Significance
Although lysosomes play a crucial role in autophagy, damaged lysosomes are eliminated by autophagy. The molecular mechanisms that recognize lysosomal damage in cells remain poorly understood, but ubiquitination is a known prerequisite for directing autophagic machinery to damaged lysosomes. FBXO27, a substrate-recognition subunit of the SCF (SKP1/CUL1/F-box protein) ubiquitin ligase complex, localizes to the cytosolic surface of endomembranes and binds glycoproteins. This paper reports that SCFFBXO27 ubiquitinates exposed glycoproteins normally sequestered on the lumenal surface of lysosomes following lysosomal damage, resulting in accelerated recruitment of autophagic machinery.
Keywords: autophagy, FBXO27, LAMP2, lysosome, ubiquitin
Abstract
Ubiquitination functions as a signal to recruit autophagic machinery to damaged organelles and induce their clearance. Here, we report the characterization of FBXO27, a glycoprotein-specific F-box protein that is part of the SCF (SKP1/CUL1/F-box protein) ubiquitin ligase complex, and demonstrate that SCFFBXO27 ubiquitinates glycoproteins in damaged lysosomes to regulate autophagic machinery recruitment. Unlike F-box proteins in other SCF complexes, FBXO27 is subject to N-myristoylation, which localizes it to membranes, allowing it to accumulate rapidly around damaged lysosomes. We also screened for proteins that are ubiquitinated upon lysosomal damage, and identified two SNARE proteins, VAMP3 and VAMP7, and five lysosomal proteins, LAMP1, LAMP2, GNS, PSAP, and TMEM192. Ubiquitination of all glycoproteins identified in this screen increased upon FBXO27 overexpression. We found that the lysosomal protein LAMP2, which is ubiquitinated preferentially on lysosomal damage, enhances autophagic machinery recruitment to damaged lysosomes. Thus, we propose that SCFFBXO27 ubiquitinates glycoproteins exposed upon lysosomal damage to induce lysophagy.
Most cell-surface proteins and secretory proteins are glycoproteins modified with highly diverse Asn (N)-linked oligosaccharides (N-glycans) that have been implicated in various extracellular processes. N-glycosylation of proteins occurs cotranslationally in the endoplasmic reticulum (ER), and glycoproteins transit from the ER through the Golgi, where N-glycans are trimmed and remodeled before they are delivered to their final destinations. This process results in N-glycans facing either out of cells or on the luminal side of organelles in the secretory pathway, including the ER, Golgi complex, lysosomes, and endosomes. Thus, most molecules that recognize N-glycoproteins are also present in these organelles or on the cell surface. However, a subset of glycosidases or lectins are found either in the cytosol or in nuclei. Peptide-N-glycanase (PNGase) and glycoprotein-specific ubiquitin ligases are cytosolic, and function in the ER-associated degradation pathway in which misfolded glycoproteins are translocated from the ER to the cytosol for degradation by the ubiquitin-proteasome system (1–4). In another case, galectin 8, a cytosolic β-galactoside–binding lectin, detects bacterial invasion by binding host glycans exposed on damaged endosomes and lysosomes, and activates xenophagy through the recruitment autophagy receptor NDP52 (5). Thus, the appearance of N-glycan–modified proteins in the cytosol likely signals the presence of unwanted proteins or organelles.
F-box proteins are receptors for substrates in the SCF (SKP1-CUL1-F-box protein-RBX1) ubiquitin ligase complex. F-box proteins belong to a large protein family comprised of ≈70 proteins in humans. Among these are FBXO2 and FBXO6, which bind to glycoproteins modified with high-mannose N-glycans found in the ER (3, 4). Biochemical and structural analyses indicate that FBXO2 recognizes the innermost chitobiose structure in N-glycans to direct unfolded glycoproteins to the ER-associated degradation pathway (6, 7). Furthermore, we found that FBXO27, as well as FBXO2 and FBXO6, bind to glycoproteins modified with high-mannose N-glycans (8). However, FBXO27 has also been reported to bind some glycoproteins modified with complex-type N-glycans (9), suggesting that the SCF complex containing FBXO27 has the ability to ubiquitinate glycoproteins that have transited from the ER to other organelles or the plasma membrane. If FBXO27 recognizes complex-type N-glycans, there must be a mechanism through which glycoproteins from these other organelles become exposed to cytoplasmic SCFFBXO27.
Emerging evidence suggests that selective autophagy is used to degrade aggregated proteins, invading pathogens, and damaged or surplus organelles, such as peroxisomes, mitochondria, lysosomes, the nucleus, and the ER (10, 11). In these systems, ubiquitination often plays a pivotal role in directing the autophagic machinery to substrates (12). It has been shown that ubiquitin-positive inclusion bodies are degraded by autophagy via p62, an adaptor protein linking ubiquitin to LC3, an autophagosome marker (13). Furthermore, a key signaling step during mitophagy involves ubiquitination of outer membrane proteins of damaged mitochondria by the ubiquitin ligase Parkin, which is recruited from the cytoplasm by PINK1-mediated phosphorylation of ubiquitin (14). In another example, autophagy specifically targets invading bacteria to restrict their growth after ubiquitination (15). Furthermore, damaged lysosomes are selectively sequestered by autophagosomes through a ubiquitin-dependent mechanism called lysophagy (16, 17). Although which proteins and how they are ubiquitinated during xenophagy and lysophagy, before autophagy, remain largely unknown, it is plausible that luminal proteins in organelles are exposed to the cytosol following membrane rupture, which may subsequently lead to their ubiquitination.
Here, we demonstrated that FBXO27 is efficiently recruited to lysosomes upon damage, and regulates subsequent accumulation of autophagic machinery. We analyzed proteins ubiquitinated upon lysosomal damage by using the trypsin-resistant tandem ubiquitin-binding entity (TR-TUBE) method, which we recently developed, to identify ubiquitin ligase substrates (18), and found that SCFFBXO27 ubiquitinates luminal sites on the lysosomal protein LAMP2.
Results
FBXO27 Recognizes N-Glycoproteins and Is Targeted to Membranes via N-Myristoylation.
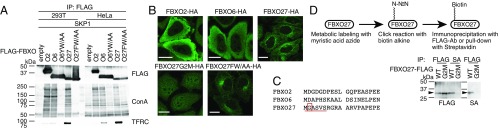
Previous work showed that FBXO2 binds to the innermost chitobiose in high-mannose N-glycans, and that the mutation of Y279 and W280 prevent binding (6). To determine whether FBXO27 and FBXO6 also recognize the chitobiose, we expressed FLAG-tagged wild-type versions of these proteins, as well as versions containing mutations in residues corresponding to Y279 and W280 in FBXO2, in 293T, or HeLa cells. We analyzed proteins immunoprecipitated using anti-FLAG antibodies on lectin blots probed with ConA, which recognizes high-mannose N-glycans (Fig. 1A). As we expected, ConA-reactive glycoproteins were not detected in coimmunoprecipitates with either mutant (FBXO6YW/AA or FBXO27FW/AA). Furthermore, we verified that FBXO27 can interact with transferrin receptor (TFRC), an endosomal glycoprotein modified with complex-type N-glycans, as previously reported (9), suggesting that FBXO27 recognizes various N-glycan structures.
Fig. 1.
FBXO27 is localized to membranes via N-myristoylation. (A) FBXO27 binds to glycoproteins modified with high-mannose N-glycans and TFRC. 293T and HeLa cells were transfected with plasmids encoding either only the FLAG tag, or FLAG-tagged FBXO2 (O2), FBXO6 (O6), FBXO6Y241A/W242A (O6YW/AA), FBXO27(O27), or FBXO27F262A/W263A (O27FW/AA), in combination with SKP1. Whole-cell lysates (WCL) were subjected to immunoprecipitation with an anti-FLAG antibody, and analyzed by blotting with the indicated antibodies and ConA lectin. (B) Localization of three glycoprotein-recognizing F-box proteins and FBXO27 mutants. HeLa cells stably expressing FBXO2-HA, FBXO6-HA, FBXO27-HA, the N-myristoylation defective mutant (FBXO27G2M-HA), or glycoprotein-binding–defective mutant (FBXO27FW/AA-HA) were subjected to immunostaining with anti-HA antibody. (Scale bars, 20 μm.) (C) The amino acid sequence of the N-terminal region of each F-box protein. The residues that constitute the N-myristoylation consensus sequence are aligned, and the predicted myristoylated Gly is boxed in red. (D) Detection of N-myristoylation in FBXO27. 293T cells expressing FBXO27-FLAG or FBXO27G2M-FLAG were incubated in the presence of myristic acid azide. Cell lysates were reacted with biotin alkyne, and FLAG-tagged proteins and biotin-labeled proteins were isolated with either anti-FLAG antibody or SA-agarose. Captured proteins were blotted with anti-FLAG antibody and SA-HRP. Arrows show the positions of FBXO27-FLAG.
We went on to examine the cellular localization of FBXO27 and observed its localization to membranes, unlike FBXO2 and FBXO6 (Fig. 1B). Although FBXO2 and FBXO6 were mainly recovered in the cytosolic fraction, FBXO27 was included in the 100,000 × g pellet fraction (Fig. S1A). We identified a putative N-terminal myristoylation motif (MGXXXS) in FBXO27 (Fig. 1C). N-myristoylation is the covalent attachment of myristic acid to the N-terminal Gly after the first Met has been removed. To determine whether the Gly confers myristoylation on FBXO27, we used a click chemistry-based approach (Fig. 1D). We expressed either wild-type FBXO27-FLAG or its G2M mutant, which we predicted to be unmyristoylated. We subsequently incubated the cells with myristic acid-azide, and generated cell lysates that were subjected to the Click-iT reaction for in vitro biotinylation. FBXO27-FLAG proteins and biotinylated proteins were isolated from the biotinylated lysate by using either anti-FLAG antibody or streptavidin (SA) beads, and were analyzed by blotting. Wild-type FBXO27 was biotinylated but not the G2M mutant, indicating that FBXO27 is N-myristoylated in cells.
Fig. S1.
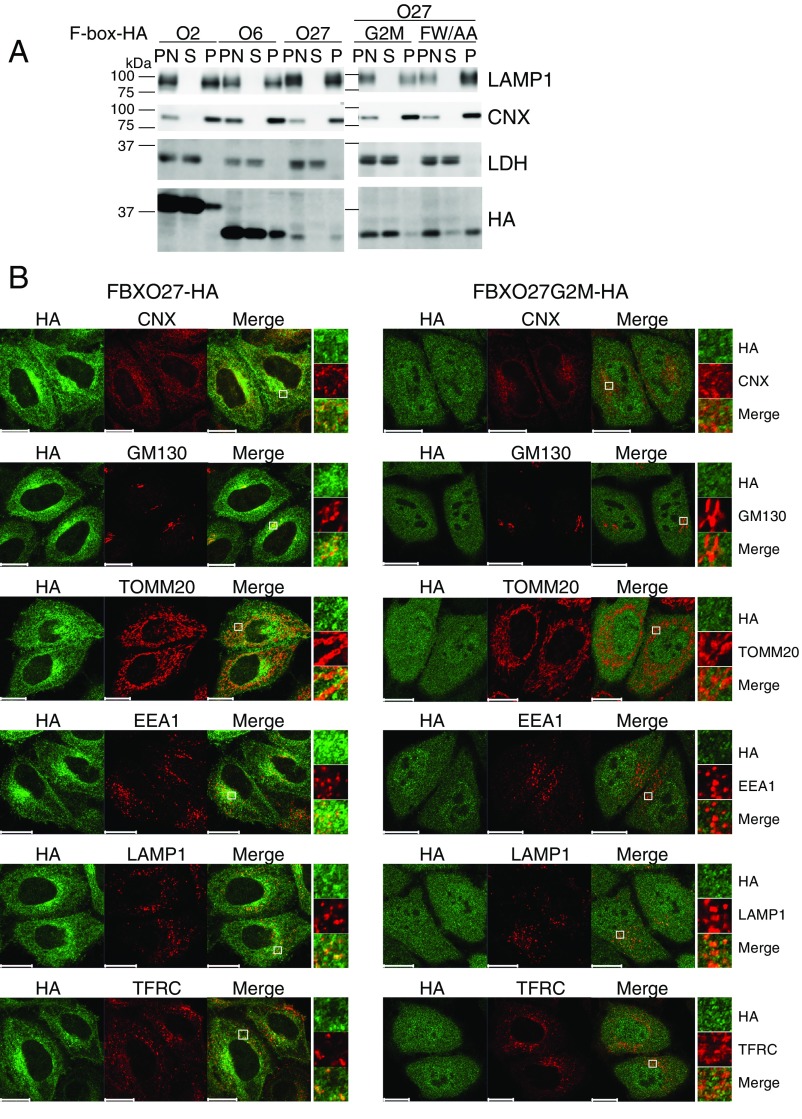
Subcellular localization of FBXO27. (A) Cell homogenates prepared from HeLa cells stably expressing C-terminally HA-tagged F-box protein using in Fig. 1B were fractionated by centrifugation. An equivalent volume of each fraction was subjected to immunoblotting with the indicated antibodies. P, pellet; PN, postnuclear supernatant; S, supernatant. (B) HeLa cells stably expressing FBXO27-HA or FBXO27G2M-HA (green) were subjected to immunostaining using the following antibodies: anticalnexin (CNX; ER marker), anti-GM130 (Golgi marker), anti-TOMM20 (mitochondria marker), anti-EEA1 (early endosome marker), anti-LAMP1 (lysosome marker), and anti-TFRC (recycling endosome marker). (Scale bars, 20 μm.) In each column, the square regions on the Left are highlighted on the Right. (Magnification, 5x.)
To determine whether the membranous localization of FBXO27 is a result of its N-myristoylation or to its ability to bind to N-glycans, we proceeded to compare the localization of the G2M mutant and the glycoprotein-binding defective FW/AA mutant (Fig. 1B and Fig. S1A). Although the membranous localization was retained in the FW/AA mutant, the G2M mutant protein was detected in whole cells and recovered in cytosolic fraction, indicating that FBXO27 is targeted to membranes via N-myristoylation. We further characterized the subcellular localization of FBXO27 by staining with antibodies for organelle marker proteins (Fig. S1B). Interestingly, FBXO27 did not colocalize with TOMM20, a protein marker of mitochondria, but colocalized with organelle marker proteins in the membrane trafficking pathway: that is, the ER, Golgi, endosomes, and lysosomes, where glycoproteins reside.
Membrane Targeting and Glycoprotein Binding Activity of FBXO27 Are Important for Its Recruitment of Damaged Lysosomes.
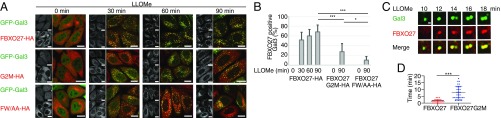
Because FBXO27 is found on the cytoplasmic surface of membranes surrounding organelles, it cannot bind to glycoproteins found on the luminal side of these organelles under normal conditions. However, if membrane integrity is impaired, SCFFBXO27 may be able to ubiquitinate glycoproteins in the ruptured organelle. For example, a diverse range of substances, such as silica, bacterial/viral toxins, and β-amyloid, can all impair lysosomal membranes in vivo. Thus, we next examined the behavior of FBXO27 following disruption of lysosomal membranes using the lysosomotropic compound l-leucyl-l-leucine methyl ester (LLOMe) (19). To monitor lysosome rupture, we used cells that express GFP-galectin3 (GFP-Gal3) (16). Gal3 is a galactose-binding lectin found in both the cytoplasm and nucleus in a static state that forms puncta within ruptured lysosomes following LLOMe treatment. We determined the rate of formation of FBXO27+ Gal3 puncta (Fig. 2 A and B) and FBXO27+ lysosomes (LAMP1) (Fig. S2 A and B) at indicated time points following LLOMe treatment. We also determined the time-course of FBXO27 recruitment by performing dual-color live-cell imaging analysis on cells that express GFP-Gal3 and FBXO27-Halo (Movies S1–S3).
Fig. 2.
Membrane targeting of and glycoprotein binding by FBXO27 are significant for its recruitment to damaged lysosomes. (A) HeLa cells stably expressing GFP-Gal3 and either FBXO27-HA or its mutants were treated with LLOMe for the indicated time, and were subjected to immunostaining with anti-HA (red) antibody. (Scale bars, 20 μm.) (B) Quantification of FBXO27 translocation to GFP-Gal3 puncta under LLOMe treatment. The data represent means + SD. Over 30 cells were counted for each experiment (n = 3). Statistical significance was determined using one-way ANOVA with posttesting according to Tukey’s test. ***P < 0.001, *P < 0.03. (C) HeLa cells expressing GFP-Gal3 and FBXO27-Halo were labeled with HaloTag TMR. After adding LLOMe, live cells were observed at 2-min intervals by fluorescence microscopy. (Scale bar, 1 μm.) (D) The time after FBXO27-Halo or FBXO27 G2M-Halo colocalization to GFP-Gal3 puncta was measured for at least 30 distinct instances. Statistical analysis was performed by Student’s unpaired test. ***P < 0.001.
Fig. S2.
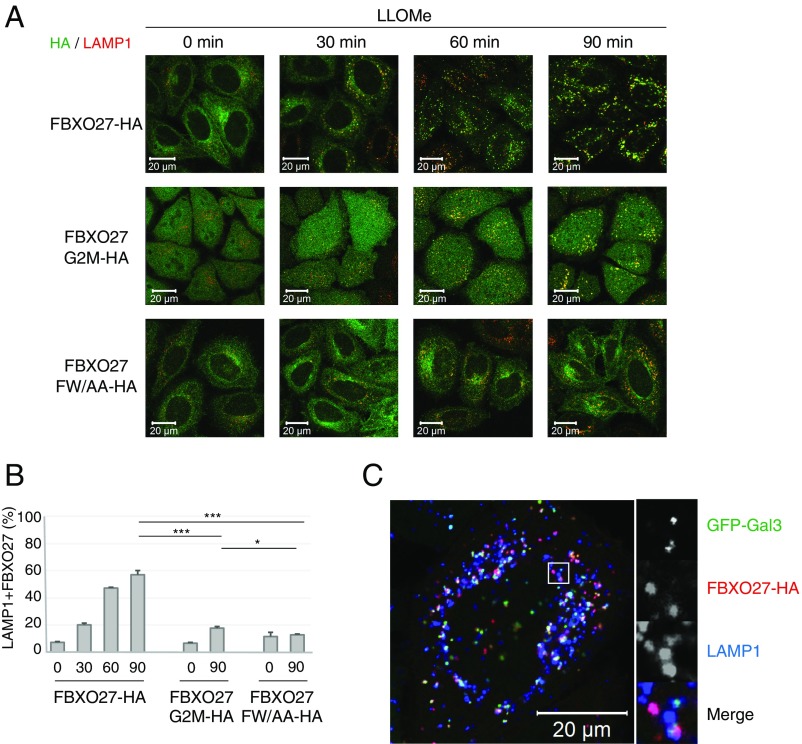
FBXO27 is recruited to damaged lysosomes. (A) HeLa cells stably expressing either FBXO27-HA or its mutants were treated with 1 mM LLOMe for the indicated time, and were subjected to immunostaining with anti-HA (green) and anti-LAMP1 (red) antibodies. (Scale bars, 20 μm.) (B) Quantification of FBXO27 translocation to lysosomes following LLOMe treatment. The error bars represent +SD from three independent replicates. Over 30 cells were counted in each replicate. Statistical significance was evaluated using one-way ANOVA with posttesting according to Tukey’s test. ***P < 0.001, *P < 0.05. (C) HeLa cells stably expressing GFP-Gal3 and FBXO27-HA were treated with 1 mM LLOMe for 60 min before being subjected to immunostaining with anti-HA (red) and anti-LAMP1 (blue) antibodies. (Scale bar, 20 mm.) The square region on the Left is highlighted on the Right. (Magnification, 3x.)
Wild-type FBXO27 started to move to damaged lysosomes soon after LLOMe addition (Fig. 2 and Movie S1). Although the newly formed Gal3 puncta immediately colocalized with FBXO27 (Fig. 2 C and D), some FBXO27+ lysosomes without Gal3 were also observed (Fig. S2C). This observation suggests that the recruitment of FBXO27 to damaged lysosomes is as effective as for Gal3. In contrast, the FW/AA mutant did not move to damaged lysosomes even 90 min after LLOMe treatment. The G2M mutant, which can bind to glycoproteins, also migrated to lysosomes following LLOMe treatment but the efficiency of its recruitment from the cytosol to lysosomes was noticeably lower compared with wild-type FBXO27 (Fig. 2 B and D). These results indicate that both membrane targeting by N-myristoylation and glycoprotein-binding activity in FBXO27 are strategies for the effective trapping of glycoproteins that appear in the cytosol following membrane rupture.
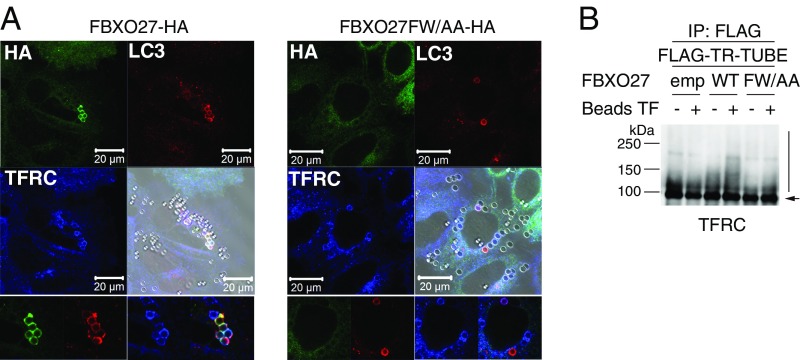
Latex beads coated with transfection reagents induce endosome damage and are targeted by LC3+ autophagosomes after being endocytosed into the cell (15, 20). We further determined whether FBXO27 was recruited to damaged endosomes in bead-transfected cells (Fig. S3A). In cells that express wild-type FBXO27, some prominent FBXO27-rings were observed surrounding endocytosed beads, and these rings colocalized with LC3. Further, ubiquitination of TFRC, a glycoprotein found in endosomes, was accelerated by bead-transfection in cells expressing FBXO27 (Fig. S3B). These results suggest that FBXO27 is recruited to damaged organelles in which glycoproteins become accessible to FBXO27 and ubiquitinates glycoproteins.
Fig. S3.
FBXO27 is recruited to the damaged endosomes in bead-transfected cells. (A) FBXO27 is recruited to ruptured endosomes. HeLa cells stably expressing FBXO27-HA or its glycoprotein-binding defective mutant (FBXO27FW/AA-HA) were transfected with Effectene-coated latex beads. After 3 h, cells were subjected to immunostaining with anti-HA (green), anti-LC3 (red), and anti-TFRC (blue) antibodies. Bright-field images are merged with immunfluoresence. (Scale bars, 20 μm.) (Bottom) LC3+ endosomes are shown. (Magnification, 2x.) (B) SCFFBXO27 ubiquitinates TFRC in ruptured endosomes. 293T cells (−) and cells stably expressing either FBXO27-HA (WT) or its mutant (FW/AA) were transfected with a plasmid encoding FLAG–TR-TUBE. Forty-five hours after transfection, cells were transfected with Effectene-coated latex beads (Beads TF). Cell lysates generated 48 h posttransfection were immunoprecipitated with anti-FLAG antibody, and the immunoprecipitates were analyzed by immunoblotting with anti-TFRC antibody. Vertical bar and arrow denote the positions of ubiquitinated and unmodified TFRC, respectively. Although wild-type FBXO27 was recruited to damaged LC3+ endosomes and enhanced TFRC ubiquitination, some LC3+ endosomes and TFRC ubiquitination were observed in FW/AA-expressing cells. These results suggest that other ubiquitin ligases are also involved in autophagy of damaged endosomes.
SCFFBXO27 Ubiquitinates Glycoproteins in Damaged Lysosomes.
Ubiquitination plays important roles in selective autophagy, including mitophagy, xenophagy, and lysophagy (12). However, it remains unclear which proteins are ubiquitinated during lysophagy. Therefore, we attempted to identify proteins ubiquitinated in response to LLOMe treatment using a dual-enrichment strategy. This strategy uses the TR-TUBE and diGly antibody that recognizes the ubiquitin remnant motif exposed following tryptic digestion of ubiquitinated proteins, in combination with mass spectrometry (18). TR-TUBE effectively interacts with ubiquitinated substrates in cells, allowing for detection and isolation of ubiquitinated proteins. To this end, 293T cells that express FLAG–TR-TUBE transiently were either treated with LLOMe or left untreated. The dual-enriched ubiquitinated peptides generated from both treated and untreated cells were subjected to MS analysis. In addition, to identify proteins whose ubiquitination was triggered by SCFFBXO27, we expressed FLAG–TR-TUBE together with FBXO27 in the presence of LLOMe.
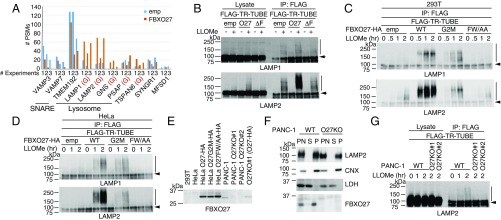
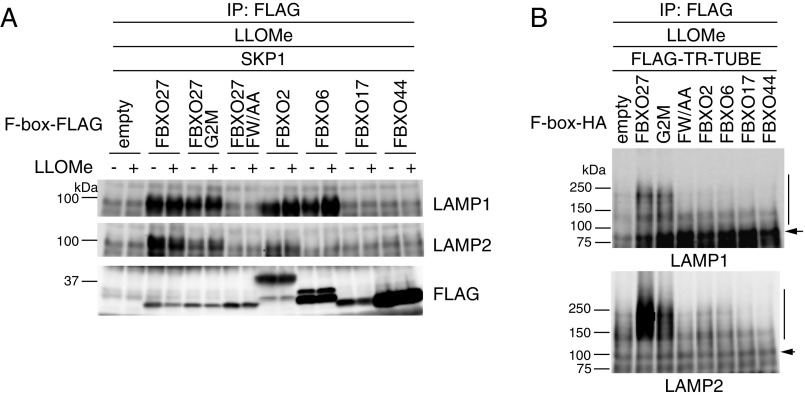
From three independent MS analyses, we identified proteins whose ubiquitination was observed only in the presence of LLOMe (Fig. 3A). By using the number of peptide spectrum matches (PSMs) as a semiquantitative index, TMEM192, a lysosomal transmembrane protein, and two v/R-SNARE proteins, VAMP3 and VAMP7, were identified as prominent ubiquitinated proteins. However, ubiquitination of these proteins was not stimulated by FBXO27 expression because they are not glycoproteins. TSPAN6, SYNGR1, and MFSD1 are reported to be cell-surface proteins, but they were also ubiquitinated in a reproducible manner upon LLOMe treatment. For lysosomal glycoproteins that were ubiquitinated in response to LLOMe treatment (LAMP1, LAMP2, GNS, TSPAN6, and PSAP), all ubiquitination sites were found on their luminal sides (Table S1), and the number of PSMs was increased by FBXO27 expression (Fig. 3A). In particular, the number of PSMs we identified on LAMP1 and LAMP2 were notably increased in FBXO27-expressing cells in the presence of LLOMe, and some ubiquitination sites were observed specifically in the FBXO27-expressing cells. Although FBXO27 expression increased the number of PSMs identified on both LAMP1 and LAMP2, immunoblot analyses using TR-TUBE showed that SCFFBXO27 seemed to ubiquitinate LAMP2 to a greater extent (Fig. 3B). Although both FBXO2 and FBXO6, as well as FBXO27, could bind to LAMP1 effectively, expressing FBXO2 or FBXO6 did not promote the ubiquitination of LAMP1 in the presence of LLOMe (Fig. S4).
Fig. 3.
SCFFBXO27 ubiquitinates glycoproteins in damaged lysosomes. (A) Identification of proteins ubiquitinated following LLOMe treatment. 293T cells were transfected with plasmids encoding FLAG–TR-TUBE with or without FBXO27, and cells were treated with or without LLOMe for 3 h before harvesting. The ubiquitinated proteins were identified by the dual-enrichment method using anti-FLAG and anti-diGly antibodies, followed by MS analysis. Total PSM numbers for identified proteins in LLOMe-treated cells expressing either FLAG tag alone (blue bars) or FBXO27 (red bars) with FLAG–TR-TUBE are shown. Glycoproteins are shown in red (G). All of the ubiquitination sites in the indicated proteins are shown in Table S1. (B) Ubiquitination assay for LAMP1 and LAMP2. Cells were transfected with plasmids encoding FLAG–TR-TUBE and either empty vector control, FBXO27 (O27), or its dominant-negative mutant (ΔF). Following transfection, cells were either treated with LLOMe or left untreated for 3 h before harvesting. Cell lysates were immunoprecipitated with anti-FLAG antibody, and analyzed by immunoblotting. Vertical bars and arrows denote the positions of ubiquitinated and unmodified LAMPs, respectively. (C and D) Time course of ubiquitination of LAMP1 and LAMP2 by FBXO27 and its mutants following LLOMe treatment in 293T (C) and HeLa (D) cells. Cells expressing FBXO27-HA (WT), FBXO27G2M-HA (G2M), or FBXO27FW/AA-HA (FW/AA) were transfected with a plasmid that encodes FLAG–TR-TUBE, and subsequently treated with LLOMe for the indicated times before harvesting. Cell lysates were immunoprecipitated with anti-FLAG antibody and analyzed by immunoblotting. (E) FBXO27 expression in cells used in this study. Lysate containing 30 μg of protein was analyzed by immunoblotting. (F) Homogenates made from either PANC-1 cells or PANC-1 cells with FBXO27 knocked out were fractionated. An equal volume of each fraction was subjected to immunoblotting. P, pellet; PN, postnuclear supernatant; S, supernatant. (G) Ubiquitination assay for LAMP2. Either PANC-1 or FBXO27 KO PANC-1 cells were transfected with a plasmid that encodes FLAG–TR-TUBE. Following transfection, cells were either treated with LLOMe or left untreated for the indicated times before harvesting. Cell lysates were subject to immunoprecipitation with anti-FLAG antibody, and analyzed by immunoblotting. Vertical bars and arrows denote the positions of ubiquitinated and unmodified LAMP2, respectively.
Table S1.
Ubiquitination sites in proteins ubiquitinated by LLOMe treatment
| Accession | Annotation | Identified peptide sequence | Ubiquitination site | N-glycosylation site | |
| Present study | Reported by MS | ||||
| P11279 | LAMP1 | ALQATVGNSYKCNAEEHVR K11(GG) C12(MT) | K337 | K319, K337 | N37, N45, N62, N76, N84, |
| EIKTVESITDIR K3(GG) | K137 (O27) | N103, N107, N121, N130, | |||
| VTKAFSVNIFK K3(GG) | K348 | N165, N181, N223, N228, | |||
| ADIDKKYR K6(GG) | K152 (O27) | N241, N249, N293, N332 | |||
| P13473 | LAMP2 | YLDFVFAVKNENR K9(GG) | K289 | N32, N38, N49, N58, N75, N101, N123, | |
| YETTNKTYK K6(GG) | K59 (O27) | N179, N229, N242m N257, N275, N300, | |||
| N307, N317, N356 | |||||
| P15586 | GNS | SWQKIQEPNTFPAILR K4(GG) | K125 | K125 | N111, N117, N183, N198, N210, |
| NKNFNIHGTNK K2(GG) | K257 (O27) | N279, N317, N362, N387, | |||
| N405, N422, N449, N480 | |||||
| O43657 | TSPAN6 | DWTDTNYYSEKGFPK K11(GG) | K171 | K128, K131, K171, K179 | N134 |
| SCCKLEDCTPQR C2(MT), C3(MT) K4(GG) C8(MT) | K179 | ||||
| P07602 | PSAP | KLVGYLDR K1 (GG) | K415 | K152, K303 | |
| EICALVGFCDEVK C3(MT), C9(MT), K13(GG) | K276 | K323, K449 | N80, N101, N215, N332, N426 | ||
| QEILAALEKGCSFLPDPYQK K9(GG),C11(MT) | K439 | ||||
| O43759 | SYNGR1 | MEGGAYGAGKAGGAFDPYTLVR M1(Ac, Ox), K10(GG) | K10 | K10 | |
| Q8IY95 | TMEM192 | TISSLEEIVEKQGDTIEYLK K11(GG), K20(GG) | K237, K246 | K201, K211, K237, K246, K254 | |
| AKPEPDILEEEKIYAYPSNITSETGFR K2(GG), K12(GG) | K201, K211 | (cytoplasmic) | |||
| HNALLSKR K7(GG) | K254 | ||||
| Q9H3U5 | MFSD1 | EQGKTGEVIK K4(GG) | K249 | K249, K255, K460 | |
| QREEIKFSHTE K6(GG) | K460 | (cytoplasmic) | |||
| Q15836 | VAMP3 | ADALQAGASQFETSAAKLK K17(GG) | K66 | K35, K42, K66, K68, K77 | |
| VNVDKVLER K5 (GG) | K35 | (cytoplasmic) | |||
| DQKLSELDDR K3(GG) | K42 | ||||
| ADALQAGASQFETSAAKLK K19(GG) | K68 | ||||
| P51809 | VAMP7 | VMETQAQVDELKGIMVR M2(Ox), K12(GG) | K137 | K125, K137, K160, K172 | |
| LELLIDKTENLVDSSVTFK K7(GG) | K160 | (cytoplasmic) | |||
| TENLVDSSVTFKTTSR K12(GG) | K172 | ||||
| GLDKVMETQAQVDELK K4(GG) | K125 | ||||
| AQTALPYAMNSEFSSVLAAQLKHHSENK K22(GG) | K115 | ||||
Fig. S4.
Binding and ubiquitination activities of FBXO27-related F-box proteins to LAMP1 and LAMP2. (A) Glycoprotein-specific F-box proteins bind to LAMP1 and LAMP2. 293T cells were transfected with plasmids encoding either only FLAG-tag alone, or one of the following C-terminally FLAG-tagged proteins, FBXO27, FBXO27G2M, FBXO27FW/AA, FBXO2, FBXO6, FBXO17, or FBXO44, in combination with SKP1, and cells were treated with or without LLOMe for 2 h before harvesting. WCLs were subjected to immunoprecipitation with an anti-FLAG antibody, and analyzed by immunoblotting with the indicated antibodies. (B) Ubiquitination assay for LAMP1 and LAMP2. 293T cells were transfected with plasmids encoding FLAG–TR-TUBE and either empty vector control, FBXO27, FBXO27G2M, FBXO27FW/AA, FBXO2, FBXO6, FBXO17, or FBXO44, and subsequently treated with LLOMe for 2 h before harvesting. Cell lysates were immunoprecipitated with anti-FLAG antibody, and analyzed by immunoblotting. Vertical bars and arrows denote the positions of ubiquitinated and unmodified LAMPs, respectively.
Next, we compared the time-course of LAMP1 and LAMP2 ubiquitination among 293T cells that express wild-type FBXO27 and its mutants following LLOMe treatment (Fig. 3C). In cells expressing wild-type FBXO27, ubiquitination started immediately after LLOMe addition and plateaued 1 h later. Moreover, LAMP2 was more effectively ubiquitinated than LAMP1. Compared with cells expressing wild-type FBXO27, the rate of ubiquitination, especially of LAMP2, was low in cells expressing G2M, a result consistent with the different rates of recruitment of wild-type FBXO27 and G2M to damaged lysosomes (Fig. 2). Furthermore, wild-type FBXO27 binds LAMP2 more effectively than the G2M mutant (Fig. S4A). To determine whether the difference in LAMP1 and LAMP2 ubiquitination efficiency is a result of differences in their cellular abundance, we analyzed ubiquitination rates in HeLa cells, which contain three times more LAMP1 than LAMP2 (21). Similar to results from 293T cells, LAMP2 was more effectively ubiquitinated compared LAMP1 in HeLa cells expressing wild-type FBXO27, whereas the rate of LAMP1 ubiquitination was almost the same in cells expressing either FBXO27 or the G2M mutant (Fig. 3D). These results suggest that SCFFBXO27 ubiquitinates LAMP2 more effectively than LAMP1, regardless of its relative abundance in cells.
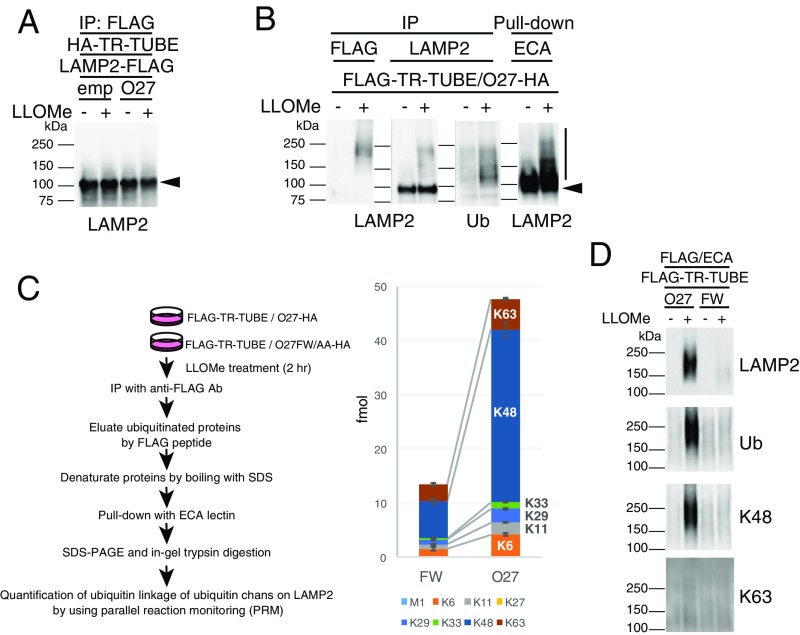
We next examined the ubiquitin chain type found on LAMP2 modified by SCFFBXO27. Because exogenously expressed LAMP2 was not ubiquitinated by SCFFBXO27 (Fig. S5A), we used Erythrina Cristagalli (ECA) lectin to enrich for endogenous LAMP2 (Fig. S5B). By using TR-TUBE and ECA lectin, we enriched for LAMP2 ubiquitinated by SCFFBXO27 to quantify its ubiquitin linkage based on the parallel reaction monitoring (PRM) method using standard absolute quantification (AQUA) peptide as internal spikes (22) (Fig. S5C). Compared with LAMP2 from cells expressing the FW/AA mutant, K48 chains on LAMP2 were increased in cells expressing FBXO27. We further assessed the ubiquitination pattern on LAMP2 by using linkage-specific antiubiquitin antibodies (Fig. S5D). These results indicated that SCFFBXO27 generates K48-linked ubiquitin chains on LAMP2 in damaged lysosomes.
Fig. S5.
SCFFBXO27 forms K48-linked ubiquitin chains on LAMP2. (A) Exogenously expressed LAMP2 fails to be detected ubiquitination by SCFFBXO27 in damaged lysosomes. 293T cells were transfected with plasmids encoding HA–TR-TUBE, LAMP2-FLAG, and either empty vector control (emp) or FBXO27 (O27). After transfection, cells were either treated with LLOMe or untreated for 2 h before harvesting. Cell lysates were immunoprecipitated with anti-FLAG antibody, and analyzed by immunoblotting. Arrowhead denotes the position of LAMP2-FLAG. (B) Confirmation of ubiquitination of isolated endogenous LAMP2 in damaged lysosomes. 293T cells were transfected with plasmids encoding FLAG–TR-TUBE and FBXO27-HA. Cells were either treated with LLOMe or left untreated for 2 h before harvesting. Cell lysates were immunoprecipitated with anti-FLAG or anti-LAMP2 antibodies, or pull-down with ECA lectin-agarose. Vertical bar and arrowhead denote the position of ubiquitinated and unmodified LAMP2, respectively. (C) Ubiquitination levels on LAMP2 by ubiquitin-PRM. 293T cells were transfected with plasmids encoding FLAG–TR-TUBE and either FBXO27-HA (O27) or FBXO27FW/AA-HA mutant (FW). After transfection, cells were treated with LLOMe for 2 h before harvesting. Cell lysates were immunoprecipitated with anti-FLAG antibody and the eluted proteins from immunoprecipitates were denatured by boiling in 2% SDS. After adding Nonidet P-40 to counteract to SDS, LAMP2 protein was enriched by using ECA lectin. Resultant ubiquitinated LAMP2 fractions were subjected to SDS/PAGE followed by in-gel trypsinization. Ubiquitin AQUA peptides were spiked in the extracted peptides and treated with H2O2 for oxidation of Met-containing pepetides. The samples were analyzed by LC-MS/MS in targeted MS/MS mode. Summary of LAMP2 ubiquitination levels determined by ubiquitin-RRM are shown (Right); mean ± SD, n = 3 biological replicates. (D) 293T cells were transfected with plasmids encoding FLAG–TR-TUBE and either FBXO27 (O27) or FBXO27FW/AA(FW). After transfection, cells were treated with or without LLOMe for 2 h before harvesting. Ubiquitinated LAMP2 was enriched as described in C, and analyzed by immunoblots with anti-LAMP2, anti-Ubiquitin (Ub: P4D1), anti-K48 chain specific (Apu2), and anti-K63 chain-specific (Apu3) antibodies.
FBXO27 Expression Enhances Recruitment of Autophagic Machinery to Damaged Lysosomes.
FBXO27 expression is restricted to several tissues, including the brain. Expression was not detected in 293T cells and HeLa cells by immunoblotting. We therefore searched for cell lines that express FBXO27 endogenously using the Body Atlas database in the NEXTBIO search engine. We identified the pancreatic adenocarcinoma cell line PANC-1 as a candidate, and verified FBXO27 expression in PANC-1 cells (Fig. 3E). Endogenous FBXO27 was also recovered in the membrane fraction, suggesting that it binds to endomembranes via N-myristoylation (Fig. 3F).
We next examined the ubiquitination of LAMP1 and LAMP2 during LLOMe treatment in PANC-1 cells. To assess ubiquitination by endogenous FBXO27, we first established FBXO27 knockout PANC-1 cells using the CRSPR/Cas9 method. Although we were unable to detect LAMP1 ubiquitination, LAMP2 was ubiquitinated after LLOMe treatment in a time-dependent manner in wild-type cells (Fig. 3G). In contrast, LLOMe treatment did not induce LAMP2 ubiquitination in FBXO27 KO cells, indicating that LAMP2 in damaged lysosomes is ubiquitinated by endogenous SCFFBXO27.
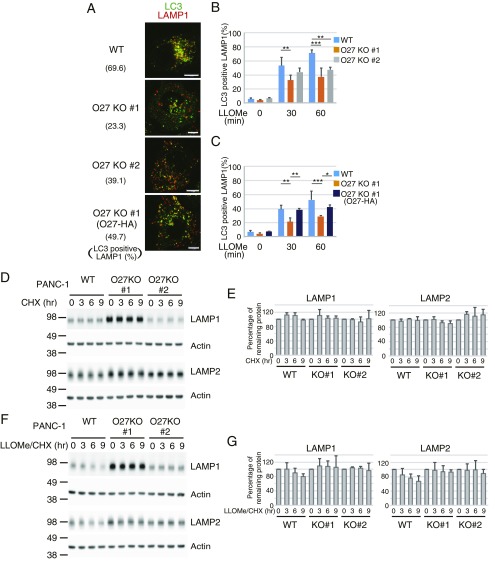
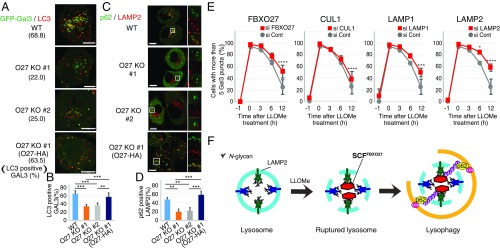
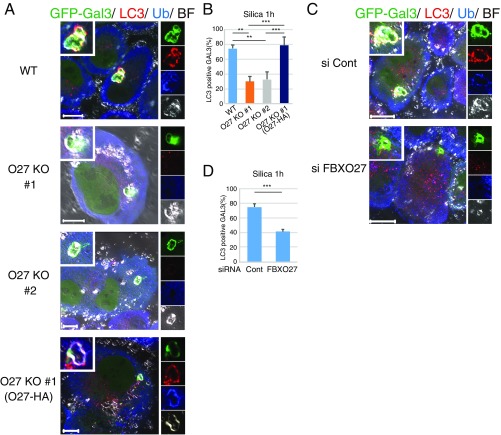
To determine whether FBXO27 expression affects the recruitment of autophagic machinery to damaged lysosomes, we measured the accumulation of LC3+ lysosomes at indicated time points following LLOMe treatment (Fig. S6 A–C). We used LAMP1 as a lysosomal marker, and observed that LC3 colocalized with ∼50% of LAMP1 puncta in wild-type PANC-1 cells 30 min after LLOMe addition. Colocalization was significantly reduced in LLOMe-treated FBXO27 KO PANC-1 cells (Fig. S6B), and this reduction was suppressed by FBXO27 expression (Fig. S6C). We also measured the colocalization of LC3 and GFP-Gal3, and found that LC3 colocalized with more than half the Gal3 puncta 30 min after adding LLOMe to transfected wild-type PANC-1 cells expressing GFP-Gal3 (Fig. 4 A and B). In contrast, the proportion of LC3+ puncta was significantly lower in FBXO27 KO cells, suggesting that SCFFBXO27 is required for proper targeting of LC3 to disrupted lysosomes. Supporting this idea, the reduced colocalization of LC3 and GFP-Gal3 in FBXO27 KO cells was rescued by transgenic FBXO27 expression. Endogenous p62, an autophagy receptor, also colocalized with LAMP2 in wild-type PANC-1 cells that had been treated with LLOMe for 30 min, and this colocalization was significantly reduced in FBXO27 KO cells (Fig. 4 C and D).
Fig. S6.
LAMP2 ubiquitination by SCFFBXO27 in damaged lysosomes recruits autophagic machinery and degrades lysosomal proteins. (A–C) Time-course of recruitment of LC3 to lysosomes following LLOMe treatment in PANC-1 cells. PANC-1, FBXO27 KO PANC-1 (O27 KO #1 and #2), or FBXO27-expressing FBXO27 KO PANC-1 [O27KO #1 (O27-HA)] cells were treated with LLOMe for either 60 min (A) or for the indicated times (B and C), and were subjected to immunostaining with anti-LC3 (green) and anti-LAMP1 (red) antibodies. (Scale bars, 10 μm.) The data represent means + SD. Over 30 cells were counted (n = 3). Statistical significance was evaluated using two-way ANOVA with posttesting according to Bonferroni’s test. ***P < 0.001, **P < 0.01, *P < 0.03. (D–G) Cycloheximide chase analyses of LAMP1 and LAMP2 in wild-type and FBXO27 KO PANC-1 cells. Cells were treated with 2 μg/mL cycloheximide (CHX) alone (D and E) or in combination with 1 mM LLOMe (F and G) for the indicated times. Cell lysates were analyzed by immunoblotting. The relative LAMP1, LAMP2, and actin (as a loading control) intensities were determined using ImageQuant TL. Data are presented as means + SD of three independent experiments.
Fig. 4.
LAMP2 ubiquitination by SCFFBXO27 in damaged lysosomes recruits autophagic machinery. (A and B) LC3 is recruited to damaged lysosomes in a FBXO27-dependent manner. Cells expressing GFP-Gal3 were treated with LLOMe for 30 min, and were subjected to immunostaining with anti-LC3 (red) antibody. (Scale bars, 10 μm.) (C and D) Colocalization of p62 with LAMP2 upon lysosomal damage requires FBXO27. Cells were treated with LLOMe for 30 min, and immunostained with anti-p62 (green) and anti-LAMP2 (red) antibodies. (Scale bars, 10 μm.) In B and D The data represent means + SD. Over 30 cells were counted (n = 3). Statistical significance was evaluated using one-way ANOVA with posttesting according to Tukey’s test. ***P < 0.001, **P < 0.01. (E) Quantification of the percentage of cells with more than five GFP-Gal3 puncta per cell for each condition. PANC-1 cells stably expressing GFP-Gal3 were treated with siRNA against luciferase (Cont), FBXO27, CUL1, LAMP1, or LAMP2. Cells were subsequently treated with LLOMe for 1 h. After LLOMe washout, cells were fixed at the indicated times and stained with DAPI. The number of Gal3 puncta per cell was counted in over 250 cells (n = 3). The data represent means ± SD. Statistical significance was evaluated using two-way ANOVA with posttesting according to Bonferroni’s test. ****P < 0.0001, ***P < 0.001, *P < 0.03. (F) Schematic diagram of LAMP2 ubiquitination by SCFFBXO27 in damaged lysosomes for recruitment of autophagic machinery.
To determine if FBXO27-dependent LC3 recruitment to lysosomes can be triggered by agents other than LLOMe, we treated cells with silica, another lysosome-damaging agent (16). In this case, most Gal3 puncta colocalized with LC3 and ubiquitin in PANC-1 cells within an hour of silica addition (Fig. S7). Similar to LLOMe results, the rate of LC3+ Gal3 puncta formation was significantly reduced in both FBXO27 KO cells and when FBXO27 was depleted by siRNA treatment. Thus, the recruitment of autophagic machinery to lysosomes by SCFFBXO27 is likely because of lysosomal damage, rather than an artifact of LLOMe treatment.
Fig. S7.
Recruitment of LC3 to GFP-Gal3+ damaged lysosomes treated with silica in a FBXO27-dependent manner. PANC-1 or PANC-1 mutant cells stably expressing GFP-Gal3 were treated with 250 μg/mL silica for 1 h (A and B). PANC-1 cells expressing GFP-Gal3 were treated with siRNA against either luciferase (Cont) or FBXO27. Forty-eight hours after siRNA transfection, cells were treated with 250 μg/mL silica for 1 h (C and D). Cells were subjected to immunocytochemistry using anti-LC3 and antiubiquitin (A and C). (Scale bars, 20 μm.) The data represent means + SD. All GFP-Gal3 puncta that appeared in silica-treated cells on each dish were counted (n = 3). Statistical significance was evaluated using one-way ANOVA with posttesting according to Tukey’s test (B), or unpaired t test (D). ***P < 0.001, **P < 0.01. (A and C) [Magnification: A, Inset and Right: WT, 1.3×; O27KO #1, 1.4×; O27KO #2, 1.3×; O27KO #1 (O27-HA), 2.4×. C, Inset and Right: si Cont, 1.4×; si FBXO27, 1.6×.]
In the absence of lysosomal damage, LAMP1 and LAMP2 proteins are relatively stable in both wild-type and FBXO27 KO PANC-1 cells (Fig. S6 D and E). However, after addition of LLOMe, we observe a gradual degradation of these proteins in wild-type, but not FBXO27 KO, PANC-1 cells (Fig. S6 F and G). These results are consistent with a model where exposed glycoproteins, including LAMP1 and LAMP2, recruit SCFFBXO27. SCFFBXO27 ubiquitinates these glycoproteins, causing subsequent recruitment of autophagic machinery, leading to degradation of these proteins during lysophagy.
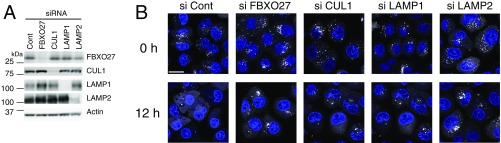
To examine whether FBXO27 regulates clearance of damaged lysosomes, we next monitored clearance of GFP-Gal3 puncta at various time points after LLOMe washout, in wild-type PANC-1 cells and in PANC-1 cells with FBXO27, CUL1, LAMP1, or LAMP2 knocked down. In control cells, the proportion of cells with more than five Gal3 puncta gradually dropped over time after LLOMe washout. Although 100% of cells had more than five Gal3 puncta immediately after LLOMe treatment, this number fell to 25% 12 h after washout. In contrast, in cells depleted of FBXO27, CUL1, LAMP1, or LAMP2 by siRNA, a significant delay in clearance of Gal3 puncta was observed (Fig. 4E and Fig. S8). These results suggest that ubiquitination of lysosomal glycoproteins by SCFFBXO27 accelerates recruitment of autophagic machinery to damaged lysosomes, leading to accelerated clearance.
Fig. S8.
Delay in the recovery of damaged lysosomes by knocking down the E3 ligase or possible ubiquitination targets. (A) Depletion efficiency of siRNA used in B was confirmed in PANC-1 cells stably expressing GFP-Gal3 by immunoblots of total lysates using the indicated antibodies. (B) PANC-1 cells stably expressing GFP-Gal3 were treated with siRNA against either luciferase (Cont), FBXO27, CUL1, LAMP1, or LAMP2. Forty-eight hours after siRNA transfection, cells were subsequently treated with LLOMe for 1 h. After LLOMe washout, cells were fixed at the indicated times and stained with DAPI. (Scale bar, 20 μm.)
Discussion
A diverse range of substances can rupture endosomal and lysosomal membranes in vivo, and the resulting damaged organelles are eliminated by a type of autophagy called organellophagy. Similar to PINK1/Parkin-mediated mitophagy, ubiquitination of damaged endosomes or lysosomes is involved in damage recognition and in recruiting autophagic machinery. However, the mechanisms underlying such ubiquitination are unclear. In this study, we showed that membrane-bound FBXO27 is rapidly recruited to ruptured lysosomes following LLOMe treatment. Lysosomal membrane rupture renders LAMP2 accessible to recognition by FBXO27 and ubiquitination by SCFFBXO27. Furthermore, SCFFBXO27-mediated ubiquitination of LAMP2 in damaged lysosomes enhances the recruitment of autophagic machinery, such as p62 and LC3 (Fig. 4F).
Although no ubiquitination sites on LAMP2 have been referenced in the proteomic database, SCFFBXO27 clearly ubiquitinated LAMP2 upon LLOMe treatment (Fig. 3). LAMP1 and LAMP2, major lysosomal membrane proteins that show 37% identity in humans, are highly glycosylated. It is unclear why LAMP2 is more effectively ubiquitinated by SCFFBXO27; however, because the efficiency of LAMP1 and LAMP2 ubiquitination is similar in cells that express the G2M mutant, the membrane targeting of FBXO27 appears to be important for LAMP2 targeting. All three glycoprotein-specific F-box proteins—FBXO2, FBXO6, and FBXO27—recognize the innermost chitobiose structure found in all N-glycans and can bind to LAMP1, but only FBXO27 is N-myristoylated and ubiquitinates LAMP1 and LAMP2 (Fig. S4). This modification may affect substrate-recognition, as well as substrate recruitment efficiency.
Knockdown of FBXO27, CUL1, LAMP1, or LAMP2 slows down, but does not abolish lysophagy (Fig. 4E), indicating that other mechanisms inducing lysophagy exist in PANC-1 cells. Consistent with this finding, Chauhan et al. reported that TRIM16, a RING-type ubiquitin ligase, recognizes endomembrane damage in cooperation with Gal-3 and ULK1, and adds K63-linked ubiquitin to the lysosomal membrane (23). Because TRIM16 is expressed globally in cells, it is likely that both TRIM16-dependent and FBXO27-dependent lysophagy pathways are operating in PANC-1 cells.
Although previous reports, including Chauhan et al., demonstrate that K63-linked ubiquitin chains are important for selective autophagy including lysophagy (23, 24), our data indicate that K48-linked ubiquitination is also important. Although the role in lysophagy at first seems inconsistent with reports implicating K48-linkages in proteasomal degradation, a recent study suggests that pathway choice between proteasomal degradation and selective autophagy is largely regulated by the oligomerization state of ubiquitin receptors, rather than specific linkage type (25).
FBXO27 is expressed at high levels in specific tissues, including the nervous system, suggesting that tissues with high energy requirements may require the FBXO27-dependent pathway in addition to K63 pathways to accelerate clearance upon lysosomal damage. Many lines of evidence indicate that abnormal protein accumulation causes neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis. Under these pathological situations, several factors can disrupt lysosomal membrane integrity directly and induce rapid lysosome-dependent cell death (26, 27). In addition to lysosomal rupture, these pathologies may induce yet unknown damage to other organelles, and it is possible that FBXO27 migrates to the damaged organelles and causes the subsequent ubiquitination of glycoproteins. Because the appearance of glycoproteins in the cytosol is aberrant, monitoring by cytosolic ubiquitin ligases is a likely cellular defense system against disease. Further studies to identify physiological substrates for FBXO27 in neuronal cells may help to advance our understanding of still unknown pathways leading to glycoprotein appearance in the cytosol.
Materials and Methods
Myristoylation Assay.
Cells were treated with 40 μM myristic acid azide (Thermo Fisher Scientific) for 12 h before lysis. Cell lysates were subjected to the Click-iT reaction for biotinylation by using the Click-iT protein reaction kit (Thermo Fisher Scientific).
Immunofluorescence and Microscopy.
Cells were cultured in glass-bottomed 35-mm dishes, fixed with 4% paraformaldehyde dissolved in PBS for 10 min, and permeabilized with 50 μg/mL digitonin in PBS for 10 min. Fixed cells were first stained with the primary antibodies described in SI Materials and Methods, and subsequently stained with the following secondary antibodies: mouse, rat, or rabbit conjugated to either Alexa Fluor-488, -568, or -633 (Life Technologies). Samples were mounted and observed with a laser-scanning confocal microscope (LSM710; Carl Zeiss) with a Plan-Apochromat 63× NA 1.4 oil differential interference contrast objective lens.
Additional information is provided in SI Materials and Methods.
SI Materials and Methods
Reagents and Antibodies.
LLOMe and silicon dioxide (S5631) were purchased from Sigma-Aldrich. ConA-HRP was from J-Oilmilles. Agarose-bound ECA lectin was purchased from Vector Laboratories. The following antibodies were used for immunoblotting: mouse anti-FLAG (DDDDK: 1:2,000, clone FLA2; MBL International), rabbit anti-TFRC (1:500; Sigma-Aldrich), mouse anti-HA (1:1,000, clone 16B12; Covance), mouse anti-LAMP1 (1:200, clone H4A3; Santa Cruz Biotechnology), mouse anti-LAMP2 (1:200, clone H4B4; Santa Cruz Biotechnology), rabbit polyclonal anti-CNX (Cell Signaling), goat polyclonal anti-LDH (1:500; Abcam), mouse antiubiquitin (1:500, P4D1; Santa Cruz Biotechnology), rabbit anti-FBXO27 (1:2,000; Proteintech), and mouse anti-actin (1:1,000, C4; Millipore). The following antibodies were used for immunostaining: mouse monoclonal anti-HA (clone 16B12; Covance); rat monoclonal anti-HA (clone 3F10; Roche), rabbit polyclonal anti-CNX (Cell Signaling), rabbit anti-GM130 (Abcam), rabbit anti-TOMM20 (Santa Cruz Biotechnology), rabbit anti-EEA1 (Cell Signaling), mouse anti-LAMP1 (clone H4A3; Santa Cruz Biotechnology), mouse anti-TFRC (TfR, clone H68.4; Thermo Fisher Scientific), rabbit anti-LC3 (MBL International), mouse antimultiubiquitin (FK2; MBL International), and rabbit anti-p62 (MBL International).
Cell Culture, Plasmid Construction, Transfections, and Generation of Stable Cell Lines.
The 293T, HeLa, and PANC-1 cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 5 U/mL penicillin, and 50 U/mL streptomycin in a 5% CO2 incubator at 37 °C. Plasmids expressing FLAG-tagged FBXO2, FBXO6, and FBXO27, untagged SKP1, FLAG–TR-TUBE, and GFP-Gal3 have been described previously (8, 16, 18). Mutations, including FBXO6 Y241A/W242A, FBXO27 F262A/W263A, and FBXO27G2M were generated by site-directed mutagenesis using the QuikChange Lightning Kit (Agilent Technologies). Plasmids for expressing C-terminally Halo-tagged FBXO27 and its mutants were generated by cDNA fragment of FBXO27 fused to XhoI/NotI fragment derived from pFC14K HaloTag CMV Flexi Vector (Promega) into pcDNA3. For transient transfection of plasmids, polyethylenimine (Polysciences) was used with 293T cells, Lipofectamine with Plus reagent (Thermo Fisher Scientific) was used with HeLa cells, and Lipofectamine LTX with Plus reagent (Thermo Fisher Scientific) was used with PANC-1 cells. Stable cell lines were established by recombinant retrovirus infection as follows. C-terminally HA-tagged F-box protein or its mutants was subcloned into the EcoRI site of the pBABE-puro vector (Addgene plasmid 1764) by using the In-fusion HD cloning kit (Takara Bio). Vector particles were produced in 293T cells by cotransfection with Gag-Pol (Addgene plasmid 14887), VSV-G (Addgene plasmid 8454), and a retrovirus plasmid (pBABE-puro/FBXO2-HA, pBABE-puro/FBXO6-HA, pBABE-puro/FBXO27-HA, pBABE-puro/FBXO27G2M-HA, pBABE-puro/FBXO27F262A/W263A-HA, or pMRX-GFP-Gal3) by using Lipofectamine LTX. Twelve hours after transfection, the medium was changed to a fresh medium and cells were further incubated for 24 h. The viral supernatants were used to infect either HeLa, 293T, or PANC-1 cells in the presence of 8 μg/mL polybrene (Sigma-Aldrich). Stable transfectants were selected for in medium containing 1 μg/mL puromycin.
Immunoprecipitation and Immunoblotting.
The 293T cells were harvested for immunoprecipitation experiments 48 h after transfection, whereas HeLa and PANC-1 cells were harvested 24 h after transfection. Harvested cells were lysed in TBS-N [10 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40] containing a protease inhibitor mixture (Roche) on ice, and lysates were cleared by centrifugation at 20,000 × g for 20 min at 4 °C. The supernatants (WCL) were immunoprecipitated with anti-FLAG antibody-conjugated agarose (anti-DDDDK, FLA-2 clone; MBL International) or anti-HA antibody-conjugated agarose (HA-7; Sigma-Aldrich) for 1 h at 4 °C. The beads were subsequently washed five times with 1 mL TBS-N and immune complexes were eluted by heating in SDS/PAGE sample buffer. In the experiment, the results of which are shown in Fig. 1, immune complexes were eluted using the FLAG peptide (Sigma-Aldrich). Immunoprecipitates and cell lysates were subjected to immunoblot analyses as previously described (3).
Cell Fractionation.
HeLa cells stably expressing F-box protein or PANC-1 cells were washed with PBS, collected by centrifugation, and homogenized in homogenization buffer (20 mM Hepes pH 7.4, 250 mM sucrose, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2) containing a protease inhibitor mixture by repeated shearing 25 times through a 25-gauge needle mounted on a 1-mL syringe on ice. After centrifugation at 1,000 × g for 10 min, the postnuclear supernatants were further centrifuged at 100,000 × g for 60 min to generate supernatant and pellet fractions. An equal volume of each fraction was subjected to immunoblot analysis.
Myristoylation Assay.
The 293T cells were transfected with a plasmid encoding FBXO27-FLAG or FBXO27 G2M-FLAG. Thirty-six hours after transfection, cells were treated with 40 μM myristic acid azide (Thermo Fisher Scientific) for 12 h. Cells were washed twice thereafter with PBS and were lysed with TBS-T [10 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1% Triton-X] containing protease inhibitor mixture on ice, and lysates were centrifuged at 20,000 × g for 20 min at 4 °C. Lysate concentrations were adjusted to 4 mg/mL with TBS-T, and 150 μL of WCL were used for the Click-iT reaction using the Click-iT protein reaction kit (Thermo Fisher Scientific) using 40 μM biotin alkyne. After the free biotin alkyne was removed using Amicon Ultra 0.5-mL centrifugal filters 3K (Millipore), the resultant supernatants were pulled down with streptavidin (SA)-agarose (Sigma-Aldrich) for 1 h at 4 °C. Immunoprecipitation with anti-FLAG was performed on the reacted lysates. The pull-down products and immune complexes were eluted by heating in SDS/PAGE sample buffer, and were subjected to immunoblot analyses by anti-FLAG antibody and HRP-conjugated SA (Pierce).
Generation of CRISPR/Cas9-Mediated Gene KO Cell Lines.
To generate FBXO27-KO Panc1 cells, CRISPR target sites were selected from the exon including the F-box domain using the CRISPR design tool made by the Zhang laboratory (crispr.mit.edu/). The selected 19 bp of target sequences (5′-CGAATAAGGTTGCGTCCGA-3′ and 5′-GCGCCCGAGCAGCGTGCGC-3′) adjacent to the protospacer adjacent motif site (NGG) was linked into the primers [forward primer: 5′-TTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG(N)19-3′ and reverse primer: 5′-GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC(N)19C-3′]. These primers were annealed and extended using Phusion polymerase (Thermo Fisher Scientific), and inserted into the AflII site of the gRNA cloning vector (Addgene plasmid 41824) by the In-fusion HD cloning kit (Takara Bio). PANC-1 cells were cotransfected with a gRNA, CRISPR nuclease (hCas9, Addgene plasmid 41815), and pMXs-puro (Cell Biolabs) plasmids by Lipofectamine LTX (Thermo Fisher Scientific). Twenty-four hours after transfection, the medium was changed to a fresh medium containing 1 μg/mL puromycin and cells were further incubated for 48 h. Each puromycin-resistant cell was plated into 96-well plates for single colony isolation. Genomic DNA was isolated from single colonies. PCR amplification of the target site was carried out, and the PCR products were analyzed by sequencing. The gene knockout clones were confirmed by immunoblotting.
Ubiquitinated Protein Identification by MS.
For ubiquitinated protein identification by MS, 1.3 × 106 293T cells were cultured in a 10-cm culture dish for 1 d, and were transfected with plasmids encoding FLAG–TR-TUBE in combination with empty, FBXO27 FBXO27ΔF. The cells were harvested 48 h after transfection, and 2 mL of WCL (∼1.5 mg/mL) was incubated with 25 mL of anti-FLAG antibody conjugated agarose beads (anti-DDDDK, FLA-2 clone; MBL International) for 1 h at 4 °C. After the agarose beads were washed five times with 1.5 mL TBS-N and twice with 1.5 mL 50 mM ammonium bicarbonate (AMBIC), bead-bound proteins were eluted three times with 25 μL of 200 µg/mL FLAG peptide (Sigma-Aldrich) in 50 mM AMBIC. Proteins were reduced in 5 mM TCEP for 30 min at 50 °C before being alkylated with 10 mM methyl methanethiosulfonate (MMTS) for 10 min at room temperature. Next, alkylated proteins were digested overnight at 37 °C with 1 μg of trypsin (Promega). Following tryptic digestion, ubiquitinated peptides were enriched using the PTMScan ubiquitin remnant motif (K-ε-GG) kit (Cell Signaling). Briefly, 20 μL of 10× IAP buffer (0.5 M Mops pH 7.2, 0.1 M Na2HPO4, 0.5 M NaCl) and 120 μL of 2× protease inhibitor mixture (Roche) were added to the tryptic digested product (60 μL), and incubated for 2 h at 4 °C with 10 μL of ubiquitin branch motif immunoaffinity beads. Next, the beads were washed twice with 250 μL 1× IAP buffer, and three times with 250 μL distilled water, before eluting bound peptides with 3 × 20 μL 0.15% TFA. The eluted peptides were desalted using GL-Tip SDB and GL-Tip GC (GL Science) before LC-MS analysis. Enriched ubiquitinated peptides were analyzed by nanoflow ultra-HPLC (Easy nLC; Thermo Fisher Scientific) coupled to a Q Exactive mass spectrometer and then searched with Proteome Discoverer software (v1.3; Thermo Fisher Scientific), as described previously (18).
Quantification of Ubiquitin Linkage of Ubiquitin Chains Using MS.
LC-MS/MS analyses and quantification of peptides by PRM were performed essentially as previously described (22). Enriched ubiquitinated LAMP2 fraction were separated by SDS/PAGE on 4–12% NuPAGE Bis⋅Tris gels with Mes buffer (Life Technologies), and the gels were stained with Bio-Safe Coomassie (Bio-Rad). Gels were washed in Milli-Q water, and the portions containing proteins >100 kDa were excised. The gel pieces were washed in 50 mM AMBIC/30% acetonitrile (ACN) for 1 h, with 50 mM AMBIC/50% acetonitrile (ACN) for 1 h, and subsequently with 100% ACN for 5 min. The dehydrated gels were incubated with 20 ng/μL trypsin in 50 mM AMBIC, 5% ACN overnight at 37 °C. Following tryptic digestion, the peptides were extracted with 0.1% TFA, 50% ACN, and subsequently with 0.1% TFA, 70% ACN. Extracted peptides were combined and concentrated to 15.5 μL by SpeedVac. The concentrated peptides were mixed with 50 fmol ubiquitin AQUA peptide mix [Custom AQUA peptide (Sigma-Aldrich), or AQUA Utimate grade (Thermo Fisher Scientific)], 0.1% TFA, 0.005% H2O2 and incubated overnight at 4 °C to oxidize methionine. For PRM analysis, the Q Exactive mass spectrometer was operated in targeted MS2 mode, and LC was conducted with a 90-min gradient. Data were processed using the PinPoint software (v1.3; Thermo Fisher Scientific). Fragment ions used for quantification were previously reported (28).
Bead Transfection.
Bead transfections were performed as previously reported (20). Transfection reagent-coated beads were prepared by mixing latex beads (NH2-17145-5; PolySciences) with Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions, except that bead suspension was used in place of DNA solution. The bead mixture was further mixed with growth medium, and added to cells by replacing the medium. One hour after bead transfection, the cells were washed twice with fresh medium, and incubated for an additional 3 h.
Immunofluorescence and Microscopy.
Cells were cultured onto glass-bottomed 35-mm dishes, fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 50 μg/mL digitonin in PBS for 10 min, and stained with the primary antibodies described above and the following secondary antibodies: mouse, rat, and/or rabbit Alexa Fluor-488, -568, and -633 (Life Technologies; 1:2,000 dilution). Samples were mounted using Slow Fade Gold and observed with a laser-scanning confocal microscope (LSM710; Carl Zeiss) with a Plan-Apochromat 63× NA 1.4 oil differential interference contrast objective lens. The images were adjusted using Photoshop CC (Adobe). Colocalization of FBXO27-HA/GFP-Gal3, FBXO27-HA/LAMP1, LC3/GFP-Gal3, or LC3/LAMP1 was analyzed by ZEN imaging software (Carl Zeiss). For time-lapse imaging, HeLa cells expressing FBXO27-Halo and GFP-Gal3 were labeled with HaloTag TMR (Promega) for 15 min. Images were captured with a laser-scanning confocal microscope system.
For counting GFP-Gal3 puncta, PANC-1 cells stably expressing GFP-Gal3 were transfected with each siRNA by Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific). The following siRNAs were used for knockdown experiments; Luciferase (Thermo Fisher Scientific 12935146), FBXO27 (Dharmacon M-018558), CUL1 (Santa Cruz Biotechnology; sc-35126), LAMP1 (Santa Cruz Biotechnology; sc-29389), and LAMP2 (Santa Cruz Biotechnology; sc-29390). Forty-eight hours after transfection, cells were treated with 1 mM LLOMe for 1 h, washed twice, and cultured in fresh growth medium for 3, 6, or 12 h. Cells were subsequently fixed and stained with DAPI. The number of Gal3 puncta was determined with Meta Imaging Series software v7.8 (Molecular Devices).
Supplementary Material
Acknowledgments
We thank Dr. S. Yamaoka (Tokyo Medical and Dental University) for providing pMRX-IRES-puro, Dr. K. Yamano for helpful discussions, and Dr. J. Horiuch for critical reading of the manuscript. This work was supported by the Japan Ministry of Education, Culture, Sports, Science, and Technology/Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP16K07705 (to Y.Y.), JP24112008 (to Y.S.), JP15H01196 (to N.M.), and JP2600014 (to K.T.); a JSPS PRESTO grant (to N.M.); and the Takeda Science Foundation (Y.S., N.M., and K.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702615114/-/DCSupplemental.
References
- 1.Hirsch C, Blom D, Ploegh HL. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 2003;22:1036–1046. doi: 10.1093/emboj/cdg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim I, et al. The Png1-Rad23 complex regulates glycoprotein turnover. J Cell Biol. 2006;172:211–219. doi: 10.1083/jcb.200507149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida Y, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida Y, et al. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J Biol Chem. 2003;278:43877–43884. doi: 10.1074/jbc.M304157200. [DOI] [PubMed] [Google Scholar]
- 5.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima T, et al. Structural basis of sugar-recognizing ubiquitin ligase. Nat Struct Mol Biol. 2004;11:365–370. doi: 10.1038/nsmb732. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida Y, Murakami A, Iwai K, Tanaka K. A neural-specific F-box protein Fbs1 functions as a chaperone suppressing glycoprotein aggregation. J Biol Chem. 2007;282:7137–7144. doi: 10.1074/jbc.M611168200. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida Y, Murakami A, Tanaka K. Skp1 stabilizes the conformation of F-box proteins. Biochem Biophys Res Commun. 2011;410:24–28. doi: 10.1016/j.bbrc.2011.05.098. [DOI] [PubMed] [Google Scholar]
- 9.Glenn KA, Nelson RF, Wen HM, Mallinger AJ, Paulson HL. Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J Biol Chem. 2008;283:12717–12729. doi: 10.1074/jbc.M709508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto K. Organellophagy: Eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205:435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: An orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–316. doi: 10.15252/embr.201541486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maejima I, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y, et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci USA. 2015;112:4630–4635. doi: 10.1073/pnas.1422313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiele DL, Lipsky PE. Mechanism of L-leucyl-L-leucine methyl ester-mediated killing of cytotoxic lymphocytes: Dependence on a lysosomal thiol protease, dipeptidyl peptidase I, that is enriched in these cells. Proc Natl Acad Sci USA. 1990;87:83–87. doi: 10.1073/pnas.87.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi S, et al. Artificial induction of autophagy around polystyrene beads in nonphagocytic cells. Autophagy. 2010;6:36–45. doi: 10.4161/auto.6.1.10324. [DOI] [PubMed] [Google Scholar]
- 21.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya H, Tanaka K, Saeki Y. The parallel reaction monitoring method contributes to a highly sensitive polyubiquitin chain quantification. Biochem Biophys Res Commun. 2013;436:223–229. doi: 10.1016/j.bbrc.2013.05.080. [DOI] [PubMed] [Google Scholar]
- 23.Chauhan S, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papadopoulos C, et al. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu K, den Brave F, Jentsch S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat Cell Biol. 2017;19:732–739. doi: 10.1038/ncb3531. [DOI] [PubMed] [Google Scholar]
- 26.Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27:6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 27.Ghavami S, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Ohtake F, Saeki Y, Ishido S, Kanno J, Tanaka K. The K48-K63 branched ubiquitin chain regulates NF-κB signaling. Mol Cell. 2016;64:251–266. doi: 10.1016/j.molcel.2016.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.