Significance
Nearly 90% of lethal antibiotic-resistant infections in the United States are caused by Gram-positive pathogens, with Staphylococcus aureus accounting for more than one-half of these. Antibiotic resistance is often encoded by plasmids and integrative elements that are exchanged between bacteria through conjugative DNA transfer. During conjugation, a relaxase protein binds, nicks, and covalently attaches to the 5′-end of the DNA, guiding it to the recipient cell, where it restores its circular closed form. We show that relaxase MobM from the promiscuous plasmid pMV158 uses a hitherto unseen mechanism for DNA nicking/closing that is based on the formation of a protein-DNA phosphoramidate adduct. Moreover, our analysis reveals that MobM-like histidine relaxases account for 85% of all relaxases in S. aureus isolates.
Keywords: histidine relaxase, antibiotic resistance, horizontal gene transfer, X-ray structure, Staphylococcus aureus
Abstract
Relaxases are metal-dependent nucleases that break and join DNA for the initiation and completion of conjugative bacterial gene transfer. Conjugation is the main process through which antibiotic resistance spreads among bacteria, with multidrug-resistant staphylococci and streptococci infections posing major threats to human health. The MOBV family of relaxases accounts for approximately 85% of all relaxases found in Staphylococcus aureus isolates. Here, we present six structures of the MOBV relaxase MobM from the promiscuous plasmid pMV158 in complex with several origin of transfer DNA fragments. A combined structural, biochemical, and computational approach reveals that MobM follows a previously uncharacterized histidine/metal-dependent DNA processing mechanism, which involves the formation of a covalent phosphoramidate histidine-DNA adduct for cell-to-cell transfer. We discuss how the chemical features of the high-energy phosphorus-nitrogen bond shape the dominant position of MOBV histidine relaxases among small promiscuous plasmids and their preference toward Gram-positive bacteria.
Acquisition of exogenous genetic material by bacteria is achieved via conjugative DNA transfer of mobile genetic elements, such as plasmids and especially integrative and conjugative elements and integrative and mobilizable elements (1). Such processes of horizontal gene transfer (HGT) are considered a strong driving force in bacterial evolution and in the ability of bacteria to colonize different niches (2). In addition to permitting the rapid evolution of the bacterial pangenome, HGT is involved in the acquisition of genetic traits that may confer selective advantages to the recipient bacteria, including antibiotic resistance (3). This is particularly important when resistance genes encoded by mobile elements are spread explosively among bacteria in hospitals, posing a serious threat to public health systems (www.cdc.gov/drugresistance/threat-report-2013; www.who.int/drugresistance/documents/surveillancereport/en/). Thus, the so-called mobilome (4) participates in the spread of antibiotic resistance, which is expected to cause 10 million casualties annually by 2050, and the consequent huge economic burden (amr-review.org/sites/default/files/Report-52.15.pdf). This has generated a unanimous call for new approaches to deal with infectious diseases caused by pathogenic bacteria (5).
A main performer in HGT is the protein relaxase, a topoisomerase-like enzyme that cleaves supercoiled plasmid DNA in a strand- and sequence-specific manner and ligates it after cell-to-cell transfer. Relaxases start DNA transfer by conjugation on recognition of their target DNA, the origin of transfer (oriT), on which they mediate generation of a hairpin-loop structure that leaves the dinucleotide to be cleaved (the nic site) in a single-stranded (ss) DNA configuration (Fig. 1B) (6, 7). On the oriT, the relaxase assembles with other proteins participating in HGT, namely the coupling protein and the proteins that constitute the type IV secretion system (T4SS). To date, relaxase-mediated nucleophilic attack at the nic site has been shown to generate a covalent linkage between a tyrosine residue and the 5′-phosphate DNA of the cleaved dinucleotide (8). This reaction leaves a free 3′-hydroxyl end that serves as primer for DNA replication by conjugative rolling-circle replication (RCR) (9, 10). The current model for conjugation hypothesizes that the covalent phosphotyrosine DNA-relaxase complex is pumped to the recipient cell by means of the coupling protein and the T4SS (11, 12). In the recipient cell, as shown for small mobilizable plasmids that replicate by the RCR mechanism, the transferred ssDNA is converted into double-stranded (ds) DNA molecules by replication from a lagging strand origin (13), followed by a second relaxase-mediated reaction to close the newly synthesized strand and supercoiling of the dsDNA by the recipient gyrase (9).
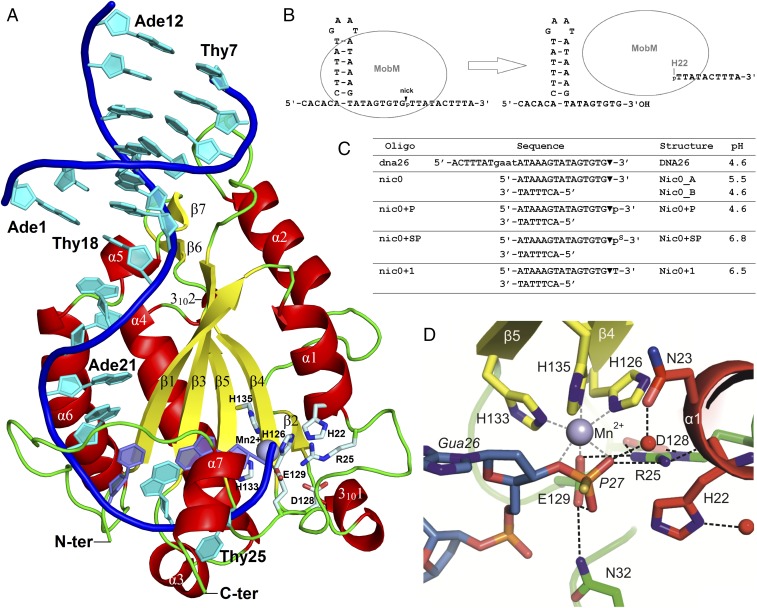
Fig. 1.
Structure of the MobM-DNA complex. (A) Crystal structure of the N-terminal MobM relaxase domain bound to the nic0+SP oligonucleotide containing the scissile phosphate (Nic0+SP structure). (B) Scheme of oriT processing by MobM. (C) Oligonucleotides used for crystallization, structure names, and pH. (D) Active site details.
On the basis of homologies along the first 300 residues, relaxases were classified into six families: MOBF, MOBQ, MOBP, MOBV, MOBH, and MOBC (14). The first four families include “classical” plasmid-encoded conjugative relaxases that belong to the superfamily of HUH (His-hydrophobic-His) endonucleases (8, 14). These endonucleases are characterized by the presence of the HUH motif (two His residues used for metal coordination, separated by a hydrophobic residue) and the Y motif containing either one (Y1) or two (Y2) active site Tyr residues, which deliver the nucleophilic hydroxyl group(s) for endonuclease-recombinase reactions (8). HUH endonucleases depend on a single metal ion for activation of the scissile phosphate for DNA cleavage and formation of a transient protein-5′-DNA covalent adduct, and finally for the joining of the nicked DNA ends. Crystal structures of members of the Y1 and Y2 relaxase families are known: from the Y1-MOBQ relaxases, MobA_pR1162 (15) and NES_pLW043 (16), and from the Y2-MOBF relaxases, TrwC_pR388 (17, 18), TraI_pF (19), and TraI_pCU1 (20).
Biochemical and biophysical, but not structural, studies have been performed on relaxases encoded by small plasmids, most of them belonging to the MOBV family (also termed MOBPre) (14, 21). So far, the best-characterized relaxase encoded by these plasmids is MobM, from the promiscuous plasmid pMV158, which has become the prototype for the MOBV1 subfamily of relaxases (14, 21). To advance our knowledge of these proteins, we undertook a structural characterization of the relaxase domain of MobM (MobMN199), a construct that retains DNA-binding and relaxase activities (7, 22). The different crystal structures solved in complex with oriT DNA sequences reveal that, unexpectedly, MOBV relaxases use a histidine (H22 of MobM), not a tyrosine, as catalytic residue. Prompted by this finding, which implies a hitherto undescribed metal-dependent mechanism for DNA cleavage, protein-DNA adduct formation, and DNA ligation, we dissected the active site mechanism by mutating all of the residues involved and tested the functional effect. In addition, we performed a computational study to describe the catalytic mechanism at the atomistic scale. Interestingly, the theoretical calculations show that a conserved glutamate (E129 of MobM) serves as (i) a general acid by protonating the 3′ oxygen-leaving group in the nicking reaction (while the phosphodiester bond is broken by H22), and (ii) a general base abstracting a proton from the 3′-hydroxyl nucleophilic end in the DNA-joining reaction (phosphoramidate bond breaking). Our results unravel a singular catalytic mechanism for the metal ion-dependent role of the conserved histidine residue present in all MOBV relaxases, thereby highlighting the role of these proteins in the spread of the thousands of small mobile genetic elements found in many pathogenic Gram-positive bacteria, especially streptococci, and staphylococci.
Results
Abundance and Features of Relaxases of the MOBV Family.
An in-depth search of the bacterial mobilome [Mob_Pre search in the National Center for Biotechnology Information’s Conserved Domain Database (NCBI CDD); www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml] retrieved more than 5,830 relaxases of the MOBV family (SI Appendix, Fig. S1 and Table S1). The majority belonged to Firmicutes phylum (5,765 hits), and the top-listed species were encountered in life-threatening bacteria, including Staphylococcus aureus (3,432 hits), Streptococcus agalactiae (453 hits), Enterococcus faecalis (200 hits), and Enterococcus faecium (109 hits). Genomic data for some pathogenic bacteria, such as S. aureus, are overrepresented in the database, and thus a high number of hits was obtained. However, strikingly, MOBV members account for approximately 85% of all conjugative relaxases reported in the NCBI CDD for S. aureus (SI Appendix, Table S1). Moreover, comparing the number of MOBV relaxases found in S. aureus (the NCBI CDD data) with the number of S. aureus genomes and plasmids available in the NCBI Genome resource revealed that up to 72% of all sequenced isolates of S. aureus contain MOBv relaxases (vs. 13% with other types of relaxases and ∼15% without a relaxase discovered to date; SI Appendix, Table S2). Similar analyses for other bacteria revealed that MOBv relaxases are frequent in the genomes of Enterococcus faecium (68%), E. faecalis (45%), and S. agalactiae (56%) and very rare in Streptococcus pneumoniae (3%) and Streptococcus pyogenes (3%). Furthermore, a NCBI Nucleotide BLAST search on the sequence of plasmid pMV158 oriT (oriTpMV158; 41 nucleotides) revealed more than 100 almost-identical sequences, found mainly in S. aureus genomes (SI Appendix, Table S3). This figure could be much higher if we consider that MobM can cleave sequences that share ∼70% identity with the oriTpMV158 (23).
With the advent of inexpensive genomic sequencing, most MOBV relaxases have been found to be encoded in bacterial genomes within various integrative elements; however, they were traditionally found mainly in RCR plasmids, MobM being representative of the entire family (21). Depending on homologies at their N200 residues, these relaxases are grouped into two main subfamilies, represented by plasmids pMV158 (MOBV1) and pBBR1 (MOBV2) (14). A further search for plasmids harboring relaxases of the MOBV1 subfamily retrieved a high number of mobile elements ranging from 2.7 to 30 kb, one-half of them encoding antibiotic resistance genes or other virulence traits (21). The genetic organization of the mobilization region was similar in all members: (i) the oriT located upstream and close to the mob gene, and (ii) the nic site in the same strand as the oriT (21). In the case of the MOBV relaxases, the following three conserved motifs, from the N terminus to the C terminus, were described (21): motif I, H(N/D)(Q/E)R, of no assigned function; motif II, NY(D/E)L, in which the Y residue (Y44 of MobM) was proposed as the catalytic one (24); and motif III, HxDExTPHMH, which contains the HUH metal-coordination signature of all relaxases (8). Curiously, residue Y44 of MobM is not conserved in all members of the MOBV family (14). Furthermore, single mutants of all seven tyrosine residues of the MobpBBR1 relaxase (a member of the MOBV family) do not substantially alter the plasmid mobilization frequencies. This observation does not support a “classical” single tyrosine-mediated initiation of conjugation of this particular plasmid (25). The foregoing findings prompted us to tackle the structural features of the MobM relaxase in complex with its target DNA.
General MobM Structure.
Six crystal structures of the MobMN199 relaxase (residues 2–196) (7), in complex with various DNA oligonucleotides, were determined to 1.9- to 3.1-Å resolution (SI Appendix, Table S4). The experimental phases were obtained by the SAD method, using a selenomethonine protein derivative dataset. The overall structure of MobMN199 shows that it has the relaxase α/β plate fold, which is reminiscent of the palm domain of the DNA polymerase Klenow fragment (Fig. 1). It consists of a central β sheet formed by five antiparallel β-strands (with topology β1, β3, β5, β4, and β2) flanked on each side by a pair of α-helices, α1/α2 and α4/α6. The active site is located on one face of the β sheet. The histidine triad (H126 and the HUH motifs H133 and H135) that coordinates the manganese ion required by the protein (7) is located on strands β4 and β5, whereas the catalytic histidine (H22; see below) is located at helix α1 (Fig. 1). The extended α6-α7 loop and the last α-helix (helix α7) create a thumb (region 5) that wraps around the ssDNA substrate and directs it into the active site (Figs. 1 and 2). On the opposite side, the active site is flanked by a β-turn located on the α1–310 loop (region 2) and, from the bottom, by the β4-β5 loop, which is partially ordered in a β-turn motif. Finally, the α1 helix (region 1), which includes the catalytic H22, closes the active site from the top. Regarding the MobM elements that bind the dsDNA, the α2-β3 loop (region 3) interacts with the minor groove of the DNA hairpin, and the fragment composed of the β6-β7 hairpin, the α5 helix, and the α5-α6 loop (region 4) interact with major groove. Details of these interactions are described in MobM–DNA Interactions below.
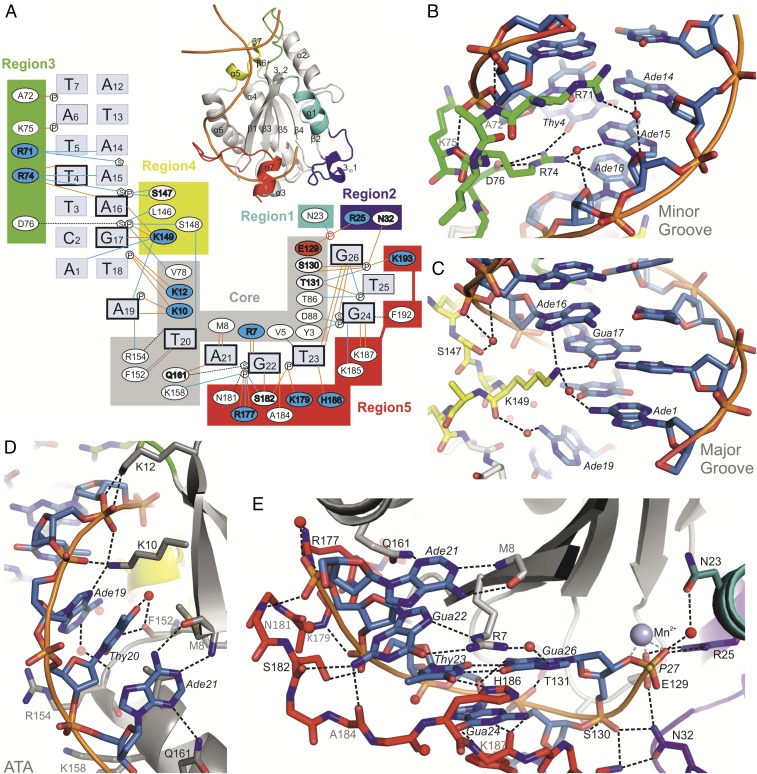
Fig. 2.
MobM–DNA interactions. (A) Scheme representing the MobM–DNA interactions in the Nic0+SP complex structure. Brown lines indicate hydrogen bonds, blue lines indicate water-mediated hydrogen bonds, and black dashed lines indicate van der Waals contacts. Residues that form side chain-DNA hydrogen bonds are indicated in bold (positively and negatively charged residues are shown on blue and red background, respectively). Nucleotides that form nucleobase-protein hydrogen bonds and the Thy23-Gua26 wobble base pair are indicated in bold type. (B) Minor groove binding by MobM region 3 (loop α2-β3). (C) Major groove binding by MobM region 4 (helix α5). (D) ATA bulge binding by the MobM core region (strand β1 and helix α6). (E) MobM-ssDNA binding by the core region, region 5 (loop α6-α7 and helix α7), region 2 (loop α1–3101), and region1 (helix α1). Residues that form side chain-DNA hydrogen bonds are labeled in black. Residues that form main chain-DNA hydrogen bonds are labeled in gray.
DNA Structure.
The oriTpMV158 is relatively complex, containing three inverted repeats (IR1, IR2, and IR3) that can generate mutually exclusive DNA hairpins (7, 21) (SI Appendix, Fig. S3C). The IR3 sequence spans 31 nucleotides upstream of the nic site and is formed by extension of the IR1 hairpin (18 nucleotides) by a 3-base ATA bulge (IR3 right arm) and a 5-bp IR3 base. The formation of the IR1 or IR3 hairpin excludes formation of the IR2 hairpin, the function of which remains unknown.
The DNA oligonucleotides used for crystallization trials were chosen based on previous work that determined the minimal substrate for MobM binding, namely the IR3 sequence without the first five nucleotides (7). We crystallized two types of DNA substrates mimicking the IR1/IR3 hairpin in complex with MobM (Fig. 1C). One type consisted of a continuous 26-base oligonucleotide upstream from the nic site which covers a sequence of the IR1 hairpin extended by 8 bases of the IR3 right arm (DNA26 oligonucleotide; bases 6–31 of oriTpMV158, here numbered 1–26; Fig. 1C). The second type consisted of two annealed oligonucleotides of the IR1 left arm and the IR3 right arm, resulting in a DNA molecule that lacks the 4-base loop of the IR1/IR3 hairpin (the nic series of oligonucleotides; bases 6–12 and 17–32 of oriTpMV158; numbered 1–7 and 12–27). The DNA molecules used for protein-DNA complex crystallization either (i) terminate at the 3′-oxygen before the cleavage site (dna26 and nic0 oligonucleotides), which mimics the 5′-side DNA cleavage product (and later serves as a substrate in the DNA-rejoining reaction), or (ii) include the scissile phosphate (nic0+P, nic0+SP, and nic0+1 oligonucleotides), which represents the cleavage substrate. In both types of MobM-DNA structure, the DNA helix of the hairpin stem adopts the B-form: however, the sequence includes a small A-tract (A14–A16) that differs from the canonical B-DNA (Fig. 2). A-tracts have a compressed minor groove, high-propeller twisted A:T base pairs, and bifurcated H-bonds at the major groove (26). This conformation is related to DNA recognition at the stem by the β-turn RxD/N motif (see MobM–DNA Interactions below).
The IR1 downstream sequence forms an extended single-stranded structure, with the exception of a Thy23-Gua26 wobble base pair that holds the ends of a U-like turn and directs the scissile phosphate into the active site of the protein (the Nic series of structures). In the DNA26 structure, bases of the hairpin loop of one protein-DNA complex enter the active site of a neighboring complex (SI Appendix, Fig. S2C) and form noncanonical base pairs with DNA bases of that neighboring complex (wobble Thy23-Gua8′ and sheared Gua24-Ade9′ base pairs); this configuration displaces the metal ion and the DNA substrate from the active site (SI Appendix, Fig. S2D). Intriguingly, in both types of MobM-DNA structures, the guanine that forms the wobble Thy-Gua base pair superimposes perfectly (Gua26 in the Nic series of structures, and Gua8′ in the DNA26 structure), thereby suggesting a preferred binding site for guanine in this region of the MobM-DNA complex.
MobM–DNA Interactions.
The specific recognition of oriTpMV158 by MobM is achieved through the formation of nine protein side chain-nucleobase hydrogen bonds between seven protein residues (R7, K10, R71, R74, K149, Q161, and H186) and eight DNA bases (Thy4, Ade6, Ade16, Gua17, Ade19, Ade21, Gua22, and Thy23) (Fig. 2). Interactions with nucleobases also include seven hydrogen bonds between the protein backbone (M8, F152, S182, A184, and K187) and four bases (Thy20, Ade21, Gua22, and Gua24). The remaining protein–DNA interactions include 14 hydrogen bonds between 11 side chains of MobM and seven sugar-phosphate moieties of the DNA backbone, five hydrogen bonds between protein and DNA backbones, and a number of van der Waals contacts (Fig. 2).
In all of the structures solved herein, MobM binds the DNA hairpin through a long track of positively charged residues that stack out from the protein surface (SI Appendix, Fig. S2F). The specific interactions within the hairpin stem region are limited to four bases interacting with the conserved R71, R74, and K149 residues (Fig. 2 and SI Appendix, Fig. S3A; note that the R71–Ade6 interaction is visible only in the DNA26 structure, which has the full DNA hairpin). In the R71 to R74 stretch, which is part of the α2-β3 loop, the protein backbone extends along the minor groove, with main chain amide-to-phosphate backbone interactions. This conformation allows the deep penetration of the two arginines in the minor groove, which narrows down to 6.8 Ǻ (O4′–O4′ distance), sandwiching the R74 guanidinium moiety between opposite sugars across the groove. The side chain of R74 is stabilized inside the minor groove by the side chain of D76, with residues 74–77 forming a β-turn. This feature (β-turn RxD/N motif) appears to be a characteristic DNA-binding motif that is crucial for specific recognition of the oriT hairpin stem (see below and SI Appendix, Fig. S4).
Whereas specific recognition of the IR1 hairpin stem is mediated by only three residues, R71, R74, and K149, which contact only a few bases, a different situation arises for the ssDNA bases downstream IR1, namely Ade19-Thy20-Ade21 of the ATA bulge and Gua22-Thy23-Gua24-Thy25-Gua26 of the right arm of the IR3 stem (IR3-R). Two kinks are observed in the DNA backbone between Thy18-Ade19 and Thy20-Ade21 nucleotides of the ATA bulge, which disrupts stacking of the respective bases (Fig. 2D and SI Appendix, Fig. S3B). The C-terminal thumb folds over the DNA backbone at the Gua22 and Thy23 bases, interacting with each phosphate or nucleobase until reaching the scissile phosphate (Fig. 2 A and D and SI Appendix, Fig. S3B). With the exception of Thy25, which is solvent-exposed, all bases turn toward the protein and are in fact embedded into the protein surface, whereas the phosphate backbone turns outward toward the solvent (Fig. 2 A, D, and E).
We highlight the role of conserved H186 here because its nitrogen ND1 atom forms a hydrogen bond with oxygen O4 of Thy23, thereby stabilizing the wobble base pair Thy23:Gua26. Moreover, the imidazole ring of H186 stacks with the guanidinium group of R7, which itself forms hydrogen bonds with Gua22 (Fig. 2E and SI Appendix, Fig. S3B). The last protein–nucleobase specific interactions are found between the Gua24 base and K187 peptide bond; however, a number of hydrogen bonds between protein residues and the DNA backbone are also found in this region. In addition, the Gua24 base is involved in a stacking interaction with F192, which appears to be relevant given the fully conserved ring character of the residues (Phe, Tyr, or His) at that position (SI Appendix, Fig. S3D). Finally, the formation of a Thy23-Gua26 wobble base pair leads to positioning of the scissile phosphate in the center of the MobM active site (Fig. 2 A and E).
Structural Comparison of MobM and Other Relaxases.
Despite very low sequence homology (9–13% sequence identity for structurally superimposable residues), structural superimposition of the structure of MobM on that of other relaxases—TrwC_pR388, TraI_F, and TraI_pCU1 from the MOBF family and MobA_pR1162 and NES_pLW043 from the MOBQ family—demonstrate general structural similarity among these proteins (2.9–4.1 Å Cα rmsd for structurally superimposable residues) (SI Appendix, Fig. S4A). Nonetheless, we observe significant differences that distinguish MobM from members of the other families. In general, MobM appears to be a simplified version of the fold with smaller loops and less additional secondary structure decoration. In this respect, it is closer to the structure of the MOBQ family members MobA_pR1162 and NES_pLW043, which are also shorter than the MOBF family representatives. Notably, in MobM, the C-terminal thumb directing ssDNA to the active site (residues 175–196) adopts an extended conformation plus one α-helix (α7), whereas in MOBF relaxases, like TrwC, it is formed by a much longer stretch (residues 210–265) that adopts a mainly α-helical conformation. In MOBQ relaxases, this element lacks structural description, because the MobA structure is based on a shorter protein construct (MobA_1–184), and for the NES relaxase construct (NES_2–220) the C-terminal residues that would form a thumb (i.e., residues 196–220) are untraceable in the crystal structure. The second evident difference is that MobM (MOBV) and NES (MOBQ) lack the extensive secondary structures that interact with the tip of the DNA hairpin, in contrast to TrwC (MOBF). As a result, the tip region of the hairpin is not covered by the protein and protrudes from the complex more prominently than it does for TrwC.
Regarding similarities, all three MobM, NES, and TrwC relaxase-DNA hairpin crystal structures have a common β-turn RxD/N motif (MobM_74-76:RKD, NES_78-80:RKN, and TrwC_75-77:RQD), which in MobM is located at region 3 of the α2-β3 loop (Fig. 2 A and B and SI Appendix, Fig. S4 C and D). Within the motif, as described above, the Arg and Asp/Asn residues enter the hairpin minor groove, whereas the middle residue positions itself between the phosphates of the DNA backbone. Superimpositions of the MobM structure on TraI_F, TraI_pCU1, and MobA structures suggest that these relaxases also use this β-turn RxD/N motif to bind DNA (TraI_F_68-70: RMD; MobA_66-68: RAN) (SI Appendix, Fig. S4 C and D); however, in these cases, the comparison is impaired owing to the lack of DNA hairpins in these crystal structures. An exception is the TraI_pCU1 relaxase, which does not have the RxD/N sequence. Interestingly, TraI_pCU1 binds its cognate DNA weakly and in a sequence-independent manner, and has been suggested to rely on a second DNA-binding protein to selectively bind the pCU1oriT (20).
At the major groove of the hairpin, interactions unique to each of the relaxases are found. MobM (MOBV1 subfamily) enters the major groove using just one residue (K149) from the short four-residue α5 helix of region 4, which defines the minimal major groove-interacting element among all structurally described relaxases (SI Appendix, Fig. S4). The MOBQu relaxase NES places R151, N154, and Y156 from an eight-residue β hairpin at the major groove (SI Appendix, Fig. S4C). In the MOBQ1 relaxase MobA, K161 superposes to MobM K149 and presumably would interact in major groove. K161 belongs to an 11-residue insertion that includes R143, which structurally superposes to MobM R71 of the minor groove-interacting region 3 (SI Appendix, Fig. S4C). The MOBF1.1 relaxase TrwC and the MOBF1.2 relaxase TraI_F also have a single Lys residue (K181 and K179, respectively) equivalent to MobM K149. However, the dsDNA-containing crystal structure of TrwC shows that MOBF relaxases use additional residues (TrwC R128 and K130; SI Appendix, Fig. S4D). These derive from an extensive β-hairpin structure that enters the major groove at the opposite face of the DNA from where the RxD/N motif enters the DNA minor groove. This area of the major groove is solvent-exposed in the MobM complex. Thus, MobM seems to have the minimal structural determinants for hairpin stem recognition.
Active Site Architecture.
In all Nic0 structures of MobM, residues H126, H133, H135, and E129 coordinate the Mn2+ ion with octahedral geometry (Figs. 1D and 3), as was unequivocally confirmed by anomalous diffraction (SI Appendix, Fig. S2E). In Nic0_A, Nic0_B, Nic0+P, and Nic0+SP structures, the fifth Mn2+ ion ligand is invariably the O3′ oxygen of Gua26, and the sixth ligand is the scissile phosphate oxygen (Nic0+P and Nic0+SP), the catalytic H22 (partial occupancy in Nic0_A and full occupancy in Nic0_B), or a water molecule (partial occupancy in Nic0_B).
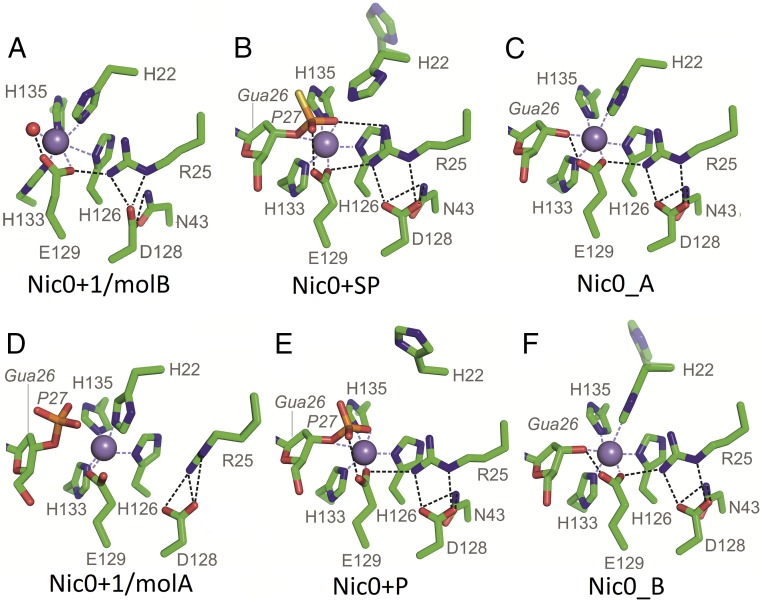
Fig. 3.
Active site snapshots. (A) Nic0+1/molB–DNA-free active site: H22 ligates Mn2+. (B) Nic0+SP active site with the DNA substrate (up to the scissile phosphate, here as a thiophosphate). H22 is in the proper position for catalysis, in-line with the scissile bond atoms; R25 and E129 interact with the scissile phosphate. (C) Nic0_A active site with the 5′ DNA product/ligation substrate. H22 ligates Mn2+; E129 interacts with the Mn2+-bound DNA O3′ of Gua26. (D) Nic0+1/molA active site with the DNA substrate placed in a nonproductive manner. The scissile phosphate does not interact with the Mn2+ ion, and H22 ligates Mn2+. A number of interactions among E129, D128, R25 (partially disordered), and N43 (disordered) are broken. (E) Nic0+P low-pH active site with the DNA substrate; as in B, but H22 protonation favors the Out position of this side chain. (F) Nic0_B low-pH active site with the 5′ DNA product/ligation substrate; as in C, but H22 protonation favors the partial Out position of this side chain. H22 minor occupancy rotamers in C and E are shown faded.
The asymmetric unit of the Nic0+1 crystal contains two protein-DNA complexes that represent two distinct states: (i) a DNA-free active site (complex B in the crystal asymmetric unit, Nic0+1/molB; Fig. 3A) and (ii) an active site in which the Gua26 O3′ atom and the scissile phosphate OP1 atom are located a considerable distance from the metal ion (3.6 and 4.2Ǻ, respectively; note that corresponding distances equal 2.7 and 2.1 Ǻ in the Nic0+P structure and 2.2 and 2.3 Ǻ in the Nic0+SP structure; complex A in the crystal asymmetric unit, Nic0+1/molA; Fig. 3D). In both complexes of the Nic0+1 structure, the metal ion attracts H22 for the interaction instead, thereby showing that in the absence of the DNA-scissile phosphate ligand, the metal ion ligates the catalytic H22 (Fig. 3 A and D). In complex B, the DNA electron density map ends at phosphate 25, indicating that the last three bases are disordered and most likely do not enter the active site. In this case, the Thy23-Gua26 wobble base pair is not formed, and the position of the Gua26 3′ oxygen (O3′) is occupied by a water molecule. In complex A, despite the wobble base pair formation and the placement of the DNA substrate in the active site, the Gua26 3′-oxygen and the scissile phosphate are shifted away from the center of the active site.
In all Nic structures, except the Nic0+1/molA structure, a group of fully conserved residues is engaged in the formation of a hydrogen bond network (Figs. 1D and 3), in which (i) D128 contacts N43 and R25; (ii) R25 interacts with the H22 peptide bond, the scissile phosphate, and the metal-ligating E129; and (iii) depending on the structure, E129 interacts with the scissile phosphate OP1 oxygen (active site with the cleavage substrate, Nic0+P and Nic0+SP structures; Fig. 3 B and E), the Gua26 O3′ oxygen (active site with the cleavage product/ligation substrate, the Nic0_A and Nic0_B structures; Fig. 3 C and F), or the metal-bound water molecule, which superimposes with the Gua26 O3′ oxygen (DNA-free active site, Nic0+1/molB structure; Fig. 3D). The Nic0+SP structure accommodates an additional contact for E129, a hydrogen bond between E129 and N32 (Fig. 1D). In the Nic0+1/molA structure, this extensive array of contacts is limited to only one: the R25–D128 interaction. This is due to displacement of the R25 side chain toward a location that in other structures would create clashes with N43 (Fig. 3D).
Mobility of the Catalytic Histidine.
Depending on the crystal structure, the catalytic H22 side chain occupies three distinct positions: the In position (metal-bound, with the NE2 nitrogen atom 2.2–2.8 Ǻ away from the Mn2+ ion; Fig. 3 A, C, D, and F), the Intermediate position (NE2 nitrogen 6.2 Ǻ away from the Mn2+ ion and 4.0 Ǻ away from the scissile phosphate phosphorous atom; Fig. 3B) and the Out position (H22 side chain far from the Mn2+ ion and the scissile phosphate; Fig. 3 B, E, and F). For structures that were determined at pH >4.6, the catalytic histidine side chain is placed in the metal-bound In position in two settings: (i) in the DNA-free and the DNA-misplaced active site structures (Nic0+1/molB and Nic0+1/molA, respectively; Fig. 3 A and D), and (ii) in the presence of the 5′ cleavage product (Gua26 3′OH) in the active site (Nic0_A; Fig. 3C). The foregoing two cases show that H22 ligates this ion in the absence of the interaction of the scissile phosphate with the metal ion. Conversely, ligation of the scissile phosphate to the active site metal ion causes the catalytic H22 to move away from the metal-bound In position to the Intermediate and Out positions as can be seen in the Nic0+SP structure, which has the H22 side chain in two alternative conformations: the Intermediate position, with major occupancy, and the Out position, with minor occupancy (Fig. 3). Similarly, protonation of the H22 imidazole ring at pH 4.6 induces the histidine side chain to move away from the metal ion, as can be observed when comparing higher and lower pH structures containing either the 5′ cleavage product (the In position at pH 5.5 vs. the In/Out positions at pH 4.6 for Nic0_A and Nic0_B structures, respectively; Fig. 3 C and F) or the cleavage substrate (the Intermediate/Out positions at pH 6.8 vs. the Out position at pH 4.6 for Nic0+SP and Nic0+P structures, respectively; Fig. 3 B and E). In fact, the relaxation activity of the MobM plasmid is highest at pH 6.5, which is slightly above the histidine imidazole pKa (6.0), and decreases steeply below pH 6.0, dropping by 30% at pH 4.6 (23). Taken together, these findings show that the H22 side chain has freedom of movement and that its interaction with the metal ion depends on the stage of the reaction.
One Metal-Ion Catalysis with a Histidine as a Nucleophile.
Nucleases cleave phosphodiester bonds through a general acid-base catalysis, where the general base activates the nucleophile by deprotonation and the general acid facilitates product formation by protonation of the leaving group (27, 28). This reaction goes typically through three stages: (i) nucleophilic attack, (ii) formation of a highly negative pentacovalent transition state, and (iii) breakage of the scissile bond. The mechanism requires that the nucleophile, the phosphorus atom, and the leaving group be in-line. In the case of metal ion-dependent DNA nucleases, metal ion B, which resides on the leaving group side and is common for one- and two-metal ion nucleases, stabilizes the pentacovalent transition state and, in certain coordination geometries, can promote a nucleophilic attack by destabilizing the enzyme-substrate complex (29). The enzymes that cleave by a two-metal ion mechanism use the second metal ion (metal ion A) to activate a catalytic water molecule that acts as a nucleophile, as well as to stabilize the transition state. In the one-metal ion enzymes, the activation role of the second metal ion is performed by a protein residue, and the catalytic water molecule can be replaced by a serine or a tyrosine hydroxyl group (28, 30). The latter was the case for the first relaxase described (17, 18), as well for other relaxases analyzed thereafter.
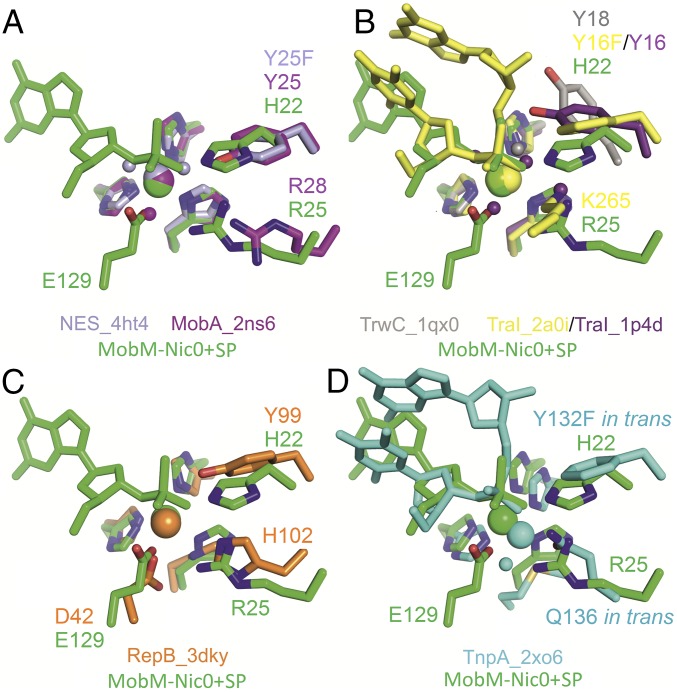
In the case of the relaxase MobM, the protein-DNA structures show that a constellation of amino acids is required for the formation of the cleavage-competent active site that is best reflected in the Nic0+SP structure. In this structure, the Mn2+ ion interacts with both the scissile phosphate OP1 oxygen atom (2.2 Ǻ distance) and the Gua26 O3′ oxygen atom (2.3 Ǻ distance). Therefore, this ion is well positioned to contribute to the stabilization of the electron-rich pentacovalent transition state. Importantly, the catalytic H22 is in the proper location for the nucleophilic in-line attack on the phosphorus atom (H22 NE2 nitrogen being 4.0 Ǻ away) and superposes with the position of the catalytic tyrosines in the crystal structures of other HUH endonucleases (MOBQ relaxases, MOBF relaxases, RepB replication initiator, and TnpA transposase; Fig. 4). In the case of superposition to MobA, which is the most similar to MobM among the structurally described relaxases, the MobM H22 NE2 nitrogen atom superposes perfectly with the MobA catalytic Y25 hydroxyl oxygen atom (Fig. 4A).
Fig. 4.
MobM Nic0+SP structure active site superimposition with other HUH endonucleases. (A) MOBQ family of one-tyrosine (Y1) relaxases MobA_pR1162 and NES_pLW1043. (B) MOBF family of two-tyrosine (Y2) relaxases TraI_pF and TrwC_pR388. (C) Replication initiation protein RepB_pMV158. (D) Transposase TnpA_Dra2 of bacterial insertion sequence ISDra2.
The foregoing findings reveal MobM as a metal-dependent nuclease that uses a histidine nitrogen atom for the nucleophilic attack on the DNA substrate. Moreover, in contrast to all known DNA breaking-joining enzymes, MobM uses a histidine nitrogen atom instead of an oxygen atom of a tyrosine/serine residue, thus operating through a phosphohistidine adduct. Known examples of nucleases belonging to the phospholipase D (PLD) superfamily of phosphodiesterases hold catalytic histidines and operate through a transient phosphohistidine intermediate adduct. However, these nucleases bear no sequence or structure relationship with relaxases, have separate domains for substrate recognition and for cleavage, are metal ion-independent enzymes, and do not support DNA ligation (28). Importantly, the acid dissociation constant of histidine is the closest to the cytoplasmic pH among known nuclease nucleophiles (His pKa = 6; Tyr pKa = 10; ribose 2′OH pKa = 12–14; Ser pKa = 13; H2O pKa = 16) (28). Thus, activation of the histidine imidazole group by deprotonation is readily achieved, and no auxiliary residue appears to be required for its activation. In contrast, other relaxases that harbor a catalytic tyrosine, such as TrwC, appear to have a residue that closely interacts with it (an Asp in TrwC) to facilitate proton abstraction.
In the MobM Nic0-SP structure, the metal-bound phosphate OP1 oxygen atom interacts with the R25 NH1 nitrogen atom, taking a position not far from K265 of TraI (19) (Fig. 4B), which has been proposed to act as the equivalent of the second metal ion (30). Similarly, in yeast type II topoisomerase, the conserved R781 residue contacts and stabilizes the covalent phosphotyrosine moiety of the protein-DNA adduct (31) and mutation of this arginine to a residue other than lysine dramatically reduces DNA cleavage and relaxation activity (32). In MobM, R25 is fixed in its position at the active site by a double-H bond interaction with D128.
In the MobM Nic0+SP structure, the second oxygen atom of the scissile phosphate, OP2, shares a hydrogen bond with the E129 OE1 oxygen, which further interacts with the N32 NE2 nitrogen, whereas the E129 OE2 oxygen ligates the metal ion and contacts the R25 NH2 nitrogen (Figs. 1D and 3B). Use of a carboxylic residue as fourth protein ligand for metal ion ligation has not yet been described for any relaxase. Intriguingly, the MobM genetic companion on pMV158, the RepB replication initiation protein (which is not a relaxase although it belongs to the HUH superfamily), in addition to the histidine triad, uses D42 for the Mn2+ ion ligation (Fig. 4C). However, its role other than metal ion coordination has not been tested (33). Interestingly, a hydrogen bond between E129 and the DNA 3′OH end in the Nic0_A and Nic0_B structures suggests that E129 acts as a general base in Gua26 3′OH activation for the nucleophilic attack on the H22-DNA adduct during the DNA ligation reaction, as discussed in Theoretical Study of the Catalytic Mechanism below.
Mutational Analysis.
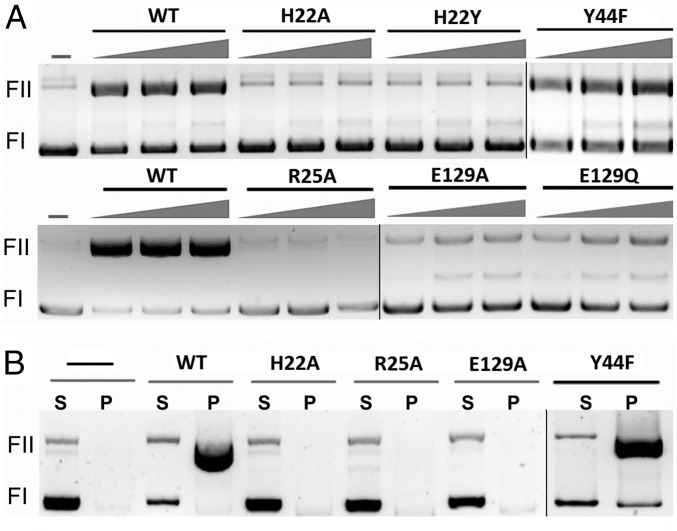
To confirm the information derived from the crystal structures, several mutations designed to change key residues of MobM were constructed, and the resulting mutated proteins were purified. The following mutations were done: (i) H22 was changed to either Ala (H22A) or Tyr (H22Y); (ii) R25 was changed to Ala (R25A); (iii) E129 was changed to either Ala (E129A) or to Gln (E129Q); and (iv) Y44 was changed to phenylalanine (Y44F), the latter because Y44 has been proposed to be the catalytic residue (24). The purified mutant proteins were tested for their ability to relax supercoiled plasmid DNA (nicking reaction) or to generate relaxosomes in vitro. This latter assay is based on the selective precipitation of DNA-protein covalent complexes by SDS and KCl (34). In both cases, pMV158 DNA was treated with the purified proteins, and the reaction products were analyzed on 1% agarose gels. The assays performed with the H22A, H22Y, and R25A mutants confirmed the essential role of these residues: the wild-type MobM protein generated ∼60% open circle (oc) relaxation products, whereas no significant oc forms were observed for any of these mutant proteins (Fig. 5A). Equivalent results were found when the generation of relaxosomes was analyzed: protein-DNA adducts were readily detected in the wild type, but not in the H22 or R25 mutants. This observation demonstrates the crucial role of amino acids of Motif I in the cleavage and generation of stable DNA-protein adducts (Fig. 5B). Relaxation assays done with the Y44F mutant protein showed that it behaved like the wild-type MobM, thus also confirming the results of crystal structures. Finally, the E129A and E129Q mutants showed a slight (if any) residual relaxation activity (Fig. 5A). Neither of the two E129 mutants was able to generate relaxosomes (Fig. 5B), thus emphasizing the significance of E129 for formation of the covalent phosphohistidine adduct (Theoretical Study of the Catalytic Mechanism below). Indeed, besides metal coordination, E129 interacts with the scissile phosphate and R25, the latter in turn contacting D128. We conclude that R25 of MobM contributes to anchoring the scissile phosphate and to stabilizing the transition state and the phosphohistidine adduct, as indicated by our calculations (SI Appendix, Fig. S5D).
Fig. 5.
In vitro activity of MobM mutant proteins. MobM nucleolytic activity by plasmid relaxation assay (A) and covalent adduct formation and stabilization by protein-DNA pull-down assay (B). MobM activity is shown as change of plasmid supercoiled (Fl) form to open circle (Fll) form. P, pellet (protein and protein-DNA fraction); S, supernatant (DNA fraction). After nicking, the relaxase remains covalently bound to plasmid DNA.
The in vivo assays were performed by constructing pMV158-derivative plasmids harboring either the H22A or the Y44F mutations. These plasmids have a single mutation in the mobM gene (i.e., a single amino acid change, H22A or Y44F, in the entire 495-residue MobM protein). Mobilization assays were done under conditions previously described (35) using S. pneumoniae cells harboring the wild-type or mutated plasmids as donors and either pneumococcal or E. faecalis cells as recipients. The results showed that whereas mobilization frequencies of 2.5 ± 0.2 × 10−4 (average of five experiments) were found for pMV158 and pMV158Y44F plasmids, no transconjugant (frequency <1.5 × 10−10, i.e., the experimental detection limit) was rescued in the five independent experiments performed with pMV158H22A (SI Appendix, Table S5). Taken together, the results from the crystal structure analysis and the functional assays demonstrate that the H22 residue is essential for the activity of the protein, thereby supporting its role as the nucleophilic residue in the DNA cleavage reaction.
Theoretical Study of the Catalytic Mechanism.
We performed theoretical calculations including classical molecular dynamics (MD) simulations and quantum mechanics/molecular mechanics (QM/MM) free energy calculations to describe the catalytic mechanism performed by MobM. Two MobM-DNA crystal structures were used for MD simulations: one in which the H22 NE2 nitrogen is positioned in-line for the nucleophilic attack on the reaction center (i.e., phosphorus atom of the scissile P-O bond) from the opposite site of the O3′ leaving group, i.e., the Nic0+SP structure, and the other in which H22 ligates the Mn2+ ion and the aforementioned atoms are not in-line (the Nic0+1/molA structure). We were able to model the covalent complex between H22 and the DNA substrate showing the expected inverted configuration of the phosphorus atom for the former (SI Appendix, Fig. S5B) but not for the latter, for which modeling of the nucleolytic reaction failed (SI Appendix, Fig. S5C). Moreover, we observed that the formation of the covalent intermediate was possible only when the O3′ leaving group atom was assumed to be protonated.
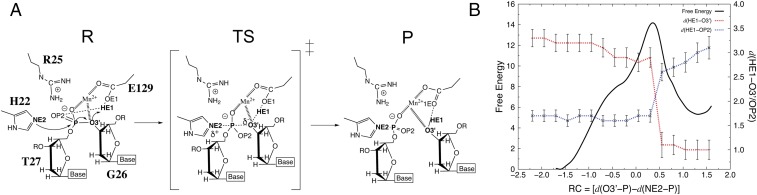
The proposed mechanism of the histidine-mediated attack to the phosphodiester bond, along with the corresponding free energy profile, is depicted in Fig. 6. The reaction occurs through an associative SN2-like mechanism involving a pentacoordinated phosphorus transition state (TS; SI Appendix, Fig. S5B), just as described in the theoretical study of the CheA histidine kinase (36). Moreover, E129 would protonate the O3′ atom, thus playing the role of a general acid in the catalytic mechanism (Fig. 6A). This glutamate is hydrogen-bonded to the OP2 of the target phosphate group in the precatalytic structure (Fig. 6 A and B), as also suggested by visual inspection of the MobM Nic0+SP structure. During catalysis, the HE2 proton from E129 reorients to form a hydrogen bond with its final acceptor atom (i.e., O3′) to finally be transferred (Fig. 6 A and B). It should be noted that in our modeling study, this proton-transfer process occurs spontaneously (i.e., it was not explicitly considered in the reaction coordinates; Fig. 6B), thereby supporting the hypothesis of E129 acting as the general acid in the nucleolytic reaction. Moreover, the proton transfer process lags behind the nucleophilic attack and is concomitant with the drop in energy observed immediately after the maximum in the free energy profile (Fig. 6B).
Fig. 6.
Theoretical calculations. The proposed mechanism of nucleolytic reaction catalyzed by MobM (A) and the corresponding QM/MM free energy surface (B). The distance relative to the protonation of the O3′ leaving group atom by E129 is depicted. P, products; R, reactants; RC, reaction coordinate; TS, transition state. Free energy is in kilocalories per mole, and distances, including RC, are in Ångstroms.
The computed free energy barrier and reaction energies were 14.2 and 5.3 kcal/mol, respectively (Fig. 6B). Moreover, consistent with the very low (if any) residual activity observed for the E129A and E129Q mutants (Fig. 5A), we were not able to find a catalytic pathway for these mutants.
Finally, our calculations also support the electrostatic stabilizing role of R25 and, accordingly, the observed inactivity of the mutant R25A (SI Appendix, Fig. S5D).
Phosphoramidate Bond Chemistry and Consequences for pMV158 Lifestyle.
The phosphoramidate bond in phosphohistidine is thermodynamically less stable than the phosphoester bond in phosphotyrosine (37). Free phosphohistidine has a half-life of seconds in acidic solution and of a few minutes at pH 7. Such lability would impair the transfer of the protein-DNA adduct across the cell membranes to the receiving cell. However, in proteins, the stability of the P-N bond is influenced by the protein environment, in particular by the side chains surrounding the phosphohistidine at the active site (37, 38). For instance, a hydrogen bond to the nonphosphorylated imidazole nitrogen would make the phosphoramidate bond significantly less labile. There are two partially conserved residues, N23 and H30, that could establish a hydrogen bond with the H22 ND1 nitrogen once the adduct is formed. N23 is the residue adjacent to the catalytic histidine, and already makes a hydrogen bond to the H22 ND1 in the Nic0_B structure. H30 is located in a flexible loop after helix α1, where the catalytic histidine is located. This loop is disordered and could not be traced in most of our structures, except in the Nic0+SP structure, where H30 ND1 is located 4.4 Ǻ away from the H22 ND1 nitrogen, whereas a water molecule bridges the interaction. A structure of the MobM-His22-DNA adduct could clarify if these residues (or other) are critical in regulating the stability of the P-N bond. Unfortunately, so far we have not been able to crystallize such an adduct.
The physiological consequences of the use of a highly transferable phosphohistidine versus highly stable phosphotyrosine bond for the DNA transfer process in MOBV relaxases remain to be unveiled. However, we propose that MOBV1 relaxases lost the need for a tyrosine hydroxyl and evolved to use a different group for substrate attack because of the lowest pKa (6.0) of histidine among known nucleophiles, thus making histidine independent from deprotonating/activating auxiliary residues, which are, however, required for other nucleophiles (tyrosine pKa ∼10–11). From a physiological standpoint, we must consider that most MOBV relaxases are encoded by Firmicutes with low G+C% content, and that many of these bacteria tend to strongly acidify the media (to below pH 5.5), perhaps as a mechanism through which to outcompete other bacterial species. Furthermore, because these small plasmids (average size ∼5 kb) (21) lack partitioning systems, they need to achieve the average copy number (∼30 copies per genome equivalent) before cell division so that newborn cells should receive sufficient plasmid copies to guarantee stable inheritance (39). Having an easy-to-break relaxase-DNA covalent link would confer a selective advantage, allowing rapid reconstitution of dsDNA-replicating molecules. In this regard, it is worth mentioning that the RCR-replicase protein of plasmid pMV158 (RepB), although a tyrosine HUH endonuclease, generates unusually transient and difficult-to-capture protein-DNA intermediates (40).
An interesting difference between tyrosine and histidine relaxases is that the former is found in both Gram-positive and Gram-negative bacteria, whereas the newly described histidine relaxase, like MobM, appears to be found almost exclusively in the Gram-positive Firmicutes phylum (SI Appendix, Fig. S1). It has been suggested that relaxases of the MOBV family have been evolutionary selected by small/mobilizable plasmids (41). We can relate this idea to the high energy of the P-N bond between the scissile phosphate and the catalytic histidine, which makes the phosphate group readily transferable to other groups (37). In such a scenario, the use of a catalytic histidine could facilitate the DNA closing reaction; on the other hand, it also would limit the size of the DNA cargo, given that the P-N bond is prone to breakage if the DNA transfer takes too long. MOBV/Mob_Pre relaxases also are found in long plasmids, as in the case of closely related multidrug resistance plasmids pSK41 (46.4 kb) and pGO1 plasmid (54 kb), the latter being the prototype for class III staphylococcal plasmids (42). However, in these multidrug resistance/multirelaxase plasmids, which use MOBQ-type relaxases for conjugative transfer, the MOBV relaxase-encoding gene is located (together with aminoglycoside and bleomycin resistance genes) within a transposable 5-kb cassette-like module flanked by the IS257-mediated recombination elements. Whether they support alternative conjugative transfer events of the 5-kb integrant or the full plasmids remains to be explored.
Another characteristic chemical feature of the phosphoramidate bond is its acidic liability (37). The existence of MOBV/Mob_Pre nucleases almost exclusively in Gram-positive bacteria might be related to the fact that there is no maintenance of the pH homeostasis in the periplasm of Gram-negative bacteria; i.e., the periplasm pH matches that of the external environment (43). Therefore, conjugal DNA transfer mediated by a His relaxase in Gram-negative bacteria in an acidic environment could lead to the P-N bond exposure to low pH and breakage of the relaxase-DNA complex during the transfer through the T4SS spanning the periplasm. In contrast, in Gram-positive bacteria, conjugation is mediated by a cell wall hydrolase (44), which allows intimate cell-to-cell contacts without the need for exposure of the transferred DNA to extracellular low pH.
Conclusion
The work presented here provides a structure-function analysis of a representative member of the MOBV1 family of relaxases, for which no structure was known. Our combined X-ray crystallography, protein-DNA biochemistry, in vivo and computational approach has allowed us to describe a metal-dependent histidine nucleolytic catalysis, represented by the MobM relaxase, which is encoded by a promiscuous plasmid actively involved in the spread of antibiotic resistance. Homologs of MobM are found in many plasmids and other mobile genetic elements of pathogenic bacteria, including S. aureus (SI Appendix, Tables S1 and S2). MobM is a metal-dependent nuclease that uses histidine nitrogen for the nucleophilic attack on the scissile phosphate. Furthermore, in contrast to other DNA-processing enzymes, MobM is a histidine relaxase, a DNA-breaking and -joining enzyme, that operates through a phosphorus-nitrogen protein-DNA adduct for cell-to-cell DNA transfer.
Materials and Methods
Detailed descriptions of protein purification, relaxation and mobilization assays, crystallography, and theoretical calculations are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. T. Chan for the construction of plasmid pMV158H22A, L. Rodríguez for help with protein purification, the staff at the Platform for Automated Crystallization, and D. Aparicio and L. Martinelli for collecting the Nic0+1 diffraction data. The Institute for Research in Biomedicine Barcelona is the recipient of a Severo Ochoa Award of Excellence from the Spanish Ministry of Economy and Competitiveness (MINECO). The Molecular Biology Institute, Barcelona Spanish Research Council’s Structural Biology Unit is the recipient of a Maria de Maeztu Award of Excellence from MINECO. This work was supported by grants from MINECO (BFU2008-02372/BMC, CSD-2006-23, BFU2011-22588, CSD-2008/00013, BIO2013-49148-C2-2-R, BFU2014-53550-P, BIO2015-69085-REDC, and BIO2015-64802), the Generalitat de Catalunya (2014-SGR134 and 2014-SGR1530), and the European Commission [project no. 260644, ERC_SimDNA, and the H2020 program (MuG and BioExcel Projects)]. R.P. and H.G. received a La Caixa/Institute for Research in Biomedicine Barcelona International PhD and Juan de La Cierva fellowship, respectively. M.O. is a Catalan Institution for Research and Advanced Studies Academia researcher.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4LVI, 4LVJ, 4LVK, 4LVL, 4LVM, and 5N2Q).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702971114/-/DCSupplemental.
References
- 1.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol Rev. 2014;38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak RAF, Waldor MK. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 3.Lanza VF, Tedim AP, Martinez JL, Baquero F, Coque TM. The plasmidome of Firmicutes: Impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr. 2015;3:PLAS-0039-2014. doi: 10.1128/microbiolspec.PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- 4.Siefert JL. Defining the mobilome. Methods Mol Biol. 2009;532:13–27. doi: 10.1007/978-1-60327-853-9_2. [DOI] [PubMed] [Google Scholar]
- 5.Chan WT, Balsa D, Espinosa M. One cannot rule them all: Are bacterial toxins-antitoxins druggable? FEMS Microbiol Rev. 2015;39:522–540. doi: 10.1093/femsre/fuv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabezón E, Ripoll-Rozada J, Peña A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzo-Díaz F, et al. The MobM relaxase domain of plasmid pMV158: Thermal stability and activity upon Mn2+ and specific DNA binding. Nucleic Acids Res. 2011;39:4315–4329. doi: 10.1093/nar/gkr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler M, et al. Breaking and joining single-stranded DNA: The HUH endonuclease superfamily. Nat Rev Microbiol. 2013;11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinosa M. Plasmids as models for studying macromolecular interactions: The pMV158 paradigm. Res Microbiol. 2013;164:199–204. doi: 10.1016/j.resmic.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Gomis-Rüth FX, Coll M. Cut and move: Protein machinery for DNA processing in bacterial conjugation. Curr Opin Struct Biol. 2006;16:744–752. doi: 10.1016/j.sbi.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Gomis-Rüth FX, Solà M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 12.Llosa M, Gomis-Rüth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: A two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 13.Kramer MG, Khan SA, Espinosa M. Plasmid rolling circle replication: Identification of the RNA polymerase-directed primer RNA and requirement for DNA polymerase I for lagging strand synthesis. EMBO J. 1997;16:5784–5795. doi: 10.1093/emboj/16.18.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 15.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. The structure of the minimal relaxase domain of MobA at 2.1 Å resolution. J Mol Biol. 2007;366:165–178. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards JS, et al. Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus. Proc Natl Acad Sci USA. 2013;110:2804–2809. doi: 10.1073/pnas.1219701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boer R, et al. Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Guasch A, et al. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 19.Larkin C, et al. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Nash RP, Habibi S, Cheng Y, Lujan SA, Redinbo MR. The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1. Nucleic Acids Res. 2010;38:5929–5943. doi: 10.1093/nar/gkq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzo-Díaz F, Fernández-López C, Garcillán-Barcia MP, Espinosa M. Bringing them together: Plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective. Plasmid. 2014;74:15–31. doi: 10.1016/j.plasmid.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-López C, et al. Functional properties and structural requirements of the plasmid pMV158-encoded MobM relaxase domain. J Bacteriol. 2013;195:3000–3008. doi: 10.1128/JB.02264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-López C, et al. Nicking activity of the pMV158 MobM relaxase on cognate and heterologous origins of transfer. Plasmid. 2013;70:120–130. doi: 10.1016/j.plasmid.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 24.de Antonio C, Farías ME, de Lacoba MG, Espinosa M. Features of the plasmid pMV158-encoded MobM, a protein involved in its mobilization. J Mol Biol. 2004;335:733–743. doi: 10.1016/j.jmb.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Szpirer CY, Faelen M, Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coll M, Frederick CA, Wang AH, Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci USA. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W. Nucleases: Diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada Y, et al. Assignment of functional amino acids around the active site of human DNA topoisomerase IIalpha. J Biol Chem. 2000;275:24630–24638. doi: 10.1074/jbc.M003243200. [DOI] [PubMed] [Google Scholar]
- 33.Boer DR, et al. Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains. EMBO J. 2009;28:1666–1678. doi: 10.1038/emboj.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trask DK, DiDonato JA, Muller MT. Rapid detection and isolation of covalent DNA/protein complexes: Application to topoisomerase I and II. EMBO J. 1984;3:671–676. doi: 10.1002/j.1460-2075.1984.tb01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenzo-Díaz F, Espinosa M. Large-scale filter mating assay for intra- and inter-specific conjugal transfer of the promiscuous plasmid pMV158 in Gram-positive bacteria. Plasmid. 2009;61:65–70. doi: 10.1016/j.plasmid.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Shi T, et al. Mechanism for the autophosphorylation of CheA histidine kinase: QM/MM calculations. J Phys Chem B. 2011;115:11895–11901. doi: 10.1021/jp203968d. [DOI] [PubMed] [Google Scholar]
- 37.Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 38.Puttick J, Baker EN, Delbaere LT. Histidine phosphorylation in biological systems. Biochim Biophys Acta. 2008;1784:100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Novick RP. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 40.Moscoso M, Eritja R, Espinosa M. Initiation of replication of plasmid pMV158: Mechanisms of DNA strand-transfer reactions mediated by the initiator RepB protein. J Mol Biol. 1997;268:840–856. doi: 10.1006/jmbi.1997.1012. [DOI] [PubMed] [Google Scholar]
- 41.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caryl JA, O’Neill AJ. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the Staphylococci. Plasmid. 2009;62:35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. 2009. Cytoplasmic pH measurement and homeostasis in Bacteria and Archaea. Advances in Microbial Physiology, ed Robert KP (Academic Press, New York), Vol 55, pp 23–33. [DOI] [PubMed]
- 44.Arends K, et al. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J Bacteriol. 2013;195:4436–4444. doi: 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.