Significance
Naturally split inteins are important tools in chemical biology and protein engineering, as they provide a rapid and bioorthogonal means to link two polypeptides, termed exteins, together in a near-traceless manner. However, their use is currently limited by sequence constraints imposed by these extein residues. The engineered split inteins reported in this work, NpuGEP and CfaGEP, demonstrate a marked enhancement in extein tolerance, offering greater promiscuity for splicing nonnative sequences. As such, they are shown to improve two important applications of naturally split inteins: protein cyclization and the chemical tailoring of native chromatin. We expect these promiscuous inteins to find broad use in other applications of split inteins that involve the construction of proteins with well-defined sequences.
Keywords: chemical biology, protein engineering, intein splicing, protein semisynthesis
Abstract
The protein trans-splicing (PTS) activity of naturally split inteins has found widespread use in chemical biology and biotechnology. However, currently used naturally split inteins suffer from an “extein dependence,” whereby residues surrounding the splice junction strongly affect splicing efficiency, limiting the general applicability of many PTS-based methods. To address this, we describe a mechanism-guided protein engineering approach that imbues ultrafast DnaE split inteins with minimal extein dependence. The resulting “promiscuous” inteins are shown to be superior reagents for protein cyclization and protein semisynthesis, with the latter illustrated through the modification of native cellular chromatin. The promiscuous inteins reported here thus improve the applicability of existing PTS methods and should enable future efforts to engineer promiscuity into other naturally split inteins.
An intein is an intervening protein domain that undergoes a unique posttranslational autoprocessing event, termed protein splicing. In this spontaneous process, the intein excises itself from the host protein and, in the process, ligates together the flanking N- and C-terminal residues (exteins) to form a native peptide bond (SI Appendix, Fig. S1A) (1, 2). Although inteins are most frequently found as a contiguous domain, some exist in a naturally split form. In this case, the two fragments are expressed as separate polypeptides and must associate before splicing takes place, so-called protein trans-splicing (SI Appendix, Fig. S1B). Unlike many well-characterized contiguous inteins that splice slowly, several naturally split inteins demonstrate rapid splicing kinetics (3–6). Indeed, the discovery of these ultrafast split inteins has enabled the development of numerous tools for both synthetic and biological applications (1).
A major caveat to splicing-based methods is that all characterized inteins exhibit a sequence preference at extein residues adjacent to the splice site. In addition to a mandatory catalytic Cys, Ser, or Thr residue at position +1 (i.e., the first residue within the C-extein), there is a bias for residues resembling the proximal N- and C-extein sequence found in the native insertion site. Deviation from this preferred sequence context leads to a marked reduction in splicing activity, limiting the applicability of protein trans-splicing (PTS)-based methods (3, 7–11). Accordingly, there is a need for split inteins whose activities are minimally affected by local sequence environment. Although efforts have previously been made to engineer promiscuous inteins (12, 13), these have not focused on naturally split inteins, which have superior fragment association and splicing kinetics (4–6).
Here, we report engineered versions of naturally split inteins that possess greatly improved extein tolerance. Guided by our understanding of active site interactions critical for efficient protein splicing, we carried out targeted saturation mutagenesis of an ultrafast split intein and then used a cell-based selection system to identify mutants that are able to support efficient splicing in a wide range of local extein contexts. The “promiscuity” of these mutant inteins was verified through a series of in vitro kinetic measurements. NMR spectroscopy and molecular dynamics (MD) simulations indicate that the mutations lead to localized structural changes in the intein active site that account for the improved extein tolerance. The mutant inteins are shown to be superior reagents for two important PTS applications: protein cyclization and protein semisynthesis, with the latter illustrated through the installation of a posttranslational modification into a histone within native cellular chromatin.
Results
Design of a Promiscuous DnaE Intein.
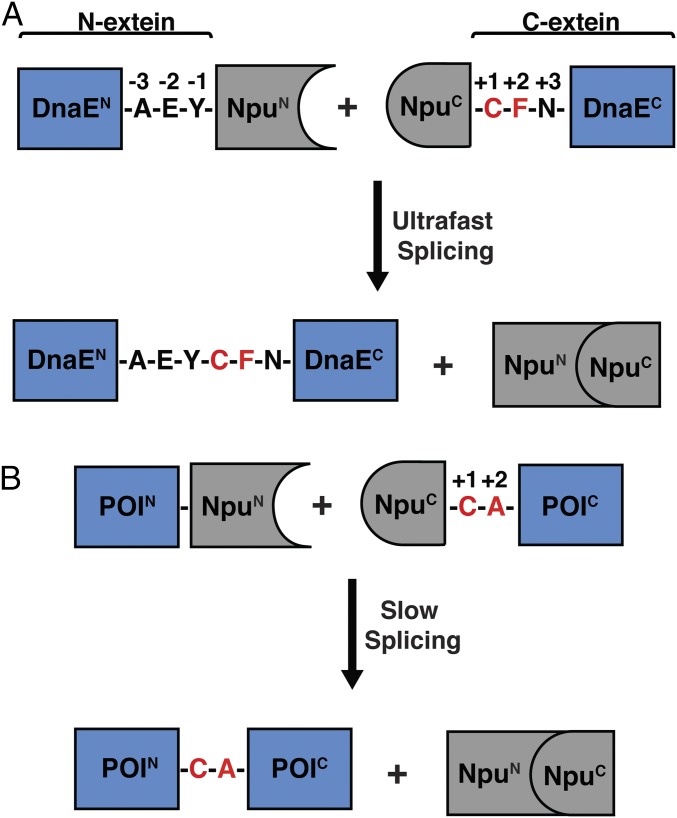
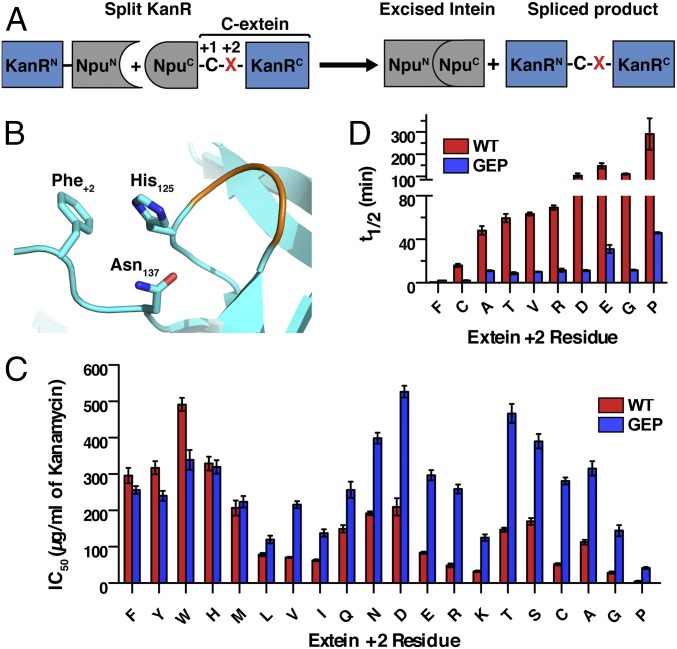
The most commonly used family of naturally split inteins is found embedded within the catalytic subunit of DNA polymerase III (DnaE) in many species of cyanobacteria (Fig. 1A) (14). These DnaE inteins, such as the well-studied and frequently used Nostoc punctiforme (Npu) split intein, show minimal sensitivity to the nature of N-extein residues (4, 9). Extein sequence preferences are largely confined to the catalytic cysteine at the +1 position and large hydrophobic residues that are preferred at the +2 position of the C-extein (9). Mutating the native Phe+2 residue in Npu (t1/2 for splicing = 0.9 min) to a less bulky residue such as Ala leads to a marked decrease in splicing rate (t1/2 = 50 min; Fig. 1B) (10). This +2 C-extein residue sensitivity is caused by loss of a stabilizing interaction between Phe+2 and His125, a key catalytic residue in the last step of protein splicing, involving the cyclization of Asn137 (10, 15, 16). Less bulky +2 residues lead to a more dynamic His125 side chain, populating additional conformations that are not conducive to catalysis. We hypothesized that engineering the loop on which His125 is located (residues 122–124) could favorably alter His125 conformational dynamics and potentially limit the effect of the +2 residue on splicing kinetics, thereby generating a more promiscuous intein (Fig. 2 A and B).
Fig. 1.
Protein trans-splicing of the Npu DnaE split intein. (A) Schematic depicting the ultrafast trans-splicing reaction of the Npu DnaE split intein under its native extein context. Npu is shown embedded within the α subunit of DnaE, flanked by its native extein residues (“AEY” at the N-extein and “CFN” at the C-extein). The +1 and +2 positions of the C-extein are highlighted in red. (B) Schematic depicting the splicing reaction of the Npu DnaE intein embedded within a target protein of interest (POI). The reaction is shown in the context of an Ala+2 C-extein residue, which is unfavorable for splicing activity.
Fig. 2.
Engineering promiscuous splicing activity into the Npu DnaE split intein. (A) Schematic showing a PTS-dependent E. coli selection system. The kanamycin resistance protein, KanR, is split and fused to N- and C-intein fragments (NpuN and NpuC). The +2 C-extein residue (red X) can be varied in the system. (B) Depiction of the Npu active site (instantaneous structure from simulation; SI Appendix) highlighting the interaction among His125, Asn137, and Phe+2 (sticks), as well as the His125 loop (orange). (C) IC50 values for kanamycin resistance in E. coli for NpuWT (red) and NpuGEP (blue) across all +2 C-extein residues (mean ± SE, n = 3). (D) In vitro splicing half-lives of NpuWT (red) and NpuGEP (blue) with indicated +2 C-extein residues (mean ± SD, n = 3). Values for NpuWT Ala+2 and NpuWT Phe+2 are from previously reported data (10, 18).
To implement this loop engineering, we used a previously reported selection assay that couples splicing activity to antibiotic resistance in Escherichia coli (16). This involves intein-mediated reconstitution of a split version of the aminoglycoside phosphotransferase (KanR) protein (Fig. 2A). We carried out saturation mutagenesis on the His125 loop (residues 122–124) of Npu and selected for kanamycin-resistant clones in the presence of the unfavorable Gly+2 (SI Appendix, Fig. S2). The majority of the colonies that were observed under selective pressure contained a GXP motif at residues 122–124 (GRP, GEP, or GPP, in place of the native ERD sequence; SI Appendix, Table S1), with the loop sequence GEP imparting the greatest improvement in splicing activity in vitro in the presence of a +2 glycine reside (SI Appendix, Fig. S3). Remarkably, the GEP mutation (NpuGEP) leads to a significant increase in kanamycin resistance for every unfavorable C-extein +2 residue in the E. coli selection assay (Fig. 2C). In vitro analysis of this mutation demonstrated an increase in splicing rate for Ala+2 (fivefold), Gly+2 (10-fold), Arg+2 (10-fold), Asp+2 (10-fold), Thr+2 (sevenfold), Cys+2 (eightfold), Val+2 (sixfold), Glu+2 (fivefold), and Pro+2 (sixfold), along with a slight decrease for Phe+2 (threefold), with all reactions achieving greater than 85% conversion (Fig. 2D and SI Appendix, Table S2).
Structural Effects of the GEP Loop Mutation.
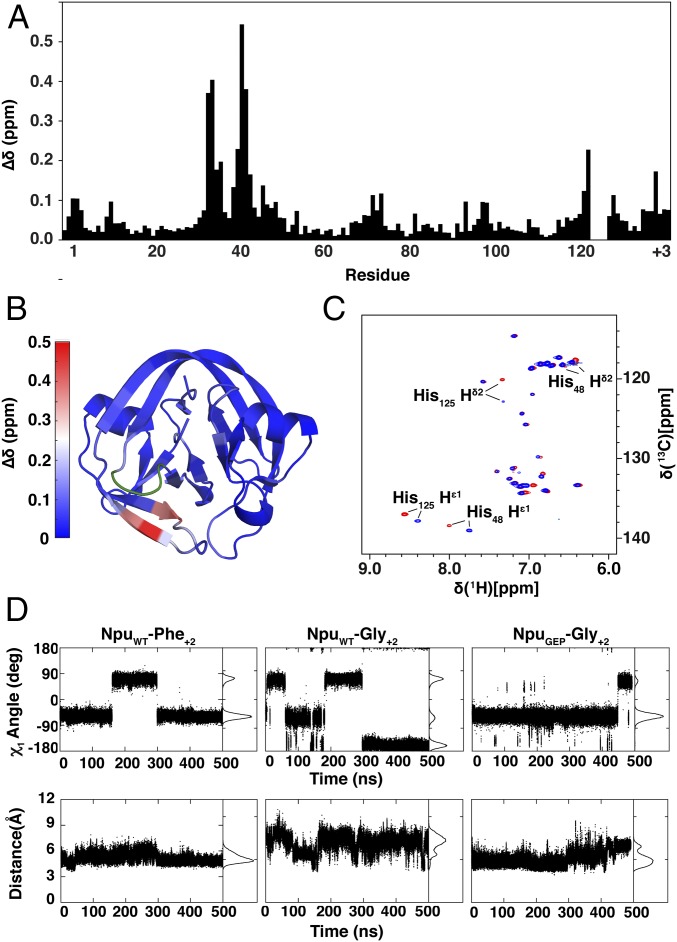
To understand the structural effect of the GEP loop mutation, artificially fused Npu intein fragments containing the WT or GEP loop and either Phe+2 or Gly+2 exteins were expressed in 15N, 13C isotopically enriched media, purified, and analyzed by NMR spectroscopy (SI Appendix, Fig. S4). To enable structural studies, splicing was inactivated through both C1A and N137A mutations. The majority of backbone resonances were assigned for all four constructs, and the chemical shift perturbations (∆δ) caused by the loop mutation were calculated for each residue and mapped onto the Npu structure (Fig. 3 A and B and SI Appendix, Figs. S5 and S6) (17). This shows that the greatest values of ∆δ are present in residues structurally proximal to the loop, with modest perturbations elsewhere. Furthermore, changes in chemical shift were also observed for the aromatic side chain protons of His125 (Fig. 3C and SI Appendix, Fig. S7), the catalytic residue targeted by the loop engineering. The highly localized changes in chemical shifts in the NMR studies demonstrate the surgical nature of the loop engineering, which leaves the majority of the residues and overall structure of the protein minimally affected.
Fig. 3.
Structural and dynamic effects of the loop mutation. (A) Differences in backbone chemical shifts between NpuWT and NpuGEP in the context of a Gly+2 extein. The weighted average chemical shift perturbation (∆δ) was calculated for each residue. His125, whose backbone could not be assigned, and residues 122–124 of the His125 loop, which were mutated, were not calculated. (B) The ∆δ values from A are shown mapped onto the crystal structure of the fused Npu intein (PDB ID: 4KL5). Residues 32, 33, 40, and 41 are depicted in shades of red corresponding to the heat map key, whereas residues 122–125 are depicted in green. (C) 1H [13C]-HSQC spectra of fused NpuWT (red) and NpuGEP (blue) inteins in the context of a Gly+2 extein. (D, Top) His125 χ1 angles calculated from individual MD simulation trajectories for NpuWT-Phe+2, NpuWT-Gly+2, and NpuGEP-Gly+2 split intein complexes. (Bottom) Distance between His125Cβ and Asn137Cβ atoms calculated from respective MD simulation trajectories.
To better elucidate the effect of the GEP loop mutation, MD simulations were carried out with Npu split intein complexes bearing the aforementioned exteins and loop mutations. Previous MD simulations demonstrated that the F+2A mutation in the C-extein resulted in a less constrained His125 side chain that sampled a catalytically unfavorable χ1 dihedral angle distribution, as well as an increased distance from the Asn137 side chain (10). In the presence of Gly+2, similarly unfavorable conformational heterogeneity for His125 was observed (Fig. 3D). Inclusion of the GEP loop mutation with Gly+2, however, constrains the His125 side chain χ1 dihedral angle and distance to Asn137 to values similar to those observed for the WT intein with Phe+2 (Fig. 3D and SI Appendix, Fig. S8). This restriction of His125 side chain conformational dynamics offers one potential explanation for the improved activity of the GEP loop mutant.
The CfaGEP Intein Improves Split Intein Mediated Protein Cyclization.
We wondered whether the GEP loop would also improve promiscuity of other members of the DnaE split intein family. Recently, we reported a consensus DnaE intein (Cfa) that possesses exceptional thermal and chaotropic stability, as well as robust yields during recombinant protein expression (18). Engineering the GEP loop mutation into Cfa (CfaGEP) also resulted in increased promiscuity at the +2 position of the C-extein in the kanamycin resistance assay (SI Appendix, Fig. S9), indicating that the precision loop engineering used for Npu is applicable to related split inteins.
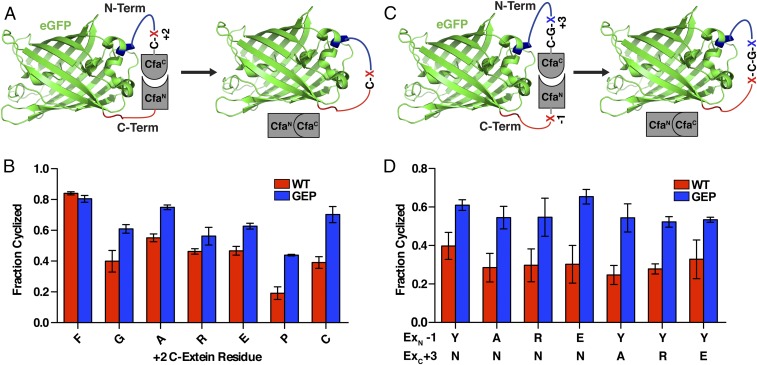
Improving split intein promiscuity should, in turn, improve the efficacy of many PTS-based methods. To demonstrate its utility, we applied CfaGEP to the split intein mediated circular ligation of peptides and proteins (SICLOPPS) (19–22). In SICLOPPS, N- and C-intein fragments are appended to the C- and N-terminus of a peptide (or protein), respectively. Spontaneous intramolecular splicing leads to the generation of a cyclic product (Fig. 4A). Because the system is genetically encoded, the sequence of the insert can be easily randomized and coupled to a cell-based selection or screen to identify cyclic peptide inhibitors of enzymes or protein–protein interactions (19, 21–23). For example, a SICLOPPS library was previously combined with an orthogonal aminoacyl-tRNA synthetase/tRNACUA pair in E. coli to evolve inhibitors of HIV protease (24). Cellular applications of protein splicing can, however, be particularly sensitive to extein mutations because of the presence of intracellular thiols, which intercept and cleave the thioester intermediates of slow-splicing inteins (SI Appendix, Fig. S10). Thus, the +2 C-extein dependency is likely to bias the sequence diversity of the libraries used in SICLOPPS screens. Illustrating this point, cyclization of the enhanced green fluorescent protein (eGFP), using a WT Cfa split intein (CfaWT) SICLOPPS system, was found to be highly sensitive to the identity of the +2 residue (Fig. 4 A and B). In contrast, use of the more promiscuous CfaGEP split intein led to improved yields of cyclized product in all unfavorable +2 contexts. Furthermore, CfaGEP maintains this improved cyclization activity even when the −1 and +3 extein positions are varied (Fig. 4 C and D). Notably, similar results were observed for the cyclization of a small ubiquitin-like modifier (SUMO)-containing construct (SI Appendix, Fig. S11), demonstrating that the improved cyclization activity associated with the CfaGEP split intein is not protein-dependent.
Fig. 4.
eGFP Cyclization with the CfaGEP split intein. (A) Schematic depicting the use of SICLOPPS for the cyclization of eGFP in E. coli with variable residues at the +2 C-extein position (red X). (B) Fraction of cyclized eGFP formed after overnight expression in E. coli for CfaWT and CfaGEP with the indicated +2 C-extein residue (mean ± SD, n = 3). (C) Schematic depicting the use of SICLOPPS for the cyclization of eGFP in E. coli with variable residues at the +3 C-extein position (blue X) and −1 N-extein position (red X). (D) Fraction of cyclized eGFP formed after overnight expression in E. coli for CfaWT and CfaGEP with the indicated +3 C-extein and −1 N-extein residues (mean ± SD, n = 3).
Semisynthesis of Chromatin in Nucleo, Using the CfaGEP Intein.
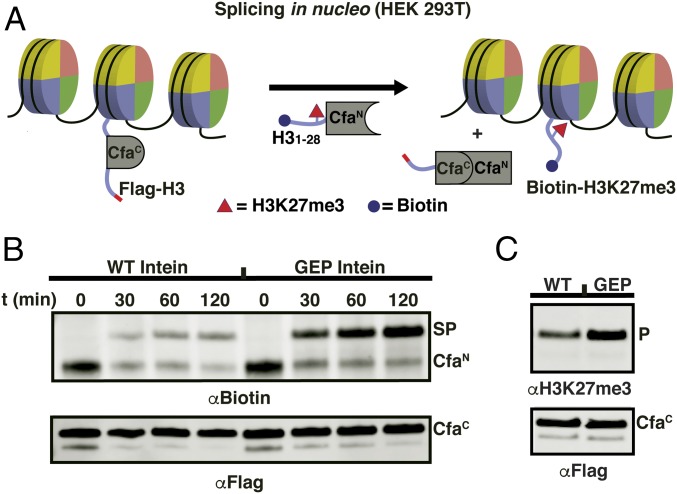
Recently, split inteins have been used to chemically modify cellular chromatin (25). This in nucleo protein semisynthesis strategy provides a means to validate in vitro observations of histone biochemistry in the context of a native cellular chromatin (26). So far, these investigations have been restricted to histone H2B, specifically involving the introduction of posttranslational modifications within the C-terminal region of this protein. We wondered whether the improved activity of the CfaGEP system might permit access to other regions of chromatin, and in particular, the N-terminal tail of histone H3, where many posttranslational modifications involved in gene regulation are clustered (27). With this in mind, we designed a PTS route that would allow synthetic access to the first 28 amino acids of H3, essentially the entire tail of the histone (SI Appendix, Fig. S12). Notably, this design places a proline at the critical +2 position of the C-extein (i.e., the histone), which is an unfavorable residue for WT DnaE inteins. Initial studies revealed poor incorporation of the requisite truncated histone–intein fusion construct (CfaCWT-H329–135) into native chromatin of HEK293T cells, possibly as a result of removal of recognition sequences required for nuclear localization and/or an inability to be recognized by histone chaperones (SI Appendix, Fig. S13) (28). This problem was solved by fusing the missing 28 residues of H3 to the N terminus of CfaC, in effect embedding the intein fragment within the histone (Fig. 5A). Note that the appended H3 fragment is removed, along with CfaC after PTS. Importantly, this insertion strategy worked for both the WT and mutant versions of CfaC, thereby allowing us to directly compare their PTS activities in this chromatin context (SI Appendix, Fig. S13). Accordingly, isolated nuclei were exposed to a semisynthetic protein containing an N-terminally biotinylated histone H3 fragment with a trimethylated lysine at position 27 fused to CfaN (biotin-H31–28K27me3-CfaN), which, on splicing, generates a modified version of full-length histone H3 (Fig. 5A). Gratifyingly, we found that the CfaGEP split intein system supports much more robust PTS compared with the WT intein (Fig. 5B), with an approximately fourfold increase in spliced product based on densitometry analysis of the immunoblot. As expected, the reaction led to an increase in the levels of the H3K27me3 modification on chromatin (Fig. 5C). Thus, use of the promiscuous CfaGEP intein in conjunction with the aforementioned intein insertion strategy allows chemical tailoring of a critical region of the H3 tail in a temporally controlled fashion.
Fig. 5.
In nucleo semisynthesis of chromatin with the CfaGEP split intein. (A) Schematic of in nucleo protein splicing on histone H3 in chromatin. Nucleosomes are depicted as discs. Extracted nuclei from mammalian cells containing transfected Flag-H31–28-CfaC-H329–135 (WT or GEP loop sequence) are treated with semisynthetic Biotin-H31–28K27me3-CfaN (spliced product = Biotin-H3K27me3). (B) Western blot analysis of in nucleo splicing reactions on histone H3 as depicted in A. PTS reactions were quenched at the indicated times with 80 mM iodoacetamide. (Top) αBiotin analysis of in nucleo splicing. CfaN, starting material (Biotin-H31–28K27me3-CfaN); SP, spliced product (Biotin-H3K27me3). (Bottom) αFlag Western blot analysis of in nucleo splicing reactions. CfaC, starting material (Flag-H31–28-CfaC-H329–135). In the case of the CfaGEP intein, the majority of CfaN starting material is converted to SP. For the CfaWT intein, in contrast, competing thiolysis (SI Appendix, Fig. S10) of the CfaN dominates, resulting in a loss of biotin signal. (C) Western blot analysis comparing in nucleo splicing yield of the CfaWT (WT) and CfaGEP (GEP) inteins (2 h, 37 °C). (Top) αH3K27me3 Western blot analysis. P, H3K27me3. (Bottom) αFlag Western blot analysis. CfaC, Flag-H31–28-CfaC-H329–135.
Discussion
Naturally, split inteins have evolved their activity in the context of a particular protein insertion site, which results in some nonnative extein residues having reduced rates of protein splicing. This extein dependence of splicing activity has been a major impediment to the applicability of split intein-based technologies, as it limits the availability of protein split sites amenable to PTS (1). Through targeted mutagenesis of a key loop of the Npu DnaE intein, we identified a mutated sequence (ERD to GEP at residues 122–124) that imparts broad improvements to the splicing activity with nonnative extein residues, overcoming limitations imposed by the +2 C-extein residue. In addition, we have shown through NMR spectroscopy that this loop engineering approach acts in a surgical manner by only affecting the chemical environment of adjacent residues, leaving the majority of the protein unperturbed. Furthermore, MD simulations indicate that the GEP mutation could enable favorable conformational dynamics of the catalytic His125 in the context of the unfavorable Gly+2 extein. Thus, the tuning of key catalytic residues within the active site through local changes in sequence, as seen with His125, can be an effective means to enable intein promiscuity.
Identifying the His125 loop as a target for mutagenesis was enabled by the detailed biochemical and structural characterization of extein dependency within the Npu DnaE intein (9, 10, 16). We expect that this strategy of thorough mechanistic characterization followed by targeted mutagenesis may similarly be applied to recently identified split inteins that possess splicing activity that is complementary to the DnaE family (6, 29). The ultrafast gp41-1, gp41-8, IMPDH-1, and NrdJ-1 inteins are especially appealing targets for this strategy because they use a serine at the +1 position of the C-extein, which makes them applicable in proteins incompatible with the Cys+1 requirement of DnaE inteins.
The greatest value of inteins lies in their use as tools for chemical biology and protein engineering. We demonstrated that the CfaGEP intein improves the scope of two such applications: protein cyclization and the in nucleo semisynthesis of chemically tailored chromatin (22, 25). The improved cyclization enabled by CfaGEP may be further extended to a SICLOPPS library and selection system to identify cyclic peptides that bind or inhibit a target enzyme (23). Furthermore, the greater splicing yields demonstrated for histone semisynthesis with the CfaGEP intein in nucleo could be applied both to histones and other cellular proteins in live cells (30, 31). Beyond the applications demonstrated in this study, we would expect the engineered GEP loop mutation to also improve many other uses of naturally split inteins, including the generation of segmentally labeled proteins for NMR spectroscopy (32, 33) and the production of recombinant proteins that would otherwise be incompatible with cellular expression systems (34). Thus, the promiscuous inteins reported in this study should expand the breadth of proteins accessible to PTS-based technologies.
Methods
A detailed description of all materials, equipment, and methods used in this study can be found in the SI Appendix. An abridged description is presented here.
Antibiotic Selection of His125 Loop Library.
The saturation mutagenesis library was transformed into DH5α competent cells and plated on LB-Agar plates containing 100 µg/mL ampicillin (Amp) and 0, 10, 20, 30, 40, or 50 µg/mL kanamycin (18 h, 37 °C). Colonies were observed on plates containing up to 30 µg/mL kanamycin. They were then isolated and sequenced to identify the residues present in the 122–124 loop region (SI Appendix, Table S1).
E. coli KanR Assay.
Splicing assays in which intein activity was coupled to kanamycin resistance in E. coli were performed as previously described (16, 18).
Recombinant Protein Production.
All recombinant proteins described in this study were expressed in Rosetta (DE3) E. coli cells (3 h, 37 °C, or 16 h, 18 °C). Isotopically enriched proteins were grown in 1 L M9 medium supplemented with [U-13C] d-glucose and 15NH4Cl as the sole carbon and nitrogen sources. The proteins were then isolated by Ni-NTA affinity purification. For N-inteins and fused inteins, the proteins were further purified by size exclusion chromatography. For C-inteins, the proteins were ligated to tripeptides and purified by preparative RP-HPLC. For all proteins described in this study, the purified products were analyzed by RP-HPLC and electrospray ionization (ESI)-MS.
In Vitro Splicing Assays.
In vitro splicing reactions were carried out as previously described (10). Equal volumes of N-inteins (15 µM) and C-inteins (10 µM) were mixed and incubated (30 °C). Individual times were quenched with 8 M guanidine hydrochloride, 4% TFA (reaction:quencher, 3:1 vol/vol) and analyzed by either RP-HPLC or ESI-MS. Peaks corresponding to the starting material, branched intermediate, and spliced product were identified, normalized, and fit to the analytical solution of the coupled differential rate equation for the three-state kinetic splicing model. The mean and SD of three independent replicates are reported.
NMR Spectroscopy.
NMR spectroscopy was performed at 37 °C in field strengths of 600 or 900 MHz on the uniformly 13C, 15N isotopically enriched proteins (400 µM). The data were then processed, and backbone chemical shifts were assigned using 1H{15N}-HSQC, HNCA, HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO experiments (35). Aromatic side chain assignments were obtained using ct-1H{13C}-HSQC, ct-13C-resolved [1H,1H]-NOESY, (HB)CB(CGCD)HD, and (HB)CB(CGCDCE)HE experiments (36). The chemical shift perturbation values were then calculated and represented as a heat map on the crystal structure of Npu DnaE (PDB ID: 4kl5) (17).
MD Simulations.
The MD simulations were carried out as previously described (10).
Cyclization of eGFP and SUMO.
As described earlier, proteins were expressed and then purified with Ni-NTA affinity beads. They were next analyzed by RP-HPLC and ESI-MS. In addition, the percentage of cyclized product was determined by SDS/PAGE, as peaks were identified that correspond to starting material, N-terminal cleavage, linear product, and cyclized product. These peaks were then normalized, and the average of three independent replicates is reported.
Semisynthesis of Biotin-H31–28K27me3-CfaN.
The biotin-H31–28K27me3 hydrazide peptide was synthesized by Fmoc-based solid phase peptide synthesis on a 2-chlorotrityl chloride resin. The resin was prepared as previously described (37), and the synthesis followed standard Fmoc solid phase peptide synthesis protocols. After cleavage from the resin, the peptide was purified by preparative RP-HPLC and analyzed by analytical RP-HPLC and ESI-MS. The purified peptide hydrazide was then converted to a thioester, as previously described (37), and ligated to an expressed Cys-CfaN protein under native conditions. The ligated Biotin-H31–28K27me3-CfaN product was further purified by FPLC, and then analyzed by RP-HPLC and ESI-MS.
In Nucleo Modification of Chromatin.
Plasmids were transfected into HEK 293T cells (107), using lipofectamine 2000, following the manufacturer’s instructions. Cells were harvested after 24 h, and nuclei were isolated as previously described (25). Splicing reactions were carried out in delivery buffer (20 mM Hepes, 1.5 mM magnesium chloride, 150 mM potassium chloride, 1 mM DTT, 1 mg/mL BSA, 1 mM ATP, protease inhibitors at pH 7.6) by addition of Biotin-H31–28K27me3-CfaN (0.25 µM), with time points quenched at the indicated times with iodoacetamide (80 mM). The samples were separated by SDS/PAGE and analyzed by Western blot, blotting against αFlag and αBiotin (Fig. 4B). For the αH3K27me3 blot (Fig. 4C), samples were quenched after 2 h, separated by SDS/PAGE, and analyzed by Western blot (αFlag and αH3K27me3).
Supplementary Material
Acknowledgments
We thank the members of the T.W.M. laboratory, especially Dr. Glen Liszczak, Dr. Robert Thompson, and Dr. Antony Burton for valuable discussions. This work was supported by the National Institutes of Health (Grants R37-GM086868, R01-GM107047, and S10 OD016305-01A1), a National Science Foundation Graduate Research Fellowship (Grant No. DGE-1148900), and Extreme Science and Engineering Discovery Environment (XSEDE) resources supported by the National Science Foundation Grant ACI-1053575.
Footnotes
Conflict of interest statement: A provisional patent application has been submitted that includes aspects of this work.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 26979, 26980, 26981, and 26984).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701083114/-/DCSupplemental.
References
- 1.Shah NH, Muir TW. Inteins: Nature’s gift to protein chemists. Chem Sci (Camb) 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novikova O, Topilina N, Belfort M. Enigmatic distribution, evolution, and function of inteins. J Biol Chem. 2014;289:14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwai H, Züger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Zettler J, Schütz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Shah NH, Dann GP, Vila-Perelló M, Liu Z, Muir TW. Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein engineering. J Am Chem Soc. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvajal-Vallejos P, Pallissé R, Mootz HD, Schmidt SR. Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J Biol Chem. 2012;287:28686–28696. doi: 10.1074/jbc.M112.372680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amitai G, Callahan BP, Stanger MJ, Belfort G, Belfort M. Modulation of intein activity by its neighboring extein substrates. Proc Natl Acad Sci USA. 2009;106:11005–11010. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong S, Williams KS, Wotkowicz C, Xu MQ. Modulation of protein splicing of the Saccharomyces cerevisiae vacuolar membrane ATPase intein. J Biol Chem. 1998;273:10567–10577. doi: 10.1074/jbc.273.17.10567. [DOI] [PubMed] [Google Scholar]
- 9.Cheriyan M, Pedamallu CS, Tori K, Perler F. Faster protein splicing with the Nostoc punctiforme DnaE intein using non-native extein residues. J Biol Chem. 2013;288:6202–6211. doi: 10.1074/jbc.M112.433094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah NH, Eryilmaz E, Cowburn D, Muir TW. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J Am Chem Soc. 2013;135:5839–5847. doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27:110–114, 116, 118–120. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 12.Appleby-Tagoe JH, et al. Highly efficient and more general cis- and trans-splicing inteins through sequential directed evolution. J Biol Chem. 2011;286:34440–34447. doi: 10.1074/jbc.M111.277350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oeemig JS, Zhou D, Kajander T, Wlodawer A, Iwaï H. NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J Mol Biol. 2012;421:85–99. doi: 10.1016/j.jmb.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspi J, Amitai G, Belenkiy O, Pietrokovski S. Distribution of split DnaE inteins in cyanobacteria. Mol Microbiol. 2003;50:1569–1577. doi: 10.1046/j.1365-2958.2003.03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun P, et al. Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing. J Mol Biol. 2005;353:1093–1105. doi: 10.1016/j.jmb.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 16.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci USA. 2009;106:10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranko AS, Oeemig JS, Kajander T, Iwaï H. Intermolecular domain swapping induces intein-mediated protein alternative splicing. Nat Chem Biol. 2013;9:616–622. doi: 10.1038/nchembio.1320. [DOI] [PubMed] [Google Scholar]
- 18.Stevens AJ, et al. Design of a split intein with exceptional protein splicing activity. J Am Chem Soc. 2016;138:2162–2165. doi: 10.1021/jacs.5b13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott CP, Abel-Santos E, Jones AD, Benkovic SJ. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem Biol. 2001;8:801–815. doi: 10.1016/s1074-5521(01)00052-7. [DOI] [PubMed] [Google Scholar]
- 21.Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc Natl Acad Sci USA. 2004;101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavassoli A, Benkovic SJ. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat Protoc. 2007;2:1126–1133. doi: 10.1038/nprot.2007.152. [DOI] [PubMed] [Google Scholar]
- 23.Lennard KR, Tavassoli A. Peptides come round: Using SICLOPPS libraries for early stage drug discovery. Chemistry. 2014;20:10608–10614. doi: 10.1002/chem.201403117. [DOI] [PubMed] [Google Scholar]
- 24.Young TS, et al. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David Y, Vila-Perelló M, Verma S, Muir TW. Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat Chem. 2015;7:394–402. doi: 10.1038/nchem.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt MT, et al. Identification of a functional hotspot on ubiquitin required for stimulation of methyltransferase activity on chromatin. Proc Natl Acad Sci USA. 2015;112:10365–10370. doi: 10.1073/pnas.1504483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soniat M, Cağatay T, Chook YM. Recognition elements in the histone H3 and H4 tails for seven different importins. J Biol Chem. 2016;291:21171–21183. doi: 10.1074/jbc.M116.730218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiel IV, Volkmann G, Pietrokovski S, Mootz HD. An atypical naturally split intein engineered for highly efficient protein labeling. Angew Chem Int Ed Engl. 2014;53:1306–1310. doi: 10.1002/anie.201307969. [DOI] [PubMed] [Google Scholar]
- 30.Giriat I, Muir TW. Protein semi-synthesis in living cells. J Am Chem Soc. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 31.Vila-Perelló M, Muir TW. Biological applications of protein splicing. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muona M, Aranko AS, Raulinaitis V, Iwaï H. Segmental isotopic labeling of multi-domain and fusion proteins by protein trans-splicing in vivo and in vitro. Nat Protoc. 2010;5:574–587. doi: 10.1038/nprot.2009.240. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Xu R, Cowburn D. Segmental isotopic labeling of proteins for nuclear magnetic resonance. Methods Enzymol. 2009;462:151–175. doi: 10.1016/S0076-6879(09)62008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing. Nucleic Acids Res. 2002;30:4864–4871. doi: 10.1093/nar/gkf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavanagh J. Protein NMR spectroscopy: Principles and practice. 2nd Ed. Academic Press; Amsterdam, Boston: 2007. p. xxv. [Google Scholar]
- 36.Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating carbon-13.beta. and proton.delta./.epsilon. chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- 37.Zheng JS, Tang S, Qi YK, Wang ZP, Liu L. Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat Protoc. 2013;8:2483–2495. doi: 10.1038/nprot.2013.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.