Significance
Data with a small number of study individuals and a large number of potential explanatory features arise particularly in genomics. Existing methods of analysis result in a single model, but other sparse choices of explanatory features may fit virtually equally well. Our primary aim is essentially a set of acceptable simple representations. The method allows the assessment of anomalies, such as nonlinearities and interactions.
Keywords: sparse effects, genomics, statistical analysis
Abstract
Data with a relatively small number of study individuals and a very large number of potential explanatory features arise particularly, but by no means only, in genomics. A powerful method of analysis, the lasso [Tibshirani R (1996) J Roy Stat Soc B 58:267–288], takes account of an assumed sparsity of effects, that is, that most of the features are nugatory. Standard criteria for model fitting, such as the method of least squares, are modified by imposing a penalty for each explanatory variable used. There results a single model, leaving open the possibility that other sparse choices of explanatory features fit virtually equally well. The method suggested in this paper aims to specify simple models that are essentially equally effective, leaving detailed interpretation to the specifics of the particular study. The method hinges on the ability to make initially a very large number of separate analyses, allowing each explanatory feature to be assessed in combination with many other such features. Further stages allow the assessment of more complex patterns such as nonlinear and interactive dependences. The method has formal similarities to so-called partially balanced incomplete block designs introduced 80 years ago [Yates F (1936) J Agric Sci 26:424–455] for the study of large-scale plant breeding trials. The emphasis in this paper is strongly on exploratory analysis; the more formal statistical properties obtained under idealized assumptions will be reported separately.
Suppose that an outcome, for example disease status or survival time, is measured on a limited number of individuals and that a large number of potential explanatory variables are available. Standard statistical methods such as least squares regression or, for binary outcomes, logistic regression need modification, essentially to take account of an assumption necessary for progress, namely of sparsity, that only a limited number of the explanatory variables have an effect. Important methods have been developed in which, for example, a least squares criterion is suitably penalized, based on the number of explanatory variables included. See, for example, ref. 1 and, for a careful account of the underlying mathematical theory, ref. 2. The outcome of such analyses is a single regression-type relation. For a very recent discussion from a different perspective and under strong assumptions, see ref. 3. The formal probabilistic behavior of the procedure in this paper under idealized conditions will be discussed in a separate paper.
This paper adopts a different, less formal, and more exploratory approach in which judgment is needed at various stages. In this the conclusion is typically that a number of different simple models fit essentially equally well and that any choice between them requires additional information, for example new or different data or subject-matter knowledge. That is, informal choices are needed at various points in the analysis. Although the choices could be reformulated into a wholly automatic procedure this has not been done here.
The combinatorial arrangements used in the method are essentially partially balanced incomplete block designs (4), in particular so-called cubic and square lattices, first developed in the context of plant breeding trials involving a very large number of varieties from which a small number are to be chosen for detailed study and agricultural use.
Outline of Method
Consider the analysis of data from independent individuals on each of which a large number, , of explanatory variables is measured together with a single outcome, . To be specific, consider analyses based on linear least squares regression. In typical applications might be roughly 100 and perhaps 1,000 or more. The idea is to begin with a large number of least squares analyses each involving a much smaller number, , of variables. The procedure in outline is as follows:
-
•
Some variables, for example, intrinsic variables such as gender, might be included in all of the regressions described below and others entered several or many times because of a prior assessment of their importance.
-
•
Arrange the variables either in a square or a cube, where preferably . Extensions to four or more dimensions are possible. We describe here the cubic case. It is immaterial if some positions in the cube are empty or if some rows, columns, and so on have more than entries, so that there is no loss of generality in the restriction of , say, to be a perfect cube.
-
•
The rows, the columns, and so forth of the cube form sets each of variables. Fit a least squares regression to each set.
-
•
From each such component analysis select a small number of variables for further study. This might be the two variables with most significant effect, or all those variables, if any, that had Student statistics exceeding some arbitrary threshold, for example the 5% point of a formal test.
-
•
Thus, each explanatory variable has been examined three times, always in the presence of a different set of explanatory variables. Those variables never selected or selected only once should, in the absence of strong prior counter evidence, be discarded. The next step depends on the number of variables remaining as selected twice or three times. If, say, is ∼100, a second phase similar to the first, probably based on representing as a square, should be used, aiming to reduce the number of potentially important explanatory variables to perhaps roughly 10 to 20.
-
•
The next phase involves more detailed analysis of the selected variables. Their correlation matrix should be calculated and for any pair of variables with a correlation exceeding, say 0.90, the corresponding scatter plot should be examined. Depending on the nature of the pair of variables, it may be decided to omit one or to replace the pair by the average of their standardized values or to proceed with both. For each of the selected variables that is not binary a regression should be fitted with a single squared term added and a probability plot produced of the corresponding statistics. Anomalous points should be checked and, for example, if necessary the corresponding variables transformed. Next, the linear by linear interactions of pairs of variable should be checked in a similar way. See, for example, ref. 5.
-
•
The final phase of the analysis is to find very small sets of variables that give adequate fit. Suppose discussion has been reduced to explanatory variables including possible interaction terms, squared terms, and so on. Provided is sufficiently small, a sensibly cautious approach is to fit all models and reject those clearly inconsistent with the data. This might be done, for example, through a likelihood ratio test against the model involving all candidate variables. It is implicit that if a model involving a subset of explanatory variables is consistent with the data, so too is a model involving any larger subset . This reduces the computational burden of the search to that of finding primitive subsets.
-
•
The computational demands of the procedure are small once the relevant code is written. Code is available from the authors upon request.
Illustration of Method
We illustrate by example how the procedure might be used and interpreted in practice, emphasizing exploratory aspects and the need for careful judgment at various stages.
Description of Data.
In a study of osteoarthritis, a set of 106 patients clinically and radiographically diagnosed with primary symptomatic osteoarthritis at multiple joint sites were selected for gene expression analysis alongside 33 healthy controls (6). Samples from each patient were subjected to transcriptional profiling using microarrays containing probes for over 48,800 genes. The raw gene expression data, scored on a positive scale, are available from the Gene Expression Omnibus under accession number GDS5363. Data on the males, one from the cases and nine from the controls, are discarded, leaving a sample of 129 females.
Analysis.
We arrange the variable indices in a hypercube and fit a linear logistic model to the log-transformed explanatory variables by maximum likelihood, using the sets of variables indexed by each dimension of the hypercube; 2,531 variables are classified as significant at the 1% level in at least three of the five analyses in which they appear. We arrange the corresponding variable indices in a hypercube and repeat the procedure twice more, successively reducing the number of potential candidate explanatory variables to 779, 66, and, finally, 17. We do not put forward our choices of significance level and the dimension of the initial hypercube as definitive; significance tests are used informally as an aid to interpretation and are calibrated to reduce the number of candidate explanatory variables to roughly 15 to 20.
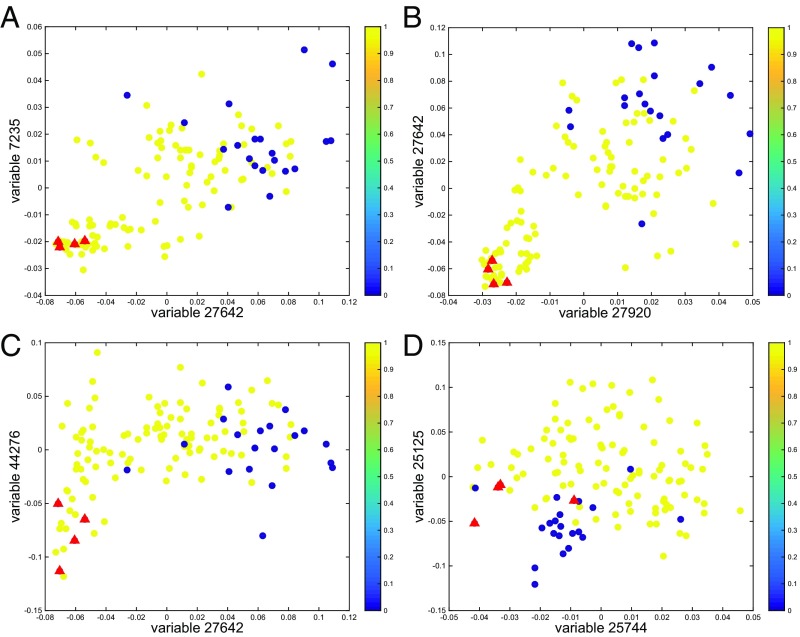
For each pair among the 17 potential candidate explanatory variables we fit a logistic model using the log-transformed variables and interaction terms between them. For all pairs of variables whose statistics exceed 2 in absolute value, scatter plots check the plausibility of the interaction. We simultaneously check whether anomalous points in different plots correspond to the same individuals. Fig. 1 displays the retained interactions and anomalous controls. The anomalous individuals are consistently anomalous across variable pairs and are discarded from the subsequent analysis. Allowing for interactions, the resulting set of candidate explanatory variables consists of 17 variables on the log scale and interactions between four pairs of them. SI Appendix, Tables S1–S15 detail many models of reasonable dimension whose fit is not significantly worse than that of the model fitted to all candidate explanatory variables, where significance is measured using an test at the 1% level.
Fig. 1.
Outcome (1 = cases, 0 = controls) as a function of the two potentially interacting variables (logarithmic scale). Four anomalous individuals in the control group are represented by red triangles.
Among the variables identified, 33385 and 46771 are identified also by ref. 6 as being highly differentially expressed between cases and controls. The biological descriptions of all variables appearing in SI Appendix, Tables S1–S15 are provided in Table 1 together with the proportion of models containing each variable.
Table 1.
Variable numbers (proportion of models in SI Appendix, Tables S1–S15 containing this variable), gene names, and biological function of the retained candidate variables
| Variable no. (occurrence rate) | Gene name | Description and biological function |
| 7235 (0.96) | ESYT2-007 | Tethers the endoplasmic reticulum to the cell membrane; plays a role in FGF signalling and may play a role in cellular lipid transport |
| 48433 (0.94) | LTBP1 | Latent transforming growth factor beta binding protein; diseases associated with LTBP1 include geleophysic dysplasia |
| 25125 (0.75) | PRR5L | Associates with the mTORC2 complex that regulates cellular processes including survival and organization of the cytoskeleton |
| 29679 (0.61) | — | mRNA |
| 48415 (0.61) | RP11-542K23.10 | RNA gene |
| 25744 (0.61) | NDEL1 | Plays a role in multiple processes including cytoskeletal organization, cell signaling and neuron migration, outgrowth, and maintenance |
| 27642 (0.53) | SRFBP1 | Serum response factor binding protein; may play a role in biosynthesis and/or processing of SLC2A4 in adipose cells |
| 45991 (0.33) | MAZ | MYC-associated zinc-finger protein |
| 36409 (0.31) | SERTAD1 | Stimulates E2F1/TFDP1 transcriptional activity |
| 48549 (0.29) | COL9A2 | Collagen type IX alpha 2 chain; mutations in this gene are associated with multiple epiphyseal dysplasia |
| 44276 (0.27) | GLS | Plays an essential role in generating energy for metabolism |
| 33385 (0.26) | LFNG | Encodes evolutionarily conserved glycosyltransferases; mutations in this gene have been associated with autosomal recessive spondylocostal dysostosis 3 |
| 37443 (0.22) | WDR20 | Regulates the activity of the USP12-UAF1 deubiquitinating enzyme complex |
| 46771 (0.19) | PLAGL2 | Zinc-finger protein that recognizes DNA and/or RNA |
| 27920 (0.18) | ANKRD24 | Protein coding gene |
| 25470 (0.14) | SPEN | Encodes a hormone inducible transcriptional repressor |
| 11643 (0.08) | NAT10 | Protein coding gene with numerous biological functions |
Gene function information was obtained from GeneCards, www.genecards.org.
There are compact messages to be extracted from SI Appendix, Tables S1–S15. Of all models specified, 96% involve the variable 7235 (ESYT2-007) and 94% involve the variable 48433 (LTBP1); 78% of models not involving variable 48433 instead contain variable 48549 (COL9A2). In fact, only 1% of all models involve neither 48433 nor 48549. It is notable, given the nature of osteoarthritis, that ESYT2-007 plays a role in fibroblast growth factor signaling essential for bone development and that mutations in this gene have been associated with various congenital bone diseases (7). LTBP1 has been associated with geleophysic dysplasia, an inherited condition characterized by abnormalities involving the bones and joints. Mutations in COL9A2 have been associated with multiple epiphyseal dysplasia, a disorder of cartilage and bone development. The most commonly occurring interaction term is between variables 25744 (NDEL1) and 25125 (PRR5L). We do not know whether this interaction is biologically interpretable.
For comparison, we fit a logistic model to all variables, the latter measured on a log scale. The lasso penalty is used. Although the number of variables selected by the lasso depends on the degree of penalization imposed, the smallest set of selected variables able to achieve the same negligible residual deviance as the models specified in SI Appendix, Tables S1–S15 has cardinality 9. The intersection of this set with the set of 17 variables in Table 1 is empty, although one of the nine variables, 41799, which corresponds to the gene H3F3B, is identified in ref. 6 as being highly differentially expressed between cases and controls. The discrepancy is attributable to the fact that many of the representations detailed define separating hyperplanes, achieving arbitrarily good fit for arbitrarily large regression coefficients. Because the norm penalty of the lasso does not admit such solutions, a lasso model of the same dimension as any of those presented makes classification errors and has worse fit. Incidentally, note that the lasso was conceived as an approximation to another subsets selection estimator (8), which unfortunately is computationally infeasible.
Conclusion
The approach here is that if there are alternative reasonable explanations of the data one should aim initially to specify as many as is feasible. This view is in contraposition to that implicit in the use of the lasso (9) and similar methods, from each of which there results a single model. Specification of reasonable alternative explanations requires judgment, in particular in the assessment of anomalies, such as nonlinearities and interactions. Here we have used significance tests as an informal guide. The essence of our approach is exploratory, leaving full interpretation to detailed subject-matter discussion.
Supplementary Material
Acknowledgments
This work was supported by an Engineering and Physical Sciences Research Council Fellowship (to H.S.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703764114/-/DCSupplemental.
References
- 1.Hastie T, Tibshirani R, Wainright M. Statistical Learning with Sparsity: The Lasso and Generalizations. CRC; Boca Raton, FL: 2015. [Google Scholar]
- 2.van de Geer S. Estimation and Testing Under Sparsity. Springer; Cham, Switzerland: 2016. [Google Scholar]
- 3.Martin R, Mess R, Walker S. Empirical Bayes posterior concentration in sparse high-dimensional linear models. Bernoulli. 2017;23:1822–1847. [Google Scholar]
- 4.Yates F. A new method of arranging variety trials involving a large number of varieties. J Agric Sci. 1936;26:424–455. [Google Scholar]
- 5.Cox DR, Wermuth N. Tests of linearity, multivariate normality and the adequacy of linear scores. J Appl Stat. 1994;43:347–355. [Google Scholar]
- 6.Ramos YFM, et al. Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann Rheum Dis. 2014;73:1844–1853. doi: 10.1136/annrheumdis-2013-203405. [DOI] [PubMed] [Google Scholar]
- 7.Su N, Jin M, Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: Learning from mouse models. Bone Res. 2014;2:14003. doi: 10.1038/boneres.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donoho D. For most large underdetermined systems of equations, the minimal -norm near-solution approximates the sparsest near-solution. Commun Pure Appl Math. 2006;59:907–934. [Google Scholar]
- 9.Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Stat Soc B. 1996;58:267–288. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.