Abstract
In eukaryotes, ribosome assembly is a highly complex process that involves more than 200 assembly factors that ensure the folding, modification and processing of the different rRNA species as well as the timely association of ribosomal proteins. One of these factors, Mpp10 associates with Imp3 and Imp4 to form a complex that is essential for the normal production of the 18S rRNA. Here we report the crystal structure of a complex between Imp4 and a short helical element of Mpp10 to a resolution of 1.88 Å. Furthermore, we extend the interaction network of Mpp10 and characterize two novel interactions. Mpp10 is able to bind the ribosome biogenesis factor Utp3/Sas10 through two conserved motifs in its N-terminal region. In addition, Mpp10 interacts with the ribosomal protein S5/uS7 using a short stretch within an acidic loop region. Thus, our findings reveal that Mpp10 provides a platform for the simultaneous interaction with multiple proteins in the 90S pre-ribosome.
Introduction
Ribosomes are ancient molecular machines essential for cell growth and viability. Eukaryotic ribosome assembly is a complex process that extends from the nucleolus to the cytoplasm and requires the assembly of 4 different ribosomal RNAs (rRNAs) with 79 ribosomal proteins (r-proteins). It is a highly coordinated and ordered multistep process that starts in the nucleolus with transcription of a long polycistronic transcript (called 35S pre-rRNA in yeast) by RNA polymerase I. This precursor carries the respective RNAs from both the 40S (18S rRNA) and the 60S subunits (5.8S and 25S rRNA) that are separated by the internal transcribed spacers 1 and 2 (ITS1 and ITS2), and flanked by the 5′ and 3′ external transcribed spacers (5′-ETS and 3′-ETS). These spacer sequences are removed during ribosome assembly. RNA polymerase III is responsible for the transcription of the 5S rRNA precursor that will become part of the large ribosomal subunit [1]. Ribosome biogenesis factors, early binding r-proteins and small nucleolar ribonucleoprotein particles (snoRNPs) associate co-transcriptionally with the nascent 35S pre-rRNA originating a huge macromolecular complex, termed 90S pre-ribosome or small-subunit processome [2, 3]. The 90S particle is composed of approximately 70 assembly factors that form stable sub-complexes, including the UTP-A/tUTP, UTP-B, UTP-C, Mpp10-Imp3-Imp4, Bms1-Rcl1, and U3 snoRNP modules, which associate in a sequential and hierarchical manner with the nascent pre-rRNA [4–6]. Within the 90S particle a series of initial cleavages occur in the 5’-ETS of the rRNA at sites A0, A1 followed by cleavage in ITS1 at site A2, yielding the 20S (precursor to 18S) and 27S pre-rRNAs (precursor to 5.8S and 25S), respectively, thereby separating the maturation pathways of the small and large subunit.
UTP-A and UTP-B are multi-protein complexes, which consist predominantly of WD40 (β-propeller) and α-helical domains that have been suggested to possess a structural role in early ribosome biogenesis steps. Recent cryo-EM structures of the 90S particle provided a first structural view into its molecular architecture, revealing that UTP-A and UTP-B form a mold-like scaffold in which the nascent 18S pre-rRNA is folded, modified and processed [7–9]. The U3 snoRNP, composed of the U3 snoRNA, the box C/D proteins and U3-specific factor Rrp9, is essential for both correct folding of the pre-rRNA as well as processing events at sites A0, A1 and A2 [10–15]. U3 snoRNP is strategically positioned at the center of the particle, allowing the formation of heteroduplexes between the U3 snoRNA and the 5’-ETS and several regions within the 18S pre-RNA [7, 10, 13, 16]. Base pairing between the U3 snoRNA and the 18S rRNA has been suggested to be involved in preventing premature formation of the central pseudoknot, an important architectural feature at the center of the mature 40S subunit [11].
The Mpp10 complex, consisting of the long disordered protein Mpp10, the uS4-like factor Imp3, and the Brix-domain protein Imp4 [17] was identified at the center of the 90S pre-ribosome adjacent to the UTP-B module and close to the 5’ region of the U3 snoRNA [7–9]. Mpp10, and its bona fide binding partners Imp3 and Imp4 have been shown to be involved in chaperoning U3::pre-18S rRNA hybrid formation by destabilization of the intramolecular box A/A’ helix of the U3 snoRNA and stabilization of the U3-hybridized state [18].
In this study, we sought to further investigate the role of the Mpp10 complex in ribosome biogenesis. We further extended Mpp10’s factor network by identifying two new binding partners, the biogenesis factor Utp3/Sas10 and the ribosomal protein Rps5/uS7 and mapped the respective regions within Mpp10 required for these interactions. Furthermore, we present the crystal structure of the Imp4-Mpp10 complex from the thermophilic fungus Chaetomium thermophilum at 1.88 Å resolution and analyze it in the context of previously published cryo-EM density of the 90S particle.
Material and methods
Yeast strains, plasmids and plasmid constructions
Complete lists of the plasmids and Saccharomyces cerevisiae strains used in this study are presented in S1 and S2 Tables, respectively. Yeast manipulations were carried out according to standard procedures [19]. Gene disruption and C-terminal tagging were performed as previously described [20, 21].
Yeast-2-hybrid assay
Yeast-2-hybrid analysis was carried out as previously described [22]. The C. thermophilum ORFs and truncations were cloned into appropriate vectors that allow expression of these proteins as N-terminal fusion proteins carrying either the transcription-activating domain (AD) or DNA-binding domain (BD) of the yeast Gal4 transcription factor. Both plasmids (LEU2 and TRP1 marker, respectively) were co-transformed into the haploid reporter yeast strain PJ69-4, which allows detection of weak (HIS3 reporter) and strong interactions (ADE2 reporter). Colonies were spotted onto SDC-Leu-Trp (spotting control), SDC-Trp-Leu-His (weak interactions) and SDC-Trp-Leu-Ade plates (strong interactions), respectively, and grown for 3–4 days at 30°C. The Y2H vector combination SV40 (AD) and p53 (BD) served as positive interaction control and also to detect potential self-activation of the investigated construct.
Tandem affinity purification from S. cerevisiae and C. thermophilum
Tandem affinity purification of FTpA-tagged (Flag-TEV-proteinA) yeast bait proteins or split-tag tandem-affinity purification of co-expressed C. thermophilum proteins in yeast, were performed in a TAP buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 5% v/v glycerol, 0.1% v/v NP-40, and 1 mM DTT as previously described [23, 24]. Pellets were thawed and resuspended in ~25 ml TAP buffer supplemented with protease inhibitor mix FY (Serva). Lysis of yeast cells was carried out by adding ~25 ml cold glass beads to the cell suspension and using a bead beater (Pulverisette, Fritsch, Germany). The lysate was first pre-cleared from the glass beads and then centrifuged (Beckman Coulter centrifuges, rotor JA-25.50, 17,000 rpm for 20 min at 4°C). The resulting supernatant was used for tandem affinity purification. In the first step, the supernatant was incubated with ~300μl slurry of pre-equilibrated IgG Sepharose beads (IgG SepharoseTM Fast Flow, Amersham Bioscience) for at least ~2 hours on a turning wheel at 4°C. After binding, IgG beads were once batch-washed in TAP buffer and then with another 15 ml (without protease inhibitors) using a Mobicol minispin column (MoBiTec, Germany). For TEV protease cleavage, TAP buffer (containing 1 mM DTT) supplemented with an aliquot of self-made recombinant HIS-TEV protease (from a 1–5 mg/ml -80°C frozen stock solution) was added to the IgG beads and incubated for another 90 min on a turning wheel at 16°C. The collected TEV eluate was transferred to a new Mobicol minispin column, containing ~50μl slurry of pre-equilibrated Flag M2 agarose beads (anti-FLAG M2 Affinity Gel, Sigma-Aldrich) and binding was performed for at least 1 hour on a turning wheel at 4°C. Flag beads were washed and co-precipitating proteins/particles were eluted by adding TAP buffer supplemented with 1.5X Flag peptide (Sigma-Aldrich).
Protein coexpression in E. coli and in vitro binding assays
Frozen pellets of E. coli cells were slowly thawed and resuspended in ~25 ml of binding buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 5% v/v glycerol, 1 mM DTT and 0.01% v/v NP-40 (IGEPAL CA-630) and lysed using a microfluidizer (Microfluidics Corp., MA, USA). The lysate was cleared by centrifugation (Beckman Coulter centrifuges, rotor JA-25.50, 17,000 rpm for 20 min at 4°C) and the supernatant was used for binding assays. For purification of GST-tagged proteins (glutathione-S-transferase tag), ~300 μl slurry of pre-equilibrated Glutathione-Sepharose (Sigma) beads was added and incubated at 4°C for 1 hour and 30 min on a turning wheel. After a washing step of the beads using a Mobicol minispin column (MoBiTec, Germany), GST-tagged proteins and the bound material were eluted by adding binding buffer supplemented with 20 mM glutathione (GSH) to the beads and incubating on a turning wheel for 20 min at room temperature. Samples were analyzed by SDS-PAGE and Coomassie staining.
Protein expression and purification for crystallization
Untagged ctImp4 (residues 76–274) and His6-ZZ-ctMpp10 (residues 433–562) were expressed separately in Rosetta 2 pLysS cells in ZYM5052 auto-induction media [25]. Cells were incubated at 37°C until OD600 reached 0.8–1.0 and then shifted to 22°C for 16–18 hours. Cells were harvested by centrifugation for 20 minutes at 4000 rpm, resuspended in lysis buffer (20 mM Na-HEPES pH 7.5, 250 mM NaCl, 40 mM Imidazole, 5 mM MgCl2) and co-lysed with a Microfluidics Fluidizer M110L (Microfluidics Corp., MA, USA). Cell debris and insoluble material were removed by centrifugation at 30000× g for 30 minutes (JA-25.50 rotor). The supernatant was applied to a 1–2 mL NiNTA column and washed with lysis buffer. The complex was eluted with lysis buffer supplemented with 250 mM Imidazole. The His6-ZZ tag on ctMpp10 was cleaved with His6-TEV protease and simultaneously dialysed against SEC buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% glycerol). His6-TEV and His6-ZZ were removed by reverse NiNTA chromatography. The flow-through containing ctImp4-ctMp10 complex was concentrated and used for size exclusion chromatography using a Superdex 75 26/60 equilibrated with SEC buffer. Fractions containing pure ctImp4-ctMpp10 complex were concentrated to 9–16 mg/mL and used for crystallization trials.
Crystallization and structure determination
Needle shaped crystals of ctImp4-ctMpp10 complex grew in conditions containing 25–30% ethylene glycol and were harvested directly without further cryo-protection. Diffraction data were collected from a single crystal at ESRF beamline ID23-2 at 100 K [26]. Data were integrated and scaled with XDS [27]. The crystals belong to the space group P62 or P64. Initial attempts to solve the structure by molecular replacement, as implemented in Phaser [28] and MOLREP [29], using Aspergillus nidulans (an) or Saccharomyces cerevisiae (sc) Rpf2-Rrs1 complex as a search model failed (PDB-IDs: 5BY8, 4XD9, 5A53 [30–32]). Subsequently we sequence adapted the search models using standard procedures as implemented in CHAINSAW, SCULPTOR [33] and MODELLER [34] and combined the resulting models with manual trimming. In addition we took into account the pseudo-symmetrical nature of the brix-fold and tried searching with two separate ‘domains’. Despite extensive trials no clear solution was found. We concluded that the low sequence identity between ctImp4 and anRpf2 and scRpf2 (both ~17%) might suggest larger structural differences between search and target model causing molecular replacement to fail. Ultimately the structure was solved by molecular replacement using the mr_rosetta [35] pipeline in PHENIX [36]. Placement of anRpf2-anRrs1 [30] in the space group P64 resulted in a log-likelihood gain (LLG) of 29.90, indicating a non-solution and similar to the results obtained with our initial molecular replacement trials. However the subsequent rebuilding and relaxation with the ROSETTA pipeline led to a better model, which improved to the LLG to 266.25, indicating a clear solution. After multiple rounds of automated building the Rfree dropped to 27%. The map clearly showed new features and allowed further manual model building. Refinement was carried out with REFMAC5 [37] from the CCP4 package [38] and PHENIX. The final structure contains one ctImp4-ctMpp10 complex in the asymmetric unit. Data collection and refinement statistics are summarized in S3 Table. Comparison of several initial MR solutions with the final model indicated that although the molecule had been correctly placed, the large RMSD (2.35 Å for 182 residues and 2.56 Å for 197 residues, anRpf2-anRrs1 and scRpf2-scRrs1 respectively) between search and target model combined with very low sequence identity resulted in very poor starting phases, preventing manual or automatic model building.
Interface residues were identified with the CONTACT program in the CCP4 suite (S4 Table) [38]. Per-residue conservation score was calculated with the ConSurf server [39].
Sucrose gradient ultracentrifugation
Flag-peptide eluates (max. 500μl) of tandem affinity purifications were further resolved by sucrose gradient ultracentrifugation. Therefore, 95% of the eluted yeast samples were loaded onto a linear 15%–40% (w/v) sucrose gradient containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 0.001% v/v NP-40, and 1 mM DTT and centrifuged for 16 hours at 129,300 xg and 4°C. Sucrose gradients were made using the Gradient Master device (BioComp Instruments). Fractions from the sucrose gradient were harvested by using the Foxy Junior® fraction collector and pooled to final fractions of ~1.2 ml. The fractions were TCA precipitated and analyzed SDS-PAGE and Coomassie staining.
Size exclusion chromatography (SEC)
Protein eluates were analyzed by size exclusion chromatography (SEC). Elution fractions were concentrated using a centricon (Amicon Ultra-4, cellulose) to the desired volume before loading on a SEC column if required. Proteins were separated on a Superdex 200 10/300 GL column attached to the Äkta Purifier System (GE Healthcare) in a buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 1% v/v glycerol, and 0.001% v/v NP-40 according to manufacturer’s recommendations. As gel filtration standard, a mix of thyroglobulin, bovine γ-globulin, chicken ovalbumin, equine myoglobin, and vitamin B12 was used (Biorad). Fractions of ~500μl were collected, TCA precipitated, and analyzed by SDS-PAGE/Coomassie staining.
Results
Utp3/Sas10 and Rps5/uS7 associate with the Mpp10 module
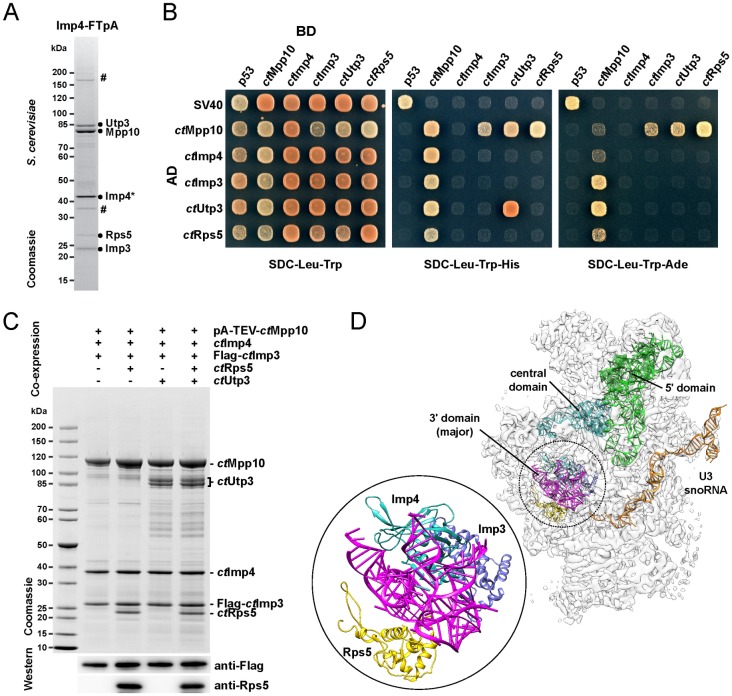
As a first step to further explore the role of the Mpp10 module in the context of ribosome biogenesis, we searched for additional factors that associate with the trimeric Mpp10/Imp3/Imp4 complex. Toward this end, the yeast chromosomal IMP4 locus was modified to encode a C-terminally Flag-TEV-proteinA-tagged Imp4 protein. Imp4-FTpA was purified from yeast via tandem-affinity purification followed by sucrose gradient ultracentrifugation. One of the top fractions obtained from the gradient yielded a pentameric complex that, in addition to Mpp10 and its bona fide binding partners Imp3 and Imp4, contained also the biogenesis factor Utp3 and the ribosomal protein Rps5 (Fig 1A). Mpp10 and Utp3, with a predicted molecular mass of 67 and 70 kDa, respectively, migrate abnormally slowly in a SDS-polyacrylamide gel, which is typical for acidic proteins [40].
Fig 1. Assembly factor network and reconstitution of the ctMpp10 module.
(A) One of the top fractions derived from 15–40% sucrose gradient ultracentrifugation containing the free pool of the yeast Mpp10 complex and associated factors, analyzed by SDS-PAGE and Coomassie staining. The sample was first tandem affinity-purified via yeast Imp4-FTpA (Mpp10 factor) and subsequently resolved by sucrose gradient ultracentrifugation. The labeled proteins were identified by mass spectrometry. Degradation products of Imp4 and dimeric version of Mpp10 are marked with a hashtag (#). (B) Systematic Yeast-2-hybrid analysis of core and associated ctMpp10 assembly factors. The indicated constructs were N-terminally fused to either GAL4 activation domain (AD) or GAL4 binding domain (BD). Yeast transformants were spotted onto SDC-Leu-Trp (permissive), SDC-Leu- Trp-His (selective), and SDC-Leu-Trp-Ade plates (selective for only strong interactions) and grown for 3 days at 30°C. (C) Recombinant co-expression and split tandem-affinity-purification (via pA-TEV-ctMpp10 and Flag-ctImp3) of the ctMpp10 core complex with stepwise addition of associated factors Utp3 and Rps5/uS7. The complexes were assembled by over-expression of indicated protein combinations in yeast (pGAL, high-copy plasmids). Shown are the corresponding Flag-peptide eluates analyzed by SDS-PAGE and Coomassie staining and Western blotting labeled bands were identified by mass spectrometry. Tags were fused to the N-terminus of the corresponding proteins. (D) Position of the ctMpp10 factors ctImp3 and ctImp4 within the 90S cryo-EM density revealing their relative position to the U3 snoRNA (orange) and pre-18S rRNA domains (green, cyan, and purple). Density map of the C. thermophilum 90S pre-ribosome (EMDB: EMD-8143) and respective fitted ctMpp10 complex members (PDB: 5JPQ) are visualized using Chimera. A magnification of the fitted models for ctImp3 (blue), ctImp4 (turquois), and ctRps5/uS7 (yellow) is shown in a circle, illustrating their close neighborhood within the particle.
U3 snoRNA-associated protein Sas10, also known as Utp3, was first identified in a screen for yeast genes that, when overexpressed, suppressed silencing of transcriptionally repressed chromatin [41]. It contains the bioinformatically defined SAS10/C1D domain, an 80 amino acid residues α-helical domain, that is present in two other yeast proteins, namely Lcp5 (another 90S factor) and Rrp47 (exosome-binding factor) [42]. The identification of Utp3 as Mpp10-associated factor is in agreement with previously published data reporting positive Yeast-2-Hybrid (Y2H) interaction between these two proteins [43, 44]. Rps5 is an essential protein located at the back of the 18S rRNA ‘head’ domain in the mature small subunit [45]. In fact, Rps5 is required for the correct assembly of other SSU head domain ribosomal proteins and therefore for the overall folding of the 3’ major domain of the mature 18S rRNA [46]. Moreover, depletion of this protein completely abolishes the nuclear export of the nascent 40S particles and causes a delay in early 18S rRNA processing events with a subsequent accumulation of early pre-rRNA species [47].
To dissect the binding properties of Utp3 and Rps5 to the Mpp10-Imp3-Imp4 complex, a systematic Y2H analysis was performed using the respective orthologs from C. thermophilum (ct) as thermophilic proteins have previously been shown to have better biochemical and biophysical characteristics in structural biology than their mesophilic counterparts [7, 48]. CtImp3, ctImp4, ctUtp3, and ctRps5 all interact with ctMpp10, whereas among these factors no interaction was detected, suggesting that Mpp10 provides a platform for its multiple binding partners (Fig 1B).
Next, we sought to reconstitute the Mpp10 module, using the C. thermophilum orthologs, by heterologous co-expression of the different members of the complex in S. cerevisiae, each under control of the inducible GAL promoter, followed by split-tag tandem affinity purification, using pA-TEV-ctMpp10 and FLAG-ctImp3 as first and second bait, respectively. We started by successfully reconstituting the core complex carrying pA-TEV-ctMpp10, FLAG-ctImp3 and untagged ctImp4. Subsequently, in addition to the core complex members, we co-expressed also ctUtp3, ctRps5 or both. In all cases, we were able to reconstitute the corresponding tetrameric and pentameric complexes. Notably, binding of ctRps5 and ctUtp3 to the Mpp10 core complex was independent from each other (Fig 1C). Mpp10 is predicted to be a rather flexible protein with large coiled-coil regions, whose exact location in the 90S has so far not been determined. Interestingly, the early-binding ribosomal protein ctRps5 is localized in the vicinity of both ctImp3 and ctImp4 in the ct90S particle, suggesting that Mpp10 might be connecting these factors in that area (Fig 1D) [7].
Taken these findings together, we could identify two additional binding partners of the Mpp10 module, the biogenesis factor Utp3 and the ribosomal protein Rps5. Furthermore, our data indicate that Mpp10 provides a shared binding platform establishing simultaneous interactions with all its binding partners.
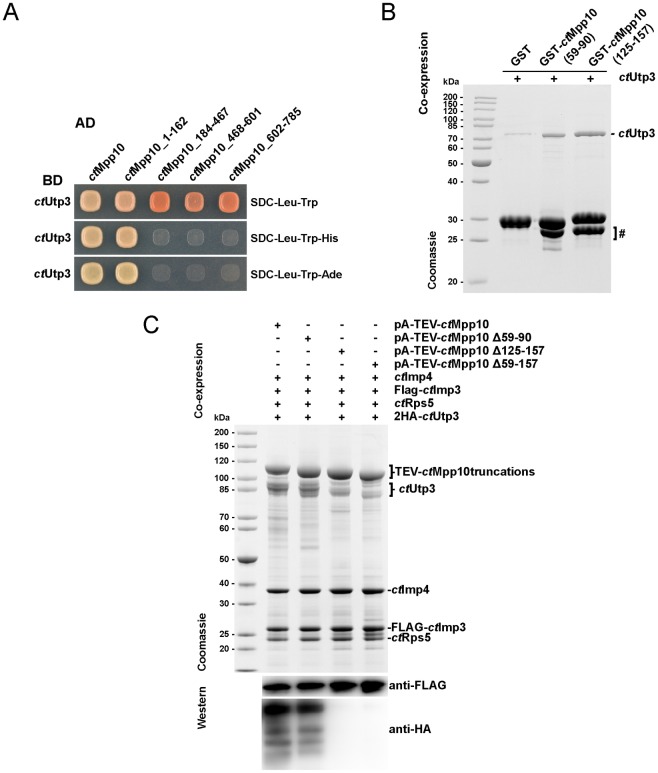
Utp3 interacts directly with the Mpp10 N-terminus
Consistent with our previous findings, the coiled-coil regions of Mpp10 have been suggested to be involved in multiple intra and/or intermolecular interactions [49]. Therefore, we set out to identify an Mpp10 minimal binding motif required for interaction with Utp3. In yeast and human the binding regions for Imp3 and Imp4 have already been reported [49, 50]. Based on these data and multiple sequence alignments and secondary structure prediction, ctMpp10 was divided into five consecutive domains: 1) an N-terminal domain (residues 1–162), 2) a long disordered acidic loop region (residues 184–467), 3) Imp4 interaction domain (residues 475–562), 4) Imp3 interaction domain (residues 612–643) and 5) a C-terminal region rich in basic residues (673–785) (S1 Fig).
To pinpoint the region of Mpp10 mediating interaction with ctUtp3, we carried out an initial Y2H assay using full-length ctUtp3 and tested binding with a series of ctMpp10 truncations. Neither the acidic loop region (residues 184–467) nor the Imp4 binding domain (residues 468–601) showed detectable binding to ctUtp3. Moreover, a fragment containing both Imp3 binding region and the C-terminus of ctMpp10 (residues 602–785) also failed to bind ctUtp3. ctUtp3 binds exclusively within the N-terminus of ctMpp10 corresponding to residues 1–162 (Fig 2A). To further narrow down ctMpp10 to a minimal motif, we tested two truncations that include two conserved hydrophobic patches present in ctMpp10 N-terminal region: ctMpp10 (59–90) and ctMpp10 (125–157) (S1 Fig). Recombinant GST, GST-ctMpp10 (59–90) and GST-ctMpp10 (125–157) were co-expressed with His-ctUtp3 in E. coli, immobilized on glutathione beads and then eluted with glutathione. Both GST-ctMpp10 (59–90) and GST-ctMpp10 (125–157), but not GST alone, were able to interact with His-ctUtp3, indicating that each motif is sufficient for Utp3 binding (Fig 2B).
Fig 2. Two hydrophobic patches within the N-terminal region mediate the direct interaction between Mpp10 and Utp3.
(A) Yeast 2-hybrid analysis of full-length ctUtp3 tested with four indicated fragments of ctMpp10. The indicated constructs were N-terminally fused to either GAL4 activation domain (AD) or GAL4 binding domain (BD). Yeast transformants were spotted onto SDC-Leu-Trp, SDC-Leu-Trp-His, and SDC-Leu-Trp-Ade plates and grown for 3 days at 30°C. (B) Recombinant GST and GST-ctMpp10 truncations were co-expressed with ctUtp3 in E. coli, and subsequently bound to glutathione resin. GSH-eluates were analyzed by SDS-PAGE followed by Coomassie staining. Labeled bands were identified by mass spectrometry. Lower bands correspond to degradation products and are marked with a hashtag (#). (C) Recombinant overexpression of the C. thermophilum Mpp10 complex components in S. cerevisiae, followed by split-tag affinity-purification using pA-TEV-ctMpp10 and Flag-ctImp3, as first and second bait, respectively. The complexes were assembled by over-expression of indicated protein combinations in yeast (pGAL, high-copy plasmids). In this case, we additionally used an N-terminally HA tagged version of ctUtp3. FLAG eluates were subject to SDS-PAGE followed by Coomassie staining and Western blot analysis using anti-HA and anti-FLAG antibodies.
To obtain further insight into the importance of these discrete motifs in binding to Utp3, we again took advantage of our reconstitution system. We then co-expressed Mpp10 constructs lacking one or both binding motifs. Interestingly, pA-TEV-ctMpp10 (Δ59–90) is still able to interact with 2HA-ctUtp3. In contrast, interaction is completely abolished upon deletion of the second minimal motif ctMpp10 (Δ125–157) and ctMpp10 (Δ59–157), suggesting that the motif corresponding to residues 125–157 motif predominantly mediates this interaction (Fig 2C).
Taken together, our findings indicate that Utp3 binds directly to the N-terminal domain of Mpp10. Two conserved motifs within ctMpp10 (residues 59–90 and 125–157) are able to mediate this interaction but only the second motif is essential for binding.
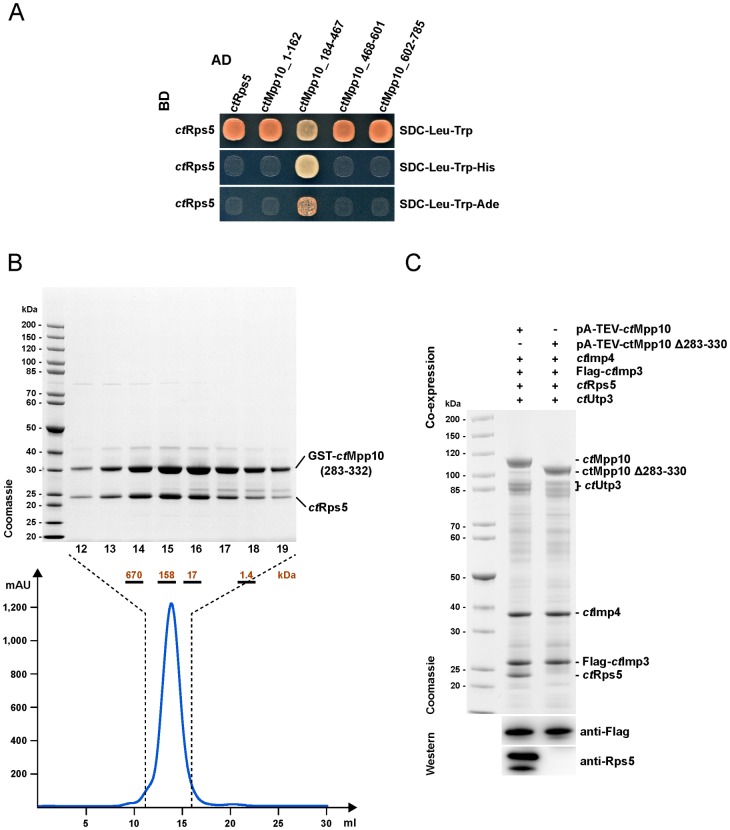
Mpp10 interaction with Rps5 requires the acidic loop region
Using an Y2H approach, similar to the one used for Utp3, we identified the acidic loop, ctMpp10 (184–467), as the region required for Mpp10 interaction with Rps5 (Fig 3A). We defined 5 different regions within the acidic loop that were tested to determine the minimal motif required for Rps5 binding by Mpp10 (S2 Fig). Y2H assays allowed the exclusion of regions 1, 4 and 5 as the binding motifs (data not shown). Two different fragments, one containing regions 2 and 3 (GST-ctMpp10 (283–332)) and other containing regions 2, 3 and 4 (GST-ctMpp10 (283–410)) were co-expressed with His-ctRps5. In both cases, we observed the formation of a complex between Mpp10 truncation and His-ctRps5 (S2 Fig). The obtained stoichiometric complex between GST-ctMpp10 (283–332) and His-ctRps5 (calculated mass of ~58 kDa) remained stable during size exclusion chromatography corroborating this solid interaction (Fig 3B). A protein size standard run on the same column suggests that this complex runs a dimer during gel filtration, most likely due to GST dimerization.
Fig 3. Identification of the minimal Rps5-binding motif within Mpp10.
(A) Yeast 2-hybrid analysis of full-length ctRps5 tested with four indicated fragments of ctMpp10. The indicated constructs were N-terminally fused to either GAL4 activation domain (AD) or GAL4 binding domain (BD). Yeast transformants were spotted onto SDC-Leu-Trp, SDC-Leu-Trp-His, and SDC-Leu-Trp-Ade plates and grown for 3 days at 30°C. (B) Size exclusion chromatography (SEC) of the reconstituted GST-ctMpp10(283–332)-ctRps5 heterodimer. Based on the elution profile recorded at 280 nm, we analyzed fractions 12–19 by SDS-PAGE and Coomassie staining. The complete elution profile (in ml) using a Superdex 200 10/300 column is shown underneath, with indication of the region corresponding to fractions 12–19. The complex was recombinantly co-expressed in E. coli, purified via GST pull-down and eluted with glutathione (GSH). Labeled bands were verified by mass spectrometry. (C) Assembly of the Mpp10 module by recombinant overexpression of the C. thermophilum counterparts, in S. cerevisiae, followed by split-tag affinity-purification using pA-TEV-ctMpp10 and Flag-ctImp3, as first and second bait respectively. The complexes were assembled by over-expression of indicated protein combinations in yeast (pGAL, high-copy plasmids). FLAG eluates were subject to SDS-PAGE followed by Coomassie staining and Western blot analysis using anti-Rps5 and anti-FLAG antibodies.
Subsequently we attempted to confirm that deletion of this region would be sufficient to abolish binding between ctRps5 and ctMpp10. While full-length ctMpp10 co-purified all members of the Mpp10 complex, ctMpp10 Δ283–330 could no longer co-purify ctRps5 (Fig 3C). For this mutant, we also observe a decrease in the levels of ctUtp3 co-purifying with the Mpp10 complex. One possible explanation is that by truncating the protein, even though not truncating the region responsible for Utp3 binding, we might be creating space constraints that affect the interaction of Mpp10 N-terminus with Utp3.
Based on these findings, we conclude that ctRps5 binds to Mpp10 within its acidic loop region and restricted the interaction site to an acidic stretch in Mpp10 corresponding to residues 283–330. Deletion of these residues specifically hampers interaction between ctMpp10 and ctRps5.
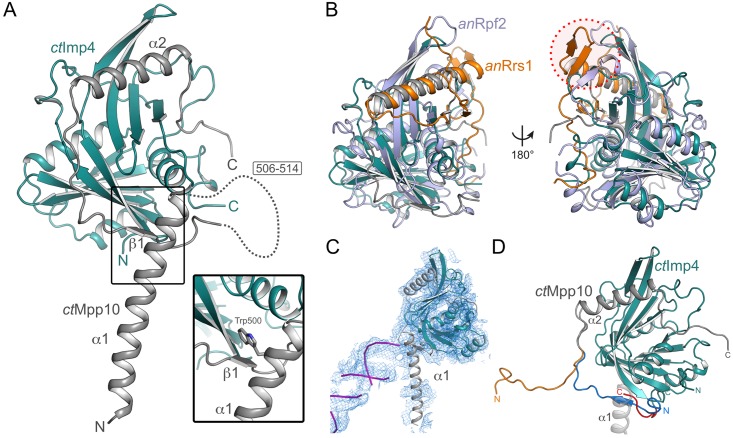
Structure of the ctImp4-ctMpp10 complex
To obtain atomic insight into the interaction network of Mpp10, we tried to crystallize complexes between Mpp10 and its cognate binding partners. We reconstituted a minimal complex of ctImp4 (residues 75–274) and ctMpp10 (residues 433–562) and determined its crystal structure to a resolution of 1.88 Å (Fig 4A). Residues 433–474 as well as 506–514 of ctMpp10 are not resolved in the crystal structure.
Fig 4. Crystal structure of ctImp4-ctMpp10 complex.
(A) Overall structure of the ctImp4-ctMpp10 complex. The BRIX domain of ctImp4 (teal) is completed by helix α2 of ctMpp10 (grey). Helix α1 of ctMpp10 interacts via Trp500 with ctImp4 (inset). For the sake of clarity ctMpp10 helix α1 is omitted in the other panels. (B) Comparison of ctImp4-ctMpp10 with anRpf2-anRrs1 (PDB-ID: 5BY8 [30]) The overall fold between the BRIX domain proteins ctImp4 (teal) and anRpf2 (light-blue) and their ligands ctMpp10 (grey) and anRrs1 (orange) is preserved. Beta-augmentation on the C-terminal sub-domain of the BRIX protein is only observed in the anRpf2-anRrs1 complex (red circle). (C) Rigid body docking of the ctImp4-ctMpp10 complex into the 5.1 Å map of the yeast 90S particle [8]. While the BRIX domain of ctImp4 (teal) and ctMpp10 helix α2 (grey) are completely covered by electron density, helix α1 of ctMpp10 is not. This suggests that in context of the 90S, this helix occupies another location. The tip of helix α1 is occupied by a RNA double helix (purple) in the yeast 90S density. (D) Alternative modeling of the termini of ctImp4 and ctMpp10 based on the crystal structure and cryo-EM density. In the crystal structure (grey/blue), the N-terminus (residues 515–530) of our ctMpp10 construct interacts with ctImp4, whereas when modeled based on the cryo-EM density it extends away (orange) from ctImp4. Likewise the C-terminus of ctImp4 can be extended and modeled (red) into the now remaining cavity between ctMpp10 helix α1 (grey ribbon) and ctImp4.
The overall fold of the ctImp4-ctMpp10 complex is very similar to that of Rpf2-Rrs1 from Aspergillus nidulans (an) and Saccharomyces cerevisiae (sc), despite the rather low sequence identity of ~17%. CtImp4 exhibits the typical BRIX fold [30–32, 51], comprised of two anti-codon binding domains, of which one is completed by the brix-ligand, by an α-helix provided by the brix-ligand, ctMpp10 in this case (S1 Fig). This helix contains several conserved charged (Glu536, Glue542 and Arg548) as well as hydrophobic residues (Ile545 and Ile549) (S1 Fig). There are several local structural differences when compared to Rpf2-Rrs1, leading to a RMSD of 2.35 Å for 182 residues (A. nidulans, Fig 4B), 2.56 Å for 197 residues (S. cerevisiae, S3 Fig). Furthermore, there is no beta-augmentation on the C-terminal subdomain of ctImp4 as observed in Rpf2-Rrs1, limiting the domain complementation to helix α2 (S3B Fig). In addition, ctMpp10 also makes several contacts with the N-terminal subdomain of ctImp4. Interaction at the N-terminal subdomain includes the short beta-strand β1, which is sandwiched between ctImp4 and helix α1 of ctMpp10. A striking difference to the Rpf2-Rrs1 complex is the presence of a long α-helix (α1) at the N-terminus of our ctMpp10 construct, which interacts via a tryptophan residue (Trp500) with ctImp4 at the interface between the two subdomains of ctImp4 (Fig 4A, inset). In order to understand if the position of ctMpp10 helix α1 represents a physiologically relevant conformation or is stabilized by crystal packing, we performed rigid body docking of our crystal structure into the previously published cryo-EM density of the 90S particle from C. thermophilum [7] as well as S. cerevisiae [8, 9]. While the completed Brix domain results in an excellent fit to the density, there is no corresponding density for helix α1 of ctMpp10 in any of the 90S pre-ribosomal particles [7–9]. In fact, the density at this position in the 90S belongs to a RNA double helix and has been partially modeled previously ([9] and Fig 4C). Furthermore, we could trace 7 residues of the ctImp4 C-terminus (which had been omitted for crystallization) within the 5.1 Å cryo-EM map of the yeast 90S particle. A tryptophan (Trp282) residue at the very C-terminus of ctImp4 would locate to the same position as the tryptophan of helix α1 of ctMpp10 (Fig 4D), suggesting that the current conformation may be the result of truncation and crystal packing. Further comparison of the cryo-EM map and our crystal structure suggests that ctMpp10 residues 515–530, locate towards other proteins within the 90S particle and do not participate in beta-augmentation with ctImp4 (Fig 4D and S3C Fig).
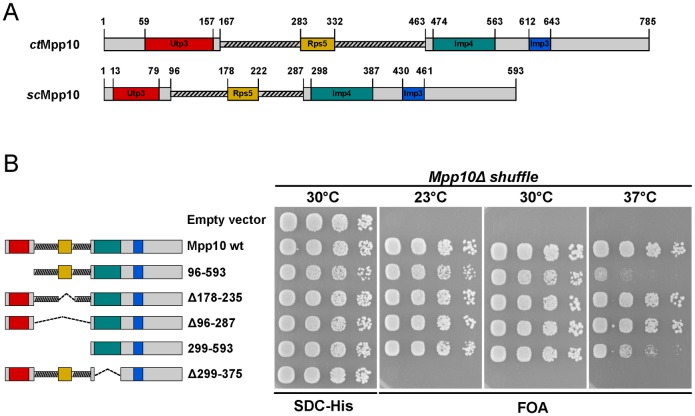
In vivo analysis of binding-deficient mpp10 mutants
To assess the impact of these interactions in vivo, we turned to the Saccharomyces cerevisiae system and used an mpp10Δ shuffle strain for complementation of the otherwise non-viable mpp10Δ (null) mutant. Based on our previous data, we located the equivalent regions in S. cerevisiae necessary for binding Utp3, Rps5 and Imp4 and deleted them (Fig 5A). A mutant containing a truncation of scMpp10 N-terminal region (scMpp10 96–593), the region responsible for binding Utp3, displays a slow growth phenotype, especially at 37°C (Fig 5B). Interestingly, a previously reported mpp10-3 mutant, consisting of a 46 amino acids N-terminal truncation, displays no growth phenotype at any of the tested temperatures [52]. This truncation is lacking our reported Utp3 minimal binding site 1 that we have found to be sufficient, but not required for binding Utp3 (Fig 2C and S1 Fig), providing an explanation for the lack of a growth defect. Interestingly, the mpp10 mutant lacking the Rps5 binding region, was viable displaying a normal growth (scMpp10 (Δ178–235)), suggesting that the interaction between Rps5 and Mpp10 is not essential. This was also observed for a mutant with the complete deletion of the acidic loop, scMpp10 (Δ96–287). Intriguingly, deletion of both N-terminal and acidic loop regions (scMpp10 (299–593)) partially rescued the ts phenotype of the scMpp10 (96–593) mutant at 37°C. Furthermore, deletion of the Imp4 binding region (scMpp10 (Δ299–375)) exhibited lethality in all of the tested temperatures (Fig 5B). Indeed, Mpp10 has been reported to be essential for the stability of both Imp3 and Imp4 presumably by binding them [53].
Fig 5. Growth analysis of strains with different Mpp10 domain truncations.
(A) Overall domain organization for both ctMpp10 and scMpp10 including the identified minimal binding motifs for ctUtp3, ctRps5, ctImp4, and the published ctImp3 [50]. (B) Growth analysis of yeast mpp10Δ shuffle strain complemented with the indicated Mpp10 constructs under TEF1 promoter control. The mpp10Δ shuffle strain was transformed with HIS3 plasmids carrying wild-type MPP10 and truncations. Subsequently, the URA3-MPP10 shuffle plasmid was shuffled out on SDC+5-FOA plates. Transformants were spotted in 6-fold serial dilutions onto SDC-His or SDC+FOA. Plates were incubated at the indicated temperatures for 3 days.
Discussion
The heterotrimeric Mpp10-Imp3-Imp4 complex is an essential 90S module, which is localized at the core of the 90S pre-ribosome adjacent to the UTP-B complex. Imp3 and Imp4, which have been implicated in controlling the formation of pre-rRNA::U3 RNA hybrids during 90S maturation [18], were localized within the ct90S cryo-EM map based on their homology to ribosomal protein uS4 and the Brix-domain, respectively. Recent reports have found small helical segments associated with both Imp3 and Imp4 that have been suggested to be Mpp10 [7–9]. Our crystal structure reveals a long α-helix (α1, residues 475–505) of ctMpp10, which is anchored via Trp500 to ctImp4 and is not part of the canonical helix (α2) that docks with the BRIX fold. Unambiguous assignment of this helix to the helical densities observed in cryo-EM 90S densities will however require higher resolution. In addition to the known interaction with Imp3 and Imp4, in this study, we have identified Utp3/Sas10 and ribosomal protein Rps5/uS7 as additional binding partners of Mpp10 and mapped their respective binding regions within Mpp10. In the 90S particle, ribosomal protein S5 has been found to bind in the vicinity of both Imp3 and Imp4. However, Utp3, presumably a rather flexible protein, could not be located in any of the 90S structures published to date, although SDS/PAGE and mass spectrometry have shown that it is a component of the particle [7–9]. Based on our data we hypothesize that Utp3 contacts Mpp10’s N-terminal region. We propose that Mpp10, positioned at the 90S core, provides an assembly scaffold with linear interaction motifs for the different members of this pentameric module.
Ribosome biogenesis is a tightly regulated process, during which ribosomal proteins are synthesized in the cytoplasm and then imported into the nucleolus where they associate with pre-ribosomal particles. However, many ribosomal proteins have been shown or suggested to be highly unstable and prone to aggregation. Furthermore, they constitute one of the most common ubiquitinated substrates in the cell, being readily subject to degradation by the ubiquitin-proteasome system when in an unbound state in the cell [54–56]. Several different reports have unveiled the existence of chaperones responsible for ensuring the correct and timely incorporation of the ribosomal [57–61]. Similarly, Mpp10 may represent one of these dedicated chaperones, ensuring the correct incorporation of Rps5 into the particle. However, deletion of the Rps5 binding motif of Mpp10 does not display a growth defect suggesting a non-essential role for this interaction. Indeed, several of these dedicated chaperones are non-essential and there are alternative and redundant pathways and factors involved in the incorporation of the ribosomal proteins into the nascent 90S pre-ribosome [57–62]. Interestingly, the cryo-EM structure of the ct90S particle suggests that Rps5 has additional physical contacts to the Emg1 homodimer and a so far UTP-A unassigned beta-propeller that could also facilitate Rps5 recruitment [7]. Alternatively, association of Rps5 with its canonical rRNA binding site within the pre-40S head domain might already be sufficient for its incorporation. Notably, Mpp10 possesses a putative nuclear localization sequence (NLS) within its C-terminal region, and it has been shown that formation of a ternary complex between Mpp10, Imp3 and Imp4 is essential for their nucleolar localization [49, 63]. One interesting possibility is that Mpp10 could additionally mediate the active import of Rps5 into the nucleus. In agreement with this idea, we used a non-radioactive pulse-chase analyses combined with affinity-purification, and found that newly synthesized scMpp10 co-precipitated with Srp1/Kap60, a karyopherin that mediates the nuclear import of cargos, together with Kap95 [64]. This interaction was lost upon deletion of Mpp10’s C-terminal region, where the putative NLS is localized (data not shown).
Interestingly deletion of Mpp10’s Utp3 binding motif is deleterious to the cell, suggesting a putative functional significance for this interaction. Mpp10 complex association with the 90S particle depends on previous incorporation of the UTP-B complex [5, 6, 65]. Furthermore, Utp3 has been suggested to be the link between the Mpp10 module and the UTP-B complex. Indeed, Y2H analysis revealed interactions between Utp3 and the members of UTP-B complex Utp6 and Utp21 [43] and we have confirmed the interaction between Utp3 and Mpp10 even pinpointing two regions in the Mpp10 N-terminal domain able to bind Utp3. Several possibilities arise for the possible function of the observed interaction. Similarly to what we proposed for Rps5, Mpp10 interaction might be important also for nuclear import and/or proper incorporation of Utp3 into the 90S particle. Additionally, this interaction could have a functional role in the context of the 90S particle. One possibility is that Utp3, being a rather flexible protein and the bridging factor between the two 90S modules could potentially ensure its proper structural arrangement towards each other. This could contribute for positioning the Mpp10 complex in the vicinity of the area where it mediates the formation of the U3::pre-18SRNA heteroduplexes. Future EM studies might help to reveal where Utp3 is sitting and might shed some more light into its function.
Supporting information
Indicated is the overall Mpp10 domain organization with an N-terminal domain, indicated by a red line below the alignment, with two conserved hydrophobic motifs required for Utp3 binding (inside a dashed line box). Next, we pinpointed the acidic loop, indicated in yellow that contains the Rps5 binding motif (inside a dashed line box), followed by the Imp4 binding region, indicated in green. Then, the Imp3 binding domain based on what was previously described in [50] in blue, followed by the C-terminal region rich in basic residues, in grey. Helix α1 and α2 of ctMpp10 are drawn above the alignment, strictly conserved residues in α2 are marked with red dots. Aligned proteins were chosen based on homology groups obtained for Mpp10 using Homologene. Alignments were performed with T-coffee and visualized with Jalview. Proteins from different species were used: Chaetomium thermophilum, Saccharomyces cerevisiae, Kluyveromyces lactis, Eremothecium gossypii, Schizosaccharomyces pombe, Homo sapiens, Pan troglodytes, Macaca mulatta, Canis lupus familiaris, Bos Taurus, Mus musculus, Rattus norvegicus, Xenopus tropicalis and Danio rerio.
(TIF)
(A) Recombinant GST and GST ctMpp10 truncations were co-expressed ctRps5 in E.coli, and subsequently bound to glutathione resin. GSH-eluates were analyzed by SDS-PAGE followed by Coomassie staining. Labeled bands were identified by mass spectrometry. (B) Scheme of the different regions of the acidic loop that were tested for binding.
(TIF)
(A) Comparison of ctImp4-ctMpp10 with scRpf2-scRrs1 (PDB-ID: 5A53 [32]). The BRIX fold is conserved between ctImp4 (teal) and scRpf2 (light-brown). The BRIX-ligands, ctMpp10 (grey) and scRrs1 (lime-green) occupy the same position via a single α-helix. No β-augmentation is observed for ctMpp10 (red circle). (B) Structure of ctImp4 can be divided into sub-domains, the N- (teal) and C-terminal half (cyan). (C) Comparison of ctImp4 (teal) and the archeal Imp4-like protein Mil (orange). While ctImp4 is completed by ctMpp10 helix α2, no such ligand is known for Mil. (D) Modeling of the ctMpp10 N-terminus (orange) based on the yeast 90S cryo-EM density suggests an interaction with Bms1 (blue).
(TIF)
(PDF)
(PDF)
(PDF)
(XLSX)
Acknowledgments
We thank Sungmin Eu for his work during his rotation internship, as well as Jochen Bassler for sharing plasmids. We thank Jürgen Kopp and Claudia Siegmann from the BZH/Cluster of Excellence: CellNetworks crystallization platform. We thank Jeremy Sloan for data collection and also acknowledge access to the beamlines at the European Synchrotron Radiation Facility (ESRF) in Grenoble and the support of the beamline scientists.
Data Availability
Coordinates and structure factors are deposited in the RCSB protein data bank (PDB) with the accession number 5O9E.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (SFB638, Z4 to I.S. and HU363/15-1 to E.H. and the Leibniz programme to I.S.); Cluster of Excellence: CellNetworks (EcTOP1 to I.S. and E.H.). I.S. and E.H. are investigators of the Cluster of Excellence: CellNetworks. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Philippsen P, Thomas M, Kramer RA, Davis RW. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978;123(3):387–404. [DOI] [PubMed] [Google Scholar]
- 2.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417(6892):967–70. doi: 10.1038/nature00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell. 2002;10(1):105–15. [DOI] [PubMed] [Google Scholar]
- 4.Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol. 2015;22(11):920–3. doi: 10.1038/nsmb.3111 [DOI] [PubMed] [Google Scholar]
- 5.Perez-Fernandez J, Martin-Marcos P, Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011;39(18):8105–21. doi: 10.1093/nar/gkr508 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Wu C, Cai G, Chen S, Ye K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev. 2016;30(6):718–32. doi: 10.1101/gad.274688.115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Kos-Braun I, et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell. 2016;166(2):380–93. doi: 10.1016/j.cell.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 8.Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017;6 doi: 10.7554/eLife.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaker-Margot M, Barandun J, Hunziker M, Klinge S. Architecture of the yeast small subunit processome. Science. 2017;355(6321). doi: 10.1126/science.aal1880 [DOI] [PubMed] [Google Scholar]
- 10.Dutca LM, Gallagher JE, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39(12):5164–80. doi: 10.1093/nar/gkr044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes JM, Ares M Jr., Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10(13):4231–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5' external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22(23):5139–47. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11(4):1531–42. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14(17):4350–6. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5' end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19(9):6012–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marmier-Gourrier N, Clery A, Schlotter F, Senty-Segault V, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5'-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res. 2011;39(22):9731–45. doi: 10.1093/nar/gkr675 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19(8):5441–52. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerczei T, Shah BN, Manzo AJ, Walter NG, Correll CC. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. J Mol Biol. 2009;390(5):991–1006. doi: 10.1016/j.jmb.2009.05.072 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- 20.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21(11):947–62. doi: 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- 21.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–36. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thoms M, Thomson E, Bassler J, Gnadig M, Griesel S, Hurt E. The Exosome Is Recruited to RNA Substrates through Specific Adaptor Proteins. Cell. 2015;162(5):1029–38. doi: 10.1016/j.cell.2015.07.060 [DOI] [PubMed] [Google Scholar]
- 24.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–2. [DOI] [PubMed] [Google Scholar]
- 25.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–34. [DOI] [PubMed] [Google Scholar]
- 26.Flot D, Mairs T, Giraud T, Guijarro M, Lesourd M, Rey V, et al. The ID23-2 structural biology microfocus beamline at the ESRF. J Synchrotron Radiat. 2010;17(1):107–18. doi: 10.1107/S0909049509041168 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–32. doi: 10.1107/S0907444909047337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–74. doi: 10.1107/S0021889807021206 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–5. doi: 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 30.Kharde S, Calvino FR, Gumiero A, Wild K, Sinning I. The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res. 2015;43(14):7083–95. doi: 10.1093/nar/gkv640 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano N, Kato K, Nakamura A, Komoda K, Tanaka I, Yao M. Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res. 2015;43(9):4746–57. doi: 10.1093/nar/gkv305 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, et al. Chaperoning 5S RNA assembly. Genes Dev. 2015;29(13):1432–46. doi: 10.1101/gad.260349.115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):303–12. doi: 10.1107/S0907444910051218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2016;54:5 6 1–5 6 37. doi: 10.1002/cpbi.3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terwilliger TC, Dimaio F, Read RJ, Baker D, Bunkoczi G, Adams PD, et al. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012;13(2):81–90. doi: 10.1007/s10969-012-9129-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–67. doi: 10.1107/S0907444912001308 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–67. doi: 10.1107/S0907444911001314 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–42. doi: 10.1107/S0907444910045749 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344–50. doi: 10.1093/nar/gkw408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan Y, Zhu Q, Huang D, Zhao S, Jan Lo L, Peng J. An equation to estimate the difference between theoretically predicted and SDS PAGE-displayed molecular weights for an acidic peptide. Sci Rep. 2015;5:13370 doi: 10.1038/srep13370 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamakaka RT, Rine J. Sir- and silencer-independent disruption of silencing in Saccharomyces by Sas10p. Genetics. 1998;149(2):903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell P. Rrp47 and the function of the Sas10/C1D domain. Biochem Soc Trans. 2010;38(4):1088–92. doi: 10.1042/BST0381088 [DOI] [PubMed] [Google Scholar]
- 43.Charette JM, Baserga SJ. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010;16(11):2156–69. doi: 10.1261/rna.2359810 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen YC, Wang HJ, Jauh GY. Dual Role of a SAS10/C1D Family Protein in Ribosomal RNA Gene Expression and Processing Is Essential for Reproduction in Arabidopsis thaliana. PLoS Genet. 2016;12(10):e1006408 doi: 10.1371/journal.pgen.1006408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–9. doi: 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 46.Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28(3):446–57. doi: 10.1016/j.molcel.2007.09.029 [DOI] [PubMed] [Google Scholar]
- 47.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20(2):263–75. doi: 10.1016/j.molcel.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 48.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146(2):277–89. doi: 10.1016/j.cell.2011.06.039 [DOI] [PubMed] [Google Scholar]
- 49.Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, et al. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31(7):1877–87. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng S, Ye K. Purification, crystallization and preliminary X-ray diffraction analysis of Imp3 in complex with an Mpp10 peptide involved in yeast ribosome biogenesis. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 7):918–21. doi: 10.1107/S2053230X14010905 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng CL, Waterman D, Koonin EV, Antson AA, Ortiz-Lombardia M. Crystal structure of Mil (Mth680): internal duplication and similarity between the Imp4/Brix domain and the anticodon-binding domain of class IIa aminoacyl-tRNA synthetases. EMBO Rep. 2005;6(2):140–6. doi: 10.1038/sj.embor.7400328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SJ, Baserga SJ. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci U S A. 1997;94(25):13536–41. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol Cell Biol. 2002;22(20):7258–67. doi: 10.1128/MCB.22.20.7258-7267.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol Biol Cell. 2016;27(17):2642–52. doi: 10.1091/mbc.E16-05-0290 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorenstein C, Warner JR. Synthesis and turnover of ribosomal proteins in the absence of 60S subunit assembly in Saccharomyces cerevisiae. Mol Gen Genet. 1977;157(3):327–32. [DOI] [PubMed] [Google Scholar]
- 56.Warner JR. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol. 1977;115(3):315–33. [DOI] [PubMed] [Google Scholar]
- 57.Stelter P, Huber FM, Kunze R, Flemming D, Hoelz A, Hurt E. Coordinated Ribosomal L4 Protein Assembly into the Pre-Ribosome Is Regulated by Its Eukaryote-Specific Extension. Mol Cell. 2015;58(5):854–62. doi: 10.1016/j.molcel.2015.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338(6107):666–71. doi: 10.1126/science.1226960 [DOI] [PubMed] [Google Scholar]
- 59.Koch B, Mitterer V, Niederhauser J, Stanborough T, Murat G, Rechberger G, et al. Yar1 protects the ribosomal protein Rps3 from aggregation. J Biol Chem. 2012;287(26):21806–15. doi: 10.1074/jbc.M112.365791 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schutz S, Fischer U, Altvater M, Nerurkar P, Pena C, Gerber M, et al. A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly. Elife. 2014;3:e03473 doi: 10.7554/eLife.03473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat Commun. 2015;6:7494 doi: 10.1038/ncomms8494 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol. 2010;189(1):57–68. doi: 10.1083/jcb.200910074 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westendorf JM, Konstantinov KN, Wormsley S, Shu MD, Matsumoto-Taniura N, Pirollet F, et al. M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol Biol Cell. 1998;9(2):437–49. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270(28):16499–502. [DOI] [PubMed] [Google Scholar]
- 65.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem. 2004;279(36):37385–97. doi: 10.1074/jbc.M404909200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Indicated is the overall Mpp10 domain organization with an N-terminal domain, indicated by a red line below the alignment, with two conserved hydrophobic motifs required for Utp3 binding (inside a dashed line box). Next, we pinpointed the acidic loop, indicated in yellow that contains the Rps5 binding motif (inside a dashed line box), followed by the Imp4 binding region, indicated in green. Then, the Imp3 binding domain based on what was previously described in [50] in blue, followed by the C-terminal region rich in basic residues, in grey. Helix α1 and α2 of ctMpp10 are drawn above the alignment, strictly conserved residues in α2 are marked with red dots. Aligned proteins were chosen based on homology groups obtained for Mpp10 using Homologene. Alignments were performed with T-coffee and visualized with Jalview. Proteins from different species were used: Chaetomium thermophilum, Saccharomyces cerevisiae, Kluyveromyces lactis, Eremothecium gossypii, Schizosaccharomyces pombe, Homo sapiens, Pan troglodytes, Macaca mulatta, Canis lupus familiaris, Bos Taurus, Mus musculus, Rattus norvegicus, Xenopus tropicalis and Danio rerio.
(TIF)
(A) Recombinant GST and GST ctMpp10 truncations were co-expressed ctRps5 in E.coli, and subsequently bound to glutathione resin. GSH-eluates were analyzed by SDS-PAGE followed by Coomassie staining. Labeled bands were identified by mass spectrometry. (B) Scheme of the different regions of the acidic loop that were tested for binding.
(TIF)
(A) Comparison of ctImp4-ctMpp10 with scRpf2-scRrs1 (PDB-ID: 5A53 [32]). The BRIX fold is conserved between ctImp4 (teal) and scRpf2 (light-brown). The BRIX-ligands, ctMpp10 (grey) and scRrs1 (lime-green) occupy the same position via a single α-helix. No β-augmentation is observed for ctMpp10 (red circle). (B) Structure of ctImp4 can be divided into sub-domains, the N- (teal) and C-terminal half (cyan). (C) Comparison of ctImp4 (teal) and the archeal Imp4-like protein Mil (orange). While ctImp4 is completed by ctMpp10 helix α2, no such ligand is known for Mil. (D) Modeling of the ctMpp10 N-terminus (orange) based on the yeast 90S cryo-EM density suggests an interaction with Bms1 (blue).
(TIF)
(PDF)
(PDF)
(PDF)
(XLSX)
Data Availability Statement
Coordinates and structure factors are deposited in the RCSB protein data bank (PDB) with the accession number 5O9E.