Significance
Endogenization of retroviruses has occurred multiple times in the course of vertebrate evolution, with the captured retroviral envelope syncytins playing a role in placentation in mammals, including marsupials. Here, we identify an endogenous retroviral envelope protein with unprecedented properties, including a specific cleavage process resulting in the shedding of its extracellular moiety in the human blood circulation. This protein is conserved in all simians—with a homologous protein found in marsupials—with a “stemness” expression in embryonic and reprogrammed stem cells, as well as in the placenta and some human tumors, especially ovarian tumors. This protein could constitute a versatile marker—and possibly an effector—of specific cellular states and being shed, be immunodetected in the blood.
Keywords: HERV, endogenous retrovirus, envelope protein, placenta, development, stem cells, tumors
Abstract
Capture of retroviral envelope genes is likely to have played a role in the emergence of placental mammals, with evidence for multiple, reiterated, and independent capture events occurring in mammals, and be responsible for the diversity of present day placental structures. Here, we uncover a full-length endogenous retrovirus envelope protein, dubbed HEMO [human endogenous MER34 (medium-reiteration-frequency-family-34) ORF], with unprecedented characteristics, because it is actively shed in the blood circulation in humans via specific cleavage of the precursor envelope protein upstream of the transmembrane domain. At variance with previously identified retroviral envelope genes, its encoding gene is found to be transcribed from a unique CpG-rich promoter not related to a retroviral LTR, with sites of expression including the placenta as well as other tissues and rather unexpectedly, stem cells as well as reprogrammed induced pluripotent stem cells (iPSCs), where the protein can also be detected. We provide evidence that the associated retroviral capture event most probably occurred >100 Mya before the split of Laurasiatheria and Euarchontoglires, with the identified retroviral envelope gene encoding a full-length protein in all simians under purifying selection and with similar shedding capacity. Finally, a comprehensive screen of the expression of the gene discloses high transcript levels in several tumor tissues, such as germ cell, breast, and ovarian tumors, with in the latter case, evidence for a histotype dependence and specific protein expression in clear-cell carcinoma. Altogether, the identified protein could constitute a “stemness marker” of the normal cell and a possible target for immunotherapeutic approaches in tumors.
Endogenous retroviral sequences represent ∼8% of the human genome. These sequences [called human endogenous retroviruses (HERVs)] share strong similarities with present day retroviruses and are the proviral remnants of ancestral germ-line infections by active retroviruses, which have thereafter been transmitted in a Mendelian manner (1–3). The >30,000 proviral copies found in the human genome can be grouped into about 80 distinct families, with most of these elements being nonprotein-coding because of the accumulation of mutations, insertions, deletions, and/or truncations (4, 5). However, some retroviral genes have retained a coding capacity, and some of them have even been diverted by remote primate ancestors for a physiological role. The so-called “syncytins,” namely syncytin-1 and -2 in humans, are retroviral envelope (env) genes captured 25 and 40 Mya, respectively, with a full-length protein-coding sequence, a fusogenic activity, and strong placental expression (6–9). These genes have been shown to be involved in placenta formation, with their fusogenic activity contributing to the formation of the syncytiotrophoblast (ST) at the maternofetal interface as a result of the syncytin-mediated cell–cell fusion of the underlying mononucleated cytotrophoblasts (CTs). Syncytins were, thereafter, identified in all placental mammals where they have been searched for, and their unambiguous role in placentation was shown via the generation and characterization of KO mice (10, 11). Syncytins are also present in marsupials, where they are expressed in a short-lived placenta that is very transiently formed (a few days) before the embryo pursues its development in an external pouch (12).
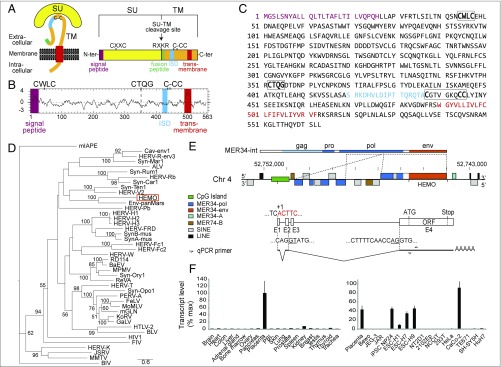
Previous systematic searches for genes encoding endogenous retroviral Env proteins within the human genome have led to the identification of 18 genes with a full-length coding sequence (among which are syncytin-1 and -2) (13, 14). These analyses have been performed using methods based on the search for characteristic motifs carried by retroviral Envs (Fig. 1), which include, from the N terminus to the C terminus, a signal peptide; a furin cleavage site (R-X-R/K-R) between the surface (SU) and transmembrane (TM) subunits, with the latter carrying additional signatures including an immunosuppressive domain (ISD; 17-aa motif), which is also found in most oncoretroviruses; a characteristic C-(X)5–7-C motif; and a transmembrane hydrophobic domain anchoring the Env protein in the cell or virion membrane (4, 15).
Fig. 1.
Characterization of the human HEMO Env retroviral protein and the HEMO env gene. (A) Schematic representation of a canonical retroviral Env protein delineating the SU and TM subunits. The furin cleavage site (consensus: R-X-R/K-R) between the two subunits, the C-X-X-C motif involved in SU-TM interaction, the hydrophobic signal peptide (purple), the fusion peptide (green), the transmembrane domain (red), and the putative ISD (blue) along with the conserved C-X5–7-CC motif (C-CC) are indicated. Adapted from ref. 38, copyright (2007) National Academy of Sciences. (B) Hydrophobicity profile of HEMO Env. The canonical structural features highlighted in A are positioned and shown in the color code used in A. The mutated furin site (CTQG) is shown as a dotted line. (C) Amino acid sequence of the HEMO Env protein with the same color code. (D) Retroviral Env protein-based phylogenetic tree with the identified HEMO-Env protein. The maximum likelihood tree was constructed using the full-length SU-TM amino acid sequences from HERV Envs (including an HERV-K consensus), all previously identified syncytins, and a series of endogenous and infectious retroviruses. The lengths of the horizontal branches are proportional to the average numbers of amino acid substitutions per site (scale bar in the lower right), and the percentage bootstrap values obtained from 1,000 replicates are indicated at the nodes. (E) Schematic representation of the HEMO gene locus on chromosome 4 (4q12; with the GRCh38 assembly coordinates of the Genome Reference Consortium). (Top) MER34-int consensus (Repbase) with putative gag, pro, and pol retroviral ORFs indicated according to consensus amino acid sequences. Dotted lines delineate parts of the MER34 sequences found in the HEMO locus. (Middle) The HEMO gene locus (10 kb) is located between the RASL11B gene (∼120 kb 5′) and the USP46 gene (∼120 kb 3′). HEMO env ORF is shown as an orange box, and repetitive sequences identified on the Dfam.org website are shown as different colored boxes, with the sense sequences above and antisense sequences below the line. Of note, the gene is part of an MER34 provirus that has kept only degenerate pol sequences (mostly in opposite orientation), a truncated putative 3′ LTR (MER34-A), and no 5′ LTR. No other MER34 sequences are found 100 kb apart from the gene. A CpG island (chromosome 4:52750911–52751703), detected by the EMBOSS-newcpgreport software, is indicated as a green box. (Bottom) Intron–exon structure predicted from the National Center for Biotechnology Information and RNA transcripts: exons found in placental RNA, as determined by RACE experiments, are indicated with the main E1-E2-E4 spliced env subgenomic transcript below. Nucleotide sequences of the start site (ACTTC...; red) and large intron splice sites for the HEMO env ORF are depicted; arrows specify qRT-PCR primers (Table S3). (F) Real-time qRT-PCR analysis of the HEMO transcripts in a panel of 20 human tissues and 16 human cell lines. Transcript levels are expressed as percentage of maximum and were normalized relative to the amount of housekeeping genes (SI Methods). Placenta values are the means of 12 samples from first trimester pregnancies, and other tissues are from a commercial panel (Zyagen).
Less stringent methods based on BLAST searches using large panels of retroviral Env proteins, including the increasing number of newly identified ERV genes from other animals, led us to identify a gene encoding a full-length retroviral Env protein with unprecedented characteristics. This Env protein gene—dubbed HEMO [human endogenous MER34 (medium-reiteration-frequency-family-34) ORF]—is the oldest captured full-length env gene identified to date in humans, because it entered the genome of a mammalian ancestor more than 100 Mya. The HEMO protein is released in the human blood circulation via a specific shedding process closely related to that observed for the Ebola filovirus, and it is highly expressed by stem cells and also, by the placenta resulting in an enhanced concentration in the blood of pregnant women. It is also expressed in some human tumors, thus providing a marker for a pathological state as well as, possibly, a target for immunotherapies.
Results
Identification of HEMO, an HERV Gene Encoding a Full-Length Env Protein.
The most recent human genome sequence release (GRCh38 Genome Reference Consortium Human reference 38, December 2013) was screened for the presence of genes encoding ERV Env proteins by a BLAST search for ORFs (from the Met start codon to the stop codon) > 400 aa using a selected series of 42 Env sequences representative of both infectious retrovirus and ERV families, including all of the previously identified syncytins (SI Methods). It yielded 45 Env-encoding ORFs, which could be, for all except one, grouped by clustalW alignments into already known HERV Env families (among which 24 Env-encoding ORFs for HERV-K, and 20 Env-encoding ORFs belonging to the set of 12 previously described HERV Envs) (Table S1). However, an unrelated env gene (HEMO) can be identified (Fig. 1) with a 563-aa ORF displaying some—but not all—of the characteristic features of a full-length retroviral Env protein, namely an N-terminal signal peptide; a CWLC motif in the putative SU subunit; and in the TM subunit, an ISD domain, a C-X6-CC motif, and a 23-aa hydrophobic transmembrane domain followed by a C-terminal cytoplasmic tail. Of note, the putative HEMO protein lacks a clearly identified furin cleavage site (CTQG instead of the canonical R-X-R/K-R) as well as an adjacent hydrophobic fusion peptide (Fig. 1B). The HEMO sequence was incorporated into the Env phylogenetic tree shown in Fig. 1D containing 42 retroviral envelope amino acid sequences used for the genomic screen. Fig. 1D shows that the sequence most closely related to the HEMO protein is Env-panMars encoded by a conserved, ancestrally captured retroviral env gene found in all marsupials, which has a premature stop codon upstream of the transmembrane domain (12).
Table S1.
Endogenous retroviral envelope protein-related sequences (ORF > 400 aa) in the human genome
| Name | Length, aa | Coordinates |
| ENV GAMMA type | ||
| EnvW | 538 | chr7:92468768–92470381 (REV) |
| EnvW-like | 475 | chrX:107052509–107053933 (REV) |
| EnvW-like | 472 | chr20:55351277–55352692) |
| EnvW-like | 468 | chr4:72926505–72927908 (REV) |
| EnvFRD | 538 | chr6:11103697–11105310 (REV) |
| EnvERV3 | 604 | chr7:64991215–64993026 (REV) |
| EnvERV3-like | 406 | chrX:52569735–52570952 (REV) |
| EnvE | 428 | chr19:20748111–20749394 (REV) |
| EnvV1 | 477 | chr19:53014091–53015521) |
| EnvV2 | 535 | chr19:53049252–53050856) |
| EnvH1 | 584 | chr2:165708193–165709944 (REV) |
| EnvH2 | 563 | chr3:166823237–166824925 (REV) |
| EnvH3 | 555 | chr2:154872220–154873884) |
| EnvH-like | 474 | chrX:72228564–72229985) |
| EnvPb | 665 | chr14:92622888–92624882 (REV) |
| EnvRb | 514 | chr3:16770303–16771844) |
| EnvFc1 | 584 | chrX:97847263–97849014) |
| EnvFc2 | 527 | chr7:153409531–153411111 (REV) |
| EnvT | 626 | chr19:20369432–20371309) |
| EnvT-like | 427 | chr14:106197668–106198948 (REV) |
| EnvHEMO | 563 | chr4:52743832–52745520 (REV) |
| ENV BETA type | ||
| Env-K–like | 412 | chr16:10418516–10419751 (REV) |
| Env-K–like | 439 | chr1:242457592–242458908) |
| Env-K–like | 475 | chr5:34462280–34463704 (REV) |
| Env-K–like | 482 | chr16:2661368–2662813 (REV) |
| Env-K–like | 487 | chr11:118722384–118723844 (REV) |
| Env-K–like | 550 | chr16:34413088–34414737 (REV) |
| Env-K–like | 550 | chr16:34997093–34998742) |
| Env-K–like | 560 | chr1:160697328–160699007) |
| Env-K–like | 588 | chr1:75380770–75382533) |
| Env-K–like | 597 | chr3:113025711–113027501 (REV) |
| Env-K–like | 658 | chr12:105311338–105313311) |
| Env-K–like | 661 | chr11:101701507–101703489) |
| Env-K–like | 687 | chr2:129962883–129964943 (REV) |
| Env-K–like | 698 | chr12:58328384–58330477 (REV) |
| Env-K–like | 698 | chr6:77717862–77719955 (REV) |
| Env-K–like | 699 | chr7:4583351–4585447 (REV) |
| Env-K–like | 699 | chr7:4591855–4593951 (REV) |
| Env-K–like | 699 | chr8:7498800–7500896 (REV) |
| Env-K–like | 699 | chr19:27638542–27640638 (REV) |
| Env-K–like | 738 | chr3:101696851–101699064) |
| Env-K–like | 885 | chr3:185564943–185567597 (REV) |
| Env-K–like | 930 | chr5:156659966–156662755 (REV) |
| Env-K–like | 1,171 | chr22:18943415–18946927) |
| Env-K–like | 1,375 | chr1: 155627591–155631715 (REV) |
Finally, BLAST analysis of the human genome indicates that the HEMO gene is part of a very old degenerate multigenic family known as medium reiteration frequency family 34 (MER34; first described in ref. 16). In this family, an internal consensus sequence with a Gag-Pro-Pol-Env retroviral structure (MER34-int) and LTR-MER34 sequences have been described and reported in RepBase (17). Genomic BLAST with the MER34-int consensus sequence could not detect any full-length putative ORFs for the gag or pol genes. Among the env sequences of the MER34 family scattered in the human genome (20 copies with >200-bp homology identified by BLAST) (Table S2), HEMO is clearly an outlier (1,692 bp/563 aa), with all of the other sequences containing numerous stop codons, short interspersed nuclear elements (SINE) or long interspersed nuclear elements (LINE) insertions, and no ORF longer than 147 aa.
Table S2.
MER34-related env sequences in the human genome
| Chromosome | Extracted sequences* | Maximum ORF bp/aa |
| 2 | 162084066–162086565 (rev) | 195/64 |
| 2 | 110061551–110064050 | 201/66 |
| 2 | 110307369–110309868 (rev) | 195/64 |
| 2 | 208368352–208370851 | 279/93 |
| 3 | 83422568–83425067 (rev) | 213/70 |
| 4 | 52743421–52745920 (rev) | 1,692/563 |
| 6 | 24704890–24713439 (rev) | 168/55 |
| 7 | 123922822–123925321 (rev) | 156/51 |
| 8 | 59785528–59788027 | 297/98 |
| 8 | 88712235–88714734 | 339/112 |
| 9 | 11114167–11116166 | 243/80 |
| 11 | 102483445–102485944 | 249/82 |
| 12 | 54733146–54735645 | 189/62 |
| 14 | 70462086–70464585 | 150/49 |
| 14 | 70237764–70240263 (rev) | 228/75 |
| 14 | 53023713–53026212 | 369/122 |
| 15 | 5078981–5081480 (rev) | 387/128 |
| 22 | 23932542–23935041 | 444/147 |
| 22 | 23938277–23940776 (rev) | 324/107 |
| X | 43460042–43462541 | 111/36 |
The HEMO gene is on chromosome 4, in bold.
Corresponds to genomic sequences sorted out by BLAST with the MER34-env consensus (Repbase MER34-int; base pairs 6,555–8,207) and >200-bp homology.
The HEMO Gene Locus and Transcription Profile.
The HEMO gene is located on chromosome 4q12 between the RASL11B and USP46 genes at about 120 kb from each gene (Fig. 9). Close examination of the HEMO env gene locus (10 kb) by BLAST comparison with the RepBase MER34-int consensus (17) reveals only remnants of the retroviral pol gene in a complex scrambled structure (Fig. 1E), with part of it being in reverse orientation and further disrupted by numerous SINE insertions. The locus organization indicates low selection pressure for the proviral non-env genes, such as often observed in the previously characterized loci harboring captured envs.
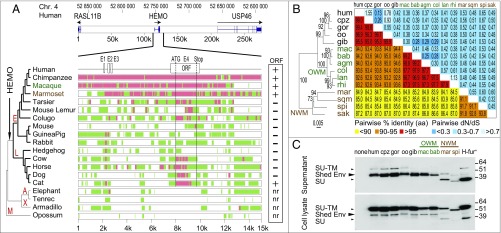
Fig. 9.
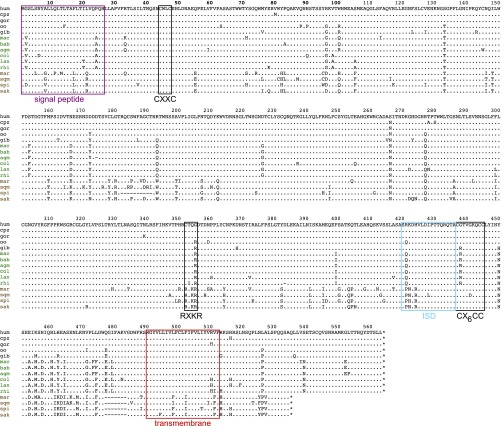
Sequence conservation and purifying selection of the HEMO gene in simians. (A) Syntenic conservation of the HEMO locus in mammalian species. The genomic locus of the HEMO gene on human chromosome 4 along with the surrounding RASL11B and USP46 genes (275 kb apart; genomic coordinates listed in Table S4) was recovered from the UCSC Genome Browser together with the syntenic loci of the indicated mammals from five major clades [Euarchontoglires (E), Laurasiatherians (L), Afrotherians (A), Xenarthres (X), and Marsupials M)]; exons and sense of transcription (arrows) are indicated. Exons of the HEMO gene (E1–E4) are shown on an enlarged view of the 15-kb HEMO locus together with the homology of the syntenic loci (analyzed using the MultiPipMaker alignment-building tool). Regions with significant homology as defined by the BLASTZ software (60) are shown as green boxes, and highly conserved regions (more than 100 bp without a gap displaying at least 70% identity) are shown as red boxes. Sequences with (+) or without (−) a full-length HEMO ORF are indicated on the right. nr, not relevant. (B) Purifying selection in simians. HEMO-based maximum likelihood phylogenetic tree was determined using nucleotide alignment of the HEMO genes (listed in Table S5 and Dataset S1). The horizontal branch length and scale indicate the percentage of nucleotide substitutions. Percentage bootstrap values obtained from 1,000 replicates are indicated at the nodes. Double-entry table for the pairwise percentage of amino acid sequence identity (lower triangle) and the pairwise value of dN/dS (upper triangle) between the HEMO gene from the various simian species listed on the phylogenetic tree to the left and listed in the same order in abbreviated form at the top. A color code is provided for both series of values. (C) Conservation of HEMO shedding in simians illustrated by Western blot analysis of 293T cells transfected with expression vectors for the indicated simian HEMO genes or the human HEMO mutant with a consensus furin site (H-fur+). Cell lysates and supernatants were harvested and treated with PNGase F before Western blot analysis with the polyclonal anti-HEMO antibody. The entire SU-TM HEMO protein is the main form observed in cell lysates, whereas the shed and the free SU form (for the NWM genes with a furin site and the H-fur+ mutant) are mainly observed in the supernatants. agm, African green monkey; bab, baboon; col, Angolan black-and-white colobus; cpz, chimpanzee; gib, gibbon; gor, gorilla; hum, human; lan, langur; mac, macaque; mar, marmoset; NWM, New World monkey; oo, orangutan; OWM, Old World monkey; rhi, golden snub-nosed monkey; sak, saki monkey; spi, spider monkey; sqm, squirrel monkey.
A quantitative RT-PCR (qRT-PCR) analysis using primers within the identified ORF and RNAs from a panel of human tissues and cell lines (Fig. 1F) shows that HEMO is expressed at a high level in the placenta. It is also significantly expressed in the kidney but at a lower level. In cell lines, expression of the HEMO gene looks heterogeneous, except for its systematic expression in stem cells [embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs)]. Quite unexpectedly, there is an absence of detectable transcripts in several placental choriocarcinoma cell lines (BeWo, JEG-3, and JAR) as well as a series of embryonal carcinoma (NT2D1, 2102Ep, and NCCIT) and tumor cell lines (but see the CaCo-2 colon adenocarcinoma).
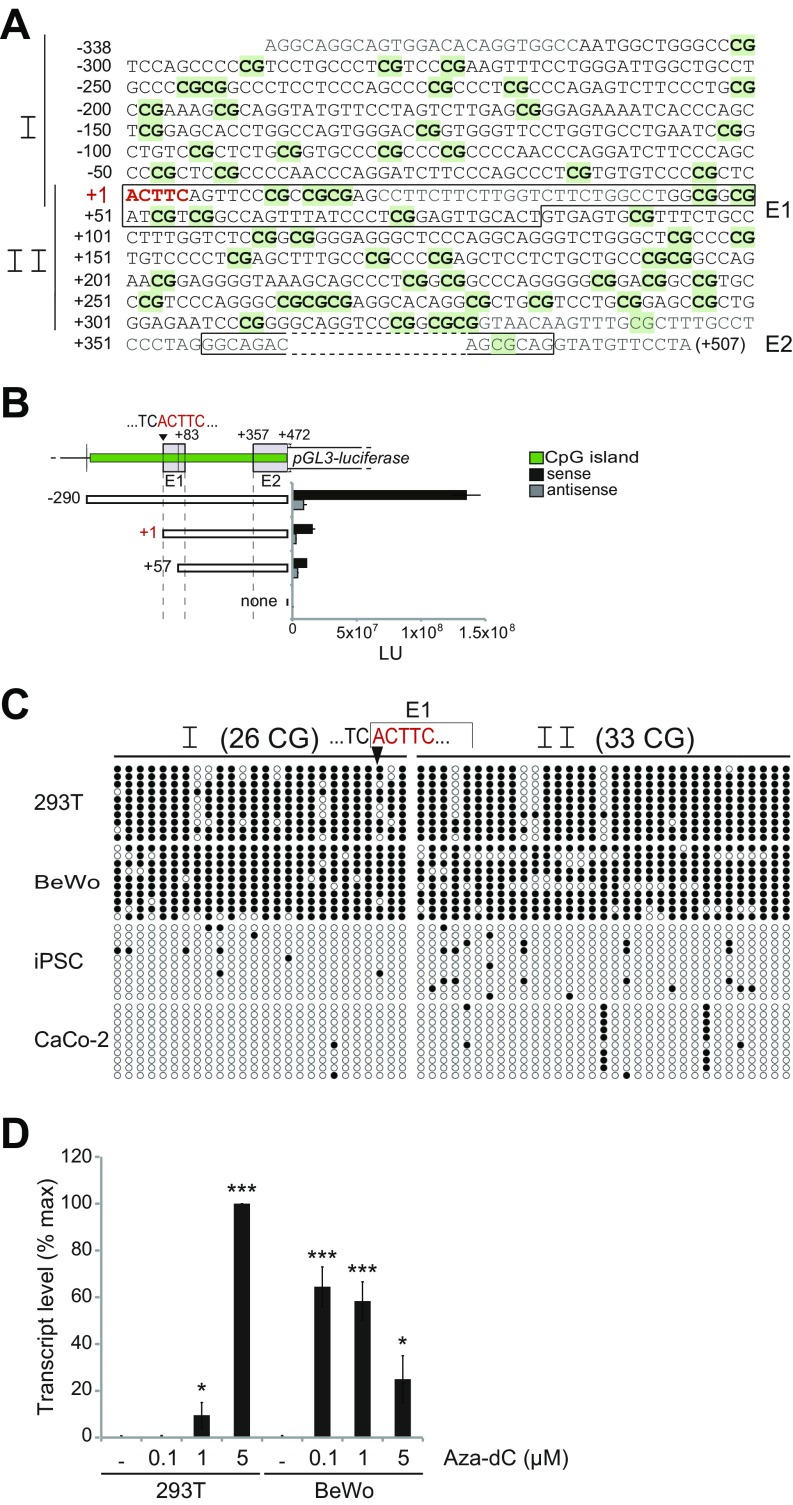
The structure of the HEMO env transcripts was determined by RACE-PCR analysis of Env-encoding transcripts from the placenta. It allowed the identification of multiply-spliced transcripts, with the intron boundaries corresponding to donor/acceptor splice sites predicted from the genomic sequence and as classically observed for retroviral env genes, a functional acceptor site located close to the env ATG start site. Interestingly, the transcript 3′ end falls within an identifiable MER34 LTR, as expected for a retroviral transcript. However, the transcription start site, located ∼5 kb 5′ to the env gene, does not correspond to any identifiable LTR structure. Rather, the sequence associated with the transcript start site is located in a CpG-rich domain (Fig. 1E and Fig. S1A) and most probably corresponds to a cellular promoter unrelated to any retroviral element. The transcript 5′ end (i.e., tc|ACTTC) falls within a canonical RNA Polymerase II Core Promoter Initiator Motif [yy|ANWYY (18)].
Fig. S1.
Characterization of the HEMO env gene promoter. (A) CpG island promoter sequence around the transcription start site (+1; ACTTC in red; position 52.751.425 on chromosome 4 of the GRCh38 assembly), with CG dinucleotides in green highlighted. Exon1 and exon2 are boxed. Nucleotide sequences in gray represent primer sequences used for amplification of the two fragments I and II (vertical bars on the left) and analyzed after bisulfite treatment (C). Primers listed in Table S3 are bisulfite-converted sequences. (B) Luciferase assay of the CpG island promoter. (Upper) Schematic representation of the promoter-luciferase constructs with the CpG island (green) containing exons E1 and E2. E2 was shortened at its 3′ end, 28 bp upstream of the donor splice site, to limit splicing out of the luciferase gene. (Lower) Promoter sequences in each pGL3 construct are indicated as white boxes, with coordinates relative to the +1 transcription start site of the gene. Control (none) corresponds to the basic pGL3 vector, with no inserted sequence. Promoter activity, expressed in light unit (LU), was determined using the Luciferase reporter assay in lysates from 293T cells transfected with the pGL3 vectors. The plotted data are the average from three independent experiments. (C) Methylation status of the HEMO promoter region as revealed by bisulfite treatment of the genomic DNA from cell lines not expressing (293T and BeWo cells) or expressing (iPSC and CaCo-2) the HEMO gene, and PCR amplification of fragments I (containing 26 CpG) and II (containing 33 CpG) delineated. The graph represents the sequencing of 10 clones for each PCR-amplified fragment, with methylated (black circles) and unmethylated (white circles) CpG indicated. (D) Effect of DNA demethylation on expression of the HEMO gene. HEMO gene transcription levels were detected by qRT-PCR and normalized to the housekeeping gene RPLP0 in cell lines (293T, BeWo) untreated (DMSO alone) or treated with 0.1–5 μM 5-Aza-dC for 3 d. Data are presented as the mean ± SEM. Asterisks indicate values significantly different from those obtained with untreated cells (unpaired two-tailed t test). *P < 0.05; ***P < 0.001.
The CpG-rich start site-containing region (CpG island) (reviewed in ref. 19) was studied further for its promoter activity by in vitro transfection assays using luciferase reporter genes. As illustrated in Fig. S1B, a 760-bp fragment including the identified start site acts as a strong promoter in this assay (>500-fold compared with none). Lower expression is observed (10- to 50-fold compared with none) in partial deletion mutants and as expected for a CpG promoter, when placed in antisense orientation.
DNA methylation patterns of sequences surrounding the transcription start site within the identified CpG island were analyzed by bisulfite treatment. As shown in Fig. S1C, the majority of the CpGs are methylated in the HEMO-negative cell lines (293T and BeWo), whereas they are unmethylated in HEMO-expressing cell lines (iPSC and CaCo-2). To get additional insight into this dependence of the promoter activity on the CpG island methylation pattern, 5-Aza-2′-deoxycytidine (5-Aza-dC) treatment was performed on BeWo and 293T cells at doses ranging from 0.1 to 5 µM (Fig. S1D). Transcripts were detectable by qRT-PCR after a 3-d treatment at low dose for BeWo cells (0.1 µM) and higher dose for 293T cells (5 µM). Of note, the high transcript level of the HEMO gene in CaCo-2 cells was not further amplified by a similar 5-Aza-dC treatment. Altogether, these results indicate that HEMO expression is sensitive to the methylation status of the CpG promoter.
HEMO Protein Synthesis and Structure: Specific Shedding.
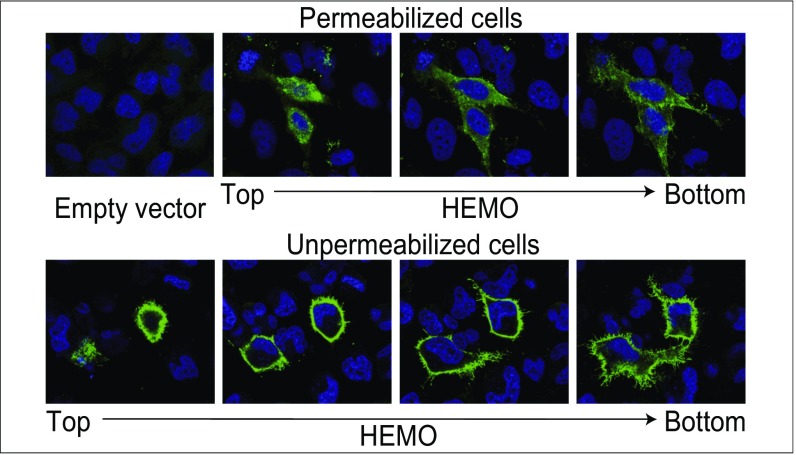
The capacity of the identified gene to produce an envelope protein was tested by introduction of the env ORF into a CMV promoter-driven expression vector and in vitro transient transfection assays. Polyclonal antibodies and mAbs were raised by immunization of mice with a recombinant protein corresponding to a 163-aa fragment of the putative SU moiety of the protein (SI Methods). As illustrated by the immunofluorescence assay shown in Fig. 2, Upper using the anti-HEMO antibodies, a strong labeling can be observed on permeabilization of the transfected cells (and not of control cells transfected with an empty vector). Furthermore, HEMO proteins can be detected at the cell surface as evidenced by the specific immunofluorescence labeling of the cell membrane of nonpermeabilized transfected HeLa cells in the successive confocal images shown in Fig. 2, Lower, consistent with HEMO being a retroviral env gene.
Fig. 2.
Immunofluorescence analysis of HEMO protein expression in transfected HeLa cells. Cells (HeLa) were transfected with the phCMV-HEMO expression vector (or an empty vector as a negative control), fixed, permeabilized (Upper) or not permeabilized (Lower), and stained for HEMO protein expression using a specific anti-HEMO polyclonal antibody (SI Methods). (Upper) Specific staining of the phCMV-HEMO transfected cells vs. empty vector transfected cells. (Lower) Successive confocal images show cell surface localization of the protein.
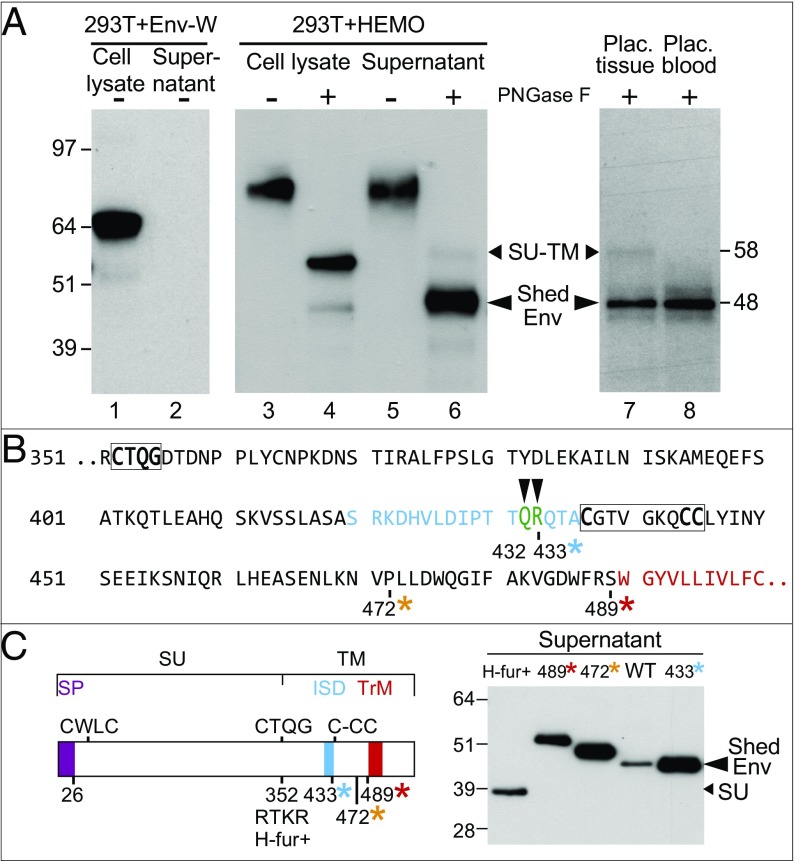
As illustrated in the Western blot of a whole-cell lysate (Fig. 3A, lane 3), transfection with the above HEMO expression vector yielded a strong band with an apparent molecular mass >80 kDa, much larger than expected for the HEMO full-length SU-TM protein (theoretical molecular mass = 61 kDa) but consistent with its glycosylation—as expected for a retroviral protein. Indeed, treatment of the cell extract with peptide N-glycosidase F (PNGase F)—to deglycosylate proteins—resolved the >80-kDa band into two bands of lower molecular mass (lane 4): a major band of ∼58 kDa and a fainter one of ∼48 kDa. The major band most probably corresponds to the full-length SU-TM protein (estimated size of 61 kDa), whereas the lower 48-kDa band has a size inconsistent with that of the sole SU subunit (estimated size 37 kDa; see below).
Fig. 3.
Characterization of the shed HEMO protein. (A) Detection of the shed HEMO Env protein by Western blot analysis. (Left) Detection of the syncytin-1 protein with the anti–Env-W polyclonal antibody (58) in the cell lysate of phCMV-Env-W transfected 293T cells. (Center and Right) Detection of the two forms of the HEMO protein (full-length SU-TM and Shed Env) with the anti-HEMO polyclonal antibody in the cell lysate and supernatant of phCMV-HEMO transfected 293T cells (Center) and first trimester placental tissue and placental blood (Right; matched representative samples from the same individual); samples were treated (+) or not (−) with PNGase F. (B) Detail of the shedding site amino acid sequence indicated by green capital letters. *Positions of the stop codons introduced in the mutants analyzed in C. (C) Migration pattern of the mutant HEMO forms analyzed as in A. (Left) Schematic representation of the HEMO protein with the same color code as in Fig. 1 and the stop codons of the generated mutants positioned together with that of the mutant with a reconstituted furin site (H-fur+; with an RTKR furin site). (Right) Supernatant of 293T cells transfected with the expression vectors for the WT and the mutant HEMO plasmids analyzed after PNGase F treatment, SDS gel electrophoresis, and Western blot as in A.
Analysis of the cell supernatants provided an unexpected answer as to the origin of the 48-kDa protein. Indeed, this 48-kDa protein turns out to be the major form in the cell supernatant (Fig. 3A, lane 6) (with PNGase F treatment of the supernatant), whereas the larger 58-kDa band observed in the whole-cell extract (Fig. 3A, lane 4) (with similar PNGase F treatment) is almost undetectable, as expected for a cell membrane-attached full-length Env protein (Fig. 3A, lane 6). This secreted 48-kDa protein is glycosylated, being observed at a much higher molecular mass in the cell supernatant without PNGase F treatment (Fig. 3A, lane 5). Altogether, these data strongly suggest that the HEMO protein, which is a transmembrane protein exported at the cell surface, can nevertheless be quantitatively released in the supernatant in the form of a protein—Shed Env—which has a molecular mass that is larger than that of the SU alone. This property, unexpected for a retroviral Env protein, is indeed not observed using the same protocols and expression vectors for syncytin-1 (HERV Env-W) used as a negative control (Fig. 3A, lanes 1 and 2).
To go further into the characterization of this shed soluble protein, we purified it from the supernatant of transfected 293T cells (SI Methods) and characterized its sequence by using mass spectrometry (MS) for the determination of both its N and C termini. As illustrated in Fig. 3B (and Fig. S2, which provides the HEMO protein sequence coverage by MS analysis of trypsin- or chymotrypsin-generated peptides), it turns out that the shed protein is truncated at its C terminus, mainly within the ISD domain, with two C-terminal sites identified with a different abundance (namely, Q432 and R433 at a 4:1 ratio). At the N terminus, the HEMO protein begins at position 27 (i.e., 2 aa after the predicted signal peptide cleavage site; using SignalP 4.1 Server software; www.cbs.dtu.dk/services/SignalP/). To confirm the MS size determination of the shed HEMO protein, several mutants were constructed by inserting stop codons before the anchoring transmembrane domain at the indicated positions (marked in Fig. 3 B and C with asterisks: 433R-stop, 472P-stop, and 489S-stop) or introducing the consensus furin site RTKR in place of the human CTQG [as in the New World monkeys (NWMs); human furin+ construct (H-fur+)] (Fig. S3). Western blot analysis of the supernatant of the HEMO mutant transfected cells then clearly showed that the wild-type (WT) deglycosylated shed HEMO protein migrates as the 433R-stop mutant as expected. In addition, the H-fur+ mutant displays a smaller 37-kDa band, consistent with the size estimated for the deglycosylated SU subunit alone. Of note, the figure also shows that the 489S- and 472P-stop mutant proteins do not comigrate with the shed WT HEMO—but are larger—indicating that, despite the fact that they still contain the shedding sequence, they have not been further processed by the shedding machinery. This absence of shedding is most probably because they are not membrane-associated (as a consequence of the premature stop codon introduced before the transmembrane domain), thus suggesting that anchoring the Env protein at the cell surface is required for an efficient processing by the shedding machinery.
Fig. S2.
MS determination of the N and C termini of the shed HEMO protein. Protein coverage of the Shed Env form, purified from the supernatant of phCMV-HEMO transfected 293T cells, is shown in green characters after trypsin proteolysis and MS characterization of the resulting peptides, or with underlined characters after chymotrypsin proteolysis (SI Methods). The HEMO N and C termini are indicated by large capital letters. The signal peptide is in purple, and the transmembrane domain is in red.
Fig. S3.
Aligned amino acid sequences of the simian HEMO proteins. The characteristic domains are delineated, with the putative proteolytic furin cleavage site (RXKR; black) between the SU and TM subunits, the signal peptide (purple) and the CWLC motif (CXXC; black) in the SU subunit, the ISD (blue), the C6XCC sequence (black), and the transmembrane domain (red) in the TM subunit. Dots indicate amino acid identity, and hyphens indicate codon deletions. Coordinates of the genomic nucleotide sequences used for aminoacid translation are listed in Table S5, except for the nucleotide sequences of gibbon, baboon, langur, spider monkey, and saki monkey, which were determined (SI Methods) and are reported in Dataset S1. Apes names are in black, OWMs names in green, and NWMs names in brown. agm, African green monkey; bab, baboon; col, Angolan black-and-white colobus; cpz, chimpanzee; gib, gibbon; gor, gorilla; hum, human; lan, langur; mac, macaque; mar, marmoset; NWM, New World monkey; oo, orangutan; OWM, Old World monkey; rhi, golden snub-nosed monkey; sak, saki monkey; spi, spider monkey; sqm, squirrel monkey.
To determine if the shed form of the HEMO protein could be observed under in vivo conditions, placental tissues (which show high transcription levels for the HEMO gene) (Fig. 1F) were recovered from first trimester legal abortions together with the local placental blood (which bathes the placental villi and can be analyzed in parallel), and proteins were extracted and deglycosylated for Western blot analyses. As shown in Fig. 3A, lane 7, the small 48-kDa band (and a very faint SU-TM 58-kDa band) can be detected in the placental tissue extract. The 48-kDa band is also detected in the placental blood, most probably corresponding to the protein secreted by the placenta. MS analysis (as above) of the 48-kDa protein in the corresponding gel bands confirmed the relevance of the immunological detection.
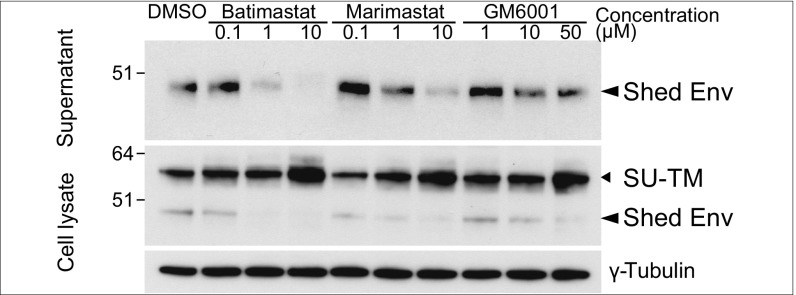
The release of a processed HEMO protein is reminiscent of what has been observed for the viral envelope protein of a completely unrelated virus (i.e., the Ebola filovirus), for which it has been further shown that cleavage was mediated by a cell-associated metalloproteinase ADAM protein (20). Accordingly, we tested whether chemical inhibitors of metalloproteinases [including the ADAM and MMP proteins (20–22)] had any effect on HEMO shedding in 293T transfected cells. As illustrated in Fig. 4, the broad-range ADAM and MMP inhibitors Batimastat and Marimastat and the MMP inhibitor GM6001 clearly inhibited HEMO release in the supernatant to various extents and in a dose-dependent manner, with visible accumulation of the nonsecreted form in the cell lysates. These experiments suggest that, in vivo, HEMO shedding could be driven by one or several metalloproteinases known to be present notably in placental cells (23–25).
Fig. 4.
Inhibition of HEMO release in the supernatant of transfected cells. Western blot analysis of cell lysate and supernatant of 293T cells transfected with the phCMV-HEMO expression vector using the polyclonal anti-HEMO antibody (SI Methods). Cells were treated for 3 d with the indicated doses of the ADAM and MMP chemical inhibitors Batismastat, Marismastat, and GM6001 or DMSO alone. Anti–γ-tubulin antibody was used as a control of cell lysate protein loading. The full-length HEMO protein (SU-TM) and the secreted form (Shed Env) are indicated by arrowheads.
HEMO Expression in Vivo: HEMO Release in the Blood Circulation of Pregnant Women.
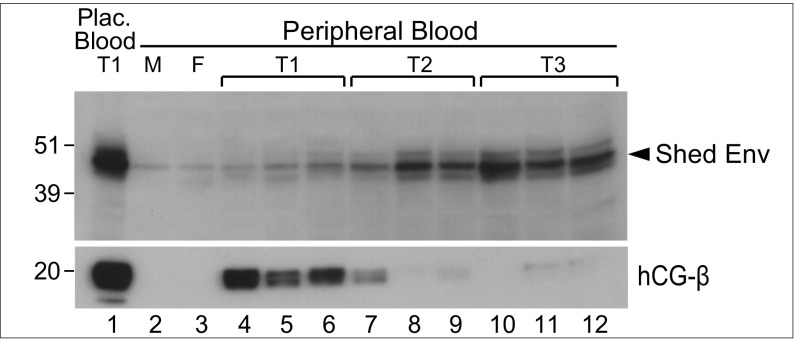
The combined results of the qRT-PCRs on the panel of human tissues shown in Fig. 1F and the shedding of the protein shown in Fig. 3 led us to hypothesize that HEMO could be detected in the blood circulation, especially in pregnant women. Sera were, therefore, collected and assayed for the presence of shed HEMO by Western blotting. Sera were treated with wheat germ agglutinin (WGA) to isolate glycosylated proteins, which were then deglycosylated. As illustrated in Fig. 5, Lower, the hCG-beta protein, which is a well-known early biomarker of pregnancy (26), shows undetectable levels in the peripheral blood of men and nonpregnant women (Fig. 5, lanes 2 and 3), whereas a very high level is observed for women in the first trimester of pregnancy (20-kDa band) (Fig. 5, lanes 4–6, T1), with a decrease at later stages (Fig. 5, lanes 7–12, T2 and T3). Remarkably, the deglycosylated shed HEMO form (48 kDa; previously identified in the placental blood) (Fig. 3A, lane 8 and Fig. 5, Upper, lane 1) can also be detected in the peripheral blood of pregnant women, beginning at a faint level in first trimester pregnancies (Fig. 5, Upper, lanes 4–12). As pregnancy proceeds, the level of HEMO protein increases very significantly, consistent with the large increase in placental mass during pregnancy. HEMO concentration at the peak can be estimated to be in the 1- to 10-nM range [by comparative Western blot analysis of serial dilutions of a purified recombinant shed HEMO protein; i.e., about one to two logs below that for hCG at the peak (T1) and for additional comparison, about the same as that for alpha-fetoprotein in the blood of pregnant women at the peak (T2)]. Of note, a faint level of shed HEMO protein can also be observed in the blood of men and nonpregnant women (Fig. 5, Upper, lanes 2 and 3), consistent with its nonnegligible expression in other organs, such as the kidney (qRT-PCR results in Fig. 1F).
Fig. 5.
Release of the HEMO protein in the peripheral blood during pregnancy. Western blot analysis of purified blood samples with the polyclonal anti-HEMO antibody (Upper) and anti–hCG-beta antibody (Lower). The shed HEMO protein is detected in the placental blood from first trimester pregnancy (T1) and from peripheral blood of men (M), nonpregnant women (F), and pregnant women from the first (T1), second (T2), and third (T3) trimesters. Each lane corresponds to distinct individuals. Bands observed at both slightly higher and lower molecular masses might correspond to minor alternatively processed/shed forms of the HEMO protein.
Identification of HEMO-Producing Cells in the Placenta.
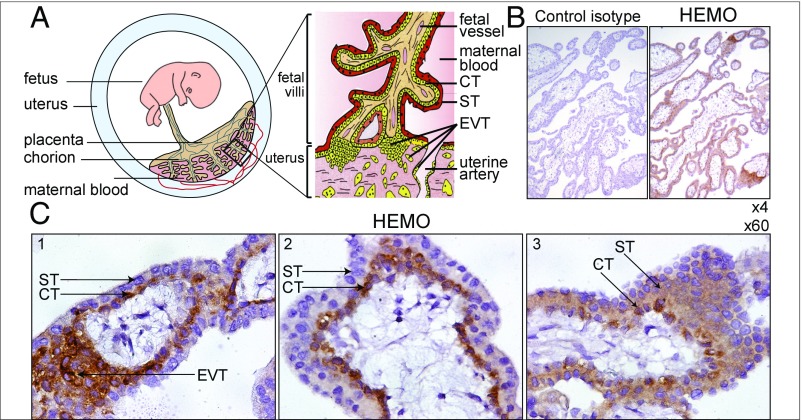
The human placenta is of the hemochorial type and characterized by the presence of fetal villi in direct contact with—and bathed by—the maternal placental blood (Fig. 6A) (9). These villi arise from the chorionic membrane—of fetal origin—and have an inner mononucleated cytotrophoblast layer (CT) underlying the surface syncytial layer, the syncytiotrophoblast (ST) (reviewed in refs. 27 and 28). The placenta invades the maternal uterine part, with anchoring villi characterized by invasive extravillous trophoblasts (EVTs).
Fig. 6.
Immunohistochemical detection of the HEMO protein in first trimester human placenta. (A) Schematic representation of the fetoplacental unit with an enlarged anchored villus bathed by maternal blood and displaying the ST layer, the underlying mononucleated CT, and the invading EVT. Adapted from ref. 59. (B) Serial sections of multiple placental villi stained with a control IgG2a mouse isotype (Left) or the anti-HEMO mAb 2F7 (Right). (Magnification: 4×.) (C) Enlarged views of placental villi showing staining of CTs (1–3), with strong staining of EVTs (1) and diffuse staining of STs (3). (Magnification: 60×.)
To localize precisely HEMO expression in the placenta, immunohistochemistry experiments were then performed on sections of first trimester placental tissues from abortion cases. As illustrated in Fig. 6 B and C, specific staining was obtained with the monoclonal anti-HEMO antibody and not with a control isotype as shown in Fig. 6B (4× magnification). In the three enlargements (60× magnification) shown in Fig. 6C, strong staining is observed in the trophoblast cells, including the villous CTs and the EVTs, suggesting that HEMO is indeed produced by these cells. More diffuse staining can be observed in the ST layer (Fig. 6C3), which is generated by CT fusion and involved in the exchanges between fetal and maternal blood.
Altogether, the immunohistochemical analyses of the placenta carried out with the above anti-HEMO antibody show strong labeling essentially at the trophoblast level and are consistent with the observed shedding of HEMO in the mother’s blood (Fig. 5).
Profile of HEMO Expression in Development.
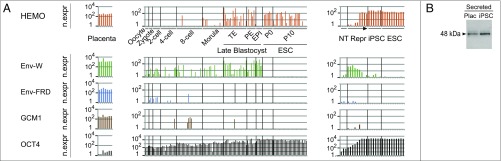
To get insight into the possible involvement of HEMO in embryonic development, we further analyzed by data mining a series of human RNA sequencing (RNA-Seq) experiments deposited at the Sequence Read Archive National Center for Biotechnology Information platform corresponding to different stages of development (29–32). Extraction of the expression profiles of a set of human genes was performed, and the results are illustrated in Fig. 7A for the HEMO, the syncytin 1 (env-W), and the syncytin 2 (env-FRD) env genes as well as specific genes expressed in either the placenta (GCM1) or stem cells (OCT4/POU5F1). For each gene of interest, read counts were verified to be equally distributed over the coding sequence (SI Methods). All three env genes (together with the placental GCM1-specific gene) are found in the RNA-Seq samples of placental tissues as expected (Fig. 7A, Left). Fig. 7A, Center clearly shows that HEMO has a wide expression profile, being expressed early in embryonic development starting at the eight-cell stage up to the late blastocyst stage and being permanently expressed in the derived ESCs from passage 0 up to passage 10. The HEMO gene RNA-Seq expression profile found in stem cells confirms the qRT-PCR results shown in Fig. 1F and is clearly different from what is observed for the two human syncytin genes: env-W, which is expressed very early in development, is completely down-regulated in the human stem cells, and env-FRD remains almost undetectable (33). Finally, RNA-Seq expression of HEMO was analyzed in the reprogramming experiments of differentiated somatic cells into iPSCs described in ref. 32, and hits reported in Fig. 7A, Right highlight the specific reprogramming of the HEMO gene—not observed with env-W and env-FRD—which parallels the expected profile of expression of the OCT4/POU5F1 transcription factor. Of note, as illustrated in Fig. 7B at the protein level, we could verify by Western blot analysis of iPSCs in culture that the HEMO gene expression unraveled above also results in the shedding of HEMO proteins, with a 48-kDa band detected in the iPSC supernatants.
Fig. 7.
Expression of the HEMO gene during development by in silico RNA-Seq analysis. (A) In silico analysis of three panels of RNA-Seq data for HEMO, syncytin-1 (env-W) and -2 (env-FRD), GCM1 (Glial Cells Missing homolog 1, a specific placenta-expressed gene), and OCT4 (highly expressed in stem cells). RNA-Seq raw data were screened with the coding part of each gene, and hits were reported in log scale per kilobase of screened sequence and after normalization with two housekeeping genes, RPLP0 and RPS6 (SI Methods). (Left) Panel of seven samples of normal placental tissues from distinct individuals at the same stage of pregnancy (29). (Center) Panel of 124 single-cell RNA-Seq of human preimplantation embryos and ESCs at the indicated stages of development or cell passage (30) (similar patterns were obtained from data in ref. 31, which covered the oocyte to morula stages). (Right) Panel of 28 RNA-Seq samples from the reprogramming (Repr.; from day 4 to day 21) of human CD34+ cells (NT) to iPSCs (six subclones) and from independent human ESC lines (32). EPI, epiblast; n.expr, normalized expression; P0, Passage 0; P10, Passage 10; PE, primitive endoderm; TE, trophectoderm. (B) Western blot analysis of WGA-purified placental blood (first trimester pregnancy) and WGA-purified supernatant of confluent iPSC cloneN (grown an extra 36 h without serum and concentrated 20×). Samples were treated with PNGase F. The shed HEMO form is detected using the polyclonal anti-HEMO antibody.
Conclusively, the HEMO gene displays a specific pattern of expression—that includes ESCs—a feature possibly linked to the “capture” of a specific CpG-rich promoter of non-LTR origin, with the bona fide production of HEMO in the form of a soluble protein from at least trophoblast and stem cells.
HEMO Expression in Tumors.
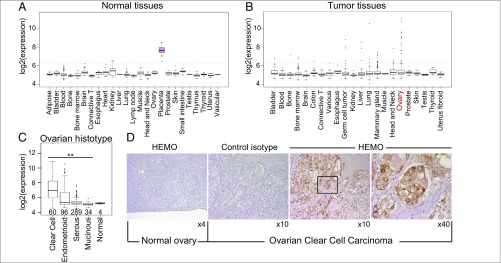
To get insight into the possible expression of the HEMO gene in human tumors, we performed an in silico analysis of microarray data using the dataset E-MTAB-62 elaborated in ref. 34, which includes 1,033 samples from normal tissues and 2,315 samples from neoplasm tissues obtained from various ArrayExpress (AE) and Gene Expression Omnibus (GEO) studies (SI Methods). In normal tissues, as expected from the qRT-PCR analysis in Fig. 1F, significant levels of expression were essentially observed in placental tissues and to a limited extent, the kidney (Fig. 8A). In several tumors, as illustrated in Fig. 8B, heterogeneity was detected among samples from the same organ (represented by the outliers plotted as black dots in Fig. 8B), with in some cases, evidence for high-level expression of the HEMO gene (for instance, in germ-line, liver, lung, or breast tumors, with the most salient heterogeneity being observed for ovary tumors). In the latter case, additional search for annotation data related to various histological types of ovarian carcinoma (35, 36) led us to correlate the highest values with specific tumor histotypes, mainly clear cell carcinoma. To enlarge this dataset, ovary tumor samples from five other GEO databases were collected and further normalized (SI Methods) together with E-MTAB-62, giving a total of 479 tumor samples. As shown in Fig. 8C, higher expression values of the gene are observed for clear cell carcinomas (60 samples) and to a lesser extent, endometrioid cancer (96 samples) samples. No clear-cut up-regulation of the HEMO gene is observed in the serous cancer histotype (289 samples; albeit with some heterogeneity) and the mucinous histotype (34 samples).
Fig. 8.
Microarray analysis of HEMO expression within normal tissues and tumor samples. (A and B) Box plot representations of normalized values obtained for HEMO gene expression extracted from the E-MTAB62 dataset (on a logarithmic scale). Original tissue categories were adjusted to group together samples from the same biological source, keeping the major groups described by the authors: normal tissues (A) and tumor samples (B). (C) Box plot representation of normalized values obtained from an enlarged ovarian tumor sample extracted as raw.CEL files from various AE and GEO studies (SI Methods). Values for normal ovarian tissues were included, as control, in the normalization process. Tumoral ovarian histotypes correspond to 60 clear cell carcinoma, 96 endometrioid, 34 mucinous, and 289 serous tumoral samples (Wilcoxon’s rank sum test). **P < 0.01. (D) Immunohistochemical analysis using the 2F7 mAb specific for the HEMO protein (or a control isotype) of formalin-fixed normal ovarian tissues (column 1) and ovarian clear cell carcinoma (column 2–4; at two magnifications).
In agreement with these transcription data, immunohistochemistry analyses of normal vs. clear cell carcinoma ovarian tissues using the anti-HEMO mAb disclose a highly specific staining of the tumoral clear cells compared with the control isotype staining (Fig. 8D).
HEMO Insertion Date and Conservation Across Mammalian Genomes.
A strong hint for a physiological role of a captured gene is its conservation in evolution and the nature of the selection to which it is subjected. Accordingly, we performed an extensive search for the HEMO gene in eutherian mammals by both in silico screening and PCR cloning and sequencing and further extended it to marsupials (the phylogenetic tree in Fig. 1D shows homology of HEMO with Env-panMars; see below). These analyses also aimed at the determination of the HEMO date of insertion into the genome of a mammalian ancestor, the determination of the coding capacity of the identified genes in the various species, and the determination of the presence of a shed HEMO protein after introduction of the cloned gene into an expression vector and transfection of 293T cells. The overall data are summarized in Fig. 9. We performed an in silico analysis of syntenic loci (coordinates listed in Table S4) by using the MultiPipMaker synteny building tool between the RASL11B and USP46 genes conserved in all mammalian genomes (each found at about 120 kb from the human HEMO gene). Focus on the 15-kb HEMO region (Fig. 9A) shows that the HEMO gene entered the genome of mammals before the radiation of Laurasiatherians (e.g., ruminants, carnivora) and Euarchontoglires (e.g., primates, rodents, lagomorphes), i.e., between 100 and 120 Mya (37), being found in neither Afrotherians (Elephant, Tenrec) nor Xenarthrans (Armadillo). It also allowed the identification of the orthologous HEMO gene in primates (and as a very degenerate sequence in rodents) and among Laurasiatherians in several species including the dog, cat, horse, and cow. Closer analysis further discloses that the HEMO gene (coordinates and sequences in Table S5 and Dataset S1) has been conserved as a full-length protein-coding sequence in all simians (Fig. S3) and unexpectedly, the cat (Fig. S4). The identified full-length HEMO ORFs show high similarities, ranging from 84 to 99% amino acid identities (Fig. 9B, lower triangle), and signs of purifying selection, with nonsynonymous to synonymous ratios (dN/dS) between all pairs of species lower than unity (mean value 0.46), except for very close species (e.g., human/chimpanzee), for which the number of mutations is not high enough to provide significant dN/dS values. For example, dN/dS values of 0.29–0.42 are observed between great apes and Old World monkeys (OWMs) (Fig. 9B, upper triangle) as expected for a host gene. These low values further fall within those that can be determined for the similarly captured primate synctin-1 (0.85) and syncytin-2 (0.28) functional env-derived genes.
Table S4.
List of genomic coordinates of the 250-kb RAS-USP46 locus
| Species | Assembly | Coordinates |
| Euarchontoglires | ||
| Human (Simian-Ape) | GRCh38/hg38, 2013 | chr4:52590972–52866835 |
| Chimpanzee-bonobo (Simian-Ape) | Max-Planck/panPan1, 2012 | JH650087:949069–1220194 |
| Rhesus macaque (Simian-OWM) | BCM Mmul_8.0.1/rheMac8, 2015 | chr5:81899847–82185019 |
| Marmoset (Simian-NWM) | WUGSC 3.2/calJac3, 2009 | chr3:140669174–140925562 |
| Tarsier (Prosimian) | Tarsius_syrichta-2.0.1/tarSyr2, 2013 | KE926088v1:194120–271011 KE938719v1:458231–525407 |
| Mouse Lemur (Prosimian) | Mouse lemur/micMur2, 2015 | KQ053245v1:1118456–1287657 |
| Colugo (Dermoptera) | G_variegatus-3.0.2 | Scaffold969: 20581–246600 |
| Mouse (Rodentia) | GRCm38/mm10, 2011 | chr5:74000038–74199471 23479169 |
| Guinea pig (Rodentia) | Broad/cavPor3, 2008 | Scaffold_24:23257653- |
| Rabbit (Lagomorpha) | Broad/oryCun2, 2009 | chrUn0056:1207035–1383532 |
| Laurasiatheria | ||
| Hedgehog (Insectivora) | EriEur2.0/eriEur2, 2012 | JH835325:6037893–6282794 |
| Cow (Ruminantia) | Bos_taurus_UMD_3.1.1/bosTau8, 2014 | chr6:69950422–70183806 |
| Horse (Perissodactyla) | Broad/equCab2, 2007 | chr3:79263527–79467858 |
| Dog (Carnivora) | Broad/CanFam3.1/canFam3, 2011 | chr13:45379782–45583929 |
| Cat (Carnivora) | ICGSC/Felis_catus_8.0/felCat8, 2014 | chrB1:164058766–164262324 |
| Afrotheria | ||
| Elephant (Proboscidae) | Broad/loxAfr3, 2009 | Scaffold_38:3843013–4119717 |
| Tenrec (Tenrecidae) | Broad/echTel2, 2012 | JH980315:5379386–5641477 |
| Xenarthra | ||
| Armadillo (Dasypodidae) | Baylor/dasNov3, 2011 | JH568349:4112648–4401060 |
| Marsupial | ||
| Opossum (Didelphimorphia) | Broad/monDom5, 2006 | chr5:173087922–173327904 |
Table S5.
List of genomic coordinates of simian and cat HEMO ORF sequences
| Species | Assembly or GenBank accession no. | Coordinates | Abbreviation |
| Human | GRCh38/hg38, 2013 | chr4: 52743829–52745520 | hum |
| Chimpanzee | CSAC 2.1.4/panTro4 | chr4: 77303213–77304904 | cpz |
| Gorilla | gorGor4.1/gorGor4, 2014 | chr4: 76069071–76070762 | gor |
| Orangutan | WUGSC 2.0.2/ponAbe2, 2007 | chr4: 67458017–67459708 | oo |
| Gibbon | GenBank MF320351 | Dataset S1 | gib |
| Macaque | BCM Mmul_8.0.1/rheMac8, 2015 | chr5: 82025079–82026770 | mac |
| Baboon | GenBank MF320353 | Dataset S1 | bab |
| African green monkey | Chlorocebus_sabeus 1.1/chlSab2, 2014 | chr7: 15740103–15741791 | agm |
| Angolan black-and-white colobus | Cang.pa_1.0 | Scf473: 133450–131759 | col |
| Langur | GenBank MF320353 | Dataset S1 | lan |
| Golden snub-nosed monkey | Isolate Xiao Hai Rrox_v1 | ENSRROG025365: 167098–168786 | rhi |
| Marmoset | WUGSC 3.2/calJac3, 2009 | chr3: 140763785–140765366 | mar |
| Squirrel monkey | Broad/saiBol1, 2011 | JH378162: 9809510–9811090 | sqm |
| Spider monkey | GenBank MF320354 | Dataset S1 | spi |
| Saki monkey | GenBank MF320355 | Dataset S1 | sak |
| Cat | ICGSC/Felis_catus_8.0/felCat8, 2014 | chrB1:164136405–164138141 | cat |
Fig. S4.
Characterization of the marsupial env-panMars gene and protein. (A) Amino acid sequence homology between marsupial env-panMars (coordinates as in ref. 12) and HEMO proteins from representative simian species and domestic cat (Table S5). Every amino acid of a marsupial sequence that is found at the same position in a simian or cat sequence is highlight in yellow. Same color code for the characteristic env domains as in Fig. 1C. cat, cat; gib, gibbon; hum, human; mac, macaque; mar, marmoset; opo, opossum; tas, Tasmanian devil; wal, wallaby. *Stop codon. (B) Detection of the HA-tagged opossum and wallaby env-panMars proteins. Western blot of cell lysates (L) and supernatants (S) from 293T cells transfected with the phCMV-empty, phCMV-Opossum-env, or phCMV-Wallaby-env expression vectors. Detection with an anti-HA antibody (Upper) and an anti–γ-tubulin antibody (Lower). (C) Structure of the env-panMars gene locus and transcripts for the opossum (Upper) and wallaby (Lower). Schematic representation of the env-panMars locus, with the env-ORF in orange and the CpG island in green. N represents uncharacterized sequences. Black arrowheads (pA) position the AATAAA polyadenylation signal sequence. Intron–exon structures are from UCSC for the opossum and were characterized by RACE PCR experiments for the wallaby (RNA from the ovaries; SI Methods). Nucleotide sequences of the start site (TTCTA for the opossum and CTTTCTA for the wallaby) and the env ORF acceptor splice site are indicated, with the dinucleotide AG (end of intron) underlined; E2-E3 intron is dotted to indicate E3 skipping in a fraction of the wallaby transcripts, as observed for the HEMO gene.
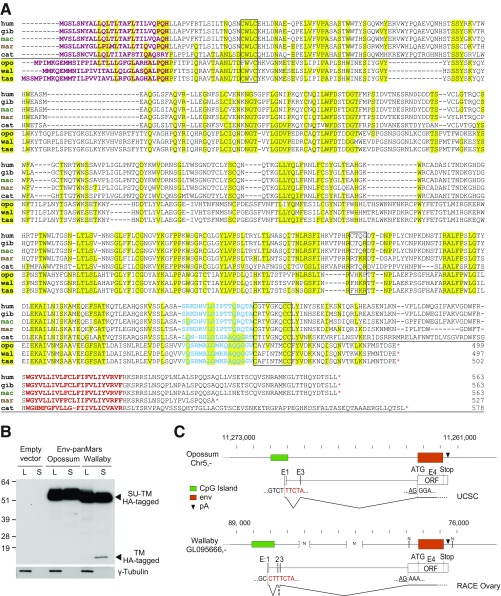
To test the conservation of the specific shedding property observed in humans, a series of simian HEMO genes were cloned, introduced into the phCMV expression vector, and tested by transfection of 293T cells as described above. As shown in Fig. 9C, the HEMO genes from all of the tested species encode a protein that can be detected with the human HEMO antibodies (yet with a lower intensity for the distant NWMs), with in all cases, evidence for protein shedding in the cell supernatant. Even in the NWM branch, where the HEMO protein has retained a functional furin site (Fig. S3), a shed form of the protein is released in the supernatant together with a smaller SU form. The smaller size observed for the spider monkey protein is consistent with a small 10-aa deletion in the 5′ part of the gene (amino acids 182–191) (Fig. S3). Accordingly, it seems that the shedding of the HEMO protein is a very well-conserved property among simians, a feature that, together with the purifying selection applying to this gene, is a hint for a possible role of this secreted protein, notably in pregnant females. Of note, the domains 3′ to the shed protein form (Fig. S3) are much less conserved at the sequence level among simians, except for the transmembrane anchoring domain that seems to be required for shedding of the HEMO protein at the cell membrane (Fig. S3).
A Related HEMO Gene in Marsupials.
To determine whether HEMO-like sequences could be present in some species where the orthologous gene could not be identified, a less stringent BLAST search was performed, which provided hits in Marsupials—but still in neither Afrotherians nor Xenarthrans. Of note, the closest env gene identified is a conserved marsupial env gene that we had previously identified to be present in all marsupials (12), namely env-panMars (the phylogenetic tree is shown in Fig. 1D). Amino acid sequence comparison of this conserved marsupial envelope protein with HEMO indicates only 20–30% similarity, but alignment of simian, cat, and marsupial (from opossum, wallaby, and Tasmanian devil) sequences (Fig. S4A) shows significant identity regions, all along the extracellular domains. The Env-panMars sequences correspond to truncated env because of a stop codon upstream of the transmembrane domain. The encoded proteins are, therefore, expected to be soluble proteins. As illustrated in Fig. S4B with HA-tagged Env-panMars proteins, the opossum and wallaby Env proteins are indeed released in the supernatant of cells transfected with the corresponding expression vectors. In the supernatant from wallaby transfected cells, a 15-kDa faint band can also be observed, which probably corresponds to the HA-tagged TM subunit produced after partial cleavage at a degenerate furin site (FHKR). No similar band is observed for the opossum [sequence at the furin site (VHKP)].
Furthermore, RACE-PCR experiments performed on wallaby RNA transcripts from ovary (Fig. S4C) locate the transcription start site within a CpG-rich region, with multiply-spliced RNAs in the promoter region as observed for the HEMO gene. In the case of the opossum, RNA-Seq data compiled in UCSC (Fig. S4C) show similar organization (with almost identical transcription start site located in a homologous CpG island and the use of the same E3 exon). Altogether, these data suggest that both the simian and marsupial env genes have a common retroviral ancestor. However, because of the long evolutionary distance between Marsupials and Eutherian mammals, which results in poor synteny data, no convincing evidence could be further obtained indicating that the marsupial gene would be the ortholog of HEMO (Fig. 9A) or of any of the noncoding copy in Table S2.
SI Methods
Database Screening and Sequence Analyses.
Retroviral endogenous env gene sequences were searched by BLAST on the human genome [GRCh38/hg38 Genome Reference Consortium Human Reference 38 (GCA_000001405.15); December 2013]. All genomic sequences containing an ORF longer than 400 aa (from start to stop codons) were extracted from the hg38 human database using the getorf program of the EMBOSS package (emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) and translated into amino acid sequences. These amino acid sequences were converted to a searchable database using formatdb (structure.usc.edu/blast/formatdb.html) and then BLASTed with blastall (structure.usc.edu/blast/blastall.html) against the SU-TM amino acid sequences of 42 retroviral envelope glycoproteins (from representative ERVs, among which are known syncytins, and infectious retroviruses) using the BLASTP program of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov). Positive envelope-containing ORFs were classified by multiple alignments of their amino acid sequences using the ClustalW protocol (www.ebi.ac.uk). ORFs consisting of highly repetitive sequences were discarded.
Maximum likelihood phylogenetic trees were constructed with RaxML 7.3.2, with bootstrap percentages computed after 1,000 replicates using the GAMMA + GTR model for the rapid bootstrapping algorithm.
Sequences were analyzed using various platforms and software: UCSC browser of the Santa Cruz University of California (https://genome.ucsc.edu/); Repbase (www.girinst.org/repbase/) (17); Repeatmasker (www.repeatmasker.org); Dfam of the University of Montana (www.dfam.org/); EMBOSS software at the Bordeaux Bioinformatics Center (CBiB) (services.cbib.u-bordeaux.fr/galaxy/), prediction servers at www.cbs.dtu.dk/services/ and www.expasy.org/, and newcpgreport for CpG island characterization at www.ebi.ac.uk/Tools/emboss/.
dN/dS ratios were obtained with the PAML program package on the PAMLX graphical user interface (version 1.2). Coordinates of the selected HEMO ORF sequences are listed in Table S5. The gibbon, baboon, spider monkey, and saki nucleotide HEMO ORF were PCR-amplified as indicated below, and the nucleotide sequences are reported in Dataset S1.
Syntenic loci were recovered for a representative number of species from the UCSC browser on a 250-kb genomic region located between two genes conserved in all species 5′ and 3′ to the HEMO locus, namely RASL11B and USP46. They were analyzed using the MultiPipMaker alignment tool (pipmaker.bx.psu.edu/pipmaker/), with the human genome sequence as a reference. Coordinates of the selected sequences are listed in Table S4.
Polyclonal and Monoclonal Anti-HEMO Antibodies.
A DNA fragment coding for 163 aa of the HEMO SU envelope subunit (amino acids 123–286) was inserted into the pET28b (Novagen) prokaryotic expression vector and expressed in BL21(DE3) bacteria. The recombinant C-terminal His-tagged protein was purified from bacteria lysates by nickel affinity chromatography. Mice immunization was performed in accordance with standard procedures. Sera containing polyclonal antibodies were recovered independently from 10 mice and tested by Western blot analyses using lysates of 293T cells transiently transfected with HEMO Env expression vector. One mouse was selected for mAb production by Agro-Bio, and one hybridoma clone was isolated (2F7; IgG2a isotype) for IgG production.
Cell Culture, 5-Aza-dC Treatment, and Metalloprotease Inhibitors.
Cells were maintained at 37 °C and 5% CO2 in DMEM for 293T (embryonic kidney), HeLa (cervix adenocarcinoma), CaCo-2 (colon adenocarcinoma), TE671 (rhabdomyosarcoma), SH-SY5Y (neuroblastoma), and HuH7 (hepatoma) human cells; RPMI Media 1640 for JAR (choriocarcinoma), 2102Ep (teratocarcinoma), and NCCIT (teratocarcinoma) human cells; and F-12K medium for BeWo (choriocarcinoma), JEG-3 (choriocarcinoma), and NTera2D1 (teratocarcinoma) human cells. All media were supplemented with 10% heat-inactivated FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin (all reagents are from Life Technology). iPSCs were grown on irradiated mouse embryonic fibroblasts (MEFs) at the Gustave Roussy iPSC Platform. When reaching confluence, cells were serum-deprived for 36 h, and supernatant was harvested, filtered (0.22-µm Millipore filters), and concentrated 20-fold on Amicon Ultra 0.5 mL (Millipore 10K).
For treatment with 5-Aza-dC (Sigma-Aldrich), 2 × 105 BeWo and 293T cells were plated in six-well dishes. Doses ranging from 0.1 to 5 μM of 5-Aza-dC were then added to the culture for 3 d, with fresh medium each day. Cells were harvested for RNA extraction 1 d later.
For treatment with metalloprotease inhibitors, 5 × 105 293T cells were seeded in six-well dishes, with 2 mL per well of culture medium. One day after seeding, cells were transiently transfected using 1.5 µg phCMV-HEMO plasmid and 4.5 µL Lipofectamine LTX (Thermo Fischer) per well. One day posttransfection, cells were incubated with culture medium supplemented with the indicated concentrations of metalloprotease inhibitors (Calbiochem): Batimastat (0.1–10 µM), Marimastat (0.1–10 µM), or GM6001 (1–50 µM). Medium with inhibitors was replenished for 2 other days, and supernatants were collected and filtered through 0.45-µm Millipore filters 1 d later. Cells were harvested the same day for protein analysis.
Luciferase Promoter Assay.
For HEMO promoter activity assay, fragments of different sizes containing the transcription start site (TSS) (+1) were PCR-amplified from human genomic DNA and cloned in sense and antisense orientation into the HindIII-NheI sites of the pGL3 Basic vector (Promega) upstream of the luciferase reporter gene [757-bp fragment: from −290 to +472; 467-bp fragment: from +1 (TSS) to +472; 408-bp fragment: from +57 to +472] [primers used are listed in Table S3, with (NNN) representing HindIII and NheI sites].
Table S3.
List of primers
| Primer names | Primer sequences |
| qRT-PCR | |
| HEMO-F1 | 5′-ACTATGGGCTCCCTTTCAAACT |
| HEMO-R1 | 5′-CATAGGAGGAAGTAGAGTGATT |
| RPLP0-F | 5′-GGCGACCTGGAAGTCCAACTA |
| RPLP0-R | 5′-CCATCAGCACCACAGCCTTC |
| G6PD-F | 5′-TGCAGATGCTGTGTCTGG |
| G6PD-R | 5′-CGTACTGGCCCAGGACC |
| RACE experiments | |
| HEMO-5′-RACE-R | 5′-CCTTGGGAGGTCCTAGTGCTAAGTGC |
| HEMO-3′-RACE-F | 5′-AAGCCACAGGAAGCTAGATTGAGATCAT |
| HEMO-R2 | 5′-GCTGTCTACTTCATCTGCTCAT |
| HEMO-R4 | 5′-CCGCAGACGTAGACAACGAA |
| HEMO-F4 | 5′-TTTCAAATAGGGCAATGAAGG |
| panMars-5′-RACE-R | 5′-CATCTGTCCTCTGGAACATCGCCCAAG |
| panMars-R2 | 5′-TCAGTTTCCATATTACCCACTT |
| panMars-R3 | 5′-CAAGGAGTGAACTGAAGTGG |
| panMars-R4 | 5′-ATTCGTCAGAACAACCCAATAG |
| Bisulfite experiments | |
| Fragment I | |
| I-F | 5′-AGGTAGGTAGTGGATATAGGTG |
| I-R | 5′-AAACCAAAAAACCAAAAAAA |
| Fragment I nested | |
| I-F2 | 5′-GTAGTGGATATAGGTGGTT |
| I-R2 | 5′-AAACCAAAAAACCAAAAAAAAAAC |
| Fragment II | |
| II-F | 5′-TTTTAATTTAGGATTTTTTTAGT |
| II-R | 5′-ATCTACCCTAAAAAAACAAA |
| Fragment II nested | |
| II-F2 | 5′-TTTTTTTTTTGGTTTTTTGG |
| II-R2 | 5′-AAAAAACAAAACRCAAACTTATTAC |
| Amplification of genomic HEMO in primate species | |
| HEMO-Ge-F-Xho | 5′-ATACATCTCGAGCATTGTCTGGAGTTTGCTTGT |
| HEMO-Ge-R-Mlu | 5′-ATACATACGCGTGGGTAAGGGTTTACAGATCAG |
| HEMO-NWM-R-Mlu | 5′-ATACATACGCGTACACCTTGGGAGGTCCTAGT |
| Amplification of promoter fragments | |
| HEMO-(-290)F | 5′-(NNN)GTCCTGCCCTCGTCCCGAAG |
| HEMO-(+1)F | 5′-(NNN)CACTTCAGTTCCCGCCGCGA |
| HEMO-(+57)F | 5′-(NNN)GCCAGTTTATCCCTCGGAGTT |
| HEMO-(472)R | 5′-(NNN)CCGCAGACGTAGACAACGAA |
| Furin site mutation | |
| HEMO-RTKR-F | 5′-CACCGCATAGACGCACCAAACGAGACACAGACA |
| HEMO-RTKR-R | 5′-TGTCTGTGTCTCGTTTGGTGCGTCTATGCGGTG |
The 293T cells were seeded in 96-well dishes with 2 × 104 cells per well. One day after seeding, cells were transfected with 100 ng DNA plasmid and 0.2 µL jetPRIME (Polyplus). Two days posttransfection, culture medium was discarded, and the activity of luciferase was detected using the Pierce Renilla-Firefly Luciferase Dual Assay Kit and the GloMax-Multi+ Luminescence Apparatus (Promega) following the manufacturer’s instructions.
Bisulfite Genomic Sequencing Analysis.
Genomic DNA from 293T, BeWo, iPSC-NP24, and CaCo-2 cells was subjected to bisulfite treatment with the EpiTect Plus DNA Bisulfite Kit (Qiagen). Two DNA fragments of the promoter region were amplified via nested PCR (two rounds of 35 cycles) with AccuPrimeTM High Fidelity polymerase (Invitrogen; Thermo Fischer), on 50–150 ng bisulfite-treated DNA using specific primers listed in Table S3. PCR products were then cloned into pGEMT-Easy vector (Promega), and a minimum of 10 clones were selected for sequencing.
Expression Vectors for the HEMO ORF from Human, Simians, and ex Vivo Assays.
The HEMO ORFs from human and selected simians (Fig. S3) were PCR-amplified from the corresponding genomic DNAs using the Phusion DNA Polymerase (Thermo Scientific), with a unique forward primer caused by high conservation 5′ to the ATG codon (hemoGe-F-Xho), and one of two reverse primers (hemoGe-R-Mlu or a specific NWM monkey hemoNWM-R-Mlu primer) (Table S3). PCR products were directly sequenced (BigDye Terminator v3.1; Thermo Fischer). The amplified HEMO gene fragments were then cloned into the XhoI and MluI sites of the phCMV-G expression vector (GenBank accession no. AJ318514) for transfection experiments. Premature stop codon HEMO mutants (Fig. 3) were constructed by inserting a TGA stop codon in a reverse primer used to PCR-amplify the indicated fragments from phCMV-HEMO and recloning as above. Substitution of the CTQG sequence by the consensus furin site RTKR (as in the NWM HEMO genes) was performed by site-directed mutagenesis with multiple PCR reactions.
HEMO protein production and release were assayed using 5 × 105 293T cells transfected with 1.5 μg phCMV-HEMO plasmid and 7.5 μL Fugene 6 (Promega) in six-well dishes. Cell media were replaced 12 h posttransfection by serum-free media. Forty-eight hours posttransfection, supernatant and cells were collected. Supernatants were filtered (0.45-μm Millipore filters) and stored at −80 °C. For cell lysates, samples were solubilized in RIPA buffer (150 mM NaCl, 25 mM Tris⋅HCl, pH 7.6, 0.1% SDS, 1% sodium deoxycholate; Thermo Scientific) with 1× Protease and Phosphatase Inhibitor Mixture (Thermo Scientific), centrifuged (14,000 × g for 20 min to eliminate debris), and stored at −80 °C before testing.
Immunofluorescence and Immunohistochemistry Assays.
For HEMO immunofluorescence assays, HeLa cells were grown on glass coverslips and transiently transfected with the phCMV-HEMO expression vector or a control empty vector (500 ng) and 1.5 µL Lipofectamine LTX (Thermo Fischer) per well of 12-well dishes. Forty-eight hours posttransfection, cells were fixed in 4% paraformaldehyde, permeabilized or not with 0.2% Triton X100, and stained with the mouse anti-HEMO polyclonal antibody (see above) and an Alexa Fluor 488-conjugated anti-mouse secondary antibody (Molecular Probes). Nuclei were stained in blue with DAPI (Sigma-Aldrich). Observations were made under a Leica TCS SP8 MP confocal microscope.
For immunohistochemistry assays, freshly collected placental tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Sections (4 μm) were stained with H&E and safran. Paraffin sections were processed for heat-induced antigen retrieval (Tris⋅EDTA, pH 9; Abcam) and incubated overnight with the monoclonal mouse anti-HEMO (2F7) antibody (1/10 dilution) or a control IgG2a isotype. Staining was visualized by using the peroxidase/diaminobenzidine Mouse PowerVision kit (ImmunoVision Technologies).
Western Blot Analyses, WGA Purification, and PNGase F Treatment.
Samples, cell supernatants, or cell lysates were analyzed by SDS/PAGE on gradient precast gels (NuPAGE Novex 4–12% Bis⋅Tris gels; Life Technologies) and transferred onto nitrocellulose membranes using a semidry transfer system. After blocking in PBS containing 0.1% Tween-20 and 5% nonfat milk, membranes were incubated overnight at 4 °C with primary antibodies [anti-HEMO mouse polyclonal antibody, 1/5,000; anti-CGB/hCG-beta rabbit polyclonal antibody (Abgent), 1/100,000; anti–γ-tubulin mouse mAb (Sigma-Aldrich), 1/1,000; anti–Env-W rabbit polyclonal antibody (from ref. 58), 1:500], washed three times, and then, incubated with species-appropriate HRP-conjugated secondary antibodies for 45 min at room temperature. Proteins were detected by using an enhanced chemiluminescence system (ECL; Pierce).
When specified, glycoproteins were first extracted from placental tissue or sera using the lectin WGA kit (Thermo Scientific); 600 μL whole-protein extracts were prepared according to the manufacturers’ guidelines and eluted in 200 μL elution buffer. When specified, samples were treated with PNGase F (NEB Biolabs) before SDS/PAGE.
MS Characterization of the N and C Termini of the HEMO Protein.
To get sufficient amounts of HEMO protein for MS characterization, 293T cells (four 10-cm dishes with 3 × 106 cells per dish) were transfected with the phCMV-HEMO expression vector (from human) in DMEM–FCS medium (10 μg per plate). Medium was replaced by serum-free DMEM 2 d later, and supernatants were recovered after 2 more days. Total secreted proteins were concentrated about 60-fold using Vivaspin 20 [Sartorius; molecular-weight cutoffs (MWCOs) of 30 kDa]. Glycoproteins from the concentrated extract were recovered using the WGA kit, eluted in 200 µL, and loaded on a 4–12% NuPAGE gel. The 80-kDa part of the acrylamide gel was excised, and proteins were eluted in a dialysis bag electrophoretically. Proteins were again concentrated using Amicon Ultra Centrifugal Filters (Ultracel-50K), treated with PNGase, and reloaded on a 4–12% NuPAGE gel for an additional purification step. The main band (seen on Coomassie Blue staining and corresponding to the shed 48-kDa HEMO protein) was excised and subjected independently to different enzymatic digestions (Trypsin, Chymotrypsin). The shed HEMO protein-associated fragments were characterized by the IMAGIF platform of Gif-sur-Yvette by nanoscale liquid chromatography coupled to tandem MS (nanoLC–MS/MS) analyses with a TripleTOF 4600 mass spectrometer (AB Sciex), thus allowing the determination of the N and C termini of the protein.
RNA, qRT-PCR, and RACE Experiments.
Total RNAs from human tissues and cells were either purchased from Zyagen or isolated using the RNAeasy Isolation Kit (Qiagen) according to the manufacturer’s instructions and treated with DnaseI (Ambion). Reverse transcription was performed with 1 μg RNA using the MLV reverse transcriptase (Applied Biosystems). qRT-PCR was carried out with an ABI Prism 7000 sequence detection system with 5 μL diluted (1:20) cDNA in a final volume of 25 μL by using SYBR green PCR master mix (Qiagen) and specific primers (listed in Table S3). Primers were selected to match at 100% to the target gene only, with dissociation curves only disclosing a single peak. Transcript levels were normalized relative to the amount of a housekeeping gene (RPLP0 or G6PD) mRNA. Samples were assayed in triplicate; 5′ RACE and 3′ RACE were performed with 100 ng DNase-treated RNA using the SMARter RACE cDNA Amplification Kit (Clontech) and the primers listed in Table S3.
RNA-Seq Data Mining.
RNA-Seq raw data were downloaded from the NCBI Sequence Read Archive with accession numbers SRP011546 (GSE36552), ERP003613 (PRJEB4337), and SRP042153 (GSE57866). RNA-Seq raw data were aligned with TopHat2 (v2.0.14) to a custom gene database of interest, including some retroviral envelope and housekeeping genes, with the following parameters: “–read-mismatches 0-g 1–no-coverage-search.” Uniquely mapped reads were selected using SAMtools (v0.1.19) for additional analysis. Only hits with exact matches were counted to avoid detection of other analogous ERV genes. Read counts were normalized by the length of the gene (after merging in kilobases) and the read counts of two housekeeping genes (RPLP0 and RPS6) and log-transformed. Specific transcripts of the gene (absence of read counts in intronic and flanking sequences and presence of split RNA-Seq reads corresponding to specific splice junctions) were also verified by BLAST on the NCBI Trace Archive Nucleotide BLAST platform. For each gene of interest, read counts were verified to be equally distributed over the coding sequence on the Integrative Genomics Viewer visualization tool (software.broadinstitute.org/software/igv).
Microarray Data Mining.
To get insight into the expression profile of the HEMO gene in normal and tumoral human tissues, an in silico analysis of microarray (Affymetrix U133A) data was performed using the dataset E-MTAB-62 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-62/files/), which includes 1,033 samples from normal tissues and 2,315 neoplasm samples obtained from various AE and GEO studies (34). We first checked that none of 25-nt probes of the Affymetrix array that were expected to hybridize with the HEMO transcript (100% identity) could possibly target any other genomic sequence: actually, a BLAST search for each probe does not reveal any matching for more than 17 over 25 nt, except for one where a match was found—in addition to HEMO—over 18 and 20 nt for two genomic sequences (unrelated to MER34 and further annotated as being untranscribed). We have further checked the specificity of the microarray signal by analysis of U133A Affymetrix array data of various tissues and cell lines for which our own data using qRT-PCR (and in some instances, immunohistochemistry) were available: for instance, a positive signal was verified in microarray experiments reported for whole placenta, EVTs, and villous CTs, whereas no signal was detected in microarray experiments from choriocarcinoma cell lines (JEG-3 and BeWo) and other known negative tissues. The dataset E-MTAB-62 was downloaded as processed expression data. Statistical significance was assessed using Wilcoxon’s rank sum test. For larger panel analyses, additional ovarian cancer datasets (AE E-GEOD-63885, E-GEOD-30311, E-GEOD-54809, E-GEOD-6008, and E-GEOD-14764 from https://www.ebi.ac.uk/arrayexpress) were preprocessed using the “expresso” function of the affy package (v1.48.0), with the following parameters: robust multiarray average for background correction, keeping only the perfect match probes (“pmonly”), and quantile normalization. Both “medianpolish” and “avgdiff” were applied as summarization methods to have normalized values in both log2-transformed values and probe set intensities log2-transformed values. After data preprocessing, the expression values of the HEMO gene were extracted and plotted with R 3.2.3 (www.r-project.org). The ovarian cancer datasets were merged using the inSilicoMerging R package (version 1.14), applying COMBAT as batch effect correction method.
Discussion
Here, we have identified an endogenous retroviral envelope gene, HEMO, with a full-length protein-coding sequence conserved in simians, including humans, and an unprecedented characteristic feature for a retroviral envelope because it is shed and released in the extracellular medium, being found at a detectable level in the blood of pregnant women. Several retroviral envelope gene “captures” have been reported among most mammalian species, and in a number of cases, these genes were shown to be syncytins (i.e., genes playing a role in placentation), with the canonical immunosuppressive and fusogenic properties inherited from their ancestral retroviral progenitors being involved in a physiological function of benefit to the host (38) (reviewed in refs. 9 and 39). The presently identified HEMO gene shares some of the properties of syncytins but is different, because it is shed in the extracellular environment with no evidence for fusogenic activity. In addition, its pattern of expression is not strictly restricted to the placenta—although it is the organ where its expression is highest. However, its conservation in evolution with characteristic features of a coopted gene (i.e., evidence for purifying selection) together with the identification of a closely related retroviral env gene captured and conserved in the remote marsupial clade (which diverged from eutherian mammals more than 150 Mya) sharing with HEMO a CpG-rich promoter and the capacity of its protein product to be released in the extracellular medium [in that case because of a stop codon located just upstream of the transmembrane domain of the TM subunit (12)] constitute a strong hint for a potential physiological role in simians (see below).
The identified retroviral env gene belongs to a poorly characterized and moderately reiterated ERV family, namely the MER34 family, with only highly degenerated elements (5, 16, 17). Analysis of the structure of the genomic locus where HEMO can be identified only reveals traces of an ancestral provirus, with a highly rearranged gene organization. Of note, an LTR structure is only barely detectable 3′ to HEMO, and the 5′ LTR is no longer present. Actually, RACE-PCR analysis of the HEMO transcripts reveals a transcription start site within a CpG-rich domain unrelated to an LTR but clearly possessing a promoter activity as shown by transfection of reporter plasmids—with the promoter in both orientations—in cells in culture. This unusual promoter is most probably responsible for the specific pattern of expression of the HEMO gene, which is found to be active in a series of stem cells ex vivo as well as in vivo very early in the developing embryo. The encoded protein itself has some unusual features, because it no longer possesses a furin cleavage site (although a functional one can still be shown for the HEMO ortholog present within the NWM genome) and more importantly, because it is specifically cleaved at the cell membrane via a metalloproteinase-mediated processing that results in the shedding of its ectodomain into the extracellular medium —observed for all simians, including NWMs. Shedding is a process that has not been reported previously for a retroviral envelope, although such a process is used by the cellular machinery for a series of cellular genes (e.g., Notch, TNF-alpha) involved, for instance, in signaling, cell mobility, and migration (reviewed in refs. 21, 22, and 40). Of note, a closely related molecular event also takes place in the case of the Ebola filovirus envelope protein, which is, in part, shed in the cell medium by a specific ADAM-mediated cleavage upstream of the transmembrane domain (20, 41). In that case as well, the shed protein is detected in the blood and anticipated to play a critical role in the associated pathology either by exerting a decoy effect on anti-Env antibodies or even through direct immune activation and increased vascular permeability in the infected individuals (42). The presently observed shedding of the HEMO retroviral envelope protein de facto makes a link between unrelated viruses (e.g., a filovirus and a retrovirus), with a possible convergent evolution for the triggering of a systemic effect via a shedding process.
A still unresolved question concerns the possible role of HEMO in human physiology and/or pathology. Because of (i) the high level of purifying selection acting on the gene in simians; (ii) the conservation in marsupials of a gene transcribed from a similar promoter type and encoding a protein closely related in both sequence and mature protein extracellular localization (both proteins are released in the supernatant by shedding for HEMO and because of absence of a transmembrane-anchoring domain for the marsupial protein); (iii) the rather uncommon profile of expression in development; and (iv) the massive shedding by the placenta of the protein into the blood, it can be anticipated that HEMO fulfils a role, most probably in pregnancy. Among the possible roles that can be hypothesized, a protective effect against infection by—still to be identified—viruses and/or retroviruses would be relevant. Such protective effects could be mediated by classical “interference” via the sequestration of the receptor for the incoming virus, an effect that could be further enhanced by the release of the HEMO protein in the blood circulation and direct targeting of such receptors (reviewed in refs. 43 and 44). Alternately, HEMO might possess a cytokine- or hormone-like activity, with a possible role in pregnancy still to be uncovered. An effect of HEMO in development should also be considered, taking into consideration that its expression is observed as early as at the eight-cell stage and persists at all of the subsequent embryonic stages. Of note, other ERVs—including HERV-H and HERV-K—have related profiles of expression, and abundant HERV-H RNA was recently shown to be a marker of cell “stemness” in humans and possibly play a role—via transcriptional effects and/or specific ERV-driven transcripts—in the maintenance of pluripotency in human stem cells (45–49) (reviewed in refs. 50–52). In the case of HEMO, which unambiguously encodes a retroviral envelope protein that can further be detected, its expression might not only be a stemness marker as for the above multicopy ERVs, but its encoded protein might also constitute—as the OCT4, SOX2, KLF4, or MYC “reprogramming” factors (53)—a molecular effector of pluripotency per se. Finally, we could unravel HEMO gene expression in a series of human tumors and show HEMO protein expression in ovarian tumors. Additional immunological analyses based on a large number of tumors and control tissues will have to be performed to definitely correlate HEMO protein expression with specific tumor histotypes (54, 55) (reviewed in ref. 56 for other retroviral Env expressed in tumors) and assess whether this protein can be considered as a reliable marker of a given tumoral state and tentatively, a possible target for immunotherapeutic approaches.
Experiments are now in progress to identify the cellular interacting partners of the HEMO protein, with the hope that their identification will allow a definite characterization of HEMO functions in vivo in both normal development and the onset of pathological processes.
Methods
Biological Samples.
First trimester human placenta tissues were obtained from legal elective terminations of pregnancy (gestational age 8–12 wk) with parent’s written informed consent from the Department of Obstetrics and Gynecology at the Cochin Hospital. Blood samples from pregnant (11–18 wk of amenorrhea) and nonpregnant (before ovulation induction hormonal therapy) women were from Labo Eylau with MTA protocol MTA2015-45. Male blood samples were from the Etablissement Français du Sang with agreement 15EFS018.
Ovary tissue samples were from the Biological Resources Centre and the Department of Laboratory Medicine and Pathology of the Gustave Roussy Institute (Research Agreement RT09916).
RNAs from human ESCs (H1, H7, and H9) were from U1170-INSERM of the Gustave Roussy Institute. iPSCs (reprogrammed CD34+ human cells at passage 24) and their supernatants were from the iPSC Platform of the Gustave Roussy Institute. The source of nonhuman primate genomic DNA is in ref. 57 [except for gibbon (from the European Collection of Authenticated Cell Cultures) and spider monkey (from Coriell Institute)], and the source of wallaby RNA is in ref. 12.
Ethics Statement.
All human samples were obtained with written informed consent. Experiments were approved by the Ethics Committee of the Gustave Roussy Institute. This study was carried out in strict accordance with the French and European laws and regulations regarding Animal Experimentation (Directive 86/609/EEC regarding the protection of animals used for experimental and other scientific purposes).
Other methods are as in ref. 12, and they are detailed in SI Methods.
Supplementary Material
Acknowledgments
We thank David Cornu and Laïla Sago (Paris-Saclay Proteomic Platform, Institut Biologie Integrative Cellule, Gif sur Yvette) and Emilie Cochet and Vasily Ogryzko (Proteomic Platform of the Gustave Roussy Institute) for MS analyses; Larissa Lordier (Gustave Roussy iPSC Platform) for the gift of iPSCs and supernatant; Antonio di Stefano and Nathalie Balayn (UMR 1170, Gustave Roussy) for the gift of ESCs RNAs; and Mélanie Polrot (Gustave Roussy Animal Care Facilities) for assistance in mice handling. We also thank Jean-Michel Teboul (Port-Royal Maternity Hospital) for providing placental tissues; Jean-Yves Scoazec, Catherine Genestie, and Christine Machavoine for the gift of tumor tissues (Gustave Roussy, Centre de Ressources Biologiques); and Martine Bacry (Laboratoire Eylau) for serum samples. We thank Philippe Dessen, Guillaume Meurice, and Bastien Job (Bioinformatic Platform, Gustave Roussy) for helpful bioinformatics assistance and discussion; Anne Dupressoir and Jérome Salmon for fruitful discussions; and Christian Lavialle for critical reading of the manuscript. This work was supported by the CNRS and grants to T.H. from the Ligue Nationale contre Le Cancer and the Agence Nationale de la Recherche (ANR “RETRO-PLACENTA”).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MF320351–MF320355).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702204114/-/DCSupplemental.
References
- 1.Feschotte C, Gilbert C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 2.Dewannieux M, Heidmann T. Endogenous retroviruses: Acquisition, amplification and taming of genome invaders. Curr Opin Virol. 2013;3:646–656. doi: 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Mager DL, Stoye JP. Mammalian endogenous retroviruses. Microbiol Spectr. 2015;3:MDNA3-0009-2014. doi: 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 4.de Parseval N, Heidmann T. Human endogenous retroviruses: From infectious elements to human genes. Cytogenet Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 5.Vargiu L, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 7.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavialle C, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA. 2011;108:E1164–E1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis G, et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci USA. 2015;112:E487–E496. doi: 10.1073/pnas.1417000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henzy JE, Johnson WE. Pushing the endogenous envelope. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120506. doi: 10.1098/rstb.2012.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tóth G, Jurka J. Repetitive DNA in and around translocation breakpoints of the Philadelphia chromosome. Gene. 1994;140:285–288. doi: 10.1016/0378-1119(94)90559-2. [DOI] [PubMed] [Google Scholar]
- 17.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 18.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 19.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolnik O, et al. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 2004;23:2175–2184. doi: 10.1038/sj.emboj.7600219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okazaki I, Nabeshima K. Introduction: MMPs, ADAMs/ADAMTSs research products to achieve big dream. Anticancer Agents Med Chem. 2012;12:688–706. doi: 10.2174/187152012802650200. [DOI] [PubMed] [Google Scholar]
- 22.Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139:3693–3709. doi: 10.1242/dev.076398. [DOI] [PubMed] [Google Scholar]
- 23.Pollheimer J, Fock V, Knöfler M. Review: The ADAM metalloproteinases - novel regulators of trophoblast invasion? Placenta. 2014;35:S57–S63. doi: 10.1016/j.placenta.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Aghababaei M, Beristain AG. The Elsevier Trophoblast Research Award Lecture: Importance of metzincin proteases in trophoblast biology and placental development: A focus on ADAM12. Placenta. 2015;36:S11–S19. doi: 10.1016/j.placenta.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Majali-Martinez A, et al. Placental membrane-type metalloproteinases (MT-MMPs): Key players in pregnancy. Cell Adhes Migr. 2016;10:136–146. doi: 10.1080/19336918.2015.1110671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole LA. New discoveries on the biology and detection of human chorionic gonadotropin. Reprod Biol Endocrinol. 2009;7:8. doi: 10.1186/1477-7827-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bischof P, Irminger-Finger I. The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol. 2005;37:1–16. doi: 10.1016/j.biocel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Maltepe E, Fisher SJ. Placenta: The forgotten organ. Annu Rev Cell Dev Biol. 2015;31:523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- 29.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 30.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 31.Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedli M, et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soygur B, Moore H. Expression of Syncytin 1 (HERV-W), in the preimplantation human blastocyst, embryonic stem cells and trophoblast cells derived in vitro. Hum Reprod. 2016;31:1455–1461. doi: 10.1093/humrep/dew097. [DOI] [PubMed] [Google Scholar]
- 34.Lukk M, et al. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho KR, Shih IeM. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurman RJ, Shih IeM. The dualistic model of ovarian carcinogenesis: Revisited, revised, and expanded. Am J Pathol. 2016;186:733–747. doi: 10.1016/j.ajpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 38.Mangeney M, et al. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denner J. Expression and function of endogenous retroviruses in the placenta. APMIS. 2016;124:31–43. doi: 10.1111/apm.12474. [DOI] [PubMed] [Google Scholar]
- 40.Jones JC, Rustagi S, Dempsey PJ. ADAM Proteases and Gastrointestinal Function. Annu Rev Physiol. 2016;78:243–276. doi: 10.1146/annurev-physiol-021014-071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolnik O, et al. Shedding of ebola virus surface glycoprotein is a mechanism of self-regulation of cellular cytotoxicity and has a direct effect on virus infectivity. J Infect Dis. 2015;212:S322–S328. doi: 10.1093/infdis/jiv268. [DOI] [PubMed] [Google Scholar]
- 42.Escudero-Pérez B, Volchkova VA, Dolnik O, Lawrence P, Volchkov VE. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 2014;10:e1004509. doi: 10.1371/journal.ppat.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nethe M, Berkhout B, van der Kuyl AC. Retroviral superinfection resistance. Retrovirology. 2005;2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malfavon-Borja R, Feschotte C. Fighting fire with fire: Endogenous retrovirus envelopes as restriction factors. J Virol. 2015;89:4047–4050. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fort A, et al. FANTOM Consortium Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 48.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Göke J, et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: War and peace. Mol Cell Biol. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 52.Izsvák Z, Wang J, Singh M, Mager DL, Hurst LD. Pluripotency and the endogenous retrovirus HERVH: Conflict or serendipity? BioEssays. 2016;38:109–117. doi: 10.1002/bies.201500096. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: A decade of progress. Nat Rev Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang-Johanning F, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 55.Wang-Johanning F, et al. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer. 2014;134:587–595. doi: 10.1002/ijc.28389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strissel PL, et al. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget. 2012;3:1204–1219. doi: 10.18632/oncotarget.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esnault C, Cornelis G, Heidmann O, Heidmann T. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013;9:e1003400. doi: 10.1371/journal.pgen.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malassiné A, et al. Expression of the fusogenic HERV-FRD Env glycoprotein (syncytin 2) in human placenta is restricted to villous cytotrophoblastic cells. Placenta. 2007;28:185–191. doi: 10.1016/j.placenta.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Malassiné A, et al. Human endogenous retrovirus-FRD envelope protein (syncytin 2) expression in normal and trisomy 21-affected placenta. Retrovirology. 2008;5:6. doi: 10.1186/1742-4690-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.