Abstract
While atherosclerotic cardiovascular disease (ASCVD) is known to be common among modern people exposed to various risk factors, recent paleopathological studies have shown that it affected ancient populations much more frequently than expected. In 2010, we investigated a 17th century Korean female mummy with presumptive ASCVD signs. Although the resulting report was a rare and invaluable conjecture on the disease status of an ancient East Asian population, the diagnosis had been based only on anatomical and radiological techniques, and so could not confirm the existence of ASCVD in the mummy. In the present study, we thus performed a paleogenetic analysis to supplement the previous conventional diagnosis of ASCVD. In aDNA extracted from the same Korean mummy, we identified the risk alleles of seven different SNPs (rs5351, rs10757274, rs2383206, rs2383207, rs10757278, rs4380028 and rs1333049) that had already been revealed to be the major risk loci of ASCVD in East Asian populations. The reliability of this study could be enhanced by cross-validation using two different analyses: Sanger and SNaPshot techniques. We were able to establish that the 17th century Korean female had a strong genetic predisposition to increased risk of ASCVD. The current paleogenetic diagnosis, the first of its kind outside Europe, re-confirms its utility as an adjunct modality for confirmatory diagnosis of ancient ASCVD.
Introduction
Analyses on human remains from archaeological sites are very useful for obtaining knowledge on the health and disease statuses of our ancestors [1–3]. Especially in the case of mummies, as their preservation status is far better than those of other types of archaeological corpses, paleopathological data attained from them tend to be much more valuable [4–6]. In addition to conventional diagnostic tools such as anatomical or radiological techniques, recently developed genetic analysis has deepened our understanding of past diseases from another perspective [7–9].
In South Korea, there have been a number of reports on mummies recovered from 15th-to-18th century Joseon tombs. During the past 10 years, those scientific studies have provided us with significant data for comprehensive understanding of Joseon peoples’ health and disease statuses, which knowledge could not easily be attained by historical and archaeological investigations [6,10–23]. Particularly by ancient DNA (aDNA) analysis on Korean mummy samples [24–32], the genetic backgrounds of specific diseases prevailing in the past could be traced.
In 2010, during a computed tomography (CT) examination, we discovered multiple aortic calcifications in a 17th century Korean female mummy (nicknamed Mungyeong). We suspected that the calcifications represented atherosclerosis of the aortic wall. This radiological finding was confirmed by subsequent autopsy in which multiple signs of aortic atherosclerosis were found once again. The mummified female must have suffered from coronary artery disease (CAD), as intimal thickening was also discovered in the left anterior descending (LAD) artery. Actually, this is a rare report of remnant atherosclerotic cardiovascular disease (ASCVD) in ancient human remains of East Asia [18].
Despite its academic implications, however, we also admit that the previous report was not sufficient to capture the full aspects of ancient ASCVD. Actually, a genetic predisposition to cardiovascular disease had already been proven in the paleogenetic study on “the Iceman,” a 5,300-year-old chalcolithic mummy [33,34]. This study showed, for the first time, that genetic analysis can be very successful in diagnosing ancient atherosclerosis. However, as this is only report of its kind to date, it is difficult to be sure that the same technique could also be applied to the diagnosis of similar cases discovered in any countries or continents.
The purpose of this study therefore was to determine whether paleogenetic analysis can also be useful for cases such as that of the above-noted 17th century Korean mummy. More specifically, we attempted to determine if paleogenetic investigation by analysis of single-nucleotide polymorphism (SNP) can be an adjunct modality to anatomical or radiological analyses for confirmatory diagnosis of ASCVD in ancient human remains.
Materials and methods
Archeological information
In April 2010, a middle-aged female mummy (repository number #278, JDHS Collection of Seoul National University College of Medicine, Seoul, South Korea) was discovered within a Joseon tomb in Mungyeong County, South Korea (Fig 1). The approximate time of burial, as estimated by a tree-ring test of the coffin wood, was the 1560s CE. The anatomical and radiological findings observed in this case are summarized in our previous report [18].
Fig 1. Mungyeong mummy examined in this study.

In brief, aortic calcifications were found on CT images. These findings were confirmed to be atheromatous plaques on autopsy. We also found other signs suggestive of atherosclerosis, such as ulcerated plaques, ruptured hemorrhages, and intimal thickening in which the necrotic core was covered by the fibrous cap. In the LAD, we also discovered intimal thickening and protrusion into the lumen, a definite sign of CAD [18]. We strongly suspected that the Mungyeong mummy had suffered from ASCVD.
aDNA extraction
This study was declared by the Institutional Review Board (IRB) of Seoul National University Hospital as an exempt (IRB no. 2013–004). It was conducted in accordance with the Vermillion Accord on Human Remains, World Archaeological Congress (South Dakota, 1989).
The brain tissue of the Mungyeong mummy was subjected to aDNA analysis. The sample (0.5–1 g) was incubated in 4 mL of lysis buffer (EDTA 50 mM, pH 8.0; 1 mg/mL of proteinase K; SDS 1%; 0.1M DTT) at 56°C for 24 hr. Total DNA was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) and then treated with chloroform/isoamyl alcohol (24:1). DNA isolation and purification was performed using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany). The purified DNA was eluted in 50 μl of EB buffer (QIAGEN) [26,27].
Polymerase Chain Reaction (PCR), cloning and sequencing
We selected 10 SNPs (rs5351, rs10757274, rs2383206, rs2383207, rs10757278, rs6903956, rs4380028, rs10953541, rs974819 and rs1333049) linked with atherosclerosis, CAD or myocardial infarction (MI) reported for European, South Asian, Chinese, Japanese and South Korean populations [33,35–43]. The relevant information on the selected SNPs is summarized in Table 1.
Table 1. PCR primer sets and SNP analysis results by Sanger sequencing.
| Related Disease | dbSNP | Primer sequence (5' → 3') | Product size (bp) | AT(℃) | SNP | Risk allele | Allele | Primer reference | |
|---|---|---|---|---|---|---|---|---|---|
| Atherosclerosis | rs53511 | Forward | TCA TCC CTA TAG TTT TAC AAG ACA GC | 74 | 58 | A/G | A | 10A | Keller et al., 2014 |
| Reverse | ATG GCC AAT GGC AAG CAG A | ||||||||
| CAD/CA/CHD | rs107572742,3,4 | Forward | CCC CCG TGG GTC AAA TCT AAG | 82 | 58 | A/G | G | 9G,1A | Keller et al., 2014 |
| Reverse | AGA ATT CCC TAC CCC TAT CTC CTA TCT | ||||||||
| CAD/CA | rs23832063,4 | Forward | TAC TAT CCT GGT TGC CCC TTC TGT C | 78 | 58 | A/G | G | 9G | Keller et al., 2014 |
| Reverse | GGT TCA GGA TTC AGG CCA TCT TG | ||||||||
| CA/MI | rs23832074 | Forward | GAT ACT TAG CCC TTG GGA CC | 121 | 58 | A/G | G | 10G | this study |
| Reverse | TTG GGC AGC TCT TTT CAT AC | ||||||||
| CA/MI | rs107572784 | Forward | GAC AGG GCT GTG GGA CAA GTC | 83 | 58 | A/G | G | 10G | this study |
| Reverse | AGA GAA GGA GAA ACT ACT CTG TC | ||||||||
| CAD/CA/MI | rs69039565,6 | Forward | GGG GAA CAG AGA GAG ATT CCA TC | 127 | 58 | A/G | A | 8G | this study |
| Reverse | GCA CCC ACT TCA ACA CTT GG | ||||||||
| CAD | rs43800287 | Forward | ACT GGG GCA TGG AAA GGT TA | 116 | 58 | C/T | C | 9C | this study |
| Reverse | AGA GGC AGT GAT ATG GAG CG | ||||||||
| CAD | rs109535417 | Forward | GTG TGC CTC TTG AGG ATA AAG C | 150 | 58 | C/T | C | 9T | this study |
| Reverse | AGG GTT CTG TTT CCT GGT CC | ||||||||
| CAD | rs9748198,9 | Forward | ACA CCA TGG ACA AAG AGA AAA | 163 | 53 | C/T | T | 10C | this study |
| Reverse | TGT ATG TAT AAG CAG GGG ATA ACT | ||||||||
| CAD/CA/CAC | rs13330499,10 | Forward | CTG CTT CAT ATT CCA ACT TGT GT | 139 | 56 | C/G | C | 8C | this study |
| Reverse | TTG CTT ACC TCT GCG AGT G | ||||||||
CAD, coronary artery disease; CA, Coronary atherosclerosis; CHD, coronary heart disease; MI, Myocardial infarction; AT, annealing temperature. SNP, single nucleotide polymorphism; AT, annealing temperature. SNP references
1Yasuda et al., 2007
2Xie et al. 2011
3Keller et al., 2012
4Shen et al., 2008
5Wang et al., 2011
6Guo et al. 2012
7C4D genetics consortium, 2011
8Zhou et al., 2012
9Dechamethakun et al., 2014
10Hinohara et al., 2008.
Ten μl of aDNA extracted from the brain-tissue sample was treated with 1.0 unit of uracil-DNA-glycosylase (New England Biolabs, MA, USA) for 30 min at 37°C. Then, aDNA (40 ng) was amplified by PCR in a 20 μl reaction mixture containing a 200 μM dNTP mixture (iNtRON Biotechnology, Seoul, Korea), 1 mg/ml of BSA (New England Biolabs, MA, USA), 10 pmol of each primer (Integrated DNA Technologies, IA, USA), 1X GeneAmp® gold buffer, 2 mM MgCl2, and 2 units of Amplitaq gold DNA polymerase (Applied Biosystems, CA, USA).
The PCR conditions were as follows: pre-denaturation at 95°C for 10 min, 50 cycles of denaturation at 95°C for 30 sec, annealing at 53–58°C for 30 sec, extension at 72°C for 30 sec, and final extension at 72°C for 10 min. The PCR products were separated on 2.5% agarose gel (Invitrogen, Carlsbad, CA, USA) and then stained with ethidium bromide. Also, negative controls (extraction controls) were applied to gel electrophoresis. We photographed the results with a Vilber Lourmat ETX-20M equipped with Biocapt software (Vilber Lourmat, Collégien, France).
Cloning and sequencing were performed for amplified PCR products of the expected sizes. After the DNA of the amplified bands was extracted using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany), bacterial transformation was completed using the pGEM-T Easy Vector system (Promega, WI, USA). Bacteria transformed by the DNA products were grown on an agar plate containing ampicillin (100ug/ml), 0.5 mM IPTG and X-GAL (40 ug/μl) for 12 -14hr.
The selected colonies were grown in LB media for 12–16 hr, after which plasmid was harvested using the QIAprep spin miniprep kit (QIAGEN, Hilden, Germany). Sequencing was performed on each strand using the ABI Prism BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA) and the 3730xl Automated Sequencer (Applied Biosystems, CA, USA). The consensus sequences were determined using Alignment Explorer implemented in MEGA7 [44]. The SNPs obtained from Sanger sequencing were observed and compared with the risk alleles of each locus.
SNaPshot analysis
We used 10 single-base-extension (SBE) primers for SNP analysis (Table 2) with each of the PCR products (1–3μl). The SBE reactions were carried out using a SNaPshot™ Kit (Applied Biosystems, CA, USA) and a PTC-200 DNA Engine (Bio-Rad Laboratories, CA, USA), following the manufacturer’s instructions. The thermal cycling conditions were as follows: 25 cycles of denaturation at 96°C for 10 sec; annealing at 50°C for 5 sec; extension at 60°C for 30 sec. The amplicons were purified with 1.0 unit of shrimp alkaline phosphatase (SAP) (USB Corporation, OH, USA), incubated at 37°C for 45 min, and heat-inactivated at 80°C for 15 min. The reactants were analyzed by an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, CA, USA) and GeneMapper®ID software, v3.2.1 (Applied Biosystems, CA, USA). The SNP scoring at each locus was confirmed by repeated SNaPshot analysis. The Sanger sequencing and SNaPshot experiments were conducted independently in different laboratories, and the results were compared.
Table 2. SNP analysis of Mungyeong mummy DNA by SNaPshot Kit.
| dbSNP | SBE Primer sequence (5' → 3') | SNP | Risk allele | Allele (PCR1) | Allele (PCR2) |
| rs5351 | TAC AAG ACA GCA AAA GAT TGG TGG CT | A/G | A | A | A |
| *rs10757274 | TCT ATC TAG TGA ATT TCA ATT ATG TC | A/G | G | G | G |
| *rs2383206 | TTC AGG ATT CAG GCC ATC TTG CAA A | A/G | G | G | G |
| rs2383207 | TTT TTT ACT CCT GTT CGG ATC CCT TC | A/G | G | G | G |
| *rs10757278 | CTG TCT TGA TTC TGC ATC GCT GC | A/G | G | G | G |
| *rs6903956 | TTG GGG GAC CAA CCT TAA GTA ATA A | A/G | A | G | G |
| rs4380028 | GGC ATG GAA AGG TTA AGT AAC TTG | C/T | C | C | C |
| rs10953541 | TAT GGG TAC CTA AGT ATT AGC AGC A | C/T | C | T | T |
| *rs974819 | TCT CCA AAC ATG AAA ATA AAA CAG TA | C/T | T | C | C |
| rs1333049 | CAT ACT AAT CAT ATG ATC AAC AGT T | C/G | C | C | C |
*Reverse direction
Authenticity of this study
In the course of the sampling and lab work, we wore sterilized protection gloves, masks, gowns, and head caps. Our aDNA lab facilities had been set up according to the protocol of Hofreiter et al. [45]. The rooms for aDNA extraction and PCR preparation, respectively, were physically separated from our main PCR lab. The DNA extraction/PCR preparation rooms were each equipped with UV irradiation, isolated ventilation, and a laminated flow hood. Every procedure in this study was performed according to the authentic aDNA analysis criteria suggested by Hofreiter et al. [45].
We also tried to see whether the modern DNA might contaminate the ancient samples used in this study. The mtDNA profiles of all of the researchers involved were determined with the permission of the Institutional Review Board of Seoul National University (H-0909-049-295).
After obtainment of the mtDNA control region sequences from the mummy and researcher samples [46–48], 40 ng of aDNA was mixed with premix solution containing 1X AmpliTaq Gold® 360 Master Mix (Life Technologies, CA, USA) and 10 pmol of each primer (Integrated DNA Technology, IA, USA). The current PCR conditions and primer sequences were the same as in our previous study [28, 49]. The PCR products, as isolated using the Qiagen gel extraction kit (QIAGEN, Hilden, Germany), were sequenced by the ABI Prism® 3100 Genetic Analyzer (Applied Biosystems, CA, USA) using the ABI Prism® BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA). The obtained sequences were aligned using Alignment Explorer implemented in MEGA7 [44] for determination of the mummy’s and researchers’ mtDNA consensus sequences, which subsequently were compared to rule out any possibility of contamination.
Results
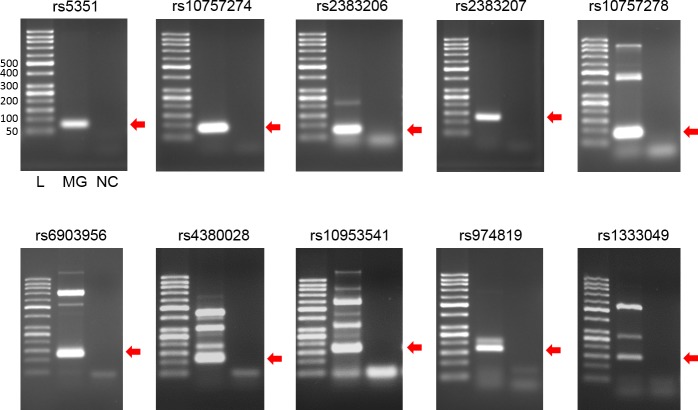
In the gel electrophoresis of the PCR amplicons for the 10 different SNPs, we found amplified bands of the expected sizes at each target locus. Meanwhile, there were no bands detectable for the negative control samples (Fig 2). For the amplified DNA, cloning and sequencing analyses were then performed. From each DNA amplicon of the target loci, between 8 and 10 clone sequences were obtained (S1 File). By multiple-sequence alignment using the ClustalW program implemented in MEGA7 [44], we genotyped each target SNP.
Fig 2. PCR amplification of DNA extracted from Mungyeong mummy sample.
Amplicons associated with each SNP site were observed (indicated by arrows). L, 50bp ladder; MG, Mungyeong mummy; NC, negative extraction control.
The SNP genotypes obtained from the consensus sequencing results were screened to determine the presence of risk alleles. Among the SNPs examined, the risk alleles of atherosclerosis were found in seven different SNPs (rs5351, rs10757274, rs2383206, rs2383207, rs10757278, rs4380028 and rs1333049). Specifically, homozygous genotypes were identified at rs5351 (AA), rs2383206 (GG), rs2383207 (GG), rs10757278 (GG), rs4380028 (CC), and rs1333049 (CC) (Table 1). In the case of rs10757274, the SNP genotype determined by Sanger sequencing was 9G, 1A. No risk alleles were found in three SNPs (rs6903956, rs974819, rs10953541) even though they also had been established as atherosclerosis-related loci for CAD and MI.
To determine the reliability of our Sanger sequencing results, we re-confirmed them by SNaPshot analysis, this time at a different laboratory. In the results, most of the SNP genotypes thus obtained were identical to those obtained by the earlier Sanger sequencing results. Additionally, rs10757274 was confirmed to be homozygous genotype (GG) (Table 2, Fig 3).
Fig 3. The genotyping results analyzed by the SNaPshot kit were identical to those of Sanger sequencing.
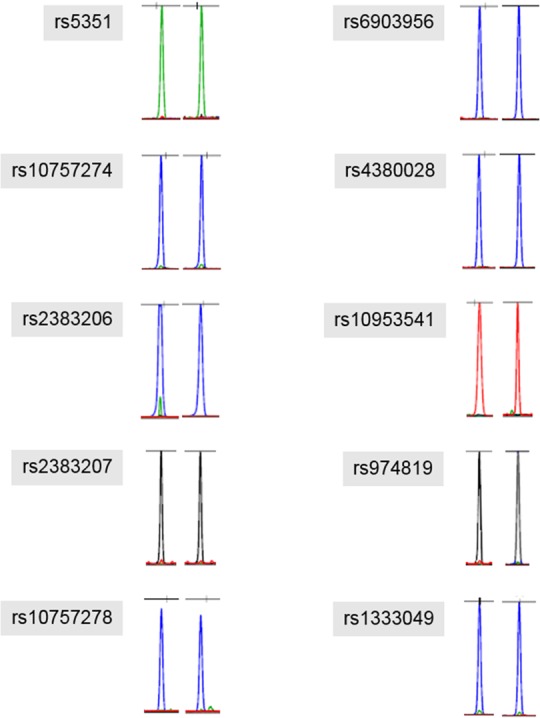
Some of the results were shown as antisense allele because SBE primer was made in a reverse direction.
To rule out any possibility of sample contamination by modern DNA, we compared the haplotypes of the mtDNA control region as obtained from the Joseon mummy and the researchers’ samples. As no identical sequences were found between them, it is highly likely that the obtained data were endogenous (Table 3, Fig 4, S2 File).
Table 3. Comparison of mtDNA hypervariable region from Mungyeong mummy and researcher samples.
| Subjects | Hypervariable region | |||
|---|---|---|---|---|
| HVI (15991–16390) | HVII (034–369) | HVIII (423–548) | ||
| Mungyeong mummy | Lab Aa | 16223T 16362C | 73G 263G 315.1C | 489C |
| Lab Bb | 16223T 16362C | 73G 263G 315.1C | 489C | |
| Researchers | 1 | 16172C 16174T 16223T 16362C | 73G 263G 309.1C 315.1C | |
| 2 | 16183C 16189C 16220C 16254G 16298C 16362C | 73G 249d 263G 315.1C | ||
| 3 | 16129A 16182C 16183C 16189C 16232A 16249C 16304C 16311C 16344T | 73G 152C 249d 263G 315.1C | ||
acloning
bdirect sequencing
Fig 4. PCR amplification for mtDNA hypervariable region of Mungyeong mummy.
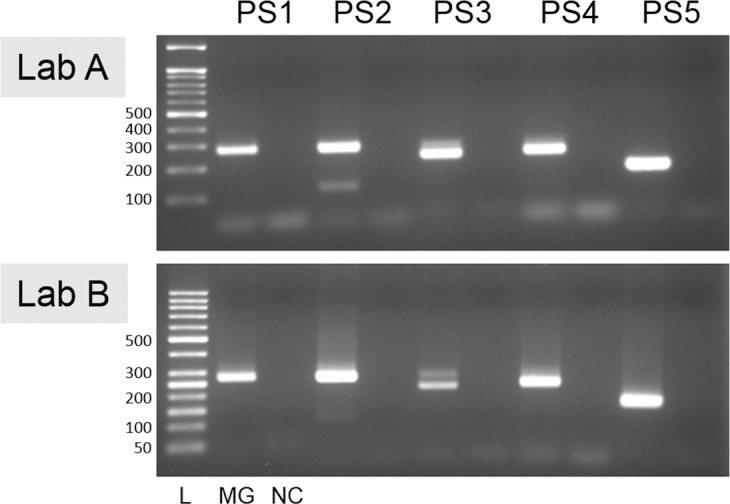
Electrophoresis of Lab A and B showed the same amplified bands (PS1, 267 bp; PS2, 267 bp; PS3, 226 bp; PS4, 235bp; PS5, 167bp). L, 50bp ladder; MG, Mungyeong mummy; NC, negative extraction control.
Discussion
Atherosclerosis is a pathological state characterized by the thickening and protrusion of intima into the vascular lumen. Although atherosclerosis lesions are benign fatty streaks that do not initially interfere with blood flow, they progress to atheromatous plaque, narrowing the vascular lumen, and eventually inducing fatal diseases such as CAD and its acute complication, MI [50,51]. This is generally called ASCVD, the leading cause of morbidity and mortality in many countries [52].
It is common to think that ASCVD is of concern only for modern people with their various risk factors including hypertension, diabetes, smoking, high caloric diet, and hyperlipidemia, among others [52,53]. However, genetic factors must not be ignored either, as many CAD and atherosclerosis cases have been correlated with the genetic predisposition of the patient [35,54,55]. Thus, in any attempts to determine the health statuses of ancient individuals based on mummified remains, we must not rule out ASCVD as a candidate cause of death either.
In this sense, recent studies on several-hundred-to-thousand-year-old mummies are significant to concerned researchers. Through examinations of actual mummy cases, ASCVD has in fact been common to many individuals in the past. For instance, based on mummy studies, Thompson et al. [56] established that certain ancient individuals suffered from atherosclerosis while alive even though they would have been significantly less exposed to risk factors than we are today. By multiple vascular calcifications on radiological images, atherosclerosis and arteriosclerosis also has been proven a common disease among the ancient Egyptians [57,58]. Pathological signs of atherosclerosis were found in an 18th century Aleutian female mummy [59] and an Italian Renaissance mummy [60] as well. All of this is not even to mention our previous report on 17th century Mungyeong mummy that also showed definitive signs of atherosclerosis and CAD [18]. Taking all of this evidence together, we know that ASCVD was a disease not infrequently seen in the ancient world.
Although pioneering studies have contributed greatly to the proper understanding of ASCVD in history, we must also consider the technical limitations of the anatomical and radiological modalities employed. This is in fact a serious issue, given that the samples examined typically are too old for confirmation by ordinary biological-investigative means. Therefore, ancient cases such as the present Mungyeong mummy, though showing multiple signs of atherosclerosis and CAD on CT or autopsy, still require novel diagnostics for confirmation.
In this regard, the recent genomic “Iceman” study is very significant to us. In their examination of this 5,300-year-old chalcolithic mummy, Keller et al. [33] found multiple vascular calcifications suggestive of atherosclerosis on CT images. Not satisfied with this evidence, they performed an aDNA analysis in which they also discovered the risk alleles of the SNPs linked to atherosclerosis and CAD. Their detailed sequencing identified the homozygous risk alleles of SNPs rs10757274 and rs2383206, both strong predictors of CAD [33,34]. Their report represents the first successful demonstration of the utility of genetic studies to the diagnosis of ancient atherosclerosis.
The Iceman study, however, is the sole report of its kind, which fact leaves the paleogenetic study of ancient CAD on an unstable basis. Especially for East Asian individuals or populations, which are genetically distant from the Iceman, there is absolutely no report to date on any paleogenetic analysis of ancient ASCVD. Of course, as greater numbers of genetic (aDNA) diagnoses of ancient ASCVD for chronologically and geographically diverse samples become available, the more accurate the information on ASCVD in history will be [34].
In the current study, we thus analyzed ASCVD-related SNPs to determine if any genetic predisposition could be detected in our 17th century Joseon mummy. Among 10 different ASCVD-related SNPs, we eventually found risk alleles in seven SNPs (rs10757274, rs2383206, rs5351, rs2383207, rs10757278, rs4380028, and rs1333049) in Sanger sequencing. These results were verified by SNaPshot analysis. Meanwhile, no risk alleles were identified in SNPs rs6903956, rs10953541 or rs974819.
As noted above, the SNPs examined in this study had already been revealed to be the major risk loci for ASCVD in modern East Asian populations. Four of these (rs10757274, rs2383206, rs2383207, and rs10757278), in chromosome region 9p21, are known to be strongly associated with CAD or MI in various human populations worldwide [36,61–68]. Briefly, the allele GG of rs10757274 has been repeatedly confirmed as a major risk locus for CAD and MI in both Chinese (odd ratio (OR) = 1.37, confidence interval (CI) = 1.31–1.43, p = 7.56E-45) and Korean (OR = 1.29, CI = 1.06–1.58, p = 0.010) populations [37,69]. The association of rs2383206 with CAD and MI phenotypes has been reported based on Chinese (OR = 1.54, CI = 1.18–2.01, p = 0.001) and Korean studies (OR = 1.30, CI = 1.06–1.58, p = 0.024) [37, 65]. The genotype and allelic frequencies for the CAD-related SNP rs2383207 differed remarkably between case and control subjects of Han Chinese (OR = 1.52, CI = 1.13–2.04) and Korean (OR = 1.32, CI = 1.06–1.63, p = 0.001) populations [37, 42]. Researchers also have discovered the contribution of SNP rs10757278 to CAD and acute coronary syndrome (ACS) risks in Han Chinese (OR = 1.91, CI = 1.35–2.68, p = 0.00035) and Korean (OR = 1.29, CI = 1.06–1.57, p = 0.001) populations [37, 70]. In our study, the homozygous risk alleles of the above-noted four different SNPs in chromosome region 9p21 were identified in the Mungyeong mummy.
Besides the SNPs on chromosome 9p21, endothelin receptor type B-rs5351 also is known to increase the risk of atherosclerosis in Japanese males (OR of the allele AA-GG = 5.0; CI = 1.13–2.04; p = 0.0187) [35]. The relationship between rs4380028 and CAD has been reported based on a Chinese population study as well (OR = 1.17, p = 0.000667) [69]. The strong associations of rs1333049 with CAD, moreover, have been observed in both Koreans (OR = 1.19, CI = 1.02–1.38, p = 0.025) and Japanese (OR = 1.30, CI = 1.13–1.49, p = 0.00027) [36]. The Mungyeong mummy examined in the present study also was homozygous for the risk alleles of the above-noted seven SNPs confirmed to be the major risk loci for atherosclerosis, CAD, and/or MI in East Asian populations. Therefore, we could not deny the possibility that the 17th century Korean mummy had a strong genetic predisposition to increased risk of ASCVD.
Conclusions
Although radiology and autopsy are still the most useful and convenient techniques for mummy studies, they have drawbacks in terms of confirmative diagnosis of ancient diseases such as ASCVD. Indeed, ancient people must have been less exposed to the relevant risk factors than are people these days. Notwithstanding, the genetic impact on ASCVD onset cannot be ignored either. Thus, research that considers both phenotypes and genotypes is necessary in order to confirm diagnoses of ancient ASCVD.
In this study, to overcome those limitations, we attempted to determine whether paleogenetic analysis can be a useful tool supplementary to the classical ASCVD detection techniques in studies on ancient human remains. By SNP analyses of Sanger sequencing and SNaPshot techniques, we found a genetic predisposition to ASCVD in the Joseon female mummy. Our attempt to genetically diagnose the presence of ASCVD, the first report of its kind outside Europe, is a small but significant step in the effort to make paleopathological disease research more efficient and authentic.
Supporting information
(PDF)
(A) Consensus sequence of mtDNA control region for Mungyeong mummy. (B) Comparison between Revised Cambridge Reference Sequence (rCRS, GeneBank accession no. NC_012920), consensus sequence and direct sequencing result of mtDNA control region.
(PDF)
Acknowledgments
First two authors (Shin DH and Oh CS) contributed equally to this study. We are grateful to Professor Frank Rühli of University of Zurich for his helpful advice on the current study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013R1A1A2009688) and by the Seoul National University Hospital (SNUH) research fund (04-2016-0390). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aufderheide AC, Rodriguez-Martin C. The Cambridge Encyclopedia of Human Paleopathology. Cambridge University Press; Cambridge, UK: 1998. [Google Scholar]
- 2.Ortner DJ. Identification of Pathological Conditions in Human Skeletal Remains. Academic Press; 2nd edition San Diego, CA: 2003. [Google Scholar]
- 3.Baca M1, Doan K, Sobczyk M, Stankovic A, Węgleński P. Ancient DNA reveals kinship burial patterns of a pre-Columbian Andean community. BMC Genet. 2012; 13:30 doi: 10.1186/1471-2156-13-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynnerup N. Mummies. Am J Phys Anthropol. 2007; Suppl 45: 162–90. [DOI] [PubMed] [Google Scholar]
- 5.Lynnerup N. Bog bodies. Anat Rec (Hoboken). 2015; 298: 1007–1012. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y-S, Lee IS, Jung G-U, Oh CS, Yoo DS, Lee WJ, et al. Radiological Diagnosis of Congenital Diaphragmatic Hernia in 17th century Korean Mummy. PLoS ONE 2014; 9: e99779 doi: 10.1371/journal.pone.0099779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nerlich A. Paleopathology and Paleomicrobiology of Malaria. Microbiol Spectr. 2016; 4(6): PoH-0006-2015. [DOI] [PubMed] [Google Scholar]
- 8.Rühli FJ, Galassi FM, Haeusler M. Palaeopathology: Current challenges and medical impact. Clin Anat. 2016; 29: 816–822. doi: 10.1002/ca.22709 [DOI] [PubMed] [Google Scholar]
- 9.Spigelman M, Donoghue HD, Abdeen Z, Ereqat S, Sarie I, Greenblatt CL, et al. Evolutionary changes in the genome of Mycobacterium tuberculosis and the human genome from 9000 years BP until modern times. Tuberculosis (Edinb) 2015; 95: S145–149. [DOI] [PubMed] [Google Scholar]
- 10.Shin DH, Choi YH, Shin KJ, Han GR, Youn M, Kim CY, et al. Radiological analysis on a mummy from a medieval tomb in Korea. Ann Anat. 2003a; 185: 377–382. [DOI] [PubMed] [Google Scholar]
- 11.Shin DH, Youn M, Chang BS. Histological analysis on the medieval mummy in Korea. Forensic Sci Int. 2003b; 137: 172–182. [DOI] [PubMed] [Google Scholar]
- 12.Shin DH, Lim DS, Choi KJ, Oh CS, Kim MJ, Lee IS, et al. Scanning electron microscope study of ancient parasite eggs recovered from Korean mummies of the Joseon Dynasty. J Parasitol. 2009; 95: 137–145. doi: 10.1645/GE-1588.1 [DOI] [PubMed] [Google Scholar]
- 13.Shin DH, Lee IS, Kim MJ, Oh CS, Park JB, Bok GD, et al. Magnetic resonance imaging performed on a hydrated mummy of medieval Korea. J Anat. 2010; 216: 329–334. doi: 10.1111/j.1469-7580.2009.01185.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee IS, Lee EJ, Park JB, Baek SH, Oh CS, Lee SD, et al. Acute traumatic death of a 17th century general based on examination of mummified remains found in Korea. Ann Anat. 2009; 191: 309–320. doi: 10.1016/j.aanat.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 15.Lee E-J, Oh CS, Yim SG, Park JB, Kim Y-S, Shin MH, et al. Collaboration of archaeologists, historians and bioarchaeologists during removal of clothing from Korean mummy of Joseon dynasty. Int J Hist Archaeol. 2013; 17: 94–118. [Google Scholar]
- 16.Lee WJ, Yoon AY, Song MK, Wilkinson CM, Shin DH. The archaeological contribution of forensic craniofacial reconstruction to a portrait drawing of a Korean historical figure. J Archaeol Sci. 2014; 49: 228–236. [Google Scholar]
- 17.Hershkovitz I, Spigelman M, Lim D-S, Lee IS, Oh CS, May H, et al. A possible case of Cherubism in a 17th-century Korean female mummy. PLoS ONE 2014; 9:e102441 doi: 10.1371/journal.pone.0102441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MJ, Kim Y-S, Oh CS, Go J-H, Lee IS, Park W-K, et al. Anatomical Confirmation of Computed Tomography-Based Diagnosis of the Atherosclerosis Discovered in 17th Century Korean Mummy. PLoS ONE 2015; 10: e0119474 doi: 10.1371/journal.pone.0119474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y-S, Lee IS, Oh CS, Kim MJ, Cha SC, Shin DH. Calcified Pulmonary Nodules Identified in 350-Year-Old-Joseon Mummy: the First Report on Ancient Pulmonary Tuberculosis from Archaeologically Obtained Pre-modern Korean Samples. J Korean Med Sci. 2016; 31: 147–151. doi: 10.3346/jkms.2016.31.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo M, Guk SM, Kim J, Chai JY, Bok GD, Park SS, et al. Paleoparasitological report on the stool from a Medieval child mummy in Yangju, Korea. J Parasitol. 2007; 93: 589–592. doi: 10.1645/GE-905R3.1 [DOI] [PubMed] [Google Scholar]
- 21.Seo M, Shin DH, Guk SM, Oh CS, Lee EJ, Shin MH, et al. Gymnophalloides seoi eggs from the stool of a 17th century female mummy found in Hadong, Republic of Korea. J Parasitol. 2008; 94: 467–472. doi: 10.1645/GE-1365.1 [DOI] [PubMed] [Google Scholar]
- 22.Seo M, Araujo A, Reinhard K, Chae J-Y, Shin DH. Paleoparasitological studies on Mummies of the Joseon Dynasty, Korea. Korean J Parasitol. 2014a; 52: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo M, Oh CS, Chai J-Y, Jeong MS, Hong SW, Seo Y-M, et al. The Changing Pattern of Parasitic Infection among Korean Populations by Paleoparasitological Study of Joseon Dynasty Mummies. J Parasitol. 2014b; 100: 147–150. [DOI] [PubMed] [Google Scholar]
- 24.Oh CS, Seo M, Chai JY, Lee SJ, Kim MJ, Park JB, et al. Amplification and sequencing of Trichuris trichiura ancient DNA extracted from archaeological sediments. J Archaeol Sci. 2010a; 37: 1269–1273. [Google Scholar]
- 25.Oh CS, Seo M, Lim NJ, Lee SJ, Lee EJ, Lee SD, et al. Paleoparasitological report on Ascaris aDNA from an ancient East Asian sample. Mem Inst Oswaldo Cruz. 2010b; 105: 225–228. [DOI] [PubMed] [Google Scholar]
- 26.Oh CS, Lee SJ, Lee SD, Kim MJ, Kim YS, Lim DS, et al. Amplification of DNA remnants in mummified human brains from medieval Joseon tombs of Korea. Anthropol Anz. 2013; 70: 57–81. [DOI] [PubMed] [Google Scholar]
- 27.Oh CS, Koh B-J, Yoo DS, Park JB, Min SR, Kim Y-S, et al. Joseon Funerary Texts Tested Using Ancient DNA Analysis of a Korean Mummy. Anat Rec (Hoboken). 2015a; 298: 1191–1207. [DOI] [PubMed] [Google Scholar]
- 28.Oh CS, Lee SD, Kim YS, Shin DH. The Use and Effectiveness of Triple Multiplex System for Coding Region Single Nucleotide Polymorphism in Mitochondrial DNA Typing of Archaeologically Obtained Human Skeletons from Premodern Joseon Tombs of Korea. Biomed Res Int. 2015b; 2015: 850648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh CS, Seo Min, Hong JH, Chai JY, Oh SW, Park JB, et al. Ancient Mitochondrial DNA Analyses of Ascaris Eggs Discovered in Coprolites from Joseon Tomb. Korean J Parasitol. 2015c; 53: 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahila Bar-Gal G, Kim MJ, Klein A, Shin DH, Oh CS, Kim JW, et al. Tracing hepatitis B virus to the 16th century in a Korean mummy. Hepatology 2012; 56: 1671–1680. doi: 10.1002/hep.25852 [DOI] [PubMed] [Google Scholar]
- 31.Shin DH, Oh CS, Lee SJ, Lee E-J, Yim SG, Kim MJ, et al. Ectopic paragonimiasis from 400 year old female mummy of Korea. J Archaeol Sci. 2012; 39: 1103–1110. [Google Scholar]
- 32.Shin DH, Oh CS, Lee HJ, Chai JY, Lee SJ, Hong D-W, et al. Ancient DNA analysis on Clonorchis sinensis eggs remained in samples from medieval Korean mummy. J Archaeol Sci. 2013; 40: 211–216. [Google Scholar]
- 33.Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, et al. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012; 3: 698 doi: 10.1038/ncomms1701 [DOI] [PubMed] [Google Scholar]
- 34.Zink A, Wann LS, Thompson RC, Keller A, Maixner F, Allam AH, et al. Genomic correlates of atherosclerosis in ancient humans. Glob Heart 2014; 9: 203–209. doi: 10.1016/j.gheart.2014.03.2453 [DOI] [PubMed] [Google Scholar]
- 35.Yasuda H, Kamide K, Takiuchi S, Matayoshi T, Hanada H, Kada A, et al. Association of single nucleotide polymorphisms in endothelin family genes with the progression of atherosclerosis in patients with essential hypertension. J Hum Hypertens. 2007; 21: 883–892. doi: 10.1038/sj.jhh.1002234 [DOI] [PubMed] [Google Scholar]
- 36.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008; 53: 357–359. doi: 10.1007/s10038-008-0248-4 [DOI] [PubMed] [Google Scholar]
- 37.Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008; 28: 360–365. doi: 10.1161/ATVBAHA.107.157248 [DOI] [PubMed] [Google Scholar]
- 38.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011; 43: 339–344. doi: 10.1038/ng.782 [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011; 43: 345–349. doi: 10.1038/ng.783 [DOI] [PubMed] [Google Scholar]
- 40.Xie F, Chu X, Wu H, Sun W, Shen M, Yang L, et al. Replication of putative susceptibility loci from genome-wide association studies associated with coronary atherosclerosis in Chinese Han population. PLoS One. 2011; 6: e20833 doi: 10.1371/journal.pone.0020833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo CY, Gu Y, Li L, Jia EZ, Li CJ, Wang LS, et al. Association of SNP rs6903956 on chromosome 6p24.1 with angiographical characteristics of coronary atherosclerosis in a Chinese population. PLoS One 2012; 7: e43732 doi: 10.1371/journal.pone.0043732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Zhang X, He M, Cheng L, Chen Y, Hu FB, et al. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol. 2008; 28: 2085–2089. doi: 10.1161/ATVBAHA.108.176065 [DOI] [PubMed] [Google Scholar]
- 43.Dechamethakun S, Ikeda S, Arai T, Sato N, Sawabe M, Muramatsu M. Associations between the CDKN2A/B, ADTRP and PDGFD polymorphisms and the development of coronary atherosclerosis in Japanese patients. J Atheroscler Thromb. 2014; 21: 680–690. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016; 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Ancient DNA. Nat Rev Genet. 2001; 2: 353–359. doi: 10.1038/35072071 [DOI] [PubMed] [Google Scholar]
- 46.Ricaut FX, Keyser-Tracqui C, Cammaert L, Crubézy E, Ludes B. Genetic analysis and ethnic affinities from two Scytho-Siberian skeletons. Am J Phys Anthropol. 2004; 123: 351–360. doi: 10.1002/ajpa.10323 [DOI] [PubMed] [Google Scholar]
- 47.Ricaut FX, Fedoseeva A, Keyser-Tracqui C, Crubézy E, Ludes B. Ancient DNA analysis of human neolithic remains found in northeastern Siberia. Am J Phys Anthropol. 2005; 126: 458–462. doi: 10.1002/ajpa.20257 [DOI] [PubMed] [Google Scholar]
- 48.Ricaut FX, Kolodesnikov S, Keyser-Tracqui C, Alekseev AN, Crubézy E, Ludes B. Molecular genetic analysis of 400-year-old human remains found in two Yakut burial sites. Am J Phys Anthropol. 2006; 129: 55–63. doi: 10.1002/ajpa.20195 [DOI] [PubMed] [Google Scholar]
- 49.Holland MM, Huffine EF. Molecular analysis of the human mitochondrial DNA control region for forensic identity testing. Curr Protoc Hum Genet. 2001; Chapter 14: Unit 14.7. [DOI] [PubMed] [Google Scholar]
- 50.Schoen FJ, Cotran RS. The Blood Vessels In: Kumar, Cotran, Robbins, editors. Robbin's Basic Pathology. 7th ed. New Delhi: Saunders; 2002. [Google Scholar]
- 51.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006; 3: e442 doi: 10.1371/journal.pmed.0030442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKibben RA, Al Rifai M, Mathews LM, Michos ED. Primary Prevention of Atherosclerotic Cardiovascular Disease in Women. Curr Cardiovasc Risk Rep. 2016; pii: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burns DK, Kumar V. The Heart In: Kumar, Cotran, Robbins, Eds. Robbin's Basic Pathology. 7th edition New Delhi: Saunders, 2002. [Google Scholar]
- 54.Chan L, Boerwinkle E. Gene-environment interactions and gene therapy in atherosclerosis. Cardiol Rev. 1994; 2: 130–137. [Google Scholar]
- 55.Huang EW, Peng LY, Zheng JX, Wang D, Xu QY, Huang L, et al. Common Variants in Promoter of ADTRP Associate with Early-Onset Coronary Artery Disease in a Southern Han Chinese Population. PLoS One 2015; 10: e0137547 doi: 10.1371/journal.pone.0137547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson RC, Allam AH, Lombardi GP, Wann LS, Sutherland ML, Sutherland JD, et al. Atherosclerosis across 4000 years of human history: the Horus study of four ancient populations. Lancet 2013; 381: 1211–1222. doi: 10.1016/S0140-6736(13)60598-X [DOI] [PubMed] [Google Scholar]
- 57.Allam AH, Thompson RC, Wann LS, Miyamoto MI, Nur El-Din Ael-H, El-Maksoud GA, et al. Atherosclerosis in ancient Egyptian mummies: The Horus Study. J Am Coll Cardiol Cardiovasc Imaging 2011; 4:315–327. [DOI] [PubMed] [Google Scholar]
- 58.Habicht ME, Bianucci R, Buckley SA, Fletcher J, Bouwman AS, Öhrström LM, et al. Queen Nefertari, the Royal Spouse of Pharaoh Ramses II: A Multidisciplinary Investigation of the Mummified Remains Found in Her Tomb (QV66). PLoS ONE 2016; 11: e0166571 doi: 10.1371/journal.pone.0166571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmerman MR, Trinkaus E, LeMay M, Aufderheide AC, Reyman TA, Marrocco GR, et al. The paleopathology of an Aleutian mummy. Arch Pathol Lab Med. 1981; 105: 638–641. [PubMed] [Google Scholar]
- 60.Fornaciari G. Renaissance mummies in Italy. Med Secoli. 1999; 11: 85–105. [PubMed] [Google Scholar]
- 61.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007; 316: 1488–1491. doi: 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007; 316: 1491–1493. doi: 10.1126/science.1142842 [DOI] [PubMed] [Google Scholar]
- 63.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447: 661–678. doi: 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ding H, Xu Y, Wang X, Wang Q, Zhang L, Tu Y, et al. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ Cardiovasc Genet. 2009; 2: 338–346. doi: 10.1161/CIRCGENETICS.108.810226 [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Wang XF, Cheng SS, Wan XH, Cao FF, Li L, et al. Three SNPs on chromosome 9p21 confer increased risk of myocardial infarction in Chinese subjects. Atherosclerosis 2009; 207: 26–28. doi: 10.1016/j.atherosclerosis.2009.04.017 [DOI] [PubMed] [Google Scholar]
- 66.Saleheen D, Alexander M, Rasheed A, Wormser D, Soranzo N, Hammond N, et al. Association of the 9p21.3 locus with risk of first-ever myocardial infarction in Pakistanis: case-control study in South Asia and updated meta-analysis of Europeans. Arterioscler Thromb Vasc Biol. 2010; 30: 1467–1473. doi: 10.1161/ATVBAHA.109.197210 [DOI] [PubMed] [Google Scholar]
- 67.Kumar J, Yumnam S, Basu T, Ghosh A, Garg G, Karthikeyan G, et al. Association of polymorphisms in 9p21 region with CAD in North Indian population: replication of SNPs identified through GWAS. Clin Genet. 2011; 79: 588–593. doi: 10.1111/j.1399-0004.2010.01509.x [DOI] [PubMed] [Google Scholar]
- 68.Jing J, Su L, Zeng Y, Tang X, Wei J, Wang L, et al. Variants in 9p21 Predicts Severity of Coronary Artery Disease in a Chinese Han Population. Ann Hum Genet. 2016; 80: 274–281. doi: 10.1111/ahg.12163 [DOI] [PubMed] [Google Scholar]
- 69.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012; 44: 890–894. doi: 10.1038/ng.2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng Q, Yuan Y, Wang S, Sun J, Zhang T, Qi M. Polymorphisms on chromosome 9p21 confer a risk for acute coronary syndrome in a Chinese Han population. Can J Cardiol. 2013; 29: 940–944. doi: 10.1016/j.cjca.2012.11.028 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(A) Consensus sequence of mtDNA control region for Mungyeong mummy. (B) Comparison between Revised Cambridge Reference Sequence (rCRS, GeneBank accession no. NC_012920), consensus sequence and direct sequencing result of mtDNA control region.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.