with this, we isolated a distinct complex containing Mtr4 and the zinc finger protein ZFC3H1. Strikingly, knockdown of either protein not only increased pt/uaRNA levels but also led to their accumulation in the cytoplasm. Furthermore, all pt/uaRNAs examined associated with active ribosomes, but, paradoxically, this correlated with a global reduction in heavy polysomes and overall repression of translation. Our findings highlight a critical role for Mtr4/ZFC3H1 in nuclear surveillance of naturally unstable lncRNAs to prevent their accumulation, transport to the cytoplasm, and resultant disruption of protein synthesis.
RNA polymerase II is responsible for production of a large repertoire of RNAs. In addition to mRNAs, these include a variety of functional, relatively stable RNAs, such as small nuclear RNAs (snRNAs) and microRNAs (miRNAs), and a diverse set of long noncoding RNAs (lncRNAs). Many of these lncRNAs have well-documented functions in either the cytoplasm or nucleus and are also often quite stable (Chen 2016). However, a large number of lncRNAs have no known functions and can be very unstable and rapidly degraded in the nucleus. These include, for example, RNAs that are transcribed upstream of protein-coding gene promoters (Preker et al. 2008; Flynn et al. 2011) as well as RNAs transcribed, frequently bidirectionally, from transcriptional enhancers (Djebali et al. 2012; Andersson et al. 2014a). Synthesis of these RNAs often involves multiple RNA processing reactions, which are typically closely linked to their transcription. Such processing invariably involves 5′ capping, frequently splicing as well as 3′ end formation, and often cleavage and polyadenylation. It is noteworthy that polyadenylation can have either a stabilizing effect, as typically observed with mRNAs, or a destabilizing effect, as found with many nuclear lncRNAs subject to rapid decay (Beaulieu et al. 2012; Ntini et al. 2013; Bresson et al. 2015).
Two interesting classes of lncRNAs are the promoter upstream transcripts (PROMPTs) and prematurely terminated RNAs (ptRNAs). PROMPTs are transcribed in both sense and antisense directions relative to transcription start sites of protein-coding genes and are polyadenylated (Preker et al. 2008). Notably, PROMPTs that are transcribed in the antisense direction, which we specifically refer to as upstream antisense RNAs (uaRNAs) (Flynn et al. 2011), appear to be processed by mechanisms of pre-mRNA 3′ end formation that are the same as or similar to those of mRNAs (Almada et al. 2013; Ntini et al. 2013; for review, see Richard and Manley 2013). Motifs similar to those that constitute poly(A) sites (PASs) in pre-mRNAs are found at the 3′ ends of uaRNAs, and much of the same complex protein machinery that is responsible for mRNA polyadenylation (Tian and Manley 2017) functions in uaRNA 3′ end formation (Ntini et al. 2013). Notably, PASs are more enriched in the upstream antisense region compared with the downstream region, whereas U1 snRNA-binding sites (which, when bound by U1 snRNP, prevent polyadenylation at nearby PASs) (Almada et al. 2013; Ntini et al. 2013) are more frequent in the sense-coding direction. Failure of the suppression of proximal PASs results in premature cleavage and polyadenylation, giving rise to ptRNAs (Kaida et al. 2010; Berg et al. 2012). PAS-driven early termination of pt/uaRNAs is linked to rapid degradation of these RNAs by the nuclear exosome despite the fact that the RNA signals and protein factors are very similar to those used for relatively stable mRNAs (Ntini et al. 2013).
What targets unstable nuclear RNAs for rapid turnover? An interesting candidate that might link PAS-mediated 3′ processing of lncRNAs to degradation is the exosome adaptor complex NEXT (nuclear exosome targeting), which consists of the RNA helicase Mtr4, the RNA-binding protein RBM7, and the zinc knuckle protein ZCCHC8 (Lubas et al. 2011). NEXT physically associates with the nuclear exosome to facilitate turnover of various types of RNA substrates, including PROMPTs, enhancer RNAs (eRNAs), 3′ extended snRNAs/small nucleolar RNAs (snoRNAs), and replication-dependent histone-encoding transcripts (Lubas et al. 2011; Andersen et al. 2013). Mtr4, which shows a higher affinity for poly(A) relative to non-poly(A) RNA (Bernstein et al. 2008), plays an essential role in exosome activation, presumably by RNA unwinding and/or feeding RNA substrates into the exosome (Wang et al. 2008; Houseley and Tollervey 2009). The substrate recognition activity of NEXT is conferred by RBM7, which shows some preference for U-rich sequences (Andersen et al. 2013). Intriguingly, all three NEXT subunits were identified in a proteomics analysis of the purified human pre-mRNA polyadenylation complex (Shi et al. 2009), suggesting a possible role for NEXT coupled to 3′ processing. Human cells possess another exosome adaptor complex, TRAMP (Trf4–Air–Mtr4 polyadenylation complex), comprising Mtr4, the noncanonical poly(A) polymerase PAPD5, and the zinc knuckle protein ZCCHC7. TRAMP substrates, however, are thought to be restricted to nucleolar RNA targets such as ribosomal RNA (rRNA) precursors (Lubas et al. 2011).
In this study, we analyzed the polyadenylated transcriptomes of cells depleted of the individual NEXT subunits to investigate whether NEXT might affect polyadenylation generally. Unexpectedly, we found that siRNA-mediated knockdown of Mtr4, but neither of the other two NEXT subunits or a TRAMP subunit, resulted in strong and specific accumulation of ptRNAs and uaRNAs. We then examined Mtr4 complexes and interacting proteins by gel filtration followed by mass spectrometry (MS) and identified the zinc finger protein ZFC3H1 as an Mtr4 partner (see also Meola et al. 2016) also necessary for degradation of ptRNAs and uaRNAs. Unexpectedly, knockdown of Mtr4 or ZFC3H1 resulted in accumulation of polyadenylated Mtr4 targets in the cytoplasm, and the exported RNAs were bound by ribosomes. Mtr4/ZFC3H1-deficient cells also showed a surprising global reduction in heavier polysomes, suggesting that ribosomes naturally bound to mRNAs were occupied by the short ORF-containing pt/uaRNAs. Consistent with this, the Mtr4/ZFC3H1-depleted cells showed a significant specific inhibition of translation. Our findings illustrate the importance of nuclear surveillance of polyadenylated lncRNAs by Mtr4/ZFC3H1 to prevent the unwanted and deleterious transport to and accumulation of these RNAs in the cytoplasm.
Results
Mtr4 prevents accumulation of ptRNAs and uaRNAs
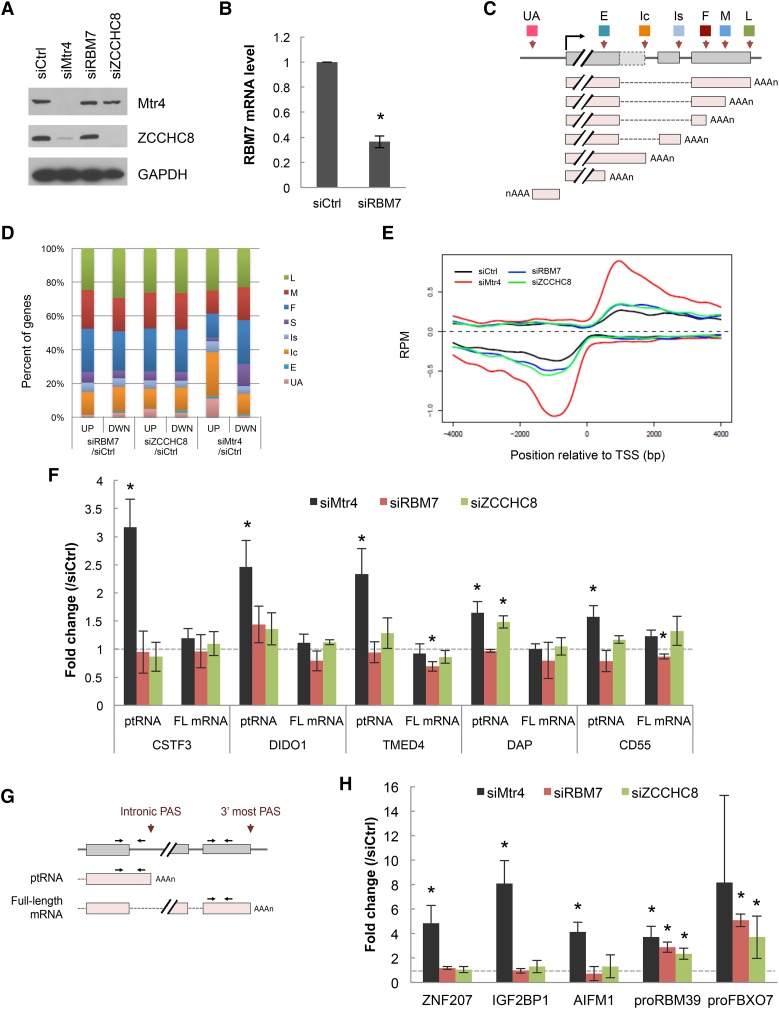
The initial aim of our experiments was to investigate the significance of the previously observed association of NEXT with the pre-mRNA polyadenylation machinery. To this end, we depleted each of the three NEXT subunits from HeLa cells with siRNAs (Fig. 1A,B) and performed 3′ region extraction and deep sequencing (3′READS) (Hoque et al. 2013) to analyze the global effects on accumulation of polyadenylated RNAs. Unexpectedly, depletion of Mtr4, but neither of the other two NEXT subunits (RBM7 or ZCCHC8), resulted in preferential accumulation of two types of transcripts: uaRNAs and ptRNAs (Fig. 1C,D). Consistent with this, metagene plots revealed that the Mtr4-depleted cells showed a sharp increase of promoter-proximal PAS usage in both sense and antisense directions, whereas depletion of RBM7 and ZCCHC8 had only minimal effects (Fig. 1E). An increase of reads corresponding to intronic PASs was observed in Mtr4 knockdown cells, while those corresponding to the 3′-most PASs, reflecting full-length mRNAs, were not affected by knockdown (Supplemental Fig. S1A), suggesting that the change was due to ptRNA stabilization as opposed to increased 3′ processing at the intronic PAS.
Figure 1.
Global analysis of poly(A)+ transcript levels following depletion of individual NEXT subunits. (A) Western blot analysis of HeLa cell extracts after 48 h of transfection of control (Ctrl), Mtr4, RBM7, or ZCCHC8 siRNA. (B) RBM7 mRNA level after 48 h of siRBM7 treatment. RBM7 mRNA was normalized to GAPDH mRNA, and the normalized levels in siCtrl-treated cells were set to 1. Bars represent mean ± SD. n = 3. An asterisk denotes significant difference from siCtrl (P < 0.05) using an unpaired Student's t-test. (C) Schematic of different transcript types analyzed: transcripts using the first (F), middle (M), or last (L) potential PAS in the 3′ untranslated region (UTR); the single (S; no 3′ UTR APA) PAS in the 3′ UTR; the intronic PAS in the composite terminal exon (Ic); the intronic PAS in the skipped terminal exon (Is); the upstream (not 3′-most) exonic PAS (E); and the upstream antisense transcripts (UA). (D) Changes in relative abundance of the indicated transcript types following knockdown of indicated NEXT subunits. The percentage of genes showing increases (UP) or decreases (DWN) of each type of transcript are indicated. False discovery rate < 0.05. (E) Metagene plots of ptRNAs and uaRNAs. Data are presented as strand-specific reads per million (RPM) at PAS positions within 4 kb upstream of or downstream from the transcription start site. (F,H) RT-qPCR [oligo(dT)-primed RT and quantitative PCR (qPCR)] analysis of select ptRNAs and corresponding full-length (FL) mRNAs (F) and uaRNAs (H) after knockdown of the individual NEXT subunits. Analysis of two representative PROMPTs—proRBM39 and proFBX07—is also shown in H. Values were normalized to GAPDH mRNA, and the normalized levels in siCtrl-treated cells were set to 1. Bars represent mean ± SD. n = 3. Asterisks denote significant difference from siCtrl (P < 0.05) using an unpaired Student's t-test. (G) Diagram of a ptRNA-producing gene and primers used for RT-qPCR. Arrows indicate the positions of primer targeting sites to analyze ptRNA and full-length mRNA.
We next performed RT-qPCR [oligo(dT)-primed RT and quantitative PCR (qPCR)] to confirm and extend the results of 3′READS. To distinguish ptRNAs and full-length mRNAs, primers were designed as shown in Figure 1G. Consistent with the 3′READS data, levels of both ptRNAs and uaRNAs were increased by Mtr4 knockdown but not by knockdown of RBM7 or ZCCHC8 (Fig. 1F,H). In contrast to the uaRNAs, increased levels of two known NEXT substrates—RBM39 and FBXO7 PROMPTs (proRBM39 and proFBXO7, respectively) (Lubas et al. 2011)—were observed in all knockdown cells (Fig. 1H). Similar results were obtained using a second Mtr4 siRNA (Supplemental Fig. S1B,C). RT-qPCR also confirmed that there were no changes in full-length mRNA levels for genes that displayed elevated ptRNA levels (Fig. 1F, FL mRNA). The other known Mtr4-containing complex, TRAMP, appears not to be involved in degradation of ptRNAs and uaRNAs, as there were no significant changes of the Mtr4 target RNAs after ZCCHC7 knockdown (Supplemental Fig. S1D,E). In contrast and as expected, the nuclear exosome is required for ptRNA and uaRNA degradation, as all tested pt/uaRNAs accumulated following codepletion of the two catalytic subunits Rrp6 and Dis3 (Supplemental Fig. S1F,G). Together, these results indicate that the exosome degrades these RNAs in an Mtr4-dependent, but NEXT- and TRAMP-independent, manner.
Identification of Mtr4-interacting proteins
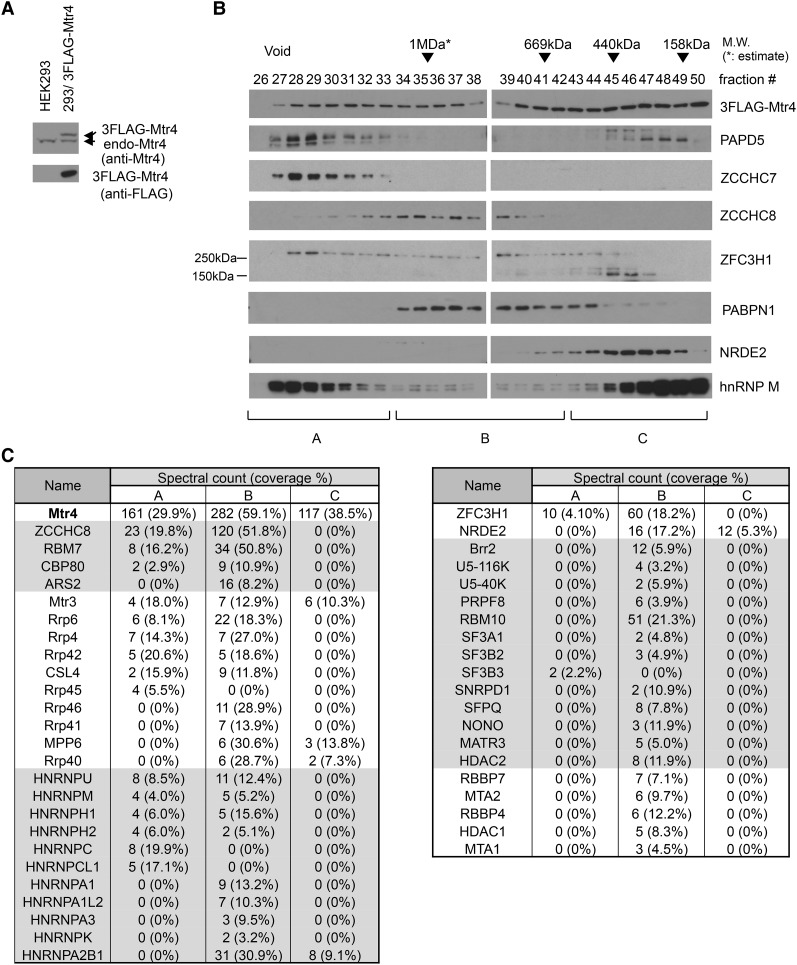
The above results suggested the possible existence of an additional Mtr4-containing protein complex that functions in ptRNA and/or uaRNA turnover. To investigate this, we prepared extracts from HEK293 cells stably expressing N-terminally 3xFlag-tagged Mtr4 (Flag-Mtr4, with expression equivalent to endogenous Mtr4) (Fig. 2A) in the presence of RNase A and performed size fractionation using Superose 6 gel filtration chromatography followed by Western blotting (Fig. 2B). Mtr4 was detected in all fractions from the void to <158 kDa in a bimodal distribution with peaks at fraction 31 and at <158 kDa, likely reflecting the existence of multiple Mtr4-containing complexes. The NEXT subunit ZCCHC8 eluted at ∼1 MDa, whereas TRAMP subunits ZCCHC7 and PAPD5 appeared mainly in the void fractions.
Figure 2.
Identification of Mtr4-interacting proteins by cofractionation and MS. (A) Western blotting analysis of HEK293 cells and HEK293 cells stably expressing 3Flag-Mtr4. (Top panel) Blotted with anti-Mtr4 antibodies. (Bottom panel) Blotted with anti-Flag antibodies. (B) Fractions from Superose 6 gel filtration 3Flag-Mtr4-expressing HEK293 cells were analyzed by Western blotting using antibodies against proteins shown at the right. Approximate molecular sizes are indicated at the top, and fractions pooled are indicated at the bottom. (C) Selected proteins copurified with 3Flag-Mtr4 in the indicated pools. Spectral counts and sequence coverage of known Mtr4-interacting partners (NEXT, exosome, and NRDE2), proteins detected as complexes (e.g., NuRD and spliceosome), and RNA processing or RNA-binding proteins are shown. A full protein list is in Supplemental Table S1.
To identify additional Flag-Mtr4-interacting proteins, we collected three Flag-Mtr4-containing pools (A–C) according to the distribution of TRAMP (ZCCHC7 and PAPD5) and NEXT (ZCCHC8). The pools were subjected to Flag immunoprecipitation, and the coimmunoprecipitated proteins were identified by MS (Fig. 2C; Supplemental Table S1). The subunits of NEXT, the exosome, and cap-binding complex (CBC), which were shown previously to associate with NEXT (Andersen et al. 2013), were detected in pools A and B. Consistent with a previous report (Nag and Steitz 2012), various splicing factors also associated with Flag-Mtr4 in pool B. Pool B also contained NuRD complex subunits, which function in histone modification (Xue et al. 1998; Zhang et al. 1998). We also found several heterogeneous nuclear ribonucleoproteins (hnRNPs)—PSF/SFPQ and p54nrb/NONO—in pool B as well as two other possibly relevant proteins: NRDE2 and ZFC3H1. Although the role of NRDE2 in human cells is unknown, the fission yeast homolog Nrl1 interacts physically with an Mtr4-like protein, Mtl1 (Lee et al. 2013; Aronica et al. 2016), and is involved in suppression of R-loop formation (Aronica et al. 2016). ZFC3H1 is a large (∼230-kDa) protein localized in the nucleus and shown to modulate IL-8 transcription (Tomita et al. 2014). ZFC3H1 is the apparent homolog of the Schizosaccharomyces pombe protein Red1, which functions in a distinct Mtl1-containing complex and, significantly, plays essential roles in the degradation of various unstable RNAs (Lee et al. 2013; Egan et al. 2014; Zhou et al. 2015).
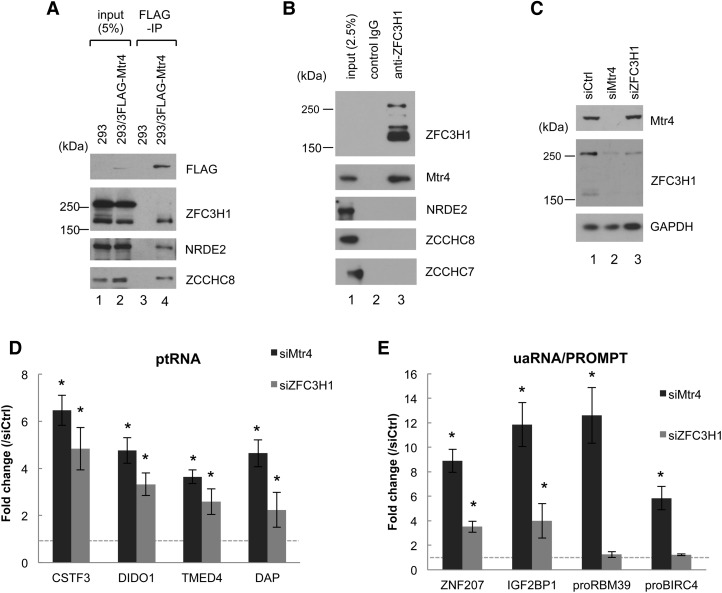
We next performed coimmunoprecipitation (co-IP) experiments to verify several of the interactions suggested by the above data. We validated RNase-resistant interactions of Mtr4 with ZFC3H1 (Fig. 3A,B), NRDE2, U5-40K, and hnRNP M (Supplemental Fig. S2). (Note that ZFC3H1 appears as two major bands of ∼250 and 150 kDa, both of which were decreased by ZFC3H1 knockdown [Figs. 3C, 6A]. The 150-kDa isoform was more efficiently immunoprecipitated with Mtr4 or ZFC3H1 antibodies [Fig. 3A], which may reflect limited epitope accessibility in the 250-kDa ZFC3H1–Mtr4 complex. The existence of a 250-kDa ZFC3H1–Mtr4 complex is supported by the observation that Mtr4 knockdown caused decreases in both the 250- and 150-kDa isoforms, likely reflecting protein destabilization [Figs. 3C, 6A]. The origin of the smaller species remains to be determined.) Importantly, neither TRAMP nor NEXT subunits were coimmunoprecipitated with ZFC3H1 or NRDE2 (Fig. 3B; Supplemental Fig. S2B,C), indicating that these proteins form complexes that are distinct from NEXT and TRAMP. In contrast, hnRNP M and U5-40K coimmunoprecipitated with ZCCHC7 and PAPD5 but not with ZCCHC8, suggesting that Mtr4 interacts with these proteins in the context of TRAMP (Supplemental Fig. S2A).
Figure 3.
Mtr4-associated ZFC3H1 is required for down-regulation of ptRNAs and uaRNAs but not NEXT substrates. (A) Cell extracts prepared from HEK293 cells and HEK293 cells stably expressing 3Flag-Mtr4 were used for immunoprecipitation with anti-Flag antibodies in the presence of benzonase and RNase A followed by Western blotting with the indicated antibodies. (B) Cell extracts prepared from HEK293 cells were used for co-IP experiments with anti-ZFC3H1 in the presence of benzonase and RNase A followed by Western blotting with antibodies against the proteins indicated at the right. (C) Western blot analysis of HeLa cell extracts after 72 h of knockdown treatment with the siRNAs indicated at the top; antibodies against the proteins are indicated at the right. (D,E) RT-qPCR analysis of the indicated ptRNAs (D) and the indicated uaRNAs and NEXT substrates proRBM39 and proBIRC4 (E) after the indicated siRNA transfections. Transcript levels were normalized to GAPDH mRNA, and the normalized levels in siCtrl-treated cells were set to 1. Bars represent mean ± SD. n = 3. Asterisks denote significant difference from siCtrl (P < 0.05) using an unpaired Student's t-test.
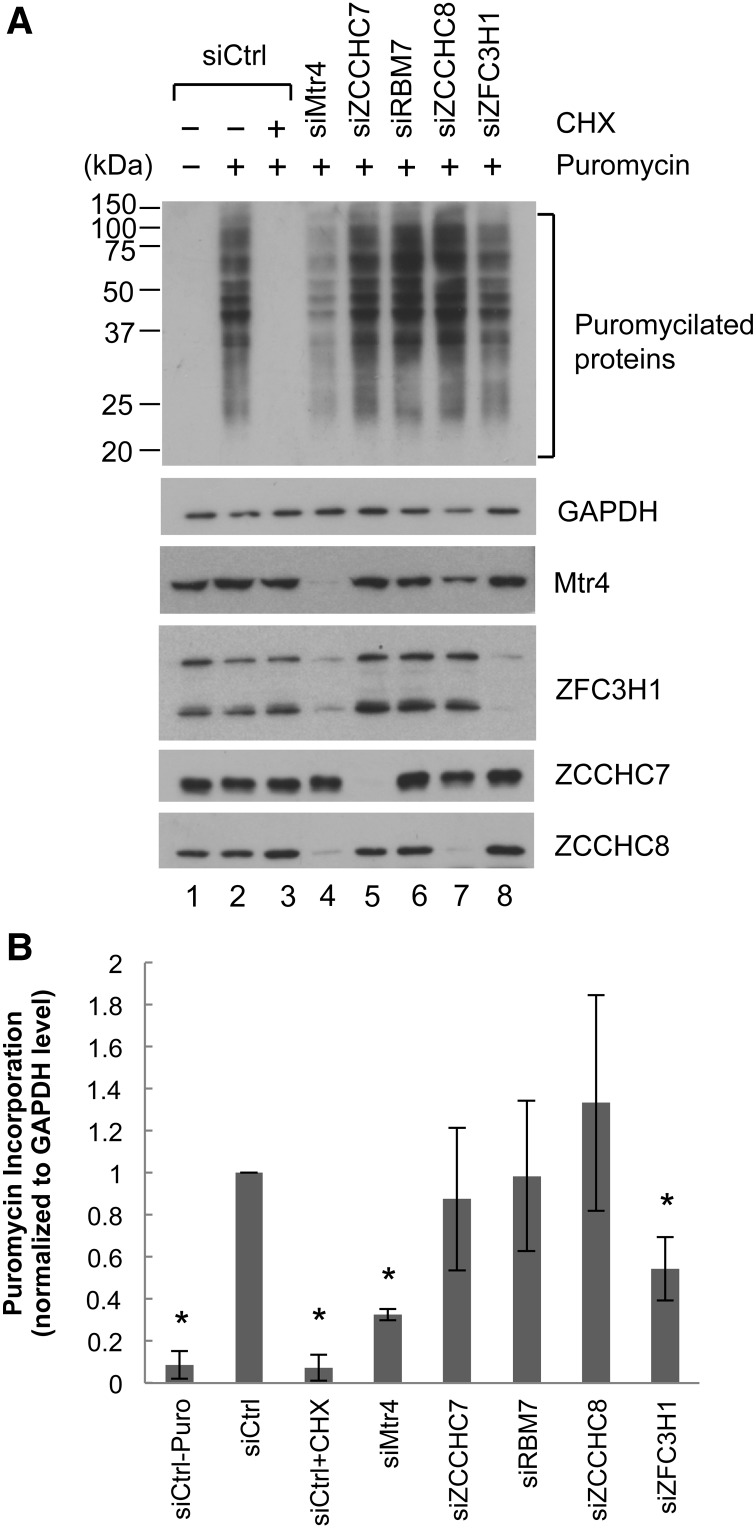
Figure 6.
Mtr4/ZFC3H1 depletion causes global reduction of translation. (A) Puromycin incorporation assay. HeLa cells transfected with the indicated siRNAs for 48 h were treated with 1 µg/mL puromycin for 30 min. (Lane 3) CHX treatment was performed 10 min prior to puromycin addition. Cell lysates were resolved by SDS-PAGE, and puromycilated proteins were detected using an anti-puromycin antibody. (B) Puromycin-incorporated protein levels as in A were quantitated using LI-COR Image Studio software and normalized by GAPDH levels. The normalized levels in lane 2 were set to 1. Bars represent mean ± SD. n = 3. Asterisks denote significant difference from lane 2 (P < 0.05) using an unpaired Student's t-test.
ZFC3H1 is required for repression of ptRNAs and uaRNAs but not NEXT substrates
Among the verified Mtr4-interacting partners described above, we decided to focus on ZFC3H1. (While this work was in progress, Meola et al. [2016] also identified ZFC3H1 as an Mtr4-interacting protein.) To determine whether ZFC3H1, like Mtr4, is required for ptRNA and uaRNA turnover, we depleted ZFC3H1 and evaluated the accumulation of these RNAs by RT-qPCR (Fig. 3C–E). Importantly, as with Mtr4 knockdown, both ptRNA and uaRNA levels were increased by ZFC3H1 knockdown (Fig. 3D,E). In contrast, ZFC3H1 knockdown, unlike NEXT knockdown, had no significant effect on the levels of two PROMPTs, proRBM39 and proBIRC4, indicating that ZFC3H1 and NEXT target distinct sets of RNA substrates (Fig. 3E).
Another protein that might be involved in pt/uaRNA degradation is the nuclear poly(A)-binding protein (PABPN1). PABPN1 interacts physically with the nuclear exosome to degrade subsets of polyadenylated lncRNA species (Beaulieu et al. 2012), and these targets include both ptRNAs (Li et al. 2015) and uaRNAs (Bresson et al. 2015; Li et al. 2015). Despite the absence of PABPN1 in our Flag-Mtr4 co-IP/MS (Fig. 2; Supplemental Table S1) and co-IP/Western blot analyses (Supplemental Fig. S3A,B), we next investigated whether PABPN1 functions similarly to Mtr4/ZFC3H1. RT-qPCR results indicate that Mtr4/ZFC3H1 and PABPN1 share RNA substrates (Fig. 3C–E; Supplemental Fig. S3C–E), although an exception was uaIGF2BP1, which was sensitive only to Mtr4/ZFC3H1 knockdown (Fig. 3E; Supplemental Fig. S3E). As with Mtr4/ZFC3H1 knockdown, PABPN1 knockdown had only minimal effect on proRBM39 and proBIRC4 levels (Supplemental Fig. S3E).
Depletion of Mtr4/ZFC3H1 causes cytoplasmic accumulation of ptRNAs and uaRNAs
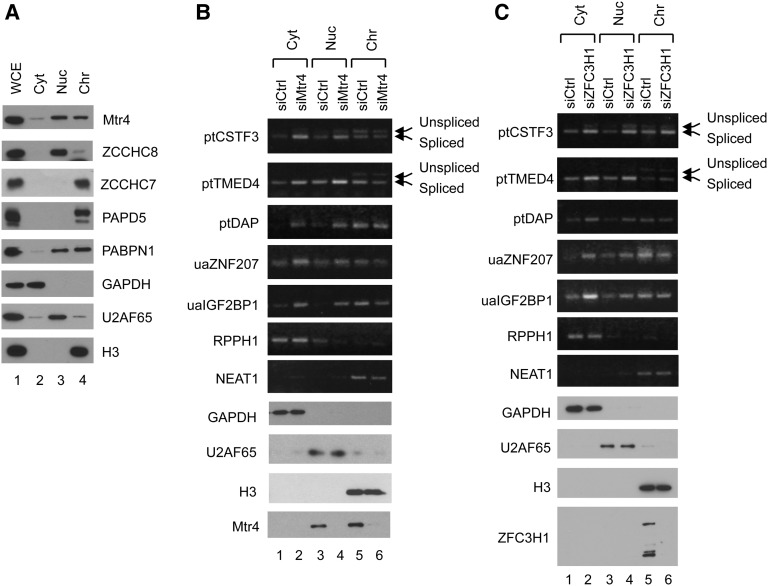
We next investigated the consequences of the increased accumulation of ptRNAs and uaRNAs caused by Mtr4/ZFC3H1 knockdown. Given that addition of a poly(A) tail can be sufficient to target mRNAs for nuclear export (Brodsky and Silver 2000; Fuke and Ohno 2008), we first investigated whether there were any changes in subcellular localization of these RNAs in knockdown cells. To this end, we separated cell compartments into cytoplasmic, nuclear-soluble, and chromatin fractions. The effectiveness of our fractionation protocol was verified by Western blotting: GAPDH, U2AF65, and histone H3 were detected predominantly in the cytoplasmic, nuclear-soluble, and chromatin fractions, respectively (Fig. 4A–C). Mtr4 was predominantly nuclear and evenly distributed in the nuclear-soluble and chromatin fractions (Fig. 4A,B), while ZFC3H1 was almost entirely in the chromatin fraction (Fig. 4C). NEXT subunit ZCCHC8 and TRAMP subunits ZCCHC7 and PAPD5 displayed different distributions; ZCCHC8 was largely nuclear-soluble, whereas ZCCHC7 and PAPD5 were exclusively in the chromatin fraction (Fig. 4A). PABPN1 was predominantly in the two nuclear fractions (Fig. 4A).
Figure 4.
Mtr4 knockdown causes cytoplasmic accumulation of stabilized ptRNAs and uaRNAs. (A) Western blotting of subcellular fractions prepared from HeLa cells. Proteins from whole-cell extract (WCE), cytoplasm (Cyt), nuclear-soluble (Nuc), and nuclear-insoluble chromatin (Chr) fractions were analyzed using antibodies directed against the proteins listed on the right. (B,C) Subcellular fractionation was performed after 72 h of siMtr4 (B) or siZFC3H1 (C) treatment, and total RNAs were isolated from each fraction, as indicated at the top. cDNA was synthesized using random or oligo(dT) primer, and the indicated transcripts (shown at the left) were analyzed by PCR. Gels were prestained with ethidium bromide (EtBr). RPPH1 and NEAT1 RNAs were amplified using random-primed RT products and served as cytoplasmic and nuclear-insoluble markers. Other RNAs were amplified using oligo(dT)-primed RT products.
To investigate ptRNA and uaRNA localization, we analyzed by RT–PCR RNA from fractions prepared as above from cells treated with Ctrl, Mtr4, or ZFC3H1 siRNAs (Fig. 4B,C). Specifically, RNA from the cytoplasmic (Fig. 4B,C, lanes 1,2), nuclear-soluble (Fig. 4B,C, lanes 3,4), and chromatin (Fig. 4B,C, lanes 5,6) fractions of knockdown cells was analyzed. Effective fractionation was verified by Western blotting of GAPDH, U2AF65, and histone H3 as above as well as by RT–PCR of a cytoplasmic lncRNA (RPPH1) and a nuclear-insoluble lncRNA (NEAT1). Moreover, unspliced forms of multiexonic ptRNAs (CSTF3 and TMED4) were enriched in the chromatin fraction, further demonstrating the validity of the fractionation. In siCtrl cells, all types of Mtr4/ZFC3H1 targets were most abundant in the chromatin fraction (Fig. 4B,C, cf. lanes 1, 3, and 5). This trend was most pronounced with the single-exonic DAP ptRNA as well as all uaRNAs, which were almost exclusively in the chromatin fraction, whereas multiexonic CSTF3 and TMED4 ptRNAs were detected in all three fractions. Most importantly, however, increased accumulation of all of the ptRNAs and uaRNAs was detected in cytoplasmic and nuclear-soluble fractions after Mtr4 or ZFC3H1 knockdown, while levels in the chromatin fraction were unchanged. These results imply that Mtr4/ZFC3H1 target transcripts, especially those with single exons, are degraded immediately after release from chromatin, and failure of this surveillance system results in significant accumulation of these RNAs in the cytoplasm.
Exported ptRNA and uaRNA associate with ribosomes
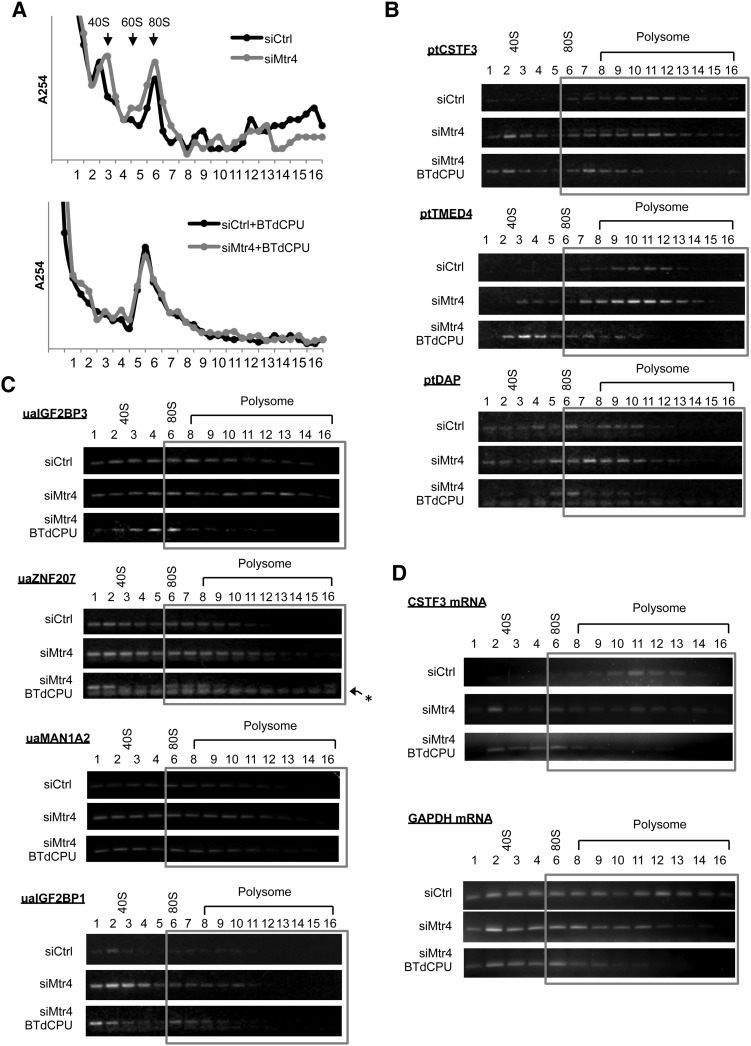
We next investigated the fate of the ua/ptRNAs that accumulate in the cytoplasm in the knockdown cells. Notably, all of these RNAs contain putative ORFs, and one possibility therefore is that they are bound by ribosomes and translated. This possibility is supported by the fact that ptRNAs and uaRNAs have very long, ∼300-nucleotide poly(A) tails, as determined by RL-PAT assays (Supplemental Fig. S4). To investigate the association of ptRNAs and uaRNAs with ribosomes, we first performed polysome fractionation by sedimenting cytoplasmic extracts prepared from siRNA-treated cells through 15%–45% sucrose gradients (Fig. 5A) and then evaluated the distribution of individual Mtr4 target transcripts by RT–PCR (Fig. 5B–D; quantitation in Supplemental Fig. S5). In Ctrl siRNA-treated cells, CSTF3 and TMED4 ptRNAs were detected mainly in polysomes, while the DAP ptRNA was found in monosomes to light polysomes (Fig. 5B). All four uaRNAs analyzed (ZNF207, IGF2BP1, IGF2BP3, and MAN1A2), although present at only low levels in the cytoplasm, were distributed between free cytosolic to light polysome fractions (Fig. 5C). While Mtr4 knockdown in general did not cause significant changes in the distribution of these RNAs, all of the ptRNAs and uaRNAs that accumulated in the cytoplasm associated with ribosomes, and the absolute amount of ribosome-bound RNA thus increased in all cases. We also analyzed the polysome profile of two full-length mRNAs (GAPDH and CSTF3), and, in contrast to the ptRNAs and uaRNAs, both shifted to lighter fractions following Mtr4 knockdown (Fig. 5D).
Figure 5.
Cytoplasmic ptRNAs and uaRNAs associate with active ribosomes but lead to reduced global translation. (A) UV absorption profiles at 254 nm of 15%–45% sucrose gradients. HeLa cells were transfected with either control siRNA (siCtrl) or Mtr4 siRNA (siMtr4) for 48 h, and cytoplasmic extracts were prepared from cells with or without 50 µM/mL BTdCPU for 3 h. (B–D) RNAs extracted from each fraction as in A were used for oligo(dT)-primed cDNA synthesis, and the indicated transcripts were analyzed by RT–PCR. Gels were prestained with EtBr. Ribosome/polysome-associated fractions are highlighted with a gray box. An asterisk marks primer dimers.
We next investigated whether the ptRNAs and uaRNAs were indeed associated with active ribosomes. To this end, we treated cells with the eIF2–GTP–tRNAiMet ternary complex inhibitor BTdCPU, which blocks formation of the preinitiation complex (Chen et al. 2011), and subjected cell extracts to sucrose gradient analysis as above. UV absorption profiles showed a sharp inhibition of translation after 3 h of BTdCPU treatment (Fig. 5A; Supplemental Fig. S6A, +BTdCPU). RT–PCR analysis of the individual Mtr4 target RNAs revealed in all cases a shift of peak positions from heavy to light fractions following BTdCPU treatment (Fig. 5B–D), providing evidence that the RNAs were associated with active polysomes. Together, our results indicate that Mtr4 target transcripts that escaped RNA surveillance in the nucleus and were exported to the cytoplasm were then bound by ribosomes and likely translated.
Despite the increased association of uaRNAs and ptRNAs with ribosomes in the Mtr4 knockdown cells, we detected an unexpected decrease in polysomes. Specifically, analysis of the UV absorption profiles revealed that 48 h of Mtr4 siRNA treatment caused a reduction in polysomes, especially in heavier polysomes (Fig. 5A; Supplemental Fig. S6A). Notably, this is consistent with the behavior of the two full-length mRNAs analyzed (see above). An even more robust reduction in polysomes was evident after 72 h of Mtr4 knockdown, again especially notable in the heavy polysome fractions (Supplemental Fig. S6B). A second Mtr4 siRNA gave similar results (Supplemental Fig. S6C). Also of note, although Mtr4-depleted cells showed reduced growth and morphological changes after 72 h, ptRNA and uaRNA levels continued to increase even after 96 h of knockdown (Supplemental Fig. S6D). These findings together indicate that normally unstable, nuclear, and hence untranslated lncRNAs associate with active ribosomes following Mtr4 knockdown, but, paradoxically, this correlates with an overall reduction in polysomes and hence, very likely, translation.
To verify that Mtr4 knockdown indeed led to a global reduction of translation and determine whether ZFC3H1 knockdown might have similar effects, we performed puromycin incorporation assays. Puromycin is a chain terminator that is incorporated into growing nascent polypeptide chains and thus can be used to label nascent polypeptides (Schmidt et al. 2009). We treated cells with low concentrations of puromycin (1 µg/mL), and translation efficiency was evaluated by detecting puromycilated nascent chains using anti-puromycin antibodies for Western blotting of whole-cell lysates (Fig. 6A, quantitation in B). Puromycin incorporation was completely blocked by pretreatment with cycloheximide (CHX) (Fig. 6A, lane 3), confirming that this method allowed us to analyze newly synthesized proteins. In agreement with the polysome fractionation data, efficiency of puromycin incorporation was substantially lower after Mtr4 or ZFC3H1 knockdown (Fig. 6A, cf. lanes 2 and 4,8). Note that siZFC3H1 treatment was less toxic than siMtr4: We observed only a small reduction in cell proliferation and slight morphological changes. Importantly, depletion of NEXT and TRAMP subunits RBM7, ZCCHC8, and ZCCHC7 did not cause a significant decrease in puromycin incorporation (Fig. 6A, lanes 5–7).
Next, we wished to address the possibility that the reduction in polysomes and translation in the Mtr4/ZFC3H1 knockdown cells might reflect another function of the proteins. Specifically, aberrant unprocessed pre-rRNAs have been observed to accumulate in Mtr4-compromised cells (Schilders et al. 2007; Tafforeau et al. 2013). We also detected pre-rRNAs by Northern blotting using probes hybridizing to 3′ extended sequences of 5.8S (ITS2) and 18S (ITS1) rRNAs in Mtr4-depleted cells (Supplemental Fig. S7A). However, Northern blotting using probes that detect both mature and unprocessed rRNAs revealed that the amount of unprocessed rRNAs relative to the mature species was extremely low: The unprocessed pre-rRNAs were detectable only when blots were overexposed such that signals for mature rRNAs were saturated and no longer in a quantitative range (Supplemental Fig. S7B, 5.8S and 18S). In addition, there were no detectable effects on the levels of mature rRNAs. Moreover, ZFC3H1 knockdown, which also caused reduced translation, did not lead to accumulation of aberrant pre-rRNAs or decrease of mature rRNAs (Supplemental Fig. S7B). These findings and other results discussed below argue strongly that the effects of Mtr4/ZFC3H1 knockdown on translation are not due to defects in ribosome assembly.
Together, our experiments revealed an unexpected role for Mtr4–ZFC3H1 in preventing global disruption of mRNA association with polysomes and subsequent translation.
Discussion
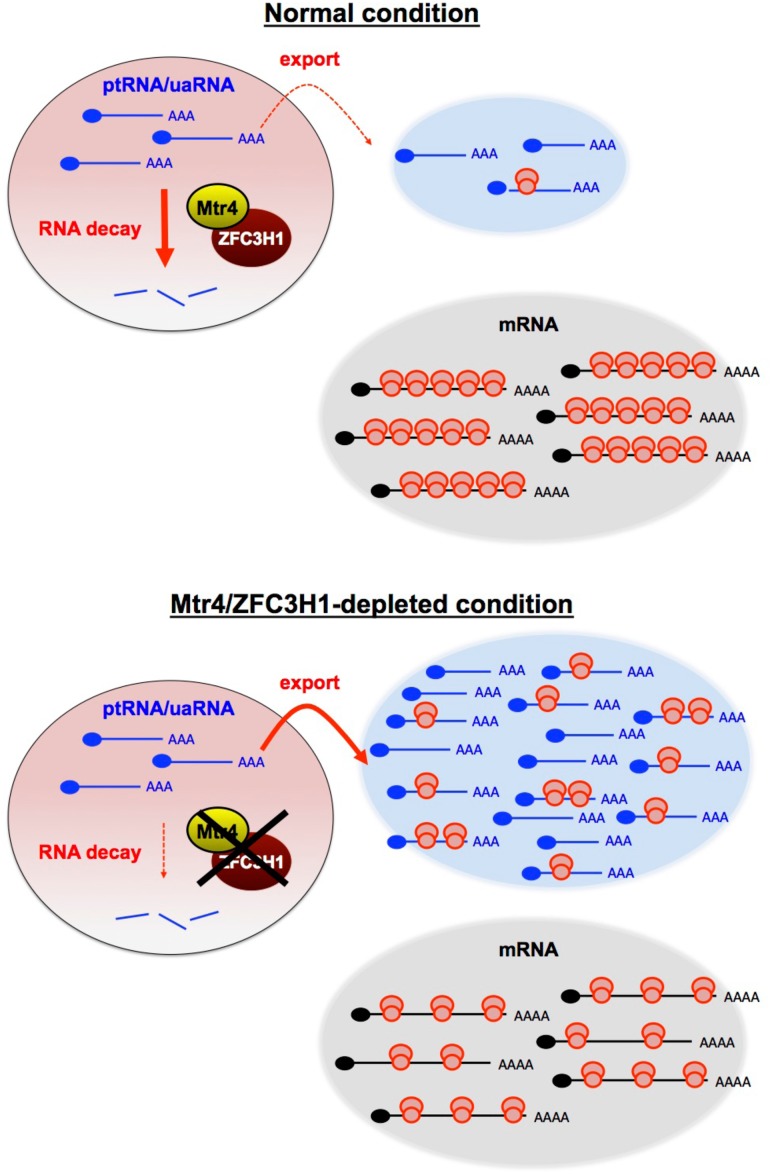
It is becoming increasingly clear that a much larger fraction of the human genome is transcribed than previously appreciated. Much of this “non-mRNA” transcription is by RNA polymerase II, and the RNAs produced are frequently subject to the processing reactions that typically give rise to mRNAs, such as splicing and polyadenylation (Jensen et al. 2013). However, unlike mRNAs, these RNAs are often retained in the nucleus and rapidly turned over. While some of the key factors in this process are known, such as the nuclear exosome and NEXT complex (Lubas et al. 2011; Meola et al. 2016; for review, see Zinder and Lima 2017), it is not well understood how and whether the degradation of these RNAs is coordinated and what the consequences might be if their turnover is prevented. In this study, we identified Mtr4 together with ZFC3H1 as a potential “master regulator” of polyadenylated lncRNA metabolism and showed that when its activity is reduced, normally unstable lncRNAs accumulate and are transported to the cytoplasm, where they appear to “swamp” ribosomes and thereby inhibit translation globally (Fig. 7). Based on these properties, we refer to the Mtr4/ZFC3H1 complex as the “polysome protector complex” (PPC).
Figure 7.
Model for the role of the Mtr4/ZFC3H1 complex in the turnover of nuclear polyadenylated transcripts and how its loss affects translation. Model depicting the impact of PPC deficiency on polyadenylated transcriptomes and global translation. Loss of the PPC results in stabilization of ptRNAs and uaRNAs, which are normally rapidly degraded in the nucleus, and these RNAs are then transported to the cytoplasm. The exported RNAs become ribosome-associated and overwhelm the translational machinery, which leads to disruption of the quantitative balance between available ribosomes and translatable RNAs. See the text for details.
ptRNAs and uaRNAs are targeted for turnover by the PPC and not by either of the other characterized Mtr4-containing complexes—NEXT or TRAMP. A large majority of these RNAs contain a PAS at their 3′ end, implying that the canonical or very similar pre-mRNA 3′ processing machinery is used for their polyadenylation. However, in contrast to mRNAs (which are generally more stable, efficiently exported to the cytoplasm, and translated), uaRNAs and ptRNAs as well as many other lncRNAs are typically rapidly degraded in the nucleus (Andersson et al. 2014b; Li et al. 2015; Schlackow et al. 2017). While neither NEXT nor TRAMP is required for ptRNA and uaRNA turnover, the nuclear exosome is. Thus, the PPC is distinct from NEXT and TRAMP, suggesting that these complexes target distinct sets of RNA substrates for degradation by the exosome. Importantly, though, only the PPC prevents accumulation and transport of lncRNAs from the nucleus that is sufficient to disrupt normal translation, as knockdown of NEXT/TRAMP subunits did not detectably affect translation.
While this work was in preparation, Meola et al. (2016) reported that Mtr4 and ZFC3H1 together with PABPN1 form a complex that preferentially degrades polyadenylated lncRNAs such as snoRNA host gene (SNHG) transcripts. They also showed that ZFC3H1 and PABPN1 knockdown resulted in the accumulation of subsets of uaRNAs and eRNAs, There are similarities as well as differences between our results and those of Meola et al. (2016). For example, in addition to uaRNAs, we identified ptRNAs as Mtr4/ZFC3H1 substrates and, notably, found that these RNAs had very long poly(A) tails and were not substrates for NEXT-mediated degradation. Also, while we did not detect an interaction between PABPN1 and Mtr4/ZFC3H1, we did observe the accumulation of most, but not all, ptRNAs and uaRNAs tested following PABPN1 knockdown. Finally and most importantly, we demonstrated that an important function of Mtr4/ZFC3H1 involves the maintenance of polysome integrity by preventing the accumulation of polyadenylated lncRNAs in the cytoplasm.
The fact that Mtr4 participates in multiple distinct complexes is reminiscent of Mtr4 proteins in fission yeast. S. pombe has two Mtr4 paralogs: Mtr4 and Mtl1. Mtr4 is a TRAMP component (Zhang et al. 2011), whereas Mtl1 forms a core complex with Red1, called MTREC (Mtl1–Red1 core) or NURS (nuclear RNA silencing) (Lee et al. 2013; Egan et al. 2014; Zhou et al. 2015). As mentioned above, Red1 is the fission yeast homolog of ZFC3H1 and is essential for exosomal decay of various RNAs, including CUTs (cryptic unstable transcripts), which are similar to uaRNAs and PROMPTs in mammals; meiotic mRNAs; and unspliced pre-mRNAs (Lee et al. 2013; Egan et al. 2014; Zhou et al. 2015). Specificity for RNA targeting by MTREC/NURS is determined by at least three distinct submodule complexes (Zhou et al. 2015). Further studies are required to determine whether the PPC also uses associated proteins to distinguish specific RNA substrates.
The global reduction of translation that we observed in PPC-depleted cells highlights the importance of rapidly degrading naturally unstable nuclear lncRNAs. Since failure of RNA surveillance by PPC depletion leads to increased accumulation of cytoplasmic polyadenylated RNAs that have the ability to recruit ribosomes, we propose that heavy polysome formation on mRNAs is hampered by “dilution of ribosomes” by the accumulated normally unstable nuclear “noncoding” RNAs in the cytoplasm (Fig. 7). Translation or ribosome binding of lncRNAs, which contain small ORFs (sORFs) and thus possibly produce micropeptides (Slavoff et al. 2013; Ruiz-Orera et al. 2014; Raj et al. 2016) has been reported in multiple species, including yeast (Ingolia et al. 2014; Smith et al. 2014), fruit flies (Dunn et al. 2013; Aspden et al. 2014), zebrafish (Chew et al. 2013; Bazzini et al. 2014), and mammals (Chew et al. 2013; Zhou et al. 2013; Ingolia et al. 2014; van Heesch et al. 2014; Fields et al. 2015; Ji et al. 2015; Calviello et al. 2016; for review, see Ingolia 2016). Moreover, a recent study provided evidence that a vast majority of monosomes is actively elongating and translating sORFs (Heyer and Moore 2016). Thus, ribosome binding/translation of some lncRNAs occurs and appears compatible with efficient cellular protein synthesis. However, our data provide evidence that when such RNAs are globally stabilized and accumulate in the cytoplasm, they “overwhelm” the system, and translation of mRNAs is repressed (Fig. 7). The fact that Mtr4/ZFC3H1 substrates have long poly(A) tails, which is in contrast to NEXT targets that largely lack poly(A) tails (Meola et al. 2016), may help explain their efficient ribosome association (Peng et al. 2008; Park et al. 2016). It is noteworthy that although eRNAs are largely nonpolyadenylated (Djebali et al. 2012; Andersson et al. 2014a), we recently identified a class of poly(A)+ eRNAs that are stabilized by Mtr4/ZFC3H1 knockdown. These eRNAs can also be transported to the cytoplasm and associate with ribosomes (K Ogami, Y Chen, and JL Manley, unpubl.), increasing the pool of lncRNAs that require surveillance by the PPC. Our results thus highlight how critical it is that such lncRNAs be degraded rapidly in the nucleus because, if they survive surveillance by the PPC, they become toxic.
Could another function of Mtr4 or ZFC3H1 be responsible for the disruption of translation that we observed? As noted above, Mtr4 is known to function in the maturation of 5.8S and 18S rRNA from cleaved rRNA precursors (de la Cruz et al. 1998; Schilders et al. 2007; Tafforeau et al. 2013). Might defects in rRNA processing contribute to reduced translation in Mtr4-deficient cells? We consider this unlikely for several reasons: First, since mature rRNAs are abundant and very stable, with half-lives that are days long (Yi et al. 1999; Defoiche et al. 2009), it would be unlikely that mature rRNA levels decrease sufficiently to affect ribosome levels and perturb translation. Our finding that the amount of unprocessed pre-rRNA that accumulated following Mtr4 knockdown was extremely small and that levels of 5.8S and 18S rRNAs were essentially unaffected is consistent with this. Second, while ZFC3H1 knockdown was shown previously to result in reductions in 47S and 45S pre-rRNAs (Tafforeau et al. 2013), our data showed no changes in downstream pre-rRNA and mature rRNA levels following ZFC3H1 knockdown. Third, unprocessed pre-5.8S rRNA in fact assembles into 60S ribosomes (Briggs et al. 1998), and the resulting immature 60S particles engage in apparently normal translation (Rodriguez-Galan et al. 2015). Indeed, polysome disassembly has not been observed in yeast or mammals under conditions that allow accumulation of aberrant pre-rRNA (Briggs et al. 1998; Strezoska et al. 2000). It is thus unlikely that defective pre-rRNA processing is responsible for the impaired polysome formation/translation that we observed in Mtr4/ZFC3H1 knockdown cells. Indeed, the fact that the excess uaRNAs and ptRNAs that accumulated in Mtr4 knockdown cells associated with active ribosomes further argues against this possibility. Mtr4-depleted cells can also accumulate mature and 3′ extended snRNA (Hrossova et al. 2015; Lubas et al. 2015) and pri-miRNA 5′ by-products (Dorweiler et al. 2014; Lubas et al. 2015). However, accumulation of these transcripts does not contribute to the decreased translation that we observed, since these RNAs are NEXT substrates (Hrossova et al. 2015; Lubas et al. 2015; K Ogami, Y Chen, M Hoque, W Li, B Tian, and JL Manley, unpubl.), and NEXT subunit knockdown had no effect on protein synthesis.
Our model implies that ribosomes must not be present in significant excess or they would otherwise be able to handle the increase in substrates produced when the PPC is depleted. Indeed, studies in yeast have suggested that ribosomes in fact are limiting for translation (Chu and von der Haar 2012; Shah et al. 2013). A similar situation likely exists in human cells. A good example is virus infection. In infected cells, there is often a competition between viral and cellular RNAs for limiting translation components. To overcome this, some viruses alter the balance of viral and cellular mRNA availability for translation by decreasing cytoplasmic cellular mRNA levels by stimulation of mRNA turnover or inhibition of mRNA export (for review, see Walsh and Mohr 2011). For example, herpes simplex virus 1 expresses the endonuclease virion host shutoff (vhs) to accelerate cellular mRNA decay, thereby preventing mRNA overload in infected cells (Dauber et al. 2014). Viral mRNAs associated with polysomes dramatically decrease in the absence of vhs, indicating that the total amount of translatable RNA needs to be regulated to ensure optimal translation of viral mRNAs.
In conclusion, we identified a complex containing the RNA helicase Mtr4 and the zinc finger protein ZFC3H1. This complex, dubbed the PPC, functions in nuclear surveillance of certain unstable polyadenylated lncRNAs to prevent their accumulation, export to the cytoplasm, and consequent disruption of protein synthesis. Our findings are significant because they provide an explanation of why so many lncRNAs are degraded in the nucleus essentially as soon as they are synthesized, as they otherwise have the potential to escape from the nucleus and overwhelm the cell's translational capacity. While it will be important in the future to learn more about the PPC (e.g., precisely how it functions and whether it can be regulated), our results have uncovered a new and unexpected function for nuclear RNA surveillance.
Materials and methods
Primers and siRNAs
All primers and siRNAs used in this study are listed in Supplemental Table S2. siRNAs against Rrp6 and Dis3 were described previously (Richard et al. 2013; Di Giammartino et al. 2014). siMtr4 s223606 was obtained from Thermo Fischer Scientific.
Cell culture and siRNA transfections
HeLa cells were cultured in DMEM supplemented with 10% FBS. The siRNAs were transfected using DharmaFECT 1 (GE Dharmacon) at 20 nM and maintained for 48, 72, or 96 h as indicated. To maintain high knockdown efficiency after 96 h, the siRNA transfection was repeated 48 h after the first transfection with half the amount of siRNA, and cells were harvested 48 h after the second transfection.
Antibodies
Mtr4 (NB100-1574), ZCCHC8 (NB100-94995), ZCCHC7 (NBP1-89175), Dis3 (H00022894-B01P), and Rrp6 (NBP1-32870) antibodies were from Novus Biologicals. ZFC3H1 (Bethyl Laboratories, A301-456A), hnRNP M (Bethyl Laboratories, A303-910A), NRDE2 (Proteintech, 24968-1-AP), U2AF65 (Sigma, U4758), histone H3 (Abcam,ab1791), and puromycin (Kerafast, 3RH11) were also used in this study. PAPD5 antibody was a generous gift from Dr. Shin-ichi Hoshino (Ogami et al. 2013).
RT-qPCR
Total RNA was isolated using TRIzol according to the manufacturer's instructions followed by DNase I digestion for 30 min at 37°C. Two micrograms of DNase I-treated RNA was reverse-transcribed with oligo(dT) primer using Maxima RT. Reactions were diluted 15 times in water, and qPCR was performed with the primers listed in Supplemental Table S1 and Power SYBR using StepOnePlus (Applied Biosystems). All data were normalized to GAPDH mRNA levels. Reagents for RT-qPCR were all from Thermo Fischer Scientific.
3′READS
Total RNA was purified from control and siRNA-treated cells. RNA integrity was analyzed by using Agilent Bioanalyzer. Samples with an RNA integrity number (RIN) >9.0 were subjected to 3′READS analysis following the protocol described in Hoque et al. (2014). Briefly, after RNA fragmentation, poly(A)+ RNA fragments were captured on magnetic beads coated with a chimeric oligonucleotide (oligo CU5T45), which contained 45 thymidines at the 5′ portion and five uridines at the 3′ portion, and subjected to RNase H digestion, which removed the bulk of the poly(A) tail and eluted RNA from beads. Eluted RNA was ligated to 5′ and 3′ adapters followed by reverse transcription, PCR amplification, and deep sequencing on an Illumina platform. 3′READS data were analyzed as described (Li et al. 2015).
Gel filtration
HEK293 or HEK293/3Flag-Mtr4 cells from 10 10-cm dishes were washed twice with PBS and lysed in lysis buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1× protease inhibitor cocktail [Biotools], 100 µg/mL RNase A) for 10 min at room temperature and 15 min on ice. Lysates were sonicated and cleared by centrifugation at 15,000 rpm in an Eppendorf centrifuge 5424 for 20 min at 4°C. The supernatants were filtered using Spin-X 0.45 µM cellulose acetate membrane (Sigma) at 16,000g for 10 min at 4°C prior to applying to a Superose 6 column. Gel filtration was performed in FPLC buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.5% NP-40) with a flow rate of 0.15 mL/min. Eluates were collected every 5 min.
MS analysis
Pooled FPLC fractions 26–33 (pool A), 34–42 (pool B), and 43–50 (pool C) were mixed with anti-Flag M2 magnetic beads for 2 h. Beads were then washed three times with wash buffer (20 mM Tris-HCl at pH 8.0, 300 mM NaCl, 0.5% NP-40, 0.5 mM EDTA). Proteins remaining on the resin were eluted using 100 µL of 150 µg/mL 3Flag peptide (ApexBio) three times and precipitated in 23% TCA and washed with cold acetone. Proteins were reduced with 5 mM Tris (2-carboxyethyl) phosphine hydrochloride (Sigma) and alkylated with 55 mM 2-chloroacetamide (Fluka Analytical). Proteins were digested for 18 h at 37°C in 2 M urea, 100 mM Tris (pH 8.5), and 1 mM CaCl2 with 2 µg of trypsin (Promega). Multidimensional protein identification technology (MudPIT) analysis was performed using an Eksigent nanoLC pump and a Thermo LTQ-Orbitrap using an in-house built electrospray stage (Wolters et al. 2001).
Protein and peptide identification and protein quantitation were done with Integrated Proteomics Pipeline (IP2; Integrated Proteomics Applications, Inc., http://www.integratedproteomics.com). Tandem mass spectra were extracted from raw files using RawConverter (He et al. 2015) and were searched against a UniProt human database with reversed sequences using ProLuCID (Peng et al. 2003; Xu et al. 2015). The search space included all fully tryptic and half-tryptic peptide candidates. Peptide candidates were filtered using DTASelect with the following parameters: -p 2 -y 1 --trypstat --extra --pI -DM 10 --DB --dm -in --brief --quiet (Tabb et al. 2002).
Co-IP
HEK293 or HEK293/3Flag-Mtr4 cells grown in two 10-cm dishes were washed twice with PBS and lysed in lysis buffer (20 mM Tris-HCl at pH 8.0, 300 mM NaCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5% NP-40, 1× protease inhibitor cocktail, 10 µg/mL RNase A, >250 U/mL benzonase) for 20 min on ice. The lysates were sonicated and then centrifuged in an Eppendorf centrifuge 5424 at 15,000 rpm for 20 min at 4°C, and supernatants were rotated with either anti-Flag M2 magnetic beads (Sigma) for 1 h or protein G beads (GE healthcare) in the presence of antibodies for 4 h. The beads were then washed three times with lysis buffer, and proteins retained on the resin were subjected to Western blot analysis.
Subcellular fractionation
Seventy-two hours after siRNA transfection, HeLa cells grown in a 10-cm dish were washed twice with PBS and collected by scraping and centrifugation. Cell pellets were resuspended in 400 µL of swelling buffer (10 mM Tris-HCl at pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 5 U of RNasin, 1× protease inhibitor cocktail) and incubated for 15 min on ice. Cells were homogenized by passing a 26-gauge needle attached to a 1-mL syringe until >90% of cells were disrupted (typically 10∼20 strokes). Half of the lysate was kept in a new tube and used as whole-cell lysate. The rest of the 200 µL of lysate was mixed with 2 µL of 10% NP-40, gently tapped, and immediately centrifuged in an Eppendorf centrifuge 5424 at 6,000 rpm for 5 min. The supernatant was kept in a new tube and used as cytoplasmic fraction. The pellet was washed once with swelling buffer, resuspended in 100 µL of glycerol buffer (20 mM Tris-HCl at pH 8.0, 75 mM NaCl, 0.5 mM EDTA, 50% glycerol, 0.85 mM DTT, 5 U of RNasin, 1× protease inhibitor cocktail) by pipetting, and then mixed with 100 µL of nucleus lysis buffer (20 mM HEPES-NaOH at pH 7.6, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1 M urea, 1% NP-40, 1 mM DTT, 5 U of RNasin, 1× protease inhibitor cocktail). The mixture was pulse-vortexed three times, incubated for 1 min on ice, and then centrifuged in an Eppendorf centrifuge 5424 at 14,000 rpm for 2 min. The supernatant was used as nuclear-soluble fraction. The pellet was washed once with a 1:1 mixture of glycerol/nucleus lysis buffer and then resuspended in 200 µL of water. RNA was extracted using TRIzol, DNase I-treated, and then reverse-transcribed with oligo(dT) or random primer using Maxima RT.
Polysome fractionation
HeLa cells were transfected with either siCtrl or siMtr4 and maintained in the same medium for the indicated times in five 10-cm dishes. On the day of harvest (∼80% confluency), cells were treated with either 50 µM BTdCPU or 100 µg/mL CHX for 3 h and 5 min, respectively, at 37°C. Cells were washed once with ice-cold PBS containing either 50 µM BTdCPU (Millipore) or 100 µg/mL CHX and then resuspended in 400 µL of polysome lysis buffer (20 mM HEPES-NaOH at pH 7.6, 150 mM NaCl, 15 mM MgCl2, 0.5% NP-40, 80 U RNasin, 1× protease inhibitor cocktail) containing either 50 µM BTdCPU or 100 µg/mL CHX. After 10 min of incubation on ice, the lysates were centrifuged at 3000 rpm for 5 min, and the supernatant was centrifuged at 15,000 rpm for 5 min in a new tube. The supernatant was loaded onto 15%–45% sucrose gradients (20 mM HEPES-NaOH at pH 7.6, 150 mM NaCl, 5 mM MgCl2, 100 µg/mL CHX) followed by centrifugation at 39,000 rpm for 90 min using an SW41Ti rotor. Fractions (200 µL each) were manually collected, and A254 was determined using a Nanodrop spectrophotometer. RNA was extracted by mixing with 1 vol of TRIzol, isopropanol-precipitated in the presence of GeneElute-LPA (Sigma), and then reverse-transcribed with oligo(dT) primer using Maxima RT. To avoid efficiency differences in RT reactions, RNA amounts were equalized by adding purified yeast RNA (Thermo Fisher Scientific). No PCR products were generated when yeast RNA RT products alone were amplified with the primers used in this study.
Puromycin incorporation assay
siRNA-transfected HeLa cells (∼50% confluent) were treated with 1 µg/mL puromycin for 30 min to label nascent polypeptides. CHX treatment was done at 10 µg/mL 10 min prior to adding puromycin. Puromycilated proteins were resolved by SDS-PAGE and detected by Western blotting using anti-puromycin antibody.
Supplementary Material
Acknowledgments
We are grateful to Shin-ichi Hoshino (Nagoya City University) for sharing the PAPD5 antibody. We also thank Rachel Giesler (Novus Biologicals) for the Mtr4, ZCCHC8, and ZCCHC7 antibodies. This work was supported by National Institutes of Health grants R01 GM28983 and R35 GM118136 to J.L.M., and R01 GM084089 to B.T. J.J.M. and J.R.Y. were supported by the National Center for Research Resources (5P41RR011823).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.302604.117.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014a. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Refsing Andersen P, Valen E, Core LJ, Bornholdt J, Boyd M, Jensen TH, Sandelin A. 2014b. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat Commun 5: 5336. [DOI] [PubMed] [Google Scholar]
- Aronica L, Kasparek T, Ruchman D, Marquez Y, Cipak L, Cipakova I, Anrather D, Mikolaskova B, Radtke M, Sarkar S, et al. 2016. The spliceosome-associated protein Nrl1 suppresses homologous recombination-dependent R-loop formation in fission yeast. Nucleic Acids Res 44: 1703–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M, Couso JP. 2014. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife 3: e03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet 8: e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J, Patterson DN, Wilson GM, Toth EA. 2008. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′ → 5′ helicase partner of the nuclear exosome. J Biol Chem 283: 4930–4942. [DOI] [PubMed] [Google Scholar]
- Bresson SM, Hunter OV, Hunter AC, Conrad NK. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet 11: e1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. 2016. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13: 165–170. [DOI] [PubMed] [Google Scholar]
- Chen LL. 2016. Linking long noncoding RNA localization and function. Trends Biochem Sci 41: 761–772. [DOI] [PubMed] [Google Scholar]
- Chen T, Ozel D, Qiao Y, Harbinski F, Chen L, Denoyelle S, He X, Zvereva N, Supko JG, Chorev M, et al. 2011. Chemical genetics identify eIF2α kinase heme-regulated inhibitor as an anticancer target. Nat Chem Biol 7: 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. 2013. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, von der Haar T. 2012. The architecture of eukaryotic translation. Nucleic Acids Res 40: 10098–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Saffran HA, Smiley JR. 2014. The herpes simplex virus 1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload. J Virol 88: 9624–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoiche J, Zhang Y, Lagneaux L, Pettengell R, Hegedus A, Willems L, Macallan DC. 2009. Measurement of ribosomal RNA turnover in vivo by use of deuterium-labeled glucose. Clin Chem 55: 1824–1833. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17: 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Li W, Ogami K, Yashinskie JJ, Hoque M, Tian B, Manley JL. 2014. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes Dev 28: 2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler JE, Ni T, Zhu J, Munroe SH, Anderson JT. 2014. Certain adenylated non-coding RNAs, including 5′ leader sequences of primary microRNA transcripts, accumulate in mouse cells following depletion of the RNA helicase MTR4. PLoS One 9: e99430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. 2014. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 20: 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. 2011. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci 108: 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke H, Ohno M. 2008. Role of poly(A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res 36: 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, Yates JR III. 2015. Extracting accurate precursor information for tandem mass spectra by RawConverter. Anal Chem 87: 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ. 2016. Redefining the translational status of 80S monosomes. Cell 164: 757–769. [DOI] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Li W, Tian B. 2014. Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods Mol Biol 1125: 119–129. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. [DOI] [PubMed] [Google Scholar]
- Hrossova D, Sikorsky T, Potesil D, Bartosovic M, Pasulka J, Zdrahal Z, Stefl R, Vanacova S. 2015. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3′-end extended forms of snRNAs. Nucleic Acids Res 43: 4236–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 165: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D. 2013. Dealing with pervasive transcription. Mol Cell 52: 473–484. [DOI] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, Struhl K. 2015. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4: e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, Balachandran V, Dhakshnamoorthy J, Taneja N, Yamanaka S, Zhou M, et al. 2013. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11: e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637. [DOI] [PubMed] [Google Scholar]
- Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, Jensen TH. 2015. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep 10: 178–192. [DOI] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen Jens S, Sandelin A, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell 64: 520–533. [DOI] [PubMed] [Google Scholar]
- Nag A, Steitz JA. 2012. Tri-snRNP-associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre-mRNA splicing. RNA Biol 9: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20: 923–928. [DOI] [PubMed] [Google Scholar]
- Ogami K, Cho R, Hoshino S. 2013. Molecular cloning and characterization of a novel isoform of the non-canonical poly(A) polymerase PAPD7. Biochem Biophys Res Commun 432: 135–140. [DOI] [PubMed] [Google Scholar]
- Park JE, Yi H, Kim Y, Chang H, Kim VN. 2016. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell 62: 462–471. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2: 43–50. [DOI] [PubMed] [Google Scholar]
- Peng J, Murray EL, Schoenberg DR. 2008. In vivo and in vitro analysis of poly(A) length effects on mRNA translation. Methods Mol Biol 419: 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Raj A, Wang SH, Shim H, Harpak A, Li YI, Engelmann B, Stephens M, Gilad Y, Pritchard JK. 2016. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife 5: e13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. 2013. How bidirectional becomes unidirectional. Nat Struct Mol Biol 20: 1022–1024. [DOI] [PubMed] [Google Scholar]
- Richard P, Feng S, Manley JL. 2013. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev 27: 2227–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Galan O, Garcia-Gomez JJ, Kressler D, de la Cruz J. 2015. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol 12: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. 2014. Long non-coding RNAs as a source of new peptides. Elife 3: e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJ. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res 35: 2564–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277. [DOI] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153: 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, Baker KE. 2014. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep 7: 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol 20: 5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell 51: 539–551. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Ieguchi K, Coin F, Kato Y, Kikuchi H, Oshima Y, Kurata S, Maru Y. 2014. ZFC3H1, a zinc finger protein, modulates IL-8 transcription by binding with celastramycin A, a potential immune suppressor. PLoS One 9: e108957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. 2014. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 15: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9: 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT. 2008. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, Yates JR III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73: 5683–5690. [DOI] [PubMed] [Google Scholar]
- Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. 2015. ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861. [DOI] [PubMed] [Google Scholar]
- Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. 1999. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol 19: 3989–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289. [DOI] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. 2011. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331: 1624–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, et al. 2013. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci 110: 15395–15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu J, Schermann G, Ohle C, Bendrin K, Sugioka-Sugiyama R, Sugiyama T, Fischer T. 2015. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat Commun 6: 7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Lima CD. 2017. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.