Abstract
Once considered as largely independent functional units, G protein-coupled receptors (GPCRs) are now recognized as having a far more diverse molecular architecture. Receptor activity-modifying proteins (RAMPs) provide a major example of proteins that interact with GPCRs to modify their function. RAMPs are able to act as pharmacological switches, chaperones, regulate signaling and/or trafficking in a receptor-dependent manner. This review covers recent discoveries in the RAMP field and summarizes the known GPCR partners and functions of RAMPs. We also discuss the first peptide-bound structures of RAMP-GPCR complexes, which give insight into the molecular mechanisms as to how RAMPs can alter the pharmacology of GPCRs and their signaling.
Keywords: Adrenomedullin, amylin, CGRP, calcium-sensing receptor, CRF, GPR30, heterodimer, GPCR, RAMP, VIP
1. INTRODUCTION
Receptor activity-modifying proteins (RAMPs) are an example of membrane-spanning accessory proteins that can alter the function of G protein-coupled receptors (GPCRs). RAMPs form a small family of 3 (in mammals) (sidebar “RAMPs from other species” near here) proteins that have substantial capacity for introducing functional diversity by interacting with GPCRs (1). First identified as chaperones that enhanced the cell surface expression of the calcitonin-like receptor (CLR) (2), RAMPs are now known to alter trafficking, signaling and pharmacology in a receptor-dependent manner (Figure 1). GPCRs are extremely important cell surface signaling proteins that are responsible for controlling numerous physiological processes and are the targets for ~30% of all medications (3). The interactions of RAMPs with GPCRs is important because they provide an elegant means for controlling their function and may provide further opportunities for drug development.
Sidebar. RAMPs from other species.
This review focuses on mammalian RAMPs but it is important to note that RAMP genes exist in a wide variety of species (98). Some very nice work has been conducted to characterize fish (including pufferfish) RAMPs. These studies include efforts to understand pharmacology and receptor trafficking, and can provide valuable insight into evolutionary mechanisms (99–101). Furthermore, a receptor activity–modifying protein (RAMP)-like triterpene glycoside receptor has been identified from a zebrafish cDNA library (102). Its functional relationship to RAMPs is not known.
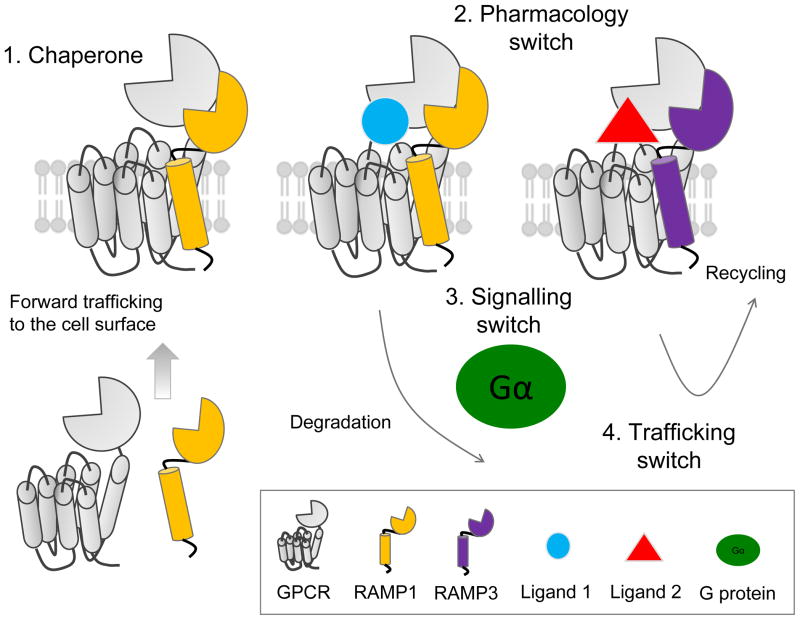
Figure 1. Summary of reported roles for RAMPs.
Four major roles that have been reported for RAMPs: 1. RAMPs enable the forward-trafficking of some GPCRs to the cell surface. 2. RAMPs can alter GPCR pharmacology, switching ligand selectivity for some GPCRs. 3. RAMPs can influence coupling to GPCR signaling pathways. 4. RAMPs may alter the trafficking pathway of some GPCRs from the cell surface, with different RAMPs controlling receptor fate through recycling or degradative pathways.
The interactions of RAMPs with the class B GPCR, CLR are the most extensively studied. In this case, RAMPs are required to chaperone CLR to the cell surface, where the RAMP/CLR complexes can act as receptors for the peptide hormones calcitonin gene-related peptide (CGRP) or adrenomedullin (AM), depending on RAMP co-expression (2). CLR/RAMP1 forms the CGRP receptor and CLR/RAMP2 or CLR/RAMP3 form the AM1 and AM2 receptors, respectively (4). RAMPs are essential for specifying CLR pharmacology, as described in section 4 of this review. There is also quite comprehensive information on interactions between the closest relative to CLR, another class B GPCR, the calcitonin receptor (CTR) and RAMPs. In this case, CTR can reach the cell surface in the absence of RAMP but CTR can drive RAMP translocation to the cell surface. The inability of RAMPs to reach the cell surface without an interacting partner has become a useful screening tool for identifying novel GPCR partners for RAMPs. It is relatively straightforward to monitor RAMP cell surface expression when co-transfected with a GPCR, compared to when transfected alone (5; 6). CTR pharmacology is altered in the presence of RAMP, such that affinity for the endocrine hormone amylin (and variably also CGRP) at this receptor is increased with each RAMP, and three amylin receptor subtypes (AMY1–3) are formed from the resulting RAMP/CTR complexes (4). This early literature on CLR, CTR and RAMP interactions has been reviewed (1). As summarized in Table 1, RAMPs are reported to interact with 9 other GPCRs. Many properties of RAMPs have only been extensively studied with CLR or CTR, and many findings still require validation and mechanistic insight (see Future Issues below).
Table 1.
Summary of RAMP partners and functional consequences
| GPCR | GPCR Class | Interacting RAMP | Consequence of interaction | Validation in vivo | Supporting references |
|---|---|---|---|---|---|
| GPR30 (an estrogen receptor) | A | RAMP3 | Receptor trafficking? | RAMP3-dependent cardioprotection | (28) |
| Calcitonin-like receptor (Adrenomedullin/CGRP receptors – CGRP, AM1, AM2) | B | RAMPs1-3 | Receptor trafficking (chaperone, internalization & recycling), pharmacology | RAMP1, RAMP2 and RAMP3 mouse models linked to adrenomedullin and CGRP biology. | (4; 34; 41; 103) |
| Calcitonin receptor (AMY1, AMY2, AMY3) | B | RAMPs1-3 | Pharmacology, modulates signaling | RAMP1 TG phenotype associated with amylin function | (4; 39) |
| PTH1 | B | RAMP2 | Unknown | Unknown | (5) |
| PTH2 | B | RAMP3 | Unknown | Unknown | (5) |
| VPAC1 | B | RAMPs1-3 | Modulates signaling | Unknown | (5) |
| VPAC2 | B | RAMPs1-3 | Modulates signaling | Unknown | (6; 25) |
| CRF1 | B | RAMP2 | Receptor trafficking (chaperone), modulates signaling | Plasma ACTH response | (6) |
| Glucagon | B | RAMP2 | Unknown | Unknown | (5) |
| Secretin | B | RAMP3 | Receptor trafficking (rescues mutant receptor) | Unknown | (71) |
| Calcium sensing receptor | C | RAMP1 and 3 | Receptor trafficking (chaperone) | Unknown | (26; 27) |
Structurally, RAMPs comprise a single transmembrane spanning domain with an extracellular N-terminal domain of ~90–100 (RAMP2 is the longer sequence) amino acids and a short intracellular C-terminal domain of ~9 amino acids (Figure 2A). Mammalian RAMP1s appear not to be glycosylated, although potential glycosylation sites are present in chick and zebrafish RAMP1. RAMP2 and RAMP3 have multiple glycosylation sites and this may be important for receptor function. For a discussion of these studies refer to (1). Although the C-termini of RAMPs are short, these regions interact with other proteins. Two basic residues in the C-terminus of RAMPs 1 and 3 appear to act as an endoplasmic retrieval signal (2). In RAMP1, these are part of a 5 residue motif (QSKRT) immediately adjacent to the plasma membrane (7). The C-terminal four amino acids of human RAMP3 (DTLL), form a type-1 PDZ1 recognition site. This motif leads to interaction with the Na+/H+ exchange regulatory factor (NHERF) or N-ethylmaleimide-sensitive factor (NSF), leading to altered receptor internalization and recycling, compared to non-PDZ motif containing RAMP1 and 2 (8; 9). Such motifs help explain the variety of contributions that RAMPs have to GPCR function (Table 1). The RAMP C-termini can also influence G protein coupling (10; 11).
Figure 2. Domain structures of RAMPs and Class B GPCRs.
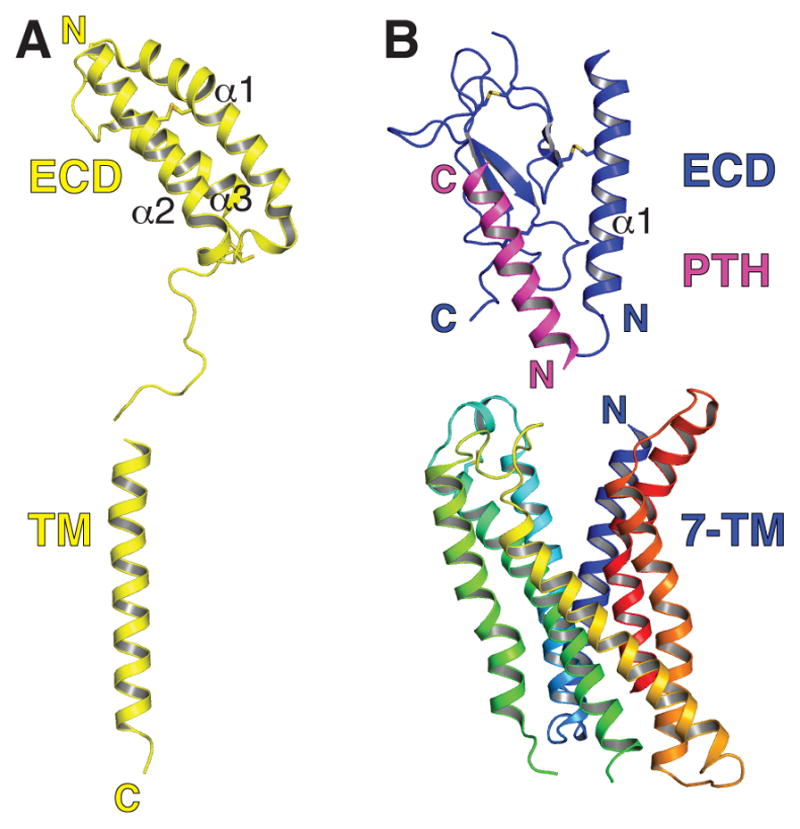
(A) RAMP domain structure shown using the ECD crystal structure of RAMP1 (PDB: 3N7P) and a generic α-helix for the TM segment. (B) Class B GPCR domain structure shown using the crystal structure of the PTH1R ECD with bound C-terminal PTH peptide fragment (PDB: 3C4M) and the peptide-free CRFR1 7-TM domain (PDB: 4K5Y). The 7-TM domain is colored ramped from blue to red for N-terminus to C-terminus. Disulfide bonds are shown as sticks in both panels. The ECD orientations with respect to the TM regions is arbitrary.
There are some very useful recent reviews on RAMPs. Volume 14 of Current Protein and Peptide Science in 2013 and a book on RAMPs both contain excellent collections of specialist reviews on the topic (12). Therefore, in this review, we will focus on the major findings on RAMP/GPCR interactions that have been discovered since the last major general review on RAMPs (1). We will provide some critical insight and will cover the identification of new GPCR partners for RAMPs, their in vivo roles and new structural information which shows how these proteins can alter receptor pharmacology.
2. NEW INSIGHTS FROM EXPRESSION STUDIES
There are two types of studies of RAMP expression that are pertinent to discuss. The first relates to reports of mRNA and protein expression and the correlation or co-localization of this expression with GPCR partners. The second relates to transfection or knockdown studies in cell models that try to identify or characterize receptor partners for RAMPs.
mRNA and protein expression
Expression analysis can be a valuable tool. PCR, RT-PCR or gene arrays, for example, can give broad oversight regarding whether RAMPs are expressed. In most cases, these studies are conducted in samples of a whole organ, rather than an individual cell type. The common result from these studies is that all three RAMPs are expressed. For example, mouse and human heart are reported to express all three RAMPs (13). Given, the spectrum of possible GPCR partners for each RAMP, this tells us nothing more than that the RAMP is expressed. Alterations in mRNA expression in disease models can be more informative. For example, decreased RAMP2 expression in parallel with an increase in RAMP3 s can potentially be informative about the relative roles of AM1 and AM2 receptors (14; 15). However, it is important to consider CLR expression at the same time in this context. What are the consequences of such changes on other GPCR partners for these RAMPs? In most examples, all RAMPs are regulated in the same direction, which does not give clues as to specific mechanisms (15). Furthermore, knowledge of mRNA expression will not necessarily translate to the expression levels of the functional unit: a specific RAMP/GPCR complex at the cell surface. A further issue, which has not been pursued since initial studies, is that RAMPs can apparently compete with one-another in cells, potentially biasing towards one functional complex over another (16).
To address deficiencies in measuring only mRNA, many groups have attempted to use commercially-available RAMP antibodies or raise their own, including one of the authors (D.L.H.). There are many more failures than successes, as is common with GPCR antibodies (17). Initiatives are in place to educate researchers about the controls required for antibody use (18) and a database in The Journal of Comparative Neurology (http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1096-9861/homepage/jcn_antibody_database.htm) provides information related to this issue. Researchers in the RAMP field should pay very careful attention to such recommendations because they are essential for generating (and publishing) reproducible results.
Very few RAMP antibodies have undergone rigorous analysis and thus, at this time we cannot recommend any commercial source. The size of a band on a western blot does not guarantee specificity, nor does pre-incubation of the antibody with the antigen. The latter only implies specificity for the antigen, which is usually a peptide sequence derived from the protein, but is not a reliable indicator that the antibody will detect the fully-folded protein, including in its native environment. Controls such as knockout tissues and cells that do not express the RAMP are necessary to be more confident regarding the validity of conclusions drawn from the use of an antibody. Many standard cell lines express endogenous RAMPs, and this should be taken in to account when designing experiments (Table 2). Most commercial suppliers do not conduct adequate validation of their antibodies. As an example of the issues, in one study, at least two bands of higher molecular weight than RAMPs themselves were detected in a western blot of samples from sham-transfected cells using a common commercial source of RAMP antibodies (19). This result clearly demonstrates that the antibodies can detect proteins other than RAMPs. The authors reported attempts to characterize the additional bands and concluded that they were artifacts and not RAMP/GPCR complexes or RAMP “homodimers”, as has been proposed to explain such observations (20). Therefore immunohistochemical experiments using these or other uncharacterized antibodies could be detecting proteins other than the RAMP itself. Such studies are impossible to interpret, without extremely rigorous controls.
Table 2. Endogenous mRNA expression (by PCR or Northern blot) of RAMPs in cell lines that are commonly used for transfection studies.
Any GPCR transfected into cells that naturally express RAMPs could potentially interact with RAMPs and influence the behavior of the GPCR. Similarly, RAMPs transfected into cells that endogenously express known partners could compete with the RAMP, influencing the outcome of the experiment. Please note that properties of cell lines can differ with passage and culture conditions. Therefore each laboratory should test their own cell stocks for RAMP expression.
| Cell line | RAMP1 | RAMP2 | RAMP3 | References |
|---|---|---|---|---|
|
| ||||
| Cos-7 | − | − | − | (104) |
| +* | +* | +* | (105) | |
| − | − | − | (27) | |
| − | + | nd | (16) | |
|
| ||||
| HEK293 | + | − | − | (27) |
|
| ||||
| HEK293S | − | − | − | (106) |
| +* | +* | +* | (6) | |
|
| ||||
| HEK293T | + | + | − | (2) |
|
| ||||
| CHO-K1 | +* | +* | +* | (6) |
|
| ||||
| CHO-P | +* | +* | +* | (105) |
−no RAMP detected, + RAMP detected, nd, no data,
RAMP detected by PCR but corresponding assays with co-transfected CLR or CTR indicated that this was insufficient to create functional AM, CGRP or amylin receptor complexes, indicating low-level endogenous RAMP expression.
Because of these problems, clear demonstration of RAMP/GPCR oligomerization in tissues has not been provided. Importantly, only a few examples show the association of a GPCR with a RAMP at the protein level through co-expression analysis in tissues with antibodies characterized by at least one of these controls. These examples are the CGRP receptor (CLR/RAMP1) and AMY1 receptor (CTR/RAMP1) (21–23). Where a RAMP is known to alter GPCR pharmacology, such as is the case of CLR and CTR/RAMP complexes, researchers should also consider the value of using different ligands to help them distinguish receptors (side bar Pharmacological Approaches for Identifying CLR/RAMP or CTR/RAMP Complexes near here).
Sidebar. Pharmacological Approaches for Identifying CLR/RAMP or CTR/RAMP Complexes.
The authors encourage readers to refer to www.guidetopharmacology.org to identify pharmacological tools that may help in distinguishing which complex mediates a particular effect. Some ligands are reasonably selective between receptors but this selectivity is usually lost at high concentrations (micromolar or higher), which is unfortunately often found in published studies. Other ligands, such as CGRP8–37 or AM22–52 alone are very poor discriminators between receptors at any concentration. We thus recommend the use of a combination of ligands wherever possible.
Identifying interactions between RAMPs and GPCRs in cell models
Experiments to identify partnerships between RAMPs and GPCRs and understand mechanisms thereof tend to rely on the use of tagged proteins. In some respects, this approach avoids the issues related to antibody specificity but it can have its own problems. For example, the addition of tags can lead to aberrant trafficking or the presence of a large protein tag (e.g., intracellular GFP C terminal tag) could impede interactions with signaling proteins (5). Therefore researchers should also conduct appropriate controls in using tagged receptors to study RAMPs. Such co-expression studies have been very valuable and have advanced the field, through the identification of new RAMP/GPCR partnerships. A few examples will be briefly described.
A range of interactions between RAMPs and class B GPCRs, other than the classic examples of CLR and CTR have been described There is no evidence for interactions between RAMPs and the GLP-1 receptor, GLP-2 receptor or GHRH receptor (5; 6) but several other interactions are summarized in Table 1. With respect to the CRF1 receptor and RAMP2, co-expression in cells increases the cell surface expression of both proteins. CRF1 receptor signaling is also affected: RAMP2 reportedly enhances Gi, Gq and G12 interactions, as well as Ca2+ responses to both CRF and urocortin (6). It is assumed that these effects are due to a direct interaction but this has not been shown.
The literature describes variable VPAC2 receptor-RAMP interactions. Christopoulos and colleagues reported that the VPAC2 receptor could not translocate any RAMP to the cell surface in the classic assay used to screen for RAMP/GPCR interactions (5). On the other hand, Wootten and colleagues detected enhanced expression of all three RAMPs with this receptor (6). This could relate to differences in the cells in which these experiments were performed. RAMP co-expression with the VPAC2 receptor produced subtle effects on basal receptor signaling; this was most clearly evident as an increase in basal signaling with RAMP1. Results of this study support others that report differential effects of RAMPs on G protein-coupling (5; 24). Another report of VPAC2 receptor/RAMP2 co-expression suggested that RAMP2 may influence peptide histidine methionine (PHM) affinity and GTP-sensitivity at this receptor (25). Although PHM and peptide histidine isoleucine (PHI) were also studied in the context of VPAC1 and VPAC2 receptor/RAMP co-expression in the other two studies (5; 6), only limited experiments were performed. It will certainly be interesting to explore these interactions further and to consider how RAMPs might influence PHM/I pharmacology and signaling.
Two reports have studied the interactions between the class C GPCR, the calcium-sensing receptor (CaSR) and RAMPs (26; 27). In both cases RAMP1 and RAMP3, but not RAMP2, appeared to enable CaSR cell surface expression. Further, Desai and colleagues used siRNA to reduce RAMP1 expression in medullary thyroid carcinoma TT cells, which naturally express mRNA for both CaSR and RAMP1. RAMP1 knockdown reduced agonist (Cinacalcet)-mediated CaSR Ca2+ signaling, showing that RAMP1 is important for CaSR receptor function in these cells (26). It will be important to determine the importance of these interactions in vivo and their relevance to the clinical use of CaSR-targeted drugs.
GPR30, an estradiol receptor, has been reported to interact with RAMP3 using BRET, immunoprecipitation and siRNA (28). The precise details of this interaction and the properties of GPR30 that are altered by this interaction are not yet clear. In vivo implications are discussed in the next section. This study is very interesting because it is the first detailed report of a class A GPCR/RAMP-interaction.
Together the studies noted above lead to the important possibility that RAMPs may interact and potentially influence not only Class B GPCRs but also GPCRs in Class A and Class C.
3. NEW INSIGHTS FROM ANIMAL MODELS
These cellular and expression studies are the first steps in demonstrating RAMP/GPCR partnerships. Such findings require validation using more complex models to define their functional relevance in physiology and pharmacology. However, as with other receptor systems that involve more than one receptor subunit, such studies are not trivial. RAMP biology is difficult to interpret because genetic intervention with a RAMP could have effects in various receptor systems (29). Nevertheless the studies that have been performed are important for documenting and validating different partnerships between RAMPs and GPCRs and to advance understanding of their roles in physiology and disease.
For example, RAMP deletion studies or transgenic models have given valuable insight into the functional roles of the receptors for the calcitonin family of peptides, including the two AM receptor subtypes and the CGRP receptor (30–33). The AM1 and AM2 receptors are derived from the interaction of CLR with RAMP2 or RAMP3, respectively. In pharmacological studies, both receptors bind the 52 amino acid peptide AM with high affinity. Although the fate of internalized AM1 and AM2 receptors differs by virtue of these different RAMPs, the roles of these receptors in vivo has been difficult to establish due to a lack of pharmacological tools or antibodies that can distinguish the effects of these receptors. AM is an important cardiovascular peptide and two groups have developed RAMP2- or RAMP3-knockout mouse models in an attempt to learn more about the receptor biology of this peptide.. Ramp3−/− mice are viable, whereas Ramp2−/− mice die in utero, implying that the two AM receptor subtypes probably have quite different functions (34; 35). Even partial loss of RAMP2 (e.g. Ramp2+/−) is deleterious, whereas a phenotype has been harder to establish even for complete loss of RAMP3. In the first reported study, Dackor and colleagues did not observe any obvious phenotypic differences in their Ramp3−/− mice up to 18 months of age, aside from a modest decrease in body weight at 9–10 months, compared to wild-type (WT) animals (34). Only when challenged by crossing Ramp3−/− with RenTgMK mice, a genetic model of AngII-mediated cardiovascular disease, did sex-dependent cardiovascular phenotypic differences emerge: renal failure and cardiac hypertrophy only in male mice (36). In contrast, lymphatic defects were evident in a separate Ramp3−/− model (35). AM has important effects in the cardiovascular and lymphatic systems, consistent with these observations in the Ramp3−/− model. However, at present it is difficult to draw firm conclusions as to the role of each AM receptor from these studies.
Several RAMP1 mouse models exist, including those with over-expression. The knockout models display a range of phenotypic differences compared to WT mice. These include blood pressure and inflammatory phenotypes (31; 37; 38). Differences in wound healing, angiogenesis and lymphangiogenesis have also been reported, each of which was suppressed in the absence of RAMP1 (32). In studies thus far, there is a clear linkage between reported actions of the peptide i.e. AM and CGRP, and the RAMP with which a cognate receptor for each peptide is formed. Thus, cardiovascular phenotypes are expected for each RAMP, in line with the activities of the peptides. However, each RAMP model might influence ligand-receptor systems that may not yet have been studied but may also contribute to the phenotypes of the mice. We will briefly describe three examples of this.
RAMP1 TG mouse models (39–41) were initially described in the context of CGRP activity in sensory nerves. However, RAMP1 forms a separate receptor for amylin with CTR, creating the AMY1 receptor. The neuronal overexpression of human RAMP1 led to sensitization to amylin. Human RAMP1 transgenic mice were lighter, leaner and had increased energy expenditure and body temperature, consistent with the classic role of amylin as an endocrine hormone that controls blood glucose and body weight (Hay et al accepted with minor revisions). Therefore, any alteration in RAMP1 expression could affect CGRP or amylin activity. As more partners for RAMP1 are identified, such as the CaSR, this becomes even more relevant (26; 27).
The second example of this is Ramp2+/− female mice, which exhibit a range of phenotypes. Many of these are recapitulated in AM and CLR-deficient animals, linking RAMP2 to AM1 receptor biology. Others are distinct, supporting a broader range of functions for RAMP2. These include endocrine phenotypes such as hyperprolactinemia, enlarged pituitary glands, accelerated mammary gland development and skeletal abnormalities (42). These types of observations yield useful hypotheses and encourage the search for novel receptor partners for RAMPs.
Finally, treatment of RenTgMK;Ramp3+/+ and RenTgMK;Ramp3−/− mice with G-1, reportedly a specific GPR30 agonist, was used to determine the in vivo relevance of the interaction between RAMP3 and GPR30 (43). Cardiovascular phenotyping revealed a reduction in cardiac hypertrophy and perivascular fibrosis that was RAMP3- and sex-dependent. Thus, cardioprotective effects of RAMP3 are linked to GPR30 as well as the AM system (36; 43).
4. NEW INSIGHTS FROM STRUCTURAL AND MOLECULAR STUDIES
Molecular understanding of GPCR pharmacology and activation mechanisms has significantly expanded with recent advances in GPCR structural studies. Crystal structures are available for nearly thirty distinct GPCRs, most of these class A receptors (44). Class B GPCR structure is less well understood, but this is beginning to change (45). Class B receptors, including those that interact with RAMPs, use two domains to bind their peptide ligands (46; 47). The C-terminal portion of the peptide ligand binds the receptor extracellular domain (ECD) to impart affinity and specificity while the N-terminal portion of the peptide binds and activates the 7 transmembrane (7-TM) domain of the receptor (Figure 2B). No structure of a full-length class B GPCR is yet available, but several structures of ligand-free and peptide-bound class B receptor ECDs (48–58), and structures of two peptide-free class B receptor 7-TM domains, those of the CRF and glucagon receptors (59; 60), are available (Figure 2B). These studies indicated that the C-terminal portion of class B GPCR peptide ligands forms an α-helix that occupies a groove in the receptor ECD. The class B 7-TM bundle is similar to that of the class A receptors except that the extracellular-facing half of the class B 7-TM bundle is more open than in a typical class A receptor, presumably to accommodate the large peptide ligand (Figure 2B). How the peptides bind the 7-TM domain and the orientation of the ECD with respect to the 7-TM domain in the intact receptors remain poorly understood.
Interactions between RAMPs and GPCRs
How RAMPs associate with class B GPCRs is best understood for CLR and CTR. Studies with chimeric proteins indicated that the RAMP and CLR/CTR ECDs play a major role in dictating ligand selectivity (61; 62). Heterologous expression and purification of a soluble 1:1 CLR:RAMP1 ECD complex (63) led to crystal structures of this complex in ligand-free and small molecule antagonist-bound states (64). Similar approaches yielded a soluble CLR:RAMP2 ECD complex and its structure in the ligand-free state (65; 66). The purified CLR:RAMP2 ECD complex behaved as a 2:2 dimer of heterodimers, but the physiological relevance of higher order oligomerization was unclear (65; 67; 68). These important structural studies revealed how the ECDs of RAMP1 and RAMP2 interact with the CLR ECD and how the antagonist drugs olcegepant and telcagepant bind the CGRP receptor ECD complex (69; 70). We recently determined the first peptide-bound crystal structures for both the CGRP and AM1 receptor ECD complexes that reveal how peptides bind and how RAMPs determine peptide selectivity as discussed below (Booe et al, in press).
Although ECD-ECD interactions are critical for RAMP modulation of CLR and CTR, the RAMP TM also interacts with the GPCR 7-TM and this interaction is likely to be most important for RAMP interactions with other class B GPCRs and with class A and class C GPCRs that lack class B GPCR-like ECDs. How the RAMP TM domain interacts with the GPCR 7-TM is best understood for the class B Secretin receptor where elegant BRET and fluorescence complementation methods were used to identify TM6 and TM7 of the Secretin receptor 7-TM as the site of contact with the RAMP3 TM (71). It is unclear if this specific interface is utilized in all RAMP-GPCR complexes.
Structural studies of peptide-bound CLR:RAMP ECD complexes
To simplify production of soluble CLR:RAMP ECD complexes for structural studies, we developed a tethered RAMP ECD-CLR ECD fusion protein approach that ensures stability of the complex and enforces 1:1 CLR:RAMP stoichiometry (68). The RAMP1 or RAMP2 ECD was covalently tethered to the CLR ECD via a flexible (Gly-Ser)x linker. The RAMP1 ECD-CLR ECD and RAMP2 ECD-CLR ECD tethered fusion proteins selectively bound CGRP and AM, respectively, but the proteins differed in their oligomeric states; the RAMP1 tethered fusion was a monomer whereas the RAMP2 tethered fusion was a dimer (68). Obtaining crystals of the peptide-bound tethered fusion proteins required refining the sequence and length of the tethers as well as identifying an amino acid substitution in the RAMP2 ECD that prevented dimerization of the RAMP2-CLR fusion, which may be an artifact, because dimerization occludes the peptide-binding site (Booe et al, in press). The RAMP1-CLR ECD fusion was crystallized in the presence of a CGRP(27–37)NH2 analog with affinity-enhancing [D31, P34, F35] substitutions (72). The RAMP2-CLR ECD fusion was crystallized in the presence of a WT C-terminal AM fragment (Figure 3). The CGRP analog-bound and AM-bound structures were refined at resolutions of 2.5 and 1.8 Å, respectively, which allowed unambiguous determination of the peptide-binding modes.
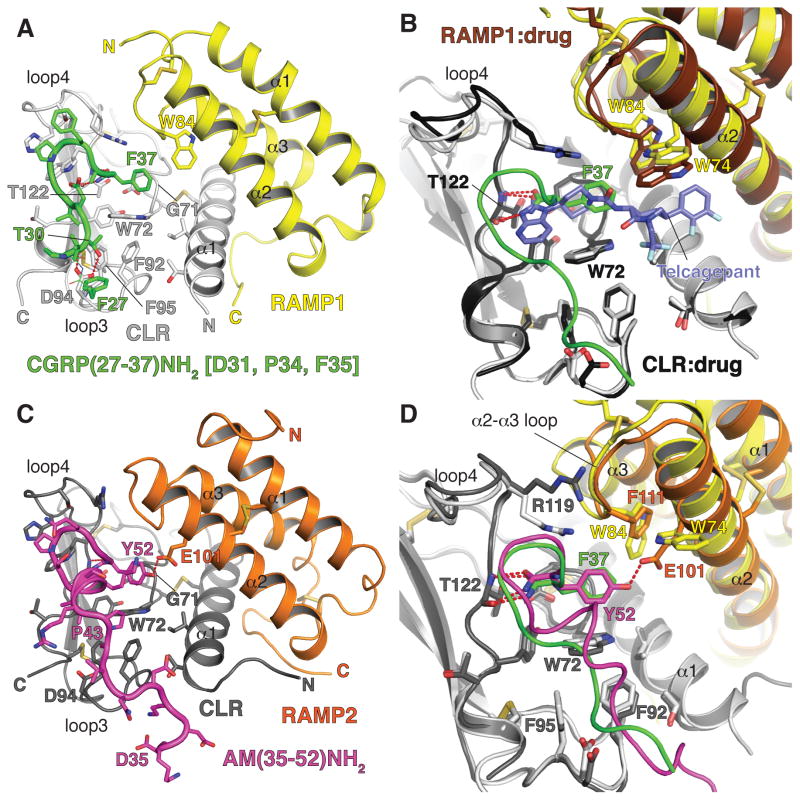
Figure 3. Structural basis for RAMP-dependent selective CGRP and AM binding to CLR.
(A) Crystal structure of the CLR:RAMP1 ECD heterodimer with bound high-affinity CGRP analog (PDB: 4RWG). The peptide is shown in cartoon representation with side chains as sticks. For clarity the mainchain atoms of peptide residues F27 and V28 are shown in lines representation. (B) Superposition of the CGRP analog-bound and small molecule antagonist-bound (PDB: 3N7R) ECD heterodimers aligned based on the CLR positions. (C) Crystal structure of the CLR:RAMP2 ECD heterodimer with bound AM (PDB: 4RWF). (D) Superposition of the CGRP analog-bound and AM-bound structures aligned based on the CLR positions.
The CGRP analog occupies a peptide-binding site that is almost entirely on CLR (Figure 3A). The peptide makes only a single critical contact to RAMP1, involving hydrophobic interaction of CGRP F37 and RAMP1 W84. Strikingly, the CGRP analog is devoid of the α-helical structure typical of class B GPCR peptide ligands. Instead, the peptide has a β-turn structure near the C-terminus that helps position the C-terminal residue so that it occupies a pocket composed of CLR W72 (known as the “Trp shelf” (64)), G71, and RAMP1 W84. The peptide C-terminal amide group hydrogen bonds with the main chain of CLR T122 at the base of loop4. The peptide C-terminal residue and amide group thus have an important “anchoring” function. Another key interaction involves hydrogen bonds between the CGRP analog T30 and CLR D94 on loop3. The structure explains antagonism of CGRP by the drugs olcegepant and telcagepant, which block anchoring of the peptide C-terminus by hydrogen bonding with the CLR T122 main chain and packing against the Trp shelf and G71 (Figure 3B).
AM occupies the same peptide-binding site on CLR as the CGRP analog and also adopts a conformation largely lacking secondary structure, other than one helical turn (Figure 3C). Like the CGRP analog, AM has a β-turn near its C-terminus that allows the AM C-terminal residue, Y52, to occupy the pocket packing against the Trp shelf and G71, and the AM C-terminal amide group hydrogen bonds with the CLR T122 main chain. Only a single contact of importance is made between AM and RAMP2, involving a hydrogen bond between the AM Y52 side chain hydroxyl and the RAMP2 E101 side chain. Prior to the β-turn, the interaction of AM with CLR differs from that of the CGRP analog. The helical turn in AM positions K46 to pack against the Trp shelf and AM Y52. AM A42 and P43 pack against a hydrophobic patch on CLR comprised of the Trp shelf, F92, and F95.
Although the lack of α-helicity in the receptor bound CGRP analog and AM peptides may seem surprising, previous NMR and mutagenesis studies of the CGRP and AM peptides indicated a lack of helicity and propensity for turn formation in their C-terminal regions (73–75). Moreover, the CT family peptide sequences are rich in helix-breaking Pro residues in their C-terminal regions. CT family peptides thus appear to bind their class B GPCR ECDs with bound conformations very different from other class B GPCR peptide ligands, although the binding site position in the CLR ECD is similar to other receptors (Booe et al., in press) (76). The CGRP analog-bound and AM-bound structures thus significantly expand our understanding of how peptides bind class B GPCRs.
Comparing the CGRP analog-bound and AM-bound structures reveals that RAMP1 and RAMP2 are distinct in their augmentation of the peptide-binding site pocket with residues that are in proximity to the peptide C-terminal residue (Figure 3D). RAMP1 W84 contacts the CGRP analog F37 phenyl ring and the equivalent RAMP2 F111 cannot make this contact. RAMP2 F111 thus “discourages” CGRP binding. RAMP2 E101 hydrogen bonds with AM Y52 and the equivalent RAMP1 W74 cannot make this contact. The lack of Glu at RAMP1 position 74 thus discourages AM binding. RAMP3 has a Trp at position 84 and a Glu at position 74; thus it is a RAMP1–2 hybrid with respect to the residues that augment the peptide-binding site pocket. RAMP3 E74 would favor AM binding by hydrogen bonding with AM Y52 and RAMP3 W84 likely explains the increased potency of CGRP at the AM2 receptor as compared to the AM1 receptor (77). The binding sites in the two receptors are further compared in Supplemental Movie S1.
Distinct RAMP binding site augmentation contributes to peptide selectivity, but there is evidence that this is insufficient to entirely account for selectivity. Despite the shared CGRP and AM binding site on CLR and only a single RAMP-mediated contact of importance involving the CGRP and AM C-terminal residues, swapping the CGRP and AM peptide C-terminal residues is insufficient to exchange their receptor binding preferences (Booe et al., in press). RAMP1 and RAMP2 elicit subtly different CLR conformations, which may alter the peptide-binding site to favor a particular peptide. The RAMP1/2 α2-α3 loop is close to the CLR R119 side chain that adopts different conformations in the two structures, which may accommodate subtle differences in the β-turn structures of CGRP and AM (Figure 3D). Subtle shifts in the positions of other CLR residues that make up the pocket may also contribute to selectivity. More work is needed to determine the extent to which such allostery contributes to selectivity, but overall the data are consistent with RAMPs determining peptide selectivity of CLR by a combination of distinct binding site augmentation and subtle effects on CLR conformation. Allosteric effects could help explain how RAMPs can influence a diverse range of GPCRs, with distinct functional consequences (Table 1).
Perhaps the most striking revelation from the new structures is that the profound effect of RAMPs on CLR pharmacology arise not from extensive RAMP-peptide contacts or dramatic RAMP-induced changes in CLR conformation, but rather from a single RAMP-peptide contact and subtle alteration of CLR. Although additional selectivity determinants may exist in portions of the receptors not addressed by the structures, the soluble ECD complexes exhibit the same peptide selectivity profiles as the intact receptors. Thus, the apparently minor effects of RAMPs can in fact greatly affect receptor function. Another important finding is that CGRP and AM occupy a shared a binding site largely on CLR, which highlights the difficulty of developing peptide analogs with selectivity for a specific GPCR:RAMP pairing. Such analogs would be valuable pharmacological tools and could also be of value as therapeutics. Despite the challenges, the new structures should help guide drug development efforts targeting GPCR:RAMP complexes.
Mutagenesis studies of CT family peptides and RAMP:GPCR complexes
The CGRP analog and AM ECD complex-binding modes observed in the crystal structures are consistent with mutagenesis studies of the peptides. Binding studies with the purified tethered RAMP1 ECD-CLR ECD fusion indicated that CGRP T30, V32, F37, and the C-terminal amide were important for receptor binding (68). Studies with intact CGRP receptor in cells similarly indicated the importance of these CGRP residues (72; 74; 78). Peptide binding studies with purified tethered RAMP2 ECD-CLR ECD (68), and binding and NMR chemical shift perturbation experiments with CLR ECD/RAMP2 ECD (66) were in agreement that AM residues P43, K46, I47, G51, Y52 were most important for receptor binding. Studies of mutant AM peptide binding to intact AM1 receptor in cells similarly identified these residues (66).
Extensive mutagenesis studies of the RAMP ECDs (79) identified RAMP1 W84 as important for CGRP function at the CGRP receptor (80) and RAMP2 F111 and E101 as important for AM function at the AM1 receptor (77). Several studies highlighted an important role for RAMP3 E74 in AM function at the AM2 receptor (77; 81; 82). These data are now explained by how these residues contact the peptide C-termini. Previous RAMP “swap” mutants highlighted the importance of Glu at position 74/101 in favoring AM binding (81; 82). RAMP1 W84 is also important for amylin and CGRP binding to the AMY1 receptor, which suggests that RAMP1 may augment a binding site on CTR that is similar to that on CLR (83). Extensive mutagenesis of the CLR ECD in the context of the CGRP and AM1 receptors fully supported the new CGRP analog and AM-bound structures (Booe et al., in press). The good agreement between the structures and mutagenesis data suggests that the peptide-bound ECD complex structures are good models for full-length agonist peptide binding to the intact receptor complexes.
Understanding of how CT family peptides bind and activate the GPCR 7-TM remains rudimentary. Nonetheless, several recent studies provided insight into this area using mutagenesis and/or chimeric approaches to explore how the CLR 7TM domain recognizes peptides. Various residues in all three extracellular loops (ECL) of CLR were important for response to CGRP in the CGRP receptor (84; 85) and ECL3 was important for responses to AM in the AM1 and AM2 receptors (86). Thr6 in the CGRP peptide, which is also conserved in AM, was shown to be important for agonist activity (87). Ultimately, structural information for the intact RAMP:CLR/CTR complexes will be required to enable mechanistic explanation of this mutagenesis data.
Activation mechanisms in RAMP-complexed GPCRs
Molecular understanding of how RAMPs may influence GPCR activation mechanisms and signaling is still in its infancy (88). Most information is available for CLR, in the AM1 and CGRP receptors. Substantial mutagenesis within the 7-TM region, C-terminal tail and helix 8 region of CLR in the CGRP receptor has revealed valuable information into how this particular receptor complex is activated (89–94). In the AM1 receptor, there is some information on the C-terminus and helix 8 regions (95; 96). At the present time, there are too few parallel studies to enable conclusions to be drawn around the role of the different RAMPs in influencing the 7-TM bundle and C-terminus. In the case of a key proline residue in TM6 of CLR, this appears to have the same important functional role in receptor activation with RAMP1, RAMP2 and RAMP3 (94). The equivalent proline in CTR is also important but this has not been looked at with each RAMP (97). Much of this information has been amassed to support a model of the 7-TM bundle of CLR with RAMP1, which is important to help interpret this volume of data (90).
5. CONCLUSIONS
RAMPs are fascinating proteins that diversify GPCR ligand binding preferences and function. Given their range of potential functions and broad expression, including in commonly used cell lines (Table 2), their influence on other GPCRs may still be underappreciated. The “Future Issues/questions List” outlines key unanswered questions in this field, and what we view are important issues that still need to be addressed.
Supplementary Material
Future Issues/questions List.
Antibodies need to be fully validated to enable characterization of RAMP/GPCR complex expression in tissues.
More specific pharmacological tools need to be developed to assist in receptor identification.
Do RAMPs compete with each other for expression in cells? More work is needed to understand this.
Some reported GPCR-RAMP interactions are only single reports and require validation and further characterizion to understand mechanisms.
Can RAMP/GPCR complexes be successfully targeted with drugs?
How do AM2, CT, and amylin selectively bind their respective RAMP/GPCR complexes?
To what extent does allostery contribute to RAMP-altered GPCR ligand selectivity?
Determination of the molecular architectures of full-length RAMP/GPCR complexes and how full-length agonist peptides bind and activate these complexes.
Are their more GPCR partners for RAMPs and other functions?
Do RAMPs partner with other classes of receptor, such as receptor tyrosine kinases?
Acronyms and Definitions List
- 7-TM
7 transmembrane
- AM
adrenomedullin
- AM1
RAMP2/CLR AM receptor subtype
- AM2
RAMP3/CLR AM receptor subtype
- AM2
AM2/intermedin peptide
- CaSR
calcium-sensing receptor
- CGRP
calcitonin gene-related peptide
- CLR
calcitonin receptor-like receptor
- CTR
calcitonin receptor
- ECD
extracellular domain
- GPCR
G protein-coupled receptor
- RAMP
receptor activity-modifying protein
Literature Cited
- 1.Hay DL, Poyner DR, Sexton PM. GPCR modulation by RAMPs. Pharmacology & Therapeutics. 2006;109:173–97. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 2.McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. NATURE. 1998;393:333–9. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 3.Garland SL. Are GPCRs still a source of new targets? Journal of biomolecular screening. 2013;18:947–66. doi: 10.1177/1087057113498418. [DOI] [PubMed] [Google Scholar]
- 4.Poyner DR, Sexton PM, Marshall I, Smith DM, Quirion R, et al. International Union of Pharmacology. XXXII. The Mammalian Calcitonin Gene-Related Peptides, Adrenomedullin, Amylin, and Calcitonin Receptors. Pharmacological reviews. 2002;54:233–46. doi: 10.1124/pr.54.2.233. [DOI] [PubMed] [Google Scholar]
- 5.Christopoulos A, Christopoulos G, Morfis M, Udawela M, Laburthe M, et al. Novel Receptor Partners and Function of Receptor Activity-modifying Proteins. J Biol Chem. 2003;278:3293–7. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- 6.Wootten D, Lindmark H, Kadmiel M, Willcockson H, Caron KM, et al. Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. British journal of pharmacology. 2013;168:822–34. doi: 10.1111/j.1476-5381.2012.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner S, Muff R, Gujer R, Fischer JA, Born W. The Transmembrane Domain of Receptor-Activity-Modifying Protein 1 Is Essential for the Functional Expression of a Calcitonin Gene-Related Peptide Receptor. Biochemistry. 2002;41:11398–404. doi: 10.1021/bi020279r. [DOI] [PubMed] [Google Scholar]
- 8.Bomberger JM, Parameswaran N, Hall CS, Aiyar N, Spielman WS. Novel Function for Receptor Activity-modifying Proteins (RAMPs) in Post-endocytic Receptor Trafficking. J Biol Chem. 2005;280:9297–307. doi: 10.1074/jbc.M413786200. [DOI] [PubMed] [Google Scholar]
- 9.Bomberger JM, Spielman WS, Hall CS, Weinman EJ, Parameswaran N. Receptor Activity-modifying Protein (RAMP) Isoform-specific Regulation of Adrenomedullin Receptor Trafficking by NHERF-1. J Biol Chem. 2005;280:23926–35. doi: 10.1074/jbc.M501751200. [DOI] [PubMed] [Google Scholar]
- 10.Udawela M, Christopoulos G, Morfis M, Christopoulos A, Ye S, et al. A Critical Role for the Short Intracellular C Terminus in Receptor Activity-Modifying Protein Function. Mol Pharmacol. 2006;70:1750–60. doi: 10.1124/mol.106.024257. [DOI] [PubMed] [Google Scholar]
- 11.Udawela M, Christopoulos G, Morfis M, Tilakaratne N, Christopoulos A, Sexton PM. The effects of C-terminal truncation of receptor activity modifying proteins on the induction of amylin receptor phenotype from human CTb receptors. Regulatory Peptides. 2008;145:65–71. doi: 10.1016/j.regpep.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Parameswaran N, Spielman W. Introduction to Ramps. In: Spielman W, Parameswaran N, editors. RAMPs. Vol. 744. Springer; US: 2012. pp. 1–11.pp. 1–11. [DOI] [PubMed] [Google Scholar]
- 13.Parameswaran N, Spielman WS. RAMPs: The past, present and future. Trends Biochem Sci. 2006;31:631–8. doi: 10.1016/j.tibs.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Nishikimi T, Yoshihara F, Kanazawa A, Okano I, Horio T, et al. Role of increased circulating and renal adrenomedullin in rats with malignant hypertension. Am J Physiol Regul Integr Comp Physiol. 2001;281:R2079–87. doi: 10.1152/ajpregu.2001.281.6.R2079. [DOI] [PubMed] [Google Scholar]
- 15.Udawela M, Hay DL, Sexton PM. The receptor activity modifying protein family of G protein coupled receptor accessory proteins. Seminars in Cell & Developmental Biology. 2004;15:299–308. doi: 10.1016/j.semcdb.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Buhlmann N, Leuthauser K, Muff R, Fischer JA, Born W. A Receptor Activity Modifying Protein (RAMP)2-Dependent Adrenomedullin Receptor Is a Calcitonin Gene-Related Peptide Receptor when Coexpressed with Human RAMP1. Endocrinology. 1999;140:2883–90. doi: 10.1210/endo.140.6.6783. [DOI] [PubMed] [Google Scholar]
- 17.Marchalant Y, Brownjohn PW, Bonnet A, Kleffmann T, Ashton JC. Validating Antibodies to the Cannabinoid CB2 Receptor: Antibody Sensitivity Is Not Evidence of Antibody Specificity. Journal of Histochemistry & Cytochemistry. 2014;62:395–404. doi: 10.1369/0022155414530995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gore AC. Editorial: Antibody Validation Requirements for Articles Published in Endocrinology. Endocrinology. 2013;154:579–80. doi: 10.1210/en.2012-2222. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Bell D, Smith LR, Zhao L, Devine AB, et al. Differential Expression of Components of the Cardiomyocyte Adrenomedullin/Intermedin Receptor System following Blood Pressure Reduction in Nitric Oxide-Deficient Hypertension. J Pharmacol Exp Ther. 2006;316:1269–81. doi: 10.1124/jpet.105.092783. [DOI] [PubMed] [Google Scholar]
- 20.Cueille C, Pidoux E, de Vernejoul M-C, Ventura-Clapier R, Garel J-M. Increased myocardial expression of RAMP1 and RAMP3 in rats with chronic heart failure. Biochemical and Biophysical Research Communications. 2002;294:340–6. doi: 10.1016/S0006-291X(02)00487-4. [DOI] [PubMed] [Google Scholar]
- 21.Eftekhari S, Salvatore CA, Calamari A, Kane SA, Tajti J, Edvinsson L. Differential distribution of calcitonin gene-related peptide and its receptor components in the human trigeminal ganglion. Neuroscience. 2010;169:683–96. doi: 10.1016/j.neuroscience.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Lennerz Jochen K, VREPCWLNNWBEFGKM Calcitonin receptor-like receptor (CLR), receptor activity-modifying protein 1 (RAMP1), and calcitonin gene-related peptide (CGRP) immunoreactivity in the rat trigeminovascular system: Differences between peripheral and central CGRP receptor distribution. The Journal of Comparative Neurology. 2008;507:1277–99. doi: 10.1002/cne.21607. [DOI] [PubMed] [Google Scholar]
- 23.Walker CS, Eftekhari S, Bower RL, Wilderman A, Insel PA, et al. A second trigeminal CGRP receptor: function and expression of the AMY1 receptor. Annals of Clinical and Translational Neurology. 2015 doi: 10.1002/acn3.197. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morfis M, Tilakaratne N, Furness SGB, Christopoulos G, Werry TD, et al. Receptor Activity-Modifying Proteins Differentially Modulate the G Protein-Coupling Efficiency of Amylin Receptors. ENDOCRINOLOGY. 2008;149:5423–31. doi: 10.1210/en.2007-1735. [DOI] [PubMed] [Google Scholar]
- 25.Muller J-M, Debaigt C, Goursaud S, Montoni A, Pineau N, et al. Unconventional binding sites and receptors for VIP and related peptides PACAP and PHI/PHM: An update. Peptides. 2007;28:1655–66. doi: 10.1016/j.peptides.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Desai AJ, Roberts DJ, Richards GO, Skerry TM. Role of Receptor Activity Modifying Protein 1 in Function of the Calcium Sensing Receptor in the Human TT Thyroid Carcinoma Cell Line. PLoS One. 2014;9:e85237. doi: 10.1371/journal.pone.0085237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouschet T, Martin S, Henley JM. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci. 2005;118:4709–20. doi: 10.1242/jcs.02598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LMF, Caron KM. G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. Journal of molecular endocrinology. 2013;51:191–202. doi: 10.1530/JME-13-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferré S, Casadó V, Devi LA, Filizola M, Jockers R, et al. G Protein–Coupled Receptor Oligomerization Revisited: Functional and Pharmacological Perspectives. Pharmacological reviews. 2014;66:413–34. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadmiel M, Fritz-Six KL, Caron KM. UNDERSTANDING RAMPs THROUGH GENETICALLY ENGINEERED MOUSE MODELS. Advances in experimental medicine and biology. 2012;744:49–60. doi: 10.1007/978-1-4614-2364-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Wetzel-Strong SE, Hua X, Tilley SL, Oswald E, et al. Deficiency of RAMP1 Attenuates Antigen-Induced Airway Hyperresponsiveness in Mice. PloS one. 2014;9:e102356. doi: 10.1371/journal.pone.0102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurashige C, Hosono K, Matsuda H, Tsujikawa K, Okamoto H, Majima M. Roles of receptor activity-modifying protein 1 in angiogenesis and lymphangiogenesis during skin wound healing in mice. The FASEB Journal. 2014;28:1237–47. doi: 10.1096/fj.13-238998. [DOI] [PubMed] [Google Scholar]
- 33.Jusek G, Reim D, Tsujikawa K, Holzmann B. Deficiency of the CGRP receptor component RAMP1 attenuates immunosuppression during the early phase of septic peritonitis. Immunobiology. 2012;217:761–7. doi: 10.1016/j.imbio.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Dackor R, Fritz-Six K, Smithies O, Caron K. Receptor Activity-modifying Proteins 2 and 3 Have Distinct Physiological Functions from Embryogenesis to Old Age. J Biol Chem. 2007;282:18094–9. doi: 10.1074/jbc.M703544200. [DOI] [PubMed] [Google Scholar]
- 35.Yamauchi A, Sakurai T, Kamiyoshi A, Ichikawa-Shindo Y, Kawate H, et al. Functional differentiation of RAMP2 and RAMP3 in their regulation of the vascular system. Journal of Molecular and Cellular Cardiology. 2014;77:73–85. doi: 10.1016/j.yjmcc.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 36.Barrick CJ, Lenhart PM, Dackor RT, Nagle E, Caron KM. Loss of receptor activity-modifying protein 3 exacerbates cardiac hypertrophy and transition to heart failure in a sex-dependent manner. Journal of Molecular and Cellular Cardiology. 2012;52:165–74. doi: 10.1016/j.yjmcc.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsujikawa K, Yayama K, Hayashi T, Matsushita H, Yamaguchi T, et al. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proceedings of the National Academy of Sciences. 2007;104:16702–7. doi: 10.1073/pnas.0705974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikami N, Sueda K, Ogitani Y, Otani I, Takatsuji M, et al. Calcitonin Gene-Related Peptide Regulates Type IV Hypersensitivity through Dendritic Cell Functions. PLoS One. 2014;9:e86367. doi: 10.1371/journal.pone.0086367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Liu X, Morgan DA, Kuburas A, Thedens DR, et al. Neuronal receptor activity-modifying protein 1 promotes energy expenditure in mice. Diabetes. 2011;60:1063–71. doi: 10.2337/db10-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabharwal R, Zhang Z, Lu Y, Abboud FM, Russo AF, Chapleau MW. Receptor activity-modifying protein 1 increases baroreflex sensitivity and attenuates Angiotensin-induced hypertension. Hypertension. 2010;55:627–35. doi: 10.1161/HYPERTENSIONAHA.109.148171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Winborn CS, Marquez de Prado B, Russo AF. Sensitization of Calcitonin Gene-Related Peptide Receptors by Receptor Activity-Modifying Protein-1 in the Trigeminal Ganglion. J Neurosci. 2007;27:2693–703. doi: 10.1523/JNEUROSCI.4542-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadmiel M, Fritz-Six K, Pacharne S, Richards GO, Li M, et al. Research Resource: Haploinsufficiency of Receptor Activity-Modifying Protein-2 (Ramp2) Causes Reduced Fertility, Hyperprolactinemia, Skeletal Abnormalities, and Endocrine Dysfunction in Mice. Molecular Endocrinology. 2011;25:1244–53. doi: 10.1210/me.2010-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LMF, Caron KM. G-protein Coupled Receptor 30 Interacts with Receptor Activity Modifying Protein 3 and Confers Sex-Dependent Cardioprotection. Journal of molecular endocrinology. 2013;51:191–202. doi: 10.1530/JME-13-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annual review of pharmacology and toxicology. 2013;53:531–56. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014;35:12–22. doi: 10.1016/j.tips.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov Today. 2005;10:417–27. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 47.Barwell J, Gingell JJ, Watkins HA, Archbold JK, Poyner DR, Hay DL. Calcitonin and calcitonin receptor-like receptors: common themes with family B GPCRs? Br J Pharmacol. 2012;166:51–65. doi: 10.1111/j.1476-5381.2011.01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grace CR, Perrin MH, Gulyas J, Digruccio MR, Cantle JP, et al. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci U S A. 2007;104:4858–63. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grace CR, Perrin MH, Gulyas J, Rivier JE, Vale WW, Riek R. NMR structure of the first extracellular domain of corticotropin releasing factor receptor 1 (ECD1-CRF-R1) complexed with a high affinity agonist. J Biol Chem. 2010;285:38580–9. doi: 10.1074/jbc.M110.121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Pioszak A, Zhang C, Swaminathan K, Xu HE. Crystal structure of the PAC1R extracellular domain unifies a consensus fold for hormone recognition by class B G-protein coupled receptors. PloS one. 2011;6:e19682. doi: 10.1371/journal.pone.0019682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pal K, Swaminathan K, Xu HE, Pioszak AA. Structural basis for hormone recognition by the Human CRFR2{alpha} G protein-coupled receptor. J Biol Chem. 2010;285:40351–61. doi: 10.1074/jbc.M110.186072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parthier C, Kleinschmidt M, Neumann P, Rudolph R, Manhart S, et al. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc Natl Acad Sci U S A. 2007;104:13942–7. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pioszak AA, Harikumar KG, Parker NR, Miller LJ, Xu HE. Dimeric arrangement of the parathyroid hormone receptor and a structural mechanism for ligand-induced dissociation. J Biol Chem. 2010;285:12435–44. doi: 10.1074/jbc.M109.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pioszak AA, Parker NR, Gardella TJ, Xu HE. Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides. J Biol Chem. 2009;284:28382–91. doi: 10.1074/jbc.M109.022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pioszak AA, Parker NR, Suino-Powell K, Xu HE. Molecular recognition of corticotropin-releasing factor by its G-protein-coupled receptor CRFR1. J Biol Chem. 2008;283:32900–12. doi: 10.1074/jbc.M805749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci U S A. 2008;105:5034–9. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Runge S, Thogersen H, Madsen K, Lau J, Rudolph R. Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J Biol Chem. 2008;283:11340–7. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- 58.Underwood CR, Garibay P, Knudsen LB, Hastrup S, Peters GH, et al. Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J Biol Chem. 2010;285:723–30. doi: 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–43. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 60.Siu FY, He M, de Graaf C, Han GW, Yang D, et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–9. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fitzsimmons TJ, Zhao X, Wank SA. The extracellular domain of receptor activity-modifying protein 1 is sufficient for calcitonin receptor-like receptor function. J Biol Chem. 2003;278:14313–20. doi: 10.1074/jbc.M211946200. [DOI] [PubMed] [Google Scholar]
- 62.Udawela M, Christopoulos G, Tilakaratne N, Christopoulos A, Albiston A, Sexton PM. Distinct receptor activity-modifying protein domains differentially modulate interaction with calcitonin receptors. Mol Pharmacol. 2006;69:1984–9. doi: 10.1124/mol.105.021915. [DOI] [PubMed] [Google Scholar]
- 63.Koth CM, Abdul-Manan N, Lepre CA, Connolly PJ, Yoo S, et al. Refolding and characterization of a soluble ectodomain complex of the calcitonin gene-related peptide receptor. Biochemistry. 2010;49:1862–72. doi: 10.1021/bi901848m. [DOI] [PubMed] [Google Scholar]
- 64.ter Haar E, Koth CM, Abdul-Manan N, Swenson L, Coll JT, et al. Crystal structure of the ectodomain complex of the CGRP receptor, a class-B GPCR, reveals the site of drug antagonism. Structure. 2010;18:1083–93. doi: 10.1016/j.str.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, et al. Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci. 2012;21:199–210. doi: 10.1002/pro.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watkins HA, Au M, Bobby R, Archbold JK, Abdul-Manan N, et al. Identification of key residues involved in adrenomedullin binding to the AM1 receptor. Br J Pharmacol. 2013;169:143–55. doi: 10.1111/bph.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hill HE, Pioszak AA. Bacterial expression and purification of a heterodimeric adrenomedullin receptor extracellular domain complex using DsbC-assisted disulfide shuffling. Protein expression and purification. 2013;88:107–13. doi: 10.1016/j.pep.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moad HE, Pioszak AA. Selective CGRP and adrenomedullin peptide binding by tethered RAMP-calcitonin receptor-like receptor extracellular domain fusion proteins. Protein Sci. 2013;22:1775–85. doi: 10.1002/pro.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Archbold JK, Flanagan JU, Watkins HA, Gingell JJ, Hay DL. Structural insights into RAMP modification of secretin family G protein-coupled receptors: implications for drug development. Trends Pharmacol Sci. 2011;32:591–600. doi: 10.1016/j.tips.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Kusano S, Yokoyama S. Ectodomain structures of the CGRP and AM receptors. Current protein & peptide science. 2013;14:375–85. doi: 10.2174/13892037113149990054. [DOI] [PubMed] [Google Scholar]
- 71.Harikumar KG, Simms J, Christopoulos G, Sexton PM, Miller LJ. Molecular Basis of Association of Receptor Activity-Modifying Protein 3 with the Family B G Protein-Coupled Secretin Receptor. Biochemistry. 2009;48:11773–85. doi: 10.1021/bi901326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rist B, Entzeroth M, Beck-Sickinger AG. From micromolar to nanomolar affinity: a systematic approach to identify the binding site of CGRP at the human calcitonin gene-related peptide 1 receptor. Journal of medicinal chemistry. 1998;41:117–23. doi: 10.1021/jm970533r. [DOI] [PubMed] [Google Scholar]
- 73.Breeze AL, Harvey TS, Bazzo R, Campbell ID. Solution structure of human calcitonin gene-related peptide by 1H NMR and distance geometry with restrained molecular dynamics. Biochemistry. 1991;30:575–82. doi: 10.1021/bi00216a036. [DOI] [PubMed] [Google Scholar]
- 74.Carpenter KA, Schmidt R, von Mentzer B, Haglund U, Roberts E, Walpole C. Turn structures in CGRP C-terminal analogues promote stable arrangements of key residue side chains. Biochemistry. 2001;40:8317–25. doi: 10.1021/bi0102860. [DOI] [PubMed] [Google Scholar]
- 75.Perez-Castells J, Martin-Santamaria S, Nieto L, Ramos A, Martinez A, et al. Structure of micelle-bound adrenomedullin: a first step toward the analysis of its interactions with receptors and small molecules. Biopolymers. 2012;97:45–53. doi: 10.1002/bip.21700. [DOI] [PubMed] [Google Scholar]
- 76.Archbold JK, Flanagan JU, Watkins HA, Gingell JJ, Hay DL. Structural insights into RAMP modification of secretin family G protein-coupled receptors: implications for drug development. Trends in pharmacological sciences. 2011;32:591–600. doi: 10.1016/j.tips.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 77.Watkins HA, Walker CS, Ly KN, Bailey RJ, Barwell J, et al. Receptor activity-modifying protein-dependent effects of mutations in the calcitonin receptor-like receptor: implications for adrenomedullin and calcitonin gene-related peptide pharmacology. Br J Pharmacol. 2014;171:772–88. doi: 10.1111/bph.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watkins HA, Rathbone DL, Barwell J, Hay DL, Poyner DR. Structure-activity relationships for alpha-calcitonin gene-related peptide. Br J Pharmacol. 2013;170:1308–22. doi: 10.1111/bph.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qi T, Hay DL. Structure-function relationships of the N-terminus of receptor activity-modifying proteins. Br J Pharmacol. 2010;159:1059–68. doi: 10.1111/j.1476-5381.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore EL, Gingell JJ, Kane SA, Hay DL, Salvatore CA. Mapping the CGRP receptor ligand binding domain: tryptophan-84 of RAMP1 is critical for agonist and antagonist binding. Biochem Biophys Res Commun. 2010;394:141–5. doi: 10.1016/j.bbrc.2010.02.131. [DOI] [PubMed] [Google Scholar]
- 81.Qi T, Christopoulos G, Bailey RJ, Christopoulos A, Sexton PM, Hay DL. Identification of N-terminal receptor activity-modifying protein residues important for calcitonin gene-related peptide, adrenomedullin, and amylin receptor function. Mol Pharmacol. 2008;74:1059–71. doi: 10.1124/mol.108.047142. [DOI] [PubMed] [Google Scholar]
- 82.Qi T, Ly K, Poyner DR, Christopoulos G, Sexton PM, Hay DL. Structure-function analysis of amino acid 74 of human RAMP1 and RAMP3 and its role in peptide interactions with adrenomedullin and calcitonin gene-related peptide receptors. Peptides. 2011;32:1060–7. doi: 10.1016/j.peptides.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 83.Gingell JJ, Qi T, Bailey RJ, Hay DL. A key role for tryptophan 84 in receptor activity-modifying protein 1 in the amylin 1 receptor. Peptides. 2010;31:1400–4. doi: 10.1016/j.peptides.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 84.Barwell J, Conner A, Poyner DR. Extracellular loops 1 and 3 and their associated transmembrane regions of the calcitonin receptor-like receptor are needed for CGRP receptor function. Biochim Biophys Acta. 2011;1813:1906–16. doi: 10.1016/j.bbamcr.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woolley MJ, Watkins HA, Taddese B, Karakullukcu ZG, Barwell J, et al. The role of ECL2 in CGRP receptor activation: a combined modelling and experimental approach. Journal of the Royal Society, Interface/the Royal Society. 2013;10:20130589. doi: 10.1098/rsif.2013.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuwasako K, Hay DL, Nagata S, Hikosaka T, Kitamura K, Kato J. The third extracellular loop of the human calcitonin receptor-like receptor is crucial for the activation of adrenomedullin signalling. Br J Pharmacol. 2012;166:137–50. doi: 10.1111/j.1476-5381.2011.01803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hay DL, Harris PW, Kowalczyk R, Brimble MA, Rathbone DL, et al. Structure-activity relationships of the N-terminus of calcitonin gene-related peptide: key roles of alanine-5 and threonine-6 in receptor activation. Br J Pharmacol. 2014;171:415–26. doi: 10.1111/bph.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barwell J, Gingell JJ, Watkins HA, Archbold JK, Poyner DR, Hay DL. Calcitonin and calcitonin receptor-like receptors: common themes with family B GPCRs? British journal of pharmacology. 2012;166:51–65. doi: 10.1111/j.1476-5381.2011.01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woolley MJ, Watkins HA, Taddese B, Karakullukcu ZG, Barwell J, et al. The role of ECL2 in CGRP receptor activation: a combined modelling and experimental approach. Journal of the Royal Society, Interface/the Royal Society. 2013;10:20130589. doi: 10.1098/rsif.2013.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vohra S, Taddese B, Conner AC, Poyner DR, Hay DL, et al. Similarity between class A and class B G-protein-coupled receptors exemplified through calcitonin gene-related peptide receptor modelling and mutagenesis studies. Journal of the Royal Society, Interface/the Royal Society. 2013;10:20120846. doi: 10.1098/rsif.2012.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Conner M, Hicks MR, Dafforn T, Knowles TJ, Ludwig C, et al. Functional and Biophysical Analysis of the C-Terminus of the CGRP-Receptor; a Family B GPCR†. Biochemistry. 2008;47:8434–44. doi: 10.1021/bi8004126. [DOI] [PubMed] [Google Scholar]
- 92.Conner AC, Simms J, Conner MT, Wootten DL, Wheatley M, Poyner DR. Diverse Functional Motifs within the Three Intracellular Loops of the CGRP1 Receptor†. Biochemistry. 2006;45:12976–85. doi: 10.1021/bi0615801. [DOI] [PubMed] [Google Scholar]
- 93.Conner AC, Simms J, Howitt SG, Wheatley M, Poyner DR. The Second Intracellular Loop of the Calcitonin Gene-related Peptide Receptor Provides Molecular Determinants for Signal Transduction and Cell Surface Expression. J Biol Chem. 2006;281:1644–51. doi: 10.1074/jbc.M510064200. [DOI] [PubMed] [Google Scholar]
- 94.Conner AC, Hay DL, Simms J, Howitt SG, Schindler M, et al. A Key Role for Transmembrane Prolines in Calcitonin Receptor-Like Receptor Agonist Binding and Signalling: Implications for Family B G-Protein-Coupled Receptors. Mol Pharmacol. 2005;67:20–31. [PubMed] [Google Scholar]
- 95.Kuwasako K, Kitamura K, Nagata S, Hikosaka T, Kato J. Structure–function analysis of helix 8 of human calcitonin receptor-like receptor within the adrenomedullin 1 receptor. Peptides. 2011;32:144–9. doi: 10.1016/j.peptides.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 96.Kuwasako K, Kitamura K, Nagata S, Hikosaka T, Kato J. Function of the cytoplasmic tail of human calcitonin receptor-like receptor in complex with receptor activity-modifying protein 2. Biochemical and Biophysical Research Communications. 2010;392:380–5. doi: 10.1016/j.bbrc.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 97.Bailey RJ, Hay DL. Agonist-dependent consequences of proline to alanine substitution in the transmembrane helices of the calcitonin receptor. Br J Pharmacol. 2007;151:678–87. doi: 10.1038/sj.bjp.0707246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benítez-Páez A. Sequence Analysis of the Receptor Activity-Modifying Proteins Family, New Putative Peptides and Structural Conformation Inference. In Silico Biology. 2006;6:467–83. [PubMed] [Google Scholar]
- 99.Nag K, Sultana N, Kato A, Dranik A, Nakamura N, et al. Ligand-induced internalization, recycling, and resensitization of adrenomedullin receptors depend not on CLR or RAMP alone but on the receptor complex as a whole. General and Comparative Endocrinology. 2015;212:156–62. doi: 10.1016/j.ygcen.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 100.Nag K, Sultana N, Hirose S. Calcitonin receptor-like receptor (CLR) influences posttranslational events of receptor activity-modifying proteins (RAMPs) Biochemical and Biophysical Research Communications. 2012;418:824–9. doi: 10.1016/j.bbrc.2012.01.116. [DOI] [PubMed] [Google Scholar]
- 101.Nag K, Kato A, Nakada T, Hoshijima K, Mistry AC, et al. Molecular and functional characterization of adrenomedullin receptors in pufferfish. 2006:R467–R78. doi: 10.1152/ajpregu.00507.2005. [DOI] [PubMed] [Google Scholar]
- 102.Cohen SP, Haack KKV, Halstead-Nussloch GE, Bernard KF, Hatt H, et al. Identification of RL-TGR, a coreceptor involved in aversive chemical signaling. Proceedings of the National Academy of Sciences. 2010;107:12339–44. doi: 10.1073/pnas.1000343107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibbons C, Dackor R, Dunworth W, Fritz-Six K, Caron KM. Receptor Activity-Modifying Proteins: RAMPing up Adrenomedullin Signaling. Mol Endocrinol. 2007;21:783–96. doi: 10.1210/me.2006-0156. [DOI] [PubMed] [Google Scholar]
- 104.Bailey RJ, Hay DL. Pharmacology of the human CGRP1 receptor in Cos 7 cells. Peptides. 2006;27:1367–75. doi: 10.1016/j.peptides.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 105.Tilakaratne N, Christopoulos G, Zumpe ET, Foord SM, Sexton PM. Amylin receptor phenotypes derived from human calcitonin receptor/RAMP coexpression exhibit pharmacological differences dependent on receptor isoform and host cell environment. In. The Journal of pharmacology and experimental therapeutics. 2000:61–72. [PubMed] [Google Scholar]
- 106.Qi T, Dong M, Watkins HA, Wootten D, Miller LJ, Hay DL. Receptor activity-modifying protein-dependent impairment of calcitonin receptor splice variant Delta(1–47)hCT((a)) function. British journal of pharmacology. 2013;168:644–57. doi: 10.1111/j.1476-5381.2012.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.