Significance
The slow delayed rectifier K+ current (IKs) determines the length of each human heartbeat because it activates after myocytes are excited and depolarize. This sensitivity to voltage, as well as dynamic regulation by hormones and second messengers, underlies the essential role of IKs in determining the duration and rhythm of cardiac action potentials. Here, we demonstrate the unexpected mechanism that establishes the voltage-dependent operation of IKs channels: SUMOylation. When native IKs channels are resident in the plasma membranes of neonatal mouse ventricular myocytes, or human channels are reconstituted in CHO cells, each of the four KCNQ1 pore-forming subunits is subject to monoSUMOylation in a manner that depends on KCNE1 accessory subunits, leading to stepwise depolarizing shifts in the activation voltage.
Keywords: KCNE1, minK, KCNQ1, KvLQT1, heart
Abstract
IKs channels open in response to depolarization of the membrane voltage during the cardiac action potential, passing potassium ions outward to repolarize ventricular myocytes and end each beat. Here, we show that the voltage required to activate IKs channels depends on their covalent modification by small ubiquitin-like modifier (SUMO) proteins. IKs channels are comprised of four KCNQ1 pore-forming subunits, two KCNE1 accessory subunits, and up to four SUMOs, one on Lys424 of each KCNQ1 subunit. Each SUMO shifts the half-maximal activation voltage (V1/2) of IKs ∼ +8 mV, producing a maximal +34-mV shift in neonatal mouse cardiac myocytes or Chinese hamster ovary (CHO) cells expressing the mouse or human subunits. Unexpectedly, channels formed without KCNE1 carry at most two SUMOs despite having four available KCNQ1-Lys424 sites. SUMOylation of KCNQ1 is KCNE1 dependent and determines the native attributes of cardiac IKs in vivo.
Cardiac IKs channels pass the slow component of the delayed rectifier potassium current that is necessary to produce normal heart rate and rhythm. Thus, IKs varies in magnitude and timing subject to neuronal and hormonal influences (1). Abnormal changes in IKs function, due to inherited mutations in KCNQ1 or KCNE1, or suppression of the current by medications, can produce long QT syndrome (LQTS) and life-threatening cardiac arrhythmias (2).
KCNQ1 (also called KV7.1 and previously KVLQT) is a classical voltage-gated potassium α-subunit with six transmembrane segments and a single pore-forming P loop. Human KCNQ1 has variants up to 676 residues in length, whereas KCNE1 (minK) has just 129 residues and a single transmembrane span (Fig. 1A). Despite its small size, KCNE1 is essential to the operation of IKs. Compared with channels formed by KCNQ1 subunits alone, KCNE1 produces a right shift in the half-maximal voltage required to activate the channel (V1/2), slows the kinetics of activation and deactivation, increases the channel unitary conductance, alters the ion selectivity of the conduction pore and, modifies its pharmacology (3–8). KCNE1 also endows IKs channel with a high affinity for PIP2 (9) and allows responsiveness to PKA-mediated phosphorylation following β-adrenergic stimulation (10). Here, we demonstrate that the V1/2 inherent to native cardiac IKs channels results from, and is modified by, KCNE1-dependent SUMOylation of KCNQ1.
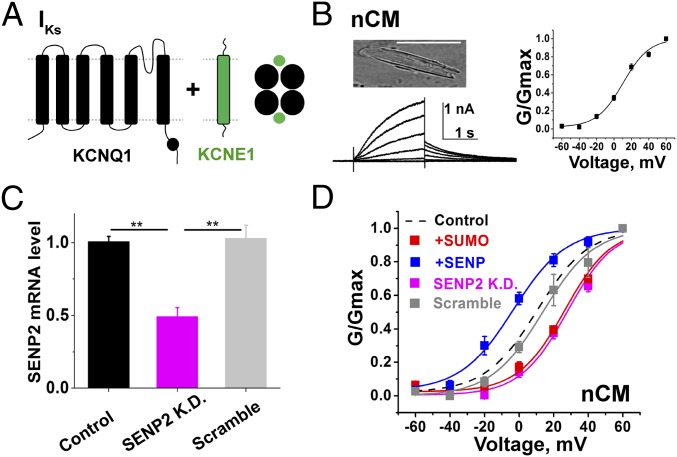
Fig. 1.
SUMO regulates IKs in neonatal mouse cardiac ventricular myocytes. nCM were identified by visualization of muscular striations and spontaneous beating. IKs was isolated with 30 µM Chromanol-293B in the bath by the protocols described in the main text and Materials and Methods; biophysical parameters are in Table 1 as mean ± SEM for n = 6–11 cells. Currents measured before application of Chromanol-293B and the Chromanol-293B–insensitive current are shown in Fig. S1. (A) Subunit topology of KCNQ1 (black) and KCNE1 (green) show the expected stoichiometry. (B, Upper Left) nCM photomicrograph. (Scale bar: 50 µm.) (B, Lower Left) Representative family of currents for native IKs. (B, Right) Representative isochronal conductance–voltage relationships for native IKs with control solution in the pipette. (C) SENP2 mRNA level measured by qPCR from groups of 5–10 eGFP-positive nCMs: untreated control (black), SENP2-shRNA knockdown (SENP2 K.D., pink), and scramble-shRNA (gray). SENP2-shRNA reduced the level of SENP2 mRNA by 51 ± 6%, compared with the scramble-shRNA, (**P < 0.01). (D) iG-V relationships for native IKs with control solution in the pipette as shown in B (black dash), 1 nM SENP2 (+SENP, blue), 1 nM SUMO2 (+SUMO, red), for nCM exposed to lentivirus expressing shRNA for SENP2 knockdown (SENP2 K.D., pink) or a scramble shRNA (scramble, gray).
SUMOylation is the enzyme-mediated linkage of one of three SUMO isoforms to the ε-amino group of specific Lys residues on a target protein (11). Present in all eukaryotic cells, the pathway was recognized to regulate the activity of nuclear transcription factors when we discovered it resident at the plasma membrane (12, 13) and to regulate the excitability of cerebellar granule neurons via SUMOylation of K2P1 and NaV1.2 (14, 15), and of hippocampal neurons via modification of KV2.1 (16).
This study was inspired by the work of Qi et al. (17), who describe partial deficiency of the deSUMOylating enzyme SENP2 in mice, showing it to produce seizures, bradycardia, and sudden death, in association with decreased M-channel currents and increased modification of KCNQ2 (KV7.2) subunits by SUMO2 in the brain. Supporting a neurogenic basis for sudden cardiac death, the authors report sinus pauses and AV conduction block following seizure activity; because they also observe increased SUMOylation of KCNQ1 in the brain by SUMO2, and others have shown that mice carrying human LQTS mutations in KCNQ1 can develop seizures and sudden death (18), we chose to study the effects of SUMOylation of KCNQ1 in the heart on the operation of IKs.
To overcome the paucity of IKs in adult mouse heart, we used a strategy for electrophysiology developed by others that allows study of the current in neonatal cardiac ventricular myocytes (nCM) (19–22). In nCM, we first used partial SENP2 knockdown and observed that the conductance–voltage relationship of native cardiac IKs was subject to tonic regulation by the SUMO pathway; the response was then characterized directly by the acute intracellular application of SUMO2 and SENP2. Reconstituting the channels in CHO cells with mouse or human KCNQ1 and KCNE1 subunits allowed identification of a single conserved SUMOylated residue—Lys424 in human KCNQ1—that determined the basis for voltage dependence of IKs channels. Total internal reflection fluorescent (TIRF) microscopy allowed counting of the number of KCNE1, KCNQ1, and SUMO2 subunits in channel complexes, and with electrophysiology, the effect of adding 1, 2, 3, and 4 SUMO2 monomers on the V1/2 of IKs activation was shown to be linearly additive. Compared with channels formed with only KCNQ1 subunits, KCNE1 incorporation to create IKs channels shifts the V1/2 by +38 mV; SUMOylation of all four sites in IKs channels yields an additional shift of +34 mV. SUMOylation was not associated with changes in the surface density or the subunit stoichiometry of IKs channels.
Results
SUMO2 and SENP2 Regulate IKs in Neonatal Mouse Cardiac Ventricular Myocytes.
Whole-cell patch-clamp recording was used to study IKs 72 h after dissociation of nCM from the harvested heart. The recording protocol was designed to isolate the slow component of the delayed rectifier potassium current, as reported by others (19–23). Briefly, a holding potential of −40 mV was used to inactivate the voltage-gated Na+ channels and 1 µM isradipine was included in the bath solution to block voltage-gated calcium channel currents. Native IKs was defined per custom as the difference current isolated by subtraction of the current measured in the presence of 30 µM Chromanol-293B, a IKs inhibitor, from the current measured before application of the drug (23–26). All currents were evoked by test pulses lasting 2 s from −60 to +60 mV in 20-mV increments, followed by 2-s tail current pulses to −40 mV and interpulse intervals of 15 s. The tail currents were used to assess isochronal conductance-voltage relationships (iG-V) and current densities, the latter after a step to +40 mV; biophysical parameters for nCM are summarized in Table 1.
Table 1.
The SUMO pathway regulates IKs in neonatal mouse cardiac ventricular myocytes
| Conditions | V1/2, mV | Current density, pA/pF | τ activation, ms | τ deactivation, ms | Zδ |
| Control | 10 ± 2 | 6.8 ± 0.4 | 583 ± 19 | 647 ± 32 | 15.1 ± 1.3 |
| SENP2 in pipette | −3.3 ± 1.4* | 12 ± 0.7* | 448 ± 13* | 877 ± 27* | 15.3 ± 1.2 |
| SUMO2 in pipette | 26 ± 2* | 3.8 ± 0.3* | 752 ± 18* | 479 ± 14* | 13.6 ± 1.6 |
| shRNA SENP2 | 28 ± 2* | 4.0 ± 0.2* | 745 ± 14* | 465 ± 15* | 13.7 ± 2.2 |
| shRNA scramble | 14 ± 2 | 6.7 ± 0.3 | 594 ± 20 | 621 ± 19 | 15.1 ± 1.5 |
IKs studied in nCMs, as per Fig. 1. V1/2 and Zδ were determined from a Boltzmann function using tail current densities at −40 mV. The tail current density was assessed by estimating the peak from a fit of the decay curve after an activating pulse at +40 mV for 2 s. The τ for activation and deactivation was determined from a single exponential fit in each experimental condition. Data are mean ± SEM with 6–11 cells. Lentivirus treated cells were studied 48 h following infection. Currents measured before application of 30 µM Chromanol-293B, and the Chromanol-293–insensitive current are show in Fig. S1. Compared with the control condition, V1/2, current density, and τ for activation and deactivation were significantly different with SENP2 or SUMO2 in the pipette, and when cells were treated with shRNA SENP2 (unpaired t test, *P < 0.01), whereas there was no significant change from the control when cells were treated with scrambled shRNA (unpaired t test, P > 0.05). Compared with the control condition, the slope factor (Zδ) was not significantly different with SENP2, SUMO2, shRNA SENP2, and scrambled shRNA (P > 0.05).
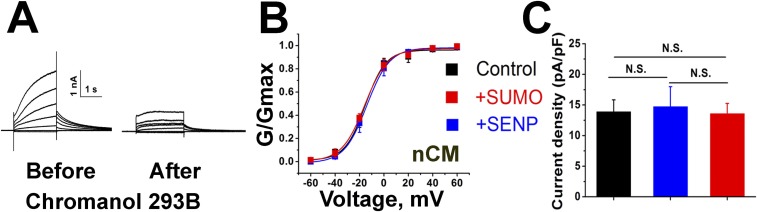
Native IKs in nCM was similar in appearance to the macroscopic, Chromanol-293B–sensitive, time-dependent, slowly activating delayed rectifier currents reported by others, including an apparent V1/2 of ∼+10 mV (Fig. 1B) (19, 20, 27–29). nCM expressing a scrambled shRNA and eGFP expressed from a lentivirus retained parameters for IKs like those in wild-type cells. In contrast, nCM expressing eGFP and shRNA directed to SENP2 showed a 51 ± 6% decrease in SENP2 mRNA by qPCR and a right shift in V1/2 to 28 ± 2 mV, decreased current density, slowed activation rates, and faster deactivation kinetics (Fig. 1 C and D and Table 1). These findings led us to hypothesize that partial knockdown of the SENP2 mRNA level could regulate the voltage-dependent activation of native IKs. To confirm the hypothesis that the right shift in the V1/2 was due to suppression of deSUMOylase activity and increased SUMOylation, we introduced 1 nM SUMO2 into the cells via the recording pipette, a manipulation that we have previously shown to produce maximal SUMOylation of potassium channels in central neurons or expressed heterologously in CHO cells (13, 16). Currents evoked in the presence of 1 nM SUMO2 had a V1/2 of 26 ± 2 mV, and manifested current densities and gating kinetics similar in magnitude to those observed in cells treated with SENP2 shRNA. In contrast, addition of 1 nM SENP2 in the pipette left-shifted the V1/2 to −3.3 ± 1.4 mV, increased the current density, speeded the kinetics of activation, and slowed the time course for deactivation. Of note, the Chromanol-293B–insensitive potassium currents in the nCM (i.e., the non-IKs currents) appeared to be unaltered by treatment with SENP2 shRNA or scrambled shRNA, or by the inclusion of SUMO2 or SENP2 in the pipette (Fig. S1).
Fig. S1.
Chromanol-293B–insensitive potassium currents in neonatal mouse cardiac ventricular myocytes are not modified by SUMO2, SENP2, or SENP2-shRNA. nCM were identified as per Fig. 1. Currents were evoked as described in the main text and studied before and after the addition of 30 µM Chromanol-293B to the bath. Data are mean ± SEM for n = 6–11 cells from each experimental condition. N.S., no statistical significance (P > 0.05). (A) A representative family of currents studied before (Left) and after (Right) the addition of 30 µM Chromanol-293B to the bath. The difference current obtained by subtraction is IKs and is shown in Fig. 1. (B) iG-V relationship for the Chromanol-insensitive component of current studied with control solution in the pipette (black), 1 nM SUMO2 (red), or 1 nM SENP2 (blue). (C) Tail current densities are not significantly different between control solution in the pipette (black), 1 nM SUMO2 (red), or 1 nM SENP2 (blue).
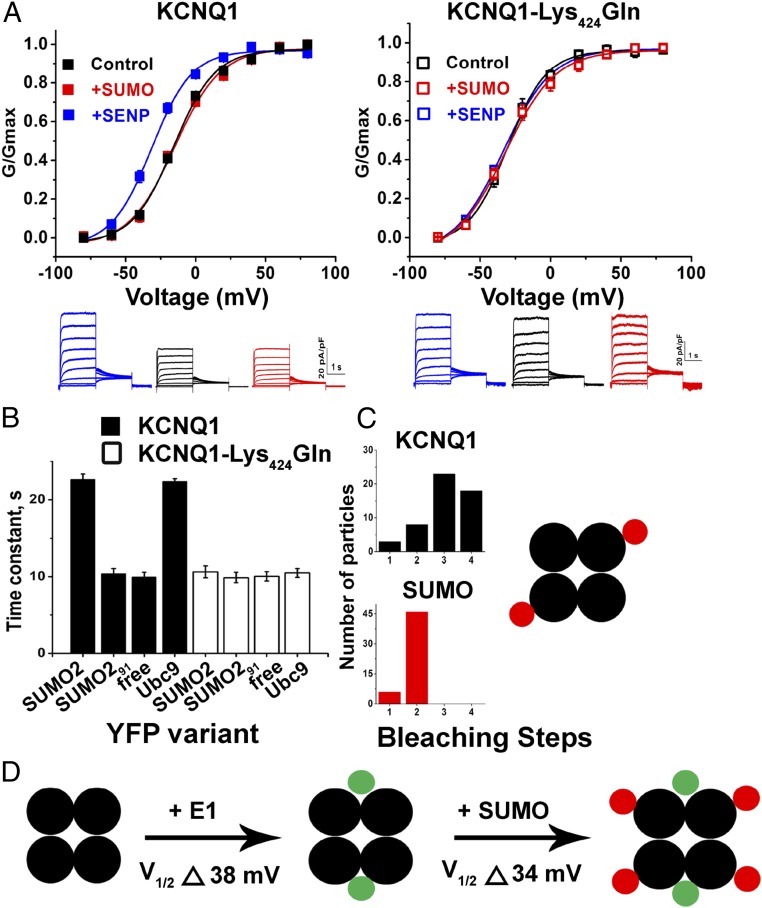
SUMO2 Regulation of Human IKs in CHO Cells Requires KCNQ1 Lys424.
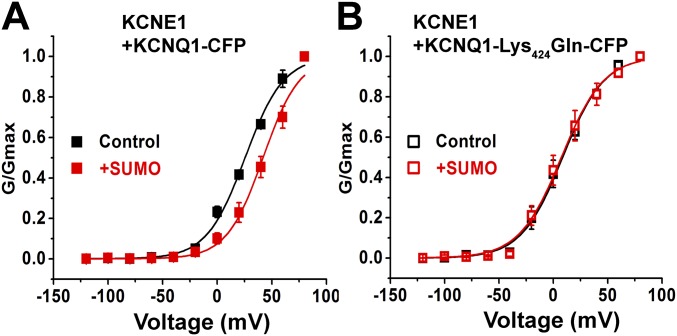
Based on our experience with other ion channels, we suspected that SUMO2 regulation of IKs would result from direct modification of the channel. To test this notion, we reconstituted the channel by expression of human clone KCNQ1 and KCNE1 subunits in CHO cells and observed that the regulatory behaviors seen in the nCM were recapitulated (Fig. 2). In accord with expectations from our prior studies of IKs in CHO cells (30), IKs channels with human subunits had a V1/2 of 25 ± 1.0 mV under control conditions; this was right-shifted to 42 ± 1 mV with 1 nM SUMO2 in the pipette and leftward to 7.6 ± 0.5 mV with 1 nM SENP2, in concert with changed current density and gating kinetics like those observed in the native cells (Table 2). Of note, 1 nM SUMO1 had the same effect on IKs in CHO cells as 1 nM SUMO2 (Table 2).
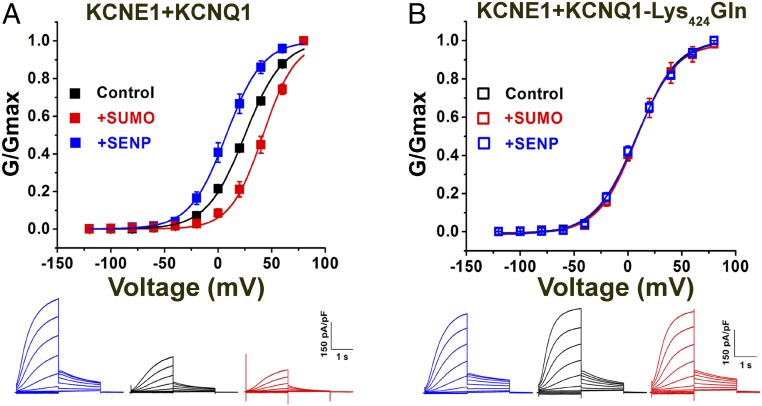
Fig. 2.
The SUMO pathway regulates human IKs in CHO cells. Human KCNQ1 and KCNE1 subunits were transfected into CHO cells, and whole-cell patch-clamp recordings were performed 24–48 h afterward, as described by the protocols in Materials and Methods. Biophysical parameters are in Table 2 (n = 6 cells, mean ± SEM). (A, Upper) iG-V relationships (filled square) for wild-type human KCNQ1 and KCNE1 with 1 nM SENP2 (+SENP, blue), control solution (black), and 1 nM SUMO2 (+SUMO, red). (A, Lower) Raw current traces for wild-type KCNQ1 and KCNE1 with 1 nM SENP2 (+SENP, blue), control solution (black), and 1 nM SUMO2 (+SUMO, red). (B, Upper) iG-V relationships for human KCNQ1-Lys424Gln and KCNE1 (open square) with 1 nM SENP2 (+SENP, blue), 1 nM SUMO2 (+SUMO, red), and control solution (black). (B, Lower) Raw current traces for KCNQ1-Lys424Gln and KCNE1, with 1 nM SENP2 (+SENP, blue), control solution (black), and 1 nM SUMO2 (+SUMO, red).
Table 2.
SUMO regulates human IKs in CHO cells
| Subunits | Pipette solution | V1/2, mV | Current density, pA/pF | τact, ms | τdeact, ms | Zδ |
| KCNE1 | Control | 25 ± 0.8 | 53 ± 6 | 510 ± 36 | 580 ± 49 | 17.9 ± 0.7 |
| KCNQ1 | ||||||
| KCNE1 | SENP2 | 7.6 ± 0.5** | 102 ± 6** | 378 ± 34* | 827 ± 36** | 17.1 ± 0.4 |
| KCNQ1 | ||||||
| KCNE1 | SUMO2 | 42 ± 1** | 28 ± 2** | 767 ± 13** | 312 ± 33** | 15.3 ± 1.2 |
| KCNQ1 | ||||||
| KCNE1 | SUMO1 | 41 ± 2** | 29 ± 1** | 776 ± 20** | 320 ± 22** | 17.7 ± 1.6 |
| KCNQ1 | ||||||
| KCNE1 | Control | 7.8 ± 0.8** | 97 ± 12** | 370 ± 39* | 833 ± 29** | 17.9 ± 0.6 |
| KCNQ1-Lys424Gln | ||||||
| KCNE1 | SENP2 | 7.6 ± 1.1** | 97 ± 13** | 376 ± 49* | 829 ± 48** | 18.8 ± 0.8 |
| KCNQ1-Lys424Gln | ||||||
| KCNE1 | SUMO2 | 7.8 ± 1.6** | 99 ± 11** | 361 ± 37* | 818 ± 35** | 17.5 ± 0.6 |
| KCNQ1-Lys424Gln | ||||||
| KCNQ1 | Control | −16 ± 1 | 6.0 ± 0.7 | 29 ± 2 | 230 ± 12 | 14.9 ± 1.1 |
| KCNQ1 | SENP2 | −32 ± 1*** | 9.2 ± 0.9*** | 30 ± 1 | 231 ± 11 | 14.9 ± 0.8 |
| KCNQ1 | SUMO2 | −14 ± 1 | 5.8 ± 0.8 | 28 ± 1 | 234 ± 12 | 15.7 ± 1.7 |
| KCNQ1-Lys424Gln | Control | −31 ± 1*** | 9.3 ± 0.6*** | 30 ± 2 | 234 ± 11 | 14.9 ± 0.9 |
| KCNQ1-Lys424Gln | SENP2 | −32 ± 1*** | 9.8 ± 1.3*** | 26 ± 3 | 241 ± 19 | 16.4 ± 1.0 |
| KCNQ1-Lys424Gln | SUMO2 | −32 ± 2*** | 9.6 ± 0.8*** | 27 ± 2 | 237 ± 17 | 17.7 ± 1.4 |
Human KCNQ1 or KCNQ1-Lys424Gln expressed in CHO cells ± KCNE1 and studied as per Fig. 2 with control solution, 1 nM SUMO2, 1 nM SUMO1, or 1 nM SENP2 in the pipette as indicated. V1/2 and Zδ were determined from a Boltzmann function using tail current densities; tail current density was assessed by estimating the peak from a fit of the decay curve after an activating pulse at +40 mV for 2 s, and the τ for activation and deactivation was determined from a single exponential fit. Data are mean ± SEM for 6–11 cells studied from each condition. Compared with KCNQ1 + KCNE1 (with control pipette solution), V1/2, tail current density, τ for activation, and deactivation were significantly different for SENP2-, SUMO2-, and SUMO1-treated conditions and also for KCNQ1-Lys424Gln + KCNE1 (with control pipette solution, SENP2, and SUMO2), unpaired t test, *P < 0.05 and **P < 0.01; whereas, the slope factor (Zδ) was not significantly different with SENP2, SUMO2, SUMO1, and KCNQ1-Lys424Gln + KCNE1 (P > 0.05). Compared with KCNQ1 (with control pipette solution), V1/2 and tail current density were significantly different for SENP2-treated conditions and also for KCNQ1-Lys424Gln (with control pipette solution, SENP2, and SUMO2), unpaired t test, ***P < 0.01, but τ for activation and deactivation was not significantly different; V1/2, tail current density and τ for activation and deactivation were not significantly different for SUMO2-treated conditions (P > 0.05); the slope factor (Zδ) was not significantly different with SENP2, SUMO2, and KCNQ1-Lys424Gln (with control pipette solution, SENP2, and SUMO2), P > 0.05.
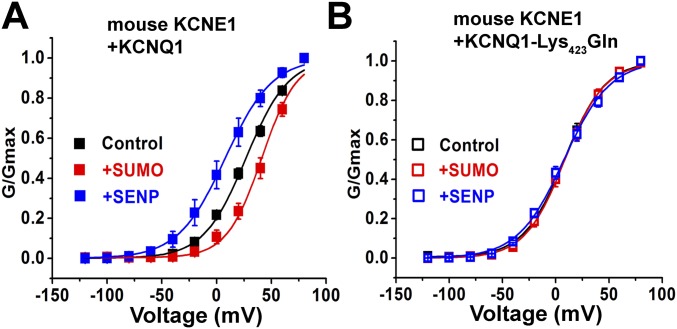
To identify potential SUMOylation site(s) in IKs channels we analyzed the primary sequences of the subunits with an algorithm (31). We found that mutating Lys424 to Gln in human KCNQ1 (KCNQ1-Lys424Gln), within the motif Phe-Lys-Leu-Asp (similar to the canonical SUMO motif ψ-K-X-E/D), produced a channel that was unresponsive to SUMO2 or SENP2, and that operated like wild-type IKs treated with SENP2, that is, a channel without linked SUMO2 (Fig. 2). Thus, IKs channels formed with KCNE1 and KCNQ1-Lys424Gln had a V1/2 of 7.8 ± 0.8, 7.8 ± 1.6, and 7.6 ± 1.1 mV, when studied under control conditions or with 1 nM SUMO2 or 1 nM SENP2 in the pipette, respectively (Table 2). IKs produced in CHO cells with mouse KCNQ1 and KCNE1 subunits responded to SUMO2 and SENP2 like their human counterparts and the analogous mutation in mouse KCNQ1 Lys423Gln, ablated responsiveness to SUMO2 and SENP2, and yielded a channel that operated like the SENP2-treated wild-type isoform (Fig. S2).
Fig. S2.
SUMO regulates mouse IKs in CHO cells. Mouse KCNQ1 and KCNE1 were transfected into CHO cells, and whole-cell patch-clamp recording was performed 24–48 h thereafter, as per Fig. 2. Data are for six cells, mean ± SEM. (A) Isochronal conductance–voltage relationships for wild-type mouse KCNQ1 and KCNE1 (filled square) with 1 nM SENP2 (+SENP, blue), control solution (black), or 1 nM SUMO2 (+SUMO, red), showed mean V1/2 values of 7.8 ± 0.8, 27 ± 1, and 41 ± 2 mV, respectively. Compared with the control condition, V1/2 is significantly different with SENP2 and SUMO2 in the pipette (unpaired t test, P < 0.01). (B) iG-V relationships for mouse KCNQ1-Lys423Gln and KCNE1 (open square) with 1 nM SENP2 (+SENP, blue), 1 nM SUMO2 (+SUMO, red), and control solution (black) with mean V1/2 values of 7.8 ± 0.9, 8.3 ± 0.6, and 8.2 ± 0.8 mV, respectively.
KCNQ1 Lys424 Is the Only Residue Modified by SUMO2 in IKs Channels.
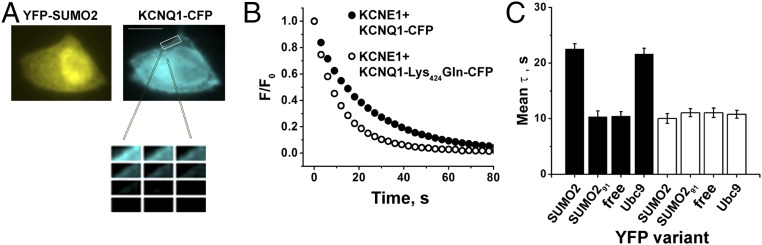
To examine interactions between IKs channels and SUMO, we measured FRET in live CHO cells between subunits tagged with CFP and YFP, a strategy we have used to study the SUMOylation of other ion channels (13–16). Measured here by the photobleaching rate of the CFP donor, FRET reports on a separation of less than 10 nm between FPs—consistent with a protein–protein interaction (Fig. 3 A and B). Tagging wild-type KCNQ1 and KCNQ1-Lys424Gln subunits with CFP did not alter their V1/2, gating kinetics, or responsiveness to SUMO2 (Fig. S3). Similarly, we have previously demonstrated that tagging the N terminus of SUMO with YFP did not alter its operation (13–16). FRET was measured between IKs channels formed with KCNQ1-CFP and YFP-SUMO2. The mean time constant for photobleaching (τ) of 22.5 ± 0.9 s was similar to the value for the interaction of KCNQ1-CFP with the SUMO ligase YFP-Ubc9 (21.6 ± 1.0 s; Fig. 3C). In contrast, KCNQ1-CFP showed τ of 10.4 ± 1.0 s with both YFP-SUMO291—a variant lacking the terminal GG motif required for covalent linkage—and free YFP (10.5 ± 0.8 s). Similarly, KCNQ1-Lys424Gln-CFP did not show FRET with YFP-SUMO2, YFP-SUMO291, free YFP, or YFP-Ubc9, the last meeting expectations because we have previously shown that the SUMO ligase requires its target Lys to bind stably to channel subunits. The lack of FRET between YFP-SUMO2 and IKs channels formed by KCNQ1-Lys424Gln-CFP argues that KCNE1 subunits are not SUMOylated.
Fig. 3.
FRET shows that KCNQ1 Lys424 is the only residue modified by SUMO2 in IKs channels. FRET was assessed in live CHO cells expressing CFP-tagged human KCNQ1 and unlabeled KCNE1 by measuring the time constant (τ) for CFP-photobleaching (donor) in the presence of YFP-tagged subunits (acceptor) and fitting with a single exponential. Data are mean τ ± SEM for 2–5 areas of the plasma membrane of 5–7 cells per group. Significant changes in τ compared with free YFP are indicated below. (A) KCNQ1-CFP (cyan) and YFP-SUMO2 (yellow) reach the cell surface. The boxed area of membrane shows donor photobleaching with continuous illumination. (Scale bar: 20 µm.) (B) Exemplar photobleaching studies show the decay of fluorescence intensity for single cells expressing YFP-SUMO2 and KCNQ1-CFP (filled) or KCNQ1-Lys424Gln-CFP (open) fit by a single exponential to give τ. (C) FRET shows assembly of KCNQ1-CFP (black bars) when expressed with untagged KCNE1 with YFP-SUMO2 and YFP-Ubc9 (τ = 22.5 ± 0.9 and 21.6 ± 1.0, respectively; P < 0.001) but not with linkage-incompetent YFP-SUMO291 or free YFP (τ = 10.4 ± 1.0 and 10.5 ± 0.8, respectively). In contrast, KCNQ1-Lys424Gln-CFP (open bars) did not show FRET with YFP-SUMO2, YFP-SUMO291, YFP-Ubc9, or free YFP (τ = 10.0 ± 0.9; 11.0 ± 0.7; 10.8 ± 0.7; and 11.0 ± 0.8, respectively).
Fig. S3.
CFP tag does not alter IKs channel biophysical properties and its responsiveness to SUMO2. CFP-tagged human KCNQ1 and KCNE1 were transfected into CHO cells and whole-cell patch-clamp recording was performed 24–48 h thereafter, as per Fig. 2. Data are for 4–6 cells, mean ± SEM. (A) Isochronal conductance–voltage relationships for human KCNQ1-CFP and KCNE1 (filled square) with control solution (black) or 1 nM SUMO2 (+SUMO, red) showed mean V1/2 values of 25 ± 1 and 42 ± 2 mV, respectively. Compared with the control condition, V1/2 is significantly different with SUMO2 in the pipette (unpaired t test, P < 0.01). (B) iG-V relationships for human KCNQ1-Lys424Gln-CFP and KCNE1 (open square) with 1 nM SUMO2 (+SUMO, red), and control solution (black) with mean V1/2 values of 7.4 ± 1.3 and 8.5 ± 1.2 mV, respectively.
IKs Channels Are Modified by SUMO2 Up to Four Times, Once on Each KCNQ1 Lys424.
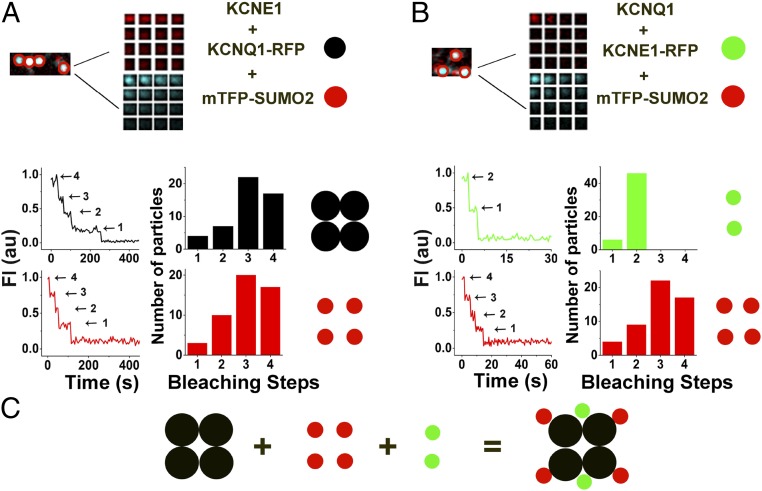
To determine the stoichiometry of SUMOylation, we counted the number of SUMO2 monomers on individual IKs channels at the surface of live CHO cells using simultaneous, two-color TIRF microscopy, and single-particle photobleaching (Fig. 4 and Table 3). We have previously shown that modification of KCNQ1 with TagRFP-T (KCNQ1-RFP) does not alter the expression or the biophysical properties of IKs channels, and that KCNQ1-RFP can be visualized at the cell surface with other subunits tagged with mTFP1 without confounding spectral overlap (30). KCNQ1-RFP and mTFP-SUMO2, in the presence of unlabeled KCNE1, were observed together at the cell membrane (Fig. 4A). In keeping with the known structure of complete channels, KCNQ1-RFP showed up to four bleaching steps, in a distribution expected (due to prebleaching and missed events) for the uniform expression of tetrameric channel assemblies (Fig. 4A). The bleaching analysis of SUMO2 shows up to four steps, consistent with one monoSUMOylation event on each KCNQ1 subunit (Fig. 4A).
Fig. 4.
IKs channels are modified by SUMO2 up to four times. Single KCNQ1-RFP or KCNQ1-Lys424Gln-RFP subunits were expressed in CHO cells with unlabeled KCNE1 and mTFP-SUMO2, or KCNE1-RFP, unlabeled KCNQ1, and mTFP-SUMO2 were expressed, as indicated, and studied by TIRFM, as described in Materials and Methods. Data represent 4–6 cells per condition, and single-particles statistical analyses are summarized in Table 3. (A, Upper) Exemplar single colocalized particles (white, encircled) observed at the surface of cells expressing KCNQ1-RFP (faux red), untagged-KCNE1, and mTFP-SUMO2 (faux cyan). The montages show simultaneous photobleaching for RFP or mTFP in the particle indicated with continuous excitation and an image every fifth frame. (A, Lower) Photobleaching time courses and histograms of photobleaching steps for colocalized particles show four KCNQ1 subunits in complexes (black) and four SUMO2 subunits in complexes (red). (B, Upper) Exemplar single colocalized particles (white, encircled) observed at the surface of cells expressing KCNE1-RFP (faux red) and untagged-KCNQ1 and mTFP-SUMO2 (faux cyan). The montages show simultaneous photobleaching for RFP or mTFP in the particle indicated with continuous excitation and an image every fifth frame. (B, Lower) Photobleaching time courses and histograms of photobleaching steps for colocalized particles show two KCNE1 subunits in complexes (green) and four SUMO2 subunits in complexes (red). (C) Cartoon presentation indicates stoichiometry: KCNQ1 subunits are black, KCNE1 subunits are green, and SUMO2 subunits are red.
Table 3.
Stoichiometry of SUMOylation of IKs
| Subunits expressed | ||||||
| Analysis | KCNE1, KCNQ1-RFP, and mTFP-SUMO2 | KCNE1-RFP, KCNQ1, and mTFP-SUMO2 | No KCNE1, KCNQ1-RFP, and mTFP-SUMO2 | |||
| Determined stoichiometry | SUMO:KCNQ1 | KCNE1:SUMO | SUMO:KCNQ1 | |||
| 4:4 | 2:4 | 2:4 | ||||
| Particles studied | 50 | 52 | 52 | |||
| Particles with | SUMO | KCNQ1 | KCNE1 | SUMO | SUMO | KCNQ1 |
| One step | 3 | 4 | 6 | 4 | 6 | 3 |
| Two steps | 10 | 7 | 46 | 9 | 46 | 8 |
| Three steps | 20 | 22 | 0 | 22 | 0 | 23 |
| Four steps | 17 | 17 | 0 | 17 | 0 | 18 |
| Confidence | 0.98 | 0.98 | 1 | 0.98 | 1 | 0.99 |
| θ | 0.76 | 0.76 | 0.95 | 0.75 | 0.95 | 0.77 |
| θ + 1 | 0.6 | 0.61 | 0.63 | 0.6 | 0.63 | 0.62 |
FP-tagged KCNQ1, SUMO2, and KCNE1 subunits were expressed in CHO cells and studied by TIRFM as described in Fig. 4. The number of photobleaching steps observed for each fluorophore in each fluorescent spot reports on the stoichiometry of the channel complex. KCNQ1 channels are tetramers and show no more than four bleaching steps when tagged with RFP (Fig. 4). Study of KCNQ1 colocalized with mTFP-SUMO2 and untagged KCNE1 reveals a 4:4 stoichiometry of KCNQ1 and SUMO2 in each IKs channel. The statistical confidence in the null hypotheses that KCNQ1 forms tetramers and that four SUMO2s assemble with each IKs channel complex was assessed to be greater than 0.98. Prebleaching and variance in quantum efficiency reduce the probability of observing each possible bleaching event (θ). θ is calculated from the value of n and the distribution of the photobleaching data per Hines (32) and as before for IKs channels (30). θ is decreased when the distribution is altered to estimate the possibility that higher numbers of missed bleaching steps, for example θ + 1, indicating that this stoichiometry is less likely.
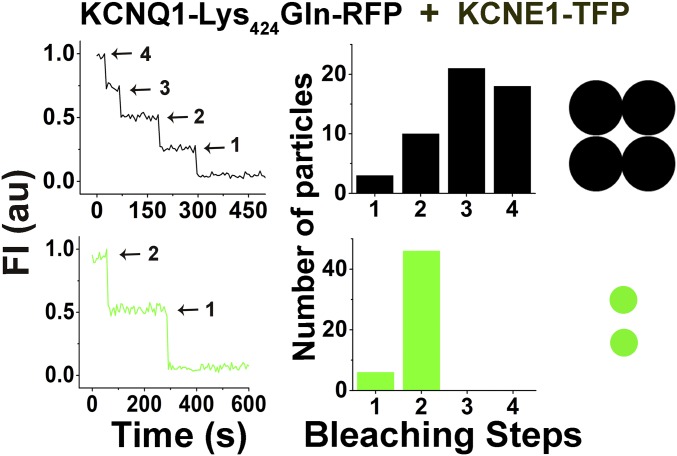
As in our recent work (30), we observed no more than two bleaching steps for KCNE1 subunits in individual IKs channels. Here, with KCNE1-RFP and mTFP-SUMO2 studied in the presence of unlabeled KCNQ1, we observed up to two steps for KCNE1 and up to four steps for SUMO2 (Fig. 4B). Based on the statistical strategy of Hines to determine the number of required particles and to estimate accuracy (32), we determine a 2:4 subunit stoichiometry for KCNE1:SUMO in fully SUMOylated single IKs channels with a confidence approaching 1 (Table 3). Of note, channels formed with KCNQ1-Lys424Gln-RFP subunits, and thus no SUMO, also had two mTFP-tagged KCNE1 subunits, indicating that the SUMOylation status of KCNQ1 did not alter the α:β subunit composition of IKs channels (Fig. S4 and Table S1).
Fig. S4.
Single IKs channels formed with KCNQ1-Lys424Gln subunits contain two KCNE1 subunits. Single KCNQ1-Lys424Gln-RFP subunits were expressed in CHO cells with KCNE1-TFP subunits to study the stoichiometry of the resultant IKs channels as per Fig. 4. Data represent five cells, and single-particle statistical analyses are reported in Table S1. Photobleaching time courses and histograms of photobleaching steps for colocalized particles indicate that there are four KCNQ1-Lys424Gln subunits (black) and two KCNE1 subunits (green) per channel complex.
Table S1.
Stoichiometry of IKs channels formed with KCNQ1-Lys424Gln subunits
| Analysis | Subunits expressed | |
| KCNE1-TFP and KCNQ1-Lys424Gln-RFP | ||
| Determined stoichiometry | KCNE1:KCNQ1 | |
| 2:4 | ||
| Particles studied | 52 | |
| Particles with | KCNE1 | KCNQ1 |
| One step | 6 | 3 |
| Two steps | 46 | 10 |
| Three steps | 0 | 21 |
| Four steps | 0 | 18 |
| Confidence | 1 | 0.98 |
| θ | 0.95 | 0.75 |
| θ + 1 | 0.63 | 0.6 |
Each SUMO2 in IKs Channels Has an Additive Effect on Voltage-Dependent Activation.
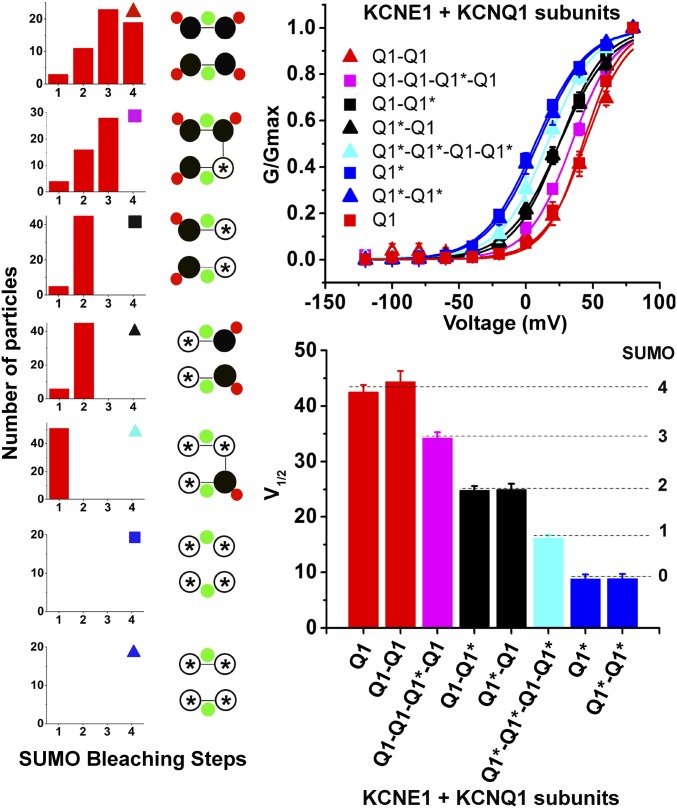
To assess the role of individual SUMO2 adducts, we studied concatemeric IKs channels that linked together KCNQ1 and/or KCNQ1-Lys424Gln subunits (indicated for this experiment here as Q1 and Q1*, respectively) to control the maximum number of SUMO2 that could link to the channel (Fig. 5). Thus, KCNE1 was expressed with Q1*-RFP or Q1*-Q1*-RFP (no SUMO sites), Q1*-Q1*-Q1-Q1*-RFP (one site), Q1-Q1*, or Q1*-Q1-RFP (two sites); Q1-Q1-Q1*-Q1-RFP (three SUMO sites); Q1-Q1-RFP or Q1-RFP (four sites). TIRF microscopy (TIRFM) analysis confirmed the enforced stoichiometry of SUMOylation (Tables S2 and S3). Whole-cell recordings showed that each SUMO2 monomer added into the IKs channels right-shifted the V1/2 by ∼+8.5 mV. This graded response accounts for the total excursion in the V1/2 of ∼30 mV (4 × ∼7.5 mV) observed when native IKs channels in nCM or IKs channels in CHO cells were studied in conditions expected to fully SUMOylate (1 nM SUMO2 in the pipette) or completely deSUMOylate (1 nM SENP2 in the pipette; Figs. 1 and 2). In addition to a change in the voltage required to activate the IKs channels, SUMOylation also evokes graded changes in gating kinetics, slowing the time course for channel activation by ∼20% and speeding deactivation by ∼20% per SUMO2 added into the complex (Table S4). For each concatemer studied, the number of SUMO2 monomers counted was equal to and not greater than the number of Lys424 residues in the channel complex, arguing against the formation of polySUMO2-chains. Analysis of the surface density of isolated and colocalized fluorescent particles revealed that mTFP-SUMO2 was not observed at the plasma membrane of CHO cells expressing KCNQ1-Lys424Gln-RFP subunits with KCNE1, indicating that KCNE1 subunits are not SUMOylated (Table S5).
Fig. 5.
Each SUMO2 in IKs channels has an additive effect on voltage-dependent activation. KCNQ1 (indicated as Q1) or KCNQ1-Lys424Gln (Q1*) subunits were linked to form RFP-tagged channels with fixed numbers of SUMO binding sites and were expressed with mTFP-SUMO2 and untagged KCNE1 in CHO cells and assessed for stoichiometry by TIRFM as per Fig. 4. Whole-cell currents were recorded as per Fig. 2. SUMO2 (1 nM) was included in the pipette solution to saturate all of the SUMO binding sites. Data represent 5–6 cells per condition and biophysical parameters and single-particle statistical analyses are summarized in Tables S1–S3. (Left) Expected SUMO2 stoichiometry and symbols (filled black circle, Q1; open black circle with an asterisk, Q1*; filled green circle, KCNE1; filled red circle, SUMO2) for indicated subunits. Photobleaching histograms for SUMO2 particles and cartoons indicating determined stoichiometry. (Upper Right) iG-V relationships for the eight channel types studied. (Lower Right) Bar graph representation for the values of determined half-maximal activation voltage for the eight channel types studied.
Table S2.
Analysis of single-particle photobleaching—TIRFM experiments with KCNQ1 and IKs dimers
| Analysis | Subunits expressed | |||||||||||
| Q1-Q1-RFP mTFP-SUMO2 | Q1-Q1-RFP KCNE1 mTFP-SUMO2 | Q1-Q1*-RFP mTFP-SUMO2 | Q1-Q1*-RFP KCNE1 mTFP-SUMO2 | Q1*-Q1-RFP mTFP-SUMO2 | Q1*-Q1-RFP KCNE1 mTFP-SUMO2 | |||||||
| Determined stoichiometry | 2:2 | 2:4 | 2:2 | 2:2 | 2:2 | 2:2 | ||||||
| Particles studied, N | 50 | 51 | 51 | 50 | 50 | 52 | ||||||
| Particles with | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 |
| One step | 4 | 5 | 4 | 6 | 6 | 5 | 5 | 4 | 5 | 5 | 6 | 5 |
| Two steps | 46 | 45 | 8 | 45 | 45 | 46 | 45 | 46 | 45 | 45 | 46 | 47 |
| Three steps | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Four steps | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Confidence | 1 | 1 | 0.98 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| θ | 0.97 | 0.96 | 0.76 | 0.95 | 0.95 | 0.96 | 0.96 | 0.97 | 0.96 | 0.96 | 0.95 | 0.96 |
| θ + 1 | 0.64 | 0.64 | 0.6 | 0.63 | 0.63 | 0.64 | 0.64 | 0.64 | 0.64 | 0.64 | 0.63 | 0.64 |
KCNQ1 (Q1) or KCNQ1-Lys424Gln (Q1*) subunits were linked to form RFP-tagged dimeric channels with fixed numbers of SUMO binding sites and were expressed with mTFP-SUMO2 and ± untagged KCNE1 in CHO cells and assessed for stoichiometry by TIRFM as per Fig. 4 and Table 3. Photobleaching and statistical parameters are described as per Table 3 and represent 50–52 particles from 4–6 cells per condition.
Table S3.
Analysis of single-particle photobleaching—TIRFM experiments with KCNQ1 and IKs concatemers
| Analysis | Subunits expressed | ||||||||||
| Q1-Q1-Q1*-Q1-RFP mTFP-SUMO2 | Q1-Q1-Q1*-Q1-RFP KCNE1 mTFP-SUMO2 | Q1*-Q1*-Q1-Q1*-RFP mTFP-SUMO2 | Q1*-Q1*-Q1-Q1*-RFP KCNE1 mTFP-SUMO2 | ||||||||
| Determined stoichiometry | 1:2 | 1:3 | 1:1 | 1:1 | |||||||
| Particles studied | 49 | 48 | 48 | 51 | |||||||
| Particles with | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 | SUMO | Q1 | |||
| One step | 5 | 49 | 4 | 48 | 48 | 48 | 51 | 51 | |||
| Two steps | 44 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | |||
| Three steps | 0 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | |||
| Four steps | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Confidence | 1 | NA | 0.99 | NA | NA | NA | NA | NA | |||
| θ | 0.96 | NA | 0.84 | NA | NA | NA | NA | NA | |||
| θ + 1 | 0.63 | NA | 0.63 | NA | NA | NA | NA | NA | |||
KCNQ1 (Q1) or KCNQ1-Lys424Gln (Q1*) subunits were linked to form RFP-tagged concatemeric channels with fixed numbers of SUMO binding sites and were expressed with mTFP-SUMO2 and ±untagged KCNE1 in CHO cells and assessed for stoichiometry by TIRFM as per Fig. 4 and Table 3. Photobleaching and statistical parameters are described as per Table 3 and represent 48–51 particles from 4–6 cells per condition. NA, not applicable.
Table S4.
Biophysical properties of IKs dimers and concatemers with SUMO2
| Subunits expressed (with KCNE1) | Current density, pA/pF | τact, ms | τdeact, ms | V1/2, mV | zδ |
| Q1-Q1 | 29 ± 1 | 754 ± 27 | 312 ± 19 | 44 ± 2 | 15.7 ± 1.8 |
| Q1-Q1* | 50 ± 2† | 500 ± 37† | 596 ± 21† | 25 ± 1† | 17.2 ± 0.7 |
| Q1*-Q1 | 52 ± 3† | 528 ± 37† | 601 ± 15† | 25 ± 1† | 19.1 ± 0.9 |
| Q1*-Q1* | 103 ± 5† | 375 ± 39† | 804 ± 19† | 8.8 ± 0.8† | 19.1 ± 0.8 |
| Q1-Q1-Q1*-Q1 | 41 ± 2† | 620 ± 12† | 469 ± 18† | 34 ± 1† | 16.3 ± 0.9 |
| Q1*-Q1*-Q1-Q1* | 75 ± 4† | 441 ± 13† | 696 ± 24† | 16 ± 1† | 17.6 ± 0.6 |
Human KCNQ1 (Q1) and KCNQ1-Lys424Gln (Q1*) subunits in dimers and tetramer constructs expressed in CHO cells with KCNE1 and studied as per Fig. 2 with 1 nM SUMO2 in the pipette. V1/2 and Zδ were determined from the same Boltzmann function as per Table 2, tail current density was assessed at −40 mV by estimating the peak from a fit of the decay curve after an activating pulse at +40 mV for 2 s and the τ for activation and deactivation was determined from fitting of a single exponential function per Table 2. Data are mean ± SEM for 6–11 cells. Compared with the control condition (wild-type KCNQ1 dimeric construct, coexpressed with KCNE1 subunit), the V1/2, tail current density, activation and deactivation time constants from all of the KCNQ1 dimeric and concatemeric constructs with variable SUMOylation sites were all statistically significantly different (†P < 0.01, unpaired t test). Compared with the control, the slope factor (Zδ) was not significantly different with all of the KCNQ1 dimers and concatemers studied (P > 0.05).
Table S5.
Colocalization and cell surface density of fluorescent particles
| Subunits expressed | Density of spots × 10−3, per µm2 | Manders’ coefficient | Determined stoichiometry SUMO:KCNQ1 or E1 | ||
| mTFP-SUMO2 alone | RFP alone | Colocalized | |||
| No KCNE1 | 17 ± 2 | 18 ± 2 | 188 ± 20 | 0.94 ± 0.01 | 2:4 |
| KCNQ1-RFP | |||||
| mTFP-SUMO2 | |||||
| KCNE1 | 48 ± 5 | 50 ± 4 | 620 ± 30 | 0.95 ± 0.01 | 4:4 |
| KCNQ1-RFP | |||||
| mTFP-SUMO2 | |||||
| No KCNE1 | NA | 205 ± 15 | NA | NA | NA |
| KCNQ1-Lys424Gln-RFP | |||||
| mTFP-SUMO2 | |||||
| KCNE1 | NA | 635 ± 43 | NA | NA | NA |
| KCNQ1-Lys424Gln-RFP | |||||
| mTFP-SUMO2 | |||||
| KCNE1-RFP | 49 ± 5 | 48 ± 6 | 628 ± 26 | 0.94 ± 0.01 | 4:2 |
| KCNQ1 | |||||
| mTFP-SUMO2 | |||||
| KCNE1-RFP | NA | 663 ± 28 | NA | NA | NA |
| KCNQ1-Lys424Gln mTFP-SUMO2 | |||||
The indicated subunits were expressed and studied as indicated in Fig. 4. The density of surface fluorescent spots either alone or colocalized and the mean Manders’ coefficient of colocalization was determined as described in Materials and Methods. The determined stoichiometry of mTFP-SUMO2 to RFP-tagged KCNQ1 or KCNE1 subunit is based on data shown in Fig. 4 and Table 3. Data are mean ± SEM for 4–6 cells studied in each group. NA, not applicable.
TIRFM was also used to show that although the current density nearly doubles when IKs channels are formed with KCNE1 and KCNQ1-Lys424Gln subunits compared with wild-type KCNE1 and KCNQ1 subunits, the increase was not associated with a change in the number of channels at the surface of live CHO cells (Table S6); thus, SUMOylation did not alter the steady-state surface expression of IKs channels.
Table S6.
Cell surface density of wild-type KCNQ1 and KCNQ1-Lys424Gln subunits (±KCNE1)
| Subunits expressed | Density of spots × 10−3, per μm2 | Manders’ coefficient | ||
| KCNE1-TFP alone | KCNQ1-RFP alone | Colocalized | ||
| No KCNE1 | NA | 194 ± 10 | NA | NA |
| KCNQ1-RFP | ||||
| KCNE1-TFP | 50 ± 4 | 44 ± 3 | 622 ± 30 | 0.94 ± 0.01 |
| KCNQ1-RFP | ||||
| No KCNE1 | NA | 192 ± 10 | NA | NA |
| KCNQ1-Lys424Gln-RFP | ||||
| KCNE1-TFP | 52 ± 4 | 45 ± 4 | 632 ± 36 | 0.94 ± 0.01 |
| KCNQ1-Lys424Gln-RFP | ||||
KCNQ1 or KCNQ1-Lys424Gln subunits with an RFP tag were expressed ±KCNE1-TFP in CHO cells. The density of surface fluorescent spots either alone, or colocalized, and the mean Manders’ coefficient of colocalization was determined as per Table S5. Data are mean ± SEM for five cells studied in each group. The numbers of single wild-type IKs channels and KCNQ1-Lys424Gln IKs channels at the surface of live CHO cells were not statistically significantly different (unpaired t test, P > 0.05); similarly, the numbers of single wild-type and KCNQ1-Lys424Gln channels were not statistically significantly different (unpaired t test, P > 0.05). NA, not applicable.
KCNE1 Determines the Valence of SUMOylation.
Unexpectedly, KCNQ1 channels, formed in the absence of KCNE1, were found to carry only two SUMO2s (Fig. 6). Whole-cell voltage-clamp recording of KCNQ1 channels with 1 nM SENP2 in the pipette showed that deSUMOylation left shifted V1/2 to −32 ± 1 mV from its control level of −16 ± 1 mV and increased the current-density. With 1 nM SUMO2 in the pipette, the V1/2 and current density remained unaltered, suggesting that KCNQ1 channels are maximally SUMOylated under control conditions (Fig. 6A). Channels formed with KCNQ1-Lys424Gln showed no change with either SUMO2 or SENP2 in the pipette, with biophysical parameters similar to those observed for KCNQ1 channels studied with 1 nM SENP2 in the recording pipette. Despite this difference from IKs channels, FRET showed that the Lys424 site was still mediating interaction with SUMO2 in a fashion that required covalent linkage and allowed assembly with the SUMO ligase, Ubc9 (Fig. 6B). In contrast to IKs channels (Fig. 4 and Fig. 5), TIRFM revealed that KCNQ1 channels formed in the absence of KCNE1 were modified at most by two SUMO2 subunits (Fig. 6C and Table 3).
Fig. 6.
In the absence of KCNE1, KCNQ1 channels carry only up to two SUMOs. KCNQ1 or KCNQ1-Lys424Gln channels were expressed in CHO cells without KCNE1 and assessed for whole-cell currents with control, 1 nM SUMO2, or SENP2 in the pipette, as per Fig. 2; FRET, as per Fig. 3; and stoichiometry by TIRFM (as per Fig. 4). Biophysical parameters and single-particle statistical analyses are summarized in Tables 2 and 3. (A, Left) iG-V relationships for wild-type human KCNQ1 channels vary with 1 nM SENP2 (+SENP, blue), control solution (black), or 1 nM SUMO2 (+SUMO, red) in the pipette, showing V1/2 of −32 ± 1 mV, −16 ± 1 mV, and −14 ± 2 mV, respectively, n = 5–6 cells each group. (A, Lower Left) Representative raw current traces for a cell with SENP2 (+SENP, blue), control solution (black), or SUMO2 (+SUMO, red). (A, Right) iG-V relationships for human KCNQ1-Lys424Gln channels with 1 nM SENP2 (+SENP, blue), 1 nM SUMO2 (+SUMO, red), and control solution (black) do not vary, showing V1/2 of −32 ± 1 mV, −32 ± 2 mV, and −31 ± 1 mV, respectively, n = 5–6 cells each group. (A, Lower Right) Representative raw current traces for a cell with SENP2 (+SENP, blue), control solution (black), or SUMO2 (+SUMO, red). (B) FRET shows assembly of KCNQ1-CFP (black bars) with YFP-SUMO2 and YFP-Ubc9 (τ = 22.7 ± 0.7 and 22.4 ± 0.4, respectively, P < 0.001) but not with linkage-incompetent YFP-SUMO291 or free YFP (τ = 10.4 ± 0.7 and 10.0 ± 0.6, respectively). In contrast, KCNQ1-Lys424Gln-CFP (open bars) did not show FRET with YFP-SUMO2, YFP-SUMO291, YFP-Ubc9, or free YFP (τ = 10.7 ± 0.8, 9.9 ± 0.7, 10.5 ± 0.6, and 10.1 ± 0.6, respectively). (C) TIRFM photobleaching shows four KCNQ1 subunits and two SUMO2 subunits in each channel complex. TIRFM single-particle statistical analysis is summarized in Table 3. (D) The cartoon indicates stoichiometry: KCNQ1 subunits are black, KCNE1 subunits are green, and SUMO2 subunits are red. The effect of KCNE1 on V1/2 without SUMOylation, inferred from study of KCNQ1-Lys424Gln channels, and when wild-type IKs channels carry four SUMO2 monomers.
Discussion
In this report, we demonstrate that SUMOylation regulates the biophysical properties of IKs channels in cardiac myocytes as well as channels formed by heterologous expression of the constituent subunits in CHO cells. IKs channels can be SUMOylated up to four times and this produces ∼34-mV right-shift in the V1/2 in nCM and CHO cells; thus, SUMOylation decreases the magnitude of repolarizing current for a given membrane potential, an excitatory effect.
We show that IKs channels have five distinct SUMOylation states and as the valence of SUMOylation increases from 0 to 4, the V1/2 of activation is right-shifted by ∼+8 mV per SUMO2, in a stepwise manner. SUMOylation also evokes graded changes in the gating kinetics of IKs channels, slowing the time course for channel activation by ∼20% and speeding deactivation by ∼20% per SUMO2 added into the complex (Table S4). TIRFM shows that the decrease in current density with SUMOylation of IKs channels is not associated with a change in the number of channels at the surface of live CHO cells (Table S6), suggesting that surface turnover of IKs channels is not SUMO dependent under the conditions of this study. Although the changes in voltage-dependent activation kinetics with SUMOylation are thus sufficient to explain the decrease in current density of IKs channels, our data do not exclude the possibility that SUMOylation has small effects on the unitary conductance.
Whereas IKs carries up to four SUMOs, KCNQ1 channels formed without KCNE1 carry a maximum of two SUMOs despite having four Lys424 binding sites (Fig. 6); perhaps this is not surprising because we observed KV2.1 channels in the absence of accessory subunits are also tetramers that carry a maximum of two SUMO monomers, on nonadjacent subunits (14). Because IKs can accommodate up to four SUMO monomers, one per Lys424, the incorporation of KCNE1 is surmised to expose either the Ubc9 binding site or the SUMO conjugation site on the two other subunits. Arguing against the proposal that highly SUMOylated KCNQ1 channels are rapidly internalized in the absence of KCNE1, we fail to detect channels with three or four SUMOs at the cell surface in single-molecule photobleaching studies despite the time resolution of our TIRF experiments (1 Hz) and the limited mobility of the channels on the surface of the mammalian cells that we study here at room temperature. It remains possible that if KCNQ1 channels are SUMOylated inside the cell, their transit to the surface might be restricted.
A number of mechanisms might be in play to shift the iG-V relationship of IKs channels. The shift in the V1/2 due to KCNE1 in the absence of SUMOylation can be inferred by study of channels formed by KCNQ1-Lys424Gln subunits and is estimated to be ∼+38 mV; our study of wild-type IKs channels shows SUMOylation to produce an excursion of up to ∼+34 mV (Fig. 6D). Because KCNE1 slows the movement of the voltage sensor domains (VSDs) in KCNQ1 to produce the slow channel activation characteristic of IKs (8, 33), SUMOylation might be anticipated to impact the energetics of this conformation change. In addition, the coupling of VSD movement and pore opening is subject to regulation, for example, via PIP2 (34), and Lys424 resides between the A and B domains in the intracellular C terminus of KCNQ1, a region associated with regulation of IKs channel function by calmodulin, phospholipids and kinases (35).
We observe macroscopic IKs current in nCM and CHO cells to operate as if the channel population carries an average of two SUMO2 monomers. This finding, and the observation that a decrease in SENP2 mRNA levels decreases IKs current in nCM, support the conclusion that SUMOylation adjusts the biophysical properties of IKs channels in the heart. Given that the biophysical properties of native IKs channels vary subject to neuronal and hormonal influences (1), it is reasonable to posit that natural regulators of IKs channels may operate via, or in concert with, the SUMO pathway. Further, regulation of the SUMO pathway by acute stressors is now well recognized in the CNS, as shown in our studies of rapid hypoxic regulation of NaV1.2 in cerebellar granule neurons (15), and it is therefore reasonable to expect it to act in a similar manner on ion channels in the heart, such as IKs.
SUMOylation of IKs decreases the repolarizing current and is thereby expected to prolong the duration of cardiac action potentials in human where it is expressed in adult ventricular myocytes; suppression of IKs is thus associated with a prolonged QT interval on the surface electrocardiogram. Whereas we study neonatal mice where IKs is prominent, in animals >6 wk of age the current declines to undetectable levels in ventricular myocytes (10, 27), which may explain why Qi et al. (17) did not observe a prolonged QT or ventricular arrhythmia with SENP2 knockdown in the adult mice they studied. These authors (17) did observe bradycardia and atrioventricular (A-V) conduction block in adult mice with SENP2 knockdown that was global or localized to the nervous system but not when it was targeted to the heart; they hypothesize that increased CNS SUMOylation can cause cardiac symptoms of neural origin as well as seizures, and our findings with neonatal cardiac myocytes do not contradict their conclusions.
The SUMO pathway has been shown to be active in slow processes in the heart, including cardiac development, regulation of metabolism, and disease progression (36). Thus, heart-specific constitutive overexpression of SENP2 in mice has been associated with major congenital defects in both the atria and ventricles of the heart (37). Whereas, chronic viral overexpression of SUMO1 has been reported to have a positive role in treating heart failure, improving cardiomyocyte intracellular calcium homeostasis via effects on SERCA2A calcium pump activity in both a pressure overload heart failure model in mice (38) and an ischemia heart failure model in swine (39), inducible overexpression of SENP1 in mice is reported to produce mitochondrial dysfunction in ventricular myocytes and attributes resembling heart failure in humans (40). That two manipulations that increase SUMOylation appear to have opposite effects on the heart is likely to reflect the multiplicity of SUMO pathway targets, stimulus-dependent changes in SUMOylation/deSUMOylation, chronic and acute changes over time, and our nascent understanding of the SUMO pathway.
In addition to the data we present here on IKs, a growing body of evidence indicates that SUMOylation regulates other cardiac ion channels, including K2P1, KV2.1, KV1.5, and TRPM4, that were studied in noncardiac tissues (or heterologous cells) but are natively expressed in the heart (12, 14, 16, 41–43). Thus, KV1.5 and KV2.1 are predominantly expressed in mouse atrial myocytes, where they contribute to the slow-repolarizing current (44), and K2P1 and TRPM4 are expressed in cardiac Purkinje fibers (45). A familial missense mutation that increases SUMOylation of TRPM4 has been associated with progressive A-V block (43). Thus, we expect SUMOylation modulates cardiac physiology in a manner that varies with localization of these SUMO-regulated proteins and activity of SUMO pathway enzymes. As we have found in our studies of central neurons, Xenopus oocytes, COS-7 cells, and CHO cells (12–15), SUMO regulation of IKs in nCM suggests that SUMO pathway enzymes will be found to be resident at the cardiac myocyte plasma membrane and not restricted to the nucleus and cytosol.
Some years ago, we described the family of subunits related to KCNE1, the mink-related peptides (46, 47), and KCNQ1 has since been shown to assemble with all of them on heterologous expression. As reviewed by Abbott et al. (48), KCNQ1 has thus far been demonstrated to assemble in vivo with KCNE1 in the heart and inner ear; with KCNE2 in the stomach, thyroid, heart, and choroid plexus; and with KCNE3 in breast, intestine, and airway epithelia. Indeed, KCNQ1 has yet to be shown to operate in the body without a KCNE subunit partner. Given that SUMO2 can modify KCNQ1 subunits in the presence of KCNE1, it is reasonable to assume that SUMO will regulate KCNQ1-containing channels with these other KCNE subunits, because the SUMO pathway is ubiquitous. In contrast, the Chromanol-293B–insensitive portion of the delayed rectifier potassium current in nCM does not appear to be regulated by the SUMO pathway. Although we did not identity its basis, others report that HERG (KV11.1) passes the Chromanol-293B–insensitive current in nCM (19, 20).
The stoichiometry of IKs channels has been an issue of debate for some years. As we have previously observed (30, 49), IKs channels studied here in CHO cells form with a fixed stoichiometry of four KCNQ1 subunits and two KCNE1 subunits, a value corroborated by some groups (50, 51) but not others (52). The data in the current study further supports the conclusion that the subunit composition of IKs channels does not vary with SUMOylation, and KCNE1 is not SUMOylated (Fig. 4 and Table S5).
We focus here on SUMO2 following up on the findings of Qi et al. (17) on M-current channels in the brain. In our hands, SUMO1 in the pipette has the same effects as SUMO2 on IKs channels in CHO cells. However, it is important to note that although SUMO1 and SUMO2 share some targets, exchanging the native cellular cytosol with pipette buffer containing 1 nM free SUMO or SENP may not faithfully reflect the behavior of the regulators expressed from their genes at low natural levels found in vivo.
Materials and Methods
Molecular Biology and Reagents.
Wild-type human KCNQ1 (NCBI ACC no. NM_000218), human KCNE1 (NP_000210.2), mouse KCNQ1 (NCBI ACC no. NM_008434), and mouse KCNE1 (NCBI ACC no. NM_008424) were handled in pRAT, an in-house vector as previously described (8, 30). Human SUMO295 and Ubc9 were amplified from a brain cDNA library (Clontech) and inserted into pMAX. Point mutations were introduced by QuikChange mutagenesis (Agilent). TagRFP-T (RFP), mTFP1 (TFP), YFP, and CFP genes were inserted in-frame with the KCNQ1, KCNE1, SUMO2, or Ubc9 where noted, as before (15, 30). SUMO2 and SENP2 were purchased from Boston Biochem. Isradipine and Chromanol-293B were purchased from Sigma-Aldrich.
Neonatal Mouse Ventricular Myocyte Primary Culture.
Neonatal ventricular myocytes were cultured from mice. Briefly, postnatal day 1–3 CD-1 mouse pups (Charles River) were decapitated and the ventricles were dissected, cut into 1-mm squares, and incubated with 0.15% Collagenase Type I (Worthington) in calcium-free, magnesium-free HBSS for 1 h at 37 °C. The tissue was triturated with a fire-polished Pasteur pipette, passed through a cell strainer (35 µm) to remove extraneous debris, and plated onto 25-mm diameter glass coverslips precoated with gelatin (0.1% in sterile water; ATCC). The cells are maintained in DMEM (Thermo Fisher) supplemented with 10% horse serum (ATCC) and 5% FBS (ATCC) and 1% penicillin and streptomycin (Thermo Fisher) at 5% CO2 and 37 °C for 72 h before experiments. Spontaneous beating and muscular striations were used to identify cells in culture as myocytes.
SENP2 Lentiviral Particle Knock-Down Assay and qPCR Analysis.
Lentiviral particles expressing GFP and an shRNA targeting mouse SENP2 or a scrambled control (OriGene Technologies) were applied to nCM for 12 h, where indicated. Cells were studied for 48 h following infection and were identified visually by the expression of GFP. Fluorescent cells (5–10 cells) were collected by glass electrodes and analyzed with the Single-Cell-to-CT Kit (Thermo Fisher Scientific) according to the manufacturer’s guidelines. In brief, the cells were lysed at room temperature and reverse transcription was performed in a thermal cycler. The products from reverse transcription were used for the real-time RT-PCR on a Rotor-Gene Q cycler (QIAGEN) according to the manufacturer’s instructions. TaqMan Gene Expression Assays were acquired from Thermo Fisher Scientific for both SENP2 and GAPDH. All reactions were done in triplicates.
Cell Culture and Heterologous Expression.
CHO-K1 cells (RRID: CVCL_0214) were purchased from ATCC, identity authenticated by cytochrome c oxidase I analysis, demonstrated to be mycoplasma free by Hoechst DNA stain and agar by culture, and maintained in F12K medium supplemented with 10% FBS (ATCC). Plasmids were transfected into cells with Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher). CHO cells for patch-clamp studies were cotransfected with eGFP for experiments performed without FP-tagged channel subunits. Experiments were performed 36–48 h posttransfection at room temperature.
Whole-Cell Voltage-Clamp Electrophysiology.
Whole-cell potassium currents were recorded with an Axopatch 200B amplifier and pCLAMP software (Molecular Devices) at filter and sampling rates of 2 and 10 kHz, respectively. CHO cells were bathed in a solution comprising, in mM: 130 NaCl, 4 KCl, 1.2 MgCl2, 2 CaCl2, and 10 Hepes; pH was adjusted to 7.4 with NaOH. Recording electrodes were fabricated from borosilicate glass (Warner) and fire polished to have a resistances of 2–4 MΩ when filled with a solution containing, in mM: 130 KCl, 1 MgCl2, 5 EGTA, 5 K2ATP, and 10 Hepes; pH was adjusted to 7.4 with KOH. CHO cells had a mean whole-cell capacitance of 14 ± 0.6 pF; series resistance was typically <5 MΩ, and the voltage error of <3 mV was not adjusted for. Cells for study were selected based on fluorescence color (free eGFP in the absence of FP-tagged subunits). Currents were evoked from CHO cells with 2-s test pulses from −120 to 80 mV from a holding potential of −80 mV, with 20-mV increments. Tail currents were recorded at −40 mV for 2 s. The interpulse interval was 15 s. Neonatal cardiac myocytes were studied in a bath solution comprising, in mM: 132 NaCl, 4.8 KCl, 2 CaCl2, 1.2 MgCl2, 10 Hepes, 5 glucose, pH 7.4, with 1 µM isradipine to block voltage-gated calcium channel currents. Pipettes were filled with a solution containing, in mM: 125 K-aspartate, 20 KCl, 10 EGTA, 10 ATP(Mg), and 10 Hepes, pH 7.3. nCM had a mean whole-cell capacitance of 30 ± 3.57 pF; series resistance was typically <5 MΩ, the voltage error of <2.5 mV was not adjusted for. Currents were evoked from nCM with 2-s test pulses from –60 to 60 mV from a holding potential of –40 mV, with 20-mV increments. Tail currents were recorded at −40 mV for 2 s. IKs was isolated by subtraction of the current measured in the presence of 30 µM Chromanol-293B, a IKs inhibitor, from the current measured before application of the drug. IKs-like currents were observed in ∼70% of nCM. Conductance (G) was calculated from the tail currents and expressed as G/Gmax as a function of the membrane voltage. G–V curves were obtained and fit to the equation as previously described (8, 30), where V is the test potential, V1/2 is the voltage of half-maximal activation, and zδ is the slope factor. Activation and deactivation kinetics were analyzed by fitting the current traces with a single-exponential equation and are reported as τact and τdeact, respectively. Current density was measured from fits of tail current decay after an activating pulse of +40 mV lasting 2 s. All of the experiments were performed at room temperature and all electrophysiological data were analyzed in pCLAMP (Molecular Devices), Excel, and OriginLab software.
Live-Cell FRET Experiments and Analysis.
FRET technique was used with live CHO cells to evaluate and validate the direct protein-protein interaction between the SUMO protein and the KCNQ1 channel, which were heterologously expressed at the cell surface. YFP-tagged SUMO2 and CFP-tagged KCNQ1 or KCNQ1-Lys424Gln was transfected, with KCNE1 where indicated. The FRET imaging data were collected using a fluorescence microscope (IX81; Olympus). CFP and YFP were excited at 458 and 514 nm, respectively, and the emission light was collected by bandpass filter of 470–500 nm and 525–575 nm, respectively (14). Images were captured using a CCD camera controlled by MetaMorph software (Molecular Devices) and analyzed with ImageJ.
TIRFM and Single-Particle Photobleaching.
Single-protein particle complexes at the surface of live CHO cells were identified and studied by TIRFM as previously described (15, 30). Briefly, the critical angle for TIRF was adjusted using a CellTIRF illuminator (Olympus) and a high numerical-aperture Apochromat objective (150×, 1.45 N.A.; Olympus) mounted on an automated fluorescence microscope (IX81; Olympus), controlled by MetaMorph software (Molecular Devices). For simultaneous illumination of two fluorophores, CellTIRF software (Olympus) was used to adjust the critical angle for each excitation wavelength to generate evanescent waves of equivalent depth (100 nm). A 561-nm laser line was used to excite RFP, and a 445-nm laser line to excite mTFP. When mTFP was studied with RFP simultaneously, emitted light signals were split by using a 520-nm dichroic mirror mounted in a Dual View adapter (Photometrics), and each wavelength was directed to one-half of a back-illuminated EM-CCD (Hamamatsu).
Analysis of Fluorescence Colocalization and Subunit Stoichiometry.
Fluorophores were photobleached by continual excitation, and data were captured as movies of 100–400 frames acquired at 1 Hz. When TFP was studied with RFP in the same cell, the data for each fluorophore were saved as separate stacks and processed in an identical manner. Misalignment of the data between stacks was corrected in ImageJ using the StackReg plugin. Next, the degree of colocalization between two fluorophores was determined by unbiased intensity correlation analysis using the Coloc2 plugin to obtain the Manders’ coefficient for each cell. For stoichiometric analysis, fluorescent spots were defined as a discrete 3 × 3 pixel region around a pixel of maximum intensity, as before (15, 30). Fluorescence is reported as the change in fluorescence normalized by the initial level of fluorescence for each trace. Statistical analyses to assess the estimated confidence with which stoichiometry can be inferred from the observed data and θ, the probability of successfully observing each possible photobleaching event were performed in RStudio as described (15, 30). The densities of colocalized and single fluorescent spots were determined following thresholding and watershed separation in ImageJ. Then, the particle number was counted in four, separate 30 × 30 pixel regions of interest for five cells per group using the Analyze plug-in.
Statistical Analysis.
Statistical analyses (unpaired t tests) were performed on biophysical parameters, cell surface density of fluorescent particles, and FRET photobleaching time-constants data using pCLAMP (Molecular Devices), Excel and OriginLab software and the statistical significance is indicated in table and figure legends. The statistical significance of TIRF photobleaching parameters was analyzed in RStudio, as before (15).
Acknowledgments
This work was supported by NIH National Heart, Lung, and Blood Institute Grant R01HL105959 (to S.A.N.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706267114/-/DCSupplemental.
References
- 1.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 2.Bohnen MS, et al. Molecular pathophysiology of congenital long QT syndrome. Physiol Rev. 2017;97:89–134. doi: 10.1152/physrev.00008.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Barhanin J, et al. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein SA, Miller C. Site-specific mutations in a minimal voltage-dependent K+ channel alter ion selectivity and open-channel block. Neuron. 1991;7:403–408. doi: 10.1016/0896-6273(91)90292-8. [DOI] [PubMed] [Google Scholar]
- 6.Sesti F, Goldstein SAN. Single-channel characteristics of wild-type IKs channels and channels formed with two minK mutants that cause long QT syndrome. J Gen Physiol. 1998;112:651–663. doi: 10.1085/jgp.112.6.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai KK, Goldstein SAN. The conduction pore of a cardiac potassium channel. Nature. 1998;391:605–608. doi: 10.1038/35416. [DOI] [PubMed] [Google Scholar]
- 8.Ruscic KJ, et al. IKs channels open slowly because KCNE1 accessory subunits slow the movement of S4 voltage sensors in KCNQ1 pore-forming subunits. Proc Natl Acad Sci USA. 2013;110:E559–E566. doi: 10.1073/pnas.1222616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. KCNE1 enhances phosphatidylinositol 4,5-bisphosphate (PIP2) sensitivity of IKs to modulate channel activity. Proc Natl Acad Sci USA. 2011;108:9095–9100. doi: 10.1073/pnas.1100872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx SO, et al. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 11.Henley JM, Craig TJ, Wilkinson KA. Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol Rev. 2014;94:1249–1285. doi: 10.1152/physrev.00008.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SAN. Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell. 2005;121:37–47. doi: 10.1016/j.cell.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Plant LD, et al. One SUMO is sufficient to silence the dimeric potassium channel K2P1. Proc Natl Acad Sci USA. 2010;107:10743–10748. doi: 10.1073/pnas.1004712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plant LD, Dowdell EJ, Dementieva IS, Marks JD, Goldstein SAN. SUMO modification of cell surface Kv2.1 potassium channels regulates the activity of rat hippocampal neurons. J Gen Physiol. 2011;137:441–454. doi: 10.1085/jgp.201110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plant LD, Marks JD, Goldstein SA. SUMOylation of NaV1.2 channels mediates the early response to acute hypoxia in central neurons. eLife. 2016 doi: 10.7554/eLife.20054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plant LD, Zuniga L, Araki D, Marks JD, Goldstein SA. SUMOylation silences heterodimeric TASK potassium channels containing K2P1 subunits in cerebellar granule neurons. Sci Signal. 2012 doi: 10.1126/scisignal.2003431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Y, et al. Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron. 2014;83:1159–1171. doi: 10.1016/j.neuron.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldman AM, et al. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Sci Transl Med. 2009 doi: 10.1126/scitranslmed.3000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuss HB, Marban E. Electrophysiological properties of neonatal mouse cardiac myocytes in primary culture. J Physiol. 1994;479:265–279. doi: 10.1113/jphysiol.1994.sp020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li RA, et al. Functional consequences of the arrhythmogenic G306R KvLQT1 K+ channel mutant probed by viral gene transfer in cardiomyocytes. J Physiol. 2001;533:127–133. doi: 10.1111/j.1469-7793.2001.0127b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D-M, et al. KCNE2 is colocalized with KCNQ1 and KCNE1 in cardiac myocytes and may function as a negative modulator of I(Ks) current amplitude in the heart. Heart Rhythm. 2006;3:1469–1480. doi: 10.1016/j.hrthm.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Terrenoire C, Clancy CE, Cormier JW, Sampson KJ, Kass RS. Autonomic control of cardiac action potentials: role of potassium channel kinetics in response to sympathetic stimulation. Circ Res. 2005;96:e25–e34. doi: 10.1161/01.RES.0000160555.58046.9a. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, et al. Biophysical properties of slow potassium channels in human embryonic stem cell derived cardiomyocytes implicate subunit stoichiometry. J Physiol. 2011;589:6093–6104. doi: 10.1113/jphysiol.2011.220863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, et al. Dynamic subunit stoichiometry confers a progressive continuum of pharmacological sensitivity by KCNQ potassium channels. Proc Natl Acad Sci USA. 2013;110:8732–8737. doi: 10.1073/pnas.1300684110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liin SI, et al. Polyunsaturated fatty acid analogs act antiarrhythmically on the cardiac IKs channel. Proc Natl Acad Sci USA. 2015;112:5714–5719. doi: 10.1073/pnas.1503488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch RF, et al. Effects of the chromanol 293B, a selective blocker of the slow, component of the delayed rectifier K+ current, on repolarization in human and guinea pig ventricular myocytes. Cardiovasc Res. 1998;38:441–450. doi: 10.1016/s0008-6363(98)00021-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Feng ZP, Kondo CS, Sheldon RS, Duff HJ. Developmental changes in the delayed rectifier K+ channels in mouse heart. Circ Res. 1996;79:79–85. doi: 10.1161/01.res.79.1.79. [DOI] [PubMed] [Google Scholar]
- 28.Ohyama H, et al. Inhibition of cardiac delayed rectifier K+ currents by an antisense oligodeoxynucleotide against IsK (minK) and over-expression of IsK mutant D77N in neonatal mouse hearts. Pflugers Arch. 2001;442:329–335. doi: 10.1007/s004240100547. [DOI] [PubMed] [Google Scholar]
- 29.Knollmann BC, et al. Isoproterenol exacerbates a long QT phenotype in Kcnq1-deficient neonatal mice: Possible roles for human-like Kcnq1 isoform 1 and slow delayed rectifier K+ current. J Pharmacol Exp Ther. 2004;310:311–318. doi: 10.1124/jpet.103.063743. [DOI] [PubMed] [Google Scholar]
- 30.Plant LD, Xiong D, Dai H, Goldstein SA. Individual IKs channels at the surface of mammalian cells contain two KCNE1 accessory subunits. Proc Natl Acad Sci USA. 2014;111:E1438–E1446. doi: 10.1073/pnas.1323548111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, et al. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;12:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hines KE. Inferring subunit stoichiometry from single molecule photobleaching. J Gen Physiol. 2013;141:737–746. doi: 10.1085/jgp.201310988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osteen JD, et al. Allosteric gating mechanism underlies the flexible gating of KCNQ1 potassium channels. Proc Natl Acad Sci USA. 2012;109:7103–7108. doi: 10.1073/pnas.1201582109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaydman MA, et al. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc Natl Acad Sci USA. 2013;110:13180–13185. doi: 10.1073/pnas.1305167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobelaim WS, et al. Competition of calcified calmodulin N lobe and PIP2 to an LQT mutation site in Kv7.1 channel. Proc Natl Acad Sci USA. 2017;114:E869–E878. doi: 10.1073/pnas.1612622114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendler L, Braun T, Müller S. The ubiquitin-like SUMO system and heart function: From development to disease. Circ Res. 2016;118:132–144. doi: 10.1161/CIRCRESAHA.115.307730. [DOI] [PubMed] [Google Scholar]
- 37.Kim EY, et al. Enhanced desumoylation in murine hearts by overexpressed SENP2 leads to congenital heart defects and cardiac dysfunction. J Mol Cell Cardiol. 2012;52:638–649. doi: 10.1016/j.yjmcc.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kho C, et al. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477:601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilemann L, et al. SUMO-1 gene transfer improves cardiac function in a large-animal model of heart failure. Sci Transl Med. 2013 doi: 10.1126/scitranslmed.3006487. [DOI] [PubMed] [Google Scholar]
- 40.Cai R, et al. Induction of SENP1 in myocardium contributes to abnormities of mitochondria and cardiomyopathy. J Mol Cell Cardiol. 2015;79:115–122. doi: 10.1016/j.yjmcc.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Dai XQ, Kolic J, Marchi P, Sipione S, Macdonald PE. SUMOylation regulates Kv2.1 and modulates pancreatic beta-cell excitability. J Cell Sci. 2009;122:775–779. doi: 10.1242/jcs.036632. [DOI] [PubMed] [Google Scholar]
- 42.Benson MD, et al. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc Natl Acad Sci USA. 2007;104:1805–1810. doi: 10.1073/pnas.0606702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruse M, et al. Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J Clin Invest. 2009;119:2737–2744. doi: 10.1172/JCI38292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bou-Abboud E, Li H, Nerbonne JM. Molecular diversity of the repolarizing voltage-gated K+ currents in mouse atrial cells. J Physiol. 2000;529:345–358. doi: 10.1111/j.1469-7793.2000.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaborit N, et al. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol. 2007;582:675–693. doi: 10.1113/jphysiol.2006.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbott GW, Goldstein SAN. A superfamily of small potassium channel subunits: Form and function of the MinK-related peptides (MiRPs) Q Rev Biophys. 1998;31:357–398. doi: 10.1017/s0033583599003467. [DOI] [PubMed] [Google Scholar]
- 47.Abbott GW, et al. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 48.Abbott GW. Biology of the KCNQ1 potassium channel. New J Sci. 2014 doi: 10.1155/2014/237431. [DOI] [Google Scholar]
- 49.Wang KW, Goldstein SAN. Subunit composition of minK potassium channels. Neuron. 1995;14:1303–1309. doi: 10.1016/0896-6273(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 50.Morin TJ, Kobertz WR. A derivatized scorpion toxin reveals the functional output of heteromeric KCNQ1-KCNE K+ channel complexes. ACS Chem Biol. 2007;2:469–473. doi: 10.1021/cb700089s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin TJ, Kobertz WR. Counting membrane-embedded KCNE beta-subunits in functioning K+ channel complexes. Proc Natl Acad Sci USA. 2008;105:1478–1482. doi: 10.1073/pnas.0710366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray CI, et al. Unnatural amino acid photo-crosslinking of the IKs channel complex demonstrates a KCNE1:KCNQ1 stoichiometry of up to 4:4. eLife. 2016 doi: 10.7554/eLife.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]