Significance
Despite intensive therapy, the cure rate for children diagnosed with high-risk neuroblastoma is still below 50%, accentuating the need for more effective therapies. Recurrent somatic mutations are relatively infrequent in neuroblastoma. We show that approximately 30% of neuroblastoma contains mutations in genes regulating Rho/Rac signaling. The mutations may be associated with activation of downstream Rho-associated kinases (ROCKs). High ROCK2 expression is associated with poor patient survival. Inhibition of ROCK activity suppresses the growth of neuroblastoma in preclinical in vivo models. ROCK inhibitors induce differentiation of neuroblastoma cells partly by glycogen synthase kinase 3β-mediated phosphorylation and degradation of MYCN proteins. These findings suggest that inhibitors of ROCK may represent a therapeutic opportunity for children with high-risk neuroblastoma.
Keywords: neuroblastoma, Rho signaling, ROCK, personalized medicine, Wnt signaling
Abstract
Neuroblastoma is a peripheral neural system tumor that originates from the neural crest and is the most common and deadly tumor of infancy. Here we show that neuroblastoma harbors frequent mutations of genes controlling the Rac/Rho signaling cascade important for proper migration and differentiation of neural crest cells during neuritogenesis. RhoA is activated in tumors from neuroblastoma patients, and elevated expression of Rho-associated kinase (ROCK)2 is associated with poor patient survival. Pharmacological or genetic inhibition of ROCK1 and 2, key molecules in Rho signaling, resulted in neuroblastoma cell differentiation and inhibition of neuroblastoma cell growth, migration, and invasion. Molecularly, ROCK inhibition induced glycogen synthase kinase 3β-dependent phosphorylation and degradation of MYCN protein. Small-molecule inhibition of ROCK suppressed MYCN-driven neuroblastoma growth in TH-MYCN homozygous transgenic mice and MYCN gene-amplified neuroblastoma xenograft growth in nude mice. Interference with Rho/Rac signaling might offer therapeutic perspectives for high-risk neuroblastoma.
Neuroblastoma is a childhood tumor of the peripheral sympathetic nervous system, causing 6% of all childhood cancers but 9% of deaths from malignant tumors in children (1, 2). Approximately half of patients present with high-risk disease characterized by unresectable primary lesions and multiple metastases (2, 3). Although survival in this group has improved, the majority of the tumors show resistance to therapy with poor patient survival despite intensive multimodal therapy, necessitating the search for new therapeutic options (2).
Compared with adult tumors, pediatric cancers exhibit significantly fewer genomic aberrations and mutations. In neuroblastoma, somatically acquired amplification of MYCN, hemizygous deletions of 1p and 11q, and gain of 17q are the most common chromosomal aberrations associated with a poor prognosis (3). Exome- and whole-genome sequencing analysis of neuroblastoma has reported an overall low somatic mutation count (12 to 18, median 15), with ALK being the most frequently mutated gene (7 to 10%) (4–6). In addition, chromothripsis, PHOX2B, and ATRX mutations have also been detected in a subset of high-risk tumors (4, 5).
Neuritogenesis is initiated during embryogenesis by a transient population of cells called the neural crest. During embryonic development, neural crest cells migrate throughout the embryo and eventually differentiate into multiple cell types, such as neurons and glial cells of the peripheral nervous system, pigment cells, fibroblasts, smooth muscle cells, and odontoblasts. The failure of neural crest cells to differentiate can result in development of cancers such as neuroblastoma and melanoma (7). A combination of Wingless (Wnt), bone morphogenetic protein, and fibroblast growth factor (FGF) signals is required to induce the formation of the neural crest and to initiate migration of neural crest cells by acquiring cell motility through epithelial–mesenchymal transition (8). The noncanonical Wnt-planar cell polarity (PCP) signaling cascade is fundamental for the migration of neural crest cells by controlling contact inhibition of locomotion between neural crest cells. PCP proteins control the activity of Rho GTPases locally by activating or inhibiting RhoA and Rac1, resulting in cells migrating away from each other upon collision (7). The stimulation of Rho signaling by PCP results in downstream activation of the serine/threonine Rho-associated coiled coil-containing protein kinases (ROCK)1 and ROCK2 (9). ROCK1 and ROCK2 phosphorylate downstream substrates such as myosin light chain and LIM kinases 1/2, which further regulate a range of cellular functions primarily through rearrangement of the actin cytoskeleton (10, 11). ROCK is dysregulated in a variety of cancers, including prostate, breast, and lung cancers, with ROCK overexpression contributing to metastasis by enhancing tumor cell invasion and motility (11).
Here we report that ROCK is a promising target for the treatment of high-risk neuroblastoma patients expressing high MYCN levels. We show that genes controlling the activity of ROCKs are frequently mutated and that high ROCK2 expression in neuroblastoma tumors corresponds to poor patient survival. Silencing or pharmacologic inhibition of ROCK induces glycogen synthase kinase (GSK)3β-mediated degradation of MYCN, neuroblastoma cell differentiation, and suppression of neuroblastoma growth in preclinical in vivo models.
Results
Neurogenesis Genes Are Frequently Mutated in Neuroblastoma.
Whole-exome and whole-genome sequencing were performed on human neuroblastoma tumors deriving from different clinical subsets together with matched germline DNA (n = 40) (Table 1) or without (n = 25) (SI Appendix, Table S1). Thirteen of the 40 neuroblastoma samples with germline control had at least one somatic mutation (32.5%) (Table 1), and 18 of the 25 neuroblastoma samples profiled contained one or more rare [<0.001% occurrence in the 1000 Genomes Project; not present in the Single Nucleotide Polymorphism database (dbSNP) or our in-house database] genetic protein-changing variants (72%) in the 177 selected genes controlling neuritogenesis (SI Appendix, Table S1). The average number of protein-changing mutations in tumors with germline controls was 19 (2 to 77), whereas 195 (range 44 to 496) protein-changing rare variants were detected in tumors without matched controls per tumor. Likewise, analysis of published and validated genomic data from 383 human neuroblastoma samples showed at least one somatic protein-changing alteration in a gene involved in neuritogenesis in 30.5% (117/383) of the samples (SI Appendix, Tables S2 and S3). For all Rho/Rac-associated genes included in the analysis, see SI Appendix, Table S4. The most commonly altered gene associated with neuritogenesis was alpha thalassemia/mental retardation gene X-linked (ATRX). However, 27.5% of the patients in our cohort contained at least one mutation or genetic aberration in genes associated with Rho/Rac signaling (11/40) other than ATRX (Table 1). The corresponding numbers in the published cohorts were 25% (97/383) (SI Appendix, Table S3).
Table 1.
Somatic mutations in Rho/Rac-associated genes in 40 human neuroblastoma tumors deriving from different clinical subsets
| Mutations in Rho/Rac-associated genes | Clinical patient characteristics | ||||||||||||
| Sample | Gene | Protein | Nucleotide | 1p del | 11q del | 17q gain | MNA | Age at diagnosis, mo | Stage INRGSS* | Risk INRG† | Outcome | Genomic profile | Sequencing |
| 1 | TENM1 | p.I2152L | c.6454A>T | No | Yes | Yes | No | 83 | L | + | DOD | 11q del | Exome |
| ATRX | Focal del exon 1–15 | ||||||||||||
| 2 | ARHGEF37 | p.V70I | c.208G>A | Yes | Yes | Yes | No | 9 | L | − | NED | 11q del | Exome |
| 3 | VAV3 | p.R330C | c.988C>T | No | Yes | Yes | No | 21 | M | + | DOD | 11q del | WGS |
| 4 | CSMD1 | p.E1766K | c.5296G>A | Yes | No | No | Yes | 41 | L | + | DOD | MNA | Exome |
| 5 | CSMD3 | — | Tandem insert exon 28–59 | No | No | No | No | 38 | M | + | DOD | OS | Exome/WGS |
| 6 | ATRX | — | Focal del exon 3–8 | No | Yes | Yes | No | 28 | M | + | DOD | 11q del | Exome |
| 7 | ATRX | — | Focal del exon 8–18 | Yes | Yes | Yes | No | 55 | M | + | NED | 11q del | WGS |
| 8 | TRIOBP | p.I1691T | c.2072T>C | No | Rearr | Yes | No | 22 | L | − | AWD | 17q gain | WGS |
| ATRX | — | Focal del exon 2–9 | |||||||||||
| TENM1 | p.E1712K | c.5134G>A | |||||||||||
| 9 | PTPRD | — | Focal gain exon 1 | Yes | No | Yes | No | 66 | M | + | NED/AWD | 17q gain | WGS |
| 10 | ARHGEF7 | — | t(13, 20)(q34;p12.2) | No | Yes | Yes | No | 61 | M | + | AWD | 11q del | WGS |
| 11 | TIAM1 | p.G719fs*5 | c.2156delG | Yes | Yes‡ | No | Yes | 41 | M | + | DOD | MNA/11q del | WGS |
| TIAM1 | p.E411Q | c.1231G>C | |||||||||||
| 12 | ATRX | — | Focal del exon 2–9 | Yes | Yes | Yes | No | 50 | M | + | PD | 11q del | WGS |
| FZD5 | p.Q472K | c.1414C>A | |||||||||||
| 13 | TENM2 | — | Tandem insert exon 1–2 | No | Yes | Yes | No | 158 | M | + | NED/AWD | 11q del | WGS |
| 14 | No | No | Yes | No | 1 | M | − | NED/AWD | 17q gain | WGS | |||
| 15 | Yes | No | Yes | Yes | 21 | M | + | AWD | MNA | WGS | |||
| 16 | Yes | Wcl11 | Yes | No | 83 | M | + | AWD | 17q gain | WGS | |||
| 17 | Yes | Yes | Yes | No | 36 | M | + | AWD | 11q del | WGS | |||
| 18 | No | No | Yes | Yes | 30 | M | + | DOD | MNA | Exome | |||
| 19 | Yes | Inter del | Wcg17 | Yes | 50 | M | + | DOD | MNA | Exome | |||
| 20 | No | Yes | Yes | No | >18 | M | + | NED | 11q del | Exome | |||
| 21 | No | Yes | No | No | 29 | M | + | DOD | 11q del | Exome | |||
| 22 | Yes | Yes | Yes | No | 35 | L | − | DOD | 11q del | Exome | |||
| 23 | Yes | No | Yes | Yes | 20 | M | + | NED/AWD | MNA | Exome | |||
| 24 | Yes | No | Yes | Yes | 37 | M | + | DOD | MNA | Exome | |||
| 25 | No | No | Wcg17 | No | 25 | L | − | NED | NO | Exome | |||
| 26 | No | No | Wcg17 | No | 1 | L | − | NED | NO | Exome | |||
| 27 | Yes | No | Yes | Yes | 13 | L | + | NED | MNA | Exome | |||
| 28 | No | No | No | No | 17 | L | − | NED | NO | Exome | |||
| 29 | No | No | Yes | Yes | N/A | N/A | + | NED/AWD | MNA | Exome | |||
| 30 | Yes | No | Yes | Yes | N/A | N/A | + | DOD | MNA | Exome | |||
| 31 | Yes | No | Yes | No | N/A | N/A | N/A | DOD | OS | Exome | |||
| 32 | No | Yes | Yes | No | 8 | M | − | AWD | 11q del | WGS | |||
| 33 | No | No | Wcg17 | No | 27 | L | − | NED | OS | WGS | |||
| 34 | Yes | No | No | No | 1 | M | − | DOD | OS | WGS | |||
| 35 | Yes | Inter del | Yes | Yes | 11 | M | + | DOD | MNA | WGS | |||
| 36 | No | No | Wcg17 | Yes | 27 | M | + | NED | MNA | WGS | |||
| 37 | No | Yes | Yes | No | 19 | M | + | NED | 11q del | WGS | |||
| 38 | Yes | Yes | Yes | No | 6 | M | − | AWD | 11q del | WGS | |||
| 39 | Yes | No | Yes | Yes | 43 | L | + | DOD | MNA | Exome/WGS | |||
| 40 | Yes | Yes | Yes | No | 29 | M | + | NED | 11q del | WGS | |||
AWD, alive with disease; del, deletion; DOD, dead of disease; insert, insertion; Inter, interstitial; L, localized; M, metastatic; MNA, MYCN amplification; N/A, not available; NED, no evidence of disease; NO, numerical only; OS, other structural; Pcl, parts of chromosome loss; PD, progressive disease; Rearr, rearrangement; Wcg, whole-chromosome gain; Wcl, whole-chromosome loss; WGS, whole-genome sequencing.
INRGSS (International Neuroblastoma Risk Group staging system) stage, modified after Monclair et al. (13); “localized” includes stage L1/L2 [International Neuroblastoma Staging System (INSS) 1, 2, and 3] and metastatic stage M (INSS 4), except for stage MS (INSS 4S).
Risk INRG, according to Cohn et al. (12); +, high risk; −, very low risk, low risk, and intermediate risk.
Patient presented with additional loss on chromosome 11p.
In our neuroblastoma cohort, 69% (27/39) were defined as high-risk disease (defined according to classification systems) (12, 13). Among the high-risk patients, 37% (10/27) had mutations in Rho/Rac-associated genes, whereas patients with Rho/Rac gene mutations were found in 25% (3/12) of patients with intermediate- and low-risk neuroblastoma (Table 1). In the patient group with mutations in genes in the Rho/Rac pathway, 9 out of 13 also had 11q deletion, a highly aggressive subgroup of neuroblastoma (Fisher’s exact test, P = 0.038) (Table 1).
ROCKs Are Expressed in Neuroblastoma and ROCK2 Is Associated with Poor Survival.
To investigate the importance of Rho signaling in neuroblastoma, we analyzed expression levels of the downstream Rho-activating kinases ROCK1 and ROCK2 in five different publicly available and validated cohorts of neuroblastoma. For ROCK1, three datasets showed a significant correlation between high expression and poor patient overall survival, one dataset displayed nonsignificant results (but with the same trend), and one dataset showed the opposite, namely that low expression of ROCK1 was significantly associated with poor overall survival. For ROCK2, four datasets showed a significant association between high expression and poor overall survival, whereas one dataset presented a nonsignificant result but with the same trend (SI Appendix, Fig. S1). In conclusion, ROCK2 was detected in neuroblastoma tumors with higher expression levels being associated with poor patient survival (SI Appendix, Fig. S1).
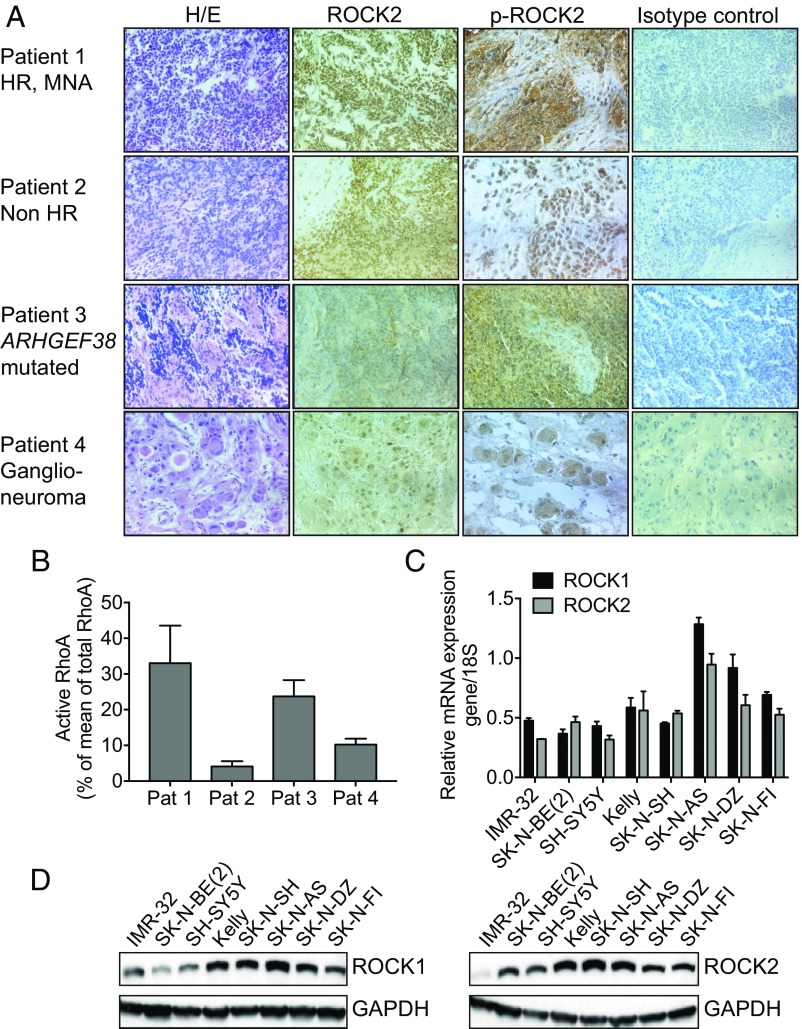
Next, we investigated the protein expression pattern of ROCK2 and an activated phosphorylated form of ROCK2 (p-Thr396) by immunohistochemistry in neuroblastoma specimens representing different clinical subgroups. This showed abundant protein levels of ROCK2 in both the cytoplasm and nucleus in neuroblastoma tumors (Fig. 1A). Immunohistochemical staining of the neural marker synaptophysin and phosphorylated ROCK2 confirmed the localization and expression of ROCK2 in neuroblastoma cells (SI Appendix, Fig. S2). The most intense positive staining for phosphorylated ROCK2 (p-Thr396) was detected in a patient with a non–MYCN-amplified tumor with a rare variant in the Rho GTPase ARHGEF38 gene (c.C1705T; p.Q569*, nonsense mutation) and in tumor cells derived from a patient with MYCN amplification (Fig. 1A). Only moderate levels of phosphorylated ROCK2 were detected in the low-risk neuroblastoma patient and in the ganglioneuroma patient samples (Fig. 1A). In line with these data, measurement of active RhoA/total RhoA in each investigated patient tumor showed a higher portion of active RhoA compared with total RhoA in the neuroblastoma samples exhibiting high ROCK2 phosphorylation (33% in patient 1 with MYCN amplification and 24% in patient 3 with Rho GTPase ARHGEF38 gene mutation) compared with tumors showing low ROCK phosphorylation (4.1% in patient 2 with non–high-risk neuroblastoma and 10% in patient 4 with ganglioneuroma) (Fig. 1B). Also, the relative level of active RhoA was higher compared with active Rac1 in the neuroblastoma sample containing ARHGEF38 mutation (patient 3, SI Appendix, Fig. S3). Similarly, real-time PCR and Western blot for ROCK1/2 in a panel of neuroblastoma cell lines, all derived from high-risk neuroblastoma patients, showed evident levels of mRNA and protein from both genes (Fig. 1 C and D).
Fig. 1.
ROCK2 is expressed in human neuroblastoma tumor samples and cell lines. (A) ROCK2 and phosphorylated ROCK2 (p-ROCK2) in tumors from four neuroblastoma patients with different clinobiological backgrounds: a high-risk (HR) tumor with MYCN amplification (MNA), non-HR tumor (numerical only genomic profile), non–MYCN-amplified tumor with a nonsense mutation in ARHGEF38, and benign ganglioneuroma tumor are shown. Images were acquired at 40× magnification except for p-ROCK2, which is at 60×. H/E, hematoxylin/eosin. (B) Quantification of active RhoA protein, expressed as percent of total RhoA in the matched patient sample, in the same four neuroblastoma patient (Pat) samples, analyzed with ELISA. Means of five determinations with SEM are displayed. (C) mRNA expression of ROCK1 and ROCK2 in neuroblastoma cell lines, assessed with real-time PCR. Data represent the mean with SD of three determinations. (D) Western blot analysis showing ROCK1 and ROCK2 protein levels in all tested neuroblastoma cell lines.
Inhibition of ROCK2 Suppresses Neuroblastoma Growth in Vitro.
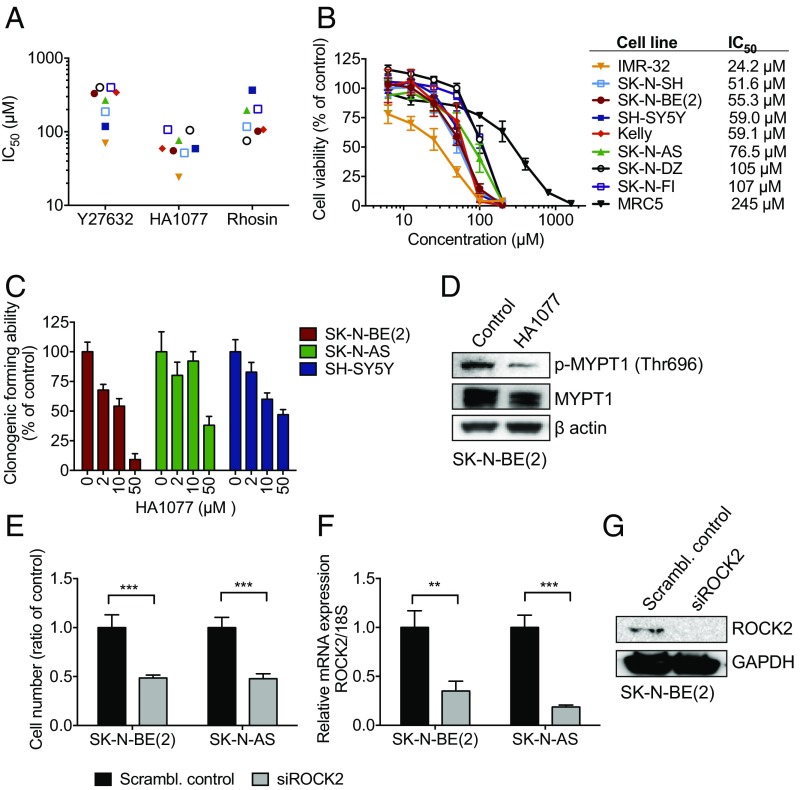
Given that ROCK proteins are expressed and activated in subsets of neuroblastoma, we investigated the effect of inhibiting ROCK activity in neuroblastoma cells. The cytotoxic effects of the two ROCK inhibitors Y27632 and HA1077 as well as a Rho inhibitor, Rhosin, were evaluated in a panel of eight human neuroblastoma cell lines with [IMR32, Kelly, SK-N-BE(2), and SK-N-DZ] or without MYCN amplification (SK-N-AS, SK-N-FI, SK-N-SH, and SH-SY5Y). HA1077, an inhibitor of both ROCK1 and 2 but with a described higher preference for ROCK2 (14), demonstrated the most effective suppression of cell growth of the three inhibitors tested (Fig. 2A) and was therefore selected for continued study. HA1077 showed a concentration-dependent decrease in cell viability after 72 h of treatment, with IC50 values ranging between 24.2 and 107 µM (Fig. 2B). The nontumorigenic fibroblast cell line, MRC5, exhibited a significantly higher IC50 of 245 µM compared with all investigated neuroblastoma cell lines. HA1077 also inhibited the tumorigenic capacity of SK-N-BE(2), SK-N-AS, and SH-SY5Y cells in a concentration-dependent manner (Fig. 2C) and suppressed protein expression of the downstream phosphorylated MYPT1 (Thr696) (Fig. 2D and SI Appendix, Fig. S4). Similarly, siRNA-mediated down-regulation of ROCK2 expression significantly suppressed the cell viability of neuroblastoma cells (Fig. 2E). The knockdown of ROCK2 was confirmed both on the mRNA and protein level (Fig. 2 F and G). To minimize the risk for possible off-target effects, we repeated the experiments using a second siRNA targeting ROCK2. Similar results on cell viability were obtained for knockdown of ROCK2 in SK-N-BE(2) cells (SI Appendix, Fig. S5).
Fig. 2.
ROCK inhibition suppresses neuroblastoma growth. (A) IC50 (µM) for the Rho/Rac signaling inhibitors Y27632, HA1077, and Rhosin in a panel of neuroblastoma cell lines after a 72-h drug exposure. The ROCK inhibitor HA1077 showed significantly lower log IC50 than the ROCK inhibitor Y27632 and the Rho inhibitor Rhosin (repeated-measures one-way ANOVA, P = 0.0067; Y27632 vs. HA1077, P = 0.001; Rhosin vs. HA1077, P = 0.0457). (B) Dose–response curves for cell viability after 72 h of HA1077 treatment in eight neuroblastoma cell lines and one fibroblast cell line, MRC5 (used as a nontumorigenic control for drug toxicity). Cell viability was determined with WST-1; IC50 values are displayed (Right). The cell lines are labeled identically in A and B. (C) Tumorigenic capacity, shown as clonogenic forming ability, was inhibited in the three tested neuroblastoma cell lines after 96 h of HA1077 exposure. (D) The expression of phosphorylated MYPT1 was decreased in neuroblastoma cells following treatment with HA1077 (48 h), as shown by Western blotting. Quantification showed a ratio for p-MYPT1/total MYPT1 of 0.41 vs. 0.15. (E) siRNA-mediated down-regulation of ROCK2 (siROCK2) expression suppressed neuroblastoma cell viability 72 h after transfection compared with scrambled control (scrambl. control) [t test; SK-N-BE(2), ***P < 0.0001; SK-N-AS, ***P = 0.0003]. (F and G) The siRNA knockdown of ROCK2 was confirmed with quantitative PCR and Western blotting [t test; SK-N-BE(2), **P = 0.047; SK-N-AS, ***P = 0.0004]. All concentrations were tested in duplicates, and the experiments were repeated at least three times; mean (A) and mean with SD (B, C, E, and F) are displayed.
ROCK2 Inhibition Induces Differentiation and Decreases Invasion in Neuroblastoma.
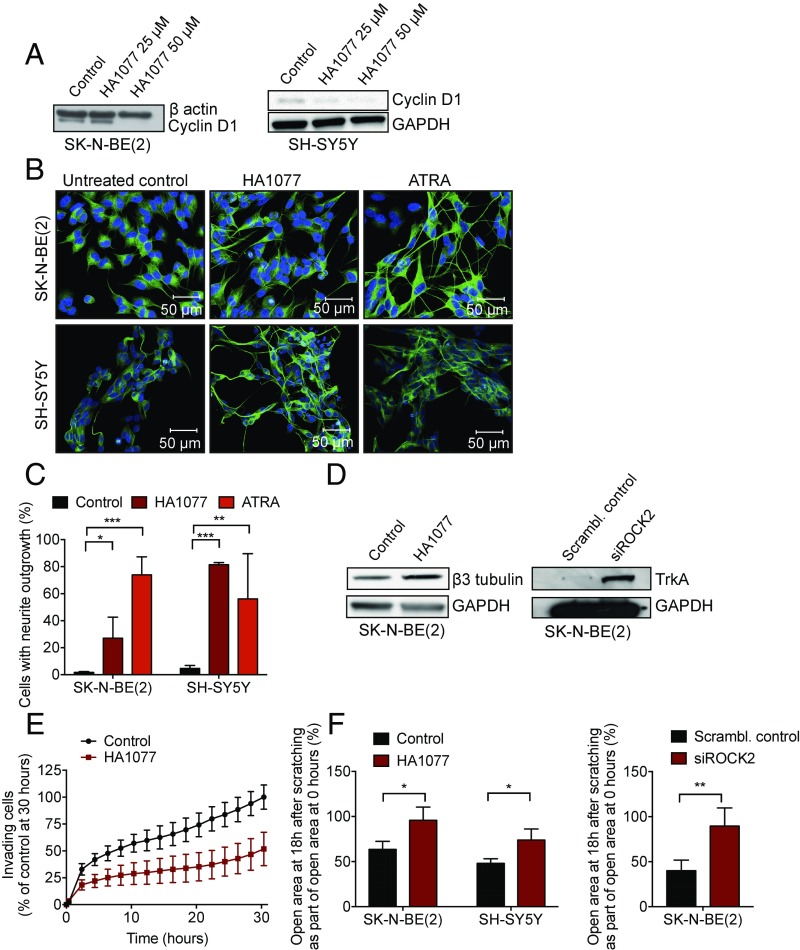
Rho kinases are important for proper cell migration and neural outgrowth (10). We therefore evaluated the effect of ROCK inhibition on neuroblastoma differentiation, migration, and invasion. HA1077 treatment reduced the level of the cell cycle-promoting cyclin D1 protein in the MYCN-amplified SK-N-BE(2) cell line as well as the non–MYCN-amplified SH-SY5Y cell line (Fig. 3A and SI Appendix, Fig. S6). Neurite outgrowth was evident after continuous incubation for 72 h with sublethal but antiproliferative concentrations of HA1077 in both SK-N-BE(2) and SH-SY5Y cells (Fig. 3B and SI Appendix, Fig. S7). The number of cells with neurite outgrowth (neurites >60 µm) was significantly higher in HA1077-treated cells compared with untreated (Fig. 3C). Up-regulation of the differentiation marker β3-tubulin was confirmed in HA1077-treated SK-N-BE(2) cells (Fig. 3D). Similarly, siRNA-mediated down-regulation of ROCK2 increased the protein expression of TrkA, another marker for neural differentiation (Fig. 3D). To investigate the effects of HA1077 on migration and invasion, we performed experiments in electronically integrated Boyden chambers as well as a wound-healing assay. HA1077 was able to reduce the invasion ability of SK-N-BE(2) toward serum, compared with untreated control in plates with electronically integrated Boyden chambers (Fig. 3E). In addition, HA1077 inhibited the rate of neuroblastoma cell migration in confluent cell monolayers, determined by the wound-healing assay (Fig. 3F). Similar results were obtained for ROCK2 siRNA-transfected SK-N-BE(2) cells (Fig. 3F).
Fig. 3.
ROCK2 inhibition induces differentiation and inhibits proliferation and invasion in neuroblastoma cell lines. (A) Treatment of SK-N-BE(2) and SH-SY5Y neuroblastoma cells with HA1077 reduced the expression of cyclin D1 protein as shown by Western blotting. Quantification with densitometry demonstrated a ratio between cyclin D1 and loading control in SK-N-BE(2) of 1 (untreated) compared with 1.06 (25 µM) and 0.26 (50 µM) in HA1077-treated cells; corresponding quantification in SH-SH5Y showed 1 compared with 0.60 and 0.48. (B) Neural differentiation of SK-N-BE(2) and SH-SY5Y in response to HA1077 (50 μM, 72 h) and retinoic acid (ATRA; 1 µM, 72 h). Cells were stained with β3-tubulin and Hoechst 33342, and images were acquired using a confocal laser-scanning microscope (Leica; TCS SP5; 20× objective). (C) The number of cells with neurite outgrowth (>60 µm) was manually counted (ANOVA with Bonferroni posttest; SK-N-BE(2), P < 0.0001; control vs. HA1077, *P = 0.033; control vs. ATRA, ***P = 0.0003; SH-SY5Y, P = 0.0002; control vs. HA1077, ***P = 0.0002; control vs. ATRA, **P = 0.0065). Means with SD are shown. (D) Up-regulation of the differentiation markers β3-tubulin in HA1077-treated cells (72 h) and TrkA after silencing of ROCK2 (96 h) compared with controls. (E) HA1077 (50 µM) repressed the invasion ability of SK-N-BE(2) cells when studied in a real-time invasion analysis in cell chambers coated with 5% Matrigel, monitored in the xCELLigence real-time cell analysis system (two-way ANOVA, P = 0.049 for treatment over 30 h; Bonferroni posttest, P < 0.05 from time point 24 h). Means with SD of three independent experiments are displayed. (F) ROCK2 inhibition, by HA1077 or siRNA knockdown, inhibited neuroblastoma migration, shown as the relative area of migrated cells 18 h after scratching measured by wound assay. Means with SD of three independent experiments are shown [t test; SK-N-BE(2), *P = 0.031; SH-SY5Y, *P = 0.028; SK-N-BE(2) siRNA, **P = 0.0054].
ROCK2 Inhibition Suppresses Neuroblastoma Growth in Vivo.
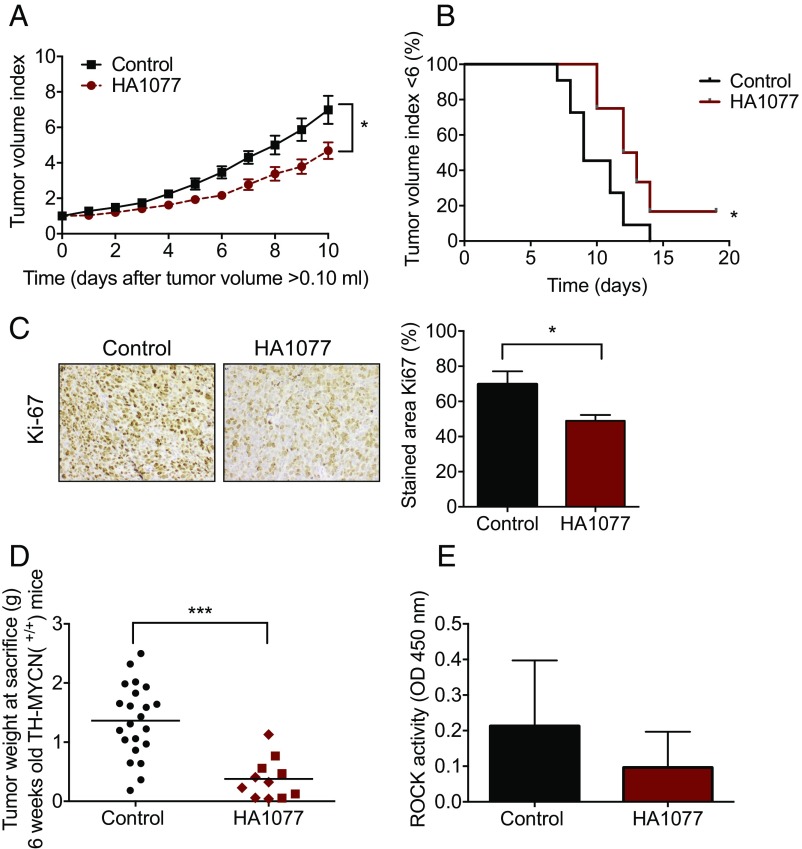
To investigate the therapeutic effects of ROCK inhibition on neuroblastoma growth, NMRI nu/nu mice carrying s.c. established SK-N-BE(2) tumors were treated with 50 mg/kg HA1077 or vehicle by daily intraperitoneal injection for 10 d. Tumor growth was significantly inhibited, as the tumor volume was reduced to 69% compared with vehicle-treated tumors (Fig. 4A). Tumor growth was also significantly delayed in HA1077-treated animals, measured as the time until the tumor reached a sixfold tumor volume (Fig. 4B). As measured by Ki-67 staining, cell proliferation was significantly inhibited after HA1077 treatment compared with untreated tumors (Fig. 4C).
Fig. 4.
HA1077 reduces neuroblastoma growth in vivo. (A) HA1077 significantly impaired the growth of established human neuroblastoma xenografts in NMRI nu/nu mice (t test, day 10, *P = 0.038; two-way ANOVA, regarding treatment, P = 0.016). Mice were engrafted with 10 × 106 SK-N-BE(2) cells s.c. and randomized to receive a daily i.p. injection of HA1077 (50 mg/kg; n = 8) or vehicle (n = 11) for 10 d, starting at the appearance of palpable tumors of ≈0.10 mm3 (mean 0.125 mm3). Means with SEM are displayed. (B) Tumor growth was significantly delayed in HA1077-treated animals, measured as time until tumor reached sixfold tumor volume (log-rank test, *P = 0.0301). (C) Cell proliferation was inhibited in tumors from mice treated with HA1077 compared with untreated controls, shown by tumor Ki-67 immunostaining (t test, *P = 0.033). Representative images and quantification, with means with SD, are shown. (D) Tumor weights from mice homozygous for the MYCN transgene (TH-MYCN+/+) treated with HA1077 [10 mg/kg (▪) or 25 mg/kg (♦); n = 11, mean 0.38 g] were significantly reduced after 10 d of treatment (starting at 4.5 wk of age) compared with matched control (n = 19; mean 1.28 g) (t test, ***P = 0.0001). Tumor weights from individual mice are shown, with lines representing the mean. (E) ROCK activity in TH-MYCN HA1077-treated tumors (n = 6) and control tumors (n = 3), measured by ROCK activity ELISA. Means with SD are presented.
Next, we evaluated the therapeutic response of HA1077 in the genetically engineered mouse model of MYCN-overexpressing neuroblastoma (TH-MYCN), which recapitulates human high-risk neuroblastoma (15, 16). Mice homozygous for the MYCN transgene (TH-MYCN+/+) were treated daily for 9 d by intraperitoneal injection of 10 or 25 mg/kg HA1077 starting at the age of 4.5 wk. Tumor growth was significantly suppressed in HA1077-treated mice compared with matched control mice; the mean tumor burden was significantly lower in treated animals (0.38 g) compared with nontreated animals (1.28 g) (Fig. 4D). No statistically significant difference in tumor growth inhibition was detected between the two doses of HA1077 tested (SI Appendix, Fig. S8). All mice gained in weight during the experiments, and no signs of toxicity were observed in the in vivo experiments (SI Appendix, Fig. S9). As measured by ELISA, ROCK activity was lower in the treated tumors compared with nontreated tumors (Fig. 4E).
Inhibition of ROCK2 Promotes MYCN Protein Degradation.
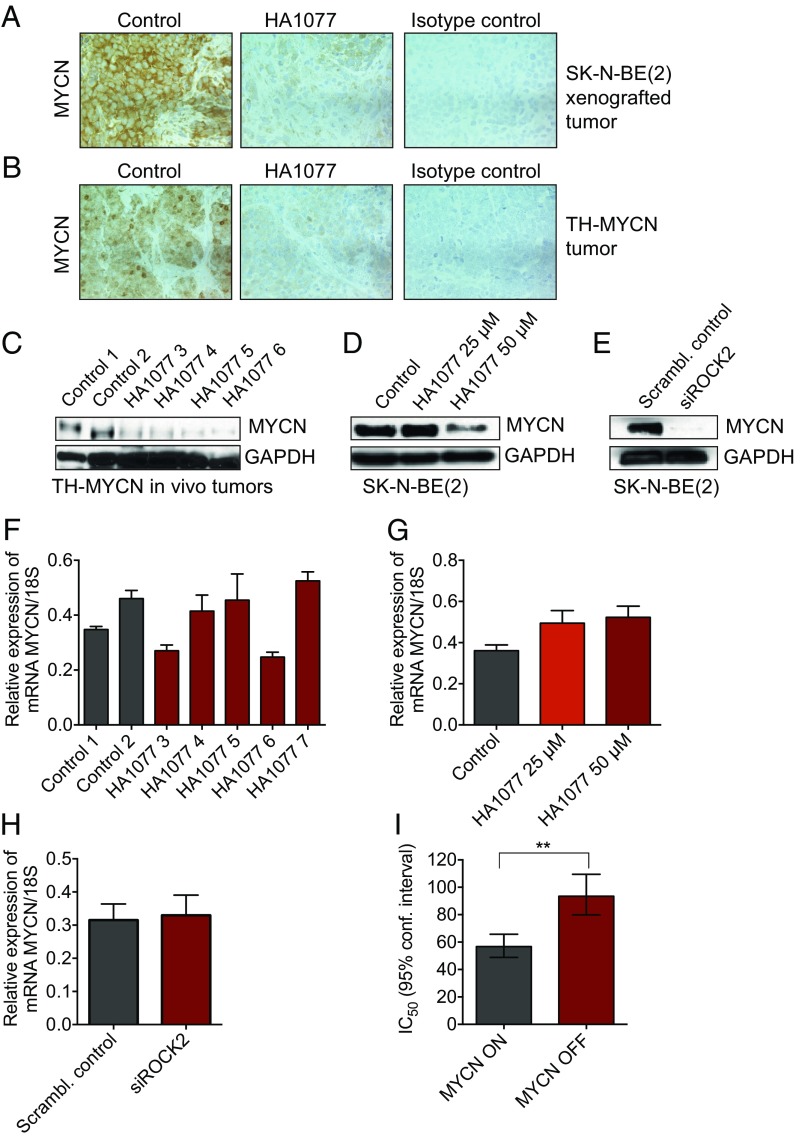
Analysis of MYCN expression in HA1077-treated mice carrying SK-N-BE(2) xenografts or tumors developed in TH-MYCN mice showed a strong reduction of MYCN protein levels compared with nontreated tumors (Fig. 5 A–C). Similarly, SK-N-BE(2) cells treated with HA1077 or transfected with ROCK2 siRNA also showed reduced expression of MYCN (Fig. 5 D and E). Also, immunohistochemistry staining of organotypic cultures derived from TH-MYCN tumors treated with HA1077 ex vivo showed reduced MYCN protein levels compared with nontreated tumors (SI Appendix, Fig. S10). However, no differences in MYCN mRNA levels were observed (Fig. 5 F–H), suggesting that ROCK inhibition regulates MYCN expression levels through posttranscriptional mechanisms. The siRNA experiments were repeated with a second siRNA targeting ROCK2 with similar results on MYCN protein and mRNA (SI Appendix, Figs. S5 and S11). Furthermore, the SH-EP MYCN-inducible Tet21N cell line, which constitutively overexpresses MYCN under the control of a tetracycline-responsive repressor element, was more sensitive to HA1077 treatment compared with non–MYCN-induced cells (Fig. 5I).
Fig. 5.
Inhibition of ROCK2 promotes degradation of MYCN protein. (A and B) Immunohistochemistry analysis of MYCN expression in HA1077-treated mice carrying SK-N-BE(2) xenografts (A) or TH-MYCN mice (B) showed reduced levels of MYCN protein levels compared with nontreated tumors. Representative images (acquired at 20× magnification) are shown, including isotype controls. (C) Western blot analysis of MYCN expression in tumors from HA1077-treated TH-MYCN mice confirmed suppression of MYCN protein levels compared with tumors from untreated controls. (D) HA1077 exposure (50 µM, 96 h) reduced MYCN protein expression in SK-N-BE(2) cells. (E) Inhibition of ROCK2 mRNA expression by siRNA suppressed MYCN protein levels (72 h). (F) Relative expression of MYCN mRNA from the same TH-MYCN tumors as in C demonstrated no differences in mRNA levels of MYCN for HA1077-treated tumors compared with tumors from untreated mice (t test, P = 0.72). (G) No decrease in MYCN mRNA levels was observed in SK-N-BE(2) cells treated with HA1077. On the contrary, mRNA MYCN levels were increased (one-way ANOVA, P = 0.015; Bonferroni posttest: control vs. HA1077, 25 µM, P = 0.033; control vs. HA1077, 50 µM, P = 0.015). (H) Inhibition of ROCK2 mRNA expression by siRNA (72 h) induced no effect on MYCN mRNA in SK-N-BE(2) neuroblastoma cells (t test, P = 0.30). Protein levels were studied with Western blot (C–E), and quantification of mRNA was assessed with real-time quantitative PCR (F–H). Data are presented as means and SD of three determinations (F–H). (I) IC50 (µM) for HA1077 was significantly lower in the SH-EP MYCN-inducible Tet21N cell line with constitutive overexpression of MYCN, compared with non–MYCN-induced cells (t test, **P = 0.003). Means with 95% confidence intervals are shown from four experiments, determined with WST-1.
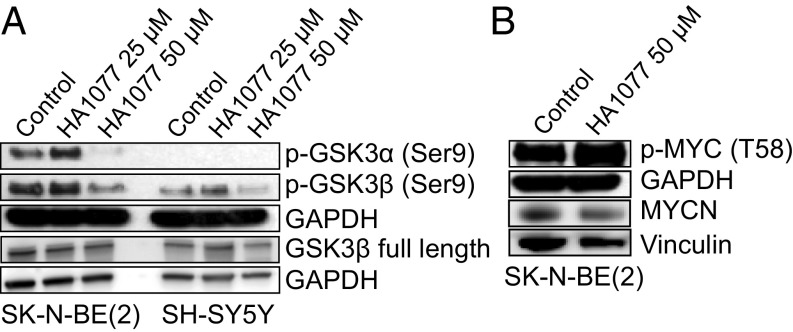
Treatment of neuroblastoma cells with HA1077 resulted in decreased GSK3β (Ser9) phosphorylation (Fig. 6A) and increased MYCN (Thr58) phosphorylation (Fig. 6B), whereas treatment with the small-molecule GSK3β inhibitors CHIR99021 or SB-216763 in combination with HA1077 resulted in increased GSK3 (Ser9) phosphorylation and increased MYCN protein levels (SI Appendix, Fig. S12). Finally, analysis of publicly available and validated gene signature data revealed significant correlation between ROCK2 and MYCN expression in neuroblastoma. Additionally, ROCK2 expression was significant higher in MYCN-amplified samples (SI Appendix, Fig. S13).
Fig. 6.
ROCK inhibition induces GSK3β-mediated phosphorylation and degradation of MYCN protein. HA1077 decreased the protein expression of phosphorylated GSK3α/β (Ser21/9) (A) and increased phosphorylated MYCN (T58) (B) in neuroblastoma cells, determined with Western blotting (72 h).
Discussion
The Rho family of GTPases is fundamental for correct polarization, locomotion, and migration of neural crest cells during embryonal development. Given the importance of Rac/Rho signaling during development, it is not surprising that several of the Rho GTPases are frequently deregulated in cancer (17, 18). Our data from whole-exome and whole-genome sequencing of primary neuroblastoma tumors showed frequent occurrence of mutations within the Rho family of GTPases and associated genes (Table 1). Recurrent mutations within Rho-family genes have been reported by others (5, 6), and analysis of our neuroblastoma cohort demonstrated that 27.5% of neuroblastoma tumors contain at least one mutation within Rho/Rac-associated genes, excluding the ATRX gene (Table 1 and SI Appendix, Table S3). Intriguingly, we found a significant accumulation of patients with an 11q deletion in the group with mutations associated with Rho/Rac signaling in our investigated cohort (Table 1). Others have shown an association between mutations in Rho/Rac genes and aggressive high-stage neuroblastomas without MYCN amplification, which is in line with our findings (5). However, we could not confirm the correlation with high-risk disease, possibly because our investigated cohort mainly consisted of high-risk patients. Mutations of Rho GTPase genes have been detected at low frequencies in various tumor types (19). Interestingly, recurrent somatic oncogenic driver mutations of Rac1 have been detected in 5 to 9% of tumor DNA from patients with melanoma, a tumor that, similar to neuroblastoma, also originates from cells within the neural crest (20, 21). This suggests that the mutations of Rho GTPases and associated genes seen in neuroblastoma and melanoma are acquired during migration and differentiation of neural crest cells. The neural crest consists of multipotent stem cells that during migration give rise to a plethora of differentiated cells, including cells of the peripheral nervous system and adrenal medulla and melanocytes. Because of the versatility and mobility of neural crest cells, they are exposed to both cell-autonomous factors and environmental cues that may lead to an inappropriate differentiation giving rise to peripheral nervous system diseases prevalent in pediatric populations (22).
Our analyses of publicly available expression arrays show that high expression of the Rac/Rho downstream signaling molecule ROCK2 is significantly correlated with high-risk neuroblastoma and poor survival (SI Appendix, Fig. S1). The correlation for high expression of ROCK1 was not as clear in the investigated datasets (SI Appendix, Fig. S1). Possible explanations of the disparity in ROCK1 and ROCK2 involvement may be differences in tissue expression and role during embryonic development, and similar differences have been considered in other malignancies (23, 24). Our data also indicate that mutations within Rho GTPase signaling may activate RhoA and increase the activity of downstream ROCK proteins, as shown by immunohistochemistry of primary neuroblastoma samples using antibodies detecting phosphorylated activated ROCK2 (Fig. 1A). Similarly, genome- and transcriptome-wide analysis of high-risk neuroblastomas showed correlation between deregulation of Rho GTPase signaling and high-risk neuroblastoma independent of MYCN gene amplification status (25). We therefore investigated the effects of inhibiting the activity of ROCK molecules in neuroblastoma using specific small-molecule inhibitors of ROCK or RNAi-mediated gene silencing. We show here that the inhibition of ROCK destabilizes MYCN proteins, induces neuroblastoma cell differentiation, and suppresses migration and invasion. The ROCK-specific inhibitor HA1077 also significantly inhibited the growth of established neuroblastoma xenograft tumors in nude mice as well as suppressed neuroblastoma growth in the MYCN-driven TH-MYCN mouse model of neuroblastoma. In preclinical in vivo models, the levels of MYCN in the tumor tissue were markedly decreased in HA1077-treated animals (Figs. 4 and 5).
One important mechanism for regulating MYCN expression is the cell cycle-dependent phosphorylation of MYCN proteins by the cell-cycle kinase cyclin B/CDK1 and GSK3β that results in Fbxw7 ubiquitin ligase-mediated proteasomal degradation (26). Our results demonstrate that inhibition of ROCK results in dephosphorylation of GSK3β (Ser9) and induction of MYCN (Thr58) phosphorylation followed by diminished MYCN protein levels. GSK3β is fundamental during neural morphogenesis through the regulation of neural polarization, axon growth, and axon branching (27). Interestingly, the activity of GSK3β is partly regulated by molecular cross-talk with ROCK through mutual protein phosphorylation (28). Hence, the mutations of Rho GTPases or associated genes that alter Rho/Rac signaling and enhance the activity of ROCK may affect the levels of MYCN proteins in neuroblastoma because active ROCK can phosphorylate and inactivate GSK3β, resulting in deregulated MYCN protein levels.
The importance of the different mutations of Rho GTPases and associated genes in neuroblastoma is not known. The majority of these mutations have the potential to activate RhoA and inhibit Rac1 (5), which is supported by RhoA activity assays presented in this study. However, further studies will be needed to verify whether these mutations are oncogenic in subsets of neuroblastoma.
Taken together, our data demonstrate that ROCK is a druggable target in neuroblastoma and that ROCK inhibitors may represent a therapeutic opportunity for neuroblastoma patients with advanced disease.
Materials and Methods
Cell Culture and Reagents.
A panel of nine human neuroblastoma cell lines, with different stage and MYCN genetic characteristics, were grown in DMEM/F12 (SH-SY5Y) or RPMI 1640 [SK-N-BE(2), SK-N-AS, SK-N-FI, SK-N-SH, SK-N-DZ, Kelly, IMR32, and Tet21N] medium supplemented with 10% FBS (Gibco), 2 mM l-glutamine, 100 IU/mL penicillin G, and 100 μg/mL streptomycin (HyClone; Thermo Scientific) in a 37 °C humidified, 5% CO2 atmosphere. Also, a fibroblast cell line, MRC5 (grown in RPMI 1640 supplemented and grown as described above), was used. All media were purchased from Gibco. Tet21N is derived from the neuroblastoma SH-EP cell line and contains a tetracycline-regulated MYCN transgene. The cells were grown as MYCN ON and MYCN were switched OFF (+doxycycline) 24 h before experiments.
The identities of the cell lines were verified by short tandem repeat genetic profiling using the AmpFLSTR Identifiler Direct PCR Amplification Kit (Applied Biosystems) in January 2014, and all cell lines were used in passages below 25. All experiments were executed in Opti-MEM (Gibco) supplemented with glutamine, streptomycin, and penicillin, with the exception of the transfection experiments, which were performed without antibiotics.
HA1077 dihydrochloride was purchased from LC Laboratories or Sigma-Aldrich; Y27632 dihydrochloride, all-trans retinoic acid, and doxycycline from Sigma-Aldrich; Rhosin from Calbiochem (Merck Millipore); CHIR99021 from Miltenyi; and SB-216763 from Sigma-Aldrich. HA1077 and Y27632 were dissolved in sterile water or 150 mM NaCl, and Rhosin, retinoic acid, doxycycline, CHIR99021, and SB-216763 were dissolved in DMSO (Sigma); further dilutions were done with Opti-MEM or PBS.
For the transfections, cells were seeded in six-well dishes, left to attach, and transfected using Lipofectamine 2000 with 20 pmol predesigned siRNAs targeting human ROCK2 (5270301; Invitrogen) and nontargeting siRNA (6568s; Invitrogen) as control or ROCK2 (locus ID 9475; SR306287) and scrambled negative control siRNA (OriGene Technologies). After 48 to 96 h, cells were subjected to further analyses.
Viability Assays.
The effects of ROCK inhibitors on cell growth were determined using a colometric formazan-based cell-viability assay (WST-1; Roche) according to the manufacturer’s description. Cells were seeded onto 96-well plates [10,000 cells per well except SK-N-BE(2), 5,000 cells per well] and incubated with drugs for 72 h. All concentrations were tested in triplicate.
To determine colony formation, 200 cells were seeded on six-well plates (Cell+; Sarstedt) in triplicate. Cells were left to attach before treatment with HA1077 for 96 h or genetic ROCK2 down-regulation. After 8 to 14 d of incubation in drug-free medium, cell cultures were washed, fixed in formaldehyde, and stained with Giemsa (Gibco). Colonies (>100 cells) with 50% plate efficiency were manually counted using a colony counter.
Western Blotting.
Protein extraction and Western blotting were performed as described earlier (29). Membranes (PVDF; Millipore) were incubated with antibodies against ROCK1 (1:1,000; 160 kDa), ROCK2 (1:1,000; 160 kDa), cyclin D1 (1:1,000; 36 kDa), TrkA (1:1,000; 140 kDa), MYCN (1:500; 62 kDa), β-actin (1:5,000; 45 kDa), GSK3β (1:1,000; 46 kDa), GSK3α/β phospho S21/S9 (1:1,000; 51 and 46 kDa), β3-tubulin (1:1,000; 55 kDa), MYPT1 (1:1,000; 140 kDa), phosphorylated MYPT1 (Thr696) (1:500; 140 kDa) (all from Cell Signaling), MYC phospho (T58/S62) (1:1,000; 57 to 65 kDa), GAPDH (1:10,000; 37 kDa), vinculin (1:10,000; 124 kDa) (all from Abcam), and MYCN (1:2,000; 62 kDa) (Santa Cruz Biotechnology). Anti-rabbit IgG, conjugated with horseradish peroxidase (Cell Signaling), was used for secondary detection and SuperSignal (Pierce) was used for chemiluminescent visualization. Quantification of blots was done with densitometry measurements in ImageJ (30).
Real-Time RT-PCR.
Total RNA was prepared from cells and tumors with the RNeasy Kit (Qiagen), followed by cDNA synthesis with the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). The mRNA expression levels of ROCK1, ROCK2, MYCN, and housekeeping genes were assessed using TaqMan (Applied Biosystems) technology on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). The TaqMan sequence-specific primers were ROCK1 (Hs01127699_m1), ROCK2 (Hs00178154_m1), MYCN (Hs00232074_m1), and 18S ribosomal RNA (Hs99999901_s1) (Applied Biosystems). Relative quantification was determined with a standard curve. All real-time RT-PCR experiments included a no-template control and were performed in triplicate.
Differentiation and Migration Studies.
To study differentiation, a morphological evaluation of neuroblastoma cells was performed after exposure to HA1077 or retinoic acid or no treatment (72 h). SK-N-BE(2) and SH-SY5Y cells were seeded on eight-well chamber slides (Nunc) [5,000 cells per well for SK-N-BE(2) and 12,500 cells per well for SH-SY5Y], left to attach, and treated with the indicated drug concentrations. After 72 h, neuroblastoma cells were fixed and immunostained with an antibody against neuron-specific β3-tubulin (1:800) followed by anti-mouse IgG Alexa Fluor 488 (1:1,000) and with Hoechst 33342 (20 µM) (Sigma). Images were taken using a confocal laser-scanning microscope (Leica; TCS SP5) and evaluated for neurite outgrowth. The number of cells with neurite outgrowth (>60 µm) was manually counted by researchers blinded to treatment, and more than 300 cells for each treatment were analyzed in randomized selected images (>5 images). In addition, neuroblastoma cells were seeded on six-well plates (75,000 cells per well), left to attach, and treated, and images were acquired with a phase-contrast microscope (Nikon; Eclipse TS100; 20× objective).
A wound assay was performed to evaluate the migration capacity after ROCK inhibition in neuroblastoma cells, or by HA1077 treatment or siRNA against ROCK2. SK-N-BE(2) and SH-SY5Y cells were seeded on six-well plates (500,000 cells per well) and left to attach for 24 h. A “wound” was made using a 200-µL pipette tip, followed by drug treatment. Images were taken at the start (t = 0) and 18 h after treatment (t = 18) using a phase-contrast microscope (Olympus; Infinity 1; 20× objective), and used for quantitative assessment of the migrated area (31). Assessment was done using automated analysis in TScratch software (32). Three independent experiments were performed with at least three images per treatment.
A cell-invasion assay was performed on 16-well cell migration/invasion plates with an 8-µm polyethylene terephthalate membrane coated with 5% Matrigel (growth factor reduced; Corning) in an xCELLigence DP instrument for real-time analysis (ACEA Biosciences). SK-N-BE(2) cells, after starvation in serum-free RPMI medium for 6 h, were seeded in the upper chamber in serum-free medium containing HA1077 (50 µM) or vehicle. The lower chamber contained complete RPMI medium (10% FCS), and cell invasion was monitored every 2 h for a period of 30 h. All treatments were performed in at least triplicate, and experiments were repeated three times.
Human Tissue Samples.
Neuroblastoma tissue from patients was obtained during surgery and stored at −80 °C. Ethical approval was obtained from the Karolinska University Hospital Research Ethics Committee (approval nos. 2009/1369-31/1 and 03-736). Informed consent for using tumor samples in scientific research was provided by parents/guardians. In accordance with the approval from the Ethics Committee the informed consent was either written or verbal, and when verbal or written assent was not obtained the decision was documented in the medical record.
Immunohistochemistry.
Formalin-fixed and paraffin-embedded (FFPE) tissue slides were deparaffinized in xylol and rehydrated in graded alcohols. For antigen retrieval, slides were boiled in a sodium citrate buffer (pH 6.0) for 10 min in a microwave oven. After blocking in 1% bovine serum albumin (BSA) for 20 min, the tissue sections were incubated with primary antibody against ROCK2 (H-85; Santa Cruz Biotechnology), ROCK2 (p-Thr396; Novus Biochemicals), or N-Myc (B8.4.B; Santa Cruz Biotechnology) diluted 1:150 in 1% PBS with 1% BSA overnight at 4 °C. Thereafter, slides were rinsed in PBS and endogenous peroxidases were blocked in 0.3% H2O2 for 10 min. As a secondary antibody, anti-rabbit horseradish peroxidase was used (Invitrogen). All slides were counterstained with hematoxylin. Matched isotype control was used as a control for nonspecific staining. The presence of activated ROCK2 in neuroblastoma cells was assessed by double immunofluorescence labeling of tissue slides using a mouse monoclonal anti-synaptophysin antibody (clone SVP-38; Sigma) and rabbit anti-ROCK2 (p-Thr396; Novus Biochemicals). For fluorescence visualization, donkey anti-mouse Alexa 594 and goat anti-rabbit Alexa 488 were used, respectively.
FFPE sections from SK-N-BE(2) xenografts were deparaffinized and rehydrated before endogenous peroxidase activity was blocked using 0.5% H2O2, and biotin was blocked using 1% BSA in Tris-buffered saline for 30 min. Tumor sections were incubated overnight at 4 °C with primary antibody against Ki-67 (1:200; Thermo Scientific). Sections were then incubated for 40 min at RT (room temperature) with a biotin-conjugated secondary antibody (anti-rabbit; 1:200; Vector Laboratories). After incubation with ABC complex (Vector Laboratories), the sections were developed for 6 min using diaminobenzidine (DAB Peroxidase Substrate Kit; Vector Laboratories) and then counterstained with Mayer hematoxylin (Histolab). Quantifications were done at 20× magnification. Sixty randomly chosen fields per slide and five slides per group were automatically quantified using Leica Qwin IM500 software, and are presented as the proportion of positive staining of the total area.
Sequencing Analyses.
DNA was extracted from frozen tumors using standard procedures and evaluated through fluorometric quantitation and DNA integrity assessment on an Agilent TapeStation prior to exome or whole-genome sequencing. Exome sequencing was performed through pair-end sequencing on Illumina platforms after enrichment with an Agilent SureSelect All Exome Enrichment Kit. Alignment against hg19 was performed using BWA (bio-bwa.sourceforge.net) with GATK local realignment followed by single-nucleotide variance calling using SNPeff (www.sourceforge.net). Whole-genome sequencing was performed on tumor DNA and corresponding constitutional DNA extracted from blood for 13 neuroblastoma patients for an average coverage of at least 60× for tumor and 30× for constitutional DNA using Illumina X Ten instrumentation located at National Genomics Infrastructure/Clinical Genomics, SciLife Laboratories. Read trimming, mapping to the human reference genome hg19, and variant calling were performed using CLC Genomics Workbench 8.0.3 software (https://www.qiagenbioinformatics.com/).
Only high-quality called variants with a minimum 10% allele frequency and a total read coverage of 10 were considered for further analysis. Variants with allele frequency above 3% in either 1000 Genomes Project (www.internationalgenome.org), Exome Aggregation Consortium (ExAC) (exac.broadinstitute.org), or National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (evs.gs.washington.edu/EVS/) were discarded as well as excluding all synonymous variants or variants in noncoding regions except those affecting canonical splice sites. Remaining variants were assessed manually through the Integrative Genomics Viewer (33) for removal of calls due to mapping artifacts and paralogs.
For tumors (exome) sequenced without corresponding DNA from constitutional tissue, a systematic filtering approach was used to identify critical variants. This was done by removal of common variants, for example, present in dbSNP version 138 or showing an allele frequency above 0.1 in either 1000 Genomes Project, ExAC, or NHLBI Exome Sequencing Project. A set of 177 genes with known function in the Rho-Rac signaling cascade, including Rho-Rac–controlling GEFs and GAPs, and other signaling molecules in the noncanonical Wnt signaling pathway was preselected for mutational analysis. Detected somatic protein-changing aberrations in these genes were compiled with all previously reported mutations in human neuroblastoma samples available in recent publications (5, 6, 34) (SI Appendix, Tables S2 and S3). Kaplan–Meier survival curves and gene correlations were produced with the R2 Genomics Analysis and Visualization Platform (r2.amc.nl).
RhoA, Rac1, and ROCK Activity Assays.
Total RhoA, active RhoA, and active Rac1 were investigated using ELISA and G-LISA kits (BK150, BK124, and BK128; Cytoskeleton). The RhoA G-LISA kit uses a plate coated with Rho-GTP–binding protein, and thus active, GTP-bound Rho in lysates will bind whereas inactive GDP-bound Rho will be washed away, and the Rac1 G-LISA plate will correspondingly bind active, GTP-bound Rac1. ROCK kinase activity was analyzed by an enzyme immunoassay measuring the phosphorylation of myosin phosphatase target subunit 1 (MYPT1) at Thr696 (STA-416; Cell Biolabs). The experiments were performed according to the manufacturer’s protocol. The total protein quantity for samples for detecting total RhoA was 30 µg, for active RhoA and active Rac1 was 50 µg, and for ROCK was 750 μg protein.
In Vivo Studies.
For the xenograft study, SK-N-BE(2) cells (10 × 106) were injected s.c. into the flanks of immunodeficient nude mice (female 5- to 6-wk-old Sca:NMRI-nu/nu, Scanbur). At palpable tumor, with a volume greater than 0.10 mm3, the mice were randomized to receive either HA1077 intraperitoneally (50 mg/kg, n = 7) or vehicle (n = 11) for 10 d. Tumors were measured daily, and tumor volume was calculated as width2 × length × 0.44. Tumor volume index was calculated as the measured tumor volume each day divided by the starting volume (day 0). The TH-MYCN animals were obtained from the Mouse Models of Human Cancers Consortium Repository as an N16 backcross to the 129X1/SvJ background and were kept as a continuous inbreeding. Genotyping, abdominal palpations, and tumor development were previously described (15). In the treatment study, homozygous mice were randomized at 4.5 wk of age to receive either no treatment (control; n = 19) or daily intraperitoneal injections with HA1077 (10 mg/kg, n = 6 or 25 mg/kg, n = 5) for 10 consecutive days, and were killed at the age of 6 wk. Some animals in the control group have been used in other published studies. Animals were maintained at a maximum of six per cage and given sterile water and food ad libitum. The animals were monitored for signs of toxicity, including weight loss. At sacrifice, tumors were dissected into smaller parts and either frozen or fixed in formaldehyde for further analysis. All animal experiments were approved by the regional ethics committee for animal research (N391/11 and N26/11), appointed and under the control of the Swedish Board of Agriculture and the Swedish Court. The animal experiments presented herein were in accordance with national regulations (SFS 1988:534, SFS 1988:539, and SFS 1988:541).
Organotypic Cultures.
Neuroblastoma tumors from TH-MYCN mice were used to create organotypic slice cultures (35). The tumor was sliced into 300-µm-thick transversal slices using a McIlwain tissue chopper (Ted Pella). The slices were moved to culture medium DMEM:F12K 1:1 supplemented with 2 mM l-glutamine, 1% penicillin-streptomycin, EGF (10 ng/mL medium; Chemicon), FGF (15 ng/mL medium; ProSpec), and B27 (1:50; Sigma) and then carefully placed on insert membranes (Millicell culture plate inserts; Millipore) on six-well plates. The membranes were coated with 0.3 mL of 0.1 mg/mL poly-d- or poly-l-lysine in advance. One milliliter of culture medium was put underneath the membrane and all fluid on top was removed. The cultures were kept in an incubator (37 °C, 5% CO2) and the culture medium was changed every second day. Control cultures always received regular culture medium, and treated slices received culture medium with 50 μM HA1077. After 7 d, slices were fixed in 4% paraformaldehyde for 1 h at 4 °C and then in 20% ice-cold methanol in PBS for 10 min. Triton X-100 (0.2%; Roche Diagnostics) and 0.1% Tween 20 (Invitrogen) in PBS was used for permeabilization for 40 min at RT, and 5% BSA (Invitrogen) with 0.05% Tween 20 in PBS was used for blocking for 2 h at RT with primary antibodies against ROCK2 (H-85) and N-MYC (B8.4.B) (Santa Cruz Biotechnology) and then diluted 1:200 in 0.05% Tween 20/PBS and incubated at 4 °C for 48 h. After a washing, slices were incubated for 1.5 h at RT with secondary antibodies (Alexa; Invitrogen) diluted 1:200 in 0.05% Tween 20/PBS. After a second washing in PBS, the slices were mounted in ProLong Gold Antifade Reagent with DAPI (Invitrogen; P36931). Stainings were analyzed with a Zeiss Axio Examiner D1 microscope (10×, 20×, and 40× water-immersion objectives) with a cooled CCD camera (Zeiss), and captured images were processed in ImageJ (1.42q; National Institutes of Health).
Statistics.
IC50 values (inhibitory concentration 50%) were determined from log concentration–effect curves in Prism (GraphPad Software) using nonlinear regression analysis. Fisher’s test was used to examine the significance of the association between the two categories. Comparison between two groups was done with the Student t test and, for comparison of three or more groups, one-way ANOVA followed by Bonferroni multiple comparisons posttest was used. Treatment groups with repeated measurements over time were compared with two-way ANOVA followed by Bonferroni multiple comparisons posttest. All tests were two-sided and carried out in Prism.
Supplementary Material
Acknowledgments
We thank the SciLifeLab/Integrative Clinical Genomics and Bioinformatics Core Facility platforms at the Sahlgrenska Academy, University of Gothenburg for assistance with the bioinformatical analysis of sequencing data. We thank Lotta Elfman and Inger Bodin for their technical assistance. This work was supported by grants from the Swedish Childhood Cancer Foundation, Swedish Cancer Foundation, Swedish Research Council, Eva och Oscars Ahréns Research Foundation, Magnus Bergvall Foundation, Mary Béve Foundation, Märta and Gunnar V. Philipson Foundation, Anna-Brita and Bo Castegren Memorial Foundation, Sigurd and Elsa Goljes Memorial Foundation, Swedish Foundation for Strategic Research (www.nnbcr.se), Karolinska Institutet, Stockholm County Council, and Cancer Research Foundations of Radiumhemmet.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706011114/-/DCSupplemental.
References
- 1.Johnsen JI, Kogner P, Albihn A, Henriksson MA. Embryonal neural tumours and cell death. Apoptosis. 2009;14:424–438. doi: 10.1007/s10495-009-0325-y. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 4.Cheung NK, et al. St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 6.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebbagh M, Borg JP. Insight into planar cell polarity. Exp Cell Res. 2014;328:284–295. doi: 10.1016/j.yexcr.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein AM, Brewer KC, Doyle AM, Nagy N, Roberts DJ. BMP signaling is necessary for neural crest cell migration and ganglion formation in the enteric nervous system. Mech Dev. 2005;122:821–833. doi: 10.1016/j.mod.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 10.Riento K, Ridley AJ. ROCKs: Multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 11.Hahmann C, Schroeter T. Rho-kinase inhibitors as therapeutics: From pan inhibition to isoform selectivity. Cell Mol Life Sci. 2010;67:171–177. doi: 10.1007/s00018-009-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohn SL, et al. INRG Task Force The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monclair T, et al. INRG Task Force The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmuson A, et al. Tumor development, growth characteristics and spectrum of genetic aberrations in the TH-MYCN mouse model of neuroblastoma. PLoS One. 2012;7:e51297. doi: 10.1371/journal.pone.0051297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alan JK, Lundquist EA. Mutationally activated Rho GTPases in cancer. Small GTPases. 2013;4:159–163. doi: 10.4161/sgtp.26530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi Y, Sipp D, Enomoto H. Tissue interactions in neural crest cell development and disease. Science. 2013;341:860–863. doi: 10.1126/science.1230717. [DOI] [PubMed] [Google Scholar]
- 23.Pinca RS, et al. Targeting ROCK2 rather than ROCK1 inhibits Ewing sarcoma malignancy. Oncol Rep. 2017;37:1387–1393. doi: 10.3892/or.2017.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thumkeo D, et al. Targeted disruption of the mouse rho-associated kinase 2 gene results in intrauterine growth retardation and fetal death. Mol Cell Biol. 2003;23:5043–5055. doi: 10.1128/MCB.23.14.5043-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stigliani S, et al. High genomic instability predicts survival in metastatic high-risk neuroblastoma. Neoplasia. 2012;14:823–832. doi: 10.1593/neo.121114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chesler L, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim WY, Snider WD. Functions of GSK-3 signaling in development of the nervous system. Front Mol Neurosci. 2011;4:44. doi: 10.3389/fnmol.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 29.Wickström M, et al. Wnt/β-catenin pathway regulates MGMT gene expression in cancer and inhibition of Wnt signalling prevents chemoresistance. Nat Commun. 2015;6:8904. doi: 10.1038/ncomms9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 32.Gebäck T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herlenius E, et al. 2012. Functional stem cell integration assessed by organotypic slice cultures. Curr Protoc Stem Cell Biol Chapter 2:Unit 2D.13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.