Significance
Toll-like receptor (TLR) signaling pathways are targeted to limit inflammation in immune cells. TLRs use adaptor proteins to drive inflammatory signaling platforms for effective microbial clearance. Here we show that MyD88 adaptor-like (MAL), an adaptor protein in TLR signaling, undergoes glutathionylation in response to LPS, driving macrophage responses to proinflammatory stimuli. We also determined the solution structure of MAL in the reduced form without disulfides, revealing a typical BB loop observed in adaptor proteins, in contrast to previously reported crystal structures. This alternate solution structure reveals the inherent flexibility of MAL, supporting the hypothesis that glutathionylation may reposition the MAL BB loop for MyD88 interaction to drive inflammation. This discovery could lead to novel approaches to target MAL glutathionylation in dysregulated TLR signaling, limiting inflammation.
Keywords: MAL/TIRAP, glutathione, Toll-like receptor, NMR spectrometry, inflammation
Abstract
MyD88 adaptor-like (MAL) is a critical protein in innate immunity, involved in signaling by several Toll-like receptors (TLRs), key pattern recognition receptors (PRRs). Crystal structures of MAL revealed a nontypical Toll/interleukin-1 receptor (TIR)-domain fold stabilized by two disulfide bridges. We therefore undertook a structural and functional analysis of the role of reactive cysteine residues in the protein. Under reducing conditions, the cysteines do not form disulfides, but under oxidizing conditions they are highly amenable to modification. The solution structure of the reduced form of the MAL TIR domain, determined by NMR spectroscopy, reveals a remarkable structural rearrangement compared with the disulfide-bonded structure, which includes the relocation of a β-strand and repositioning of the functionally important “BB-loop” region to a location more typical for TIR domains. Redox measurements by NMR further reveal that C91 has the highest redox potential of all cysteines in MAL. Indeed, mass spectrometry revealed that C91 undergoes glutathionylation in macrophages activated with the TLR4 ligand lipopolysaccharide (LPS). The C91A mutation limits MAL glutathionylation and acts as a dominant negative, blocking the interaction of MAL with its downstream target MyD88. The H92P mutation mimics the dominant-negative effects of the C91A mutation, presumably by preventing C91 glutathionylation. The MAL C91A and H92P mutants also display diminished degradation and interaction with interleukin-1 receptor-associated kinase 4 (IRAK4). We conclude that in the cell, MAL is not disulfide-bonded and requires glutathionylation of C91 for signaling.
Toll-like receptors (TLRs) are an important class of pattern recognition receptors. Upon recognition of pattern-associated molecular patterns, TLRs undergo dimerization. The consequent dimerization of the cytoplasmic Toll/interleukin-1 receptor (IL-1R)/resistance protein (TIR) domains leads to the recruitment of TIR domain-containing adaptor proteins, such as MyD88 and MyD88 adaptor-like (MAL; or TIRAP). MAL is required to recruit MyD88 to multiple TLRs, including TLR2 (1, 2), TLR4 (3), and TLR9 (4). The assembly culminates in an oligomeric assembly of kinases, including IRAK2 and IRAK4 (5, 6), termed the Myddosome, leading to NF-κB activation and inflammatory gene expression.
The crystal structures of the TIR domain of MAL (7–10) showed significant differences from the structures of other TIR domains, including MyD88 (11), IL-1 receptor accessory protein-like 1 (IL-1RAPL) (12), TLR1, TLR2 (13), and TLR10 (14). In particular, the MAL TIR-domain structures lack helix αB and feature a long AB loop situated between helix αA and β-strand βB, which includes the conserved and functionally important BB-loop proline-containing motif (15–17). Furthermore, these crystal structures contain two disulfide bonds involving residues C89–C134 and C142–C174, respectively. The presence of disulfides is unusual for a cytosolic protein and poses an intriguing possibility of redox control of MAL-mediated signaling.
Posttranslational modifications of MAL have been studied extensively (18–22); however, redox mechanisms affecting MAL have not been characterized. Glutathione (γ-l-glutamyl-l-cysteinyl-glycine; GSH), a tripeptide composed of the amino acids glutamic acid, cysteine, and glycine, is the most abundant nonprotein tripeptide and is used by the cell to maintain redox homeostasis and prevent overoxidation of cytoplasmic components (23). The cysteine thiol group (-SH) forms a mixed disulfide with GSH, which can be spontaneously or enzymatically catalyzed. GSH, however, does not act solely as an antioxidant. Recent evidence has identified that GSH can act as both a positive and negative regulator of proteins, regulating STAT3 phosphorylation (24), caspase-1 activity (25), and the anion carrier proteins mitochondrial uncoupling proteins 2 and 3 (UCP2 and UCP3) (26). GSH has also been linked to stabilization of hypoxia-inducible factor 1-α (HIF1α) under ischemic conditions (27).
Here, we present the structural and functional characterization of the role of reactive cysteine residues in MAL. We determined the solution structure of the C116A mutant of TIR domain of MAL, observing remarkable structural differences compared with the disulfide-bonded structure, including the relocation of a β-strand and repositioning of the BB-loop region to a location more typical for TIR domains. C91 is found to be the most reactive cysteine in the protein, and indeed is shown to undergo glutathionylation in macrophages. This glutathionylation is required for the interaction of MAL with MyD88 and subsequent phosphorylation and degradation by IRAK4. Jointly, our data show that the disulfide-bonded form of MAL is unlikely to be the signaling-competent form in the cell, but that modification of reactive cysteines is functionally important, in particular glutathionylation of C91 during signaling.
Results
Redox States of Cysteines in Escherichia-coli-Expressed MALTIR.
The crystal structures of the TIR domain of MAL (7–10) revealed two disulfide bonds involving residues C89–C134 and C142–C174, respectively. To analyze the redox states of cysteine residues under different redox conditions, we expressed the protein corresponding to its TIR domain (residues 79–221; MALTIR) in three different Escherichia coli (E. coli) cell types (BL-21, SHuffle, and Origami). BL-21 cells contain a reducing cytosolic environment, similar to eukaryotic cells. On the other hand, SHuffle and Origami are strains that have been engineered so that cytoplasmic reductive pathways are impaired, resulting in an oxidative cytosolic environment that promotes the formation of cytoplasmic disulfide bonds (28). MALTIR produced in all three bacterial strains yielded a similar elution profile by size-exclusion chromatography (SI Appendix, Fig. S1). Ellman’s reagent was used to estimate the average percentage of reduced cysteine residues in MALTIR (SI Appendix, Table S1). These results indicate that the cysteine residues in MALTIR are in the reduced form under reducing conditions, but are sensitive to redox modifications. The intact mass of MALTIR-expressed E. coli BL-21 cells was further analyzed using mass spectrometry, revealing the possible presence of one disulfide bond.
To analyze the role of individual cysteine residues in MALTIR, single-cysteine mutants were produced in both BL-21 and SHuffle cells (SI Appendix, Fig. S1). The results show that some cysteine residues are more prone to oxidation than others (SI Appendix, Table S2). Exposure of MALTIR to 5 mM oxidized glutathione (GSSG) resulted in a number of modifications including S-glutathionylation, dioxidation and dehydroalanine formation (SI Appendix, Table S3 and Fig. S2). Taken together, our results show that the cysteine residues of MAL are susceptible to modifications in solution.
NMR Solution Structure of MALTIRC116A Reveals Remarkable Structural Differences from the Crystal Structures.
We used NMR spectroscopy to characterize MALTIR structurally. To identify the best experimental conditions for such studies, the effects of temperature and protein concentration on the stability of MALTIR and its single-cysteine mutants were examined. The protein was less prone to precipitation at lower temperatures and lower concentrations. Remarkably, precipitated protein could be resolubilized by incubation on ice. The cysteine mutations had varying effects on the MALTIR structure, with several mutants including C91A showing very few differences and the C157A mutant showing the highest differences from the wild-type protein (SI Appendix, Tables S4 and S5 and Fig. S3). The C116A mutant protein (designated here as MALTIRC116A) was chosen for further structural studies due to high protein expression levels and as it remained monomeric for ∼12 h in solution at high concentrations. The NMR data were acquired as multiple parts that were coassembled for processing [details reported elsewhere (29)]. For the MALTIRC116A protein, 72% of all atoms could be assigned; the unassigned 28% were mostly in the disordered regions of the proteins. The calculations yielded an ensemble of 20 structures with a heavy-atom amide-backbone (NH, Cα, C′) rmsd of 1.28 Å (SI Appendix, Table S6 and Fig. S4).
Typical TIR-domain structures feature a central five-stranded parallel β-sheet (strands βA–βE) surrounded by five α-helices (αA–αE) on both sides of the sheet (30). By contrast, the crystal structures of MALTIR (7–10) revealed an absence of helix αB; the sequence corresponding to strand βB in typical TIR domain structures is instead located in the long and flexible AB loop. The solution structure of the reduced form of MALTIRC116A reveals that the secondary-structure arrangement is more consistent with a typical TIR-domain fold than with the crystal structures of MALTIR (Fig. 1, SI Appendix, Figs. S4 and S5, and Movie S1). The AB loop connecting αA and βB is short, comprising only 7 residues (109–115). This loop is followed by the strand βB (residues 116‒119), as supported by a large number of short- to medium-range interresidue NOEs (SI Appendix, Fig. S6). Because resonance assignments of the backbone residues of wild-type MALTIR could be made in this region, we were able to confirm that the secondary structures of these residues (116–119) in both the MALTIRC116A and the wild-type MALTIR protein were consistent with the formation of a β-strand. This arrangement allows for the formation of a long flexible BB loop between the strand βB and the helix αB; the loop corresponds to residues 120‒134. This loop contains many unassigned atoms, and those assigned show few long-range NOEs, suggesting inherent flexibility. The helix αB is short (comprising 5 residues), similar to MyD88TIR; for comparison, it comprises 11 residues in TLR1TIR and 9 residues in TcpBTIR, a bacterial MAL mimetic which disrupts TLR signaling.
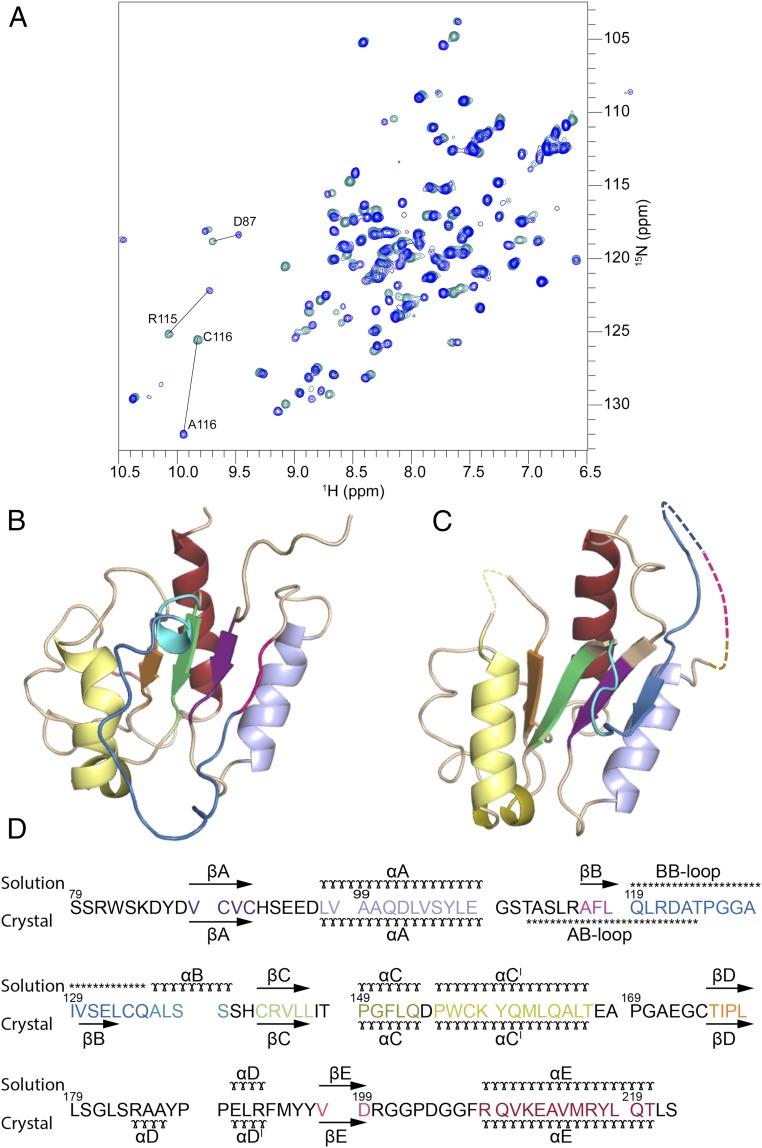
Fig. 1.
Comparison of the solution and crystal structures of MALTIR. (A) Overlay of 1H-15N-HSQC spectra from MALTIRC116A (blue) and wild-type MALTIR (cyan). Most chemical shifts of 1H and 15N are conserved between the spectra, with large shifts detected only for the mutated C116 residue, the neighboring residue R115, and additionally, residue D87. (B) Solution structure of MALTIRC116A, shown in cartoon representation. (C) Crystal structure of MALTIR (7) (PDB ID Code 2Y92) shown as in B. Regions in the crystal structure comprising residues 112–123 (AB loop) and 168–171 do not have interpretable electron density and are not included in the crystal structure model; they are shown here for illustration purposes by dotted lines. (D) Structure-based sequence alignment, showing the elements of secondary structure. The solution secondary structure is based on the CYANA calculations (60). (ɤ) α-helix, (→) β-strand, (***) loop.
The chemical shifts of the Cβ atoms of the six cysteine residues in MALTIRC116A range from 25.8 to 31.4 ppm, which indicates they are all reduced. The solution structure contains one pair of cysteine residues (C142 and C174) in close-enough proximity (5.4–6.9 Å) for possible disulfide-bond formation; however, chemical shifts of cysteine Cβ atoms indicate that they are both reduced. The comparison of NMR spectra of MALTIRC116A at pH values 7.5, 8.0, or 8.6 (SI Appendix, Table S7 and Fig. S7) suggests that the structure remains unchanged over the tested pH conditions.
MALTIR Is Sensitive to Redox Change.
Exposure of the wild-type MALTIR to redox conditions that ranged from “fully reducing” (GSH only) to “fully oxidizing” (GSSG only) and included three physiological conditions (31) of −225 mV, −199 mV, and ,190 mV, indicated that cysteines varied in reactivity. Major changes in chemical shifts were observed for the fully oxidizing condition containing only GSSG, especially for cysteine residues 89, 91, and 157 (SI Appendix, Fig. S8). Cysteine residue 91 was the most reactive, with 45% signal loss at −190 mV, compared with −225 mV, indicating that this residue had undergone oxidation to a large extent. The data suggest that the redox potential of C91 is close to −190 mV, whereas cysteines C89, 134, 142, and 172 have a redox potential <−190 mV and that C116 and C157 have redox potentials <<−190 mV. Cysteines are prone to modifications in an oxidative environment induced by inflammatory stimuli and the modifications have been shown to act as signals, for example in peroxiredoxin secretion from exosomes (32).
MAL Is Glutathionylated Endogenously.
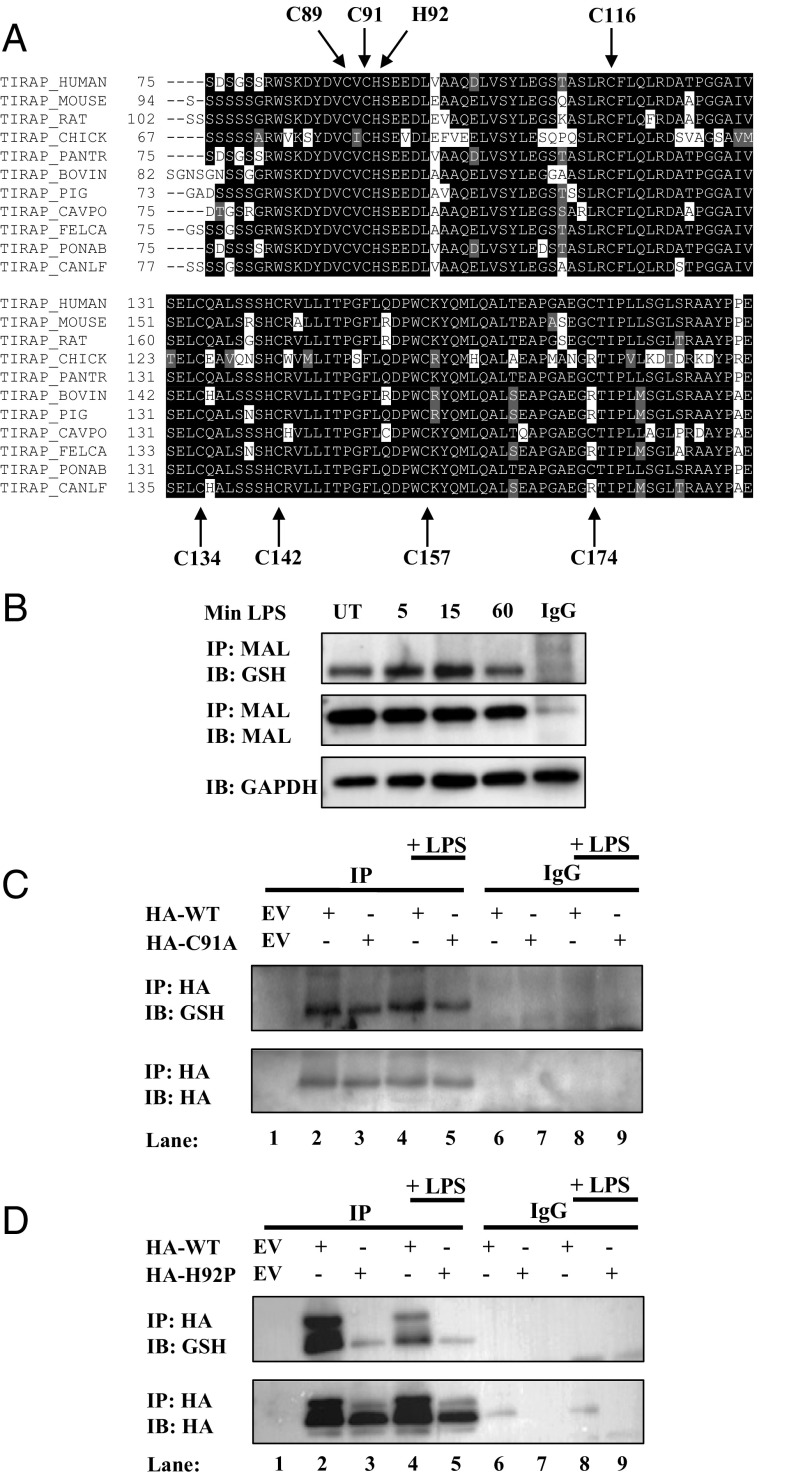
Bioinformatic analyses of MAL identified conservation of cysteine residues across most species, including human, mouse, and rabbit, except for C174, which is variable in these species (Fig. 2A). Given the reactive cysteines in MAL, we wished to test if it was glutathionylated endogenously. MAL was indeed glutathionylated basally and glutathionylation increased after 5 and 15 min of LPS treatment, and reverted to basal levels after 60 min of LPS treatment (Fig. 2B). To assess the glutathionylation site of MAL, we used cysteine-to-alanine mutants of MAL. Based on the evidence that cysteine 91 is the most reactive thiol, we chose the C91A mutant for initial functional studies. Overexpression of wild-type MAL and its C91A mutant (MALC91A) in HEK293 TLR4-MD2-CD14 cells (MTCs), followed by immunoprecipitation of MAL under nonreducing conditions, was performed to examine glutathionylation as a potential modification. To prevent deglutathionylation, we used the alkylating compound N-ethylmaleimide to modify free sulfhydryl groups. We also sought to identify if induction of TLR4 signaling by LPS would affect the potential glutathionylation status of MAL. Overexpressed wild-type MAL was found to be variably glutathionylated and underwent increased glutathionylation in response to LPS (Fig. 2C; compare lane 2 to lane 4), whereas MALC91A displayed decreased glutathionylation (Fig. 2C; compare lane 3 to lane 2), identifying that C91 is the main glutathione target in MAL.
Fig. 2.
MAL is glutathionylated on cysteine 91. (A) Sequence alignment highlighting MAL cysteine conservation across species. (B) BMDMs were treated with LPS (100 ng/mL) for 5, 15, or 60 min. Cells were lysed in low-stringency lysis buffer containing 50 mM N-ethylmaleimide. MAL was immunoprecipitated from lysate on A/G beads coated with MAL antibody and subjected to nonreducing SDS/PAGE. (C and D) MTC cells were transfected with empty vector, HA-tagged wild-type MAL, MALC91A (C), or MALH92P (D). Cells were treated with LPS (100 ng/mL) for 45 min. Cells were lysed in low-stringency lysis buffer containing 50 mM N-ethylmaleimide. MAL was immunoprecipitated from lysate on A/G beads coated with anti-HA antibody and subjected to nonreducing SDS/PAGE. The blots in B, C, and D are representative of three independent experiments.
Positively charged amino acids are known to influence cysteine susceptibility to glutathionylation. This occurs via deprotonation of neighboring cysteines, promoting cysteine thiolate anion formation, which would promote recruitment of glutathione to prevent further oxidation (33). The flanking amino acid to C91, histidine 92, was mutated to proline (H92P) to eliminate the possible positive charge, which could in turn affect C91 glutathionylation. Indeed, MAL could not be glutathionylated in MALH92P (Fig. 2D).
MAL Mutations Limit TLR2 and TLR4 Signaling.
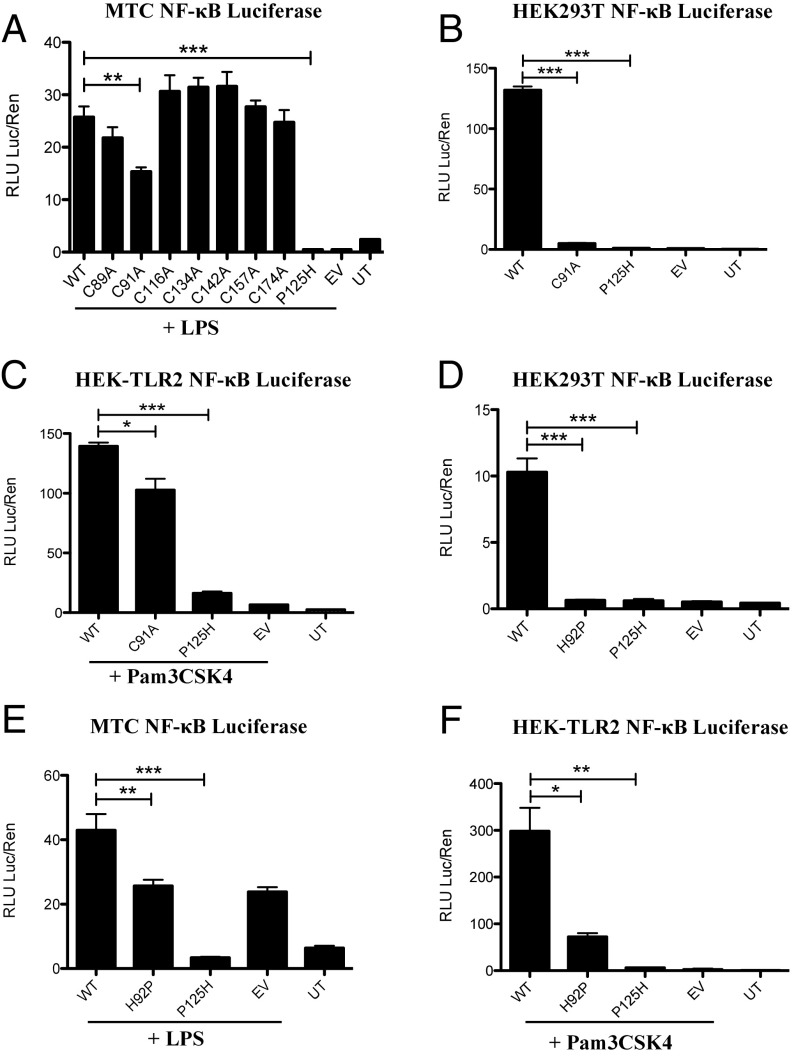
TLR pathway activation can be measured using overexpressed MAL and an NF-κB luciferase reporter construct. We initially examined the effect of each cysteine mutant on LPS-treated MTC cells. NF-κB activation was not affected in cells transfected with most cysteine-to-alanine mutants, the notable exception being MALC91A, the overexpression of which had a dominant-negative effect (Fig. 3A). MALC116A could drive NF-κB activation similar to wild-type MAL, supporting the suggestion that this mutation had no significant effect on MAL structure and function, consistent with the NMR data. To confirm the dominant-negative effect of the C91A mutation, we overexpressed MALC91A in HEK293T cells. MALC91A overexpression could not drive NF-κB activation in HEK293T cells (Fig. 3B). Transfection of MALC91A into HEK293-TLR2 cells treated with TLR2 agonist Pam3Csk4 yielded similar results to TLR4-driven NF-κB activation; however, the dominant-negative effects of MALC91A are less pronounced (Fig. 3C). Transfection with MALH92P was also unable to drive NF-κB (Fig. 3D), and acted as a dominant-negative mutant against LPS (Fig. 3E) and Pam3Csk4 (Fig. 3F). MAL P125H, which is known to act as a dominant-negative mutant, served as a control, inhibiting NF-κB activation.
Fig. 3.
Overexpression of MAL mutants limits NF-κB activation. MTC cells (A and E), HEK293T cells (B and D), and HEK-TLR2 cells (C and F) were transfected with wild-type MAL, MAL mutants, NF-κB reporter, and TK Renilla plasmids. Cells were lysed in 1× passive lysis buffer and lysates were used for NF-κB reporter assays. NF-κB activity was assayed using 1× Luciferase Assay Mix. TK Renilla fold was assayed using 1:500 dilution of coelenterazine in PBS. (A and E) MTC cells were treated with LPS (100 ng/mL) for 6 h before lysis with 1× passive lysis buffer. (C and F) HEK-TLR2 treated with Pam3Csk4 (100 ng/mL) for 6 h before lysis with 1× passive lysis buffer. Fluorescence was measured using FLUOstar Optima plate reader and gain adjustment was used to control for fluorescence. The data in A–F represent mean ± SEM of three individual experiments, each carried out in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
C91A and H92P MAL Mutants Cannot Reconstitute TLR4-Mediated IκBα Degradation.
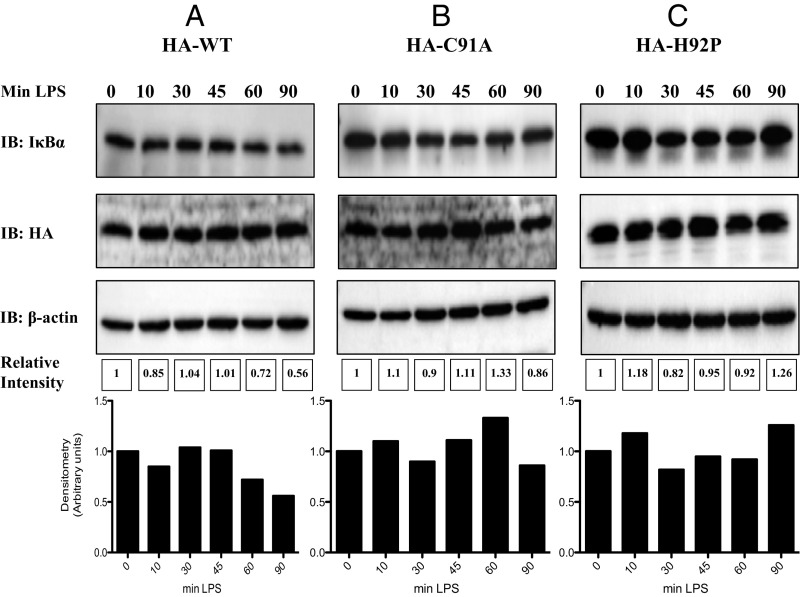
To further confirm impaired signaling by both MALC91A and MALH92P, we reconstituted MAL-deficient immortalized bone-marrow-derived macrophages (iBMDMs) and examined the degradation of the NF-κB inhibitory protein IκBα as a readout for NF-κB activation by TLR4 stimulation with LPS. The reconstitution of wild-type MAL yielded degradation of IκBα, with degradation reaching 50% at 90 min (Fig. 4A). Reconstitution of MAL-deficient iBMDMs with MALC91A failed to reconstitute TLR4 signaling, with no degradation of IκBα detected (Fig. 4B). Similarly, MALH92P did not reconstitute TLR4 signaling, with minimal degradation of IκBα (Fig. 4C).
Fig. 4.
C91A and H92P MAL mutants fail to reconstitute TLR4 signaling in MAL-deficient iBMDMs. (A) MAL-deficient iBMDMs were reconstituted with 5 μg of MAL wild-type plasmid, (B) 5 μg MALC91A, or (C) 5 μg MALH92P using the Neon electroporation system. MAL-deficient iBMDMs (1.1 × 106 cells/mL) were resuspended in 110 μL buffer R and 5 μg of wild-type MAL, MALC91A, or MALH92P. iBMDMs were immediately loaded to a 100 μL Neon tip and electroporated at 1,680 V for 20 ms and one pulse. Electroporated cells (100 μL) were placed directly into 900 μL prewarmed antibiotic-free media and incubated at 37 °C and 5% CO2 overnight. Immediately the next morning, the cells were washed with PBS, media replaced with fresh DMEM containing 10% FCS, and treated with LPS for the times indicated. Cells were lysed in 5× sample-loading buffer and subjected to SDS/PAGE. IκBα, HA-tag, and β-actin were assessed by Western blot. The blots in A–C are representative of three independent experiments. Densitometry was performed using ImageLab 5.0 using β-actin as a loading control.
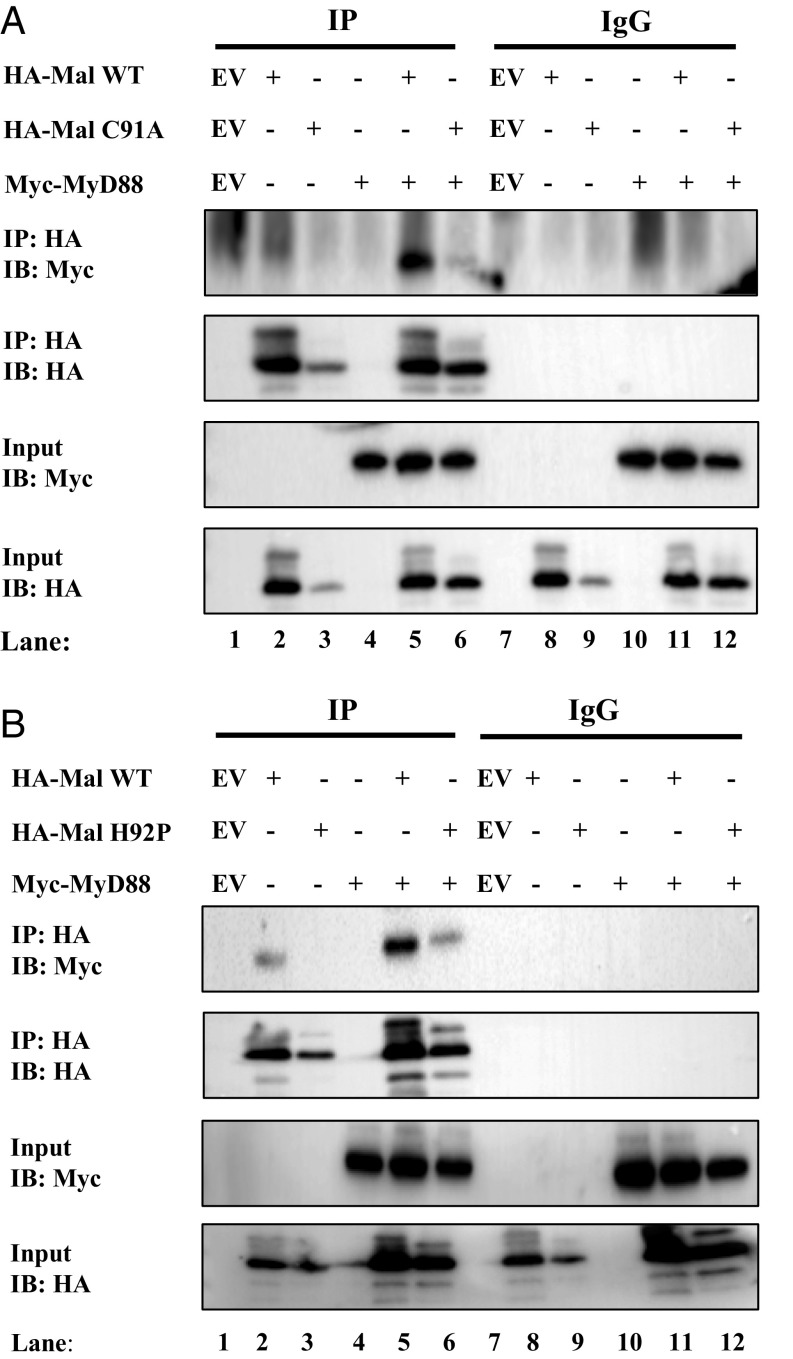
C91A and H92P MAL Mutants Cannot Interact with MyD88.
TLR stimulation induces MAL recruitment and interaction with MyD88 to drive formation of the Myddosome (4). We therefore sought to examine if MALC91A and MALH92P would be impaired in their ability to interact with MyD88. Wild-type MAL, when overexpressed, interacted with overexpressed MyD88 (Fig. 5A, lane 5, Top). MALC91A, however, did not interact with MyD88 (Fig. 5A, lane 6; compare with lane 5, Top). Lysates were examined to confirm the expression of each protein before immunoprecipitation (Fig. 5A, third and fourth panels). MALC91A appeared to express poorly when transfected (Fig. 5A, lane 3, fourth panel); however, cotransfection of MyD88 appeared to drive MALC91A expression to equal levels as wild-type MAL (Fig. 5A, lanes 5 and 6; compare with lane 2, Bottom). Similarly, when MALH92P was overexpressed, it failed to interact with MyD88, whereas wild-type MAL interacted with MyD88 (Fig. 5B, lane 6; compare with lane 5, Top). As above, lysates were examined for expression of plasmids before immunoprecipitation (Fig. 5B, third and fourth panels). These findings indicate that the C91A and H92P mutations prevent the interaction of MAL with MyD88, explaining the dominant-negative effect of these mutants.
Fig. 5.
MAL mutants C91A and H92P cannot interact with MyD88. (A and B) HEK293T cells were transfected with wild-type MAL, Myc-MyD88, EV, and (A) MALC91A or (B) MALH92P. In each transfection, the total concentration of plasmid DNA was kept constant by addition of relevant EV control. Posttransfection (48 h), the cells were lysed in low-stringency lysis buffer. Fifty μL of lysate was kept for analysis, while the remaining lysate was added to protein A/G beads coated with 1 μg of anti-HA antibody or 1 μg mouse-IgG control antibody for 2 h rotating at 4 °C. Samples were washed three times in 1 mL low-stringency lysis buffer and dried A/G beads were resuspended in 50 μL sample-loading buffer. Whole cell lysates and immunoprecipitated samples were analyzed by SDS/PAGE. Proteins were detected by immunoblot using anti-Myc and anti-HA antibodies. The results shown are representative of three independent experiments.
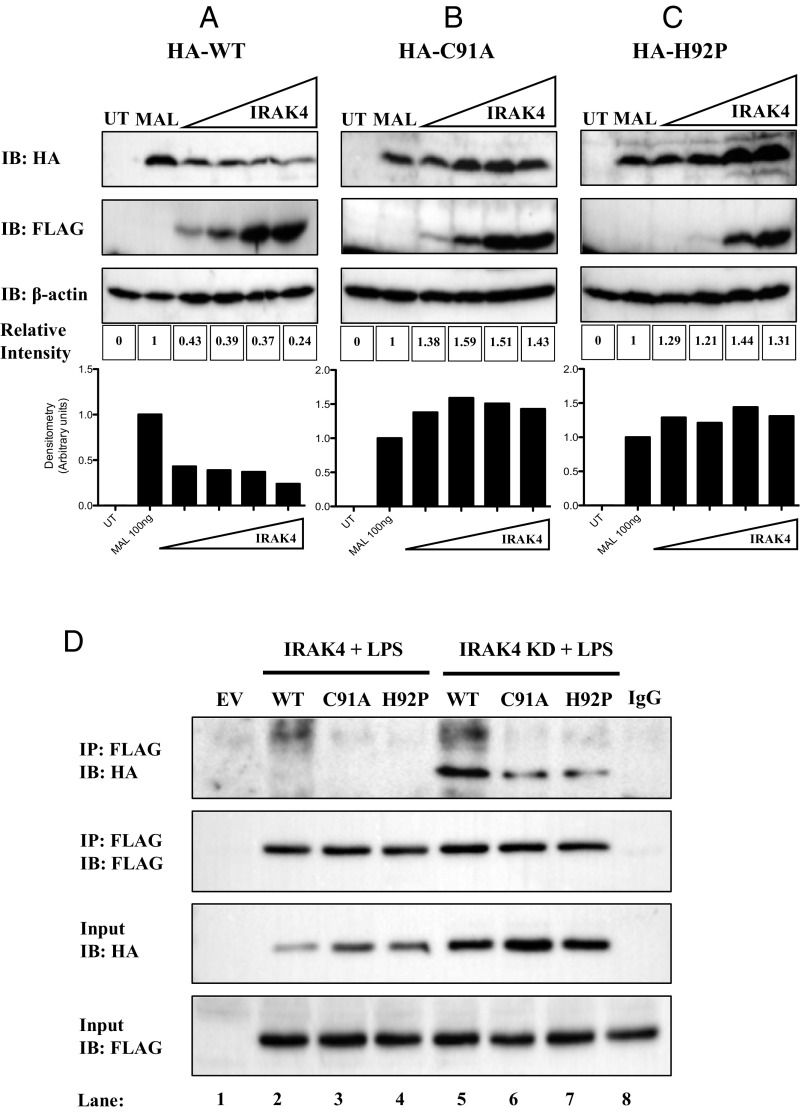
IRAK4 Cannot Degrade or Interact with C91A and H92P MAL Mutants.
It has been shown previously that IRAK1 and IRAK4 phosphorylate MAL on threonine 28, promoting MAL degradation (18, 22). We therefore sought to examine if the dominant-negative effects of MALC91A and MALH92P would affect IRAK1/4-mediated degradation of MAL. Overexpression of wild-type MAL with increasing concentrations of a plasmid encoding IRAK4 decreased MAL protein expression dose-dependently (Fig. 6A, Top). Overexpression of IRAK4 was unable to cause the degradation of MALC91A (Fig. 6B, Top). Correspondingly, overexpression of IRAK4 failed to induce degradation of MALH92P with increasing IRAK4 concentrations (Fig. 6C, Top).
Fig. 6.
IRAK4 cannot degrade or interact with C91A and H92P MAL mutants. HEK293T cells were transfected with FLAG-IRAK4, (A) wild-type MAL, (B) MALC91A, (C) MALH92P, or mock-transfected. In each transfection, the total concentration of plasmid DNA was kept constant by addition of relevant EV control. Posttransfection (24 h), the cells were lysed in 50 μL 5× sample-loading buffer. Cell lysates were analyzed by SDS/PAGE. Proteins were detected by immunoblot using anti-HA, anti-FLAG, and anti-β-actin antibodies. Densitometry was performed using ImageLab 5.0 software using β-actin as a loading control. (D) MTC cells were transfected with wild-type MAL, MALC91A, MALH92P, FLAG-IRAK4, kinase-dead FLAG-IRAK4 (IRAK4-KD), or EV. In each transfection, the total concentration of plasmid DNA was kept constant by addition of relevant EV control. Posttransfection (24 h), the cells were lysed in low-stringency lysis buffer. Fifty μL of lysate was kept for analysis, while the remaining lysate was added to protein A/G beads coated with 1 μg of anti-FLAG antibody or 1 μg mouse-IgG control antibody for 2 h rotating at 4 °C. Samples were washed three times in 1 mL low-stringency lysis buffer and dried A/G beads were resuspended in 50 μL sample-loading buffer. Whole-cell lysates and immunoprecipitated samples were analyzed by SDS/PAGE. Proteins were detected by immunoblot using anti-HA and anti-FLAG antibodies. The results shown are representative of three independent experiments.
As shown previously, the kinase-dead form (KD), but not kinase-active form of IRAK4, interacts with MAL (18). We therefore examined whether IRAK4-KD could interact with MAL mutants. As shown in Fig. 6D, wild-type MAL did not interact with IRAK4 when overexpressed (Top, lane 2), as expected, nor did the MALC91A or MALH92P mutants (Top, lanes 3 and 4, respectively). Wild-type MAL interacted strongly with IRAK4-KD (Top, lane 5); however, this interaction was attenuated with MALC91A and MALH92P (Top, lanes 6 and 7). Wild-type MAL was considerably depleted in lysates from IRAK4-transfected cells as opposed to cells transfected with IRAK4-KD (compare lane 2 to lane 5, third panel). However, the level of expression of the MALC91A and MALH92P mutants was similar between IRAK4 and IRAK4-KD (third panel; compare lanes 3 and 4 to lanes 6 and 7). Comparable levels of IRAK4 and IRAK4-KD were present before immunoprecipitation (Bottom).
Discussion
The crystal structures of the TIR domain of MAL (7–10) all show unusual features compared with other TIR-domain structures, in particular the presence of two disulfide bridges and the presence of a long flexible AB loop. We therefore aimed to characterize the TIR domain of MAL and its cysteine mutants in solution. Based on the available MAL crystal structures, the construct comprising residues 79‒221 (designated here as MALTIR) was selected for structural studies by solution NMR spectroscopy. The C116A mutant (MALTIRC116A) was found to have superior stability and behaved as a stable monomer, compared with the wild-type protein and other cysteine-to-alanine mutants. The MALTIRC116A structure was found to be representative of the wild-type MALTIR protein, based on the comparison of chemical shifts and secondary structure analysis. The structure was solved at a pH of 8.6 in a high ionic strength of 200 mM sodium chloride. However, the comparison of spectra of 15N-labeled MALTIR in the pH range of 8.6–7.5 shows that the structure is representative of a more physiological pH (SI Appendix, Fig. S6). The extreme conditions of high salt and pH yielded suitable sample concentrations for NMR analysis. These conditions, however, result in a loss of signals at rapidly exchanging backbone amide NHs and poor signal-to-noise ratio of the NMR resonances. To overcome these difficulties, an unconventional data acquisition strategy and advanced processing methods at ultrahigh fields had to be used to produce a medium-to-low-resolution solution structure of MALTIR (29).
The solution structure shows a structural rearrangement in the protein compared with the crystal structures (Fig. 1, SI Appendix, Fig. S5, and Movie S1). Consistent with other “typical” TIR-domain structures, the solution structure contains a long BB loop (residues 120–134) situated between strand βB and helix αB. The BB loop is a conserved feature among TIR domains and a number of studies have identified it to be important for mediating homotypic TIR-domain interactions (15–17, 34, 35). The striking structural differences are the result of the secondary structure arrangement between residues 111 and 139 (Fig. 1, SI Appendix, Fig. S5, and Movie S1). The crystal structures contain a flexible AB loop corresponding to residues 111–124, linking helix αB and strand βB. This rearrangement is caused by cysteine residues 89 and 134 forming a disulfide bond and shifting the location of strand βB. In the solution structure, the Cα atoms of these two cysteine residues are 11 Å apart, making disulfide-bond formation unlikely (36). The second disulfide bond found in the crystal structures involves the pair C142 and C174. In the solution structure, these residues remain in close proximity (∼6 Å); however, the chemical shifts of the Cβ-atoms from these residues suggest that the cysteine side chains are in a reduced state. In addition, these two cysteine residues are not conserved and are not likely required for the folding of the protein (7). The crystal structure of MALTIR has been used in a number of attempts to model the interactions involving this adaptor during signaling (7, 37–41); our work suggests that this structure may not be the signaling-competent form and questions the validity of the findings of these modeling studies. Our work now provides a more appropriate template structure for modeling approaches and interaction studies.
Our data point to C91 as being the most reactive cysteine in MAL and available for posttranslational modification. Indeed, we identified that MAL can be glutathionylated on this residue. MAL was basally glutathionylated in murine BMDMs, LPS transiently increased MAL glutathionylation, and the glutathionylation returned to basal levels after 60 min. Mutagenesis of C91 to alanine confirms this residue as the site of glutathionylation. Mutagenesis of the neighboring amino acid H92 also abrogated glutathionylation. MALC91A or MALH92P were unable to signal to NF-κB when overexpressed and acted as dominant-negative mutants of TLR4 signaling and could not reconstitute MAL-deficient cells. Interestingly, MALC91A showed only minor dominant-negative effects against TLR2. Previous reports suggest MAL is not required by TLR2 (42), which may explain the failure of MALC91A to exert a strong dominant-negative effect on TLR2. Finally, immunoprecipitation showed that the interaction with MyD88 of MALC91A or MALH92P was reduced, and both mutants failed to undergo degradation in response to IRAK4 overexpression. In the cell, glutathionylation of MAL on C91 might therefore be critical for MAL to signal via the Myddosome and IRAK4. As the modified residue is close to the BB loop, the modification could influence the conformation of this functionally important flexible loop.
Redox regulation of proteins is a rapidly evolving field. Increasing evidence suggests that during TLR-mediated immune activation, the cytosolic environment becomes more oxidizing. This can lead to the modification of cysteine residues on cytosolic proteins, most commonly to their oxidation by hydrogen peroxide (H2O2) species produced by NADPH (43), or by glutathionylation (44). Glutathionylation is emerging as an important posttranslational modification in a number of proteins including NF-κB (45). The glutathione transferase GSTO1-1 has been associated with the glutathionylation of cysteines in proteins in the TLR4 pathway, resulting in an on/off switch (46, 47). The precise target of GSTO1-1 in TLR4 signaling is unclear; however, the available evidence suggests that it regulates proteins early in the TLR4 signaling cascade, which could involve the cytosolic TIR domain of TLR4, MAL, or MyD88 (48). MyD88 has also been shown to be redox-regulated, with H2O2 treatment resulting in modification of MyD88 thiols (49). Glutathionylation of the redox enzyme glutathione transferase pi (GSTP) on cysteines 47 and 101 has been previously reported to alter its monomeric structure (50). Monomeric GSTP can associate with Jun N-terminal kinase (JNK) in resting cells, and dissociates from JNK under oxidative stress conditions, including H2O2 treatment, possibly by glutathionylation (51). We now contribute an example of glutathionylation of C91 in MAL, which appears to promote MyD88 and IRAK4 interaction. We identified glutathionylation of MAL as a positive regulatory mechanism in TLR4 activation, promoting Myddosome assembly for NF-κB activation.
Experimental Procedures
Biological Reagents, Mice, and Cell Culture.
HEK293T cells were obtained from the Centre for Applied Microbiology and Research, Wiltshire, UK. MTCs and HEK293-TLR2 cells were purchased from Invivogen. All experiments were carried out with prior ethical approval from the Trinity College Dublin Animal Research Ethics Committee (HPRA licence AE19136). C57BL/6 mice were killed using CO2; bone marrow was extracted and differentiated using L929 media for 6 d into primary macrophages. All cell lines and primary cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS, 1% penicillin/streptomycin, and maintained at 37 °C in a humidified atmosphere of 5% CO2. The HA-tagged MAL plasmid has been described previously (15), and site-directed mutagenesis of this plasmid was performed to obtain HA-tagged MAL mutants using QuikChange II Mutagenesis Kit (Agilent Technologies). These plasmids were used for mammalian cell transfections. Anti-TIRAP (D6M9Z), Anti-IκBα (44D4), and Anti-Myc (2276S) antibodies were purchased from Cell Signaling Technology. Anti-HA.11 antibody was purchased from Biolegend (MMS-101R). Anti-HA (H3663), anti-FLAG (F3165), and anti-β-actin antibody (Clone AC-74) were purchased from Sigma. Anti-GSH antibody was obtained from ViroGen (101-A-100). Protein A/G beads (sc-2003) and Mouse IgG (sc-2025) were purchased from Santa Cruz Biotechnology. Rabbit IgG was obtained from Millipore (2455657). GeneJuice was purchased from Novagen (70967). LPS from E. coli serotype EH100 was obtained from Enzo Life Sciences (ALX-581-010). Neon transfection kit was purchased from Bio-sciences (MPK10096). Pam3Csk4 (tlrl-pms), Blasticidin (anti-bl-1), Normocin (anti-nr-2), and Hygromycin B Gold (anti-hg-1) were obtained from Invivogen. All other reagents were obtained from Sigma unless otherwise stated.
Coimmunoprecipitation Studies.
HA-tagged MAL (3 μg) with 4 μg HA-tagged empty vector (EV) and 7 μg MAL mutants were coexpressed in HEK293T or MTC cells in 10-cm dishes. Cells were seeded at 2.5 × 105 cells per mL 24 h before transfection with GeneJuice per manufacturer’s instructions. Once 70% confluent, cells were washed with PBS and lysed in 700 μL low-stringency lysis buffer (50 mM Hepes pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Nonident P-40). Supernatants were removed and agitated for 10 min by occasional vortexing. Supernatants were pelleted at 4 °C; 50 μL of each supernatant was retained to analyze expression of input. Supernatant was then added to relevant precoupled antibody on A/G beads. Samples were incubated for 2 h rotating at 4 °C. Following incubation, the beads were washed three times with wash buffer and boiled in sample-loading buffer before being subjected to SDS/PAGE. For glutathione immunoprecipitations, 50 mM N-ethylmaleimide was added to the low-stringency lysis buffer and treated as above. After washing beads three times, sample-loading buffer without DTT was added and gently heated at 50 °C for 10 min to aid elution off beads. For endogenous immunoprecipitations, BMDMs were seeded at 1 × 106 cells per mL in 10-cm dishes and treated with the same method as glutathione immunoprecipitations.
Reporter Assays.
HEK293T, MTC, and HEK-TLR2 cells were seeded at 2 × 105 cells per mL The following day, cells were cotransfected with 20 ng NF-κB reporter plasmid and 50 ng of each respective MAL plasmid or EV using GeneJuice. In each experiment, 60 ng TK Renilla plasmid was transfected to normalize for transfection efficiency and cell death. After 24 h, reporter activity was assessed as previously described (52). Data are expressed as the mean relative light intensity folded over control pooled for three separate experiments with SEM.
Electroporation Assay.
The Neon transfection system (100 μL tip) was used for transfection. MAL-deficient iBMDMs were harvested and resuspended to a density of 1.1 × 106 cells per mL in DMEM. Cells were washed once to remove media and resuspended in 110 μL of Buffer R. Resuspended cell suspension (100 μL) was electroporated at 1,680 V for 20 ms with one pulse for optimal transfection with minimal cell death. Immediately after, electroporation cells were placed in 900 μL DMEM without antibiotics in a 12-well plate and left to adhere overnight. Cells were washed with PBS and treated with LPS for indicated times. SDS/PAGE was carried out as listed above. Densitometry was calculated using ImageLab 5.0 software (Bio-Rad).
Expression in E. coli and Purification of MALTIR and Its Cysteine Mutants.
BL-21, SHuffle (New England Biolabs) or Origami (Merck Millipore) E. coli cells were transformed by heat-shock with the pMCSG7 vector (53) encoding wild-type MALTIR (residues 79‒221) or its mutants C89A, C91A, C116A, C134A, C142A, C157A, and C174A. The cells were then grown in 50-mL starter cultures of Luria’s broth (LB) with 100 mg/L of ampicillin overnight at 37 °C. Cells were centrifuged at 800 × g and washed in M9 salts. One mL of starter culture was added to each flask containing 500 mL autoinduction media (54), with 15N-labeled ammonium chloride added for the expression of 15N-labeled protein (55), and grown while shaking at 37 °C until the optical density (OD) of each flask reached an absorbance of 0.8 at the wavelength of 600 nm. Once the OD was reached, the temperature was reduced to 20 °C for overnight protein expression. The cells were then lysed by sonication, and the protein was purified using nickel-affinity chromatography. Tobacco etch virus protease was added to the protein for an overnight incubation at 4 °C to cleave the 6-histidine tag from the protein. The protein was further purified by reeluting over the nickel-affinity column, followed by size-exclusion chromatography with a Superdex 75 gel-filtration column (GE Healthcare 26/600 S75) into a buffer consisting of 20 mM Tris⋅HCl (pH 8.6) with 200 mM NaCl.
Expression and Purification of 15N and 13C-Labeled MALTIRC116A.
The plasmid coding for the C116A mutant of MALTIR (MALTIRC116A) was transformed into E. coli BL-21 cells by heat-shock, and grown overnight in a starter culture of LB in the presence of 100 mg/L ampicillin while shaking at 37 °C. The protein was then expressed in the same media while shaking at 37 °C until the cells reached an OD of 0.7 at the wavelength of 600 nm. The sample was centrifuged at 800 × g, washed in M9 salts, and resuspended in minimal M9 media containing 13C glucose and 15N ammonium chloride until the cell density reached an OD of 0.8 at the wavelength of 600 nm. The temperature was then reduced to 20 °C and the cells were induced with 1 mM isopropyl-1-thiogalactopyrano-side for overnight expression. The protein was purified as described above.
Structural Analysis by NMR.
Double- and triple-resonance NMR experiments, including 2D 1H-15N-heteronuclear single quantum correlation (HSQC), 3D CbCacoNH, HbHacoNH, HNCaCb, HNCa, HNcoCA, HNCo, HNCa, HCcH, HCCh, 13C-HSQC-NOESY (separate spectra for the aliphatic and aromatic regions), and 15N-HSQC-NOESY (mixing time of 90 ms for all NOESY spectra), were conducted using ∼300 µM protein concentration, and 5-mm Shigemi tubes at 291 K on a 900-MHz Bruker Avance-III spectrometer equipped with a cryogenically cooled probe. All 3D experiments, excluding the NOESY spectra, were acquired using nonuniform sampling and processed using maximum entropy reconstruction as previously described (29, 56, 57). The 1H, 15N, and 13C atom frequencies were assigned to the protein using the CCPNMR software (58), followed by backbone torsion-angle prediction calculations based on chemical shifts using TALOS+ (59) and NOE calculations using CYANA 3.0 (60). The structure has been deposited in the Protein Data Bank (PDB ID code 2NDH).
The 1H and 15N chemical shifts of purified 15N-labeled MALTIR cysteine mutants were measured by 1H-15N-HSQC using a 900-MHz NMR spectrometer and analyzed with the CCPNMR software (58). The chemical shifts of each mutant were compared with the wild-type protein.
To compare the effect of pH on the structure, the 15N-labeled MALTIRC116A was divided and buffer-exchanged into three buffers, each made of 10 mM Hepes and 50 mM salt, at pH 7.5, 8.0, or 8.6. The chemical shifts of 1H and 15N were measured at each pH value and analyzed using the CCPNMR software (58).
NMR datasets were compared against each other using the Euclidean method for distance measurements, following the frequency assignment of 1H and 15N atoms in each 1H-15N-HSQC experiment using the CCPNMR software (58). Individual residues were compared by d2, where d2() = . Overall comparison between datasets was calculated using rmsd, where rmsd =.
Determination of Redox Shifts of Cysteines in Wild-Type MALTIR.
Stock solutions of GSH and GSSG were made in 10 mM Hepes buffer and adjusted to pH 7.5. Nitrogen was bubbled through both solutions to remove oxygen. A 1H spectrum of each stock was measured using a 900-MHz magnet and analyzed with the TopSpin 3.2 (Bruker) software to confirm whether the stock-solution redox species were indeed reduced or oxidized. Purified 15N-labeled wild-type MALTIR in 10 mM Hepes (pH 7.5) buffer was added to five different buffers containing varying ratios of GSH:GSSG that equate to specific electrochemical potentials (GSH only, −225 mV, −198.67 mV, −189.77 mV, and GSSG only). Electrochemical potential was calculated using the Nernst equation Eo = EoGSH/GSSG − (RT/nF) lnKeq, where EoGSH/GSSG is the standard potential of glutathione at pH 7.5 (−240 mV) (61), R is the universal gas constant (8.314 J K−1⋅m−1), T is the absolute temperature, n is the number of electrons transferred, F is the Faraday constant (9.648 104 C mol−1), and Keq is the equilibrium constant ([GSH]2/GSSG). A 1H-15N-HSQC was completed for each of the redox conditions at 298 K, using a 900-MHz NMR spectrometer, to determine the chemical shifts and intensity of the resonances from each residue. To correct for non-redox-related contributions to intensity changes (e.g., salt concentrations, etc.), the intensities were normalized to the average of a set of signals (15 peaks) from residues that were distal to any cysteine residues and neither overlapped with other peaks nor were strongly perturbed by the titration.
Ellman’s Assay.
Wild-type MALTIR was purified from three types of E. coli cells (BL-21, SHuffle, and Origami), each with different cytosolic redox environments. The protein sample from BL-21 expression was split into three conditions: with the addition of 10 mM reducing agent 1,4-DTT, with the addition of 0.03 mM hydrogen peroxide, and with no addition. Ellman’s reagent (dithiobisnitrobenzoic acid) was added to each of the five conditions before measuring the optical density at 412 nm using a spectrophotometer [NanoDrop, Thermo Scientific]. Calculations were performed based on the proportion of oxidized thiols, which correlated with the color intensity of the solution. The percent of reduced thiols in solution was used to calculate to the nearest number of reduced cysteine residues on MALTIR.
Mass Spectrometric Analysis of MAL Oxidized Using Glutathione.
Purified wild-type MALTIR from E. coli BL-21 cells was buffer-exchanged into either 10 mM Hepes (pH 7.5) or 20 mM Tris (pH 8.6). Five mM GSSG was then added to each solution, while keeping another without glutathione for control. These samples were incubated at room temperature for 1 h before subjecting each to a trypsin digest. Samples were separated using reversed-phase chromatography on a Shimadzu Prominence nanoLC system. Using a flow rate of 30 µl/min, samples were desalted on an Agilent C18 trap (0.3 × 5 mm, 5 µm) for 3 min, followed by separation on a Vydac Everest C18 (300 A, 5 µm, 150 mm × 150 µm) column at a flow rate of 1 µl/min. A gradient of 10–60% buffer B over 30 min where buffer A = 1% ACN/0.1% Formic acid (FA) and buffer B = 80% ACN/0.1% FA was used to separate peptides. Eluted peptides were directly analyzed on a TripleTof 5600 instrument (ABSciex) using a Nanospray III interface. Gas and voltage settings were adjusted as required. An MS TOF scan across m/z 350–1800 was performed for 0.5 s followed by information-dependent acquisition of up to 20 peptides across m/z 40–1800 (0.05 s per spectra). Data were converted to mgf format and searched in MASCOT accessed via the Australian Proteomics Computational Facility. Data were searched in the SwissProt database, searching all species, with trypsin as enzyme, two miscleavages, MS tolerance of 50 ppm, MS/MS tolerance of 0.1 Da. Oxidation (Met, variable) and carbamidomethylation (Cys, fixed) modifications were also included.
Intact Mass.
Intact masses were analyzed using purified wild-type MALTIR from E. coli BL-21 cells and directly measured on TripleTOF 5600 (Sciex) and Orbitrap Elite (Thermo) mass spectrometers. The presence of disulfide bonds was detected by comparing the measured mass of the protein to the theoretically calculated mass. The number of disulfide bonds in MALTIR was calculated using the theoretical mass of reduced MALTIR, which corresponded to 16,006 Da and 16,017 Da, for monoisotopic and average masses, respectively.
Supplementary Material
Acknowledgments
We thank Professor Paul Hertzog for his contributions to troubleshooting electroporation parameters of iBMDMs. The Queensland NMR Network is acknowledged for providing access to the 900-MHz spectrometer. This work was funded by Science Foundation Ireland (12/IA/1531 to L.A.J.O.) and the National Health and Medical Research Council (NHMRC Grants 1003326, 1107804, and 1071659 to B.K.). B.K. is an NHMRC Principal Research Fellow (1003325 and 1110971). M.M. is supported by an Australian Research Council Future Fellowship (FT10100925).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C.K. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the Protein Data Bank [(PDB ID code 2Y92 (MAL crystal structure) and PDB ID code 2NDH (MAL solution NMR structure)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701868114/-/DCSupplemental.
References
- 1.Yamamoto M, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 2.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 4.Bonham KS, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–716. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valkov E, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci USA. 2011;108:14879–14884. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Z, Lu J, Zhou W, Shen Y. Structural insights into TIR domain specificity of the bridging adaptor Mal in TLR4 signaling. PLoS One. 2012;7:e34202. doi: 10.1371/journal.pone.0034202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder GA, et al. Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. J Biol Chem. 2014;289:669–679. doi: 10.1074/jbc.M113.523407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo JR, Kim S, Shoelson SE, Park SY. X-ray crystallographic structure of TIR-domain from the human TIR-domain containing adaptor protein/MyD88-adaptor-like protein (TIRAP/MAL) Bull Korean Chem Soc. 2012;33:3091–3094. [Google Scholar]
- 11.Ohnishi H, et al. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc Natl Acad Sci USA. 2009;106:10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan JA, Brint EK, O’Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 14.Nyman T, et al. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 15.Dunne A, Ejdeback M, Ludidi PL, O’Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J Biol Chem. 2003;278:41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Zienkiewicz J, Hawiger J. Interactive sites in the MyD88 Toll/interleukin (IL) 1 receptor domain responsible for coupling to the IL1beta signaling pathway. J Biol Chem. 2005;280:26152–26159. doi: 10.1074/jbc.M503262200. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Z, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci USA. 2006;103:10961–10966. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunne A, et al. IRAK1 and IRAK4 promote phosphorylation, ubiquitination, and degradation of MyD88 adaptor-like (Mal) J Biol Chem. 2010;285:18276–18282. doi: 10.1074/jbc.M109.098137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray P, et al. MyD88 adapter-like (Mal) is phosphorylated by Bruton’s tyrosine kinase during TLR2 and TLR4 signal transduction. J Biol Chem. 2006;281:10489–10495. doi: 10.1074/jbc.M508892200. [DOI] [PubMed] [Google Scholar]
- 20.Ulrichts P, et al. Caspase-1 targets the TLR adaptor Mal at a crucial TIR-domain interaction site. J Cell Sci. 2010;123:256–265. doi: 10.1242/jcs.056002. [DOI] [PubMed] [Google Scholar]
- 21.Mansell A, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, et al. Membrane targeting of TIRAP is negatively regulated by phosphorylation in its phosphoinositide-binding motif. Sci Rep. 2017;7:43043. doi: 10.1038/srep43043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris D, et al. Glutathione and infection. Biochim Biophys Acta. 2013;1830:3329–3349. doi: 10.1016/j.bbagen.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Xie Y, Kole S, Precht P, Pazin MJ, Bernier M. S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology. 2009;150:1122–1131. doi: 10.1210/en.2008-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 26.Mailloux RJ, et al. Glutathionylation acts as a control switch for uncoupling proteins UCP2 and UCP3. J Biol Chem. 2011;286:21865–21875. doi: 10.1074/jbc.M111.240242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe Y, et al. Glutathione adducts induced by ischemia and deletion of glutaredoxin-1 stabilize HIF-1α and improve limb revascularization. Proc Natl Acad Sci USA. 2016;113:6011–6016. doi: 10.1073/pnas.1524198113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobstein J, et al. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb Cell Fact. 2012;11:56. doi: 10.1186/1475-2859-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miljenović T, Jia X, Lavrencic P, Kobe B, Mobli M. A non-uniform sampling approach enables studies of dilute and unstable proteins. J Biomol NMR. 2017;68:119–127. doi: 10.1007/s10858-017-0091-z. [DOI] [PubMed] [Google Scholar]
- 30.Ve T, Williams SJ, Kobe B. Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis. 2015;20:250–261. doi: 10.1007/s10495-014-1064-2. [DOI] [PubMed] [Google Scholar]
- 31.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 32.Mullen L, Hanschmann EM, Lillig CH, Herzenberg LA, Ghezzi P. Cysteine oxidation targets peroxiredoxins 1 and 2 for exosomal release through a novel mechanism of redox-dependent secretion. Mol Med. 2015;21:98–108. doi: 10.2119/molmed.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalle-Donne I, et al. Molecular mechanisms and potential clinical significance of S-glutathionylation. Antioxid Redox Signal. 2008;10:445–473. doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 34.Toshchakov VU, Basu S, Fenton MJ, Vogel SN. Differential involvement of BB loops of toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J Immunol. 2005;175:494–500. doi: 10.4049/jimmunol.175.1.494. [DOI] [PubMed] [Google Scholar]
- 35.Stack J, Bowie AG. Poxviral protein A46 antagonizes Toll-like receptor 4 signaling by targeting BB loop motifs in Toll-IL-1 receptor adaptor proteins to disrupt receptor:adaptor interactions. J Biol Chem. 2012;287:22672–22682. doi: 10.1074/jbc.M112.349225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waschütza G, et al. Engineered disulfide bonds in recombinant human interferon-gamma: The impact of the N-terminal helix A and the AB-loop on protein stability. Protein Eng. 1996;9:905–912. doi: 10.1093/protein/9.10.905. [DOI] [PubMed] [Google Scholar]
- 37.Vyncke L, et al. Reconstructing the TIR side of the Myddosome: A paradigm for TIR-TIR Interactions. Structure. 2016;24:437–447. doi: 10.1016/j.str.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Paracha RZ, et al. Structural evaluation of BTK and PKCδ mediated phosphorylation of MAL at positions Tyr86 and Tyr106. Comput Biol Chem. 2014;51:22–35. doi: 10.1016/j.compbiolchem.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Guven-Maiorov E, et al. The architecture of the TIR domain signalosome in the Toll-like receptor-4 signaling pathway. Sci Rep. 2015;5:13128. doi: 10.1038/srep13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guven-Maiorov E, Keskin O, Gursoy A, Nussinov R. A structural view of negative regulation of the Toll-like receptor-mediated inflammatory pathway. Biophys J. 2015;109:1214–1226. doi: 10.1016/j.bpj.2015.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naro C, Sette C. Dissecting a hub for immune response: Modeling the structure of MyD88. Structure. 2016;24:349–351. doi: 10.1016/j.str.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Kenny EF, et al. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J Immunol. 2009;183:3642–3651. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- 43.Ngkelo A, Meja K, Yeadon M, Adcock I, Kirkham PA. LPS induced inflammatory responses in human peripheral blood mononuclear cells is mediated through NOX4 and Giα dependent PI-3kinase signalling. J Inflamm (Lond) 2012;9:1. doi: 10.1186/1476-9255-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynaert NL, et al. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci USA. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao BC, Hsieh CW, Lin YC, Wung BS. The glutaredoxin/glutathione system modulates NF-kappaB activity by glutathionylation of p65 in cinnamaldehyde-treated endothelial cells. Toxicol Sci. 2010;116:151–163. doi: 10.1093/toxsci/kfq098. [DOI] [PubMed] [Google Scholar]
- 46.Menon D, Board PG. A role for glutathione transferase Omega 1 (GSTO1-1) in the glutathionylation cycle. J Biol Chem. 2013;288:25769–25779. doi: 10.1074/jbc.M113.487785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menon D, Coll R, O’Neill LA, Board PG. Glutathione transferase omega 1 is required for the lipopolysaccharide-stimulated induction of NADPH oxidase 1 and the production of reactive oxygen species in macrophages. Free Radic Biol Med. 2014;73:318–327. doi: 10.1016/j.freeradbiomed.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 48.Menon D, Coll R, O’Neill LA, Board PG. GSTO1-1 modulates metabolism in macrophages activated through the LPS and TLR4 pathway. J Cell Sci. 2015;128:1982–1990. doi: 10.1242/jcs.167858. [DOI] [PubMed] [Google Scholar]
- 49.Stottmeier B, Dick TP. Redox sensitivity of the MyD88 immune signaling adapter. Free Radic Biol Med. 2016;101:93–101. doi: 10.1016/j.freeradbiomed.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 50.Tew KD, et al. The role of glutathione S-transferase P in signaling pathways and S-glutathionylation in cancer. Free Radic Biol Med. 2011;51:299–313. doi: 10.1016/j.freeradbiomed.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adler V, et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jefferies CA, et al. Bruton’s tyrosine kinase is a Toll/interleukin-1 receptor domain-binding protein that participates in nuclear factor kappaB activation by Toll-like receptor 4. J Biol Chem. 2003;278:26258–26264. doi: 10.1074/jbc.M301484200. [DOI] [PubMed] [Google Scholar]
- 53.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Tyler RC, et al. Auto-induction medium for the production of [U-15N]- and [U-13C, U-15N]-labeled proteins for NMR screening and structure determination. Protein Expr Purif. 2005;40:268–278. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Mobli M. Reducing seed dependent variability of non-uniformly sampled multidimensional NMR data. J Magn Reson. 2015;256:60–69. doi: 10.1016/j.jmr.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Mobli M, Maciejewski MW, Gryk MR, Hoch JC. An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat Methods. 2007;4:467–468. doi: 10.1038/nmeth0607-467. [DOI] [PubMed] [Google Scholar]
- 58.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 59.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 61.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.