Significance
Orthomyxoviruses, including the influenza viruses A–D, all encode a matrix protein M1 that forms a continuous matrix layer underneath the viral envelope. Here we report the crystal structure of a full-length M1 from an orthomyxovirus that infects fish. Consisting of two domains connected by a flexible linker, M1 forms an infinite two-dimensional lattice in crystal through parallel packing. A cryoelectron tomography reconstruction of intact virions confirmed that the M1 lattice closely mimics the viral matrix layer, with the N domain abutting the viral envelope and the C domain pointing toward the virus interior. Our findings provide valuable insights into the mechanisms of M1 in mediating self-oligomerization, membrane association, and viral genome packaging.
Keywords: matrix protein, structure, orthomyoxovirus, ISAV, assembly
Abstract
Many enveloped viruses encode a matrix protein. In the influenza A virus, the matrix protein M1 polymerizes into a rigid protein layer underneath the viral envelope to help enforce the shape and structural integrity of intact viruses. The influenza virus M1 is also known to mediate virus budding as well as the nuclear export of the viral nucleocapsids and their subsequent packaging into nascent viral particles. Despite extensive studies on the influenza A virus M1 (FLUA-M1), only crystal structures of its N-terminal domain are available. Here we report the crystal structure of the full-length M1 from another orthomyxovirus that infects fish, the infectious salmon anemia virus (ISAV). The structure of ISAV-M1 assumes the shape of an elbow, with its N domain closely resembling that of the FLUA-M1. The C domain, which is connected to the N domain through a flexible linker, is made of four α-helices packed as a tight bundle. In the crystal, ISAV-M1 monomers form infinite 2D arrays with a network of interactions involving both the N and C domains. Results from liposome flotation assays indicated that ISAV-M1 binds membrane via electrostatic interactions that are primarily mediated by a positively charged surface loop from the N domain. Cryoelectron tomography reconstruction of intact ISA virions identified a matrix protein layer adjacent to the inner leaflet of the viral membrane. The physical dimensions of the virion-associated matrix layer are consistent with the 2D ISAV-M1 crystal lattice, suggesting that the crystal lattice is a valid model for studying M1–M1, M1–membrane, and M1–RNP interactions in the virion.
All members of the Orthomyxoviridae family, including the influenza viruses A–D, thogotovirus, and isavirus, encode a matrix protein called M1. In the influenza A virus, M1 is produced by a colinear transcript made from the gene segment 7 (1). As one of the most abundantly made viral proteins, the influenza A virus M1 plays multiple roles during the virus life cycle. Upon viral entry, the acidification of the viral interior in the endosome weakens the interaction between M1 and the viral ribonucleoprotein complexes (vRNPs), thus allowing M1-free vRNPs to be imported to the nucleus for viral RNA replication and transcription (2, 3). As infection proceeds, newly synthesized M1 enters the nucleus to mediate the nuclear export of nascent vRNPs. A “daisy-chain” complex is formed with the vRNP binding to the C-terminal domain of M1 (4) and the N-terminal domain of M1 interacting with the nuclear export protein (NEP), also called NS2. Through its nuclear export signal (NES), NEP is specifically recognized by the cellular exportin Crm1, which then facilitates the transport of vRNPs across the nuclear membrane to the cytoplasm in a RanGTP-dependent manner (5). It has been shown that M1 binding to vRNP in the nucleus is able to block mRNA transcription (4, 6). In the cytosol, M1 interacts with the cytoplasmic tails of the glycoproteins HA and NA. Such interactions promote M1 association with lipid raft membranes and the subsequent polymerization of M1, leading to the elongation of the budding virion (7, 8). Membrane-bound M1 serves as a docking site for the recruitment of the viral RNPs and may also mediate the recruitment of M2 to the site of virus budding (9, 10). Sequence variations in M1 have been found to correlate with the ability of the virus to form either filamentous or spherical particles (11). There are conflicting reports as to whether heterogeneously expressed M1 alone is sufficient for budding (7, 12) or that HA and NA are essential for assembly and budding of virus-like particles (13).
The structure of the influenza A virus M1 has been studied extensively by several research groups, but so far only structures of the N-terminal fragment are available (14–17). Whereas the M1 N domain was found to adopt essentially the same fold in all four reported structures, solution pH appeared to have some effects on its intermolecular interaction, with dimers observed at acidic pH (14) and monomers at neutral pH (15). Electron microscopy images showed that the influenza A virus matrix protein layer in intact particles is made up of rod-shaped M1 with monomers as basic building blocks (18). Electron tomography analysis of infectious influenza virus particles indicates that the matrix protein undergoes conformational changes and dissociates from viral membrane at acidic pH (19). A recent study also shows that the rigid matrix protein layer of the influenza A virus maintains the integrity and shape of the virus and organizes the virus membrane at neutral pH and even during membrane fusion (20). The crystal structures of the M1 N domains from the influenza C virus and thogotovirus have also been reported recently (21, 22).
The infectious salmon anemia virus (ISAV) belongs to the genus Isavirus and is the first orthomyxovirus detected in fish (23), causing serious disease in Atlantic salmon in several continents. Like the influenza A and B viruses, ISAV has a negative-sense RNA genome with eight gene segments that express a total of 10 polypeptides. The smallest segment, segment 8, has two overlapping colinear ORFs that respectively encode two proteins M1 and M2, with M1 being the viral matrix protein with a molecular weight of ∼22 kDa (24). As expected, ISAV M1 is expressed late during infection and accumulates in both nucleus and cytosol (25, 26). A number of findings indicate that the overall organization of the ISA virion, including the viral RNP (27), the nucleoprotein (28), and the viral fusion machinery (29), shares a high level of structural homology to those of the influenza viruses.
To better understand the structure and function of the orthomyxovirus matrix protein, here we determined the crystal structure of the full-length M1 from ISAV (ISAV-M1) at 2.6-Å resolution. The crystal structure of ISAV-M1 has an elbow shape with an N domain and a C domain. In the crystal, ISAV-M1 monomers assemble into an infinite 2D lattice with structural features resembling the membrane-associated protein matrix of the influenza A virus. Cryoelectron tomography of the intact ISA virion showed that ISAV-M1 forms a protein matrix underneath the viral envelope. Fitting the ISAV-M1 2D assembly into the averaged tomogram indicated that the 2D M1 lattice closely mimics the authentic matrix protein layer in infectious particles.
Results
Purification of ISAV-M1.
ISAV-M1 was overexpressed in Escherichia coli and purified as a monomer with a molecular weight of ∼25 kDa according to the gel filtration chromatogram (SI Appendix, Fig. S1). Similar elution profiles were obtained at both pH 4.0 and 7.5, indicating that the oligomerization behavior of ISAV-M1 in solution did not change with pH. The addition of a 6×His tag at either the N or C terminus of ISAV-M1 did not change its solution behaviors compared with the untagged version.
Crystal Structure of ISAV-M1.
Both N-terminal His-tagged and nontagged ISAV-M1 produced similarly shaped, thin-plate crystals that belong to the space group P21. Due to the presence of a pseudotranslation symmetry, heavy atom phasing using Se-Met was not successful. After many heavy atom soaking trials, the crystal structure of ISAV-M1 was eventually solved by single isomorphous replacement with anomalous scattering (SIRAS) using a gold(III) derivative. The final model, which contains three ISAV-M1 molecules, was refined to 2.6-Å resolution using a native dataset to a Rfree of 25.8% and Rwork of 19.2% (SI Appendix, Table S1 and Figs. 1 and 2). All three ISAV-M1 polypeptides are structurally ordered from beginning to end. In the crystal, ISAV-M1 assembles into infinite 2D lattices with interesting implications on M1 polymerization and membrane association (see Higher Order Organization of ISAV-M1). The basic building unit of such 2D lattices is a monomer, consistent with ISAV-M1 being a monomer in solution.
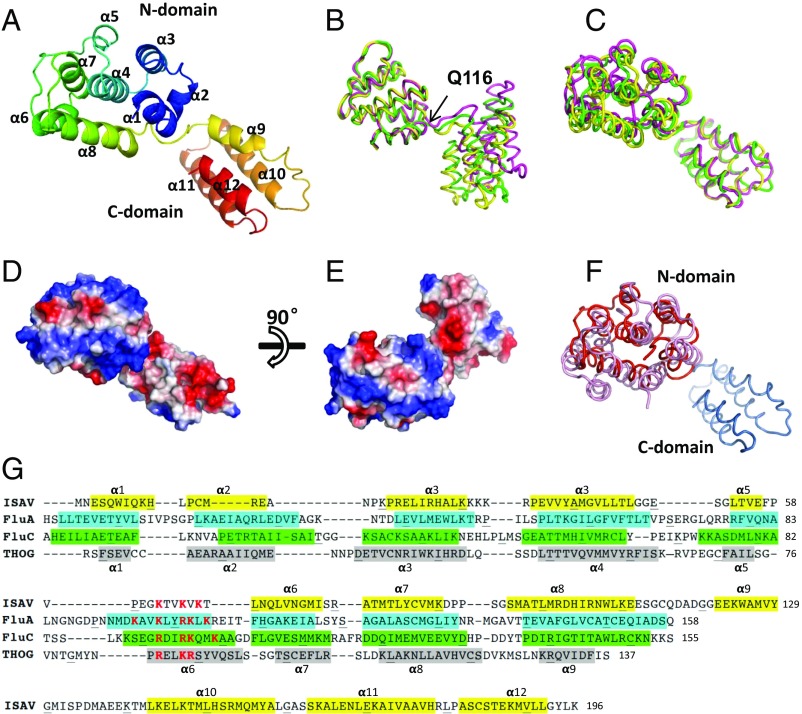
Fig. 1.
Crystal structure of the ISAV-M1. (A) ISAV-M1 crystal structure. The ribbon diagram is rainbow colored with the N terminus in blue and C terminus in red. (B) Superimposition of the three ISAV-M1 molecules related by pseudotranslation symmetry. The three molecules A–C are shown in green, magenta, and yellow, respectively. Superimposition was calculated based on the N domain. Q116 is the hinge point where the structural deviation of the C domain begins. (C) Superimposition of the three ISAV-M1 molecules based on the C domain. (D and E) Molecular surface of ISAV-M1 colored by electrostatic potential viewed from different directions. (F) ISAV-M1 superimposed onto FLUA-M1. ISAV-M1 is shown in red with FLUA-M1 shown in pink. (G) Secondary structure assignment of M1 proteins from ISAV, FluA (influenza A virus), FluC (influenza C virus), and THOG (thogotovirus). The four sequences are aligned according to structure alignment by DALI combined with some manual adjustment. Residues forming α-helices are colored (i.e., ISAV in yellow, FluA in cyan, FluC in green, and THOG in gray) and numbered. The α-helix labels on Top of the aligned sequences apply to ISAV, and the α-helix labels at the Bottom of the aligned sequences apply to FluA. For FLUA-M1, FLUC-M1, and THOG-M1, only the N-domain sequences are shown as the structures of their C domains are not yet available.
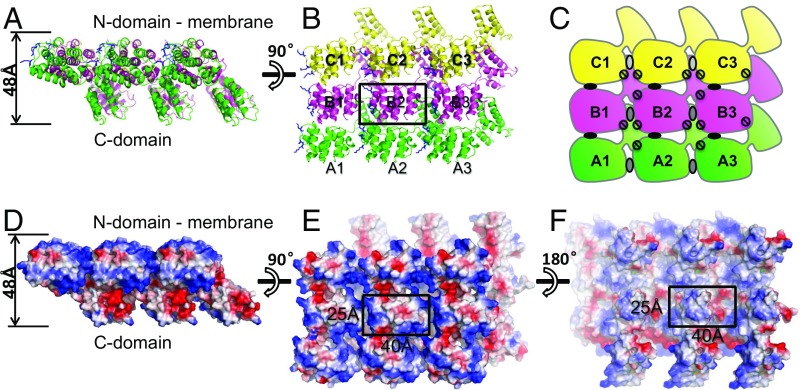
Fig. 2.
The 2D crystal lattice of ISAV-M1. (A and B) Ribbon diagram of the 3 × 3 array viewed from two different directions. The three molecules A–C related by pseudotranslation symmetry are colored in green, magenta, and yellow, respectively. Molecules A1–B1–C1 are related by crystal translation symmetry to A2–B2–C2 and A3–B3–C3. (C) Schematic drawing of the 3 × 3 array corresponding to B. Intermolecular interactions between N and N domains are highlighted by ovals (in either black or gray color), whereas interaction between N and C domains are represented by circles. (D–F) Surface representation of the 3 × 3 array viewed from three different directions. Molecules are colored by electrostatic potential with positive in blue and negative in red. A single repeating unit is highlighted in B, E, and F.
Full-length ISAV-M1 has the shape of an elbow with a dimension of 32 × 32 × 65 Å3 (Fig. 1A). It can be divided into two linear domains, the N domain (residues 1–111) and the C domain (residues 122–196), that are connected by an extended, 10-aa loop linker (residues 112–121). Each ISAV-M1 is comprised of 12 α-helices (i.e., α1–α12), with the first 8 α-helices (i.e., α1–α8) forming the N domain and the last 4 α-helices (i.e., α9–α12) making up the C domain. The N and C domains from the same molecule do not directly interact with each other.
The structure of the ISAV-M1 N domain can be described as a helical bundle with a central α-helix (i.e., α4) surrounded by five α-helices (i.e., α1, α3, α5, α7, and α8) in the shape of a barrel, which is further packed by two α-helices (i.e., α2 and α6) on the outside. Nearly all residues at the interface between the central α-helix and its five surrounding helices are hydrophobic in nature. Interaction between the two outermost α-helices and the core α-helical barrel are also largely mediated by hydrophobic residues, indicating that the entire N domain likely functions as a rigid unit during virus infection. Two cysteines (i.e., C13 and C115) lie adjacent to each other near the linker loop (SI Appendix, Fig. S2). These two cysteines form a disulfide bond in only one of the three noncrystallographic symmetry (NCS)-related molecules (i.e., molecule C). The formation of the disulfide bond keeps the linker loop closely tethered to the N domain.
In the C domain, the four α-helices α9–α12 pack closely against each other, forming an α-helical bundle that is stabilized by a hydrophobic core. A salt bridge between residues E122 and R153 from two adjacent α-helices further helps to strengthen the structural integrity of the C domain.
Comparing the structures of the three M1 molecules from the same crystallographic asymmetric unit reveals considerable structural flexibility in the loop linker (Fig. 1 B and C). For instance, superposition of the three molecules by their N domains shows that the hinge angles can vary by as much as 40°, with molecules B and C adopting the largest and smallest hinge angles, respectively (Fig. 1B, magenta vs. yellow). Major deviation in the linker conformation occurs around Q116 in the middle of the linker (Fig. 1B). The linker region from residues E112 to Q116 is held in place by both covalent (i.e., S–S bond) and noncovalent interactions with the N domain.
Comparison of ISAV-M1 to Homologous Viral Matrix Proteins.
Phylogenetic analyses of orthomyxovirus M1 proteins indicate that the six genera within the family form separate clusters, with ISAV and Thogotovirus forming the two most divergent groups (SI Appendix, Fig. S3). Using the program DALI (30) and ISAV-M1 as the reference, pairwise structure alignment to FLUA-M1, FLUC-M1 and THOG-M1 gave a Z score of 4.1 (rmsd = 3.6 Å for 90 Cα atoms), 4.0 (rmsd = 3.9 Å for 93 Cα atoms), and 5.2 (rmsd = 2.9 Å for 84 Cα atoms), respectively, indicating that these M1 proteins are evolutionarily related in the absence of apparent sequence identities (SI Appendix, Fig. S4).
The FLUA-M1 contains a total of 252 amino acid residues, which is ∼25% larger than the 196-aa ISAV-M1. Like ISAV-M1, the M1 of the influenza A virus also has a two-domain organization, with its first 165 residues making up the N domain. Eight of the nine α-helices in the FLUA-M1 N domain can be matched to those in ISAV-M1 (Fig. 1 F and G). The most notable differences are: (i) the replacement of the helix α6 in FLUA-M1 by a structured loop in ISAV-M1, and (ii) a shorter α2 in ISAV-M1 than its counterpart in FLUA-M1. The positively charged region located between α5 and α6 in ISAV-M1 is highly conserved and likely plays an important role in mediating M1 membrane association (see Functional Implications).
The crystal structure of ISAV-M1 C domain is the first reported for any orthomyxovirus M1 proteins. When it was submitted for a homology search by DALI (30), a number of diverse protein hits were identified, including the Clostridium difficile toxin glucosyltransferase domains, the diversity-generating retroelement protein bAvd, and two S23 ribosomal-like proteins Xanthomonas campestris Xcc0516 and Bacteriodes thetaiotamicron B.t_0352. Considering the different functions and surface properties of these molecules, the structural homology is likely due to convergent evolution, as the four-helix bundle found in the ISAV-M1 C domain is a common structural fold.
Higher Order Organization of ISAV-M1.
In the crystal, ISAV-M1 forms an infinite 2D sheet of a single protein layer (Fig. 2). In this 2D layer ISAV-M1 molecules are oriented in a parallel manner, with their N domains situated on top and their C-terminal domains at bottom. The top face of the sheet is smooth and nearly flat without obvious pores, whereas the bottom face has a rugged appearance due to pronounced protrusions caused by the C domain. This lattice sheet has a regular spacing of 40 × 25 Å2 and a height of 48 Å. Each ISAV-M1 molecule lies ∼45° sideways; therefore, the height of the lattice is shorter than the length of the molecule (i.e., 48 vs. 65 Å). In the crystal, multiple layers of such 2D sheets stack on top of each other in the same direction as directed by a crystallographic 21 screw axis.
To closely examine molecular interactions involved in the formation of the 2D lattice, a 3 × 3 array of nine ISAV-M1 molecules is taken from the sheet (Fig. 2). These nine molecules can be generated from the three NCS-related monomers (i.e., A–C) by simply applying crystallographic translation symmetry (Fig. 2 A–C). Using the central molecule B2 as the reference, we calculated the buried surface between B2 and all eight surrounding molecules to determine dominant intermolecular interactions in the 2D network (SI Appendix, Figs. S5 and S6). If we ignore NCS-related differences, only four of the eight pairs of molecules involve unique interactions.
Among these four pairs of molecules, the largest interface (∼2,300 Å2) is given by B2–C2 (or A2–B2), the two adjacent molecules related by the pseudotranslation symmetry (Fig. 2 B and C and SI Appendix, Fig. S5). The B2–C2 interface, which will be referred to as “interface I” in our discussion, involves the interaction between the B2 N and C domains with the C2 N domain. The second largest interface (∼400–800 Å2), which is called interface II, is observed in the B2–A1 pair (or the B2–C3 pair), which involves the interaction between the A1 C domain and the B2 N domain. A much weaker interaction (i.e., ∼300 Å2), interface III, is observed between molecules from the same row or molecules related by crystallographic translation symmetry (i.e., B2–B1 or B2–B3). This interface involves only N domains from neighboring molecules. There is no interaction at the B2–C1 or the B2–A3 interface.
Based on the interaction between B2 and its adjacent molecules, we speculate that M1 could form an elongated ribbon through interactions mediated by interface I. Such 1D ribbons could further polymerize into extended 2D sheets through interface II and interface III. Therefore, the three interfaces I–III are solely responsible for maintaining the intact 2D lattice of ISAV-M1 as seen in the crystal. Amino acid residues responsible for forming the intricate 2D crystal lattice appear to be strictly conserved among all ISAV isolates (SI Appendix, Figs. S6 and S7).
The 2D crystal lattice sheet of ISAV-M1 bears close resemblance to the influenza A virus matrix protein layer associated with the viral envelope at the particle interior (18). FLUA-M1 appears as thin rod-shaped molecules, and with one end touching the membrane, they form a regularly spaced lattice of densely packed rods with a spacing of ∼40 × 40 Å2 and a height of 60 Å (18). Under the negative staining EM, most of the FLUA-M1 thin rods also lie ∼45° sideways relative to the plane of the viral membrane. The spacing and the height of the ISAV-M1 lattice sheet are slightly smaller than that of the influenza A virus matrix protein layer, which is not surprising, considering the smaller molecular size of ISAV-M1.
ISAV-M1 Membrane Binding.
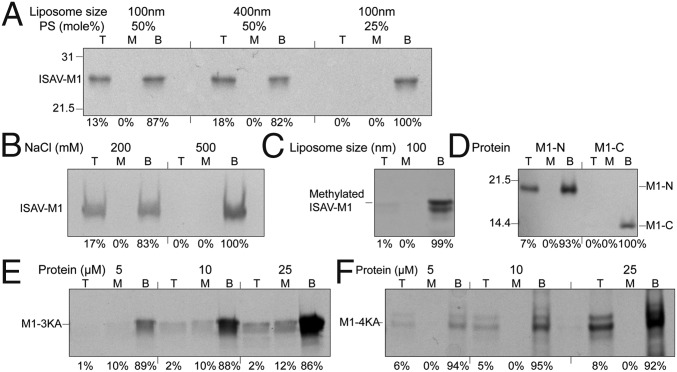
Because FLUA-M1 has been shown to bind negatively charged liposomes (18), here we tested the ability of ISAV-M1 to bind phospholipids using a discontinuous Accudenz gradient following a previously described protocol with some modifications (18). The detection of the ISAV-M1 in the top fraction confirmed the ability of ISAV-M1 to interact with phospholipids (Fig. 3A). The strength of the interaction is dependent on the size of the liposome, as 18% of the total protein was found in the top fraction, using ∼400-nM liposomes compared with 13% using ∼100-nM liposomes (Fig. 3A). The preference for larger liposomes likely reflects the effect of membrane curvature on ISAV-M1 binding. As M1 is found at the interior of the viral particle, it may prefer a concave or a flat membrane, and a membrane with too large of a convex curvature may actually inhibit binding. ISAV-M1 liposome binding also depends on the phospholipid composition: When the ratio of phosphatidylserine to phosphatidylcholine was reduced from 50:50 to 25:75, ISAV-M1 binding to liposome was completely abolished (Fig. 3A), indicating that net negative charges on the lipid are essential. Solution ionic strength also affects ISAV-M1 lipid binding, as the interaction was no longer observed in 500 mM NaCl (Fig. 3B), suggesting that ISAV-M1 lipid binding is largely mediated by electrostatic interactions. Our results were consistent with those previously reported for FLUA-M1 and also the M protein from vesicular stomatitis virus (VSV) (18, 31).
Fig. 3.
ISAV-M1 membrane association by liposome flotation. M1 protein was incubated overnight with liposomes under different conditions and floated on an Accudenz discontinuous gradient. Top (T), Middle (M), and Bottom (B) fractions of the discontinuous gradient were analyzed by SDS/PAGE and stained with Coomassie blue. Flotation assays of M1 with (A) PC:PS (50:50) 100 nm or 400 nm liposomes, and 100 nm PC:PS (75:25) liposomes; (B) 400 nm PC:PS (50:50) liposomes incubated with M1 the Tris buffer with 200 mM or 500 mM NaCl; (C) DMAB-treated M1 floated with 100 nm PC:PS (50:50) liposomes; (D) M1 N-terminal truncation (1–119) and C-terminal truncation (120–196) floated with 400 nm PC:PS (50:50) liposomes; (E) M1-3KA (K63, 66, 68A) and (F) M1-4KA (K30, 31, 32, 33A) at different concentrations floated with 400 nm PC:PS (50:50).
To confirm that positive charges on ISAV-M1 play an important role in liposome binding, the protein was subjected to lysine methylation (32). Methylated ISAV-M1 completely lost its liposome binding activity (Fig. 3C). To dissect which one of the two ISAV-M1 domains is responsible for liposome binding, we engineered two truncation mutants, ISAV-M1-N (amino acids 1–119) and ISAV-M1-C (amino acids 120–196), among which only ISAV-M1-N was capable of liposome binding (Fig. 3D). There are two clusters of positively charged residues in the ISAV-M1 N domain (amino acids 1–111): one is composed of K30–K33 located between α3 and α4, and the other is composed of K63, K66, and K68 located between α5 and α6 (SI Appendix, Fig. S8 and Fig. 2 A and B). Both clusters are situated on the top surface of the 2D lattice and well positioned to interact with an approaching phospholipid membrane. Two M1 mutants named M1-4KA and M1-3KA were subsequently generated, each containing the mutations K30A/K31A/K32A/K33A and K63A/K66A/K68A, respectively. Whereas M1-4KA retained most of its activity, M1-3KA had almost completely lost its ability to bind liposomes (Fig. 3 E and F), suggesting that the loop connecting α5 and α6 likely plays an important role in facilitating ISAV-M1 membrane recruitment and association (SI Appendix, Figs. S8 and S9).
ISAV-M1 RNA Binding.
Because FLUA-M1 was reported to nonspecifically bind ssRNA (33, 34), we measured the RNA binding affinity of the ISAV-M1 protein using fluorescence polarization assays with synthetic RNA oligos (28). Three RNA oligos of different length (i.e., 12 nt, 24 nt, and 48 nt) were used, but only modest differences were observed in their binding affinities (SI Appendix, Fig. S10A), suggesting that the RNA binding footprint on ISAV-M1 is 12 nt or shorter. When the same fluorescence polarization assay was used to quantitate ISAV-M1 RNA binding stoichiometry, it was determined that each ISAV-M1 binds to one RNA molecule (SI Appendix, Fig. S10B). Full-length ISAV-M1 and ISAV-M1-N exhibited similar RNA binding affinities (SI Appendix, Fig. S10C), but the RNA binding affinity of ISAV-M1-C was ∼40-fold weaker compared with the full-length ISAV-M1.
The RNA binding sequence of the FLUA-M1 was mapped to amino acid residues 90–111 located in the N domain (35, 36). The equivalent region in ISAV-M1 is the loop connecting α5 and α6, which was found essential for ISAV-M1 membrane association as discussed above. Interestingly, when the two mutants M1-3KA and M1-4KA were subjected to fluorescence polarization assays, little difference was observed in the RNA binding affinities of the two mutants compared with the wild-type M1 (SI Appendix, Fig. S10D), suggesting that neither one of the two charged loops plays a major role in ISAV-M1 RNA binding.
Cryoelectron Tomography of ISA Virion.
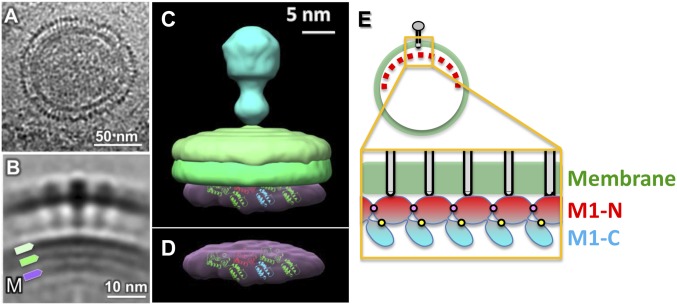
Cryoelectron tomography (cryo-ET) was used to analyze the structural organization of the matrix protein layer in the ISA virion in situ. When visualized by cryo-ET, ISA virions showed numerous mushroom-like and occasionally rod-like protruding spikes extending outward from the virion surface (Fig. 4 A and B). These protruding spikes were likely hemagglutinin esterase (HE) and fusion (F) proteins. At the base of these membrane protein molecules, we frequently observed triple-layered density features. To obtain high-resolution details of the matrix protein layer, we extracted a total of 2,600 subtomograms of envelope spikes from 10 virus tomograms. Subtomogram averaging and classification were used to analyze the envelope spike and the viral envelope as described (37). The two outermost layers had approximately equal densities with an overall thickness of ∼3.5 nm and likely corresponded to the lipid bilayer of the viral envelope (Fig. 4B). The innermost layer at the particle interior was assigned as the matrix protein lattice, which is supposedly to lie underneath the viral membrane in close association. The ISAV-M1 layer was ∼4 nm away from the inner membrane leaflet and was ∼4 nm thick (Fig. 4 A and B). A 3 × 3 2D crystal lattice of the ISAV-M1 was docked into the density of the third layer, and the results showed that the overall height of the whole molecule closely matches the thickness of the density (Fig. 4 C and D).
Fig. 4.
Cryoelectron tomography of the ISA virion. (A) Central slide from a tomogram of an ISA viral particle. (B) Averaged structure showing the ISA viral envelope. Triple-layered membrane densities are visible in A and B. (C) A 3D rendering of the averaged structure. The three layers of density are interpreted as the inner membrane leaflet, the outer membrane leaflet, and the matrix protein shell. A 3 × 3 ISAV-M1 crystal lattice is fitted into the matrix protein layer. (D) Isolated density corresponding to the matrix protein layer. (E) Schematic diagram showing M1 protein polymerization under the viral envelope. The formation of the matrix protein layer involved interactions between the M1 N and N domains (represented by pink dots) as well as the M1 N and C domains (represented by yellow dots).
Functional Implications.
The crystal structure of the ISAV-M1 reported here provides a first glimpse into the structure of a full-length matrix protein from the Orthomyxoviridae family. Each ISAV-M1 molecule assumes the shape of an elbow with two globular domains that are connected by a flexible linker. Unlike the linker sequence in FLUA-M1, which is highly susceptible to proteolytic attack (15, 38), the linker of ISAV-M1 has the sequence of 112ESGCQDADGG121 and is highly resistant to contaminating proteases, possibly due to the lack of hydrophobic or arginine/lysine residues, thus allowing us to determine its full-length crystal structure.
The N domain of ISAV-M1 is structurally homologous to the FLUA-M1 N domain with superimposable secondary structure elements (Fig. 1 F and G). Although there is no X-ray structure currently available for the C domain of FLUA-M1, it is predicted to contain four α-helices (39, 40), exactly what is observed in the ISAV-M1 structure. The high α-helical content of the FLUA-M1 C domain was also confirmed by a number of solution studies (15, 39). Additional evidence suggesting that ISAV-M1 and FLUA-M1 share similar structure and function includes: (i) The elbow-shape of ISAV-M1 closely resembles that of virion-associated FLUA-M1 molecules seen in negative-staining EM images (18); (ii) both ISAV-M1 and FLUA-M1 exist as monomers in solution (15); and (iii) ISAV-M1 demonstrates an inherent membrane association activity similar to that of the FLUA-M1 (18).
ISAV-M1 polymerizes into infinite 2D arrays in crystal, possibly as a consequence of a crowding effect due to the use of polyethylene glycol as the precipitating reagent. The requirement of both N and C domains for 2D lattice formation likely explains why similar 2D lattices have not been observed in the crystal structures of FLUA-M1, FLUC-M1, and THOG-M1 that only include the N domain. An earlier study using small angle X-ray scattering indicated that the full-length FLUA-M1 monomers coexist in solution with a small fraction of large clusters with a layered architecture (41), suggesting a key role of the C terminus of FLUA-M1 in the formation of 2D lattice sheets. Similarly, studies on FLUC-M1 also showed that membrane tubule formation requires its C domain, corroborating its essential role for FLUC-M1 polymerization (22). In vivo, the 2D M1 lattice formation is likely promoted by cell membrane, as it has been shown that FLUA-M1 and FLUC-M1 oligomer formation is strongly enhanced by membrane binding and does not require the presence of other viral proteins (22, 41).
The formation of an S–S bond between cysteine residues C13 and C115 has been observed to modulate the relative orientation of the two domains of ISAV-M1 (Fig. 1B and SI Appendix, Fig. S2). An interesting hypothesis is that the status of the S–S bond may affect ISAV-M1 polymerization/depolymerization during virus budding and/or virus entry. Variations in the oxidative status of the S–S bond may explain the difficulties we had in reproducing ISAV-M1 crystals. Many batches of purified proteins could not produce crystals, and in all cases proteins stored overnight were no longer useful in crystallization. Cysteines are not found at the equivalent positions in FLUA-M1, suggesting that FLUA-M1 may use an alternative mechanism to modulate the hinge angle between the N and C domains.
ISAV-M1 binds to liposomes in vitro with a strong preference for phosphatidylserine (PS) over phosphatidylcholine (Fig. 3). Phosphatidylserine is universally present in the inner leaflet of the plasma membrane in all multicellular organisms ranging from mammals to nematodes (42). As an important component in the cell clearance pathways, PS becomes externalized during apoptosis to signal for cell clearance. Indeed, studies of ISAV infection at 15 °C indicated that infected cells showed increased surface expression of phosphatidylserine (43). ISAV-M1 interactions with phosphatidylserine may facilitate the recruitment of M1 to the cell membrane and promote its interaction with other viral glycoproteins (i.e., HE and F) during virus assembly and budding. The N domain of ISAV-M1 is largely responsible for liposome binding, a situation similar to FLUA-M1 (4). Three basic residues (i.e., K63, K66, and K68) located on top of the 2D lattice have been found important for ISAV-M1 liposome binding, confirming our speculation that the top face of the 2D lattice directly interacts with the viral membrane and the C domain faces the virus interior. The three basic residues from ISAV-M1 are structurally aligned to a polybasic region composed of residues 95–105 in the FLUA-M1 N domain, which was also speculated to mediate membrane association (4). This polybasic region appears to be conserved in influenza C and thogotovirus as well (Fig. 1G). There is evidence that multiple polypeptide regions of FLUA-M1 are necessary to sustain membrane binding in vivo (44). Likewise, other basic residues on the top face of the ISAV-M1 2D lattice, including K31–K34, may make further contributions to allow efficient membrane recruitment of ISAV-M1 during virus morphogenesis.
FLUA-M1 interacts with RNPs to mediate RNP export from the nucleus and to facilitate RNP packaging into budding virions. However, the mechanism of M1-RNP interaction remains poorly understood. FLUA-M1 has been shown to bind NP (45), RNA (34, 36), as well as RNP (4, 35, 46). In particular, Baudin et al. demonstrated that it is the FLUA-M1 C domain, instead of the N domain, that is required for RNP binding (4). In a more recent study, it was shown that FLUA-M1 is SUMOylated at K242 from the C domain, and that the SUMOylation of M1 is required for the interaction between M1 and viral RNP (46). ISAV-M1 showed no binding activity toward purified recombinant NP in vitro. Nevertheless, the structural organization of the ISAV-M1 2D lattice suggests that ISAV-M1 C domain, which points toward the viral interior, likely mediates RNP interaction. Such interaction may require NP molecules to be arranged in the form of the double-helical RNP instead of oligomeric rings found in recombinant samples. We showed that ISAV-M1 N domain has a nonspecific ssRNA binding activity similar to FLUA-M1. Therefore, the N domain may contribute to RNP binding through its nonspecific RNA binding activity. RNA loops on the surface of RNP may extend through the gaps in between adjacent C domains in the 2D M1 lattice to reach the N domain. The ssRNA binding by the ISAV-M1 is most likely mediated by residues from the side of the N domain that are accessible from the interior of the viral particle. Since the highly conserved lysine residue K188 is found near the C terminus of ISAV-M1, it remains to be seen whether it is a potential SUMOylation site implicated in RNP binding.
As shown in Fig. 4E, results from our study support a virus assembly model involving the matrix layer. The matrix is made of a single layer of M1 molecules arranged in a 2D lattice similar to that seen in the ISAV-M1 crystal. The formation of the 2D M1 lattice requires interaction mediated by both the N and C domains, thus explaining that only full-length M1 molecules are efficiently incorporated into viral particles (45). The top face of the lattice contacts the membrane, whereas bottom face interacts with the RNPs through protrusions made of the C domain. This model also predicts that interactions between M1 and viral glycoproteins are likely mediated by the N domain as it lies adjacent to the viral envelope. Factors that either weaken M1 interaction with the membrane or M1 lattice stability would result in the loss of the matrix layer. We expect that the 2D lattice of ISAV-M1 will serve as a useful structural platform so that our understanding of biochemical activities of orthomyxovirus M1 can be unified and placed in a biologically relevant context.
Materials and Methods
Details of experimental procedures are available in SI Appendix, SI Materials and Methods. The structure of ISAV-M1 was determined by SIRAS. Membrane binding activity of ISAV-M1 was analyzed using a discontinuous Accudenz gradient with phospholipids. ISAV-M1 RNA binding was measured using fluorescence polarization assays with synthetic RNA oligos. Cryoelectron tomography was used to analyze the structural organization of the matrix protein layer in intact ISA virions.
Supplementary Material
Acknowledgments
We thank Yusong Guo and John Olson for helpful discussions. This work was supported by the Welch Foundation (Awards C-1565 to Y.J.T. and AU-1714 to J.L.), the National Institutes of Health Grant (AI077785 to Y.J.T.), the National Key Research and Development Program of China Grant (2016YFD0500205 to M.J.), the Hamill Foundation award (to Y.J.T.), and the Kresge Science Initiative Endowment Fund (Rice University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5WCO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701747114/-/DCSupplemental.
References
- 1.Lamb RA, Lai CJ. Conservation of the influenza virus membrane protein (M1) amino acid sequence and an open reading frame of RNA segment 7 encoding a second protein (M2) in H1N1 and H3N2 strains. Virology. 1981;112:746–751. doi: 10.1016/0042-6822(81)90319-6. [DOI] [PubMed] [Google Scholar]
- 2.Neumann G, Hughes MT, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin F, Petit I, Weissenhorn W, Ruigrok RW. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology. 2001;281:102–108. doi: 10.1006/viro.2000.0804. [DOI] [PubMed] [Google Scholar]
- 5.Akarsu H, et al. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2) EMBO J. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye ZP, Pal R, Fox JW, Wagner RR. Functional and antigenic domains of the matrix (M1) protein of influenza A virus. J Virol. 1987;61:239–246. doi: 10.1128/jvi.61.2.239-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gómez-Puertas P, Albo C, Pérez-Pastrana E, Vivo A, Portela A. Influenza virus matrix protein is the major driving force in virus budding. J Virol. 2000;74:11538–11547. doi: 10.1128/jvi.74.24.11538-11547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruigrok RW, Baudin F, Petit I, Weissenhorn W. Role of influenza virus M1 protein in the viral budding process. Int Congr Ser. 2001;1219:397–404. [Google Scholar]
- 9.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc Natl Acad Sci USA. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latham T, Galarza JM. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J Virol. 2001;75:6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 15.Arzt S, et al. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology. 2001;279:439–446. doi: 10.1006/viro.2000.0727. [DOI] [PubMed] [Google Scholar]
- 16.Harris A, Forouhar F, Qiu S, Sha B, Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- 17.Safo MK, et al. Crystal structures of influenza A virus matrix protein M1: Variations on a theme. PLoS One. 2014;9:e109510. doi: 10.1371/journal.pone.0109510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruigrok RW, et al. Membrane interaction of influenza virus M1 protein. Virology. 2000;267:289–298. doi: 10.1006/viro.1999.0134. [DOI] [PubMed] [Google Scholar]
- 19.Fontana J, Steven AC. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J Virol. 2013;87:5621–5628. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang M, et al. pH-dependent conformational changes of a Thogoto virus matrix protein reveal mechanisms of viral assembly and uncoating. J Gen Virol. 2016;97:2149–2156. doi: 10.1099/jgv.0.000551. [DOI] [PubMed] [Google Scholar]
- 22.Saletti D, et al. The Matrix protein M1 from influenza C virus induces tubular membrane invaginations in an in vitro cell membrane model. Sci Rep. 2017;7:40801. doi: 10.1038/srep40801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cottet L, Rivas-Aravena A, Cortez-San Martin M, Sandino AM, Spencer E. Infectious salmon anemia virus: Genetics and pathogenesis. Virus Res. 2011;155:10–19. doi: 10.1016/j.virusres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Bierin E, et al. Segment 8 encodes a structural protein of infectious salmon anaemia virus (ISAV); the co-linear transcript from Segment 7 probably encodes a non-structural or minor structural protein. Dis Aquat Organ. 2002;49:117–122. doi: 10.3354/dao049117. [DOI] [PubMed] [Google Scholar]
- 25.Ramly RB, Olsen CM, Braaen S, Rimstad E. Infectious salmon anaemia virus nuclear export protein is encoded by a spliced gene product of genomic segment 7. Virus Res. 2013;177:1–10. doi: 10.1016/j.virusres.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Falk K, Aspehaug V, Vlasak R, Endresen C. Identification and characterization of viral structural proteins of infectious salmon anemia virus. J Virol. 2004;78:3063–3071. doi: 10.1128/JVI.78.6.3063-3071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falk K, Namork E, Rimstad E, Mjaaland S, Dannevig BH. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) J Virol. 1997;71:9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng W, Olson J, Vakharia V, Tao YJ. The crystal structure and RNA-binding of an orthomyxovirus nucleoprotein. PLoS Pathog. 2013;9:e1003624. doi: 10.1371/journal.ppat.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook JD, Soto-Montoya H, Korpela MK, Lee JE. Electrostatic architecture of the infectious salmon anemia virus (ISAV) core fusion protein illustrates a carboxyl-carboxylate pH sensor. J Biol Chem. 2015;290:18495–18504. doi: 10.1074/jbc.M115.644781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakowski JJ, Petri WA, Jr, Wagner RR. Role of matrix protein in assembling the membrane of vesicular stomatitis virus: Reconstitution of matrix protein with negatively charged phospholipid vesicles. Biochemistry. 1981;20:3902–3907. doi: 10.1021/bi00516a037. [DOI] [PubMed] [Google Scholar]
- 32.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakefield L, Brownlee GG. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elster C, Larsen K, Gagnon J, Ruigrok RW, Baudin F. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J Gen Virol. 1997;78:1589–1596. doi: 10.1099/0022-1317-78-7-1589. [DOI] [PubMed] [Google Scholar]
- 35.Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. Association of influenza virus matrix protein with ribonucleoproteins. J Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada A, Miura T, Takeuchi H. Zinc- and pH-dependent conformational transition in a putative interdomain linker region of the influenza virus matrix protein M1. Biochemistry. 2003;42:1978–1984. doi: 10.1021/bi027176t. [DOI] [PubMed] [Google Scholar]
- 39.Shishkov AV, et al. The in situ spatial arrangement of the influenza A virus matrix protein M1 assessed by tritium bombardment. Proc Natl Acad Sci USA. 1999;96:7827–7830. doi: 10.1073/pnas.96.14.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shishkov A, et al. Spatial structure peculiarities of influenza A virus matrix M1 protein in an acidic solution that simulates the internal lysosomal medium. FEBS J. 2011;278:4905–4916. doi: 10.1111/j.1742-4658.2011.08392.x. [DOI] [PubMed] [Google Scholar]
- 41.Shtykova EV, et al. Structural analysis of influenza A virus matrix protein M1 and its self-assemblies at low pH. PLoS One. 2013;8:e82431. doi: 10.1371/journal.pone.0082431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klöditz K, Chen YZ, Xue D, Fadeel B. Programmed cell clearance: From nematodes to humans. Biochem Biophys Res Commun. 2017;482:491–497. doi: 10.1016/j.bbrc.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiøtz BL, Baekkevold ES, Poulsen LC, Mjaaland S, Gjøen T. Analysis of host- and strain-dependent cell death responses during infectious salmon anemia virus infection in vitro. Virol J. 2009;6:91. doi: 10.1186/1743-422X-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thaa B, Herrmann A, Veit M. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology. 2009;383:150–155. doi: 10.1016/j.virol.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Noton SL, et al. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu CY, Jeng KS, Lai MM. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol. 2011;85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.