Significance
The distal end of the eukaryotic flagellum/cilium has critical functions, yet due to its small dimensions and association of tip structures with the axoneme is rather intractable to studying. We have developed biochemical approaches to identify a cohort of proteins specific for the flagellum tip structures. We sublocalized these proteins into individual structures. Using functional studies, we elucidated how the identified proteins contribute to the function of the flagella connector, the mobile membrane junction at the tip of the trypanosome flagellum.
Keywords: flagellar distal end, trypanosome, flagella connector, axonemal capping structure, structure immunoprecipitation
Abstract
The distal end of the eukaryotic flagellum/cilium is important for axonemal growth and signaling and has distinct biomechanical properties. Specific flagellum tip structures exist, yet their composition, dynamics, and functions are largely unknown. We used biochemical approaches to identify seven constituents of the flagella connector at the tip of an assembling trypanosome flagellum and three constituents of the axonemal capping structure at the tips of both assembling and mature flagella. Both tip structures contain evolutionarily conserved as well as kinetoplastid-specific proteins, and component assembly into the structures occurs very early during flagellum extension. Localization and functional studies reveal that the flagella connector membrane junction is attached to the tips of extending microtubules of the assembling flagellum by a kinesin-15 family member. On the opposite side, a kinetoplastid-specific kinesin facilitates attachment of the junction to the microtubules in the mature flagellum. Functional studies also suggest roles of several other components and the definition of subdomains in the tip structures.
Flagella and cilia are evolutionarily conserved organelles, which function in motility, signaling, and sensing. Malfunctions of flagella/cilia have been implicated in many inherited diseases and developmental abnormalities, including primary ciliary dyskinesia, Meckel–Gruber syndrome, Bardet–Biedl syndrome, and polycystic kidney disease (1). The core microtubule-based axoneme and the proximal basal body have been the focus of much study, including their ultrastructure, protein composition, and assembly in relation to the cell cycle, cell, and tissue development. In contrast, there is a dearth of information about the distal tip of the flagellum/cilium. This is the site of assembly of components delivered by the intraflagellar transport system (2), of localization of soluble signaling molecules (3, 4), and of distinct mechanical properties critical for flagellar beat (5). These unique tip features are reflected by a very discrete ultrastructure (6–9). However, we know little about how flagellum tip structures form during flagellum assembly, how they change postaxoneme assembly, or their molecular composition. There is only a single well-characterized protein (FAP256/CEP104) known to be related to structures at the distal end of the axoneme (10, 11).
Trypanosoma brucei is a unicellular flagellate that causes sleeping sickness in humans and nagana in animals. This parasite undergoes a complex life cycle during which it switches between the mammalian host and the tsetse fly insect vector. In T. brucei the flagellum is attached along most of its length to the cell body and is essential for cell motility, morphogenesis, and cell division (12, 13). Trypanosomes offer a superb system for understanding flagellum tip functions. Not only do they exhibit the general tip features outlined above, but also, during a single cell cycle, each cell assembles a new flagellum while maintaining the existing one. Hence, both the assembling and existing tip structures can be addressed simultaneously in the same cell.
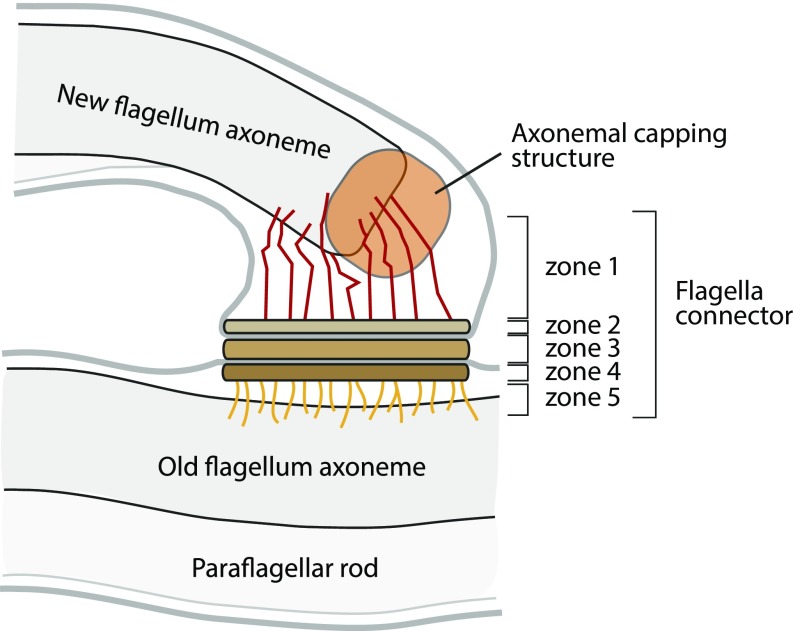
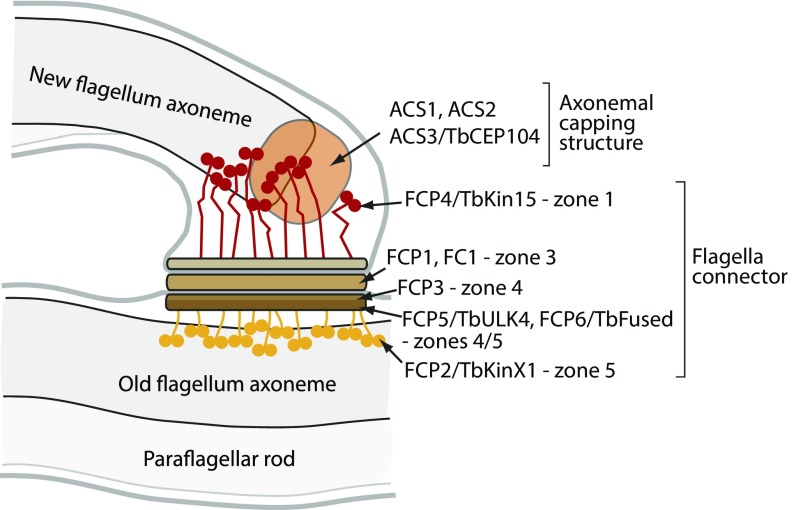
The assembling trypanosome flagellum tip has a flagella connector (FC), which is a mobile membrane junction linking it to the side of the old flagellum, an association that is implicated in inheritance of cell pattern (9, 14, 15). The FC has a complex structure and can be divided into five distinct zones (Fig. 1) (9, 15). Zone 1 in the new flagellum is composed of a filamentous network linking axonemal microtubule tips to zone 2—a discrete electron-dense layer underlying the new flagellum membrane at the membrane junction. Zone 3 comprises the new and old flagellum membranes at the junction plus the interstitial layer. Zone 4 is the discrete electron-dense layer underneath the old flagellum membrane, and zone 5 comprises a filamentous network linking zone 4 to microtubules of the old axoneme.
Fig. 1.
Schematics of the flagella connector and the new flagellum tip region.
The FC is formed very soon after formation of the transition zone (TZ), the junction between the basal body and the axoneme (9, 15). The FC then provides constant attachment of the extending new flagellum tip on the old flagellum while migrating along the old flagellum (14, 15). This migration is independent of new axoneme elongation (12, 16). The FC movement ceases when the structure reaches a “stop point” (16). Continued extension of the new flagellum is thus associated with a posterior migration of the new basal body, together with the associated kinetoplast (mitochondrial DNA), contributing to mitochondrial genome segregation (16). This robust flagella connection is severed by an unknown mechanism during the late stage of cytokinesis (15).
In addition to its relevance in the cell cycle of T. brucei, the FC is an experimentally useful structure making the trypanosome flagellum system particularly tractable for examination of the distal tip. Using this, we developed a proteomic approach that proved very powerful in identifying molecular constituents of discrete structures in the flagellum tip cytoskeleton. We identified tip proteins, some of which are specific to the tip of the new flagellum and others of which are shared between the new and old flagellum tips. We have further localized these proteins to individual tip structures. The precise localization, presence of protein domains of known functions, and, importantly, analyses of depletion phenotypes give a clear view of functions of the components and insights into redundancies in these complex cytoskeletal architectures.
Results
Identifying Flagella Connector Proteins.
To identify FC constituents, we used 2D difference gel electrophoresis and mass spectrometry (MS) to compare the protein composition of procyclic (tsetse midgut) form trypanosome flagellar cytoskeletons with those of bloodstream form trypanosomes, which do not possess an FC (15, 17). Proteins with a significantly higher abundance in the procyclic flagella were tagged with yellow fluorescence protein (YFP) and localized (18).
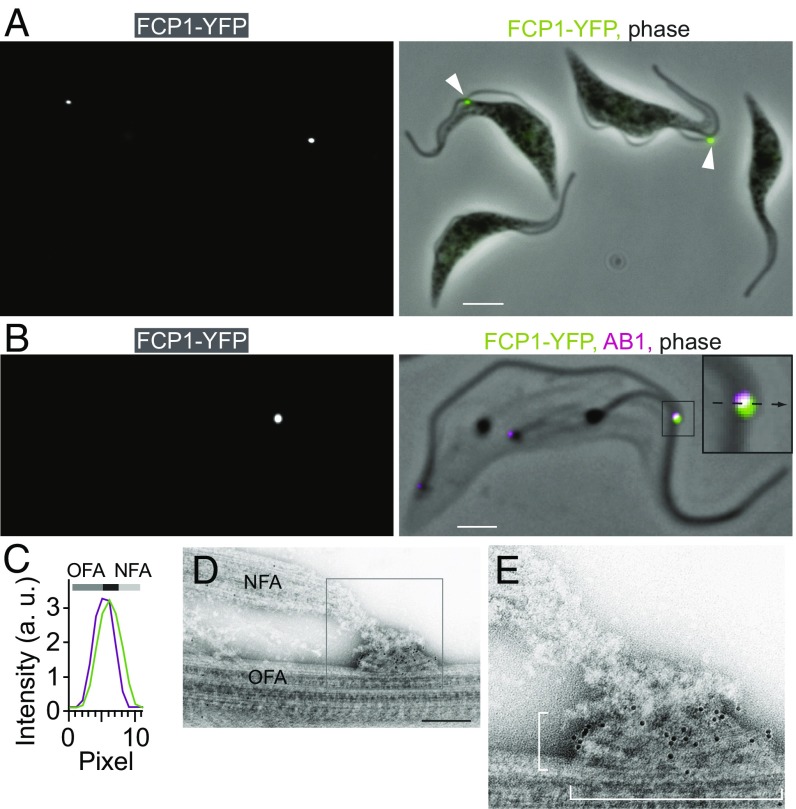
One protein, Tb927.8.940-YFP, was present as a dot at the new flagellum tip exclusively in cells with two flagella (2F cells) (Fig. 2A) (18). The signal was resistant to detergent treatment and overlapped with that of the AB1 monoclonal antibody, which marks a central component of the FC (zones 2–4) (15) (Fig. 2 B and C and SI Appendix, Table S1). To confirm the localization of Tb927.8.940-YFP to the FC and to sublocalize it, we performed immunogold labeling and transmission electron microscopy (TEM). We labeled detergent-treated negatively stained whole-mount cytoskeletons, so that many cells could be viewed and their cell-cycle status determined. In these samples, the filaments of zone 1 are clearly visible (Fig. 2 D and E). Zones 2–4 likely correspond to an ∼90-nm wide and 400-nm long tri-laminar core (TLC) (15). Following Tb927.8.940-YFP labeling, the majority of gold particles were found within the TLC (Fig. 2 D and E and SI Appendix, Table S2), which is in accord with Tb927.8.940 having two predicted transmembrane domains (for bioinformatics analysis, see SI Appendix, Table S3). We concluded that Tb927.8.940 is an FC-specific constituent and named it the flagella connector protein 1 (FCP1) (18).
Fig. 2.
Localization of FCP1 by light and electron microscopy. (A) A fluorescence image of FCP1-YFP–expressing cells (Left) merged with a phase-contrast image (Right, YFP signal in green), showing that the fusion protein localizes exclusively to the new flagellum tip in dividing 2F cells (arrowheads). (Scale bar, 5 μm.) (B) A fluorescence image of a cytoskeleton prepared from a 2F FCP1-YFP–expressing cell (Left) merged with a fluorescent image of AB1 antibody and with a phase-contrast image (Right, YFP signal in green, AB1 signal in magenta). (Inset) A higher magnification view of the new flagellum tip region. (Scale bar, 2 μm.) (C) A plot of YFP (green) and AB1 (magenta) intensities along a line running across the FC perpendicular to the old flagellum (dotted line in B Inset). The bars above the intensity plot indicate positions of the FC (black), the old flagellum axoneme (OFA, dark gray), and the new flagellum axoneme (NFA, light gray) in respect to the fluorescence signals. (D) TEM image of the new flagellum tip region in a negatively stained cytoskeleton of an FCP1-YFP–expressing cell. Immunogold labeling detecting YFP was performed. (Scale bar, 200 nm.) (E) A higher magnification image of the FC shown in D. Gold particles are concentrated in the TLC, the dimensions of which are indicated with brackets.
A Western blot with an anti-YFP antibody indicated that FCP1-YFP migrates at a lower molecular weight than predicted (∼120 kDa versus ∼260 kDa) (SI Appendix, Fig. S1), suggesting some proteolytic processing. However, proteomics provided evidence for the full-length FCP1 protein migrating at a higher molecular weight than the tagged one (SI Appendix, Fig. S1). Whether the truncated variant is a consequence of YFP tagging remains to be determined.
Structure Immunoprecipitation Approach Identifies Flagellum Tip Proteins.
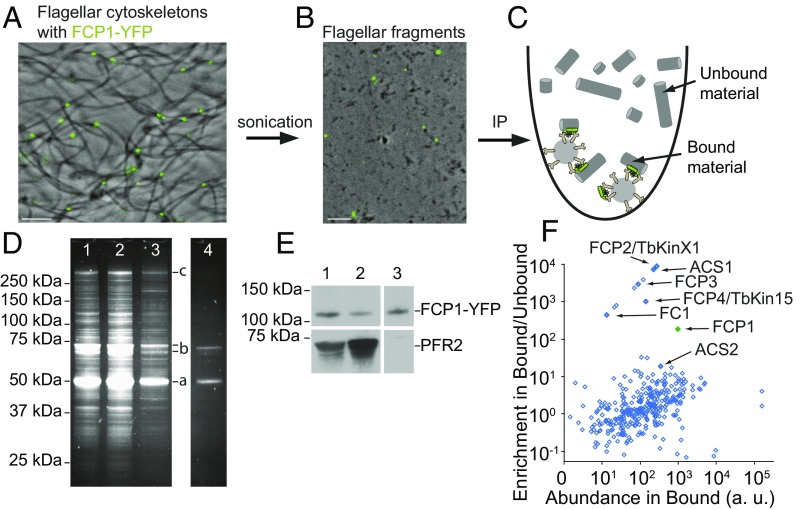
We developed a successful approach for identification of more tip proteins, which we termed structure immunoprecipitation (SIP). Flagellar cytoskeletons prepared from FCP1-YFP–expressing trypanosomes (Fig. 3A) were fragmented by sonication (Fig. 3B), and fragments containing YFP-tagged FCs were enriched by binding to anti-YFP antibody/magnetic beads (Fig. 3C). SDS/PAGE confirmed the specificity of the anti-YFP antibody interaction because the vast majority of proteins in the bound material were absent from the negative control, a similarly treated untagged cell line (Fig. 3D). The protein profile of the bound material was similar to the input material of sonicated flagellar cytoskeletons (Fig. 3D), consistent with cytoskeletal fragments rather than pure FCs being immunoprecipitated. However, Western blotting analysis confirmed the enrichment of FCP1-YFP in the bound material relative to the input (Fig. 3E), and this enrichment was even higher when comparison was made to the FC-depleted unbound material (Fig. 3E). Other FC constituents were expected to be revealed by similar enrichment patterns. In addition, due to an association of the FC with the distal end of the axoneme, the SIP approach may also reveal components of other flagellum tip structures.
Fig. 3.
Workflow of the structure immunoprecipitation approach used for identification of flagellum tip proteins. (A and B) Phase-contrast images of flagellar cytoskeletons from FCP1-YFP–expressing cells before (A) and after (B) sonication, merged with images of YFP signal (green). (Scale bars, 5 μm.) (C) Schematics showing that fragments of flagellar cytoskeletons with attached FCs are immunoprecipitated with anti-YFP magnetic beads (Bound material), whereas flagellar fragments without FCs do not interact with the beads (Unbound material). (D) An SDS/PAGE gel of the SIP experiment. (1) Input material (sonicated flagellar cytoskeletons). (2) Unbound material. (3) Bound material, which is a complex mix of flagellar proteins, including α- and β-tubulins (a), PFR1 and 2 (b), and dynein heavy chains (c). (4) Material recovered from anti-YFP beads incubated with flagellar fragments of a cell culture not expressing YFP. (E) Western blot analysis of the SIP experiment. (1) Input. (2) Unbound material. (3) Bound material. The bait FCP1-YFP was visualized using anti-YFP antibody (Top), and PFR2 was chosen as an example of an abundant flagellar protein, which is not an FC constituent (Bottom). In the bound material, the relative enrichment of FCP1-YFP (as estimated from the FCP1-YFP/PFR2 signal ratio) was 54-fold in respect to the input and 173-fold in respect to the unbound material. Note that the samples were loaded in different ratios on the SDS/PAGE gel and the Western blot. (F) A plot of the relative abundance of each protein species in the bound material (x axis) versus its relative enrichment in the bound material over the unbound material (y axis) as determined by MS. The proteins subsequently validated as localizing to the flagellum tip region are indicated. The bait protein FCP1 is in green.
To assess enrichment of protein species in the bound compared with the unbound material, we analyzed both fractions by label-free spectral index normalized quantitation MS (19, 20). Results of a single SIP experiment are summarized in Fig. 3F (see also Dataset S1, part 1). A total of 336 protein species were detected in the bound material. The bait protein FCP1 was enriched 219-fold. Ten proteins showed a higher enrichment, and a number of proteins showed a lower but significant enrichment.
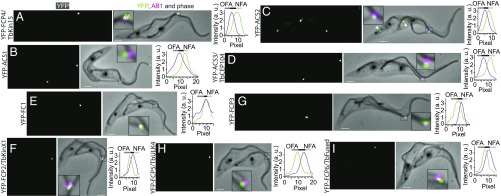
To validate these candidates, we expressed them as YFP-tagged fusion proteins in procyclic trypanosomes. Five of the top 10 most highly enriched candidates, and one from the group with a lower enrichment, localized to flagellar tips (Fig. 3F and SI Appendix, Fig. S2) with the YFP signals retained after detergent extraction (Fig. 4). This high hit rate was achieved with a single SIP experiment, showing the power of the approach. Four proteins were present exclusively at the new flagellum tip of dividing 2F cells. These proteins included Tb927.3.4960, a member of the kinetoplastid-specific kinesin-X1 clade (21), which we named FCP2/TbKinX1; Tb927.8.7540, a previously uncharacterized repetitive kinetoplastid-specific protein, FCP3; Tb927.10.890, a member of the ubiquitous kinesin-15 family (21), FCP4/TbKin15; and Tb927.11.1340, which was recently described as the FC constituent FC1 (22). In addition, two proteins without clear homology outside the Kinetoplastida, Tb927.7.6180, which we named axonemal capping structure 1 (ACS1), and Tb927.11.450, ACS2, were present at tips of both new and old flagella (Fig. 4). For bioinformatics analysis of these proteins, see SI Appendix, Table S3. The remainder of the tagged proteins localized to various structures other than flagellar tips (Dataset S1, part 1).
Fig. 4.
Proteins localizing to the new flagellum tip region. (Left and Middle) Fluorescence images of cytoskeletons prepared from cells expressing YFP-tagged proteins (Left) merged with fluorescent images of AB1 antibody staining and with phase-contrast images (Middle, YFP in green, AB1 in magenta). (Insets) A higher magnification view of the new flagellum tip region. (Scale bars, 2 μm.) (Right) Fluorescence intensities of YFP (green curves) and AB1 (magenta curves) were measured along a line running across the FC and perpendicular to the old flagellum, such as the dotted line in A, Inset. The bars above the intensity plots indicate positions of the FC (black), the old flagellum axoneme (OFA, dark gray), and the new flagellum axoneme (NFA, light gray). (A–D) Fluorescence signals of YFP-FCP4/TbKin15 (A), YFP-TbACS1 (B), YFP-ACS2 (C) and YFP-ACS3/TbCEP104 (D) were found closer to the new flagellum than the AB1 signal. Note that, in addition to the flagella tip signals, YFP-ACS2 displayed a rod-like signal alongside the flagella close to their base (arrowheads in C). (E) Fluorescent signal of YFP-FC1 often overlapped with the AB1 signal. (F–I) Fluorescent signals of YFP-FCP2/TbKinX1 (F), YFP-FCP3 (G), YFP-FCP5/TbULK4 (H), and YFP-FCP6/TbFused (I) were found closer to the old flagellum than the AB1 signal.
The 1 M NaCl incubation during flagellar cytoskeleton preparation may have solubilized less strongly associated FC constituents. We therefore developed a flagellar cytoskeleton preparation protocol using 0.2 M of NaCl and repeated the SIP experiment using FCP1-YFP as bait. All five proteins identified as specific for the new flagellum tip in the 1-M NaCl SIP experiment were again highly enriched (Dataset S1, part 2). However, a number of proteins likely to be false positives based on previous experimental or bioinformatics knowledge were also highly enriched. This may stem from an inefficient shielding of unspecific ionic interactions between solubilized proteins and axonemal microtubules at this lower salt concentration. We therefore compared our flagellum tip SIP data with SIP data from a control experiment targeting the tripartite attachment complex at the proximal end of the flagellum (23). A pattern emerged, whereby the new flagellum tip-specific proteins were detected exclusively in the bound material of the FCP1-YFP SIP (SI Appendix, Table S4). Two other previously uncharacterized proteins with an identical pattern also localized to the new flagellum tip of dividing 2F cells (SI Appendix, Fig. S2), with the YFP signals retained after detergent extraction (Fig. 4 and SI Appendix, Fig. S2). Tb927.1.2710 has no apparent homology outside the Kinetoplastida and was not studied further. Tb927.11.8150 is a member of the ubiquitous Unc-51–like kinase 4 (ULK4) family, and we refer to it as FCP5/TbULK4. The T. brucei genome contains a second gene coding for an ULK protein, Tb927.11.4470, a member of the Fused family (24). This FCP6/TbFused protein tagged with YFP was also found to localize specifically to the new flagellum tip of dividing cells (SI Appendix, Fig. S2 and Fig. 4).
Identified Proteins Localize to Different Subcompartments.
To determine the localization of the identified proteins within the tip region, we compared the position of their YFP signals with the position of the AB1 antibody signal, a marker for the middle (TLC) of the FC (15). In the following sections, we refer to the protein names for clarity, while recognizing that they are localized by YFP fusions. Unless indicated otherwise, we present data obtained using detergent-extracted cells, termed the cytoskeletons; these allow for a straightforward determination of the flagellar configuration, the cell-cycle status, and the morphology of the cellular cytoskeleton.
We observed that FCP4/TbKin15 localized between the AB1 signal and the new axoneme tip (Fig. 4A and SI Appendix, Table S1). Similarly, two proteins present at the tips of all flagella, ACS1 and ACS2, localized closer to the new axoneme tip than the AB1 signal did (Fig. 4 B and C and SI Appendix, Table S1). Tb927.10.14880, a trypanosome ortholog of FAP256/CEP104, identified bioinformatically as a reciprocal best BLAST hit for human CEP104, also displayed a similar localization pattern (Fig. 4D and SI Appendix, Table S1). We therefore named it ACS3/TbCEP104. The signal of FC1 often overlapped with that of AB1 (Fig. 4E and SI Appendix, Table S1). FCP2/TbKinX1, FCP3, FCP5/TbULK4, and FCP6/TbFused localized between the AB1 signal and the old axoneme (Fig. 4 F–I and SI Appendix, Table S1).
Immunogold EM Labeling Sublocalizes Proteins to Specific Tip Structures.
To determine more precisely the protein localization into tip structures, we performed EM immunogold labeling using anti-YFP antibodies on cell lines expressing fusion proteins.
FCP4/TbKin15 localized to the distal portion of the new axoneme (zone 1 of the FC) (Fig. 5A and SI Appendix, Table S2).
Fig. 5.
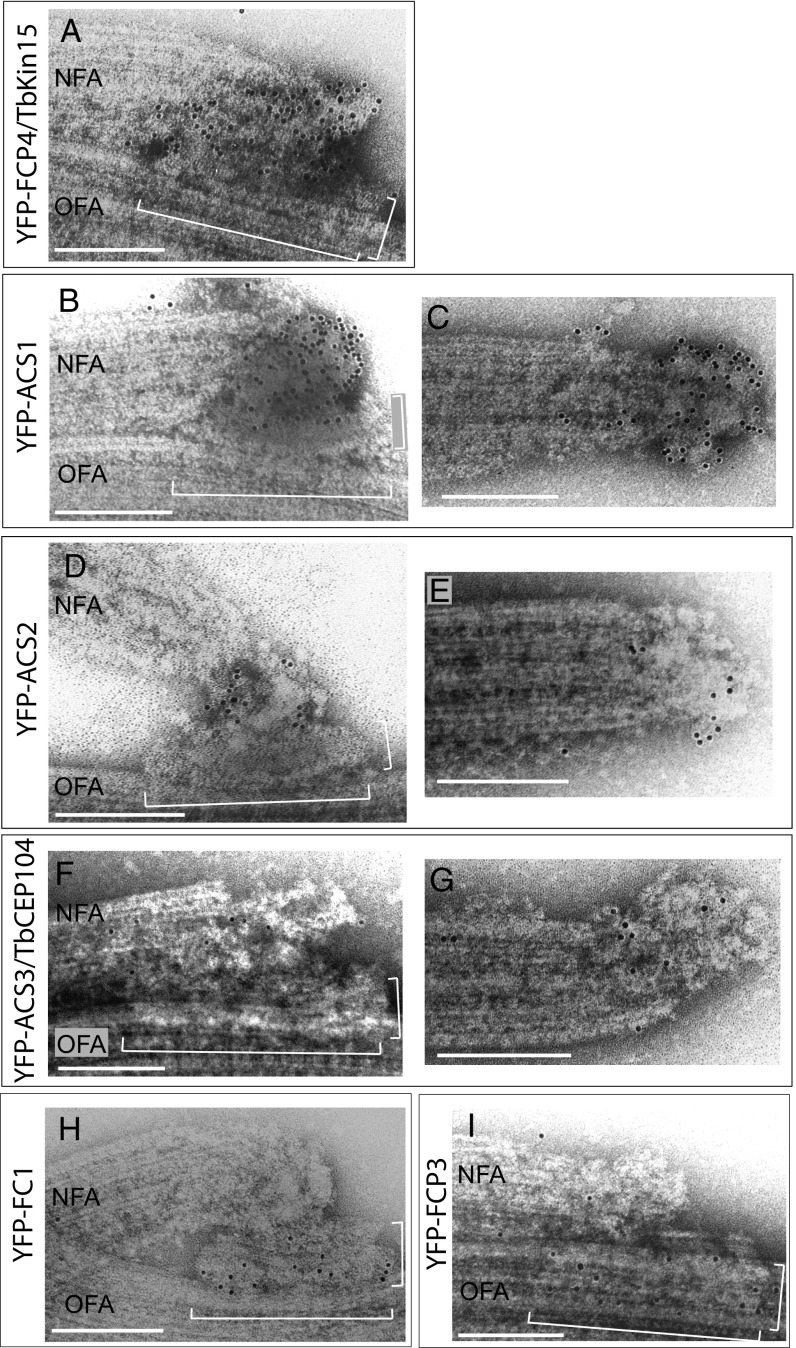
Immunogold sublocalization of identified proteins. TEM images of negatively stained cytoskeletons of cells expressing YFP-tagged proteins. YFP was detected with antibodies conjugated to 10-nm gold particles. (Scale bars, 200 nm.) (A, B, D, F, H, and I) New flagellum tip regions of the labeled cytoskeletons. The TLC is indicated with brackets. (C, E, and G) Flagellum tip regions of the labeled cytoskeletons of 1F cells. NFA, new flagellum axoneme; OFA, old flagellum axoneme.
In the new flagellum, ACS1 localized to a circular structure capping the distal end of the axoneme, the ACS (Fig. 5B and SI Appendix, Table S2). A strongly labeled ACS was also observed at tips of the old axoneme in 2F cells and of the axoneme in 1F cells (Fig. 5C). ACS2 was localized between the TLC and the tip of the new axoneme (Fig. 5D and SI Appendix, Table S2) and within the ACS of the old axoneme and the axoneme in 1F cells (Fig. 5E). Labeling for ACS3/TbCEP104 in the new flagellum tip region was weaker, making it difficult to localize it unambiguously. However, it appeared associated with the new axoneme, in particular with its tip (Fig. 5F and SI Appendix, Table S2). In nondividing 1F cells, the protein localized to the ACS (Fig. 5G).
Similar to FCP1 (Fig. 2E), FC1 and FCP3 were localized to the TLC (zones 2–4) (Fig. 5 H and I and SI Appendix, Table S2). We were unable to localize FCP2/TbKinX1, FCP5/TbULK4, and FCP6/TbFused by immunogold labeling. This may reflect inaccessibility of these proteins to the gold-conjugated antibody.
Tip Structures Assemble Early After Transition Zone Formation.
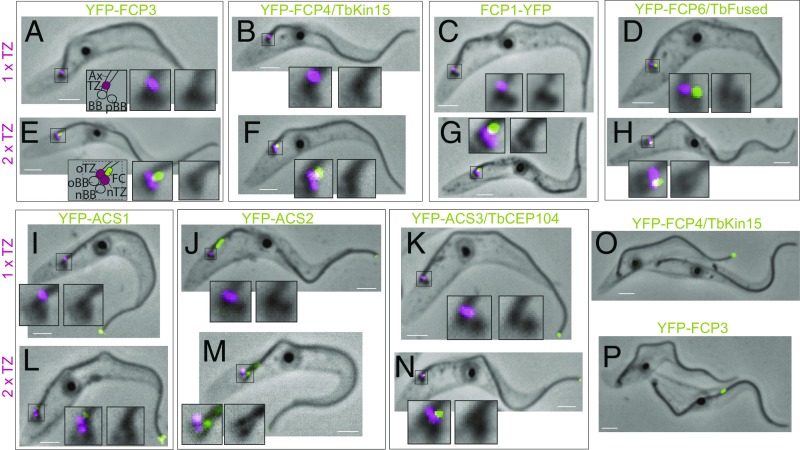
The start of new flagellum formation is one of the earliest visible events in the T. brucei cell cycle (25, 26). It initiates with maturation of the probasal body into the new basal body, followed by assembly of the new flagellum TZ, and then the flagellar axoneme (26). Using light microscopy and antibodies against the T. brucei TZ constituent protein FTZC (27), we were able to assess the timing of appearance of individual FC constituents in the new flagellum tip region.
There was no FCP3 or FCP4/TbKin15 signal observed in the basal body/probasal body region of cells with a single TZ (n = 100) (Fig. 6 A and B). In the case of FCP1, there was no signal in 94% of cells (n = 76 of 81) (Fig. 6C), with the remaining 6% of cells having a very faint signal. Finally, 20.2% (n = 44 of 217) of cells with a single TZ possessed a probasal or new basal-body–associated FCP6/TbFused signal (Fig. 6D). In 100% of cells with the second TZ recently formed on the new basal body (as judged by the absence of the new axoneme by phase-contrast imaging), FC constituents were always observed juxtaposed distal to the TZ (n > 20 for each protein shown in Fig. 6 E–H). This shows that, although some FC constituents may be present on the new basal body, the FC is fully assembled in the very earliest period after TZ formation.
Fig. 6.
Flagellum tip structure assembly and disassembly during cell cycle. (A–N) Phase-contrast images of cytoskeletons merged with the fluorescent signal of YFP-tagged proteins (green) and of the anti-FTZC antibody (magenta), a TZ marker. Insets represent an enlarged view of the basal body/probasal body region; the phase-contrast image (Right) and the merge with fluorescent images (Middle in A and E, Left in the other images) are shown. (Scale bars, 2 μm.) (A–D and I–K) Cells with a single TZ. The schematics in A indicates the positions of the axoneme (Ax), the transition zone (TZ), the basal body (BB), and the probasal body (pBB). (E–H and L–N) Cells with two TZ. The schematics in E indicates the position of the old transition zone (oTZ), the new transition zone (nTZ), the old flagellum basal body (oBB), the new flagellum basal body (nBB), and the flagella connector (FC). (O–P) Cytoskeletons of 2F cells in cytokinesis with their FCs severed. Phase-contrast images were merged with the signal of YFP (green). (Scale bars, 2 μm.) The YFP-FCP4/TbKin15 signal was retained at the tip of the new flagellum (O), and the YFP-FCP3 signal was retained on the old flagellum (P).
We observed that no ACS constituent signals were present in the basal body/probasal body region of cells with a single TZ (n > 60 cells for each protein) (Fig. 6 I–K). Upon formation of the new TZ, the signals were found juxtaposed distal to it in 71% of ACS1 (n = 10 of 14 cells), 94% of ACS2 (n = 16 of 17 cells), and 100% of ACS3/TbCEP104 (n = 16) tagged cells (Fig. 6 L–N), suggesting that the ACS is elaborated in the early period after TZ formation. The signal of ACS1 at the tip of short new flagella was low (Fig. 6L) and increased progressively as the new flagellum elongated, ultimately reaching the levels observed at the old flagellum tip (SI Appendix, Fig. S3A). The intensity of the ACS2 new flagellum tip signal was independent of the new flagellum length and twofold higher than that at the tip of the old flagellum (SI Appendix, Fig. S3B). Finally, ACS3/TbCEP104 intensities at the new and old flagellum tips were similar (SI Appendix, Fig. S3C).
Severing of the FC During Cytokinesis and Its Subsequent Disassembly.
Identification of constituents of different FC zones enabled us to examine where within the FC structure severing occurs during disconnection of the old/new flagella (15) and what happens to FC constituents thereafter.
In the rare cells undergoing cytokinesis with flagella disconnected at the FC, the FCP4/TbKin15 signal was retained at the new flagellum tip (n = 9 of 9 cells) (Fig. 6O), consistent with the protein constituting zone 1 of the FC. Furthermore, 10.4% (n = 35 of 338) of nondividing 1F cells possessed an FCP4/TbKin15 dot-like signal, invariably located at the flagellar tip (SI Appendix, Fig. S4A); these are likely daughters inheriting the new flagellum after a recent cell division event. This also implies that the protein is removed from the flagellar tip before construction of the new flagellum initiates.
In contrast, the signal of FCP3 was generally retained in the old flagellum following disconnection of the two flagella due to FC severing (7 cells versus 1 cell with the signal at the new flagellum tip) (Fig. 6P); this is consistent with the localization of FCP3 to the old flagellum (zone 4 and/or 5) (Fig. 4). The signal was positioned between 40 and 60% of the flagellum length, consistent with the position of the FC stop point (ref. 16 and our measurements of 2F cells in cytokinesis with connected flagella in SI Appendix, Fig. S4B). A small proportion of nondividing 1F cells (5.2%, n = 14 of 268 cells) also possessed a dot-like signal in a similar position (SI Appendix, Fig. S4 B and C). These cells likely inherited the old flagellum after a recent cell division event with an FC remnant remaining at the stop point for a limited period after cytokinesis. Similarly, 4% of 1F cells (n = 17 of 423 cells) possessed the FCP2/TbKinX1 signal on their flagellum, again in the position consistent with that of the FC stop point in 2F cells in cytokinesis (SI Appendix, Fig. S4B).
The behavior of the FC constituents indicated that the severing event occurred within zones 2–4. We therefore assessed the behavior of FCP1, which localizes to these zones. In the rare cells in cytokinesis with disconnected flagella, the FCP1 signal was found at the new flagellum tip (n = 3) or on the old flagellum side (n = 3) or was absent (n = 2). Thus, FC severing can occur on either side of the membrane junction. In accordance, 1F cells with the signal present either on the side (n = 9) or at the tip (n = 6) of their flagellum were present in the culture. The low frequency of these cells (< 1% of 1F cells) indicated rapid removal of the FCP1 from the FC remnant following severing.
Depletion of the FC Constituents Results in Precocious FC Severing.
We depleted individual proteins by inducible RNAi in the corresponding YFP-tagged background. Following RNAi induction for 72–96 h, the YFP signals were reduced in the majority of cells, often below a level detectable by direct fluorescence microscopy, suggesting effective knockdown (Fig. 7 A and B and SI Appendix, Fig. S5). In uninduced cultures, 2F cells with flagella not connected at the FC are rare and are restricted to a fraction of postmitotic cells undergoing cytokinesis (maximum 10% of all postmitotic cells, SI Appendix, Table S5). In contrast, depletion of four FC constituents individually—FCP1, FC1 (also reported in ref. 22), FCP2/TbKinX1, and FCP4/TbKin15—resulted in a substantial increase in cells with their new flagellum tip not in contact with the old flagellum (Fig. 7 A and B and SI Appendix, Fig. S5 and Table S5). The phenotype was observed in cells in all cell-cycle stages (SI Appendix, Table S5). It was, however, most prevalent in late cell-cycle stages, suggesting that it was caused by precocious FC severing rather than lack of FC formation.
Fig. 7.
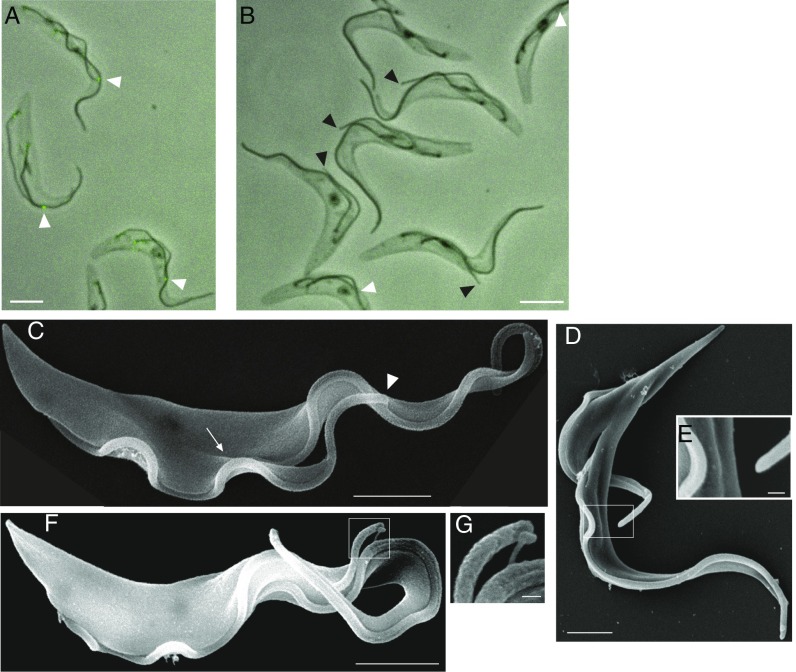
Knockdown of FCP2/TbKinX1 causes aberrant flagella connection. (A and B) Images of cytoskeletons prepared from cell cultures uninduced (A) or 96 h induced for FCP2/TbKinX1 RNAi (B). RNAi was performed in cells expressing YFP-FCP2/TbKinX1, and lack of YFP signal (green) following RNAi induction indicates efficient protein depletion. All displayed cells were precytokinesis. Black arrowheads denote tips of the new flagella not in contact with the old flagella; white arrowheads denote tips of the new flagella with functional FCs. (Scale bars, 5 μm.) (C–G) SEM images of cells with long new flagella. (Scale bars: 2 μm in C, D, and F and 200 nm in E and G.) (C) A cell in cytokinesis from a culture not induced for RNAi. The arrowhead indicates the tip of the new flagellum, which is in contact with the old flagellum; the arrow indicates the cleavage furrow. (D) A cell in late cytokinesis found in a culture not induced for RNAi. The tip of the new flagellum is not connected to the old flagellum. (E) An enlarged view of the new flagellum tip of the cell in D. (F) A precytokinesis cell with depleted FCP2/TbKinX1. The tip of the new flagellum is not in direct contact with the old flagellum. (G) An enlarged view of the new flagellum tip of the cell in F. Note that a thin tube emanating from a rectangular structure at the new flagellum tip connects it to the old flagellum.
The aberrant flagella connection was most frequent upon depletion of FCP2/TbKinX1 (SI Appendix, Table S5), occurring in 10.1% of 2F cells with a single kinetoplast and a nucleus (n = 51 of 503 cells), in 38.0% of 2F cells with two kinetoplasts and a nucleus (n = 78 of 205), in 53.8% of mitotic cells (n = 84 of 156), and in 67.3% of postmitotic cells (n = 66 of 98). However, RNAi induction had no clear impact on culture growth rates (SI Appendix, Fig. S6A), nor on the proportion of cells in different cell-cycle stages (SI Appendix, Fig. S6B). Affected postmitotic cells appeared otherwise morphologically normal (SI Appendix, Fig. S6C). Similarly, 1F cells from these cultures did not exhibit morphological abnormalities (SI Appendix, Fig. S6D).
We examined the possibility that the FC was functional in living cells, but was severed in the process of sample preparation, indicative of FC weakening. First, we imaged live cells by light microscopy. In uninduced cultures, no 2F cells with long new flagella not connected to the old ones were observed (n = 24), including cells in cytokinesis (SI Appendix, Fig. S7A). In contrast, 45.2% of cells (n = 19 of 42) with long new flagella had no visible flagella connection in RNAi-induced cultures (SI Appendix, Fig. S7B).
Next, we used scanning electron microscopy (SEM) to image whole cells fixed directly in growth medium (without any detergent). In uninduced cultures, all cells with long new flagella before cytokinesis (n = 12) and the majority of cells in cytokinesis (n = 22 of 26) retained a functional FC (Fig. 7C) with unconnected flagella observed only in cells with a very prominent cleavage furrow (Fig. 7D). In these cells, the tip of the new flagellum had no link to the old one (n = 4) (Fig. 7E). In RNAi-induced cultures, SEM confirmed the presence of 2F cells without a cleavage furrow in which the new flagellum tip was not in close contact with the old flagellum (Fig. 7F). Intriguingly, in the majority of these cells, a thin tube linked the new flagellum tip to the side of the old flagellum (n = 19 of 29) (Fig. 7F). The tube originated from a rectangular structure, which protruded above the membrane close to the new flagellum tip (Fig. 7G). It is noteworthy that the aberrant flagella connection did not affect the new flagellum positioning on the cell surface (Fig. 7F).
To address whether the presence of 2F cells with functional FCs in the RNAi-induced culture was due to an incomplete RNAi penetrance, we constructed a cell line with a knockout of both alleles coding for FCP2/TbKinX1. The proportion of 2F cells with a functional FC in both cultures was comparable (SI Appendix, Table S5), indicating that precocious FC severing likely operated in only some cells because of a complex local protein environment and functional redundancies.
Depletion of FCP3, FCP5/TbULK4, FCP6/TbFused, and ACS constituents by RNAi had no apparent effect on the FC, which was severed in a timely manner (SI Appendix, Fig. S5 and Table S5). Depletion of FCP6/TbFused, however, led to the occurrence of “double” cells, i.e., two cells connected by a thick cytoskeletal bridge at their posterior ends (SI Appendix, Fig. S8). Although of a low frequency (3% of all cells, n = 16 of 518), this phenotype is significant, because we did not observe similar cells in wild-type cultures. These cells were likely products of an uncompleted cytokinesis and resemble a phenotype observed in flagella motility mutants (28, 29).
Depletion of FC Constituents Reveals Function of Individual Proteins.
To study the cause of the aberrant flagella connection in RNAi-induced cultures, we imaged the new flagellum tip region in cytoskeletons by negative staining TEM. To exclude cells not affected by RNAi, we performed immunogold labeling for the respective YFP-tagged protein targeted by RNAi. Importantly, the vast majority of cytoskeletons in RNAi-induced cultures were devoid of gold labeling (above the background level) (Fig. 8), confirming specificity of labeling and RNAi effectiveness.
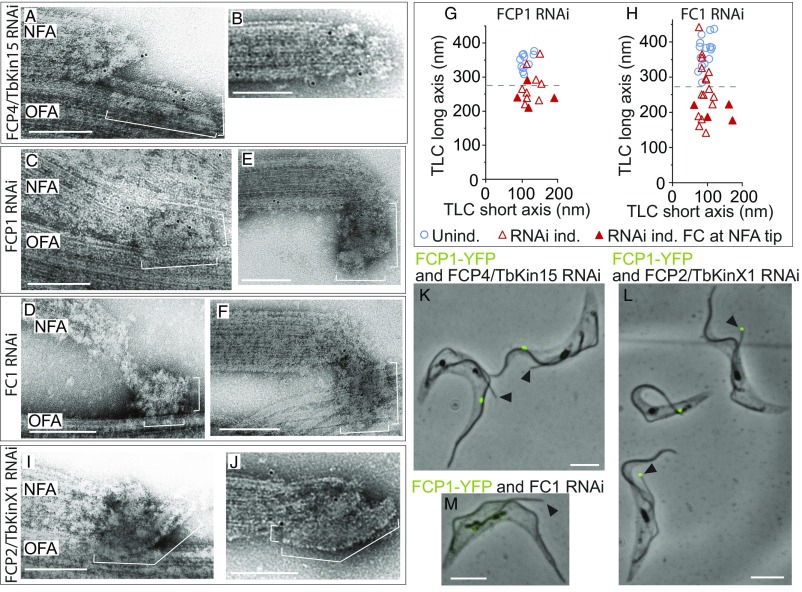
Fig. 8.
Depletion phenotypes reveal function of FC constituents. (A–F, I, and J) TEM images of cytoskeletons prepared from 2F cells depleted of individual FC constituents by RNAi. The cells originally expressed a YFP-tagged variant of the targeted protein; hence lack of specific gold anti-YFP labeling indicates efficient protein depletion. The brackets indicate long and short axes of the TLC. (Scale bars, 200 nm.) (A) The new flagellum tip region in a cell depleted of FCP4/TbKin15. The TLC is present but the filaments connecting it to the tip of the new axoneme are mostly absent. (B) The new axoneme tip not in contact with the old axoneme in a cell depleted of FCP4/TbKin15. There is no TLC-like structure attached to the axoneme tip. (C and D) The new flagellum tip region in cells depleted of FCP1 (C) or FC1 (D). The organization and dimensions of the TLC are aberrant. (E and F) The new axoneme tip not in contact with the old axoneme in cells depleted of FCP1 (E) or FC1 (F). A structure reminiscent of the aberrant TLC (in C and D) is present. (G and H) Plots of the length of the TLC long axis versus its short axis measured in cytoskeletons from cultures uninduced (blue circles) or induced (red triangles) for RNAi against FCP1 (G) or FC1 (H). Open triangles indicate FCs connected to both axonemes; full triangles indicate FCs present at the tip of the new axoneme but not in contact with the old one. Dashed lines indicate 280 nm, which is the smallest dimension of the long axis observed in uninduced cells. For both FCP1 and FC1, the TLC long axis difference between induced and uninduced cultures is statistically significant at P < 0.05. (I) The new flagellum tip region in a cell depleted of FCP2/TbKinX1. The TLC is bent, and a significant part of it is not in contact with the old axoneme. (J) In some FCP2/TbKinX1-depleted cells, a bent but otherwise structurally normal TLC is present at the new axoneme tip, but the FC is not in contact with the old axoneme. NFA, new flagellum axoneme; OFA, old flagellum axoneme. (K–M) Merges of phase-contrast images with fluorescence images (green) of cytoskeletons prepared from cell cultures expressing FCP1-YFP and depleted of FCP4/TbKin15 (K), FCP2/TbKinX1 (L), or FC1 (M). Black arrowheads denote tips of the new flagella not in contact with the old flagella. (Scale bars, 5 μm.)
Depletion of FCP4/TbKin15 caused the filaments in zone 1 linking the TLC to the new axoneme tip to become less conspicuous (Fig. 8A), whereas the structure and dimensions of the TLC were unchanged (Fig. 8A and SI Appendix, Fig. S9). In cells with unconnected axonemes (n = 11), no FC-like structures were observed either at the tip of the new axoneme (Fig. 8B) or on the old axoneme. Clearly, in the absence of the flagella connection, the position of the FC along the old axoneme cannot be easily anticipated.
Depletion of TLC-localizing FCP1 or FC1 resulted in the loss of structural organization in the TLC (Fig. 8 C and D). Its long axis was often shorter than 280 nm (Fig. 8 C, D, G, and H), which was never observed in uninduced cells (Fig. 8 G and H). Structures of a similar, reduced length were also found at tips of the new axonemes unconnected to the old ones due to FCP1 or FC1 depletion (Fig. 8 E–H).
Depletion of FCP2/TbKinX1 compromised the attachment of the TLC to the old axoneme. The TLC was either partially detached (Fig. 8I) or not in contact with the old axoneme (Fig. 8J). In these cases, the association with the new axoneme tip was typically preserved (n = 14 versus 1 cell without an FC at the new axoneme tip) (Fig. 8J). The TLC displayed its typical trilaminar organization and in most cases normal dimensions (SI Appendix, Fig. S9).
Depletion of the remaining FC and ACS constituents, which did not lead to an aberrant FC activity, had no effect on the organization and dimensions of the TLC (SI Appendix, Fig. S9) nor on its attachment to the two axonemes. Thus, an aberrant FC activity in particular RNAi-induced cultures is linked to constituent-specific changes in FC ultrastructure.
Importantly, EM imaging also revealed the function of the key mechanistic players of the FC. To test this quantitatively, we created cell lines allowing for inducible RNAi knockdown of individual proteins in the background of YFP-tagged FCP1 (a positional marker localizing between the old and the new flagellum). We observed that all 2F cells with unconnected axonemes due to depletion of FCP4/TbKin15 (n = 35) possessed the YFP-labeled membrane junction on the side of the old axoneme (Fig. 8K), consistent with this protein constituting the linkage between the junction and the tip of the new axoneme microtubules. All 2F cells with unconnected axonemes due to depletion of FCP2/TbKinX1 (n = 60) possessed the YFP-labeled junction at the tip of the new axoneme (Fig. 8L), showing that this protein attaches the junction to the side of the microtubules in the old axoneme. Interestingly, knockdown of FC1 caused the FCP1-YFP signal to diminish in all 2F cells, including cells with unconnected axonemes (n = 20) (Fig. 8M), indicating a dependency in recruitment of the two junction-localizing proteins to the FC and providing an explanation for the similarity of their depletion phenotypes.
Discussion
The SIP Approach: Identifying Constituents of Discrete Cytoskeletal Structures.
Biochemistry of higher-order cytoskeletal structures represents a considerable challenge due to their insoluble nature. Because purification of individual structures is difficult, we devised a simple biochemical approach based on their enrichment. The enrichment pattern efficiently identified FC constituents, although these proteins would have never been considered as likely candidates based on the abundance criterion typifying a purification approach. Moreover, the majority of these proteins were not detected in previous flagellar fractions; hence their substantial enrichment before MS analysis appears fundamental for their detection. We recently used a similar approach for identification of constituents of the trypanosome TZ (30). Hence, the SIP approach is highly successful for analyzing discrete protein structures and regions of the cytoskeleton of cells. It adds to the existing repertoire of techniques, such as the proximity-dependent biotinylation approach (31).
The FC Is a Membrane Junction Attached by Two Types of Kinesins to the Cytoskeleton of Two Flagella.
We identified seven constituents of the FC, including the FC1 protein (22). Defining the localization of these proteins has enabled a molecular description of the individual zones of the FC as diagrammed in Fig. 9. This information, together with bioinformatics and depletion studies, provides a mechanistic insight into the function of individual constituents in the FC.
Fig. 9.
Schematics of the new flagellum tip with the model of the FC.
FCP4/TbKin15, a member of the ubiquitous kinesin-15 family of microtubule plus-end–directed motors (32), links the plus ends of extending new axoneme microtubules to the membrane junction in zone 1. Motifs responsible for ATP binding and hydrolysis are well conserved in the motor domain of FCP4/TbKin15 (SI Appendix, Fig. S10), so the kinesin is likely capable of active motility. The force exerted by the kinesin on the microtubule ends, together with the lack of various axonemal complexes in this part of the axoneme, may explain the microtubule disorganization observed in the distal axonemal region of a growing T. brucei flagellum (33). In plants, kinesin-15s were shown to attach plus ends of dynamic microtubules to the middle region of the phragmoplast, the key structure for plant cytokinesis (34). Hence, the kinesin-15 family may have evolved specifically to provide a linkage and maintain an alignment between cellular structures and dynamic ends of microtubules.
The proteins FCP1, FC1, and FCP3 were localized to the TLC, corresponding to zones 2–4. FCP1 contains predicted membrane domains and is therefore the best candidate for a constituent of the interstitial zone 3 with its intraflagellar domains also contributing to zone 2 and/or 4. FC1 facilitates incorporation of FCP1 into the FC, possibly through direct interactions. FCP3 segregates into the old flagellum following FC severing, suggesting that it is a constituent of zone 4.
Zones 4 and/or 5 also contain two evolutionarily conserved kinases. They belong to ULK families already implicated in flagellum-related functions in animals (35, 36) with the mammalian ULK Fused demonstrated to be essential for construction of motile cilia (35). Plant orthologs of both kinases were shown to be critical for the phragmoplast organization, resembling the function of plant kinesin-15s (37, 38). Moreover, Arabidopsis Fused was shown to interact with the kinesin-15s (39), and the involved motifs are conserved in FCP6/TbFused (SI Appendix, Fig. S11), indicating that the Fused kinase–kinesin-15 module is evolutionarily ancient. ULK4 from plants is enzymatically inactive and possesses a microtubule-binding activity (37). Such an activity would be advantageous in the context of the FC. Another evolutionarily conserved kinase, polo-like kinase, was previously also shown to localize, among other structures, to the FC (40).
The kinetoplastid-specific kinesin FCP2/TbKinX1 in zone 5 attaches the junction to the old axoneme. The kinesin is also likely to contribute to the movement of the junction along the axoneme, as analysis of the FCP2/TbKinX1 sequence suggests that it is ATP- hydrolysis-competent (SI Appendix, Fig. S10). Our work shows that it is not essential for FC motility. Likewise, none of the other suggested sources of FC motility are necessary; the FC is motile in cells depleted of key intraflagellar transport system constituents, such as IFT88 (12), with the cells showing both a deficient intraflagellar transport in the old flagellum (41) and the absence of the new axoneme extension (12). A defective flagellar beating did not cause morphological phenotypes expected for an immotile FC, e.g., formation of flagellar loops (42). Possibly, several of these processes contribute to the FC function with individual processes having a more prominent role when the others fail.
We discovered that the FC is a remarkable molecular machine with its principal mechanistic components being two types of kinesins functionally cross-connected by, and operating across, a membrane junction. Due to the asymmetry of the FC the two motors fulfill fundamentally different roles. They will no doubt differ in their biochemical activities required to produce a balanced movement and render the connection between the beating flagella robust, flexible, and dynamic.
The ACS Contains Kinetoplastid-Specific and Evolutionarily Conserved Proteins.
CEP104 orthologs are present at the tips of flagella/cilia in diverse organisms, such as trypanosomes, Chlamydomonas, and mammals (this work and ref. 10), despite significant variance in morphology of the structures capping their axonemal microtubules. This points toward the involvement of CEP104 in fundamental processes that operate at flagellar tips. However, in contrast to Chlamydomonas and mammalian cells (10), depletion of ACS3/TbCEP104 in trypanosomes has no gross effect on either the presence or the length of the flagellum (SI Appendix, Fig. S5). This indicates either functional redundancies in the trypanosome ACS or an intrinsic difference between a flagellum that is maintained through many cell cycles versus one that is renewed every cell cycle.
Two other T. brucei ACS constituents that we identified, ACS1 and ACS2, are unique for Trypanosoma and for trypanosomatids, respectively. Recently, another trypanosomatid-specific protein, FLAM8, was shown to localize to the distal region of the axoneme, including tips of axonemal microtubules (43), and may also be an ACS constituent. Clearly, a subset of ACS constituents is of a restricted evolutionary distribution, explaining why comparative genomic approaches failed to identify them, and possibly accounting for the morphological variations between capping structures (44).
Tip Structure Modulation and the Cell Cycle.
The behavior of tip structure constituents implies that the tip structures are assembled very early during flagellum elongation at the onset of axoneme assembly (Fig. 6). This is consistent with EM analyses of growing cilia in other organisms (45, 46). The sequence of structure assembly may reflect an ordering of protein availability; examining the trypanosome whole-genome mRNA profiling data by Archer et al. (47) revealed that levels of mRNAs coding for tip structure constituents consistently peak before those of axonemal proteins in the cell cycle.
In trypanosomes, the new flagellum extends whereas the length of the old flagellum does not change (48, 49). Subunits for axoneme assembly must be preferentially imported into the new flagellum, requiring a mechanism that differentiates between the two flagella. We show that the new flagellum must also attract the ACS constituents and the new flagellum FC constituent FCP4/TbKin15. Moreover, a targeting mechanism for the old flagellum must also exist, providing for the selective import of FCP2/TbKinX1 and FCP3. As the material imported into a flagellum is recruited via the basal body and controlled by the TZ, proteins specific for the old or the new basal body/TZ structures are likely to account for the specificity. These proteins are currently unknown, but biochemical approaches, such as the SIP, may identify them in the future.
In cells depleted of individual FC constituents, the FC appears initially functional, making it difficult to determine the role of the structure during early stages of flagellum elongation. As these cells progress through the cell cycle, the FC deteriorates, which may be caused by forces imposed on the FC by beating flagella. This appears to be the process that displaces the membrane junction and pulls out a membrane tube when the attachment to the old axoneme has been compromised upon depletion of FCP2/TbKinX1 (Fig. 7G). Such forces perhaps underlie severing of the junction in late cytokinesis in wild-type cells, or precociously, if the junction has been weakened by depletion of its constituent, as observed in cells knocked down for FC1 (22). Notably, this aberrant or absent flagella connection does not affect the new flagellum pattern on the cell surface (this work and ref. 22), providing evidence for a dominant role of the flagellum attachment zone in flagellum positioning (50).
FC constituents are quickly removed post cytokinesis. This distinguishes them from the ACS constituents, which are always present at the axonemal tip. Therefore, there have to be mechanisms for selective removal of proteins and remodeling of the flagellar tip.
Understanding tips of flagella and cilia should be facilitated by the combination of the SIP approach and the distal tip constituents identified in this work. It is likely that studies in different organisms will give a coordinated view of the evolutionary cell biology of this important part of a fascinating cellular organelle.
Materials and Methods
Cell Growth and Preparation of Genetically Modified Cell Cultures.
Trypanosoma brucei brucei procyclic cells of the strain SmOxP927 (51) were grown at 28 °C in SDM-79 medium (Gibco) supplemented with 10% FCS (52).
YFP-tagged proteins were expressed from endogenous loci. Construction of the FCP1-YFP–expressing cell line was done as described (18). Other candidate proteins were N-terminally tagged with YFP (for details on tagging, see Dataset S1, part 3).
RNAi knockdown cell lines were prepared by cloning a gene-specific sequence into an appropriate vector (Dataset S1, part 3). The FCP2/TbKinX1 knockout cell line was prepared by inserting the sequences immediately upstream and downstream of the Tb927.3.4960 ORF into pJ1014 and pJ1015 vectors (gift of Jack Sunter, Sir William Dunn School of Pathology, University of Oxford, Oxford) (Dataset S1, part 3), which were used to knock out each of the two protein-coding alleles, respectively. Vectors or PCR products were transfected into trypanosomes following a standard protocol (53). RNAi was induced by the addition of doxycycline (0.5 μg/mL final) to the medium. Reagents were purchased from Sigma unless stated otherwise.
Two-Dimensional Difference Gel Electrophoresis Comparison of Procyclic Versus Bloodstream Flagellar Cytoskeletons.
The procedure was done as described in detail in ref. 18. Briefly, 1.5 × 108 of flagellar cytoskeletons prepared from procyclic and bloodstream cells using 1 M NaCl (see below) were labeled with a different cyDye (GE Healthcare). Samples were pooled and proteins were separated by 2D electrophoresis. Eighteen-centimeter pH3-11 nonlinear IPG strips (GE Healthcare) were used for the first dimension focusing, and second-dimension separation was performed using 10% wt/vol acrylamide/bisacrylamide SDS/PAGE. Protein spots were visualized in each spectral channel, and those of a significantly higher abundance in the procyclic sample were excised, digested with trypsin, and analyzed on a 4800 MALDI-TOF-TOF (Applied Biosystems) at the Central Proteomics Facility of Sir William Dunn School of Pathology, University of Oxford. Data were searched against a custom T. brucei protein database based on T. brucei genome version 2.2 (Tritrypdb.org).
Structure Immunoprecipitation Approach.
Flagellar cytoskeleton preparation using 1 M NaCl.
Flagellar cytoskeletons were prepared from FCP1-YFP–expressing procyclic cells using 1% Igepal CA-630 and 1 M NaCl and subsequently were fragmented by sonication. The sonicated material (input) was incubated with anti-GFP antibodies bound to Dynabeads (Thermo Fisher Scientific). The beads were pelleted, and the supernatant (the unbound material) was retained for further analysis. The bound material was eluted from the beads by incubating with a buffer containing 0.3% SDS. The fractions were analyzed with an LTQ XLOrbitrap mass spectrometer (Thermo Scientific). Peptides were identified by searching MS/MS spectra against a custom protein database based on T. brucei genome version 2.2 using the Central Proteomics Facility Pipeline (20). Data from individual slices were pooled, and protein abundance was estimated using the SINQ tool (20). For the detailed protocol, see SI Appendix.
Flagellar cytoskeleton preparation using 0.2 M NaCl.
For the FC SIP experiment, cytoskeletons were prepared from FCP1-YFP–expressing procyclic trypanosome cells as in the 1-M NaCl procedure. For the control SIP of the tripartite attachment complex, cytoskeletons were prepared from YFP-TAC102–expressing cells (54). The cytoskeletons were briefly sonicated to break cellular microtubules, which was further facilitated by incubation on ice. Resulting flagellar cytoskeleton fragments were further fragmented by additional sonication. The subsequent immunoprecipitation step and sample analysis were performed similarly to the 1-M NaCl procedure (for details, see SI Appendix).
Immunofluorescence Staining.
Procyclic cells settled on microscope slides were fixed in −20 °C methanol or, to extract cytoskeletons, incubated in 1% Igepal CA-630 and fixed. The mouse monoclonal AB1 antibody and the rabbit polyclonal anti-FTZC and anti-ClpGM6 (for the flagellum attachment zone length measurements) antibodies were used following published protocols (13, 15, 27). For more details, see SI Appendix.
Light Microscopy.
Phase-contrast and fluorescence images of fixed cells and cytoskeletons were acquired using a Leica DM5500B microscope with a Leica 100× (N.A. 1.4) HCX PL APO oil immersion objective and an Orca-ER (Hamamatsu) or a Neo 5.5 sCMOS (Andor) digital camera. The images were acquired in Leica Application Suite Software. Length measurements and quantification of fluorescence signals were performed using ImageJ software (55).
Electron Microscopy.
TEM.
Cells settled onto formvar-coated nickel mesh grids (Agar Scientific) were extracted with 1% Igepal CA-630 and labeled with anti-GFP antibody (Invitrogen A11122) and secondary goat anti-rabbit antibody conjugated with 10 nm gold (Sigma G7402). Samples were fixed in 2.5% glutaraldehyde, washed, and stained with 1% aurothioglucose (UPS Reference Standard). The grids were allowed to dry before imaging them on a Tecnai12 TEM (FEI). For more details, see SI Appendix.
SEM.
Cells were fixed in medium with 2.5% (vol/vol) glutaraldehyde for 2 h at room temperature with mild shaking (30 rpm). The fixed cells were centrifuged at 500 × g for 10 min, washed with PBS, and resuspended in PBS. The cells were settled onto coverslips, dehydrated with increasing concentrations of ethanol, critical-point-dried using an Autosamdri-815 critical point dryer (Tousimis), sputter-coated with gold using a Q150R Rotary-Pumped Sputter Coater/Carbon Coater (Quorum Technologies), and imaged using a JSM-6390 SEM (JEOL).
Supplementary Material
Acknowledgments
We thank Bungo Akiyoshi and Bill Wickstead for help with development of SIP; Samuel Dean, Jack Sunter, Richard Wheeler, and Anneliese Hoffmann for their help, Torsten Ochsenreiter and Frederic Bringaud for reagents; members of the K.G. laboratory for discussions; and Benjamin Thomas, Svenja Hester, and Gabriela Ridlova from the Central Proteomics Facility at Sir William Dunn School of Pathology for sample analysis. The EM sample preparation and imaging were done in the EM facility at the Sir William Dunn School of Pathology; we thank Errin Johnson for assistance with the EM work. Work in the K.G. laboratory is funded by the Wellcome Trust (WT066839MA and 104627/Z/14/Z). V.V. was supported by a Long Term Fellowship from the European Molecular Biology Organization and by the Czech Science Foundation (GA CR) Junior Project 16-26444Y and is a holder of the J. E. Purkyne Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, www.ebi.ac.uk/pride (dataset identifiers PXD006777 and PXD006761).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703553114/-/DCSupplemental.
References
- 1.Fliegauf M, Benzing T, Omran H. When cilia go bad: Cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saada EA, et al. Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei Flagellar membrane. Eukaryot Cell. 2014;13:1064–1076. doi: 10.1128/EC.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolley D, Gadelha C, Gull K. Evidence for a sliding-resistance at the tip of the trypanosome flagellum. Cell Motil Cytoskeleton. 2006;63:741–746. doi: 10.1002/cm.20159. [DOI] [PubMed] [Google Scholar]
- 6.Dentler WL, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sale WS, Satir P. The termination of the central microtubules from the cilia of Tetrahymena pyriformis. Cell Biol Int Rep. 1977;1:45–49. doi: 10.1016/0309-1651(77)90008-x. [DOI] [PubMed] [Google Scholar]
- 8.Dalen H. An ultrastructural study of the tracheal epithelium of the guinea-pig with special reference to the ciliary structure. J Anat. 1983;136:47–67. [PMC free article] [PubMed] [Google Scholar]
- 9.Höög JL, et al. 3D architecture of the Trypanosoma brucei flagella connector, a mobile transmembrane junction. PLoS Negl Trop Dis. 2016;10:e0004312. doi: 10.1371/journal.pntd.0004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satish Tammana TV, Tammana D, Diener DR, Rosenbaum J. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J Cell Sci. 2013;126:5018–5029. doi: 10.1242/jcs.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezabkova L, Kraatz SHW, Akhmanova A, Steinmetz MO, Kammerer RA. Biophysical and structural characterization of the centriolar protein Cep104 interaction network. J Biol Chem. 2016;291:18496–18504. doi: 10.1074/jbc.M116.739771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes P, et al. Modulation of a cytoskeletal calpain-like protein induces major transitions in trypanosome morphology. J Cell Biol. 2014;206:377–384. doi: 10.1083/jcb.201312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira-Leite FF, Sherwin T, Kohl L, Gull K. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science. 2001;294:610–612. doi: 10.1126/science.1063775. [DOI] [PubMed] [Google Scholar]
- 15.Briggs LJ, et al. The flagella connector of Trypanosoma brucei: An unusual mobile transmembrane junction. J Cell Sci. 2004;117:1641–1651. doi: 10.1242/jcs.00995. [DOI] [PubMed] [Google Scholar]
- 16.Davidge JA, et al. Trypanosome IFT mutants provide insight into the motor location for mobility of the flagella connector and flagellar membrane formation. J Cell Sci. 2006;119:3935–3943. doi: 10.1242/jcs.03203. [DOI] [PubMed] [Google Scholar]
- 17.Hughes L, Towers K, Starborg T, Gull K, Vaughan S. A cell-body groove housing the new flagellum tip suggests an adaptation of cellular morphogenesis for parasitism in the bloodstream form of Trypanosoma brucei. J Cell Sci. 2013;126:5748–5757. doi: 10.1242/jcs.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portman N. 2011. Deconstructing the trypanosome cytoskeleton: From structures to functions via components and complexes. PhD thesis (University of Oxford, UK)
- 19.Griffin NM, et al. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat Biotechnol. 2010;28:83–89. doi: 10.1038/nbt.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trudgian DC, et al. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics. 2011;11:2790–2797. doi: 10.1002/pmic.201000800. [DOI] [PubMed] [Google Scholar]
- 21.Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAllaster MR, et al. Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol Biol Cell. 2015;26:3013–3029. doi: 10.1091/mbc.E15-04-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones NG, et al. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS Pathog. 2014;10:e1003886. doi: 10.1371/journal.ppat.1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: Timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 26.Vaughan S, Gull K. Basal body structure and cell cycle-dependent biogenesis in Trypanosoma brucei. Cilia. 2016;5:5. doi: 10.1186/s13630-016-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bringaud F, et al. Characterization and disruption of a new Trypanosoma brucei repetitive flagellum protein, using double-stranded RNA inhibition. Mol Biochem Parasitol. 2000;111:283–297. doi: 10.1016/s0166-6851(00)00319-4. [DOI] [PubMed] [Google Scholar]
- 28.Branche C, et al. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci. 2006;119:3443–3455. doi: 10.1242/jcs.03078. [DOI] [PubMed] [Google Scholar]
- 29.Ralston KS, Lerner AG, Diener DR, Hill KL. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell. 2006;5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean S, Moreira-Leite F, Varga V, Gull K. Cilium transition zone proteome reveals compartmentalization and differential dynamics of ciliopathy complexes. Proc Natl Acad Sci USA. 2016;113:E5135–E5143. doi: 10.1073/pnas.1604258113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morriswood B, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryot Cell. 2013;12:356–367. doi: 10.1128/EC.00326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boleti H, Karsenti E, Vernos I. Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- 33.Höög JL, et al. Modes of flagellar assembly in Chlamydomonas reinhardtii and Trypanosoma brucei. eLife. 2014;3:e01479. doi: 10.7554/eLife.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YR, Liu B. Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr Biol. 2000;10:797–800. doi: 10.1016/s0960-9822(00)00564-9. [DOI] [PubMed] [Google Scholar]
- 35.Wilson CW, et al. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, et al. Ulk4 is essential for ciliogenesis and CSF flow. J Neurosci. 2016;36:7589–7600. doi: 10.1523/JNEUROSCI.0621-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krupnova T, et al. Microtubule-associated kinase-like protein RUNKEL needed [corrected] for cell plate expansion in Arabidopsis cytokinesis. Curr Biol. 2009;19:518–523. doi: 10.1016/j.cub.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Oh SA, et al. A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr Biol. 2005;15:2107–2111. doi: 10.1016/j.cub.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 39.Oh SA, et al. Arabidopsis fused kinase and the Kinesin-12 subfamily constitute a signalling module required for phragmoplast expansion. Plant J. 2012;72:308–319. doi: 10.1111/j.1365-313X.2012.05077.x. [DOI] [PubMed] [Google Scholar]
- 40.Ikeda KN, de Graffenried CL. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J Cell Sci. 2012;125:3173–3184. doi: 10.1242/jcs.101162. [DOI] [PubMed] [Google Scholar]
- 41.Fort C, Bonnefoy S, Kohl L, Bastin P. Intraflagellar transport is required for the maintenance of the trypanosome flagellum composition but not its length. J Cell Sci. 2016;129:3026–3041. doi: 10.1242/jcs.188227. [DOI] [PubMed] [Google Scholar]
- 42.Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 43.Subota I, et al. Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub-localization and dynamics. Mol Cell Proteomics. 2014;13:1769–1786. doi: 10.1074/mcp.M113.033357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisch C, Dupuis-Williams P. Ultrastructure of cilia and flagella: Back to the future! Biol Cell. 2011;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 45.Dentler WL. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- 46.Portman RW, LeCluyse EL, Dentler WL. Development of microtubule capping structures in ciliated epithelial cells. J Cell Sci. 1987;87:85–94. doi: 10.1242/jcs.87.1.85. [DOI] [PubMed] [Google Scholar]
- 47.Archer SK, Inchaustegui D, Queiroz R, Clayton C. The cell cycle regulated transcriptome of Trypanosoma brucei. PLoS One. 2011;6:e18425. doi: 10.1371/journal.pone.0018425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyler KM, Matthews KR, Gull K. Anisomorphic cell division by African trypanosomes. Protist. 2001;152:367–378. doi: 10.1078/1434-4610-00074. [DOI] [PubMed] [Google Scholar]
- 49.Bastin P, MacRae TH, Francis SB, Matthews KR, Gull K. Flagellar morphogenesis: Protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol Cell Biol. 1999;19:8191–8200. doi: 10.1128/mcb.19.12.8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunter JD, Varga V, Dean S, Gull K. A dynamic coordination of flagellum and cytoplasmic cytoskeleton assembly specifies cell morphogenesis in trypanosomes. J Cell Sci. 2015;128:1580–1594. doi: 10.1242/jcs.166447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poon SK, Peacock L, Gibson W, Gull K, Kelly S. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol. 2012;2:110037. doi: 10.1098/rsob.110037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brun R, Schönenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 53.McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 54.Trikin R, et al. TAC102 is a novel component of the mitochondrial genome segregation machinery in Trypanosomes. PLoS Pathog. 2016;12:e1005586. doi: 10.1371/journal.ppat.1005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.