Abstract
Efficient use of feed resources is a challenge in the pork industry because the largest variability in expenditure is attributed to the cost of fodder. Efficiency of feeding is directly related to feeding behavior. In order to identify genomic regions controlling feeding behavior and eating efficiency traits, 338 Duroc boars were used in this study. The Illumina Porcine SNP60K BeadChip was used for genotyping. Data pertaining to individual daily feed intake (DFI), total daily time spent in feeder (TPD), number of daily visits to feeder (NVD), average duration of each visit (TPV), mean feed intake per visit (FPV), mean feed intake rate (FR), and feed conversion ratio (FCR) were collected for these pigs. Despite the limited sample size, the genome-wide association study was acceptable to detect candidate regions association with feeding behavior and eating efficiency traits in pigs. We detected three genome-wide (P < 1.40E-06) and 11 suggestive (P < 2.79E-05) single nucleotide polymorphism (SNP)-trait associations. Six SNPs were located in genomic regions where quantitative trait loci (QTLs) have previously been reported for feeding behavior and eating efficiency traits in pigs. Five candidate genes (SERPINA3, MYC, LEF1, PITX2, and MAP3K14) with biochemical and physiological roles that were relevant to feeding behavior and eating efficiency were discovered proximal to significant or suggestive markers. Gene ontology analysis indicated that most of the candidate genes were involved in the development of the hypothalamus (GO:0021854, P < 0.0398). Our results provide new insights into the genetic basis of feeding behavior and eating efficiency in pigs. Furthermore, some significant SNPs identified in this study could be incorporated into artificial selection programs for Duroc-related pigs to select for increased feeding efficiency.
Introduction
Pork is a major meat resource for humans, providing over ~37% of all meat in 2012–2014[1]. The increasing demand for pork has prompted breeders around the world to significantly improve swine production. However, the cost of feed is the single largest variable for swine production, ranging from 50% to 85% of the total production cost[2]. The cost of feed can be further compounded by competition between animal agriculture, human food, and biofuel industries resulting in augmentation of the demand for grain and higher grain prices[3]. Improving feed conversion ratio (FCR) and other feeding behavioral traits is important to solve these problems.
With the development of computerized systems that record feed intake and related measurements, extensive research investigations on feeding behavior and eating efficiency have been performed[1, 4, 5]. Several studies have shown a strong correlation between the feeding behavioral traits and eating efficiency traits in livestock. For instance, Do DN et al. showed that the average daily feed intake (DFI) is positively correlated to FCR, and total time spent at feeder per day (TPD) is negatively correlated to mean feed intake rate (FR) [6]. Rauw et al. reported that faster-consuming pigs have higher levels of food intake, improved growth rates, and accumulated more body fat[7]. However, simple correlation studies between the feeding behavioral traits and eating efficiency traits could not provide a direct approach insight into the genetic determinants of control of FCR and other feeding behavioral traits.
In previous decades, quantitative trait loci (QTL) mapping was the method of choice for scientists to understand the genetics of complex traits such as FCR. With the development of genetic linkage studies, thousands of QTLs have been detected for economically important traits in livestock[8]. For example, 55 QTLs for DFI and 179 QTLs for FCR have been identified in different pig chromosomes (http://www.animalgenome.org/cgi-bin/QTLdb/SS/index). However, the biggest challenge for QTL mapping is to locate a large interval of at least 20 centimorgans (cM) in length. With the development of dense genomic markers and the significant reduction in sequencing costs, genome-wide association studies (GWAS) have proven to be a useful and powerful method in addressing this challenge in human[9] and other animal species[10, 11].
Duroc is widely used in pig production based on its excellent growth traits. It is significant for pig practitioners and even human feeding behavior researchers to identify potential genes that are associated with feeding behavior and eating efficiency. Therefore, understanding the genetic determinants controlling FCR and other feeding behavioral traits based on GWAS is crucial to improving Duroc breeding programs and enhancing their feeding efficiency. In pigs, there is an increasing number of association studies on Duroc purebred to detect SNPs associated with polygenetic traits, such as obesity-related traits[12]. Here, we conducted GWAS for feeding behavior and eating efficiency traits to identify the precise locations of QTLs for such important traits in Duroc pigs.
Materials and methods
Ethics statement
The experimental procedures used in this study met the guidelines of the Animal Care and Use Committee of the South China Agricultural University (SCAU) (Guangzhou, People’s Republic of China). The Animal Care and Use Committee of the SCAU (Approval number SCAU#0017) approved all the animal experiments described in this study.
Animals and phenotype recording
Between 2011 and 2014, a total of 338 Duroc boars (average IBS value was 0.733 ± 0.015) were collected from the Guangdong Wen’s Foodstuffs Group Co., Ltd. (Guangdong, China). All 338 boars were approximately fed under uniform feeding conditions for measurements of feed behaviors and eating efficiency traits during the fattening period (approximately 11 weeks) from 30 kg to 100 kg live weight. These pigs were group housed in half-open cement-floor pens (10 animals in each pen, with an average of 2 m2 per pig). Each animal was labeled a unique electric tag on its ear during the testing period. In the present study, both 338 boars had feeding behavior phenotypic data, and 324 had FCR phenotypic data. Both phenotypic data were collected by using the Osborne FIRE Pig Performance Testing System (Kansas, American) of Guangdong Wen’s Foodstuffs Group Co., Ltd. (Guangdong, China)[1]. The time, duration, feed consumption, and weight of each individual were recorded at every visit. Average daily feed intake (DFI) was calculated based on the total amount of recorded total feed intake divided by the number of corresponding feed days. The following feeding behavior and eating efficiency traits were defined and calculated for each boar: average DFI (kg/d), TPD (min), NVD, TPV (= TPD/NVD), FPV (kg), FR (= DFI/TPD, g/min), and FCR[4, 6].
Genotyping and quality control
Genomic DNA was extracted from ear tissues using the traditional method of phenol-chloroform and adjusted to a concentration of 50 ng/μL[13]. DNA quality was assessed by ratios of light absorption (A260/280 and A260/230) and electrophoresis. Genotyping was performed using the Porcine SNP60 Beadchip of Illumina (San Diego, CA, USA)[14], which contains 61,565 SNP markers across the entire genome. A total of 338 samples were genotyped. QC was conducted using Plink v1.07[15]. Briefly, animals with call rates of > 0.95 (MIND) and SNPs with call rates of > 0.99 (GENO), minor allele frequency > 0.01 (MAF), P value>10−6 for the HWE test were included. Moreover, all of the SNPs located on the sex chromosome and unmapped regions were excluded from the analysis. A final set of 35,791 informative SNPs from 338 pigs were used for the subsequent analyses.
GWAS
Genome-wide association analysis is a research method that is generally used to determine the correlation between high-density genetic markers and complex traits. The R package GenABEL was used to perform genome-wide association analysis under a general linear mixed model[16, 17]. The model included a random polygenic effect for which the variance-covariance matrix was proportionate to genome-wide identity-by-state[18]. The following mixed model was used to perform GWAS: Y~μ+Xb+Kw+Sc+Za+e, where Y is the vector of phenotypes; μ is the overall mean; b is the vector of fixed effects including pigsty and year-season effects; w is the vector of live weight of individuals when the measurement is completed considered as covariate; c is the vector of SNP effects; a is the vector of random additive genetic effects with a~N(0, Gσα2), where G is the genomic relationship matrix calculated from the 60K SNP makers in the Duroc population and σα2 is the polygenetic additive variance; K is the regression coefficient of live weight of individuals when the measurement is completed; and e is the vector of residual errors with e~N(0, Iσe2), where I is the identity matrix and σe2 is the residual variance. X, S, and Z are incidence matrices for b, c, and a respectively.
The Bonferroni method was used to determine the genome-wide significance threshold, in which the conventional P-value was divided by the number of tests performed[19]. According to the Bonferroni method, the genome-wide significant (significant) and chromosome-wide significant (suggestive) thresholds were P < 0.05/N and P < 1/N, respectively, where N is the number of SNPs tested in the analyses. In the present study, the significant and suggestive thresholds were 1.40E-06 (0.05/35791) and 2.79E-05 (1/35791), respectively.
Quantile-quantile plots
Because population stratification greatly impacts GWAS reliability, quantile-quantile (Q–Q) plot analysis is considered an effective way to determine the reliability of the GWAS results. In a Q–Q plot, the horizontal axis represents the expected -log10P value, and the vertical axis represents the observed -log10P value. The diagonal line represents y = x, the shaded region shows a 95% confidence interval based on a Beta distribution. An overall deviation above the diagonal identity line is generally suggestive of severe population stratification[20]. Deviations from the diagonal line indicate that either the assumed distribution is incorrect or that the sample contains values arising in some other manner, similar to that generated by true association[21]. The Q–Q plot was constructed using the R software.
Haplotype block analysis
The software Plink v1.07 [http://pngu.mgh.harvard.edu/purcell/plink/] and Haploview [http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview] were utilized for haplotype block analysis. Linkage disequilibrium blocks were defined using Haploview 4.2 based on SNPs with MAF values of > 0.05, Mendel errors of < 2, and P-value in the HWE test of < 10−3. We performed haplotype block analysis, which requires at least two suggestive SNPs in a chromosome[1, 22].
Gene ontology analysis
SNP positions from the Sus scrofa 10.2 genome version were downloaded from www.animalgenome.org/pig/. The Ensembl annotation of the Sus scrofa 10.2 genome version was employed to find genes that were nearest the significant SNPs [http://ensembl.org/Sus_scrofa/Info/Index]. To annotate significant SNP positions to previously mapped QTLs in pigs, all QTL data in pigs were downloaded from http://www.animalgenome.org/cgi-bin/QTLdb/SS/download?file=gbpSS_10.2 (accessed on April 3, 2016)[23]. Gene Ontology analysis was performed on the GO website [http://geneontology.org/][24].
Results
Phenotypes and quality control (QC) of genotypes
A summary of the statistics of the seven traits are presented in Table 1. Prior to GWAS analysis, we assessed the distribution of all phenotypes by using the Shapiro test[25]. All phenotypic data conformed to the Gaussian distribution. After QC-filtering of the genotypic data, 18,792 markers that showed low (< 1%) minor allele frequencies, 489 markers with low (< 99%) call rate, and 840 markers not within Hardy-Weinberg equilibrium (HWE) (P < 10−6) were excluded from the analysis. A total of 6,002 SNPs located on the sex chromosome and unknown chromosomal regions were thus removed. A final set of 35,791 SNPs and 338 pigs was retained for subsequent GWAS analysis. The number of markers on each chromosome and average distances between two markers after QC are presented in S1 Table. The average physical distance between two neighboring SNPs on the same chromosome was approximately 66.8 kb and ranged from 54.7 (SSC14) to 85.8 kb (SSC1).
Table 1. Descriptive statistical analysis of feeding behavior and eating efficiency in a male Duroc population.
| Trait1 | Units | N | Mean | SD | Min | Max |
|---|---|---|---|---|---|---|
| DFI | kg | 338 | 1.985 | 0.278 | 1.463 | 2.617 |
| TPD | min | 338 | 65.469 | 16.478 | 37.596 | 128.807 |
| NVD | count | 338 | 7.805 | 2.963 | 3.096 | 17.06 |
| TPV | min | 338 | 9.54 | 3.798 | 4.077 | 20.618 |
| FPV | kg | 338 | 0.295 | 0.117 | 0.107 | 0.712 |
| FR | g/min | 338 | 31.77 | 7.521 | 14.042 | 48.111 |
| FCR | kg/kg | 324 | 2.06 | 0.244 | 1.7 | 2.7 |
1DFI: Total daily feed intake, FPV: Mean feed intake per visit, FR: Mean feed intake rate, NVD: Number of visits to the feeder per day, TPD: Total time spent at feeder per day, TPV: Time spent to eat per visit, FCR: Feed conversion ratio.
Mean, standard deviation (SD), minimum (min) and maximum (max) values are presented for all of the phenotypes included in the association study (N).
Significant SNPs and haplotype block analysis
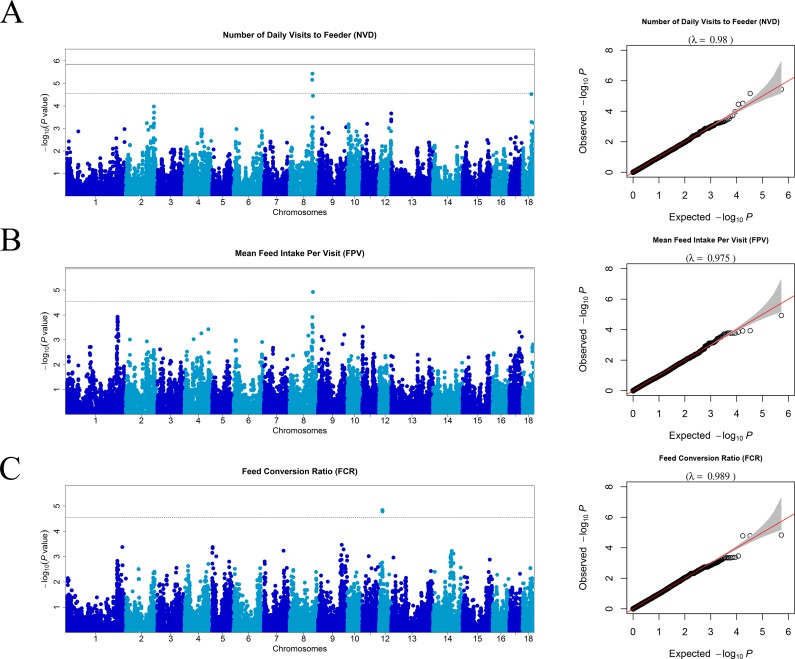
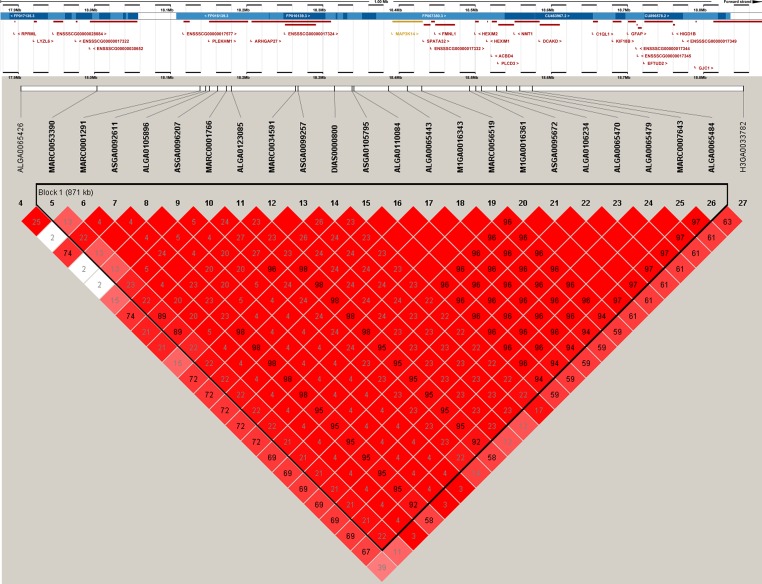
Manhattan plots of GWAS of all traits after QC and the Q–Q plots are shown in Fig 1 and S1 Fig. The average genomic inflation factors (λ) of the GWAS for all feeding behavior and feed conversion ratio traits were 0.991, ranging from 0.973 (average duration of each visit, TPV) to 1.03 (DFI), which suggests that there was little or no evidence of residual population structure effects on test statistic inflation[26]. The tag (significant and suggestive) SNPs detected by the associated test for the seven traits are shown in Table 2. In total, three significant and 11 suggestive SNPs were identified. The three significant SNPs were associated with DFI, whereas no suggestive SNP was associated with FR and TPD. The number of suggestive SNPs associated with DFI, FCR, NVD, TPV, and mean feed intake per visit (FPV) was 4, 3, 2, 1, and 1, respectively. The chromosomes and exact positions based on Sus scrofa Genebuild 10.2 as well as genes neighboring the tag SNPs are listed in Table 2. Two of 14 SNPs were located within intronic regions of known genes, and one haplotype block was detected in genomic regions affecting FCR on SSC12 (Fig 2). Approximately 22 SNPs of the haplotype block were located in SSC12 and ranged in size from 17.9 Mb to 18.8 Mb, three of these SNPs significantly affected FCR. Furthermore, 25 genes were identified within the haplotype block (Fig 2).
Fig 1. Manhattan plots of genome-wide association studies for eating efficiency and feeding behavior in male Duroc pigs.
The inserted quantile–quantile (Q–Q) plots in the right show the observed versus expected log p-values. In the Manhattan plots, negative log10 P values of the quantified SNPs were plotted against their genomic positions. Different colors indicate various chromosomes. The solid and dashed lines indicate the 5% genome-wide and chromosome-wide Bonferroni-corrected thresholds, respectively. On the vertical axis, Manhattan plot and Q-Q plot for the number of visits to the feeder per day (NVD), FPV, and feed conversion ratio (FCR), respectively.
Table 2. Tag SNPs and closest genes for feeding behavior and eating efficiency traits.
| Trait1 | SNP ID | SSC2 | Location (bp)3 | Adjusted P value4 | Nearest gene | Gene location5 | Distance/bp6 | Homo sapiens homologs |
|---|---|---|---|---|---|---|---|---|
| DFI | ASGA0036538 | 7 | 123,104,663 | 6.10E-08 | SERPINA3 | 7:123,094,888–123,101,407 | -3256 | SERPINA3 |
| DRGA0006936 | 6 | 132,729,420 | 3.90E-07 | LRRC7 | 6:132,544,632–132,612,363 | -117057 | LRRC7 | |
| DRGA0016772 | 17 | 55,848,150 | 9.00E-07 | ENSSSCG00000007456 | 17:55,754,008–55,846,536 | -1614 | - | |
| ASGA0025032 | 5 | 21,273,976 | 1.80E-06 | ENSSSCG00000021160 | 5:21,279,720–21,280,679 | +5744 | - | |
| ALGA0037698 | 6 | 152,852,810 | 4.00E-06 | PIK3R3 | 6:152,798,324–152,914,703 | Within | PIK3R3 | |
| ASGA0018273 | 4 | 11,639,918 | 1.60E-05 | ENSSSCG00000005961 | 4:11,122,206–11,124,073 | -515845 | - | |
| ASGA0018324 | 4 | 12,174,119 | 2.44E-05 | MYC | 4:12,778,444–12,783,481 | 604325 | MYC | |
| FPV | ALGA0049273 | 8 | 122,208,906 | 1.20E-05 | LEF1 | 8:122,085,975–122,204,345 | -4561 | LEF1 |
| NVD | ASGA0087256 | 8 | 119,705,102 | 3.65E-06 | PITX2 | 8:119,920,838–119,926,111 | +215736 | PITX2 |
| MARC0008579 | 8 | 118,421,256 | 6.87E-06 | AP1AR | 8:118,369,021–118,401,177 | -20079 | AP1AR | |
| TPV | ALGA0008796 | 1 | 268,986,440 | 1.92E-05 | ENSSSCG00000029421 | 1:268,964,060–268,973,843 | -22380 | - |
| FCR | MARC0053390 | 12 | 17,941,131 | 1.44E-05 | LRRC37B | 12:17,946,401–17,959,278 | +5270 | LRRC37B |
| MARC0034591 | 12 | 18,338,026 | 1.64E-05 | MAP3K14 | 12:18,391,607–18,431,381 | +53581 | MAP3K14 | |
| ASGA0099257 | 12 | 18,342,922 | 1.64E-05 | MAP3K14 | 12:18,391,607–18,431,381 | Within | MAP3K14 |
1DFI: Total daily feed intake, FPV: Mean feed intake per visit, NVD: Number of visits to the feeder per day, TPV: Time spent to eat per visit, FCR: Feed conversion ratio.
2Sus scrofa chromosome
3SNP positions in Ensembl.
4Genome-wide significant associations are underlined.
5Gene location in Ensembl.
6+/-: The SNP located in the upstream/downstream of the nearest gene; NA: not assigned.
Fig 2. The linkage disequilibrium (LD) block and candidate genes in the significantly associated region on SSC12.
LD blocks are marked with triangles. Values in boxes are LD (r2) between SNP pairs and the boxes are colored according to the standard Haploview color scheme: LOD >2 and D’ = 1, red; LOD >2 and D’<1, shades of pink/red; LOD <2 and D’ = 1, blue; LOD <2 and D’<1, white (LOD is the log of the likelihood odds ratio, a measure of confidence in the value of D’). Annotated genes in the chromosomal region retrieved from the Ensemble genome browser (www.ensembl.org/Sus_scrofa/Info/Index).
Comparison with previously mapped QTL in pigs
A total of four SNPs associated with DFI were identified within the genomic region where QTLs for feed intake traits and/or feed conversion ratio were previously been mapped in pigs (Table 3). One SNP on SSC1 associated with TPV and one locus on SSC6 that was associated with DFI was located on previously reported QTL regions for average daily gain in different pig populations. Moreover, eight SNPs were located in the genomic region where QTLs were previously detected by GWAS or linkage maps for backfat and obesity-related traits in pigs.
Table 3. Comparative mapping of tag SNPs with previous QTLs reported in the pig QTL database (as of April 3, 2016) and previous GWAS results.
| Traits1 | SNP | SSC2 | SNP position (bp)3 | Starting QTL position (bp)4 | Ending QTL position (bp)5 | QTL_ID6 | Corresponded trait in the QTL database |
|---|---|---|---|---|---|---|---|
| DFI | ASGA0036538 | 7 | 123,104,663 | 121,429,704 | 124,338,264 | 65,101 | Fat weight (total) |
| DRGA0006936 | 6 | 132,729,420 | 94,382,869 | 146,365,886 | 2,879 | Daily feed intake | |
| DRGA0016772 | 17 | 55,848,150 | 45,880,990 | 67,855,516 | 16,842 | Backfat on the last rib | |
| ASGA0025032 | 5 | 21,273,976 | 3,688,083 | 34,660,429 | 979 | Feed intake | |
| ALGA0037698 | 6 | 152,852,810 | 149,326,918 | 153,593,360 | 19,382 | Average daily gain | |
| ASGA0018273 | 4 | 11,639,918 | 7,237,639 | 12,618,993 | 5,162 | Feed conversion ratio | |
| ASGA0018324 | 4 | 12,174,119 | 7,237,639 | 12,618,993 | 5,162 | Feed conversion ratio | |
| FPV | ALGA0049273 | 8 | 122,208,906 | 108,610,930 | 139,007,531 | 17,782 | Backfat on the last rib |
| NVD | ASGA0087256 | 8 | 119,705,102 | 108,610,930 | 139,007,531 | 17,782 | Backfat on last rib |
| MARC0008579 | 8 | 118,421,256 | 108,610,930 | 139,007,531 | 17,782 | Backfat on last rib | |
| TPV | ALGA0008796 | 1 | 268,986,440 | 87,993,461 | 279,498,596 | 22,269 | Average daily gain |
| FCR | MARC0053390 | 12 | 17,941,131 | 17,226,344 | 23,672,762 | 3,967 | Backfat between the last 3rd and 4th lumbar |
| MARC0034591 | 12 | 18,338,026 | 17,226,344 | 23,672,762 | 3,967 | Backfat between the last 3rd and 4th lumbar | |
| ASGA0099257 | 12 | 18,342,922 | 17,226,344 | 23,672,762 | 3,967 | Backfat between the last 3rd and 4th lumbar |
1DFI: Total daily feed intake, FPV: Mean feed intake per visit, NVD: Number of visits to the feeder per day, TPV: Time spent to eat per visit, FCR: Feed conversion ratio.
2Sus scrofa chromosome
3SNP positions in Ensembl
4Starting position of the mapped QTL in the QTL database
5Ending position of the mapped QTL in the QTL database
6Identity of QTL in the pig QTL database or published literature
Candidate genes at significant or suggestive level
The aim of the present study was to identify and characterize genes that were related to novel feeding behavior and eating efficiency. To obtain more credible results, we attempted to reduce the number of potential genes based on biochemical and physiological roles that were relevant to feeding behavior and eating efficiency traits. Ultimately, a total of five candidate genes were obtained. Of these, serpin family A member 3 (SERPINA3), myc proto-oncogene protein (MYC) were correlated to DFI; lymphoid enhancer binding factor 1 (LEF1), which is a potential functional candidate gene, was associated with FPV; paired-like homeodomain 2 (PITX2), a potential functional candidate gene, was associated with NVD; and mitogen-activated protein kinase kinase kinase 14 (MAP3K14) was correlated with FCR. We then conducted gene ontology analysis of the five candidate genes, which indicated that most of the genes were involved in the development of the hypothalamus (GO:0021854, P< 0.0398).
Discussion
Comparison of QTLs identified in this study with those in previous studies
GWAS is a powerful tool for the genetic analysis of important traits of domestic animals. Recently, some authors have reported the results of GWAS for feeding behavior and eating efficiency in pig. For example, Do DN et al. identified 16 significant SNPs and 76 suggestive SNPs that were associated with feeding behavior traits in the Duroc population[4]. Of these SNPs, 36 SNPs were located in genome regions where QTLs for feeding behavior and/or feed intake traits were previously been reported in pigs[4]. In their study, a moderate threshold criterion was employed, in which loci with P < 5E-05 were considered as moderately genome-wide significant and those with P < 5E-04 were considered to be suggestively genome-wide significant. They finally found a large number of SNPs associated with behavior and/or feed intake traits, but this method also raised the possibility to detect false positive results. In our study, the Bonferroni correction, which is a stringent correction method, was employed to reduce the occurrence of false positive results. Compared to the results of previous studies, we detected a markedly lower number of tag SNPs; however, we believe that these SNPs provide promising markers to improve feed efficiency related traits in the Duroc population that we tested. Moreover, Guo et al. also identified six QTLs that were associated with eating efficiency and feeding behavior at suggestive and significance levels, and two QTLs were associated with more than one trait in a White Duroc × Erhualian F2 resource population[27]. Although progress has been made in identifying QTLs by GWAS, the majority of associations detected from GWAS are still for common variants. QTL caused by low-frequency or rare variants have not been efficiently identified in previous pig GWAS, especially in those using small sample size.
The present study detected a SNP (ALGA0008796) associated with TPV for average daily gain in Yorkshire pigs in SSC1[28]. Two SNPs (ASGA0018273 and ASGA0018324) associated with DFI for feed conversion ratio were identified in SSC4 of Large White×·Landrace × Leicoma pigs[29], and a SNP (ASGA0025032) associated with DFI for feed intake was localized on SSC5 of Meishan × Pietrain pigs[30]. Two SNPs (DRGA0006936 and ALGA0037698) associated with DFI were also identified in SSC6, one was situated in a region linked to DFI in Pietrain pigs[31], whereas the other was in a region related to average daily gain[32]. Some other QTLs and SNPs associated with feeding behavior were not found in regions that were previously reported to be related to feeding behavior, nevertheless, we compared our GWAS results for backfat and obesity-related traits with those of previous studies. We found that three SNPs (MARC0053390, MARC0034591 and ASGA0099257) associated with FCR on SSC12 were in previously mapped QTLs that spanned 40.2–64.7 (cM) for backfat between the last 3rd and 4th lumbar in a commercial four-way cross[33]. Moreover, two SNPs (ASGA0087256 and MARC0008579) associated with NVD and a SNP (ALGA0049273) associated with FPV on SSC8 were detected in regions for backfat on the last rib in an Iberian × Meishan F2 sow family[34]. The close proximity of the three SNPs suggests that the same gene also affects the NVD and FPV traits of backfat on the last rib. In general, six SNPs were located in genome regions where QTLs for feeding behavior and eating efficiency were previously reported in pigs.
Comparative mapping may facilitate in the validation of our results, as well as refine QTL regions and target candidate genes for complex traits such as feeding behavior. In addition, by comparing our results with those of previous QTL studies, we have determined that a large proportion of feeding behavior and eating efficiency traits located that have been previously reported. These observations are suggestive of a genetic correlation between feeding behavior and eating efficiency traits, as well as the occurrence of regional pleiotropic effects on feeding traits.
Furthermore, compared to conventional breeding, the marker-assisted selection (MAS) can speed up the breeding programs[35]. The MAS has been applied to improve reproduction rate, feed intake, growth rate and meat quality in commercial lines of pigs[36]. In this study, significant associations between polymorphisms and feeding behavior traits were revealed in the Duroc sire population. These associated variants provide novel molecular markers for the MAS to facilitate the genetic improvement of feeding behavior and eating efficiency traits in the Duroc sire population.
Potential candidate genes
Potential candidate genes for average daily feed intake
Average daily feed intake is an important feeding behavior trait, and therefore is a major ethological concern of animal nutrition workers. The most significant locus, ASGA0036538, was closest to the SERPINA3 gene (Table 2). The protein encoded by this gene is a plasma protease inhibitor and a member of the serine protease inhibitor class. Yang et al. reported that SERPINA3 promotes endometrial cancer cell growth by regulating G2/M cell cycle checkpoints and apoptosis in human[37]. Graff et al. also identified a SNP within the SERPINA3 could increase body weight significantly in human[38]. Moreover, Grubbs et al. reported that SERPINA3 may play direct and important biological roles in the pathways that control residual feed intake (RFI) in young pigs[39]. Therefore, SERPINA3 on SSC7 could be a candidate gene for DFI. The MYC gene was situated proximal to the significant SNP ASGA0018324, which is associated with DFI (Table 2). Palomero et al. determined that the MYC gene is involved in a feed forward loop transcriptional network that promotes leukemic cell growth[40]. Additionally, Malynn et al. found that MYC gene knockout mice tend to show lower weight and slower growth rate[41]. Therefore, the MYC gene on SSC4 could be a candidate gene for DFI.
The marker ALGA0037698 on SSC6, which is located on fourth intron of the PIK3R3 gene, was associated with DFI (Table 2). Previous studies have shown that the gene PIK3R3 is involved in cancer development, such as gastric cancer[42], ewing Sarcoma[43] and metastatic colorectal cancer[44]. However, no report to date has described a link between DFI and PIK3R3 in any species. In addition, there were three genes (ENSSSCG00000005961, ENSSSCG00000021160 and ENSSSCG00000007456) near the significant SNPs for DFI with unknown function. These three genes have not been studied to date and thus its function in pig has not been established. Moreover, the pig genome has not been completely annotated; therefore, additional research studies to better elucidate the association between these genes and DFI is warranted.
Potential candidate genes for FPV to the feeder
A single SNP with suggestive thresholds was associated with FPV. The lymphoid enhancer binding factor 1 (LEF1) was localized proximal to the SNP ALGA0049273 (Table 2). The protein encoded by this gene can bind to a functionally important site in the T-cell receptor-alpha enhancer, thereby conferring maximal enhancer activity. The biological functions of this gene include positive regulation of cell growth and cell proliferation in humans[45] and mice[46]. We thus inferred that the LEF1 gene increases the production of energy based on body requirements by changing its feeding behavior. Therefore, LEF1 on SSC8 could be a candidate gene for FPV.
Potential candidate genes for the number of visits to the feeder per day
The adaptor-related protein complex 1-associated regulatory protein (AP1AR) was located proximal to the suggestive SNP ASGA0087256, which is associated with NVD (Table 2). This protein is essential for adaptor protein complex 1 (AP-1)-dependent transport between the trans-Golgi network and endosomes. Diseases associated with AP1AR include gliosarcoma. No functional characterization of the gene in pigs has been conducted to date. The PITX2 gene has been localized proximal to the suggestive SNP ASGA0087256, which is associated with NVD (Table 2). The encoded protein acts as a transcription factor and regulates the expression of procollagen lysyl hydroxylase, thereby influencing terminal differentiation of somatotrophs. Kappeler et al. found that regulating somatotroph function can change the food intake behavior of rats[47]. Senescence is related to a dysfunction of the somatotroph axis. Veyrat-Durebex et al. observed that aging Lou rats exhibit a decreased capacity to adjust feeding behavior to metabolic demands[48]. We inferred that the PITX2 gene on SSC8 participates in regulating somatotroph function and could be thus a candidate gene for NVD.
Potential candidate genes for feed conversion ratio
The present study observed that the region on SSC12 showed the strongest association with FCR. The most significant SNP MARC0053390 association with FCR was located in an 871-kb LD block that comprised 25 genes (Fig 2). The leucine-rich repeat containing a 37B gene (LRRC37B) was located proximal to the top SNP, and this gene was related to human height[49]. The remaining two suggestive SNPs were also located within the 871-kb LD block, and one of these was closest to the MAP3K14 gene, whereas the other marker, MARC0059507, was located on the MAP3K14 gene and was associated with FCR (Table 2). This gene encodes mitogen-activated protein kinase kinase kinase 14, which is a serine/threonine protein-kinase. In mice, D-serine suppresses the intake of high-preference food[50]. Moreover, MAP3K14 gene knockout mice showed a reduction in the weight of the mammary fat pad. In human, the MAP3K14 gene is associated with multiple sclerosis[51], which involves motor and sensory dysfunction. Therefore, MAP3K14 on SSC12 could be a candidate gene for FCR. This is the first report that associates the LD block on SSC12 with FCR.
Potential candidate genes for other feeding traits and gene ontology analysis
The ENSSSCG00000029421 gene was located proximal to suggestive SNP ALGA0008796, which is associated with TPV. ENSSSCG00000029421 is a Sus scrofa gene, and no homologs have been identified in Homo sapiens. To the best of our knowledge, no studies on this gene have been conducted to date. Based on TPD and FR, no SNP has reached the suggestive threshold, and thus these two traits were not further investigated in the present study.
Gene ontology analysis of the five candidate genes indicated that most of the genes were involved in the development of the hypothalamus. Bouret previously reported that the hypothalamus apparently plays an essential role in controlling appetite[52]. Moreover, Yoshimatsu et al. showed that hypothalamic histamine neurons play an important role in the central regulation of feeding behavior in rats, which is mainly controlled by leptin[53]. Likewise, Peng et al. showed that the BMPR1A gene regulates the development of hypothalamic circuits that are critical to the feeding behavior of mice[54]. These results suggest that the hypothalamus serves as the linkage between the five candidate genes and feeding behavior and eating efficiency traits.
One limitation of the current study is the considerable number of false negative genetic associations. Increasing the size of the study population may potentially prevent the generation of false-positive results. Fine-mapping and the identification of causal variants should also be based on the premise that we increase the sample size. All of these studies may serve as a foundation for better understanding the genetic mechanism underlying feeding behavior and eating efficiency.
Conclusions
The present study provides a list of SNPs that are associated with feeding behavior and eating efficiency traits in pigs, and also offers valuable information on the genetic architecture and candidate genes for these traits. Fourteen significant or suggestive SNPs were detected in the Duroc sire population. Of these, two SNPs for NVD and one SNP for FPV were determined to be in close proximity on SSC8, thereby indicating a pleiotropic effect. Five candidate genes based on biochemical and physiological roles that were relevant to feeding behavior and eating efficiency were discovered closest to the significant or suggestive markers. Gene ontology analysis indicated that most of the genes were involved in hypothalamus development. The identification of several genomic regions and putative positional genes that were associated with feeding behavior and eating efficiency in the present study may contribute to marker-assisted selection in pig breeding.
Supporting information
The inserted quantile–quantile (Q–Q) plots in the right show the observed versus expected log p-values. In the Manhattan plots, negative log10 P values of the quantified SNPs were plotted against their genomic positions. Different colors indicate various chromosomes. The solid and dashed lines indicate the 5% genome-wide and chromosome-wide Bonferroni-corrected thresholds, respectively. On the vertical axis, Manhattan plot and Q-Q plot for total daily feed intake (DFI), total daily time spent at feeder per day (TPD), Time spent to eat per visit (TPV) and mean feed intake rate (FR), respectively.
(TIF)
(DOCX)
Acknowledgments
We would like to thank the Natural Science Foundation of China (31601912), the Natural Science Foundation of Guangdong Province (2016A030310447), and the Science and Technology Planning Project of Guangdong Province (2015B020231010) supported this study. We would also like to thank the Wens Foodstuffs Group Co., Ltd. for providing all phenotypic data and ear tissue samples.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors appreciate the financial support from the Natural Science Foundation of China (31601912), the Natural Science Foundation of Guangdong Province (2016A030310447), and the Applied Science and Technology Research and Development of Special Funds of Guangdong Province (2015B020231010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang K, Liu D, Hernandez-Sanchez J, Chen J, Liu C, Wu Z, et al. Genome Wide Association Analysis Reveals New Production Trait Genes in a Male Duroc Population. PLoS One. 2015;10(9):e0139207 doi: 10.1371/journal.pone.0139207 ; PubMed Central PMCID: PMCPMC4587933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young JM, Cai W, Dekkers JC. Effect of selection for residual feed intake on feeding behavior and daily feed intake patterns in Yorkshire swine. J Anim Sci. 2011;89(3):639–47. doi: 10.2527/jas.2010-2892 . [DOI] [PubMed] [Google Scholar]
- 3.Swick RA, Beveridge M, Phillips M, editors. Scientific Opportunities to Overcome the Challenge of Increasing Demand for Food and Feed. Program & Abstracts of the CGIAR Science Forum 2011: The Agriculture——Environment Nexus; 2011.
- 4.Do DN, Strathe AB, Ostersen T, Jensen J, Mark T, Kadarmideen HN. Genome-wide association study reveals genetic architecture of eating behavior in pigs and its implications for humans obesity by comparative mapping. PLoS One. 2013;8(8):e71509 doi: 10.1371/journal.pone.0071509 ; PubMed Central PMCID: PMCPMC3747221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan CA, Emmans GC, Tolkamp BJ, Kyriazakis I. Analysis of the feeding behavior of pigs using different models. Physiology & behavior. 2000;68(3):395–403. [DOI] [PubMed] [Google Scholar]
- 6.Do DN, Strathe AB, Jensen J, Mark T, Kadarmideen HN. Genetic parameters for different measures of feed efficiency and related traits in boars of three pig breeds. J Anim Sci. 2013;91(9):4069–79. doi: 10.2527/jas.2012-6197 . [DOI] [PubMed] [Google Scholar]
- 7.Rauw WM, Soler J, Tibau J, Reixach J, Gomez Raya L. Feeding time and feeding rate and its relationship with feed intake, feed efficiency, growth rate, and rate of fat deposition in growing Duroc barrows. J Anim Sci. 2006;84(12):3404–9. doi: 10.2527/jas.2006-209 . [DOI] [PubMed] [Google Scholar]
- 8.Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41(Database issue):D871–9. doi: 10.1093/nar/gks1150 ; PubMed Central PMCID: PMCPMC3531174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–69. doi: 10.1038/nrg2344 . [DOI] [PubMed] [Google Scholar]
- 10.Andersson L. Genome-wide association analysis in domestic animals: a powerful approach for genetic dissection of trait loci. Genetica. 2009;136(2):341–9. doi: 10.1007/s10709-008-9312-4 . [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Yang J, Zhou L, Ren J, Liu X, Zhang H, et al. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PLoS Genet. 2014;10(10):e1004710 doi: 10.1371/journal.pgen.1004710 ; PubMed Central PMCID: PMCPMC4207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puig-Oliveras A, Revilla M, Castello A, Fernandez AI, Folch JM, Ballester M. Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci Rep. 2016;6:31803 doi: 10.1038/srep31803 ; PubMed Central PMCID: PMCPMC4989154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan YY, Ma JW, Yuan F, Huang LB, Yang KX, Xie JP, et al. Genome-wide identification of quantitative trait loci for pork temperature, pH decline, and glycolytic potential in a large-scale White Duroc x Chinese Erhualian resource population. J Anim Sci. 2009;87(1):9–16. doi: 10.2527/jas.2008-1128 . [DOI] [PubMed] [Google Scholar]
- 14.Ramos AM, Crooijmans RPMA, Affara NA, Amaral AJ, Archibald AL, Beever JE, et al. Design of a High Density SNP Genotyping Assay in the Pig Using SNPs Identified and Characterized by Next Generation Sequencing Technology. Plos One. 2009;4(8). ARTN e6524 doi: 10.1371/journal.pone.0006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–6. doi: 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- 17.Yu JM, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–8. doi: 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- 18.Hayes BJ, Goddard ME. Technical note: Prediction of breeding values using marker-derived relationship matrices. Journal of Animal Science. 2008;86(9):2089–92. doi: 10.2527/jas.2007-0733 [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Cui J, Chazaro I, Cupples LA, Demissie S. Power and type I error rate of false discovery rate approaches in genome-wide association studies. Bmc Genet. 2005;6 Suppl 1:S134 doi: 10.1186/1471-2156-6-S1-S134 ; PubMed Central PMCID: PMCPMC1866802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong X, Liu X, Zhou L, Yang J, Yang B, Ma H, et al. Genome-wide association analysis reveals genetic loci and candidate genes for meat quality traits in Chinese Laiwu pigs. Mamm Genome. 2015;26(3–4):181–90. doi: 10.1007/s00335-015-9558-y . [DOI] [PubMed] [Google Scholar]
- 21.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 23.Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Research. 2013;41(D1):D871–D9. doi: 10.1093/nar/gks1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556 ; PubMed Central PMCID: PMCPMC3037419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Theune J. Comparison of power for the D'Agostino and the Wilk‐Shapiro test of normality for small and moderate samples. Statistica Neerlandica. 1973;27(4):163–8. [Google Scholar]
- 26.Pearson TA, Manolio TA. How to interpret a genome-wide association study. Jama-J Am Med Assoc. 2008;299(11):1335–44. doi: 10.1001/jama.299.11.1335 [DOI] [PubMed] [Google Scholar]
- 27.Guo YM, Zhang ZY, Ma JW, Ai HS, Ren J, Huang LS. A genomewide association study of feed efficiency and feeding behaviors at two fattening stages in a White Duroc x Erhualian F-2 population. Journal of Animal Science. 2015;93(4):1481–9. doi: 10.2527/jas.2014-8655 [DOI] [PubMed] [Google Scholar]
- 28.Onteru SK, Gorbach DM, Young JM, Garrick DJ, Dekkers JC, Rothschild MF. Whole Genome Association Studies of Residual Feed Intake and Related Traits in the Pig. PLoS One. 2013;8(6):e61756 doi: 10.1371/journal.pone.0061756 ; PubMed Central PMCID: PMCPMC3694077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duthie C, Simm G, Doeschl-Wilson A, Kalm E, Knap PW, Roehe R. Quantitative trait loci for chemical body composition traits in pigs and their positional associations with body tissues, growth and feed intake. Anim Genet. 2008;39(2):130–40. doi: 10.1111/j.1365-2052.2007.01689.x [DOI] [PubMed] [Google Scholar]
- 30.Lee SS, Chen Y, Moran C, Stratil A, Reiner G, Bartenschlager H, et al. Linkage and QTL mapping for Sus scrofa chromosome 5. J Anim Breed Genet. 2003;120:38–44. doi: 10.1046/j.0931-2668.2003.00422.x [Google Scholar]
- 31.Mohrmann M, Roehe R, Knap PW, Looft H, Plastow GS, Kalm E. Quantitative trait loci associated with AutoFOM grading characteristics, carcass cuts and chemical body composition during growth of Sus scrofa. Anim Genet. 2006;37(5):435–43. doi: 10.1111/j.1365-2052.2006.01492.x [DOI] [PubMed] [Google Scholar]
- 32.Li XP, Kim SW, Do KT, Ha YK, Lee YM, Yoon SH, et al. Analyses of porcine public SNPs in coding-gene regions by re-sequencing and phenotypic association studies. Mol Biol Rep. 2011;38(6):3805–20. doi: 10.1007/s11033-010-0496-1 [DOI] [PubMed] [Google Scholar]
- 33.Harmegnies N, Davin F, De Smet S, Buys N, Georges M, Coppieters W. Results of a whole-genome quantitative trait locus scan for growth, carcass composition and meat quality in a porcine four-way cross. Anim Genet. 2006;37(6):543–53. doi: 10.1111/j.1365-2052.2006.01523.x [DOI] [PubMed] [Google Scholar]
- 34.Tomas A, Ramirez O, Casellas J, Munoz G, Sanchez A, Barragan C, et al. Quantitative trait loci for fatness at growing and reproductive stages in Iberian x Meishan F-2 sows. Anim Genet. 2011;42(5):548–51. doi: 10.1111/j.1365-2052.2010.02169.x [DOI] [PubMed] [Google Scholar]
- 35.Nguyen MT, Barnes AC, Mather PB, Li YT, Lyons RE. Single nucleotide polymorphisms in the actin and crustacean hyperglycemic hormone genes and their correlation with individual growth performance in giant freshwater prawn Macrobrachium rosenbergii. Aquaculture. 2010;301(1–4):7–15. [Google Scholar]
- 36.Goddard ME, Hayes BJ. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet. 2009;10(6):381–91. doi: 10.1038/nrg2575 [DOI] [PubMed] [Google Scholar]
- 37.Yang GD, Yang XM, Lu H, Ren Y, Ma MZ, Zhu LY, et al. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Patho. 2014;7(4):1348–58. [PMC free article] [PubMed] [Google Scholar]
- 38.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, et al. Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22(17):3597–607. doi: 10.1093/hmg/ddt205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grubbs JK, Dekkers JC, Huff-Lonergan E, Tuggle CK, Lonergan SM. Identification of potential serum biomarkers to predict feed efficiency in young pigs. J Anim Sci. 2016;94(4):1482–92. doi: 10.2527/jas.2015-9692 . [DOI] [PubMed] [Google Scholar]
- 40.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48):18261–6. doi: 10.1073/pnas.0606108103 ; PubMed Central PMCID: PMCPMC1838740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malynn BA, de Alboran IM, O'Hagan RC, Bronson R, Davidson L, DePinho RA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes & development. 2000;14(11):1390–9. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y, Yap CS, et al. Genetic and bioinformatic analyses of the expression and function of PI3K regulatory subunit PIK3R3 in an Asian patient gastric cancer library. BMC Med Genomics. 2012;5:34 doi: 10.1186/1755-8794-5-34 ; PubMed Central PMCID: PMCPMC3479415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niemeyer BF, Parrish JK, Spoelstra NS, Joyal T, Richer JK, Jedlicka P. Variable expression of PIK3R3 and PTEN in Ewing Sarcoma impacts oncogenic phenotypes. PLoS One. 2015;10(1):e0116895 doi: 10.1371/journal.pone.0116895 ; PubMed Central PMCID: PMCPMC4300218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Yang X, Li C, Cao X, Luo X, Hu J. PIK3R3 induces epithelial-to-mesenchymal transition and promotes metastasis in colorectal cancer. Mol Cancer Ther. 2014;13(7):1837–47. doi: 10.1158/1535-7163.MCT-14-0049 . [DOI] [PubMed] [Google Scholar]
- 45.Reya T, O'Riordan M, Okamura R, Devaney E, Willert K, Nusse R, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13(1):15–24. [DOI] [PubMed] [Google Scholar]
- 46.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8(1):11–20. [DOI] [PubMed] [Google Scholar]
- 47.Kappeler L, Zizzari P, Grouselle D, Epelbaum J, Bluet-Pajot MT. Plasma and hypothalamic peptide-hormone levels regulating somatotroph function and energy balance in fed and fasted states: A comparative study in four strains of rats. J Neuroendocrinol. 2004;16(12):980–8. doi: 10.1111/j.1365-2826.2004.01259.x [DOI] [PubMed] [Google Scholar]
- 48.Veyrat-Durebex C, Gaudreau P, Boghossian S, Alliot J. Effects of peripheral and central administration of GHRH on feeding in aging LOU rats. Peptides. 2001;22(12):2119–26. . [DOI] [PubMed] [Google Scholar]
- 49.Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. doi: 10.1038/nature09410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki T, Kinoshita Y, Matsui S, Kakuta S, Yokota-Hashimoto H, Kinoshita K, et al. N-methyl-D-aspartate receptor coagonist D-serine suppresses intake of high-preference food. Am J Physiol-Reg I. 2015;309(5):R561–R75. doi: 10.1152/ajpregu.00083.2015 [DOI] [PubMed] [Google Scholar]
- 51.Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–9. doi: 10.1038/nature10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouret SG. Role of early hormonal and nutritional experiences in shaping feeding behavior and hypothalamic development. J Nutr. 2010;140(3):653–7. doi: 10.3945/jn.109.112433 . [DOI] [PubMed] [Google Scholar]
- 53.Yoshimatsu H, Itateyama E, Kondou S, Tajima D, Himeno K, Hidaka S, et al. Hypothalamic neuronal histamine as a target of leptin in feeding behavior. Diabetes. 1999;48(12):2286–91. [DOI] [PubMed] [Google Scholar]
- 54.Peng CY, Mukhopadhyay A, Jarrett JC, Yoshikawa K, Kessler JA. BMP receptor 1A regulates development of hypothalamic circuits critical for feeding behavior. J Neurosci. 2012;32(48):17211–24. doi: 10.1523/JNEUROSCI.2484-12.2012 ; PubMed Central PMCID: PMCPMC3589760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The inserted quantile–quantile (Q–Q) plots in the right show the observed versus expected log p-values. In the Manhattan plots, negative log10 P values of the quantified SNPs were plotted against their genomic positions. Different colors indicate various chromosomes. The solid and dashed lines indicate the 5% genome-wide and chromosome-wide Bonferroni-corrected thresholds, respectively. On the vertical axis, Manhattan plot and Q-Q plot for total daily feed intake (DFI), total daily time spent at feeder per day (TPD), Time spent to eat per visit (TPV) and mean feed intake rate (FR), respectively.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.