Significance
Successful rearrangement of the immunoglobulin locus is critical for B-cell development in the bone marrow and the function of our immune system. Here, we used a conditional gene deletion mouse strain to show that Hdac3 is required for the regulation of chromatin structure and for the productive recombination of the IgH locus, which creates immune diversity and allows B-cell maturation. Although the importance of Hdac3 deacetylase activity was recently drawn into question, we used bone marrow transplantation assays to demonstrate the requirement of Hdac3 deacetylase activity for the production of a fully recombined B-cell receptor, progenitor B-cell survival, and successful differentiation to mature B cells.
Keywords: histone deacetylase, chromatin structure, B-cell development, HDAC3, VDJ recombination
Abstract
Histone deacetylase 3 (HDAC3) is the catalytic component of NCoR/SMRT corepressor complexes that mediate the actions of transcription factors implicated in the regulation of B-cell development and function. We crossed Hdac3 conditional knockout mice with Mb1-Cre knockin animals to delete Hdac3 in early progenitor B cells. The spleens of Hdac3F/−Mb1-Cre+/− mice were virtually devoid of mature B cells, and B220+CD43+ B-cell progenitors accumulated within the bone marrow. Quantitative deep sequencing of the Ig heavy chain locus from B220+CD43+ populations identified a defect in VHDJH recombination with a severe reduction in productive rearrangements, which directly corresponded to the loss of pre-B cells from Hdac3Δ/− bone marrow. For Hdac3Δ/− B cells that did show productive VDJ rearrangement, there was significant skewing toward the incorporation of proximal VH gene segments and a corresponding reduction in distal VH gene segment use. Although transcriptional effects within these loci were modest, Hdac3Δ/− progenitor cells displayed global changes in chromatin structure that likely hindered effective distal V-DJ recombination. Reintroduction of wild-type Hdac3 restored normal B-cell development, whereas an Hdac3 point mutant lacking deacetylase activity failed to complement this defect. Thus, the deacetylase activity of Hdac3 is required for the generation of mature B cells.
Histone deacetylase 3 (Hdac3) functions as the catalytic component of NCoR/SMRT corepressor complexes that are recruited by sequence-specific transcription factors to regulate transcription through the deacetylation of both histone and nonhistone proteins (1–3). Although bulk histone acetylation is generally controlled during replication by Hdac1 and Hdac2 (4, 5), Hdac3 is required for the maintenance of heterochromatin in some tissues (6, 7). Furthermore, the ability of Hdac3 to modulate chromatin accessibility has profound effects on gene transcription, DNA replication, and DNA repair (8–13). For example, hepatocyte-specific deletion of Hdac3 resulted in global changes in histone acetylation, nucleosomal compaction, and changes in gene expression (6). Although Hdac3 has robust deacetylase activity and is proposed to mediate the activity of some class II HDACs that lack intrinsic deacetylase function (14, 15), deacetylase inactive mutants of Hdac3 appeared to partially complement the phenotype of hepatocyte-specific Hdac3-knockout mice (16). This was interesting given that the livers of Albumin-Cre-Hdac3−/− mice displayed phenotypes associated with global increases in histone acetylation, including a loss of heterochromatin (6, 9).
The adaptive immune system has provided an excellent model for the study of higher-order chromatin structure and also provides a simple genetic system in which to dissect mechanisms of action for Hdac3. Lymphocytes rely on a series of recombination-dependent genome-editing processes, such as VDJ recombination and class-switch recombination, for their development and function. These recombination events are regulated through locus accessibility and require long-range chromatin interactions (17, 18). The Rag1 and Rag2 recombinases introduce DNA double-strand breaks at recombination signal sequences, followed by processing and repair of these breaks to produce a functional Ig chain with a unique combination of one V, one D, and one J segment for the heavy chain and one V and one J segment for the light chain. Importantly, this process relies on chromatin remodeling to juxtapose VH and D segments located up to several megabases apart (18, 19). The effective completion of VDJ recombination creates a functional B-cell receptor that signals further development (20, 21). Consistently, disruption of VDJ recombination causes severe combined immunodeficiency in humans and animal models (22, 23).
Here, we crossed mice harboring a conditional Hdac3 allele to Mb1-Cre transgenic mice to define the role of Hdac3 in early B-cell development. Mb1 is expressed before the onset of VDJ recombination of the IgH locus (24), and inactivation of Hdac3 resulted in impaired B-cell development before the formation of a functional B-cell receptor (BCR). Deep sequencing of the heavy chain locus revealed a dramatic reduction in productive VDJ recombination with a particularly profound defect in distal VH gene use, suggesting that long-range recombination events were especially impaired. Although distal IgH elements remained accessible in the absence of Hdac3, an increase in micrococcal nuclease sensitivity suggested that changes in chromatin compaction may impair distal VDJ recombination resulting in failed B-cell maturation. Although previous analysis in the mouse liver implied that Hdac3-associated phenotypes may be independent of the deacetylase activity of Hdac3 (16), re-expression of an Hdac3 mutant lacking deacetylase activity failed to restore normal development of Hdac3Δ/− B cells.
Results
Hdac3 Is Required for B-Cell Survival and Maturation.
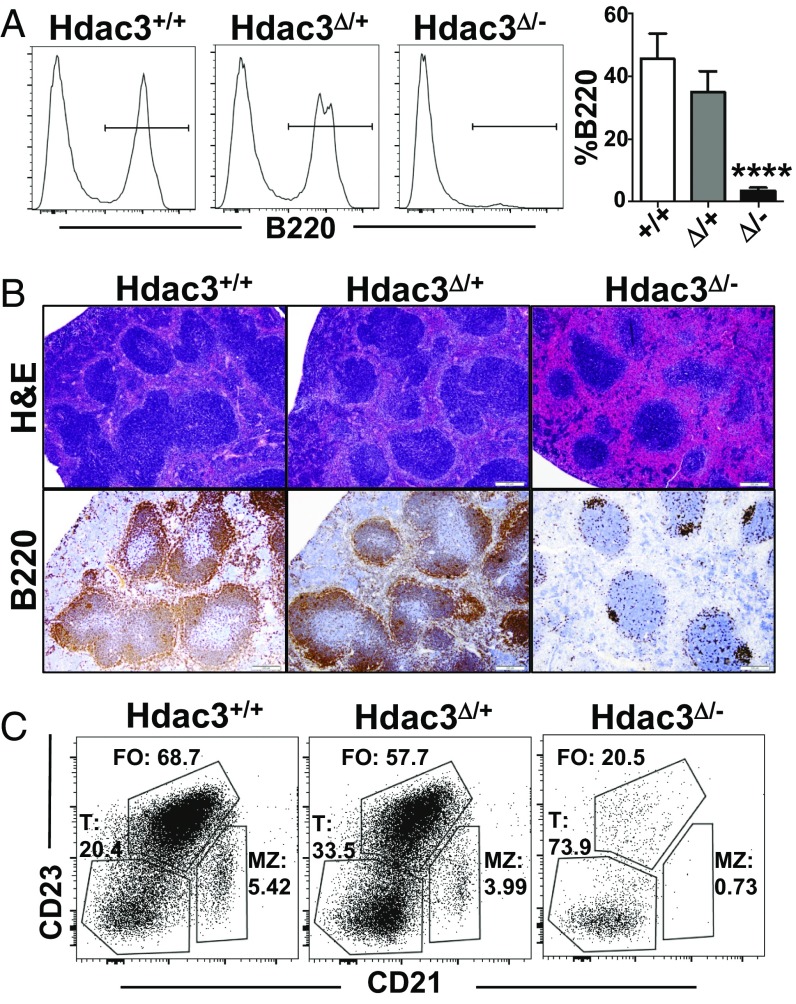
Hdac3F/− mice were crossed with Mb1-Cre transgenic mice, resulting in deletion of Hdac3 in early B-cell progenitors. Western blot analysis indicated that there was a significant reduction of Hdac3 in heterozygous B220+ B cells sorted from the bone marrow and a nearly complete loss of Hdac3 from Hdac3F/−Mb1-Cre+ B220+ B cells (Fig. S1A). This loss of Hdac3 was associated with global changes in histone acetylation as detected in Western blot analysis (Fig. S1B). Spleens isolated from Hdac3-deficient mice were noticeably smaller as well as darker in color than spleens isolated from control animals (Fig. S1C). This dark appearance is likely due to a dramatic reduction in B lymphocytes, as flow cytometry analysis revealed a complete loss of B220+ splenocytes from Hdac3-deleted animals (Fig. 1A). H&E staining of spleen sections revealed a dramatic reduction in follicle size in the absence of Hdac3 (Fig. 1B, Upper) and immunohistochemistry for B220 confirmed the paucity of B lymphocytes with only rare sporadic clusters of B cells remaining in the follicles (Fig. 1B, Lower). Flow cytometry of these residual Hdac3Δ/− B cells revealed a skewing toward less differentiated, transitional B cells (B220+CD23−CD21−) and a loss of the more mature follicular (B220+CD23+CD21low) and marginal zone B-cell populations (B220+CD23lowCD21hi) (Fig. 1C). Therefore, Mb1-Cre–mediated deletion of Hdac3 resulted in an early block to B-cell development.
Fig. S1.
(A) Western blot analysis of lysates of bone marrow B cells with the indicated genotypes shows Hdac3 loss. (B) Western blot analysis was performed from bone marrow B cells to examine global levels of histone acetylation. (C) Representative image of spleens isolated from either Hdac+/+Mb1-Cre+/− or Hdac3F/−Mb1-Cre+/− animals.
Fig. 1.
Hdac3 deletion results in loss of mature B cells. (A) FACS analysis of B220+ populations from the spleens of wild-type, heterozygous, or homozygous Hdac3-deleted mice. Graph at Right shows quantification from n = 5 (+/+), 4 (+/−), and 7 (−/−) mice. (B) Formalin-fixed paraffin-embedded spleen sections were subject to H&E staining (Upper) and immunohistochemistry for B220 (Lower). (C) FACS plots of B220+ splenic B cells to identify CD23−CD21− transitional B cells (T), CD23+CD21low follicular B cells (FO), and CD23lowCD21+ marginal zone B cells (MZ). ****P < 0.0001.
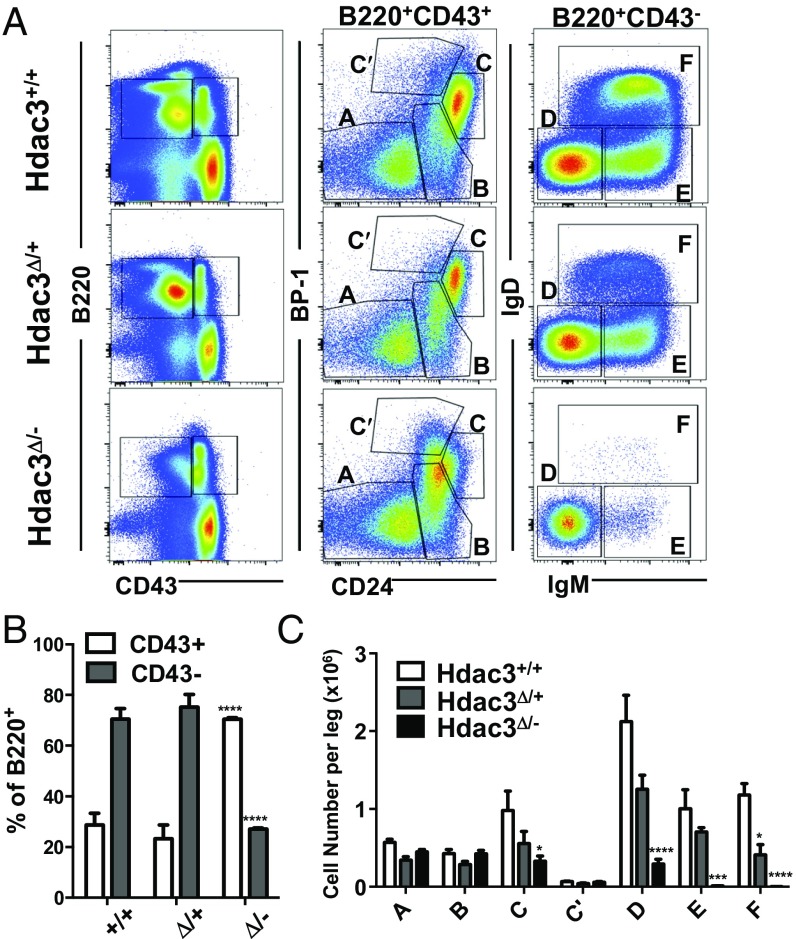
B-cell maturation in the bone marrow is intimately linked to recombination at Ig loci. Specifically, productive VDJ recombination at the heavy chain locus results in expression of a pre-BCR and transition from a CD43+ pro–B-cell to a CD43− pre–B-cell (25). Thus, whole bone marrow was segregated based on B220 and CD43 expression (Fig. 2A, Left). Compared with wild-type or heterozygous mice, Hdac3Δ/− bone marrow showed an accumulation of B220+CD43+ cells, which include the most immature B-cell populations in the marrow (Fig. 2B). We next separated B-cell populations into “Hardy fractions,” which further discriminated B220+CD43+ progenitor cells on the basis of CD24 and BP-1 expression (Fig. 2A, Center) and more mature B220+CD43− populations on the basis of surface IgM and IgD expression (Fig. 2A, Right). In the absence of Hdac3, there was a noticeable reduction in Fraction C (Fig. 2 A, Center, and C). Although some Hdac3-deficient B cells were able to develop to the B220+CD43− stage, these cells did not make it past Fraction D. The dramatic loss of Fractions E and F revealed the absence of immature/mature B-cell populations, which display surface Ig. Interestingly, heterozygous deletion of Hdac3 also resulted in a reduction of the most mature lymphocytes (Fraction F), suggesting a degree of haploinsufficiency. Therefore, in the absence of Hdac3, B-cell progenitors failed to develop into mature B cells with a fully rearranged Ig present on their cell surface.
Fig. 2.
Hdac3 is required for B-cell development within the bone marrow. (A) FACS analysis of bone marrow with anti-B220 and anti-CD43 expression (Left). B220+CD43+ early B-cell progenitors were further subdivided based on CD24 and BP-1 expression (Middle). B220+CD43− B cells were further subdivided based on surface IgM and IgD expression (Right). (B) Quantification of B cells expressing CD43 from experiment in depicted in A. (C) Quantification of Hardy Fractions A–F depicted in A. *P < 0.05, ***P < 0.001, ****P < 0.0001.
Hdac3 Loss Alters B-Cell Gene Expression.
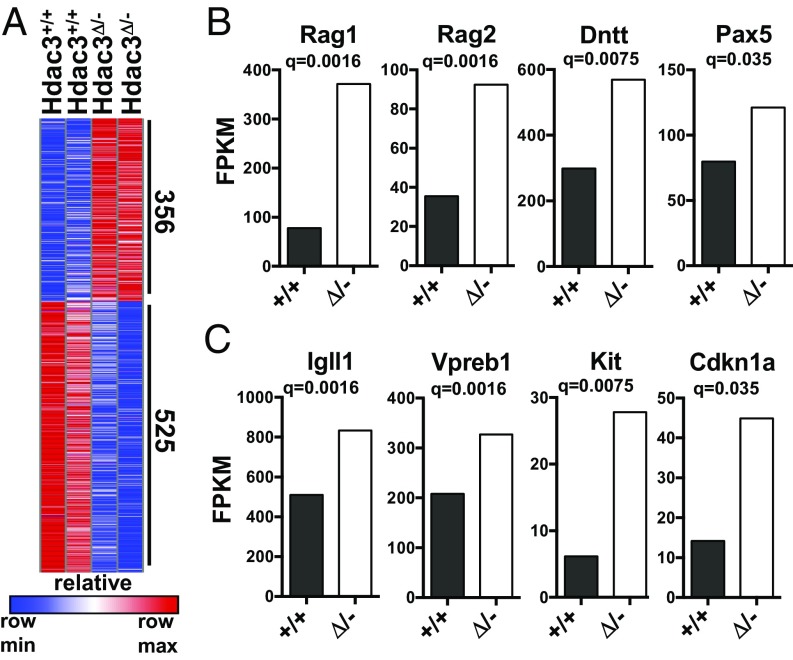
Hdac3 functions as the catalytic component of NCoR/SMRT corepressor complexes that are recruited by transcription factors to regulate gene expression (1, 3). Therefore, we sorted B220+CD43+ cells from the bone marrow of Hdac3Δ/− and control animals for RNA-seq analysis to examine transcriptional changes that might account for the aberrant B-cell development observed in the absence of Hdac3. Hdac3 deletion resulted in the increased expression of 356 genes (Fig. 3A) although slightly more genes showed down-regulation upon Hdac3 deletion (26). Among those genes significantly up-regulated upon Hdac3 loss were critical regulators of B-cell development, including Rag1, Rag2, terminal deoxynucleotidyl transferase (encoded by Dntt), and Pax5, which are required for VDJ recombination (Fig. 3B). Indeed, further junctional diversity is achieved during recombination by the addition of nucleotides by Dntt (27, 28), whereas Pax5 drives germ-line IgH transcription to maintain locus accessibility (29). Hdac3Δ/− B-cell progenitor cells also maintained continued expression of Igll1 (Igλ5) and Vpreb1 (Fig. 3C), which associate to form an Ig-like surrogate light chain assembled into the pre-BCR. Pre-BCR signaling in turn promotes silencing of Igll1 and Vpreb1 to make way for light chain rearrangement and mature BCR assembly (30, 31). Thus, elevated surrogate light chain expression in Hdac3Δ/− B-cell populations in conjunction with the elevated expression of the pro–B-cell marker Kit (Fig. 3C) provides further evidence of impaired B-cell development before the generation of a productive pre-BCR.
Fig. 3.
Gene expression changes observed upon Hdac3 loss associated with failed pre–B-cell differentiation. (A) Heatmap depicting significant gene expression changes between Hdac3+/+ and Hdac3Δ/− pro-B cells as determined by RNA-seq analysis. (B) Fragments per kilobase of transcript per million mapped reads (FPKM) values of significantly changed genes that contribute to VDJ recombination. (C) FPKM values of Igll1, Vpreb1, c-Kit, and p21.
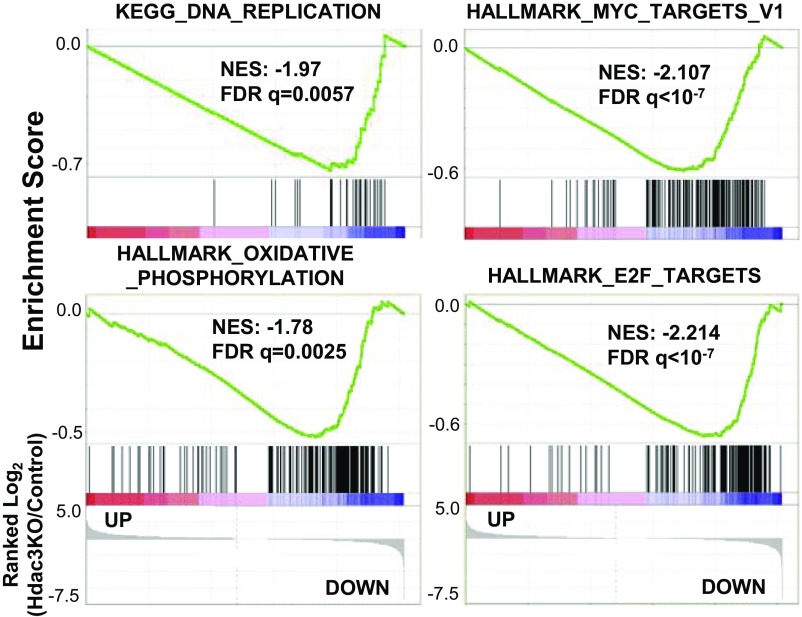
Gene set enrichment analysis did not identify any pathways with an overrepresentation of genes up-regulated upon Hdac3 loss. However, there were several signatures enriched within genes down-regulated upon Hdac3 loss including DNA replication, oxidative phosphorylation, E2F targets, and Myc-regulated genes (Fig. S2). In addition to the silencing of surrogate light chain, the pre-BCR also provides a signal to proliferate following VDJ recombination when Igμ is first made (32). The down-regulation of these gene sets suggests that Hdac3Δ/− B-cell progenitor cells were less proliferative and metabolically active than control cells, again suggesting a lack of pre-BCR signaling normally present within the B220+CD43+ population.
Fig. S2.
GSEA revealed a significant enrichment of genes down-regulated in Hdac3-deficient pro-B cells in signatures associated with cell proliferation.
Hdac3 Is Required for Productive VDJ Rearrangements.
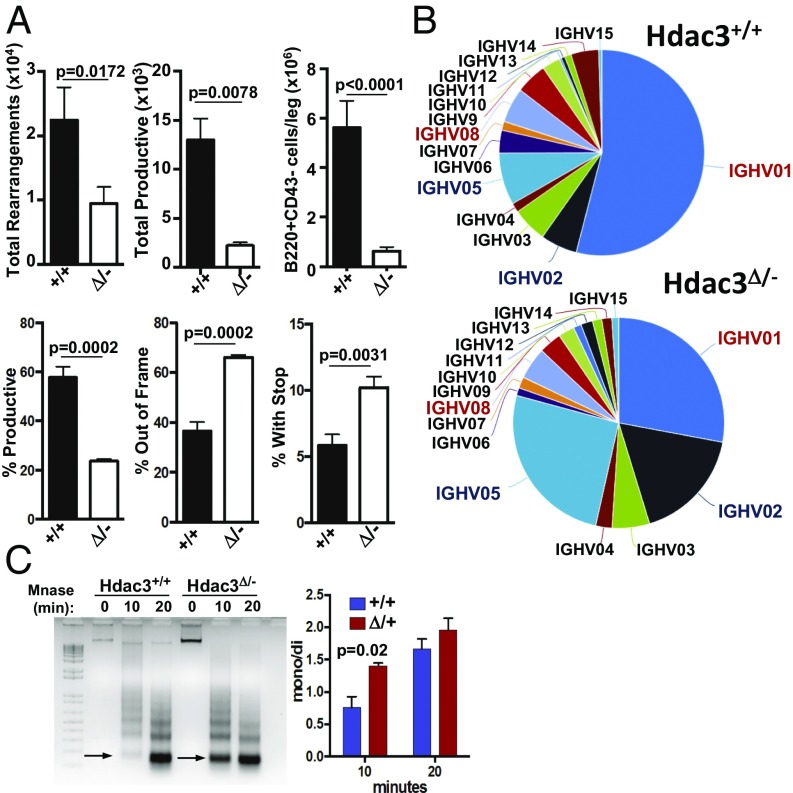
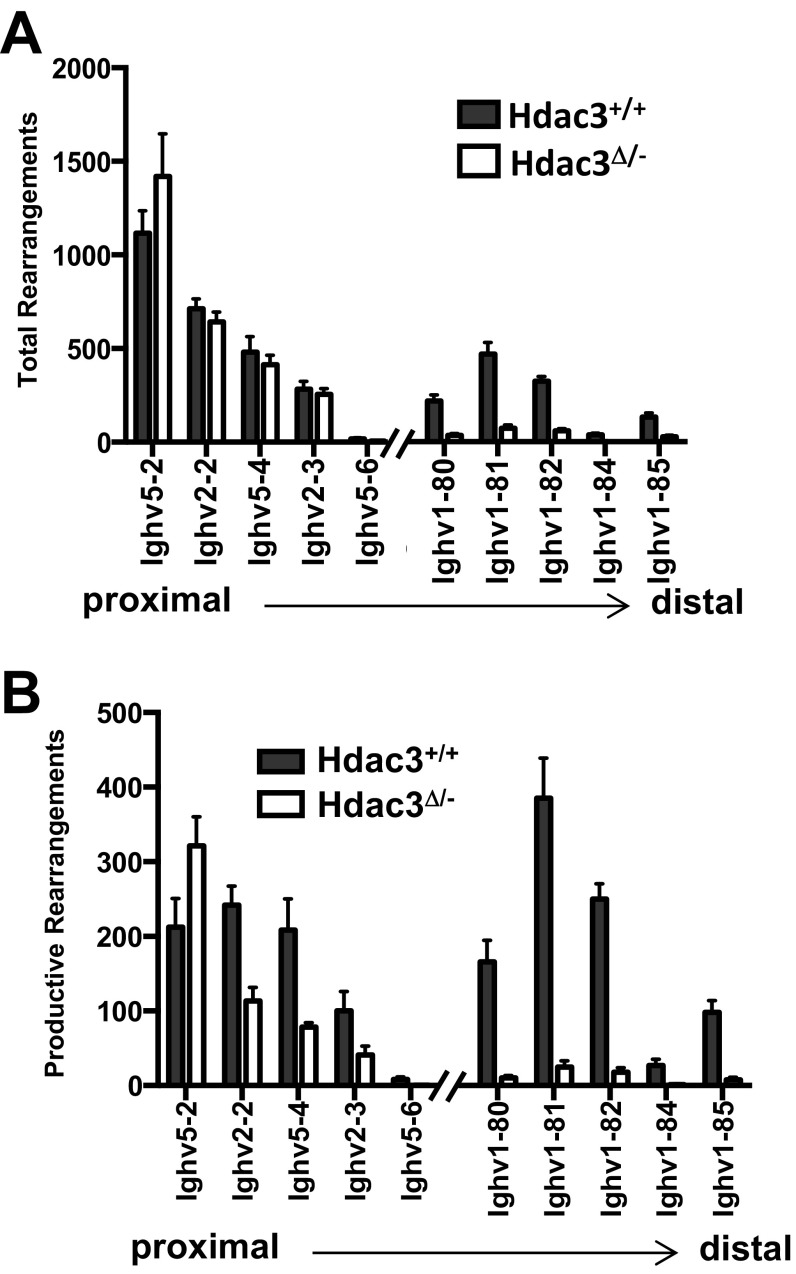
The gene expression analysis is consistent with a model in which MB1-Cre–mediated Hdac3 loss impaired B-cell development by preventing the generation of a functional BCR. Therefore, genomic DNA was isolated from B220+CD43+ bone marrow cells followed by focused deep sequencing of the heavy chain locus to quantitatively characterize VDJ rearrangements. Compared with wild-type mice, Hdac3Δ/− B cells contained significantly fewer heavy chain rearrangements (Fig. 4A, Upper). As germ-line IgH sequences are not detected by this method, a significant portion of Hdac3-deleted B cells had not rearranged the heavy chain locus, which likely accounts for the significant accumulation of CD43+ pro-B cells in the absence of Hdac3. Consistently, the reduction in total productive rearrangements closely mirrored the reduction in CD43− pre–B-cell populations observed in the absence of Hdac3 (Fig. 4A, Upper). In fact, compared with wild-type B cells, in which 60% of heavy chain rearrangements generated a productive rearrangement, only 20% of the total rearrangements were productive in the absence of Hdac3 (Fig. 4A, Lower). In particular, there were increases in rearrangements that were either out of frame or introduced a stop codon (Fig. 4A, Lower). Thus, a significant reduction in successful VDJ recombination events accounts for the dramatic reduction in CD43− B-cell populations in Hdac3Δ/− mice. In addition to the paucity of productive rearrangements, for those Hdac3Δ/− B cells that generated a functional IgH, there was significant skewing toward the incorporation of proximal V segments (IGHV05/VH7183, IGHV02/VHQ52) with a substantial reduction in the inclusion of distal variable gene families (IGHV01/VHJ558 and IGHV08/VH3609; Fig. 4B and Fig. S3 A and B).
Fig. 4.
Hdac3 deletion impairs VDJ recombination. (A) Total (Upper Left) and total productive (Upper Middle) VDJ gene rearrangements present in wild-type and Hdac3-deleted B cells as quantified by ImmunoSEQ analysis. Total numbers of B220+CD43− bone marrow cells/leg were quantified from wild-type and Hdac3Δ/− animals (Upper Right). Percentage of productive rearrangements were quantified (Lower Left), and nonproductive rearrangements were characterized as out of frame (Lower Middle) or inappropriate insertion of a stop codon (Lower Right). (B) Pie chart showing VH gene family use. Red type: two most distal gene families; blue type: two most proximal segments. (C) Micrococcal nuclease digestion of genomic DNA isolated from bone marrow B cells from Hdac3+/+Mb1+/− or Hdac3F/−Mb1+/− mice. Arrows indicate the monosome band that is increased in KO cells. The quantification at the Right shows the ratio of mono/dinucleosomes as determined from three independent experiments performed on chromatin pooled from two wild-type and two Hdac3-deleted mice.
Fig. S3.
Absolute numbers of total rearrangements (A) and absolute numbers of productive rearrangements (B) from the five most proximal and five most distal VH genes.
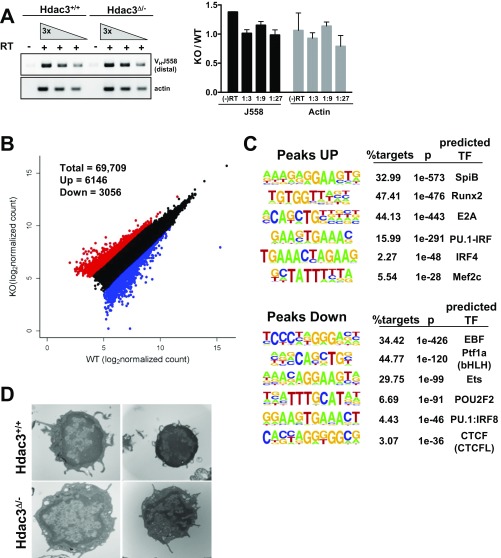
Histone acetylation controls chromatin conformation to regulate transcription, and VDJ recombination is regulated by the accessibility of the gene segments to the recombinase machinery (19). Given the reduction in distal recombination events, RT-PCR was used to detect the germ-line transcripts initiating from distal (VHJ558) gene segments. However, transcription of distal gene segments was unchanged in Hdac3Δ/− B cells (Fig. S4A), suggesting that distal gene segments remained accessible to the recombination machinery in the absence of Hdac3.
Fig. S4.
(A) RT-PCR detected germ-line transcription through the Ig heavy chain locus. Quantification of biological replicates is depicted as intensity relative to wild-type control in the graph to the Right. (B) ATAC-seq performed on 50,000 sorted B220+CD43+ B cells identified regions of chromatin accessibility. The scatterplot depicts 69,709 ATAC peaks identified in at least two of four wild-type and Hdac3−/− ATAC-seq samples. Peaks increased by at least twofold in knockouts are highlighted in red, and peaks decreased by at least twofold in knockouts relative to wild-type controls are highlighted in blue. (C) Homer-based de novo motif analysis was performed on genomic positions associated with either a gain or a loss of ATAC peaks. (D) Transmission electron microscopy of wild-type and Hdac3-deleted bone marrow B cells.
Next, we performed the assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) on sorted B220+CD43+ B cells in Hdac3−/− B cells. ATAC-seq allows the identification of nucleosome-free regions, such as those present at enhancers and promoters, based on the ability of a modified Tn5 transposase to mediate integration of sequencing adapters (33). Consistent with the efficient production of germ-line transcripts (Fig. S4A), ATAC-seq did not reveal large changes in the accessibility of the IgH locus in Hdac3-deleted pro-B cells and again suggested that the distal gene segments remained accessible in the absence of Hdac3. However, 6,146 peaks increased whereas 3,056 peaks were decreased more than twofold from a total of 69,709 peaks that were detected in at least two of four samples (Fig. S4B). Consistent with the fact that ATAC-seq identifies enhancer and promoter elements, transcription factor motif analysis of the ATAC peaks gained upon Hdac3 loss identified significant enrichment of sequences associated with a number of transcription factors with well-established roles in B-cell development including E protein, Runx, and PU.1 family members (Fig. S4C, Upper). Interestingly, both Runx1 and PU.1 are critical for early B-cell development, in part due to their ability to up-regulate the expression of another critical B-cell transcription factor, EBF1 (34, 35). However, peaks containing EBF1 consensus sequences are among the most depleted upon Hdac3 deletion (Fig. S4C, Lower), and the motif for the boundary factor CTCF was also within the peaks that were reduced.
Because ATAC-seq reports primarily on nucleosome-free regions, we extracted chromatin from B cells isolated from the bone marrow and used limited digestion with micrococcal nuclease (MNase) to assess nucleosome compaction. Chromatin from Hdac3-deficient cells proved much more sensitive to MNase digestion, with an increase in monosomes after 10 min (Fig. 4C). Thus, although Hdac3 may not be required for the establishment of nucleosome-free regions within the heavy chain locus, it may play a more general role in the regulation of nucleosome compaction. Although not quantitative, we also used transmission electron microscopy to examine nuclear architecture. In B cells lacking Hdac3, there was a reduction in the dark staining condensed chromatin present in control cells (Fig. S4D, Bottom Left). Additionally, although wild-type B cells contained large, round nuclei with very little cytoplasmic space, Hdac3-deficient B cells often displayed a collapsed and irregularly shaped nuclear architecture (Fig. S4D, Bottom Right). Thus, these studies suggest changes in chromatin structure and nucleosome compaction in Hdac3Δ/− B cells.
Hdac3 Deacetylase Activity Is Critical for Hdac3 Function.
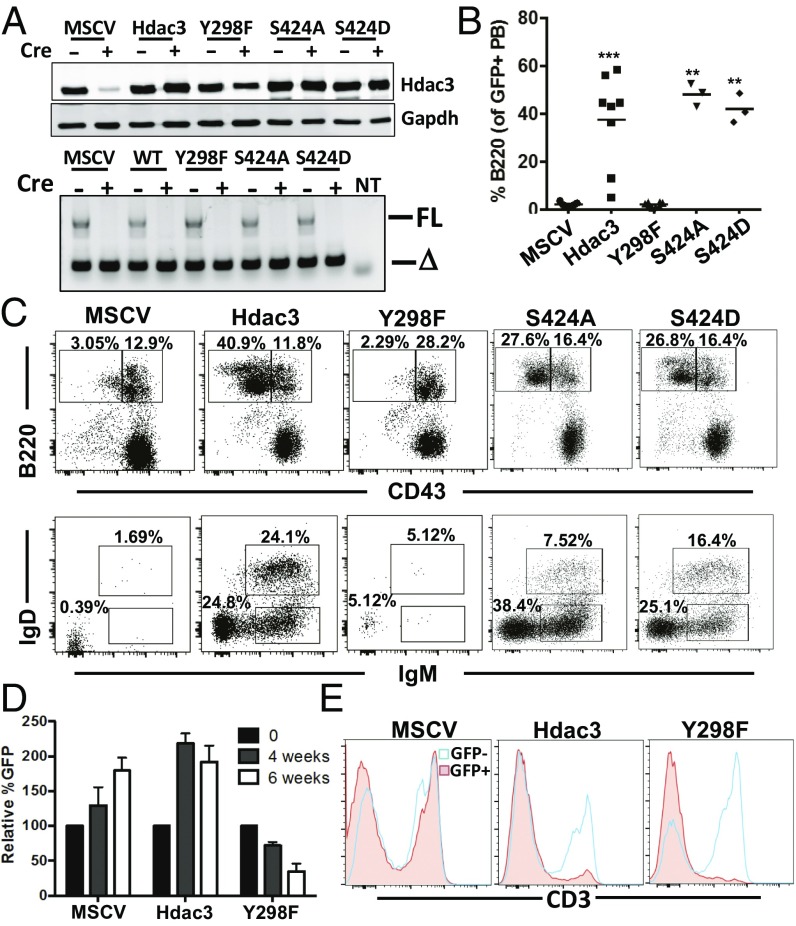
The expression of an Hdac3 deacetylase domain mutant, Hdac3Y298F, which disrupts deacetylase activity while retaining the ability to associate with NCOR1, was reported to partially complement the Hdac3−/− phenotypes observed in Hdac3-deleted livers without affecting histone acetylation (16). However, the liver model did not allow analysis at the single-cell level to assess complementation. Therefore, to understand the importance of Hdac3 deacetylase activity for B-cell development, as well as define the potential contribution of phosphorylation of Ser424 to Hdac3 regulation, we used retroviral delivery of Hdac3 mutants. We used a GFP-expressing virus to ensure analysis of only mutant-expressing cells. We first confirmed the expression of Hdac3 or mutants lacking deacetylase activity or the Ser424 phosphorylation site using a simple fibroblast system and found that the retrovirus yielded near-endogenous levels of wild-type Hdac3 or the mutants (Fig. 5A).
Fig. 5.
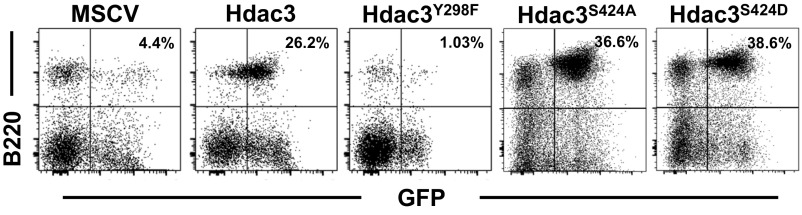
Hdac3 deactylase activity is required for B-cell development. (A) Hdac3F/− fibroblasts were infected with retrovirus-expressing wild-type Hdac3, a deacetylase dead mutant (Hdac3Y298F), or phosphorylation mutants (Hdac3S424A and Hdac3S424D) before deletion of endogenous Hdac3 by infection with adenoviral Cre recombinase. Cells were harvested 3 d following Ad-Cre infection and immunoblotted for Hdac3 (Upper) or genotyped to assess recombination at the endogenous Hdac3 locus (Lower). (B) Retrovirally transduced bone marrow was transplanted into lethally irradiated recipient mice. At 4 wk posttransplant, peripheral blood was analyzed for the presence of B220+ B cells. ***P = 0.0003, **P = 0.0001. (C) GFP+ bone marrow was FACS-analyzed to identify the presence of B220+CD43− B-cell populations (Upper). B220+CD43− populations were further analyzed for surface Ig expression (Lower). (D) GFP levels were assessed in transplanted bone marrow (time 0) and then assessed in the peripheral blood of transplanted mice 4 and 6 wk posttransplant. (E) Transplanted mice were killed at 6 wk, and GFP-positive and -negative splenic T cells were identified by cell-surface CD3 expression.
Next, we infected bone marrow isolated from Hdac3F/−Mb1-Cre mice before transplantation into lethally irradiated mice, and GFP-positive cells were monitored over time. At 4 wk posttransplant, peripheral blood was analyzed for the presence of GFP+ B cells (Fig. 5B). Expression of wild-type Hdac3 or of the two mutants that disrupt Ser424 (S424A and S424D), a residue subject to phosphorylation, was able to restore peripheral B-cell populations, indicating that Ser424 phosphorylation is dispensable in B-cell development. At the same time, mice transplanted with bone marrow expressing the deacetylase dead mutant Y298F were unable to generate mature circulating B cells (Fig. 5A). At 6 wk posttransplant, both the spleen and the bone marrow were harvested from transplanted animals, and B-cell development was analyzed. As the analysis of the peripheral blood suggested, the deacetylase dead mutant (Y298F) was unable to restore B-cell populations in the spleen as indicated by a lack of GFP+B220+ splenocytes (Fig. S5). At the same time, Hdac3Y298F failed to restore the development of CD43− pre-B cells (Fig. 5C, Upper) and failed to generate mature B-cell populations with rearranged Ig expressed on the cell surface (Fig. 5C, Lower). In contrast, cell-surface Ig expression in bone marrow B-cell populations and mature B-cell populations in the spleen was restored upon expression of either wild-type Hdac3 or the Serine424 mutants.
Fig. S5.
At 6 wk posttransplant, splenocytes derived from retrovirally transduced bone marrow were identified by GFP expression and further analyzed for B220 expression to identify splenic B-cell populations.
It is notable that, although vector control or wild-type Hdac3-expressing cells increased over time in the peripheral blood, the Hdac3Y298F mutant not only failed to restore normal B-cell development, but also Hdac3Y298F-expressing cells failed to survive over the 6-wk time frame (Fig. 5D). This suggested that the Y298F mutant had dominant negative effects in non–B-cell hematopoietic lineages in which Mb1-Cre is not expressed (such that Hdac3 was not deleted). Therefore, we used flow cytometry to examine splenic T-cell populations and found that CD3+ cells were significantly reduced in the mutant-expressing (GFP+) population compared with nontransduced (GFP−) splenic T cells (Fig. 5E). It is also worth noting that simply overexpressing wild-type Hdac3 also had an effect on T-cell accumulation, although not to the same degree as the deacetylase dead mutant. Overall, these studies indicate that deacetylase activity is a critical component of Hdac3 functions in B cells. In addition, a deacetylase dead Hdac3 mutant functions in a dominant-negative manner, likely by binding to and impairing the activity of NCoR/SMRT complexes.
Discussion
HDAC inhibitors have significant activity in lymphoma, and genetic models of the targets of these drugs continue to delineate the roles of individual Hdac proteins in lymphoid development. Deletion of Hdac1/2 in early B-cell development resulted in a complete loss of mature B cells from the spleen, whereas their deletion from mature B cells impaired proliferation following mitogenic stimulation (36). These data not only suggest that Hdac1/2 are critical for cell-cycle progression, especially through the S phase, but also suggest that the cell-cycle defects are responsible for the B-cell developmental phenotypes (36). Deletion of Hdac3 in hematopoietic stem and progenitor cells also identified key roles in cell-cycle control. However, there were also defects in the formation of the earliest lymphoid-primed multipotent progenitor cells, suggesting a more complex mechanism underlying the role of Hdac3 in lymphoid development (10). Deletion of Hdac3 in early T cells also caused a lack of cell survival and impaired differentiation, but deletion later in development caused only functional defects (37–39). Here, deletion of Hdac3 in early committed B progenitor cells uncovered unexpected roles for Hdac3 in yielding productive VDJ recombination. The dramatic reduction in the number of productive VDJ rearrangements and the skewing toward proximal VH gene segment incorporation in those rare successful recombination events suggests that Hdac3 is required for the efficiency of recombination, perhaps due to its role in the regulation of chromatin structure.
By using a quantitative sequencing-based approach for the study of VDJ recombination events in which each rearrangement detected corresponds to a single B cell within the isolated population (40), we were able to show a tight relationship between productive rearrangement and cell survival. These results imply that the loss of Hdac3Δ/− cells was due to a failure in VDJ recombination. This approach also demonstrated the skewing of recombination patterns, as the five most proximal VH gene segments showed an increased frequency of use in the absence of Hdac3, whereas the absolute number of rearrangements incorporating all but the most proximal gene segment (Ighv5-2 or VH81X) was reduced in Hdac3Δ/− B-cell populations. Importantly, recombination between Ighv5-2 and the nearest DH segment still requires the looping out of an ∼100-kb DNA fragment. Therefore, even the rearrangements using the most proximal gene segments require long-range chromatin interactions (18). However, analysis of the five most distal VH gene segments revealed a much more dramatic reduction in the absolute numbers of total and productive rearrangement events in the absence of Hdac3. Therefore, a general trend emerges whereby the greater the genomic distance between segments, the less likely that a rearrangement between them will occur in Hdac3Δ/− B cells.
Because Hdac3 is a target of drug discovery efforts, it is important to note that the deacetylase activity of Hdac3 was absolutely required to restore B-cell functions (Fig. 5 and Fig. S5), which indicates that Hdac3-selective inhibitors could modulate immune functions. Although some deacetylase-independent functions of Hdac3 were previously reported (16), the use of different model systems (liver versus B cells) and the mode of inactivation of Hdac3, such as the use of a combination of different viruses to delete and complement Hdac3 deficiency in the liver (16), may contribute to the differences in deacetylase-independent and -dependent phenotypes observed in different tissues. Hence, in our bone marrow transplantation model, we assessed the level of infection before transplant to follow the requirement for deacetylase activity on a cell-by-cell basis over a 6-wk period. By segregating the GFP+/Hdac3–re-expressing cells, we demonstrated complementation of the B-cell defect with wild-type Hdac3 and that the deacetylase dead Hdac3 mutant failed to restore B-cell development in transplanted mice. Moreover, the Hdac3Y298F mutant not only failed to complement, but also appeared to be toxic to cells, as mutant-expressing cells were selected against over time (Fig. 5). In fact, by 6 wk posttransplant all mice receiving Hdac3Y298F-expressing bone marrow had very few GFP-positive cells remaining in the peripheral blood and five of eight mice had undetectable levels of GFP-positive cells in the spleen despite high GFP+ cell numbers present at transplant. Furthermore, given that the Mb1-Cre transgene is active only in B cells, the near absence of GFP+CD3+ splenic T cells indicates that Hdac3Y298F impaired T-cell development in the presence of endogenous wild-type Hdac3, suggesting that this mutant acts in a dominant-negative fashion. These results suggest that the Hdac3Y298F mutant retained the ability to bind to NCOR and SMRT as previously reported (16) and likely disrupted their activities, as these corepressors are required for T-cell development (41). In all, our data stress the importance of Hdac3 deacetylase activity for the regulation of chromatin structure and Ig rearrangement necessary for B-cell development and indicate that inhibition of Hdac3 remains a viable therapeutic strategy.
Materials and Methods
Bone Marrow Transplantation.
Bone marrow was isolated from 8-wk-old Hdac3F/−Mb1-Cre mice and retrovirally transduced with empty vector control, wild-type Hdac3, or the indicated Hdac3 mutant as previously described (42). Following retroviral transduction, 2e5 cells were transplanted by tail-vein injection into lethally irradiated C57BL/6 mice. At 4 wk posttransplant, the peripheral blood was checked for the presence of GFP+ circulating B cells. At 6 wk posttransplant, animals were killed, and bone marrow and spleen were harvested to assess B-cell development.
Animals were housed under pathogen-free conditions and all experiments were conducted in accordance with an Institutional Animal Care and Use Committee-approved protocol under guidelines set forth by the Vanderbilt University Medical Center.
Flow Cytometry.
Single-cell suspensions were obtained from bone marrow by flushing the femur and tibia or from spleen by pressing through a 70-μM strainer. Following erythrocyte lysis (Buffer EL, Qiagen), cells were stained with noted combinations of fluorophore-conjugated antibodies in PBS + 0.5% BSA at 4 °C, washed, and acquired on a 5-laser BD LSRII. Data analysis was performed using FlowJo software (Tree Star).
RNA-Seq.
Total RNA was isolated from FACS-sorted B220+CD43+ bone marrow cells by TRIzol (Ambion) and provided to the Vanderbilt Technologies for Advanced Genomics (VANTAGE) Core Shared Resource for library construction following enrichment of polyadenylated RNAs.
Transmission Electron Microscopy.
B220+ bone marrow cells were isolated from four knockout and four wild-type mice, pelleted, and resuspended in 1 mL of fixative (2.5% glutaraldehyde in 0.1 M sodium cacodylate) at room temperature for 1 h. Fixed samples were provided to the Vanderbilt Cell Imaging Shared Resource for processing and imaging on a Philips/FEI T-12 high-resolution transmission electron microscope.
ImmunoSEQ.
VDJ recombination analysis was performed by Adaptive Biotechnologies on genomic DNA isolated from B220+CD43+ bone marrow. Briefly, libraries were constructed using bias-controlled multiplex PCR to amplify all possible IgH rearrangements (40), and high-throughput sequencing of CDR3 of the B-cell receptor was performed with spike-in of synthetic controls to allow for normalization and accurate quantification. Results were analyzed using the immunoSEQ analyzer software.
ATAC-seq.
ATAC-seq was performed on 50,000 sorted B220+CD43+ bone marrow cells as previously described (33) with a 5% spike-in of Drosophila S2 cells.
Data Sharing.
All mouse strains are available upon request, and the datasets are available in the Gene Expression Omnibus database.
SI Materials and Methods
Mice.
Mice harboring a conditional Hdac3 allele (12) were crossed to Mb1-Cre knockin animals (24) and maintained on a C57BL/6J background. Mice heterozygous for Mb1-Cre were used for experiments at 6–8 wk of age.
Genotyping.
Genotyping of the hdac3 locus was performed with the primers F-CCACTGGCTTCTCTAAGTTC and R-CCCAGGTTAGCTTTGAACTCT, which generate products associated with floxed (935 bp), wild-type (895 bp), and knockout alleles (211 bp).
Immunohistochemistry.
Ten-percent formalin-fixed spleens were paraffin-embedded and sectioned. H&E staining was performed according to standard protocols to determine splenic architecture. Briefly, deparaffinized slides were rehydrated and boiled briefly in antigen retrieval (10 mM sodium citrate, 2 mM EDTA, pH 6.0). Following cooling, endogenous peroxidases were quenched, and sections were blocked with 10% serum and incubated with 1:100 antibody overnight at 4 °C. Sections were washed and incubated with biotinylated secondary antibody before horseradish peroxidase streptavidin was added and visualization with 3,3′-diaminobenzidine (Sigma).
VDJ Germ-Line Transcription.
For germ-line transcription, total RNA was isolated from FACS-sorted B220+c-Kit+ pro-B cells as described above. RNA was pretreated with DNase before cDNA synthesis. For cDNA synthesis, 500 μg of RNA was synthesized with an iScript cDNA synthesis kit (Bio-Rad), and 500 μg was used in a no reverse transcriptase reaction. cDNA was diluted as noted in Fig. S4 and amplified with 35 cycles of PCR using a 58 °C annealing temperature and primers specific for proximal or distal germ-line transcripts. Actin served as a control.
B-Cell Isolation and Western Blotting.
B cells were purified from bone marrow using B220 microbeads (Miltenyi Biotech). Following positive selection over a MACS column, cells were lysed in RIPA buffer with protease inhibitors. Samples were sonicated, and precleared lysates were resolved by SDS/PAGE, transferred to PVDF membrane, and probed with specific antibodies.
RNA-seq Analysis.
Samples were sequenced with an Illumina HiSeq 2500 on an SR-50 run aiming for 30 million reads/sample. Preprocessed reads were aligned to the mouse transcriptome (mm10, downloaded from the University of California at Santa Cruz Genome Browser) using TopHat, and differential gene expression was determined by Cuffdiff as previously described (43). Gene set enrichment analysis (GSEA) was performed using Molecular Signatures Database v5.0 available from the Broad Institute and based on the list of expressed genes preranked by the logtwofold change in expression between Hdac3Δ/- compared with wild type (44). A false discovery rate of 0.05 was used as the cutoff to select significantly enriched pathways.
ATAC-seq Analysis.
For data analysis, adapter sequences of ATAC-seq reads were trimmed by cutadapt (cutadapt -a CTGTCTCTTATACACATCT-minimum-length 15 -paired-output) and then aligned to the mm10 + dm3 genome (boie2 -p 8 -X 2000 -q–no-mixed–no-discordant). Drosophila and mitochondrial reads were removed. Regions enriched in ATAC-seq signal were called using MACS2 with a q-value of 0.001 (callpeak -q 0.001 -nomodel -extsize 140) (45). Differential peaks in knockout vs. wild type were identified using DiffBind based on consensus peaks present in at least two samples. Significantly changed peaks were identified by q < 0.1 and at least a twofold change.
Germ-Line Transcription Primers.
The following primers were used: VH7183 F-CGGTACCAAGAASAMCCTGTWCCTGCAAATGGASC, VH7183 R-GTCTCTCCGCGCCCCCTGCTGGTCC, VHJ558 F-CGAGCTCCARCACAGCCTWCATGCSAR, VHJ558 R-CTCAGTCYCAGKGCAGYTTSCTRCT, Actinb F-GGCTGTATTCCCCTCCATCG, and Actinb R-CCAGTTGGTAACAATGCCATGT.
Antibodies.
Antibodies were used for Western blot analysis for Hdac3 (Abcam) and Actin (Sigma). Antibody binding was detected by fluorescent secondary antibody following imaging with the Odyssey system (Licor). The following flow cytometry antibodies from eBioscience were used: B220(RA3-6B2), CD23(B3B4), CD21(eBio4E3), CD43(eBioR2/60), BP-1(6C3), CD24(M1/69), IgD (11–26), and IgM(II/41). For immunohistochemistry, B220 was used (BD Biosciences-550286).
MNase Assay.
B220+ bone marrow cells were isolated, and cells were pooled from two mice, pelleted, and resuspended in 1-mL cell lysis buffer (10 mM Tris, pH 7.4, 300 mM sucrose, 3 mM CaCl2, 2 mM MgCl2, 1% Triton X-100, 5 mM DTT, protease inhbitors) for 5 min on ice. Nuclei were pelleted and resuspended in 250 μM reaction buffer (50 mM Tris, pH 8.0, 5 mM CaCl2). One microliter of nuclei were diluted in 9 μL of 1 M NaOH, and DNA concentration was determined. Forty micrograms of chromatin was used for each 50-μL reaction. A total of 1.25 μL of 1 U/μL MNase (diluted in 50% glycerol) was added to each reaction and left at room temperature for the indicated time points. Samples were moved to ice and treated with a 10× stop solution (250 mM EDTA pH 8.0, 5% SDS) for 10 min. Samples were treated with 1 μL RNase A (10 μg/μL) and 10 μL 10% SDS for 30 min at 37 °C before treating with proteinase K at 42 °C overnight. Samples were phenol chloroform-extracted, and 2.5 μg of extracted DNA was run on a 1.5% agarose gel.
Acknowledgments
We thank all the members of the S.W.H. laboratory for helpful discussions, reagents, and advice and Flow Cytometry and VANTAGE for sharing resources, services, and support. This work was supported by the T. J. Martell Foundation; the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation; National Institutes of Health Grants R01-CA109355, R01-CA164605, and R01-CA64140 (to S.W.H.); core services performed through Vanderbilt Digestive Disease Research (National Institute of Diabetes and Digestive and Kidney Diseases Grant P30DK58404); the Vanderbilt-Ingram Cancer Center (Grant NCI P30CA68485); and by NIH Grant 1S10RR028106-01A1 for the Next Generation Nucleic Acid Sequencer, housed in VANTAGE. K.R.S. was supported by Grant 5 T32 CA009582-26 and Postdoctoral Fellowship PF-13-303-01-DMC from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus database (accession no. GSE98651).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701610114/-/DCSupplemental.
References
- 1.Karagianni P, Wong J. HDAC3: Taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- 2.Yoon HG, et al. Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaskara S, et al. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin. 2013;6:27. doi: 10.1186/1756-8935-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaskara S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt HM, Pelzel HR, Schlamp CL, Nickells RW. Histone deacetylase 3 (HDAC3) plays an important role in retinal ganglion cell death after acute optic nerve injury. Mol Neurodegener. 2014;9:39. doi: 10.1186/1750-1326-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stengel KR, Hiebert SW. Class I HDACs affect DNA replication, repair, and chromatin structure: Implications for cancer therapy. Antioxid Redox Signal. 2015;23:51–65. doi: 10.1089/ars.2014.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summers AR, et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest. 2013;123:3112–3123. doi: 10.1172/JCI60806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells CE, et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One. 2013;8:e68915. doi: 10.1371/journal.pone.0068915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Z, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 15.Mihaylova MM, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 18.Subrahmanyam R, Sen R. Epigenetic features that regulate IgH locus recombination and expression. Curr Top Microbiol Immunol. 2012;356:39–63. doi: 10.1007/82_2011_153. [DOI] [PubMed] [Google Scholar]
- 19.Chaumeil J, Skok JA. A new take on v(d)j recombination: Transcription driven nuclear and chromatin reorganization in rag-mediated cleavage. Front Immunol. 2013;4:423. doi: 10.3389/fimmu.2013.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spanopoulou E, et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 21.Young F, et al. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz K, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 23.de Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 24.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer CB, et al. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Reports. 2015;13:1444–1455. doi: 10.1016/j.celrep.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: Mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 28.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 29.Hesslein DG, et al. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melchers F. The pre-B-cell receptor: Selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 31.Karasuyama H, et al. The expression of Vpre-B/lambda 5 surrogate light chain in early bone marrow precursor B cells of normal and B cell-deficient mutant mice. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 32.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 33.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209:1255–1262. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imperato MR, Cauchy P, Obier N, Bonifer C. The RUNX1-PU.1 axis in the control of hematopoiesis. Int J Hematol. 2015;101:319–329. doi: 10.1007/s12185-015-1762-8. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi T, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stengel KR, et al. Histone deacetylase 3 is required for efficient T cell development. Mol Cell Biol. 2015;35:3854–3865. doi: 10.1128/MCB.00706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu FC, et al. Histone deacetylase 3 is required for T cell maturation. J Immunol. 2015;195:1578–1590. doi: 10.4049/jimmunol.1500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thapa P, et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun. 2013;4:1582. doi: 10.1038/ncomms2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 41.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 42.Wunderlich M, Mulloy JC. Model systems for examining effects of leukemia-associated oncogenes in primary human CD34+ cells via retroviral transduction. Methods Mol Biol. 2009;538:263–285. doi: 10.1007/978-1-59745-418-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]