Significance
Ubiquitination—the covalent attachment of ubiquitin to substrates—is a posttranslational modification that regulates virtually every aspect of cellular function in eukaryotes. The final step of substrate ubiquitination requires the coordination of two types of enzymes: ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). Whereas E3s can function with different E2s, coordination between E3s has been reported only for E3s of the same class. Here we show that two distinct E2/E3 pairs (UBC-18/ARI-1 and UBC-3/CUL-1) coordinate to ubiquitinate a common substrate and regulate its steady-state levels in C. elegans. Failure to regulate the substrate's levels leads to a serious developmental defect and lethality in worms. Our work provides evidence that cross-talk between two classes of E3s and their respective dedicated E2s occurs in an organism.
Keywords: ubiquitin, RBR E3 ligase, Cullin-RING ligase, UBCH7, CDC34
Abstract
Ubiquitination, the crucial posttranslational modification that regulates the eukaryotic proteome, is carried out by a trio of enzymes, known as E1 [ubiquitin (Ub)-activating enzyme], E2 (Ub-conjugating enzyme), and E3 (Ub ligase). Although most E2s can work with any of the three mechanistically distinct classes of E3s, the E2 UBCH7 is unable to function with really interesting new gene (RING)-type E3s, thereby restricting it to homologous to E6AP C-terminus (HECT) and RING-in-between-RING (RBR) E3s. The Caenorhabditis elegans UBCH7 homolog, UBC-18, plays a critical role in developmental processes through its cooperation with the RBR E3 ARI-1 (HHARI in humans). We discovered that another E2, ubc-3, interacts genetically with ubc-18 in an unbiased genome-wide RNAi screen in C. elegans. These two E2s have nonoverlapping biochemical activities, and each is dedicated to distinct classes of E3s. UBC-3 is the ortholog of CDC34 that functions specifically with Cullin-RING E3 ligases, such as SCF (Skp1-Cullin-F-box). Our genetic and biochemical studies show that UBCH7 (UBC-18) and the RBR E3 HHARI (ARI-1) coordinate with CDC34 (UBC-3) and an SCF E3 complex to ubiquitinate a common substrate, a SKP1-related protein. We show that UBCH7/HHARI primes the substrate with a single Ub in the presence of CUL-1, and that CDC34 is required to build chains onto the Ub-primed substrate. Our study reveals that the association and coordination of two distinct E2/E3 pairs play essential roles in a developmental pathway and suggests that cooperative action among E3s is a conserved feature from worms to humans.
Protein ubiquitination is a posttranslational modification that regulates virtually every aspect of cellular function in eukaryotes. Covalent attachment of the small protein ubiquitin (Ub) to other cellular proteins can lead to a variety of outcomes, including proteasomal degradation of the substrate, changes in cellular localization, and altered enzymatic activity and protein–protein interactions (1). Protein ubiquitination is accomplished via a trio of enzymes: a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub ligase (E3). The E1 activates the C terminus of Ub and transfers activated Ub onto an E2 active-site cysteine (Cys) to generate an E2∼Ub (here “∼” denotes a thioester bond) that interacts with an E3 Ub ligase to coordinate substrate ubiquitination. A given organism has dozens of E2s and hundreds of E3s, providing an extensive foundation for diverse E2/E3 combinations to tightly regulate different substrates and their type of Ub modification. The identification of functional E2/E3/substrate relationships and their roles in vivo remains an important but underdeveloped area of investigation.
Three classes of eukaryotic E3 ligases use distinct Ub-transfer mechanisms. Members of the largest class, the RING (really interesting new gene) E3s, do not contain an active site. They bind an E2∼Ub to activate Ub transfer directly to a substrate, most often a lysine (Lys) residue. RBR (RING-in-between-RING) and HECT (homologous to E6AP C-terminus) E3s contain an active-site Cys residue to which Ub is transferred from the E2∼Ub to generate a covalent E3∼Ub thioester before transfer of Ub to a substrate. Most E2s can transfer Ub onto Cys residues (transthiolation) when bound to RBR- or HECT-type E3 or onto Lys residues (aminolysis) when bound to RING-type E3s. The E2 UBCH7 is unique in that it can perform only transthiolation reactions and thus is a dedicated E2 for RBR and HECT E3s (2).
Relatively little is known about the biological role of UBCH7. Although lacking a homolog in yeast, UBCH7 is highly conserved in worms and flies. The Caenorhabditis elegans homolog, UBC-18, is implicated in developmental processes through its cooperation with the RBR E3 ARI-1 (3). UBCH7 has been shown to regulate S-phase transition of the cell cycle in human cell culture (4); however, to date, it is mainly the multisubunit RING-type E3s known as Cullin-RING ligases (CRLs) associated with substrate ubiquitination that control cell cycle progression (5–7). Because UBCH7 is unable to work directly with RING E3s, including the CRLs, how it performs its role during regulation of the cell cycle is unclear.
C. elegans UBC-18 displays the same biochemical properties as its human counterpart and is able to work only with RBR and HECT E3s (2). Although a ubc-18 loss-of-function allele is viable, worms carrying this allele show a reduced growth rate and reduction in brood size (8). Furthermore, when ubc-18 is disrupted in combination with loss of function in lin-35, a retinoblastoma ortholog, animals arrest as first-stage larvae (L1) with a lethal defect in formation of the pharynx, the anterior portion of the worm gastrointestinal tract (8).
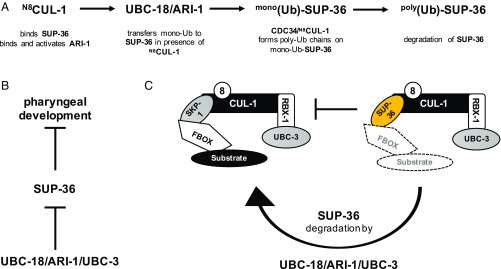
To define the biological roles of UBCH7, we conducted an RNAi screen to identify genes that interact genetically with ubc-18 in C. elegans. Our screen revealed that the C. elegans ortholog of the conserved E2 CDC34, ubc-3, acts with ubc-18 in pharyngeal development. CDC34 is considered a dedicated E2 for Skp1/Cullin/F-box (SCF)-type E3s, a subclass of CRLs. We further uncovered synthetic genetic relationships between ubc-18 and other components of SCF-type RING E3s. Given that UBC-18 is incapable of working with RING E3s and UBC-3 is a dedicated E2 for CRL E3s, these two E2s cannot substitute directly for each other. Instead, our data support a model in which UBC-18 and UBC-3 work either coordinately or in parallel pathways. In vitro, the RBR E2/E3 pair UBC-18/ARI-1 (UBCH7/HHARI) monoubiquitinates SUP-36, “priming” it as a substrate for polyubiquitination by CDC34/UBC-3. Consistent with this observation, both ubc-18 and ubc-3 regulate in vivo steady-state levels of SUP-36 protein; however, the “Ub priming” function of ARI-1 is not required for other Cullin E3s in C. elegans. Our findings complement a recent study in mammalian cell culture suggesting that RBR and CRL E3 complexes can work together (9). Furthermore, our work provides biological evidence for the existence of cross-talk between two different types of E3s, RING-and RBR-type E3s, and their respective, dedicated E2s in an organism.
Results
ubc-18 and ubc-3 Cooperate in Pharyngeal Development.
To identify genes that function with ubc-18, we performed an RNAi screen using ubc-18(ku354) mutant worms. The ku354 allele contains a substitution that changes a Glu at the E1-binding interface of UBC-18 to Lys, resulting in loss of function, likely owing to reduced charging of UBC-18 with Ub (8). Despite a smaller brood size and a slower growth rate, ubc-18(ku354) mutant worms are viable (8). In our primary screen, first larval stage (L1) worms were grown to adulthood (P0) on bacteria expressing gene-specific dsRNA clones from the genome-wide Ahringer library (10), and progeny (F1) were scored for penetrant phenotypes. We screened only clones from autosomes (a total of 14,400 library clones), a fraction of which (9.8%) failed to grow, yielding 12,982 clones that were screened, representing 67% of the total predicted genes in the C. elegans genome (10). Our primary screen identified 213 genes that caused a stronger phenotype when depleted in ubc-18(ku354) mutant animals compared with N2 wild-type (WT) controls. We retested these clones, resulting in the identification of 16 RNAi clones that robustly and reproducibly exhibited synthetic phenotypes with ubc-18 on rescreening (Table 1). Among these candidates was lin-35, which was previously shown to be synthetically lethal with ubc-18 (8), validating our screening method (Fig. 1A).
Table 1.
Novel genetic interactions with ubc-18 identified from the RNAi screen
| Gene | Synthetic phenotype | Process | Function/homology |
| ubc-3 | Lethal | Ubiquitination | E2 specific to SCF type E3; regulates G1 and G2 in cell cycle |
| skr-2 | Lethal | Ubiquitination | Skp1-related component of SCF type E3 ubiquitin ligase complex |
| skr-1 | Lethal | Ubiquitination | Skp1-related component of SCF type E3 ubiquitin ligase complex |
| egl-27 | Lethal | Chromatin regulation | MTA (human metastasis) homolog, nucleosome remodeling |
| lin-54 | Lethal | Nucleic acid regulation | DNA binding |
| Y65B4A.1 | Lethal | Nucleic acid regulation | Nucleotide binding |
| smu-2 | Lethal | Nucleic acid regulation | RNA splicing |
| emb-4 | Lethal | Nucleic acid regulation | Ortholog to human spliceosome |
| dpy-10 | Lethal | Body morphology and movement | Cuticle collagen protein |
| prx-13 | Lethal | Proteolysis | Human PEX13 homolog (endopeptidase); human Zellweger syndrome |
| ptc-1 | Lethal | Cytokinesis | Sugar modification; eggshell development |
| B0035.1 | Lethal | Unknown | ZNF207/BUGZ ortholog |
| T09A5.9 | Lethal | Unknown | PPP1R7 homolog (protein phosphatase) |
| fbf-2 | Sterile | Nucleic acid regulation | RNA binding |
| mrpl-10 | Sterile | Mitochondrial translation | Mitochondrial ribosomal protein, large subunit |
| trr-1 | Sterile | Chromatin regulation | Homolog of protein kinase in chromatin remodeling complex; synMuv |
Validated novel hits from RNAi screen using two ubc-18 loss-of-function alleles: ubc-18(ku354) and ubc-18(tm5426). Our screen also identified lin-35, previously shown to be synthetically lethal with ubc-18 (8). Process and function/homology are from Wormbase build WS254 (www.wormbase.org).
Fig. 1.
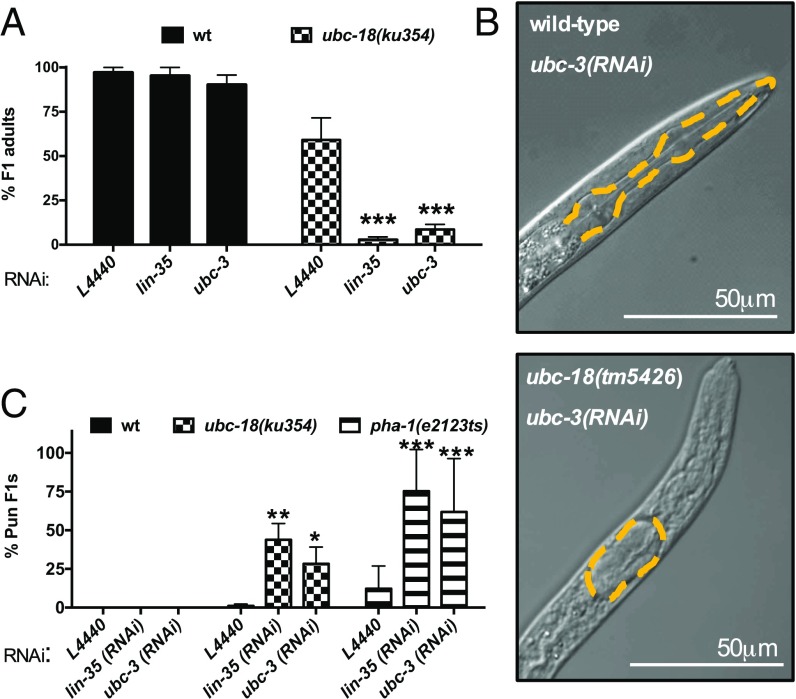
RNAi screen identified a synthetic genetic interaction between ubc-18 and ubc-3. (A) Quantitation of synthetic interaction between ubc-3 and ubc-18, measured as percent survival to adulthood. The pL4440 empty vector was included as the negative control, and lin-35 served as a positive control (8). Table 1 contains a list of other synthetic interactions identified in the RNAi screen. Error bars indicate the SD of at least three independent experiments. (B) Synthetic lethality between ubc-18(ku354) and ubc-3(RNAi) is associated with a Pharynx unattached (Pun) phenotype. In each image, the pharynx basement membrane is traced with the yellow dashed line. (Top) Wild-type L1 grown on ubc-3(RNAi). (Bottom) ubc-18(tm5426); ubc-3(RNAi) L1. (C) Quantification of the synthetic Pun phenotype. The percentage of Pun L1 observed in animals grown on the indicated RNAi for ubc-18(ku354) (checkered bars) and pha-1(e2123) mutant L1s (striped bars) is shown. The N2 WT control (wt, black bars) did not produce Pun progeny under any of the experimental conditions. Error bars indicate the SD of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 relative to L4440 controls. The absence of asterisks implies no significant difference relative to L4440 controls.
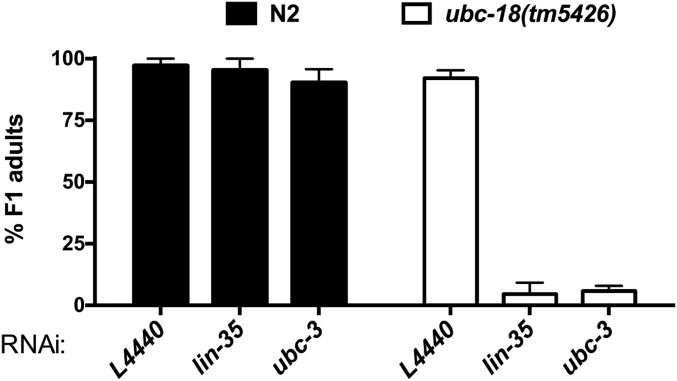
We found that among the confirmed hits, depletion of ubc-3, an E2 Ub-conjugating enzyme homologous to mammalian CDC34, was lethal in the ubc-18 mutant animals (Fig. 1A). To validate the synthetic interaction observed between ubc-18 and ubc-3, we tested another allele of ubc-18, tm5426. The tm5426 allele is a 308-bp deletion beginning in the second exon and extending into the third intron, and is a predicted molecular null. We found that ubc-18(tm5426) animals arrested as L1 larvae when grown on ubc-3(RNAi), similar to ubc-18(ku354) mutant animals (Fig. 1A and Fig. S1). We conclude that the synthetic lethal interaction between ubc-18 and ubc-3 is robust and statistically significant.
Fig. S1.
Synthetic genetic interactions between ubc-18(tm5426), and ubc-3(RNAi). Viability of animals grown on ubc-3(RNAi) was assessed for WT (N2, black bars) and ubc-18(tm5246) animals as in Fig. 1B. Error bars indicate the SD of at least three independent experiments.
Of the remaining 15 genes identified, 12 are synthetically lethal and 3 are synthetically sterile with ubc-18 (Table 1). The products of those genes include components of the ubiquitination machinery, metabolic enzymes, and nuclear proteins involved in transcription or splicing. We were especially interested in the interaction of ubc-18 with ubc-3. Based on known properties of the mammalian homologs, it is highly unlikely that ubc-3 and ubc-18 act redundantly with the same E3. The UBC-3 homolog in humans, CDC34, is a dedicated E2 for CRLs, multisubunit RING-type E3s that target cellular substrates for proteasomal degradation (7, 11). The mammalian homolog of UBC-18, UBCH7, is biochemically restricted to function only with RBR and HECT E3s and cannot function with CRLs (2). Thus, a synthetic interaction between these two E2s suggests either the presence of parallel pathways or coordination between an RBR/HECT E3 and a RING-type CRL—a relationship between two different classes of E3s that has not been reported in a biological system.
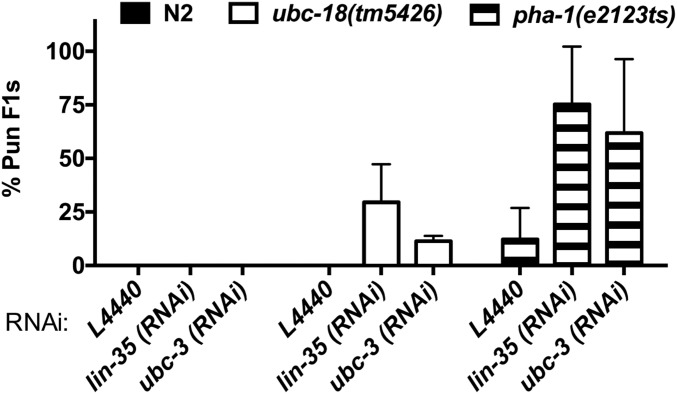
We noted that ubc-18 mutant animals grown on ubc-3(RNAi) arrest as L1, reminiscent of the reported synthetic phenotype between ubc-18 and lin-35, in which simultaneous inactivation of ubc-18 and lin-35 results in the Pharynx unattached (Pun) phenotype (8). Pun animals have a defect in pharyngeal morphogenesis in which the anterior end of the pharynx does not attach properly to the buccal cavity. As a result, Pun animals are unable to feed, arrest as L1, and consequently die (8). We considered the possibility that the synthetic interaction between ubc-18 and ubc-3 also results in a Pun phenotype. Indeed, examination of ubc-18; ubc-3(RNAi) animals revealed a significant fraction of Pun L1s (Fig. 1 B and C and Fig. S2). Although we noted variability in the penetrance of this phenotype among experiments, we never observed the Pun phenotype in WT animals, consistent with previous reports (8, 12) (Fig. 1C and Fig. S2). Although we did find a low frequency of Pun larvae (1–2%) for ubc-18(ku354) mutant animals grown on the control RNAi food, the penetrance was much greater in the ubc-3(RNAi) animals (Fig. 1C). These data suggest that ubc-3 also acts in the genetic network controlling pharyngeal morphogenesis.
Fig. S2.
Fraction of Pun L1, as in Fig. 1C. The percentage of Pun F1 progeny from animals grown on the indicated RNAi for ubc-18(tm5426) (white bars) and pha-1(e2123ts) (striped bars) is shown. Experiments with pha-1(e2123ts) were conducted at 15 °C. The isogenic wild-type control (N2; black bars) did not form Pun progeny under any of the experimental conditions. Error bars indicate the SD of at least three independent experiments.
We used additional genetic tools to test the hypothesis that ubc-3 acts with ubc-18 in pharyngeal morphogenesis. We first asked whether ubc-3 is synthetically lethal with pha-1, a gene that acts redundantly with class B synthetic multivulva (synMuv) genes to regulate pharyngeal development (12). We found a high penetrance of the Pun phenotype in pha-1(e2123ts) mutant animals grown on ubc-3(RNAi) plates at the permissive temperature of 15 °C (Fig. 1C and Fig. S2), similar to the findings in ubc-18(RNAi); pha-1(e2123ts) animals (12). We also found that disruption of sup-35, sup-36, or sup-37, which are known to suppress the synthetic genetic interaction between ubc-18 and lin-35 (3, 8), also suppressed the ubc-18/ubc-3 synthetic interaction (Table 2). Taken together, our data show that ubc-3 acts with ubc-18, lin-35, and pha-1 in the previously described genetic network that regulates pharyngeal development.
Table 2.
Viability of animals to adulthood for each genotype
| Genotype | % viable (n) |
| ubc-18(tm5426) | 92 (470) |
| ubc-18(tm5426); lin-35(RNAi) | 5 (603) |
| ubc-18(tm5426); ubc-3(RNAi) | 6 (853) |
| ubc-18(tm5426); sup-35(RNAi); ubc-3(RNAi) | 96 (161) |
| ubc-18(tm5426); sup-36(e2217); ubc-3(RNAi) | 100 (171) |
| ubc-18(tm5426); sup-37(e2215); ubc-3(RNAi) | 100 (263) |
| pha-1(e2123ts) | 75 (135) |
| pha-1(e2123ts); lin-35(RNAi) | 0.8 (69) |
| pha-1(e2123ts); ubc-3(RNAi) | 0 (69) |
| pha-1(e2123ts); sup-35(e2223); ubc-3(RNAi) | 99.7 (696) |
| pha-1(e2123ts); sup-36(e2217); ubc-3(RNAi) | 87.9 (555) |
| pha-1(e2123ts); sup-37(e2215); ubc-3(RNAi) | 97.3 (802) |
Temperature-sensitive pha-1 strains were assayed at the permissive temperature (15 °C). Data are aggregated from three independent experiments.
Genetic Interactions Between ubc-18 and Genes of the SCF E3 Ubiquitination Machinery.
The observed genetic interaction between ubc-18 and ubc-3 suggests that both a CRL E3 complex and an RBR/HECT E3 are involved in pharyngeal development. Consistent with this idea, our genome-wide RNAi screen also identified the SKP1-like CRL components skr-1 and skr-2 as synthetically lethal with ubc-18 (Table 1 and Fig. S3). SKP1 proteins are adaptors that connect F-box proteins to the CUL-1 scaffold in the SCF subclass of CRLs E3 complexes (13). Our screen did not recover genes coding for Cullin or F-box proteins, however. To assess the roles of other SCF components or other Cullins, we tested an additional set of CRL components for genetic interactions with ubc-18 by RNAi.
Fig. S3.
Synthetic genetic interactions with skr-1/2. Viability of WT, N2 (black bars), ubc-18(ku354), and ubc-18(ku354); sup-36(e2217) grown on the indicated RNAi was measured. Error bars indicate the SD of at least three independent experiments.
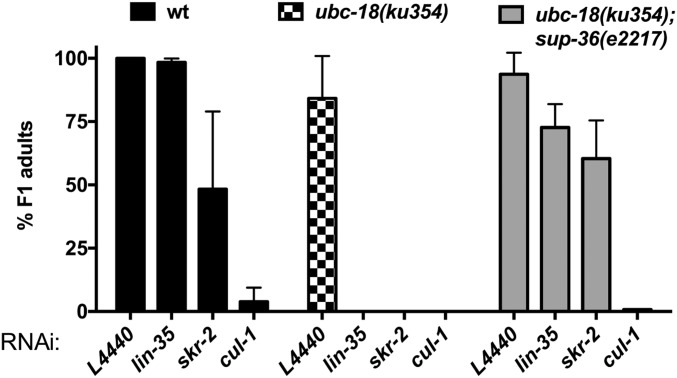
The C. elegans genome includes more than 20 SKP1-related genes, 6 Cullin-RING E3s, and more than 300 F-box genes (14–17). The candidate genes that we tested included five Cullins (cul-1, -2, -4, -5, -6) and SKP1-related (skr) and F-box genes chosen based on published genetic and physical interactions with members of the pharyngeal attachment pathway (18–21). Consistent with previous reports (22), we found that only 25% of WT animals grown on cul-1(RNAi) survived to adulthood, which excluded cul-1(RNAi) from our primary screen. However, we found that lethality was more penetrant for ubc-18 mutant animals grown on cul-1(RNAi) (Fig. 2A). We did not observe genetic interactions between ubc-18 and cul-4, -5, or -6(RNAi) (Fig. 2A).
Fig. 2.
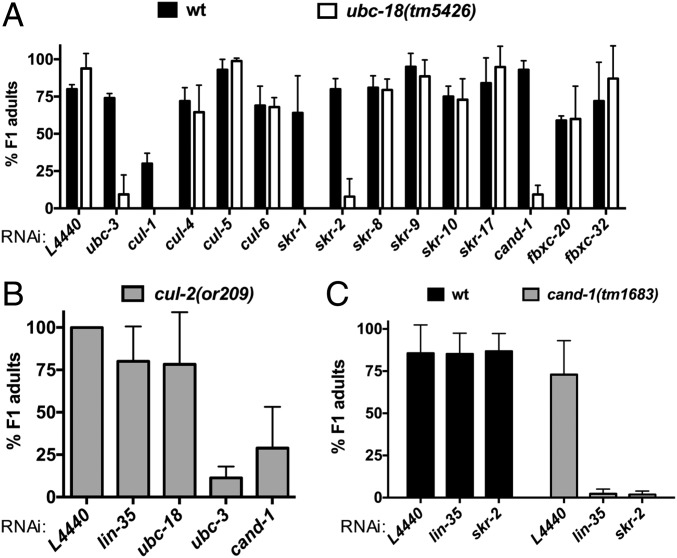
ubc-18 interacts synthetically with genes encoding SCF E3 ubiquitination machinery. (A) Quantification of synthetic lethal interactions with ubc-18. The percentage of F1 progeny of RNAi-fed animals that survive to adulthood is shown for WT (wt) and ubc-18(tm5426) strains (black and white bars, respectively). The rrf-3(pk1426) mutant served as the wild-type control. This mutant is wild type at the ubc-18 locus and reported to be RNAi-sensitive (49). Error bars are the SD between technical replicates in one representative experiment. The candidate screen was performed only once for genes that were not affected. Synthetic interactions with skr-1/2, cul-1, and cand-1 were observed in at least three independent experiments. (B) ubc-18 does not genetically interact with cul-2(or209ts). The percentage of F1 progeny of RNAi-fed animals that survived to adulthood is shown for cul-2(or209ts). Error bars are the SD between two independent experiments. (C) cand-1 genetically interacts with lin-35 and skr-2/1. The percentage of F1 progeny viable to adulthood is shown for N2 WT (wt, black bars) and cand-1(tm1683) mutant animals (gray bars) grown on each RNAi. Error bars represent the SD of at least three independent experiments.
To test for genetic interactions with cul-2, we used the temperature-sensitive cul-2(or209) allele (23). Although ubc-3(RNAi) was synthetically lethal with cul-2(or209), even at the permissive temperature, we did not observe a synthetic phenotype with either lin-35 or ubc-18 (Fig. 2B). This result suggests that although UBC-3 can function as an E2 for CUL-2 ligases in worms, this function is unrelated to the function of UBC-3 with UBC-18. Of the six SKP1-related genes tested, only skr-1/2 was synthetically lethal with ubc-18 (Fig. 2A and Fig. S3). Depletion of skr-10 or skr-17 did not lead to lethality in ubc-18 mutant animals, although these genes have been reported to have synthetic genetic interactions with lin-35 that result in larval lethality (21). We also did not observe genetic interactions with skr-8 or skr-9, which suppress the synthetic lethality of ubc-18; lin-35 animals (19). Based on these findings, we conclude that ubc-18 works with ubc-3, cul-1, and skr-1/2 in pharyngeal development.
Owing to the large number of F-box proteins in worms, we considered it unlikely that RNAi of a single F-box would recapitulate the loss of function of UBC-3, which works with many different SKR/CUL-1/F-box combinations. Thus, we used RNAi against Cullin-associated NEDD8-dissociated protein 1 (cand-1) to perturb F-box function on a wider scale. CAND-1 is required for the exchange of SKP1/F-box modules (and their substrates) on and off a SCF complex (24–26), and its depletion impacts known downstream outputs of SCF E3s in worms (27). We found that RNAi depletion of cand-1 in WT animals had no phenotype, but significantly reduced survival in ubc-18 mutant animals (Fig. 2A). Moreover, although cand-1 deletion mutants are viable, the additional knockdown of lin-35 or skr-1/2 is lethal (Fig. 2C). Overall, our data are consistent with a model in which both the RBR/HECT-specific E2 ubc-18 and the SCF-associated E2 ubc-3 play critical roles in pharyngeal development in worms.
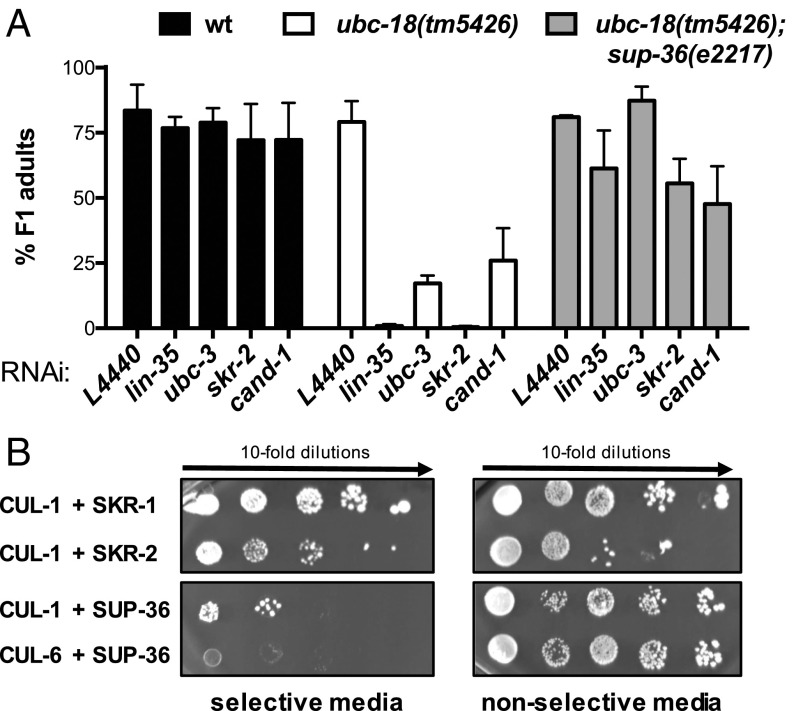
Similar to genetic interactions between ubc-18 and other components of pharynx development, the synthetic interactions between ubc-18 and SCF components ubc-3, skr-1/2, and cand-1 are each suppressed by a sup-36 null allele (19) (Fig. 3A, Table 2, and Fig. S3). Based on sequence similarity, sup-36 is predicted to encode a SKP1-like protein (19). The single SKP1 gene product in humans binds simultaneously to SCF subunits CUL-1 and F-box proteins. SUP-36 was previously found to bind to several F-box proteins (FBXC-20, FBXC-32, and FBXC-53) in yeast two-hybrid (Y2H) experiments (18–20), supporting the notion that it has SKP1-like properties. To further test the assignment of SUP-36 as a SKP1-like protein, we tested for its ability to interact with Cullins in a Y2H assay, and observed a physical interaction between SUP-36 and CUL-1 (Fig. 3B). Thus, SUP-36 can bind to both CUL-1 and F-box proteins, consistent with it being part of an SCF complex. However, the fact that a sup-36 null allele suppresses the synthetic phenotype of ubc-3 and ubc-18 indicates that these two E2s act to oppose the function of SUP-36.
Fig. 3.
SUP-36 has SKP1-like properties. (A) The sup-36(e2217) null allele suppresses the synthetic genetic interactions between ubc-18 and SCF components. (B) Yeast two-hybrid experiments with either CUL-1 or CUL-6 fused to the Gal4 DNA-binding domain and SKR-1, SKR-2, or SUP-36 fused to the Gal4 DNA-activating domain. (Left) Growth on selective media (−His, −Leu, −Trp) indicates interactions of the two proteins tested. As shown previously, CUL-1 binds to both SKR-1 and SKR-2. SUP-36 binds to CUL-1 (Top), but not to CUL-6 (Bottom). (Right) Yeast were also spotted on nonselective media (−Leu, −Trp) to control for growth.
UBC-18/ARI-1 and UBC-3/CUL-1 Regulate SUP-36 Protein Levels.
A simple explanation for why loss of SUP-36 suppresses the synthetic lethality between ubc-18 and ubc-3 is that SUP-36 is a ubiquitination target of UBC-18 and UBC-3 (Fig. 3A, Table 2, and Fig. S3). Indeed, the E2/RBR E3 pair UBC-18/ARI-1 has been reported to regulate protein levels of SUP-36 in worms (19), providing indirect evidence that SUP-36 may be a substrate of UBC-18/ARI-1. This observation is puzzling, however, because the human homolog of ARI-1 (HHARI) cannot build poly-Ub chains that are normally considered to target a substrate to the proteasome (9) (Fig. S4). On the other hand, the human ortholog of UBC-3 (CDC34) together with the E3 CUL-1 does generate poly-Ub chains on substrates that lead to proteasomal degradation (28–30). These known biochemical activities, together with our genetic data, led us to hypothesize that SUP-36 might be a common substrate of UBC-18/ARI-1 and UBC-3/CUL-1. To test this possibility, we measured SUP-36 levels in embryos using a SUP-36::GFP reporter. As reported previously, SUP-36::GFP levels are increased in worms grown on ubc-18(RNAi) and ari-1(RNAi), indicating that UBC-18/ARI-1 directly or indirectly controls SUP-36 protein levels (19) (Fig. 4 A and B and Fig. S5A). We found that depletion of ubc-3 by RNAi results in increased SUP-36::GFP levels (Fig. 4 A and B and Fig. S5A), providing evidence that UBC-3 is also involved in the degradation of SUP-36. Consistent with our hypothesis that SUP-36 might be a common substrate for UBC-18/ARI-1 and UBC-3/CUL-1, SUP-36::GFP levels were increased even further when worms were grown on ubc-18(RNAi); ubc-3(RNAi) compared with on either ubc-18(RNAi) or ubc-3(RNAi) alone (Fig. 4 A and B and Fig. S5A).
Fig. S4.
HHARIRBR does not form poly-Ub chains. Autoubiquitination assays in which the GST-tagged E3 (HHARI) served as a proxy substrate were performed with either WT Ub (Left) or Lys-less Ub (Right). The construct used, GST-HHARIRBR, lacks the Ariadne domain and thus is constitutively active (32). Lys-less Ub is unable to form Ub-chains; therefore, the higher molecular weight bands observed are numerous multiple mono-Ub modifications. Products were visualized by Western blot analysis against the GST-tag on HHARI.
Fig. 4.
SUP-36 protein levels are regulated by UBCH7/HHARI (UBC-18/ARI-1) and CDC34/CUL-1 (UBC-3/CUL-1). (A) Representative images for SUP-36::GFP expression levels measured in comma to twofold embryos under indicated RNAi conditions. (B) Quantification of embryos in the comma to twofold stage [L4440 vector, n = 27; ubc-18(RNAi), n = 45; ubc-3(RNAi), n = 67; ubc-18(RNAi);ubc-3(RNAi), n = 95]. Boxplots show quartiles, with mean indicated by the thick black line, and whiskers show 95% confidence intervals. Statistical significance was determined via Welch’s unequal variance t tests. Measurements for embryos in early embryogenesis and twofold to threefold stage are provided in Fig. S5A. (C) In vitro immunoprecipitation (IP) using recombinant T7-SUP-36 (IP: T7) and recombinant, untagged N8-CUL-1. Results were visualized by Western blot analysis with indicated antibodies. Replicates are shown in Fig. S5B. (D) In vitro ubiquitination of SUP-36. Each reaction contained T7-SUP-36 and other reaction components as indicated. All time points are post-ATP addition, except lane 1 (0 min). Longer time courses are shown in Fig. S6A. (E) The T7-SUP-36 in vitro ubiquitination assay was performed using Lys-less Ub, which is unable to form chains. Unmodified SUP-36 and single-ubiquitinated and double-ubiquitinated T7-SUP-36 are indicated by arrows. All time points are 10 min after ATP addition; longer time courses are shown in Fig. S6 A and B.
Fig. S5.
(A) SUP-36::GFP expression levels were measured in early embryogenesis and twofold to threefold embryos under indicated RNAi conditions. (Left) Quantification of SUP-36 expression in early embryogenesis. L4440 vector control, n = 15; ubc-18(RNAi), n = 19; ubc-3(RNAi), n = 41; ubc-18(RNAi);ubc-3(RNAi), n = 32. (Right) Quantification of SUP-36 levels at the twofold to threefold stage. L4440 vector control, n = 30; ubc-18(RNAi), n = 65; ubc-3(RNAi), n = 72; ubc-18(RNAi);ubc-3(RNAi), n = 60. Statistical significance was determined using Welch’s unequal variance t test, with P < 0.05. (B) Three replicates of in vitro IP using recombinant T7-SUP-36 (IP: T7) and recombinant, untagged N8-CUL-1. The SUP-36N109K/F110K double mutant at the predicted SUP-36/CUL-1 interface retains binding to N8-CUL-1, in contrast to results reported for the analogous mutation in human SKP1 (13). Results were visualized by Western blot analysis with the indicated antibodies. The intensities of elution bands were quantified using ImageJ. Ratios of intensities (for either CUL-1 or SUP-36, as indicated) in mutant SUP-36 elutions compared with WT SUP-36 elutions are shown. (C, Left) A SUP-36 homology model was generated using Phyre2 (50) and then overlaid with the structure of human SKP1 bound to CUL-1 (Protein Data Bank ID code 1LDK; SUP-36 is shown in red, SKP1 in yellow, and CUL-1 in blue). SUP-36 residues N109 and F110 overlay closely with N108/Y110 of human SKP1 at the SKP1/CUL-1 interface (13). (C, Right) Region of clustal omega sequence alignment containing SUP-36 residues N109 and F110 (red) and the aligned sequences of (worm) SKR-1 and (human) SKP-1.
The foregoing observations suggest that the two E2s work in parallel and/or coordinately to regulate protein levels of SUP-36 (Fig. 4 A and B and Fig. S5A). The distinctly different intrinsic biochemical activities of UBC-18 and UBC-3 suggest a model in which UBC-18 (with its E3 ARI-1) modifies SUP-36 with a single Ub onto which UBC-3 (with its E3 CUL-1) can extend poly-Ub chains (Table S1). To test this hypothesis, we performed biochemical experiments to assess the activities of UBC-18 and UBC-3 in SUP-36 ubiquitination. We used human homologs for UBC-18 (UBCH7), UBC-3 (CDC34), ARI-1 (HHARI), and neddylated CUL-1 (N8-CUL-1), because we have previously demonstrated that combined recombinant worm/human enzymes exhibit biochemical activity, consistent with their high sequence similarity (2). We confirmed that human N8-CUL-1 binds to SUP-36 directly by in vitro pull-down experiments (Fig. 4C and Fig. S5B), corroborating the interaction between SUP-36 and CUL-1 identified by the Y2H assay (Fig. 3B). Because SUP-36 is a predicted SKP1 family member, we examined whether mutations in SUP-36 at the predicted SUP-36/CUL-1 interface (N109K/F110K; Fig. S5C) disrupt binding to N8-CUL-1, as has been observed for the analogous mutation in human SKP1 (13). SUP-36N109K/F110K still bound to N8-CUL-1 in our assay (Fig. S5B), suggesting that it binds differently from a canonical SKP1, or that it binds more tightly and is not effectively disrupted by the interface mutations.
Table S1.
Summary of known functions of proteins relevant to this study
| C. elegans (human) | Type of protein | Binding partners | Functions | Effects on SUP-36 protein levels |
| UBC-18 (UBCH7) | E2 Ub-conjugating enzyme | ARI-1/ HHARI | Transfers Ub onto Cys residues (RBR/HECT active sites), but not Lys | Increased (RNAi) |
| Can only function with RBR/HECT-type E3s | ||||
| UBC-3 (CDC-34) | E2 Ub-conjugating enzyme | CUL1 | Able to build K48-linked chains with Cullin E3 ligases | Increased (RNAi) |
| ARI-1 (HHARI) | RBR E3 ligase | UBC-18/ | Contains active site Cys, transfers mono-Ub only | Increased (RNAi) |
| UBCH7 | Activated by binding to neddylated CUL1 | |||
| High affinity for UBC-18/UBCH7 | ||||
| CUL-1/RBX-1 (CUL1/RBX1) | RING E3 ligase | CDC34 | Forms complexes with RBX-1 (RING domain) and SKP1/SKR1 | Unknown |
| HHARI | Known to function with CDC34 to build poly-Ub chains | |||
| SKP1 | Activates ARI-1/HHARI through physical interactions | |||
| SUP-36* | ||||
| SKR-1/2 (SKP1) | Scaffold for CUL-1/CUL1 | CUL1 | Acts as scaffold protein between CUL-1/CUL1 and Fbox proteins | Unknown |
| Fbox proteins | • | |||
| SUP-36 (???) | SKP1-like | CUL-1/ | Inhibits pharyngeal morphogenesis | Unknown |
| CUL1* | Molecular function unknown |
New physical interactions reported in this study.
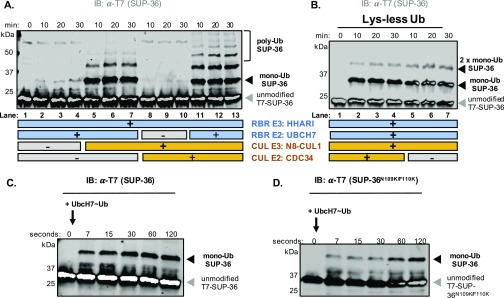
We performed biochemical assays using purified recombinant proteins, and product formation was evaluated by Western blot analysis (Fig. 4D). By itself, neither of the two functional E2/E3 pairs, UBCH7/HHARI or CDC34/N8-CUL-1, modified SUP-36 to a detectable degree (Fig. 4D, lanes 2/3 and 4/5, respectively); however, adding N8-CUL-1 to reactions with UBCH7/HHARI led to the generation of SUP-36 modified by one or two Ubs (lanes 8/9). In contrast, adding HHARI to reactions with CDC34/N8-CUL-1 did not produce any detectable modification of SUP-36 (lanes 10/11). Only when all four enzymes were present did we observe poly-Ub chain formation on SUP-36 (lanes 12/13). We suspected that the products that we observed in the absence of CDC34 were monoubiquitinated SUP-36. To test this possibility, we used Lys-less Ub, which is unable to support the synthesis of poly-Ub chains (Fig. 4E). Consistent with mono-Ub formation, reactions containing all four components and Lys-less Ub gave rise to a similar modification pattern as that seen in reactions using WT Ub that lack CDC34 (Fig. 4D, lanes 8/9 and Fig. S6A, lanes 5–7). Taken together, these data indicate that CDC34 (UBC-3), but not UBCH7 (UBC-18), is capable of poly-Ub chain formation on SUP-36.
Fig. S6.
(A) SUP-36 in vitro ubiquitination assay similar to the assay shown in Fig. 4D, but with longer time points displayed. Each reaction contained T7-SUP-36 and other reaction components as indicated below the gel. All time points are after ATP addition, except lane 1 (0 min). In the presence of CUL-1, UBCH7/HHARI modify SUP-36 with one and two single Ubs (lanes 5–7), whereas no product is detected when CDC34 is the sole E2 (lanes 8–10). All four components (UBCH7/HHARI and CDC34/CUL-1) are required for the formation of poly-Ub chains on SUP-36 (lanes 11–13). (B) Longer time points of the Lys-less Ub assay shown in Fig. 4E. SUP-36 is modified with one or two single Ubs in the presence of all four components. The same pattern is observed when CDC34 is omitted, confirming that CDC34 is required for Ub chain formation (in the presence of WT Ub), and that the multiple SUP-36 bands seen in Fig. 4D (lanes 8–9) and A (lanes 5–7) arise from addition of multiple mono-Ubs. Unmodified SUP-36 and mono- and double-Ub T7-SUP-36 are indicated by arrows. (C) Precharged UBCH7∼Ub was added to reactions containing HHARI, N8-CUL-1, and T7-SUP-36. The time points shown are after UBCH7∼Ub addition. (The 0 s time point was taken immediately before UBCH7∼Ub addition.) The results were visualized by Western blot analysis against T7 (SUP-36). (D) The same assay as in C, but using the SUP-36N109K/F110K mutant at the predicted SUP-36/CUL-1 interface.
The biochemical results imply that polyubiquitination of SUP-36 is a multistep process that requires two E2s. We propose that UBCH7 (UBC-18) acts to attach the initial Ub to form mono-Ub-SUP-36. Given that UBCH7 (UBC-18) alone is unable to modify substrate Lys residues directly (2), UBCH7 (UBC-18) ubiquitinates SUP-36 with its partner HHARI (ARI-1), an RBR E3 that specifically transfers mono-Ub moieties (9) (Fig. S4). A second E2 and its associated E3 must then build a poly-Ub chain on mono-Ub-SUP-36. In vitro, this second step can be carried out by CDC34 (UBC-3) and N8-CUL-1.
In principle, the two-step modification of SUP-36 could be accomplished either through the action of two distinct E2/E3 complexes or coordinated through a larger multienzyme complex. Evidence exists in support of a multienzyme complex; a physical interaction between human HHARI and N8-CUL-1 has been reported, and HHARI ligase activity is known to be activated by N8-CUL-1 (9, 31). Our reconstituted assay system involving a bona fide substrate whose ubiquitination depends on both E2s lends further support to this model. First, recombinant UBCH7 (UBC-18)/HHARI (ARI-1) did not modify SUP-36 unless N8-CUL-1 was present as well (Fig. 4D, lanes 8/9 compared with lanes 2/3). This observation can be explained by N8-CUL-1 binding and activating autoinhibited HHARI, as reported recently (9, 31, 32). Second, N8-CUL-1 bound directly to SUP-36 (Fig. 4C), providing a potential substrate-binding site. Importantly, we did not observe the reverse situation, in which HHARI activates N8-CUL-1 to modify SUP-36. Thus, binding of both HHARI (ARI-1) and SUP-36 to N8-CUL-1 is not sufficient for N8-CUL-1/CDC34 (UBC-3) to transfer Ub directly to SUP-36 without the function of UBCH7 (UBC-18).
Taken together, our genetic data and biochemical assay results support the idea that UBCH7 (UBC-18)/HHARI (ARI-1) carries out monoubiquitination of SUP-36 in a N8-CUL-1–dependent manner, and that CDC34 (UBC-3)/CUL-1 polyubiquitinate SUP-36 to negatively regulate SUP-36 protein levels, thereby opposing its function. Substrate modification by a CUL-1 complex typically requires a substrate to bind to an F-box protein that binds to the Cullin scaffold through SKP1. In contrast, our data suggest that a SKP1-like protein, SUP-36, behaves like a substrate of CUL-1 both in vitro and in vivo. Notably, our biochemical reactions were carried out in the absence of either (human) SKP1 or an F-box protein. To rule out nonspecific modification of SUP-36 by UBCH7/CUL-1/N8-CUL-1, we performed a biochemical assay analogous to those previously published for canonical Cullin substrate ubiquitination in the presence of UBCH7/HHARI, in which precharged UBCH7∼Ub is added to initiate reactions (9). In these reactions (Fig. S6C), we observed monoubiquitination of SUP-36 in the presence of UBCH7/CUL-1/N8-CUL-1 within seconds, implying that SUP-36 modification is unlikely to result from random collisions. In agreement with our finding that SUP-36N109K/F110K is still able to bind N8-CUL-1 (Fig. S5B), we also observed monoubiquitination of the mutant SUP-36N109K/F110K (Fig. S6D), supporting the notion that the SUP-36/CUL-1 interaction is at least partially noncanonical.
Discussion
Here we provide biological evidence of collaboration between two different types of E3s, RING-and RBR-type E3s, and their respective, dedicated E2s in an organism. Although our results are in some ways reminiscent of those from a recent study in mammalian cell culture suggesting that RBR and CRL E3 complexes can work together (9), there are some notable distinctions. Our findings indicate that the “Ub priming” function of ARI-1 is not required for all CUL-1 substrates or for other Cullin E3s in C. elegans. This conclusion is supported by the fact that a ubc-18 null allele is viable, yet cul-1(RNAi) substantially reduces survival. In addition to CUL-1, HHARI reportedly can form complexes with CUL-2, -3, and -4A (9, 31); however, we did not observe synthetic interactions between ubc-18 and cul-4 or cul-5 by RNAi (Fig. 2A) or between ubc-18 and cul-2(or209). In contrast, ubc-3(RNAi) was synthetically lethal with cul-2(or209) (Fig. 2B), corroborating that ubc-3 function with Cullins extends beyond the involvement of ubc-18. We conclude that UBCH7 (UBC-18) and its E3 HHARI (ARI-1) function coordinately with UBC-3 (CDC34)/CUL-1 to ubiquitinate some common substrates, including SUP-36, whose protein levels must be regulated to ensure proper pharyngeal development.
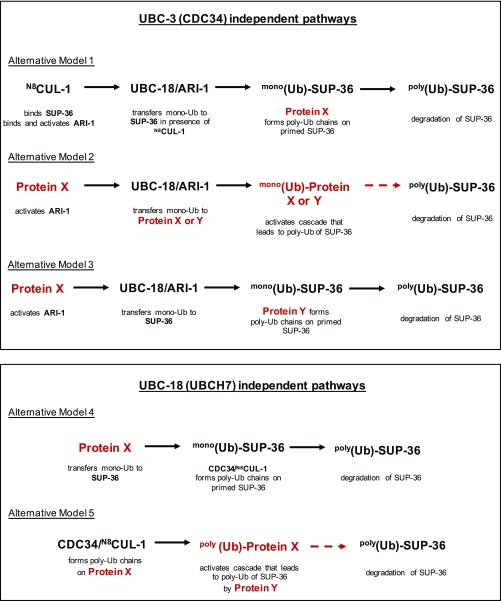
Taken alone, the genetics data would seem to imply that parallel pathways, each involving one of the E2s, regulate SUP-36; however, in combination with the biochemical results, both parallel and sequential pathways can be proposed that are consistent with the data (Fig. S7). We favor a model in which UBC-18 (UBCH7) and UBC-3 (CDC34), along with their dedicated E3s, the RBR E3 ARI-1 (HHARI) and the RING-type E3 CUL-1, work sequentially to polyubiquitinate SUP-36 (Fig. 5A). The sequential model is consistent with the known biochemical activities of the enzymes involved (Table S1). Although the synthetic genetic interactions that we observed could be interpreted as evidence for UBC-18 and UBC-3 acting in parallel pathways, there is precedent for synthetic genetic interactions between members of a complex, especially in components of essential complexes or when hypomorphic alleles are tested (33). In principle, stepwise modification of SUP-36 by UBCH7/HHARI and CDC34/CUL-1 could be carried out by two separate E2/E3 complexes; however, several lines of evidence indicate that this is not the case. First, a direct interaction between the two E3s is known to occur and to lead to activation of HHARI (31, 32), indicating that a three-enzyme complex carries out the first modification reaction. Second, SUP-36 binds directly to CUL-1 (Figs. 3B and 4C and Fig. S5B), suggesting that CUL-1 may serve to recruit and bind a substrate for HHARI (ARI-1) as well as for itself. Together with our experimental data, these observations suggest that a large complex containing both UBC-18/ARI-1 and UBC-3/CUL-1 may direct the sequential monoubiquitination and polyubiquitination of SUP-36 in vivo.
Fig. S7.
Several potential parallel pathways that are consistent with genetic data presented herein. We cannot rule out the formal possibility that another E3 with polyubiquitinating activity may perform the elongation step following the action of UBC18/ARI-1 on SUP-36, or that UBC-18 and UBC-3 use their different functions to act in parallel pathways. Combinations of alternative models 1–3 with either model 4 or 5 also would be consistent with the genetic results.
Fig. 5.
Proposed model for UBC-18/UBC-3-dependent ubiquitination of SUP-36. (A) A sequential model in which UBC-18/ARI-1 acts coordinately with UBC-3/CUL-1 to regulate the proteasomal degradation of SUP-36 is consistent with genetic and biochemical data (this work and others). In the model, CUL-1 binds SUP-36 and binds and activates ARI-1, which, together with UBC-18, monoubiquitinates SUP-36. The E2 UBC-3, with its E3 CUL-1, then builds poly-Ub chains using mono-Ub-SUP-36 as its substrate, thereby promoting the degradation of SUP-36. (B) SUP-36 protein levels increase when ubc-18, ari-1, or ubc-3 is depleted by RNAi (this work and ref. 19), and SUP-36 levels are even higher when both ubc-18 and ubc-3 are depleted, leading to inhibition of pharyngeal morphogenesis (this work). (C) A model in which SUP-36 inhibits formation of a canonical SKR-1/CUL-1 complex, and this inhibition is released when SUP-36 is ubiquitinated by UBC-18/ARI-1 and UBC-3/CUL-1, leading to its degradation, is consistent with these observations: (i) a functional SKR-1/CUL-1 complex is required for pharyngeal morphogenesis; (ii) the synthetic phenotype of ubc-18;skr-1/2(RNAi) can be rescued by a sup-36 null allele; and (iii) the SKP1-like SUP-36 can bind to CUL-1, and its degradation is dependent on UBC-18/ARI-1/UBC-3.
Another possibility consistent with our genetic and biochemical data is that other E2s can partially substitute for the functions of UBC-18 and UBC-3 in vivo. One candidate for this action is ubc-2, the worm ortholog of UBCH5, which is active with both HHARI and CUL-1 and has been proposed to perform the priming step for (human) SCF E3s (2, 34). We were not able to test this possibility, because ubc-2(RNAi) is embryonic lethal (35), precluding its testing with ubc-18 or ubc-3. Nevertheless, our data indicate that no E2 can effectively compensate for the simultaneous knockdown of both ubc-18 and ubc-3. Our original screen did not any identify any E3s, suggesting UBCH7/HHARI as the primary machinery for generating polyubiquitin chains on SUP-36. However, the limits of RNAi preclude us from formally ruling out the possibility that other factors are involved as well. Several potential parallel pathways are shown in Fig. S7.
Our findings also reveal some intriguing, possibly unconventional properties of SCF E3 ligases. Although yeast and humans each contain a single gene encoding for the substrate adaptor subunit SKP1, there are more than 20 Skp1-related (skr) genes predicted in C. elegans (15). Presumably, each of these may bind to a subset of the F-box proteins in worms to provide a large diversity of substrate-binding modules. Although not identified as a Skp1-like protein based on sequence alone, SUP-36 has been suggested to have Skp1-like properties based on its predicted protein sequence and ability to bind F-box proteins (18–20). Our data further support the identification of SUP-36 as Skp1-like by demonstrating its direct binding to CUL-1 (Figs. 3B and 4C and Fig. S5B). Paradoxically, whereas SUP-36 can bind to both F-box proteins and CUL-1, we show that it also acts as a substrate for a CUL-1 complex during pharyngeal morphogenesis. To our knowledge, neither yeast nor human Skp1 has been identified as a substrate for SCF-dependent ubiquitination. We also found that ubc-18 and skr-1/2 exhibit a synthetic phenotype that is rescued by a sup-36 null allele (Fig. 3A and Fig. S3), indicating that a functional, canonical SKP-1/CUL-1 complex is required for proper pharyngeal development and reduction of its function can be counteracted by removing SUP-36. Thus, we speculate that SUP-36 may act as an inhibitor of SCF complexes in pharyngeal morphogenesis (Fig. 5 B and C). Whether that role requires an F-box protein, and whether such an F-box protein could stabilize an inhibitory SUP-36/CUL-1 complex or signal for SUP-36 degradation, are open questions for future studies.
Previously reported evidence indicates that SUP-36 inhibits pharyngeal development with two other proteins, SUP-35 and SUP-37, possibly through the function of a large macromolecular complex (19). Consistent with that idea, both sup-35 and sup-37 mutant alleles also rescue the ubc-18; ubc-3(RNAi) synthetic lethal phenotype (Table 2). Notably, SUP-36 has been shown to bind to a microtubule-associated protein PTL-1, and SUP-35 has been shown to bind directly to microtubules (19). Taken together, the data suggest that SUP-35 and SUP-36 work together in pharyngeal morphogenesis, and that SUP-36 may have roles independent of its Skp1-like function. It is worth noting here that the single Skp1-like gene in yeast, SKP-1, is known to perform both SCF-dependent and SCF-independent functions, the latter as part of the kinetochore (36, 37). In contrast, the sole documented function of the single human SKP1 gene product is to act as an adaptor connecting F-box proteins to CUL-1. Whether human SKP1 has lost its non-SCF functions, or whether there is a functional ortholog to SUP-36, remains to be discovered.
Coordination among E3s is an emerging theme for tight regulation of protein ubiquitination. A study published while the present paper was in preparation demonstrated that depletion of HHARI in cell culture leads to accumulation of known CRL substrates, and that some CRL substrates are primed with Ub by HHARI/UBCH7 followed by poly-Ub chain formation by CDC34/CRL in vitro (9). Although our genetic and biochemical evidence complements those findings by showing that the orthologs of these enzymes regulate the levels of a common substrate in worms and work together to ubiquitinate that common substrate in vitro, our genetic data also imply that the ability of the E2/E3 pairs to work coordinately is not critical for all biological CUL-1–dependent processes. Which substrates are common among the RBR E2/E3 and CRL E2/E3 pairs, how they are selected, and the biological importance of the coordinated modification are all key questions to be addressed in the future.
Materials and Methods
Strains and Maintenance.
Worms were cultured according to standard methods on nematode growth medium plates seeded with OP50 bacteria at 20 °C, except for temperature-sensitive strains, which were maintained at 15 °C (38). The strains used in this study are listed in Table S2. Double-mutant strains were constructed by standard techniques, and alleles were genotyped by PCR using primer sequences shown in Table S3.
Table S2.
C. elegans strains used in this study
| Strain | Genotype | Source |
| N2 | Wild type | Bristol isolate, from CGC |
| WY34 | ubc-18(ku354) | CGC |
| DLM37 | ubc-18(tm5426) | NBRP strain, outcrossed to N2 (this work) |
| GE24 | pha-1(e2123) | CGC |
| DLM70 | ubc-18(tm5426); sup-35(e2223) | DLM37 crossed to GE346 (this work) |
| MT14541 | sup-36(e2217) | David Fay, University of Wyoming |
| DLM72 | ubc-18(tm5426); sup-36(e2217) | DLM37 crossed to MT14541 (this work) |
| DLM74 | ubc-18(tm5426); sup-37(e2215) | DLM37 crossed to GE347 (this work) |
| GE346 | pha-1(e2123); sup-35(e2223); dpy-18 (e499) | CGC |
| DLM71 | pha-1(e2123); sup-36(e2217) | GE24 crossed to MT14541 (this work) |
| GE337 | pha-1(e2123); sup-37(e2215); dpy-18 (e499) | CGC |
| DLM76 | cand-1 (tm1683) | NBRP strain outcrossed to N2 (this work) |
| EU640 | cul-2(or209) | CGC |
| WY1001 | fdEx239(rol-6gf; SUP-36::GFP) | D. Fay |
CGC, Caenorhabditis Genetics Center; NBRP, National Bioresource Project.
Table S3.
Primers for genotyping and cloning Y2H constructs
| Gene (allele) | Forward | Reverse | PCR product |
| Genotyping | |||
| ubc-18 (ku354) | cagaaggtgagaaaatcatg | ggctccttatcctgaaaatt | 200 bp, ku354 allele introduces Hpy188I site |
| ubc-18 (tm5426) | cgagaatgttgagtgcgagg | agtgacagcaaggccatcat | 430 bp (WT), 120 bp (mutant) |
| pha1 (e2123) | gctttctctgctgaaacc | gcgcactactgaatcagagt | 900 bp, e2123 introduces AccI site |
| sup-35 (e2223) | atgctctttggcctcttctg | acgtctccaaagcaaaagcg | 480 bp, e2223 removes BseI site |
| sup-36 (e2217) | cgcgcaatactcatagagtttc | aatccatccgtcttcctctg | 750 bp (WT) or no band (mutant) |
| sup-37 (e2215) | gaaagcccatggcagaattcatccagatatgatggag | gggtgagagcgcagatgcgagtattc | 105 bp, allele removes DpnI site |
| cand-1 (tm1683) | cgcatcatgtcagtaatcgg | caaaaaaggctcatttgggc | 875 bp (WT), or 250 bp (mutant) |
| Y2H constructs | |||
| cul-1 | atgacaaatagcgaaccgagaagaatgacg | agagagcgcaaaatgaagcc | |
| cul-6 | atgagcatcgaagcagtgtggggaac | agccagtaattttatgccaaatactc | |
| skr-1 | cgattcgaattcatggctgatcaaaagaaag | ctgaagggatccttaatcctcgcaccagg | |
| sup-36 | cttccgcgaattcatgccatcttgcgagatcaaattcg | cgaggggaggatccctattggacattctgg | |
RNAi Knockdown and Screening.
RNAi knockdown of target genes was performed as described previously (10). All RNAi clones were obtained in HT115 bacteria from the Ahringer library. The empty vector pL4440 served as a negative control in these experiments. lin-35(RNAi) served as a positive control for synthetic lethality with ubc-18(ku354) mutant animals. The ku354 allele encodes a nonconservative missense mutation (E10K) in the E1-binding surface and acts as a strong loss-of-function allele (8). unc-45 and pos-1, which generate strong uncoordinated (unc)/sterile (ste) and embryonic lethal (emb) phenotypes, respectively, were used to gauge the strength of the RNAi effect in specific experiments. For double-RNAi experiments, animals were fed a 1:1 mixture of fresh, saturated overnight cultures of both RNAi strains.
For the primary screen, 10 P0 worms were grown on each indicated RNAi clone from L1 in 24-well plates. In each well, 15 μL of saturated bacterial culture was seeded onto solid agar. After 5 d, the F1 progeny were examined for ste, emb, or larval arrest (lva) phenotypes. Bacterial clones that caused a phenotype in the ubc-18(ku354) mutant strain but not in WT animals were retested three times to detect those with robust synthetic interactions. If both WT and ubc-18 mutants showed identical phenotypes, these RNAi were excluded from further analysis. Of the 12,982 clones from the Ahringer library that were screened, 213 were selected for secondary screening. To quantitate RNAi phenotypes and for the secondary screen, timed egg-laying assays were performed using ubc-18(tm5426) animals. Synchronized populations of L1 larvae were grown to gravid adulthood on RNAi, then 10 RNAi-fed adults were transferred to fresh RNAi plates and allowed to lay eggs for up to 4 h. The adults were removed, and progeny were scored within the next 72 h.
To quantitate synthetic phenotypes, the fraction of embryos that developed to fertile adults was calculated. Embryos were counted immediately after the egg-laying period, and the number of adults were counted after 3 d or when control animals on L4440 RNAi food had developed to gravid adults. To quantitate the synthetic Pun phenotype, animals were rinsed off plates at 48 h after the timed egg-laying assay, allowed to settle by gravity, and then resuspended in M9 with 25 mM sodium azide. The fraction of Pun L1 larvae was assessed using a Nikon 90i compound microscope with Nomarsky optics and a 40× oil-immersion objective.
The statistical significance in quantitative analyses of synthetic lethality (Fig. 1A and Fig. S1) was assessed using two-way ANOVA with Bonferroni posttest on data from three separate biological replicates, each consisting of three technical replicates. The statistical significance of synthetic Pun phenotypes (Fig. 1C) was assessed similarly by two-way ANOVA on data from at least two separate biological replicates, each consisting of at least two technical replicates. For all experiments, data were graphically plotted and statistical analyses performed with GraphPad Prism 5.
SUP-36::GFP Measurements.
The C. elegans strains used in this study were maintained using standard procedures as described previously (39), using the WY1001 strain [fdEx239(rol-6gf; SUP-36::GFP)]. RNAi used standard feeding protocols (40) for the following targets: empty vector (pDF129.36), ubc-3 (Geneservice library), ubc-18 (pDF67), and ubc-3+ubc-18 (mixed 1:1). All RNAi clones were confirmed via sequencing. SUP-36::GFP–positive L4 hermaphrodites were plated on appropriate RNAi plates (empty vector, ubc-3, ubc-18, or ubc-3+ubc-18). Embryos were collected the next day via gentle suction with a microfluidic aspirator tube and placed on a 3% agar pad affixed to a glass slide. Quantitative fluorescent imaging was performed as described previously (19) with several modifications. In brief, embryos were imaged with a Nikon Eclipse microscope using OpenLab version 5.0.2, with an identical exposure time and microscope setup for all images. Differential interference contrast images were obtained for each sample before fluorescent measurement to confirm the embryonic stage and ensure adequate depth of focus. Embryos from each strain were staged into three distinct age groups based on developmental morphology: early morphogenesis, comma to twofold, and twofold to threefold. Fluorescence was quantified using Fiji ImageJ software (41, 42). The integrated fluorescence density was determined for each embryo by subtracting the background fluorescence from the calculated integrated density. Mean fluorescence was subsequently calculated for each worm strain and developmental group for comparison of SUP-36 expression levels. Statistical significance was determined using Welch’s unequal variance t test using R version 3.3.2 (43), with the significance level set at P < 0.05.
Y2H Assays.
To construct Y2H fusion constructs, the full coding sequence of each gene was amplified by PCR from cDNA. Primer sequences are listed in Table S3. To generate cDNA, mRNA was isolated from mixed-stage C. elegans using TRIzol (Thermo Fisher Scientific) following the manufacturer’s recommended protocol, and cDNA was synthesized from 5 μg total RNA using SuperScript III Reverse Transcriptase (Invitrogen). The PCR primers were designed to include restriction enzyme sites, and the PCR products were digested with the appropriate restriction enzyme (New England BioLabs) and ligated into plasmids to generate protein fusions with the Gal4-activation domain (GAD) or Gal4 DNA-binding domain (GBD). Plasmids pGAD-C1 (SKR-1 and SUP-36) and pGBD-C1 (CUL-1 and CUL-6) (44) were used in this study. Final plasmids were sequenced to verify that the fusion constructs were correct. Y2H assays were performed as described previously (45). Serial dilutions of yeast cells expressing fusions with GAD and GBD fusions were spotted onto selective media (-His, -Leu, -Trp) to assess interactions and onto nonselective media (-Leu, -Trp) with histidine to control for spotting efficiency.
Recombinant Proteins Used for in Vitro Ubiquitination Assays.
The following constructs, coded for full-length and human proteins unless indicated otherwise, were used in this study: E1, wheat E1, HA-UB-WT, HA-UB-K0 (Lys-less), UBCH7, CDC34, neddylated-(N8)-CUL-113-776/RBX-1116-108, T7-SUP-36, HHARI, and GST-HHARI-RBR177-395.
Expression and Purification of Recombinant Proteins.
Proteins were expressed and purified as described previously (2, 13, 32, 46–48). Full-length SUP-36 was cloned into a pet28a expression vector in-frame with an N-terminal His-T7 tag. Mutations were introduced using Quickchange. SUP-36 was expressed in Escherichia coli (BL21 DE3 cells) grown in LB and induced with 200 μM isopropyl β-d-1-thiogalactopyranoside at 16 °C for 18–20 h. SUP-36 was initially purified from cell lysate using Ni2+-affinity chromatography, followed by cleavage of the His-tag with thrombin (Sigma-Aldrich). After thrombin capture (p-aminobenzamidine-agarose; Sigma-Aldrich), further Ni2+-affinity chromatography was performed to capture uncleaved product, followed by size exclusion chromatography (SEC) in 25 mM NaPO4 and 150 mM NaCl, pH 7.0.
Immunoprecipitation Assays.
Protein G Dynabeads (Novex) were incubated with T7 antibody (Novagen; 69522) in 25 mM NaPO4, 150 mM NaCl pH 7.0, and 0.1% Tween 20 for 30 min and then washed twice. Then 2 µM N8-CUL-1 was mixed with 2 µM T7-SUP-36 for 15 min, followed by incubation with the Dynabead-antibody complex for 20 min. Samples were washed three times with 25 mM NaPO4, 150 mM NaCl pH 7.0, and 0.1% Tween 20. Bound proteins were eluted with SDS/PAGE reducing buffer. Samples were separated by SDS/PAGE and analyzed by Western blot analysis for CUL-1 (MyBioSource; MBS 821982) and T7 (EMD Millipore; AB3790). All binding was performed at room temperature.
Ubiquitination Assays.
E3 autoubiquitination assays.
Here 0.5 μM wheat E1, 20 μM Ub, 2 μM UBCH7, and 2 μM GST-HHARIRBR were incubated at 37 °C in 25 mM NaPO4 and 150 mM NaCl, pH 7.0. Reactions were initiated by the addition of 10 mM ATP and quenched with SDS/PAGE reducing buffer. Time points were run on SDS/PAGE gel and visualized by Western blot analysis for GST (HHARI).
SUP-36 ubiquitination assays.
Here 0.5 μM human E1, 20 μM Ub, 2 μM UBCH7, 2 μM CDC34, 1 μM HHARI, 1 μM N8-CUL-1/RBX-1, and 2 μM T7-SUP-36 were incubated as indicated in each assay at 37 °C in 25 mM NaPO4 and 150 mM NaCl, pH 7.0. Reactions were initiated by the addition of 10 mM ATP and quenched with SDS/PAGE reducing buffer. Time points were run on SDS/PAGE gel and visualized by Western blot analysis for T7 (Millipore; AB3790).
UBCH7∼UB Discharge Assays.
UBCH7 was charged with HA-UB as described previously (48), and UBCH7∼UB was purified by SEC at 4 °C. Then 2 μM N8-CUL-1/RBX-1, 2 μM HHARI, and 3 μM T7-SUP-36 (WT or N109K/F110K) were premixed in 25 mM NaPO4 and 150 mM NaCl, pH 7.0. Reactions were performed at 37 °C and initiated on the addition of 5 μM purified UBCH7∼UB immediately after taking a 0 s time point. Each time point was quenched with SDS/PAGE reducing buffer and run on SDS/PAGE gel, followed by Western blot analysis for T7 (EMD Millipore; AB3790).
Acknowledgments
We thank N. Zheng and Haibin Mao for their generous gift of purified recombinant neddylated-CUL-1/RBX-1 and CDC34; R. Gardner for plasmids and assistance with yeast two-hybrid experiments; and M. Stewart for a critical reading of the manuscript and insightful discussions. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health’s Office of Research Infrastructure Programs (Grant P40 OD010440). This work was supported by the National Institute of General Medical Sciences (Grant R01 GM088055 to R.E.K., R01 GM066868 and P20 GM103432 to D.S.F., and National Research Service Award 5T32 GM007270 to K.K.D.), the National Institute on Aging (Grant R01 AG044378 to D.L.M.), and the U.W. Hurd Fellowship Fund (K.K.D.). D.L.M. is a New Scholar in Aging at the Ellison Medical Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705060114/-/DCSupplemental.
References
- 1.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani K, Fay DS. A mechanistic basis for the coordinated regulation of pharyngeal morphogenesis in Caenorhabditis elegans by LIN-35/Rb and UBC-18-ARI-1. PLoS Genet. 2009;5:e1000510. doi: 10.1371/journal.pgen.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitcomb EA, Taylor A. Ubiquitin control of S phase: A new role for the ubiquitin conjugating enzyme, UbcH7. Cell Div. 2009;4:17. doi: 10.1186/1747-1028-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaugule VK, Walden H. Specificity and disease in the ubiquitin system. Biochem Soc Trans. 2016;44:212–227. doi: 10.1042/BST20150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng N, Wang Z, Wei W. Ubiquitination-mediated degradation of cell cycle-related proteins by F-box proteins. Int J Biochem Cell Biol. 2016;73:99–110. doi: 10.1016/j.biocel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Fay DS, Large E, Han M, Darland M. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development. 2003;130:3319–3330. doi: 10.1242/dev.00561. [DOI] [PubMed] [Google Scholar]
- 9.Scott DC, et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166:1198–1214 e1124. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 11.Schwob E, Böhm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 12.Fay DS, et al. The coordinate regulation of pharyngeal development in C. elegans by lin-35/Rb, pha-1, and ubc-18. Dev Biol. 2004;271:11–25. doi: 10.1016/j.ydbio.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 14.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nayak S, et al. The Caenorhabditis elegans Skp1-related gene family: Diverse functions in cell proliferation, morphogenesis, and meiosis. Curr Biol. 2002;12:277–287. doi: 10.1016/s0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]
- 16.Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 17.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polley SR, et al. Implicating SCF complexes in organogenesis in Caenorhabditis elegans. Genetics. 2014;196:211–223. doi: 10.1534/genetics.113.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceron J, et al. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol. 2007;7:30. doi: 10.1186/1471-213X-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka A, et al. Multiple Skp1-related proteins in Caenorhabditis elegans: Diverse patterns of interaction with Cullins and F-box proteins. Curr Biol. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- 23.Merlet J, Pintard L. Role of the CRL2(LRR-1) E3 ubiquitin-ligase in the development of the germline in C. elegans. Worm. 2013;2:e25716. doi: 10.4161/worm.25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 25.Wu S, et al. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce NW, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosu DR, et al. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Dev Biol. 2010;346:113–126. doi: 10.1016/j.ydbio.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodge A, Mendenhall M. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol Gen Genet. 1999;262:55–64. doi: 10.1007/s004380051059. [DOI] [PubMed] [Google Scholar]
- 29.Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell. 2003;11:1435–1444. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 30.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 31.Kelsall IR, et al. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 2013;32:2848–2860. doi: 10.1038/emboj.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duda DM, et al. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: Autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 34.Wu K, Kovacev J, Pan ZQ. Priming and extending: A UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhen M, Schein JE, Baillie DL, Candido EP. An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in the nematode Caenorhabditis elegans. EMBO J. 1996;15:3229–3237. [PMC free article] [PubMed] [Google Scholar]
- 36.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai C, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 38.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stiernagle T. Maintenance of C. elegans. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahringer J, editor. WormBook. 2006. Reverse genetics. [DOI] [Google Scholar]
- 41.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 44.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbaum JC, et al. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17:1221–1235. doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dove KK, et al. Structural Studies of HHARI/UbcH7∼Ub reveal unique E2∼Ub conformational restriction by RBR RING1. Structure. 2017;25:890–900. doi: 10.1016/j.str.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 50.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]