Significance
Approximately 34% of triple-negative breast cancer (TNBC) patients relapse with local recurrence or metastasis within 5 y of radiation and chemotherapy treatment. There are currently no targeted therapies to treat TNBC. This study offers therapeutic targets for blocking TNBC metastasis: cell-surface antigen CUB-domain containing protein 1 (CDCP1) and proteins in the lipid metabolism pathway. CDCP1 regulates lipid metabolism by reducing cytoplasmic lipid droplet abundance, stimulating fatty acid oxidation and oxidative phosphorylation. This metabolic pathway likely contributes to the energy production required for cell migration and metastasis of TNBC, and represents a potential therapeutic target.
Keywords: CDCP1, lipid droplets, FAO, metastasis, TNBC
Abstract
Triple-negative breast cancer (TNBC) is notoriously aggressive with high metastatic potential, which has recently been linked to high rates of fatty acid oxidation (FAO). Here we report the mechanism of lipid metabolism dysregulation in TNBC through the prometastatic protein, CUB-domain containing protein 1 (CDCP1). We show that a “low-lipid” phenotype is characteristic of breast cancer cells compared with normal breast epithelial cells and negatively correlates with invasiveness in 3D culture. Using coherent anti-Stokes Raman scattering and two-photon excited fluorescence microscopy, we show that CDCP1 depletes lipids from cytoplasmic lipid droplets (LDs) through reduced acyl-CoA production and increased lipid utilization in the mitochondria through FAO, fueling oxidative phosphorylation. These findings are supported by CDCP1’s interaction with and inhibition of acyl CoA-synthetase ligase (ACSL) activity. Importantly, CDCP1 knockdown increases LD abundance and reduces TNBC 2D migration in vitro, which can be partially rescued by the ACSL inhibitor, Triacsin C. Furthermore, CDCP1 knockdown reduced 3D invasion, which can be rescued by ACSL3 co-knockdown. In vivo, inhibiting CDCP1 activity with an engineered blocking fragment (extracellular portion of cleaved CDCP1) lead to increased LD abundance in primary tumors, decreased metastasis, and increased ACSL activity in two animal models of TNBC. Finally, TNBC lung metastases have lower LD abundance than their corresponding primary tumors, indicating that LD abundance in primary tumor might serve as a prognostic marker for metastatic potential. Our studies have important implications for the development of TNBC therapeutics to specifically block CDCP1-driven FAO and oxidative phosphorylation, which contribute to TNBC migration and metastasis.
The transmembrane glycoprotein, CUB-domain containing protein 1 (CDCP1), is a driver of migration and invasion in multiple forms of carcinoma, including renal (1, 2), ovarian (3, 4), prostate (5), pancreatic (6, 7), colon (8–12), and triple-negative breast (TNBC) (13, 14) carcinomas, among others. Furthermore, CDCP1’s role in tumor metastasis was confirmed in vivo in lung (15, 16), ovarian (17), prostate (5), and colon (9) cancers. Although CDCP1’s role in TNBC metastasis has not been established to date, high CDCP1 expression has been validated as a prognostic marker of poor survival in TNBC when combined with positive node status (18).
Our understanding of CDCP1’s upstream regulators, its mechanism of activation, and downstream signaling continues to expand (recently reviewed in ref. 19). Studies by others (5) and our group (14) have demonstrated that CDCP1 cleavage is necessary for its activation, and recently, we have further shown that cleavage stimulates CDCP1 homodimerization (14). Homodimeric CDCP1 stimulates phosphorylation of protein kinase C δ (PKCδ) by Src kinase, leading to migration and invasion of TNBC cells in vitro (14). Adding to our knowledge of CDCP1’s downstream signaling, we report that CDCP1 regulates lipid metabolism in TNBC by reducing cytoplasmic lipid droplet (LD) abundance and promoting fatty acid oxidation (FAO) in TNBC cells.
Breast tissue is known to be a dynamic site of lipid metabolism (20), and LD abundance changes dramatically during development (21, 22), menstruation, pregnancy, and lactation (23). LDs are formed by budding off of the endoplasmic reticulum (ER) and store fatty acids (FAs) in the form of diacyl- and triacylglycerol (DAG and TAG, respectively) and cholesterol in the form of cholesteryl esters. LDs interact with a multitude of organelles (i.e., peroxisomes, autophagosomes, and mitochondria) to shuttle FAs for incorporation into cell membranes, posttranslational modifications, and FAO (24). In order for long-chain FAs to enter any of these pathways, they must first be activated to become acyl-CoAs. Long-chain (12–20 carbons) FAs are activated by typical acyl-CoA synthetase ligase (ACSL) enzyme family members, including members 1, 3, 4, and 5 (25). Acyl-CoAs may enter FAO, which yields acetyl-CoAs used in the tricarboxylic acid cycle, generating NADH, which is used for oxidative phosphorylation (OxPhos) to ultimately produce adenosine triphosphate (ATP). Acyl-CoAs can also enter anabolic pathways and the shuttling of acyl-CoAs into either anabolic or catabolic pathways depends significantly on their localization within the cells (25, 26). Although there is conflicting data of the respective role and cell-type specificity of each ACSL in catabolic and anabolic pathways, ACSL3 has mainly been implicated in lipid anabolism (lipid storage) in mouse adipocytes and primate kidney cells (27, 28).
Dysregulated lipid metabolism and FAO have recently been linked to breast cancer progression. Photoacoustic imaging of the mouse mammary tumor virus–polyoma middle T (MMTV-PyMT)–driven breast cancer model demonstrated that transformation of normal breast tissue to invasive carcinoma resulted in a progressive decrease of cytoplasmic LD abundance (29). In line with this finding, pharmacological activation of peroxisome proliferator-activated receptor-γ caused accumulation of LDs in breast cancer cells and decreased their proliferation (30). Furthermore, Src (31) and Myc (32) oncogenes were recently reported to contribute to lipid metabolism dysregulation in TNBC. Src kinase has been reported to stimulate FAO in a positive feedback loop, and pharmacological and genetic inhibition of FAO blocked TNBC metastasis in vivo (31). Myc has also been reported to stimulate FAO and pharmacological inhibition of FAO blocked tumor growth of Myc-driven TNBC tumors (32). These data indicate that lipid metabolism represents a potential therapeutic target in TNBC.
Lipid metabolism dysregulation has also been reported in other types of cancer. Pharmacological inhibition of Myc in neuroblastoma increased LD abundance, accompanied by growth arrest and apoptosis in vitro and increased animal survival in vivo (33). High-passage prostate cancer cells (comparable to metastatic cells) have lower LD abundance than their low-passage (comparable to primary tumor) counterparts (34). In a spontaneous mouse model of pancreatic cancer, the synthesis of acyl-CoA–modified lipids was found to be the most down-regulated metabolic pathway compared with normal pancreatic tissue (35). These data indicate that dysregulated lipid metabolism is characteristic of progression of multiple forms of cancer.
Here, we used coherent anti-Stokes Raman scattering (CARS) and two-photon excited fluorescence (TPEF) microscopy methods to assess TNBC metabolism. CARS microscopy detects the C-H bond in FAs, allowing quantification of the LD abundance (36). TPEF microscopy detects fluorescence from reduced NADH and oxidized flavin adenine dinucleotide (FAD). The relative ratio of the two fluorophores is defined as the optical redox ratio (ORR) (37), which we have previously correlated with the OxPhos rate (38). Previously, we found that CDCP1 is overactivated in TNBC compared with normal breast epithelial cells (14). Here we demonstrate that CDCP1 dysregulates lipid metabolism in TNBC by inactivating ACSLs, reducing LD abundance, and stimulating FAO, which increases OxPhos. This process results in a “low-lipid” phenotype, a high migratory ability in vitro, and metastasis in vivo. These findings provide insight into CDCP1 as a regulator of LD abundance and FAO. Because CDCP1 is a druggable target (5, 14), these findings have important implications for TNBC therapeutic targeting and development of prognostic markers.
Results
CDCP1 Lowers Intracellular LD Abundance in Breast Cancer Cells.
We have previously identified CDCP1 protein as a master-regulator of migration and invasion in TNBC (14). To get insight into the signaling pathways it regulates, we conducted mass spectrometric (MS) analysis of proteins interacting with CDCP1. We overexpressed CDCP1 with a C-terminal Flag-tag in HEK 293T cells and conducted coimmunoprecipitation (co-IP). Unexpectedly, we found that CDCP1 interacts with multiple proteins involved in metabolism based on PANTHER analysis (www.pantherdb.org/) (SI Appendix, Fig. S1). Among these proteins was ACSL3, involved in lipid metabolism.
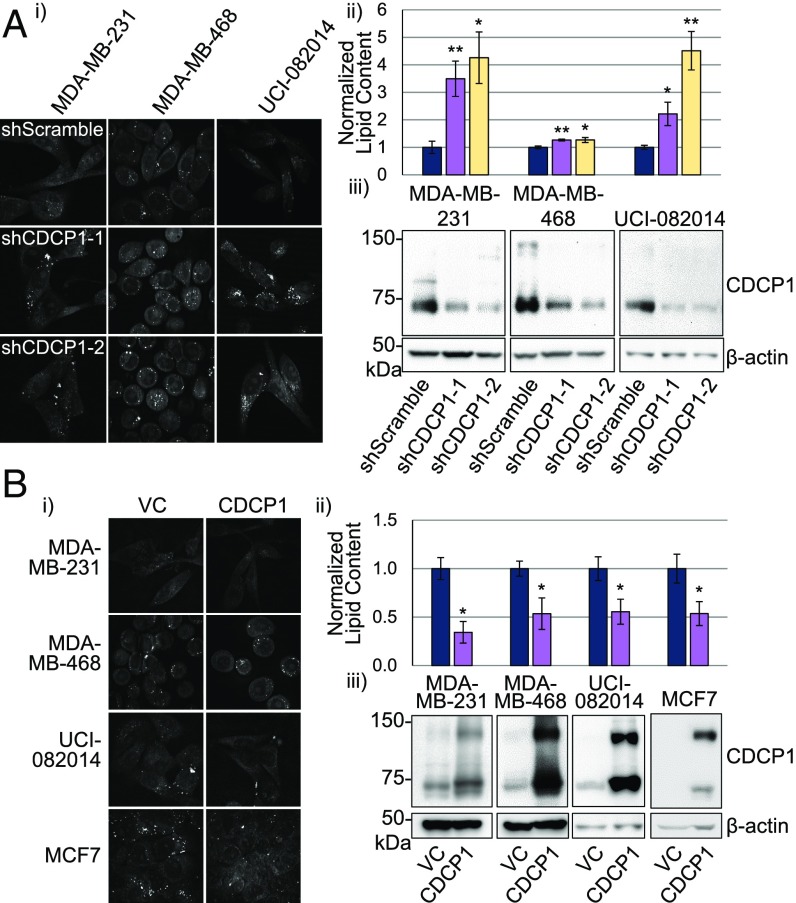
To assess CDCP1’s role in lipid metabolism, we first analyzed the effect of CDCP1 knockdown on LD abundance in three TNBC cell lines: MDA-MB-231, MDA-MB-468, and UCI-082014 (14), using two different shRNAs. We found that knocking down CDCP1 expression in all three cell lines increased LD content as analyzed by CARS microscopy (Fig. 1A). Second, we overexpressed CDCP1 in MDA-MB-231, MDA-MB-468, UCI-082014, and MCF7 cells and found that overexpression of CDCP1 decreased LD content of these cells (Fig. 1B). These results demonstrate that CDCP1 decreases LD abundance in breast cancer cells.
Fig. 1.
CDCP1 lowers intracellular LD abundance of breast cancer cells. (A) CDCP1 knockdown increases LD abundance in three TNBC cell lines: (i) representative 40× CARS images; (ii) quantitation of CARS; (iii) Western blot confirming CDCP1 knockdown. (B) CDCP1 overexpression decreases LD abundance in three TNBC cell lines and ER+ MCF7 breast cancer cell line not expressing endogenous CDCP1: (i) representative 40× CARS images; (ii) quantitation of CARS; (iii) Western blot confirming CDCP1 overexpression. n = 3 for A and B. P values analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs; *P < 0.05, **P < 0.01. shScr, shScramble; shC1 & 2, shCDCP1-1 and 2.
We further analyzed which lipid types were affected by CDCP1 expression in MDA-MB-231 and MDA-MB-468 cells using gas chromatography and found that multiple long-chain (16 and 18 carbons) FAs had lower abundance in shScramble-transduced control cells compared with shCDCP1-transduced cells (SI Appendix, Fig. S2).
CDCP1 Expression Correlates with Breast Cancer Cell Invasiveness and Causes Appearance of Low-Lipid Broken Spheroids in 3D Culture.
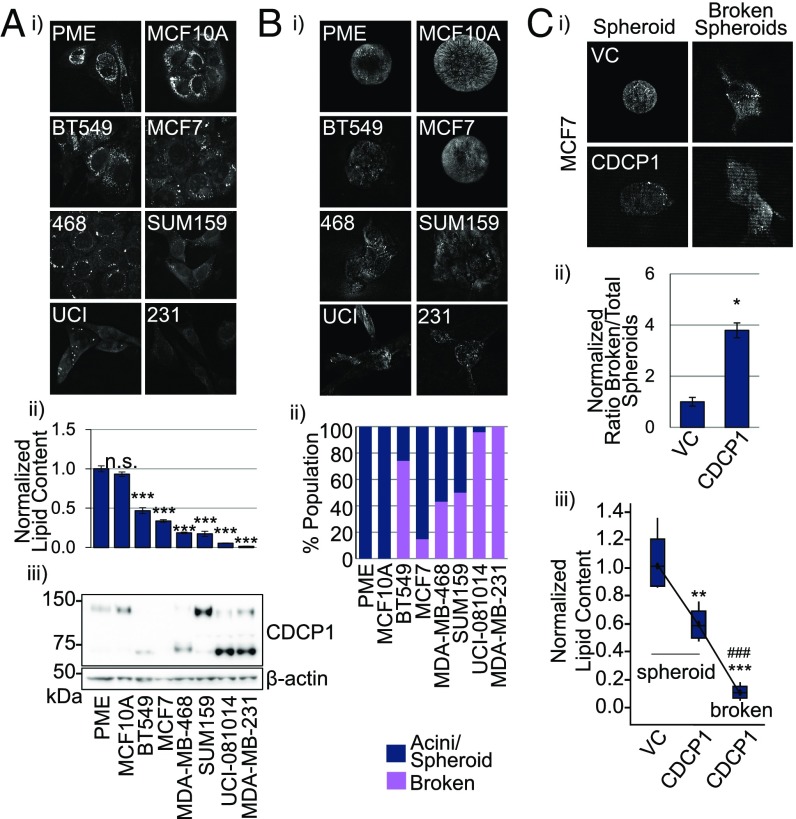
Because primary mammary epithelium cells (PME) were reported to have abundant LDs (30), we sought to compare their LD content to breast cancer cells. We found that PME and a nonmalignant breast cell line, MCF10A, had at least a twofold higher LD content than a panel of six breast cancer cell lines as analyzed by CARS microscopy (Fig. 2 A, i and ii).
Fig. 2.
Expression of cleaved CDCP1 isoform correlates with breast cancer cell invasiveness and causes appearance of low-lipid broken acini in 3D culture. (A) The LD abundance is lower in breast cancer cells compared with PME and nontumorigenic breast epithelial (MCF10A) cells in 2D culture. (i) Representative 40× CARS images; (ii) quantitation of CARS, statistics based on comparison with PME; (iii) Western blot of CDCP1 expression. (B) Lower LD abundance correlates with invasiveness in 3D culture. (i) Representative 40× CARS images of breast cancer cell lines in 3D culture; (ii) quantitation of intact acini (PME and MCF10A), intact spheroids and broken spheroids (breast cancer cell lines). (C) CDCP1 overexpression in MCF7 cells promotes invasion observed as broken spheroids in 3D culture, which have lower LD content than intact spheroids. (i) Representative 40× CARS images; (ii) quantitation of the ratio of broken spheroids to total spheroids in VC- and CDCP1-transduced MCF7 cells; (iii) quantitation of LD abundance in spheroids formed by VC- and CDCP1-transduced MCF7 and broken spheroids formed by CDCP1-transduced MCF7 cells. n = 3 in A, ii and C, iii. P values analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs; *P < 0.05, **P < 0.01, ***P < 0.001 compared with VC; ###P < 0.001 compared with CDCP1 spheroids.
We found that CDCP1 was expressed in five of six breast cancer cell lines in the panel, as well as in PME and MCF10A (Fig. 2 A, iii). Studies by others (5) and our group (14) have shown that CDCP1 cleavage leads to its activation to stimulate migration, invasion, and metastasis. Importantly, CDCP1 appears to be mostly full-length in PME and MCF10A and cleaved in five of five breast cancer cell lines that express CDCP1. Although CDCP1 is mostly full-length in SUM159 cells, the abundance of cleaved CDCP1 is still higher than in PME and MCF10A. Accordingly, all of the breast cancer cell lines formed irregular acini structures, spheroids, in 3D culture and invaded the surrounding matrix compared with the normal hollow acini formed by PME and MCF10A cells (Fig. 2B). Moreover, when we overexpressed CDCP1 in MCF7 cells (lacking endogenous CDCP1), they became more invasive and formed more broken spheroids (Fig. 2 C, i and ii). Importantly, we observed heterogeneity in lipid content in 3D culture: intact spheroids had higher lipid content than broken spheroids (Fig. 2 C, iii). These data suggest a link between lipid droplet abundance and invasiveness, both of which are regulated by CDCP1.
CDCP1 Interacts with ACSL Family Members Involved in Lipid Metabolism.
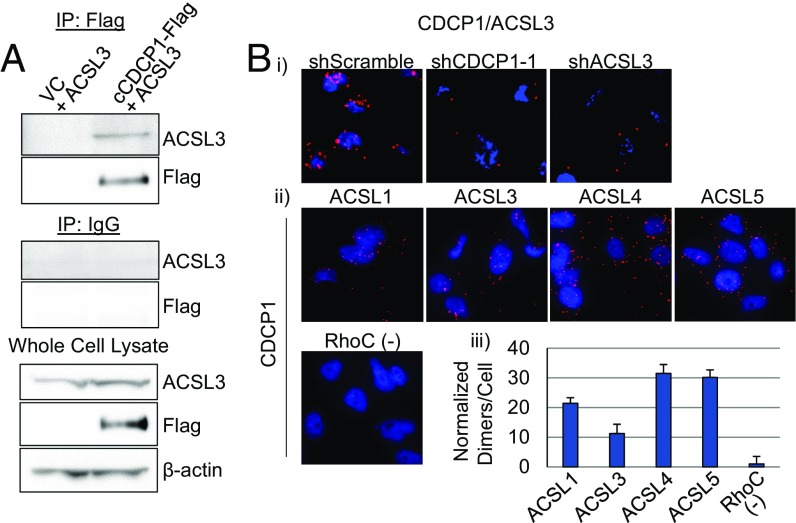
To investigate the molecular mechanism of the LD regulation by CDCP1, we decided to validate the interaction of CDCP1 with ACSL3 found by MS. CDCP1 and ACSL3 interact in HEK 293T cells overexpressing cCDCP1-Flag and ACSL3, as shown by co-IP (Fig. 3A). We further found that CDCP1 interacts with ACSL3 endogenously in MDA-MB-231 cells by proximity ligation assay (PLA) using shCDCP1-1 and shACSL3 as negative controls (Fig. 3 B, i).
Fig. 3.
CDCP1 interacts with ACSL family of proteins involved in lipid metabolism. (A) CDCP1 coimmunoprecipitates with ACSL3 in HEK 293T cells overexpressing ACSL3 and Flag-tagged cleaved CDCP1. (B) 40× images of PLA conducted in MDA-MB-231 cells demonstrating that (i) CDCP1 interacts with ACSL3 endogenously (negative controls: shCDCP1-1– and shACSL3-transduced cells); (ii) CDCP1 interacts with ACSL family members endogenously; negative control: anti-CDCP1 + anti-RhoC antibody pair, based on no reports of CDCP1 and RhoC interaction; and (iii) quantitation of PLA in ii.
ACSL3 belongs to an ACSL protein family, including typical members 1, 3, 4, and 5 (26, 39). Thus, we conducted additional PLA assays and found that CDCP1 interacts with ACSL1, -3, -4, and -5 (Fig. 3 B, ii and iii) (RhoC antibody was used as a negative control). All ACSL antibodies and RhoC antibody used for PLA were verified to work for immunofluorescence (SI Appendix, Fig. S3).
CDCP1 Regulates ACSL Activity.
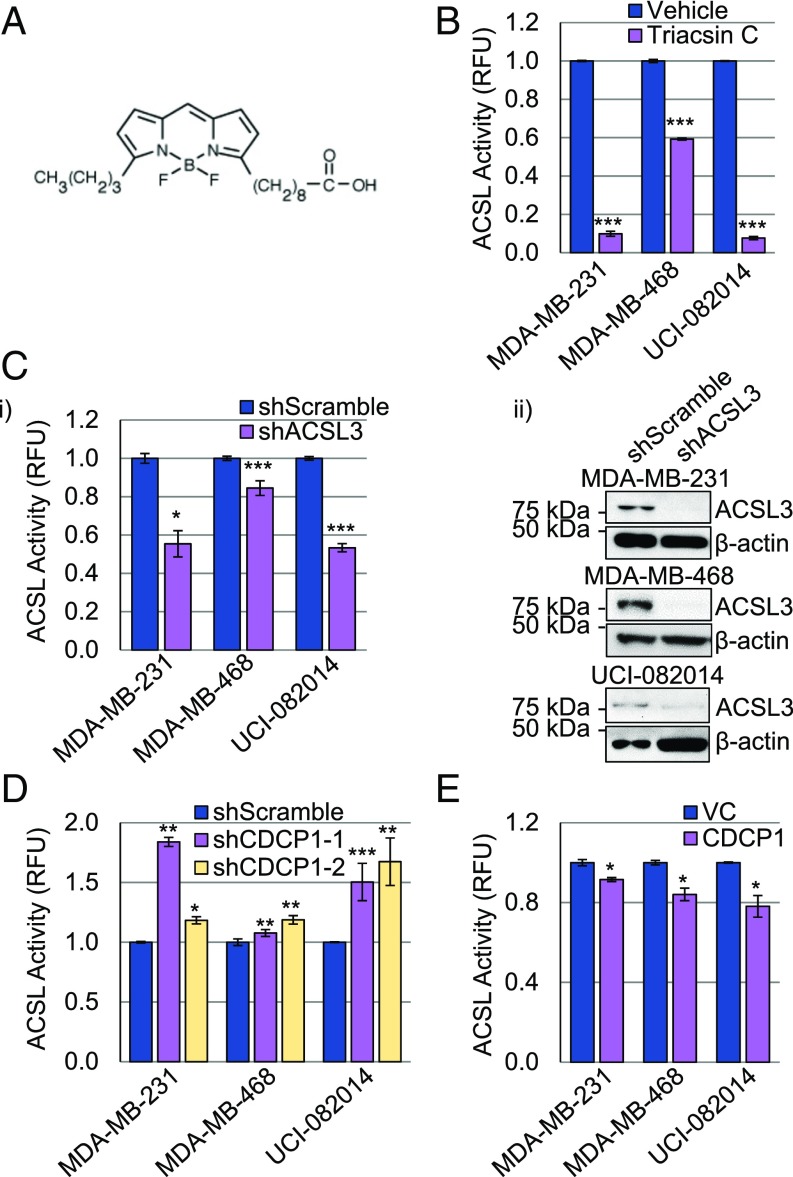
The CDCP1/ACSL interaction suggested that CDCP1 is an ACSL regulator. We analyzed the CDCP1’s effect on ACSL activity using a previously established assay (40–43) based on midchain BODIPY labeled palmitate (structure in Fig. 4A). First, we validated the assay in TNBC cell lines treated with a pan-ACSL inhibitor, Triacsin C (Fig. 4B), and shACSL3-transduced TNBC cell lines (Fig. 4C). In both cases, we observed a decrease in ACSL activity compared with vehicle-treated cells and shScramble-transduced cells. Second, we analyzed ACSL activity in the same TNBC cell lines with manipulated CDCP1 expression. We found that ACSL activity was increased on average by 40% in shCDCP1-transduced cells compared with shScramble-transduced cells (Fig. 4D). Accordingly, ACSL activity was decreased on average by 15% in CDCP1 overexpressing cells compared with vector-control (VC) cells (Fig. 4E). CDCP1 expression was assessed by Western blot in Fig. 1 A, iii and Fig. 1 B, iii. These data suggest that CDCP1-mediated reduction of ACSL activity leads to decreased acyl-CoA production and storage in LDs.
Fig. 4.
CDCP1 regulates ACSL activity. (A) Structure of BODIPY used for ACSL activity assays. (B and C) Validation of ACSL activity assay in TNBC cells. (B) Triacsin C (5 μM) reduces ACSL activity compared with DMSO vehicle. (C, i) ACSL3 knockdown reduces ACSL activity; (ii) Western blot confirming ACSL3 knockdown. (D) CDCP1 knockdown increases ACSL activity in TNBC cells. (E) CDCP1 overexpression decreases ACSL activity in TNBC cells. P values analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs; *P < 0.05, **P < 0.01, ***P < 0.001. Quantitation is the average of an n ≥ 3 for each panel. RFU, relative fluorescence units.
Low-Lipid Content Favors Promigratory Phenotype of Breast Cancer Cells.
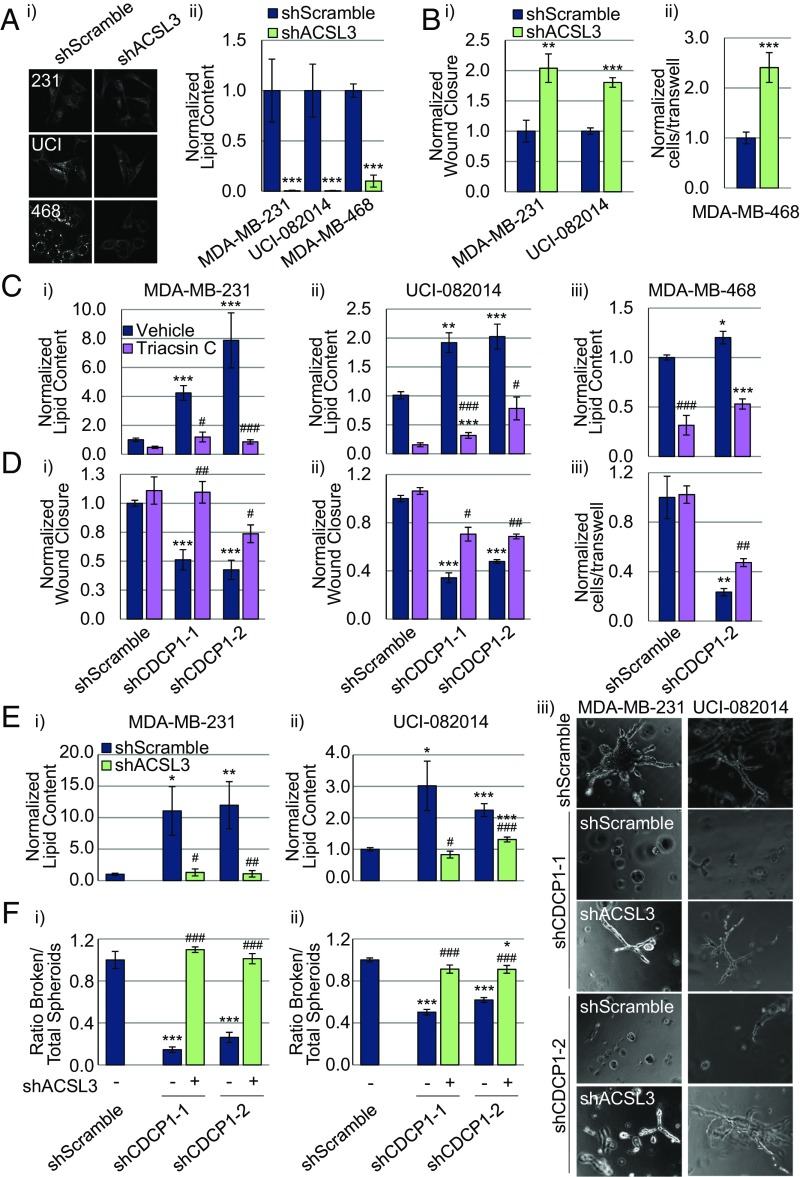
We have shown that CDCP1 decreases cytoplasmic LD abundance and stimulates invasiveness in TNBC cells, and previously we have shown that CDCP1 stimulates TNBC cell migration (14). Our findings that CDCP1 interacts with ACSLs and negatively regulates their activity led us to investigate the effect of ACSL expression on migration. Consistent with the above data, we found that knocking down ACSL3 expression reduced LD abundance (Fig. 5A), increased TNBC cell migration (Fig. 5B) and invasion (SI Appendix, Fig. S4A), and lung metastasis of MDA-MB-231 cells injected into the mammary fat pad of Rag2−/− mice (SI Appendix, Fig. S4B). We further found that treatment of shCDCP1-transduced TNBC cells with pan-ACSL inhibitor, Triacsin C, reduced the LD abundance to the level of shScramble-transduced cells (Fig. 5C) and partially rescued the migration defects caused by CDCP1 knockdown (Fig. 5D and SI Appendix, Fig. S5). The data acquired with genetic (shACSL3) and pharmacologic (Triacsin C) inhibition of ACSL activity generated consistent results.
Fig. 5.
Low-lipid content favors promigratory phenotype of TNBC. (A) ACSL3 knockdown reduces LD abundance in TNBC cell lines: (i) representative 40× CARS images; (ii) quantitation of CARS. (B) ACSL3 knockdown increases migration of MDA-MB-231 and UCI-082014 (i) and MDA-MB-468 (ii) cells. (C and D) CDCP1 knockdown increases LD abundance (C) and reduces migration (D) in MDA-MB-231 (i), UCI-082014 (ii), and MDA-MB-468 cells (iii), and 5 μM Triacsin C treatment for 16 h (lowering lipids) partially rescues migration of shCDCP1-transduced cells. (E and F) Increase in LD abundance (E) and reduction in invasion (F) by CDCP1 knockdown can be rescued by co-knockdown of ACSL3. (E) Quantitation of LD abundance measured by Oil Red O (ORO) staining. (F) Quantitation of the ratio of broken/total spheroids in MDA-MB-231 (i) and UCI-082014 (ii) 3D cultures. (iii) Representative 20× phase-contrast images of 3D MDA-MB-231 and UCI-082014 cultures. n = 3 for A–F. P values analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs. *P < 0.05, **P < 0.01, ***P < 0.001 compared with respective vehicle-treated shScramble cells; #P < 0.05, ##P < 0.01, ###P < 0.001 compared with respective vehicle-treated shCDCP1 cells.
Interestingly, we observed distinct effects of ACSL3 and CDCP1 knockdowns on cell proliferation: ACSL3 knockdown decreased proliferation of MDA-MB-231, MDA-MB-468, and UCI-082014 cells (SI Appendix, Fig. S6A), whereas CDCP1 knockdown did not change proliferation (SI Appendix, Fig. S6B), consistent with previous reports for cell types other than TNBC (44, 45). Because CDCP1 is known to regulate multiple pathways in addition to ACSL (1, 5, 13, 14, 46, 47), it is likely that these override the effect of the CDCP1/ACSL axis on cell proliferation.
To further investigate the contribution of the ACSL pathway to CDCP1’s effect on LD abundance and invasion, we simultaneously knocked down ACSL3 and CDCP1 in MDA-MB-231 and UCI-082014 cells (see SI Appendix, Fig. S7A for knockdown validation) and conducted 3D assays (similar to Fig. 2). We found that the increase in LD abundance seen by knocking down CDCP1 was rescued by co-knockdown of ACSL3 in MDA-MB-231 and UCI-082014 cells (Fig. 5E and SI Appendix, Fig. S7B). Furthermore, we found that the decrease in ORR seen by knocking down CDCP1 was partially rescued by co-knockdown of ACSL3 (SI Appendix, Fig. S7C). Upon culturing the cells in 3D, we found that knocking down CDCP1 reduced invasiveness and growth of the cells [consistent with our previous findings (14)], and that co-knocking down ACSL3 rescued invasiveness of the cells and did not rescue the reduced growth (Fig. 5F). These data link inhibition of ACSL activity occurring downstream of CDCP1 to decreased LD abundance, and increased migration and invasion.
CDCP1 Increases FAO in Breast Cancer Cells.
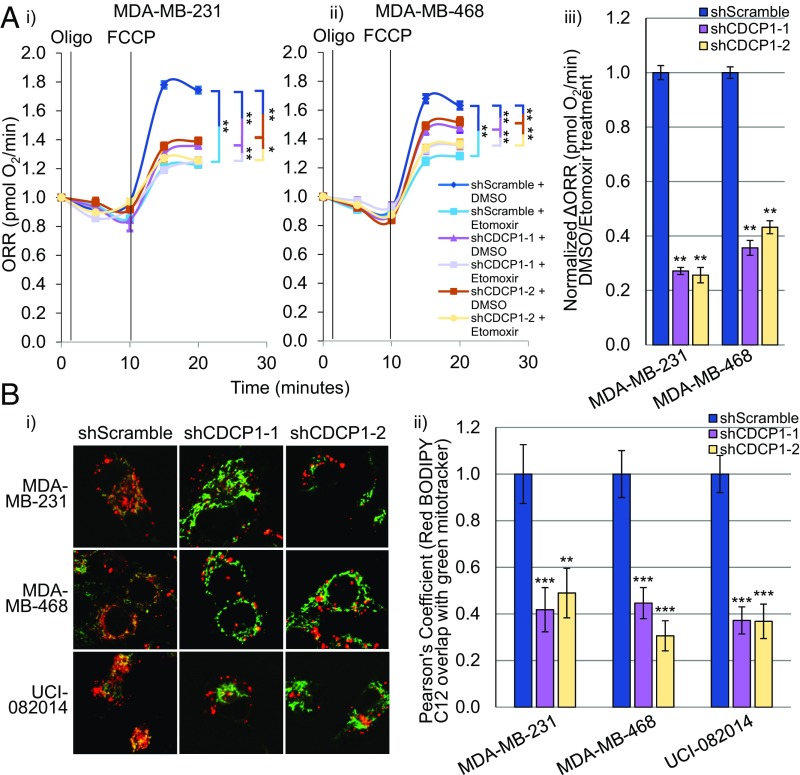
Interestingly, recent studies conducted in TNBC report two opposing disturbances in lipid metabolism: down-regulation of activators of lipid synthesis, along with up-regulation of activators of FAO (32, 33). We next sought to determine if the low-lipid phenotype caused by CDCP1 is a result of increased lipid utilization by FAO, in addition to decreased lipid activation for storage in LDs. First, we found that shCDCP1-transduced MDA-MB-231, MDA-MB-468, and UCI-082014 cells had lower levels of OxPhos than shScramble-transduced cells measured by ORR (SI Appendix, Fig. S8A). Second, to assess the contribution of FAO in OxPhos, we measured ORR after cell treatment with DMSO vehicle or FAO inhibitor Etomoxir (48, 49). We found that the ΔORR between DMSO- and Etomoxir-treated cells was lower in shCDCP1-transduced than in shScramble-transduced cells (Fig. 6A), indicating that FAO rates decrease upon CDCP1 knockdown, lowering OxPhos dependent on the products of FAO. Third, to assess if the CDCP1/ACSL axis contributes to OxPhos, we treated TNBC cells with Triacsin C and found that OxPhos increased as measured by ORR (SI Appendix, Fig. S8B). We verified that Triacsin C treatment increased FAO in TNBC cells (SI Appendix, Fig. S8C) using the same analysis as in Fig. 6 A, iii. Furthermore, we found that the decrease in ORR and FAO in MDA-MB-231 cells by CDCP1 knockdown was rescued by Triacsin C treatment (SI Appendix, Fig. S8 A and D). Finally, treatment of TNBC cells with Etomoxir to inhibit FAO increased LD abundance (SI Appendix, Fig. S9A) and reduced migration (SI Appendix, Fig. S9B) of TNBC cell lines, consistent with previous studies (31–33). These data indicate that CDCP1 contributes to a low-lipid TNBC phenotype by increasing FAO and OxPhos and that ACSL inhibition by CDCP1 increases FAO and OxPhos. These data are in line with reports on CDCP1 (1–14), FAO (31), and OxPhos (50–52) promoting cell migration.
Fig. 6.
CDCP1 knockdown reduces FAO in TNBC cells. (A) CDCP1 knockdown reduces FAO as measured by ORR in MDA-MB-231 (i) and MDA-MB-468 (ii) cells. Oligomycin (Oligo, ATP synthase inhibitor) followed by carbonyl cyanide-4-phenylhydrazone (FCCP, ATP synthesis uncoupler) treatment allowed measurement of maximum respiration potential. Etomoxir (FAO inhibitor, blocking transport of FAs into the mitochondria) treatment allowed measurement of FAO fueled by FAs already present in mitochondria. (iii) shCDCP1-transduced cells have lower ΔORR [calculated as 1 (DMSO-treated ORR/Etomoxir-treated ORR) and then all data were normalized to shScramble control, which was set to 1] than shScramble-transduced cells, indicating that CDCP1 knockdown reduces lipid stores in the mitochondria available for FAO. (B) Representative 63× images (i) and quantitation (ii) of colocalization of red BODIPY C12 (lipids) with green MitoTracker (mitochondria), indicating that less lipids colocalize with the mitochondria 16 h after BODIPY C12 addition. n = 3 for A and B. P values analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs; *P < 0.05, **P < 0.01, ***P < 0.001.
We also assessed the differences in lipid transport to mitochondria between shScramble- and shCDCP1-transduced cells, as described in Rambold et al. (53). We pulsed TNBC cell lines with red C12 BODIPY (structure in SI Appendix, Fig. S10A) for 16 h and then costained with green MitoTracker to visualize mitochondria. Colocalization quantitated by Pearson’s coefficient of red (lipid) and green (mitochondria) staining indicated that CDCP1 knockdown reduced transport of lipids into the mitochondria (Fig. 6B). We verified that mitochondrial mass did not differ in shScramble- and shCDCP1-transduced cells (SI Appendix, Fig. S10B).
Extracellular Portion of Cleaved CDCP1 Blocks CDCP1 Dimerization in Vivo, Which Is Accompanied by High LD Abundance and Reduced Metastasis.
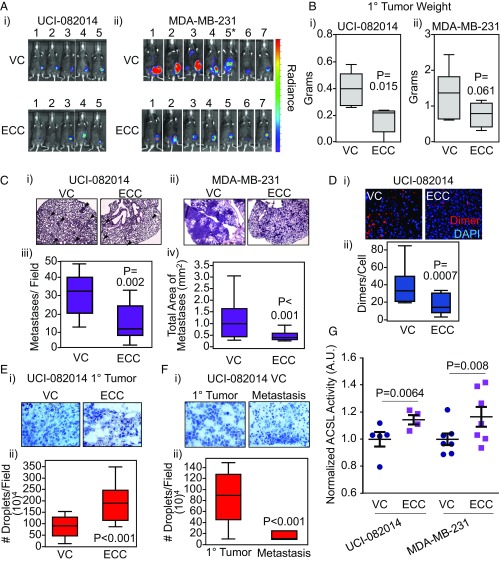
We previously demonstrated that CDCP1 cleavage and dimerization are important for driving TNBC migration and invasion (14). Moreover, we were able to block CDCP1 dimerization and activity by the expression of the secreted extracellular portion of cleaved CDCP1 (extracellular portion of cleaved CDCP1, ECC) (14). Thus, we hypothesized that in vivo ECC would block TNBC metastasis and increase the LD abundance in primary tumors. First, we found that LD abundance was higher in ECC-transduced UCI-082014 and MDA-MB-231 cells than in VC-transduced cells in vitro (SI Appendix, Fig. S11A). Second, we implanted VC- or ECC-firefly luciferase-labeled UCI-082014 and MDA-MB-231 cells to fat pads of Rag2−/− mice. Five (UCI-081014) to 8 (MDA-MB-231) weeks after cell implantations, mice were imaged and then killed (Fig. 7A). We found that ECC inhibited the primary tumor growth in the UCI-082014–based model (Fig. 7 B, i). Although the same tendency was observed in the MDA-MB-231–based model (Fig. 7 B, ii), the data did not reach statistical significance (P = 0.0613). Furthermore, ECC inhibited metastasis to the lungs in both mouse models (Fig. 7C). We verified that ECC was expressed by Western blot (SI Appendix, Fig. S11B) and inhibited CDCP1 dimerization as assessed by PLA in the primary tumors (Fig. 7D and SI Appendix, Fig. S11C). As expected, we found that ECC increased the intracellular LD abundance in the primary tumors (Fig. 7E and SI Appendix, Fig. S11D). Importantly, we also found that the lung metastases of the VC-transduced cells had lower LD abundance than their corresponding primary tumors (Fig. 7F), providing further support to the low-lipid phenotype supporting metastasis. Finally, we found that ACSL activity was higher in ECC-expressing UCI-082014 and MDA-MB-231 tumors compared with VC control tumors (Fig. 7G).
Fig. 7.
Expression of a CDCP1-blocking fragment, ECC, increases LD abundance and reduces metastasis. (A) Representative images of mice with luciferase-labeled UCI-082014 (i) or MDA-MB-231 (ii) tumors at week 5 and 8 posttumor implantation, respectively. *VC-5 killed 2 wk early because of extensive disease progression and visibly reduced quality of life. (B) ECC reduces primary (1°) tumor growth of UCI-082014 (i) and trends toward reducing 1° tumor growth of MDA-MB-231 (ii) cells implanted into the fat pad of Rag2−/− mice. (C) ECC expression reduces lung metastasis of UCI-082014 and MDA-MB-231 tumors. (i and ii) Representative 4× images of H&E-stained lungs from mice implanted with UCI-082014 (i), metastases are shown by arrowheads, and MDA-MB-231 (ii) cells. (iii and iv) Quantitation of UCI-082014 (iii) and MDA-MB-231 (iv) metastasis. (D) ECC reduces CDCP1 dimer formation in 1° tumors as assessed by PLA. Representative 20× images (i) and quantitation (ii) of PLA. (E) ECC increases LD abundance in 1° tumors. Representative 20× images (i) and quantitation (ii) of Oil Red O (ORO) staining. (F) VC-transduced 1° tumors have higher LD abundance than the corresponding metastatic nodules. Representative 20× images (i) and quantitation (ii) of ORO staining. (G) ECC stimulates ACSL activity in 1° tumors. Quantitation of ACSL activity in VC- and ECC-transduced UCI-082014 and MDA-MB-231 primary tumor tissue. Each point represents an individual tumor. P values (as indicated) analyzed by one-way ANOVA with multiple comparison post hoc t test and error bars represent SEMs. A.U., arbitrary units.
In summary, our data indicate that CDCP1 promotes TNBC metastasis by reducing LD abundance, promoting lipid accumulation in the mitochondria for FAO to fuel OxPhos, and promoting cell migration. CDCP1 regulates those processes, in part, by suppressing ACSL activity. As a result, TNBC tumors have a low-lipid phenotype.
Discussion
CDCP1 function-blocking antibodies have demonstrated effectiveness at inhibiting tumor growth (54) and metastasis (5) in vivo. We have previously shown that the CDCP1 function-blocking fragment, ECC, inhibits CDCP1 dimerization and activation in vitro (14) and here show its efficacy in vivo in two animal models of TNBC. Our data support targeting CDCP1 in TNBC to block metastasis and provide insight into the mechanism of CDCP1-induced metastasis. We demonstrate that CDCP1 regulates lipid metabolism, reducing LD abundance and stimulating FAO. Products of FAO, in turn, stimulate OxPhos, which contributes to TNBC migration and metastasis. Our data also show that the CDCP1/ACSL axis contributes to dysregulated lipid metabolism.
Importantly, the contributions of FAO (31, 32) and OxPhos (50) to TNBC migration and metastasis and OxPhos to metastasis of other cancers (55, 56) have been recently documented, supporting our proposed mechanism. Accordingly, pharmacological and genetic inhibitors of carnitine palmitoyltransferases blocked lipid transport to mitochondria, FAO, OxPhos, and metastasis of TNBC cells (31, 32). Recently, Myc overexpression has been shown to drive FAO in TNBC (32, 33). All of the cancer cell lines used in this study overexpress Myc compared with MCF10A cells (SI Appendix, Fig. S12). Importantly, MCF7 cells, which do not express endogenous CDCP1, overexpress Myc, which might explain their CDCP1-independent low-lipid phenotype (Fig. 2A). Because both Myc and CDCP1 are overexpressed in TNBC cell lines used in this report, it is worth investigating if Myc and CDCP1 pathways contribute to TNBC lipid metabolism independently or if there is cross-talk. Interestingly, several studies have shown that proteins involved in the CDCP1 pathway, Src (47) and PKCδ (1), are involved in lipid metabolism as well. FAO and OxPhos activate Src, which in turn induces OxPhos in TNBC (31), and OxPhos activates PKCδ (57). Taken together, these data suggest that targeting lipid metabolism in general, and CDCP1 in particular, are viable therapeutic strategies to inhibit FAO, OxPhos, and metastasis of TNBC.
In this study, we suggest that, through FAO and subsequent OxPhos, lipids provide energy contributing to migration and metastasis. However, although the NADH and FADH2 of FAO are generally shuttled into OxPhos, the acetyl-CoA produced from FAO can enter many different pathways, including the tricarboxylic acid cycle, which feeds into OxPhos, posttranslational modifications, and biosynthetic pathways. In addition, we envision other mechanisms linking lipid metabolism to migration and metastasis. For example, LDs appear to be not merely inert structures for lipid storage, but dynamic signaling organelles (58, 59). The literature supports the cross-talk between cellular LDs and membrane lipid rafts (60–62), where multiple proteins involved in migration, including integrins (63, 64) and CDCP1 (54, 65), reside. On the other hand, the biophysical properties of high-lipid vs. low-lipid cells remain unknown and await investigation. Thus, our study opens an intriguing research direction into dissecting the mechanism of LD contribution to cancer cell migration and metastasis, which will be instrumental in revealing new pathways for therapeutic targeting of TNBC.
Here we show that CDCP1 promotes FAO in normoxia. Our observations put CDCP1 at the crossroads of metabolic pathways, as we (1, 14) and others (2) have previously shown that CDCP1 activity increases under hypoxia, recognized to favor the metabolic shift from OxPhos to glycolysis (66). CDCP1 activity increases through its cleavage and phosphorylation, likely through activation of Src kinase (2). Because most TNBC tumors have multiple hypoxic areas (67), it raises the question of regional CDCP1 activation status and coordination of FAO/OxPhos and glycolysis. Further investigation is needed to establish the role of CDCP1 in FAO under a range of oxygen tensions and the role of lipids supplied by the microenvironment vs. being endogenously synthetized.
In this study, we concentrated on CDCP1-driven TNBC metastasis, and it is worth investigating whether our data translate to other types of breast cancer. The only non-TNBC cell line used in this study was MCF7, which is ER+, and although CDCP1 overexpression leads to increased invasiveness in 3D and decreased LD abundance, these cells do not endogenously express CDCP1. It is important to keep in mind that the role of CDCP1 in HER2+ breast cancer was addressed just on the level of primary tumor growth, not metastasis, generating controversial results. Homozygous germline CDCP1 knockout in a MMTV-PyMT spontaneous mouse model lead to increased tumor growth rate (68). However, in a BT474-based mouse model, CDCP1 overexpression led to increased tumor growth (44). Furthermore, high coexpression of CDCP1 and HER2 was correlated with decreased patient survival (44). It will be important to investigate the regulation of lipid metabolism by CDCP1 across breast cancer subtypes.
In this study, we used two animal models of TNBC. Whereas ECC reduced primary tumor growth in a UCI-082014–based animal model, it only trended toward reducing primary tumor growth in a MDA-MB-231–based model. However, in both models ECC reduced metastasis. Although both MDA-MB-231 and UCI-082014 are triple-negative, they have different genetic backgrounds, which likely contribute to the degree of their dependency on the CDCP1 pathway for tumor growth. In this respect, we recently established UCI-082014 from a primary tumor specimen and they are still to be characterized for driver mutations; MDA-MB-231 harbor mutations in BRAF and KRAS, which are potent inducers of tumor growth (69, 70). In addition, UCI-082014 cell implantations generate large primary tumors, which micrometastasize to the lungs; MDA-MB-231 cell implantations generate rather small primary tumors, which at the same time effectively macrometastasize. This difference in the two tumor models’ dynamics also likely contributes to the observed primary tumor growth phenotype. Because CDCP1 represents a potential therapeutic target, it will be important to determine the genetic and epigenetic factors informing on the effect of CDCP1 blockage on the primary tumor growth. Accordingly, ECC-based therapeutic could be potentially used in a neo-adjuvant or adjuvant setting.
To date, the ACSL activity was predominantly studied in the liver, where it does not affect FAO. This finding is highlighted by a study that found that the pan-ACSL inhibitor, Triacsin C, preferentially inhibited TAG synthesis and did not affect FAO (71). Furthermore, although both ACSL1 and ACSL5 have been reported to localize at the mitochondrial and the LD membranes (72) and ACSL1 has been reported to up-regulate genes involved in FAO (73), they do not affect the FAO rate (74, 75). Based on our data, ACSLs promote lipid storage in LDs (Fig. 5A and SI Appendix, Fig. S7B), inhibit OxPhos (SI Appendix, Fig. S7C), and inhibit FAO (SI Appendix, Fig. S8C) in TNBC. Considering that ACSL products, acyl-CoAs, are competed for by enzymes involved in LD storage, incorporation into lipid membranes, posttranslational modifications, and FAO (25, 76), our data that ACSL activity inhibits FAO are consistent with a choice to direct acyl-CoAs toward storage in LDs. Accordingly, when ACSL activity is blocked, acyl-CoAs become limited and may preferentially enter FAO, and when ACSL activity is induced, acyl-CoAs become abundant and may preferentially enter LDs for storage. Additional studies are needed to investigate the regulatory mechanism, as well as the cell-type–specific roles of ACSLs in lipid metabolism. Because CDCP1 has a large effect on LD abundance and a modest effect on ACSL activity, we expect that CDCP1 interacts with and regulates additional metabolic proteins that are yet to be uncovered.
Interestingly, ACSL3 was recently reported to promote lung cancer growth in a KRASG12D-driven mouse model, although its contribution to metastasis was not investigated (77). Our in vitro data show that ACSL3 increases TNBC cell proliferation and at the same time decreases TNBC migration (Fig. 5 and SI Appendix, Fig. S6). Our in vivo data show that ACSL3 reduces metastasis (SI Appendix, Fig. S4).
The observation that VC-transduced UCI-082014 generated metastases with lower LD abundance than the corresponding primary tumors suggests that cells in primary tumors with low LD abundance effectively migrate and metastasize and keep the low LD abundance phenotype at metastatic sites. Importantly, this finding raises the question of TNBC tumor heterogeneity in terms of CDCP1 expression and activity, as well as CDCP1-dependent and -independent regulation of LD abundance. Thus, the frequency of cells with high CDCP1 activity and low LD abundance in primary tumors might represent a prognostic marker for metastatic potential. This observation also implies that the average CDCP1 expression and activity should be higher at metastatic sites than in corresponding primary tumors. Accordingly, Alajati et al. (44) reported that CDCP1 expression is higher in HER2+ patient breast cancer metastases than the primary tumors. It is important to note that although our data support the mechanism where low LD abundance stems from CDCP1 overexpression and lipid FAO, thus depleting LDs, the dynamics of lipid metabolism in TNBC tumor tissue is largely unknown. The low LD abundance phenotype can arise from low-lipid synthesis and high-lipid consumption. In both cases the amount of lipids detected in the tumor tissue will decrease. Adding to complexity are the additional cell types present in the tumor microenvironment, including adipocytes, potentially capable of supplying lipids to breast cancer cells for FAO. Supporting the importance of lipid FAO for metastasis are the results from Park et al. (31), which show that Etomoxir, blocking mitochondrial lipid consumption, does block metastasis. Further studies are needed to clarify those important considerations.
In conclusion, our data establish CDCP1 as a master-regulator of lipid metabolism in TNBC, lowering LD abundance and increasing FAO, leading to a low-lipid phenotype of TNBC. This signaling pathway is partially regulated by the CDCP1/ACSL axis. Because of the interconnection of metabolic pathways, increased FAO leads to increased OxPhos, contributing to migration and metastasis. As a result, CDCP1 represents a potential therapeutic target for TNBC management, along with other components of the lipid metabolism pathway. Thus, our findings have important implications for therapeutic targeting and development of prognostic markers of TNBC.
Materials and Methods
CARS Microscopy.
A 76-MHz mode-locked Nd:vanadate laser provides a beam at 1,064 nm functioning as the Stokes beam and a second harmonic generated beam at 532 nm to pump an optical parametric oscillator. The pump beam generated by the optical parametric oscillator is spatially and temporally overlapped with the Stokes beam and sent to the microscope. The two beams are focused on the cells through a 60×, 1.2 numerical aperture water objective lens. The generated CARS signals are collected through the condenser and focused onto a Hamamatsu photomultiplier tube with a 650 ± 50-nm bandpass filter in front.
TPEF Microscopy.
A scanning laser TPEF microscope was used to measure the metabolic states of the cell lines. The laser was set to 740 nm to excite NADH (480 ± 50-nm bandpass filter) and tuned to 900 nm to excite FAD+ (530 ± 50-nm bandpass filter). We used 20 mW for 740 nm and 40 mW for 900 nm with a 6-µs pixel dwell time. The laser power was kept constant and the response of the photo multiplier tubes was calibrated with a reference sample (0.02 μM fluorescein solution at pH 7.0). The images of NADH and FAD+ of the same cells were taken sequentially and were used to calculate ORR, defined as FAD+/(NADH + FAD+).
FAO Analysis.
Two dishes for each experimental group were prepared. On each imaging dish, 80,000 cancer cells were plated and cultured in full-growth medium (DMEM, 10% FBS, 25 mM glucose, 2 mM GlutaMAX, 1% penicillin/streptomycin) for 24 h to allow cell attachment. The growth medium was exchanged to substrate-limited medium (DMEM, 0.5 mM glucose, 1 mM GlutaMAX, 0.5 mM carnitine, and 1% FBS) a day before the experiment. The cancer cells were switched to basal FAO medium (DMEM, 2.5 mM glucose, 0.5 mM carnitine, and 5 mM Hepes) 45 min before the experiment. Etomoxir was added to the medium 15 min before the experiment at a final concentration of 40 µM. In each imaging dish, three locations were randomly chosen. The baseline ORRs were measured at time 0 before Oligo addition. The optical redox changes were monitored every 5 min for a total of two measurements. Then, 5 µL of FCCP was added and the ORRs were measured at 15 min and 20 min. Images were taken using TPEF Microscopy (see above). All ORR measurements were done in the absence of exogenous lipid sources.
Virus Production and Infection of Target Cells.
Detailed protocol is described in Razorenova et al. (78). HEK 293T cells were transfected with each lentiviral plasmid along with packaging plasmids, pVSVG and ΔR8.2. Virus-containing media were transferred to target TNBC cells, which were selected in antibiotic-containing media for a minimum of 1 wk.
Western Blotting.
The Western blotting protocol was adapted from Razorenova et al. (79). Forty micrograms of protein were used per lane. Antibodies used are listed in SI Appendix, Table S1.
Scratch Assays.
A detailed protocol of scratch assays is described in Wright et al. (14). For scratch assays, 400,000 MDA-MB-231 or UCI-082014 cells were plated per well of a six-well plate. Twenty-four hours later, cells were treated with DMSO control or 5 μM Triacsin C for 16 h before assay. Cells were allowed to migrate for 6 h.
Transwell Migration.
A detailed protocol of assay and quantitation is described in Thompson et al. (80). MDA-MB-468 cells were treated with DMSO control or 5 μM Triacsin C for 16 h before assay. Cells were allowed to migrate for 16 h.
In Vivo Experiments.
All animal research in this study was approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. UCI-082014 and MDA-MB-231 cells were transduced with VC or pLM-CMV-ECC-His (14). Rag2−/− mice were orthotopically injected in the fourth mammary fat pad with 106 cells mixed with Matrigel. Before being killed at 5 wk for the UCI-082014–based model and at 8 wk for the MDA-MB-231–based model, mice were intraperitoneally injected with 150 mg/kg d-luciferin and imaged for 2 min on large-binning using the IVIS lumina imager. Mice were then killed, and tumors and lungs were collected for analysis. UCI-082014 metastases were quantitated as the average number of metastases per field in three to five images taken from three nonserial sections per lung per mouse. MDA-MB-231 metastases were quantitated as the total area of metastases per field because of the metastatic burden being too large to quantitate individual metastases. Quantitation of Oil Red O staining was done using ImageJ to quantitate the number of lipid droplets per cell. Quantitation of the PLA assay was done by manually counting the number of dimers per cell.
All other material and methods can be found in SI Appendix, Supplemental Material and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Kaiser for MDA-MB-231 and MDA-MB-468 cell lines; Dr. Wang for SUM159 and BT549 cell lines; Dr. Dai for MCF10A cell line; Dr. Verma for pVSVG and ΔR8.2 lentivirus packaging plasmids; Dr. Chumakov for the pLM-CMV lentiviral vector; Dr. Boiko for Rag2−/− mice; Jeffrey Kim at the University of California, Irvine Pathology for Oil Red O staining; Dr. Ghassemian at the University of California, San Diego Biomolecular and Proteomics Mass Spectrometry Facility for analysis of CDCP1-binding partners; Carla Harris and Dr. Swift at the Vanderbilt Hormone Assay & Analytical Services Core for gas chromatography quantitative analysis of intracellular lipid types; and Dr. Fan for critical reading of the manuscript. This work was supported by Concern Foundation Award CF-204722 (to O.V.R.); National Cancer Institute Grant R01CA142989 (to B.J.T.); National Institute of Biomedical Imaging and Bioengineering Grant P41EB015890 (to B.J.T.); an Arnold and Mabel Beckman Foundation award (to B.J.T.); and National Cancer Institute Grant F31CA196226 (to H.J.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703791114/-/DCSupplemental.
References
- 1.Razorenova OV, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKCdelta-driven migration. Proc Natl Acad Sci USA. 2011;108:1931–1936. doi: 10.1073/pnas.1011777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerling BM, et al. Identification of CDCP1 as a hypoxia-inducible factor 2α (HIF-2α) target gene that is associated with survival in clear cell renal cell carcinoma patients. Proc Natl Acad Sci USA. 2013;110:3483–3488. doi: 10.1073/pnas.1222435110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, et al. Elevated CDCP1 predicts poor patient outcome and mediates ovarian clear cell carcinoma by promoting tumor spheroid formation, cell migration and chemoresistance. Oncogene. 2016;35:468–478. doi: 10.1038/onc.2015.101. [DOI] [PubMed] [Google Scholar]
- 4.Vlad C, Kubelac P, Onisim A, Irimie A, Achimas-Cadariu P. The role of CDCP1 (CUB domain-containing protein 1) and ADAM12 (a disintegrin and metalloproteinase 12) in ovarian cancer. J BUON. 2015;20:673–679. [PubMed] [Google Scholar]
- 5.Casar B, et al. Blocking of CDCP1 cleavage in vivo prevents Akt-dependent survival and inhibits metastatic colonization through PARP1-mediated apoptosis of cancer cells. Oncogene. 2012;31:3924–3938. doi: 10.1038/onc.2011.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura S, Hamada S, Masamune A, Satoh K, Shimosegawa T. CUB-domain containing protein 1 represses the epithelial phenotype of pancreatic cancer cells. Exp Cell Res. 2014;321:209–218. doi: 10.1016/j.yexcr.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Miyazawa Y, et al. CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res. 2010;70:5136–5146. doi: 10.1158/0008-5472.CAN-10-0220. [DOI] [PubMed] [Google Scholar]
- 8.Orchard-Webb DJ, Lee TC, Cook GP, Blair GE. CUB domain containing protein 1 (CDCP1) modulates adhesion and motility in colon cancer cells. BMC Cancer. 2014;14:754. doi: 10.1186/1471-2407-14-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao W, et al. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology. 2013;145:636–46.e5. doi: 10.1053/j.gastro.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 10.Perry SE, et al. Expression of the CUB domain containing protein 1 (CDCP1) gene in colorectal tumour cells. FEBS Lett. 2007;581:1137–1142. doi: 10.1016/j.febslet.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 11.Scherl-Mostageer M, et al. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene. 2001;20:4402–4408. doi: 10.1038/sj.onc.1204566. [DOI] [PubMed] [Google Scholar]
- 12.Yousef I, et al. Infrared spectral signatures of CDCP1-induced effects in colon carcinoma cells. Analyst (Lond) 2011;136:5162–5168. doi: 10.1039/c1an15437k. [DOI] [PubMed] [Google Scholar]
- 13.Law ME, et al. Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene. 2013;32:1316–1329. doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright HJ, et al. CDCP1 cleavage is necessary for homodimerization-induced migration of triple-negative breast cancer. Oncogene. 2016;35:4762–4772. doi: 10.1038/onc.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu KL, et al. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci Rep. 2015;5:16426. doi: 10.1038/srep16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uekita T, et al. CUB domain-containing protein 1 is a novel regulator of anoikis resistance in lung adenocarcinoma. Mol Cell Biol. 2007;27:7649–7660. doi: 10.1128/MCB.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrington BS, et al. Cell line and patient-derived xenograft models reveal elevated CDCP1 as a target in high-grade serous ovarian cancer. Br J Cancer. 2016;114:417–426. doi: 10.1038/bjc.2015.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turdo F, et al. CDCP1 is a novel marker of the most aggressive human triple-negative breast cancers. Oncotarget. 2016;7:69649–69665. doi: 10.18632/oncotarget.11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright HJ, Police AM, Razorenova OV. Targeting CDCP1 dimerization in triple-negative breast cancer. Cell Cycle. 2016;15:2385–2386. doi: 10.1080/15384101.2016.1204849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6:1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell TD, et al. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: roles of adipophilin and lipid metabolism. J Lipid Res. 2007;48:1463–1475. doi: 10.1194/jlr.M600474-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Russell TD, et al. Adipophilin regulates maturation of cytoplasmic lipid droplets and alveolae in differentiating mammary glands. J Cell Sci. 2011;124:3247–3253. doi: 10.1242/jcs.082974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: Use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2003;8:287–307. doi: 10.1023/b:jomg.0000010030.73983.57. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F, Du G. Dysregulated lipid metabolism in cancer. World J Biol Chem. 2012;3:167–174. doi: 10.4331/wjbc.v3.i8.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grevengoed TJ, Klett EL, Coleman RA. Acyl-CoA metabolism and partitioning. Annu Rev Nutr. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mashek DG, Li LO, Coleman RA. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2007;2:465–476. doi: 10.2217/17460875.2.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 28.Poppelreuther M, et al. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J Lipid Res. 2012;53:888–900. doi: 10.1194/jlr.M024562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson KE, Bachawal SV, Tian L, Willmann JK. Multiparametric spectroscopic photoacoustic imaging of breast cancer development in a transgenic mouse model. Theranostics. 2014;4:1062–1071. doi: 10.7150/thno.9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uray IP, Rodenberg JM, Bissonnette RP, Brown PH, Mancini MA. Cancer-preventive rexinoid modulates neutral lipid contents of mammary epithelial cells through a peroxisome proliferator-activated receptor γ-dependent mechanism. Mol Pharmacol. 2012;81:228–238. doi: 10.1124/mol.111.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JH, et al. Fatty acid oxidation-driven Src links mitochondrial energy reprogramming and oncogenic properties in triple-negative breast cancer. Cell Reports. 2016;14:2154–2165. doi: 10.1016/j.celrep.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camarda R, et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zirath H, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci USA. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue S, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillaumond F, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2015;112:2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suhalim JL, et al. Effect of desiccating stress on mouse meibomian gland function. Ocul Surf. 2014;12:59–68. doi: 10.1016/j.jtos.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice WL, Kaplan DL, Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One. 2010;5:e10075. doi: 10.1371/journal.pone.0010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou J, et al. Correlating two-photon excited fluorescence imaging of breast cancer cellular redox state with seahorse flux analysis of normalized cellular oxygen consumption. J Biomed Opt. 2016;21:60503. doi: 10.1117/1.JBO.21.6.060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandal S, Khan P, Li L, Davie JR. Metabolomics and transcriptional responses in estrogen receptor positive breast cancer cells. In: Gunduz M, Gunduz E, editors. Breast Cancer—Carcinogenesis, Cell Growth and Signalling Pathways. In Tech, Rijeka; Croatia: 2011. pp. 257–284. [Google Scholar]
- 40.Bu SY, Mashek MT, Mashek DG. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem. 2009;284:30474–30483. doi: 10.1074/jbc.M109.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Transcriptional activation of hepatic ACSL3 and ACSL5 by oncostatin m reduces hypertriglyceridemia through enhanced beta-oxidation. Arterioscler Thromb Vasc Biol. 2007;27:2198–2205. doi: 10.1161/ATVBAHA.107.148429. [DOI] [PubMed] [Google Scholar]
- 42.Nchoutmboube JA, et al. Increased long chain acyl-Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog. 2013;9:e1003401. doi: 10.1371/journal.ppat.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soupene E, Dinh NP, Siliakus M, Kuypers FA. Activity of the acyl-CoA synthetase ACSL6 isoforms: Role of the fatty acid Gate-domains. BMC Biochem. 2010;11:18. doi: 10.1186/1471-2091-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alajati A, et al. Interaction of CDCP1 with HER2 enhances HER2-driven tumorigenesis and promotes trastuzumab resistance in breast cancer. Cell Reports. 2015;11:564–576. doi: 10.1016/j.celrep.2015.03.044. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda JI, et al. Epigenetic regulation of the expression of the novel stem cell marker CDCP1 in cancer cells. J Pathol. 2006;210:75–84. doi: 10.1002/path.2026. [DOI] [PubMed] [Google Scholar]
- 46.Casar B, et al. In vivo cleaved CDCP1 promotes early tumor dissemination via complexing with activated β1 integrin and induction of FAK/PI3K/Akt motility signaling. Oncogene. 2014;33:255–268. doi: 10.1038/onc.2012.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benes CH, Poulogiannis G, Cantley LC, Soltoff SP. The SRC-associated protein CUB Domain-Containing Protein-1 regulates adhesion and motility. Oncogene. 2011;31:653–663. doi: 10.1038/onc.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H, et al. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro-oncol. 2016;19:43–54. doi: 10.1093/neuonc/now128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003, 1–15; erratum in Nat Cell Bio (2014) 16:1125. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caino MC, et al. Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J Clin Invest. 2013;123:2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues MF, et al. Enhanced OXPHOS, glutaminolysis and β-oxidation constitute the metastatic phenotype of melanoma cells. Biochem J. 2016;473:703–715. doi: 10.1042/BJ20150645. [DOI] [PubMed] [Google Scholar]
- 53.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kollmorgen G, et al. Antibody mediated CDCP1 degradation as mode of action for cancer targeted therapy. Mol Oncol. 2013;7:1142–1151. doi: 10.1016/j.molonc.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu CL, et al. Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition—A Warburg-reversing effect. PLoS One. 2015;10:e0121046. doi: 10.1371/journal.pone.0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Reports. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 57.Rybin VO, et al. Stimulus-specific differences in protein kinase C delta localization and activation mechanisms in cardiomyocytes. J Biol Chem. 2004;279:19350–19361. doi: 10.1074/jbc.M311096200. [DOI] [PubMed] [Google Scholar]
- 58.Martin S, Parton RG. Lipid droplets: A unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 59.Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Galbiati F, Razani B, Lisanti MP. Emerging themes in lipid rafts and caveolae. Cell. 2001;106:403–411. doi: 10.1016/s0092-8674(01)00472-x. [DOI] [PubMed] [Google Scholar]
- 61.van Meer G. Caveolin, cholesterol, and lipid droplets? J Cell Biol. 2001;152:F29–F34. doi: 10.1083/jcb.152.5.f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown DA. Lipid droplets: Proteins floating on a pool of fat. Curr Biol. 2001;11:R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 63.Krauss K, Altevogt P. Integrin leukocyte function-associated antigen-1-mediated cell binding can be activated by clustering of membrane rafts. J Biol Chem. 1999;274:36921–36927. doi: 10.1074/jbc.274.52.36921. [DOI] [PubMed] [Google Scholar]
- 64.Gagnoux-Palacios L, et al. Compartmentalization of integrin alpha6beta4 signaling in lipid rafts. J Cell Biol. 2003;162:1189–1196. doi: 10.1083/jcb.200305006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyazawa Y, et al. CDCP1 regulates the function of MT1-MMP and invadopodia-mediated invasion of cancer cells. Mol Cancer Res. 2013;11:628–637. doi: 10.1158/1541-7786.MCR-12-0544. [DOI] [PubMed] [Google Scholar]
- 66.Masson N, Ratcliffe PJ. Hypoxia signaling pathways in cancer metabolism: The importance of co-selecting interconnected physiological pathways. Cancer Metab. 2014;2:3. doi: 10.1186/2049-3002-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernardi R, Gianni L. Hallmarks of triple negative breast cancer emerging at last? Cell Res. 2014;24:904–905. doi: 10.1038/cr.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spassov DS, Wong CH, Wong SY, Reiter JF, Moasser MM. Trask loss enhances tumorigenic growth by liberating integrin signaling and growth factor receptor cross-talk in unanchored cells. Cancer Res. 2013;73:1168–1179. doi: 10.1158/0008-5472.CAN-12-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muoio DM, Lewin TM, Wiedmer P, Coleman RA. Acyl-CoAs are functionally channeled in liver: potential role of acyl-CoA synthetase. Am J Physiol Endocrinol Metab. 2000;279:E1366–E1373. doi: 10.1152/ajpendo.2000.279.6.E1366. [DOI] [PubMed] [Google Scholar]
- 72.Yan S, et al. Long-chain acyl-CoA synthetase in fatty acid metabolism involved in liver and other diseases: An update. World J Gastroenterol. 2015;21:3492–3498. doi: 10.3748/wjg.v21.i12.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 74.Parkes HA, et al. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am J Physiol Endocrinol Metab. 2006;291:E737–E744. doi: 10.1152/ajpendo.00112.2006. [DOI] [PubMed] [Google Scholar]
- 75.Lewin TM, Kim JH, Granger DA, Vance JE, Coleman RA. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 76.Ellis JM, Bowman CE, Wolfgang MJ. Metabolic and tissue-specific regulation of acyl-CoA metabolism. PLoS One. 2015;10:e0116587. doi: 10.1371/journal.pone.0116587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Padanad MS, et al. Fatty acid oxidation mediated by Acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Reports. 2016;16:1614–1628. doi: 10.1016/j.celrep.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Razorenova OV, Ivanov AV, Budanov AV, Chumakov PM. Virus-based reporter systems for monitoring transcriptional activity of hypoxia-inducible factor 1. Gene. 2005;350:89–98. doi: 10.1016/j.gene.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Razorenova OV, et al. The apoptosis repressor with a CARD domain (ARC) gene is a direct hypoxia-inducible factor 1 target gene and promotes survival and proliferation of VHL-deficient renal cancer cells. Mol Cell Biol. 2014;34:739–751. doi: 10.1128/MCB.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thompson JM, et al. Rho-associated kinase 1 inhibition is synthetically lethal with von Hippel-Lindau deficiency in clear cell renal cell carcinoma. Oncogene. 2017;36:1080–1089. doi: 10.1038/onc.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.