Summary
Coordinated expression of mitochondrial and nuclear genes is required to maintain proper mitochondrial function. However, the precise mechanisms that ensure this coordination are not well defined. We find that signaling from mitochondria to the nucleus is influenced by mammalian target of rapamycin (mTOR) activity via changes in autophagy and p62/SQSTM1 turnover. Reducing mTOR activity increases autophagic flux, enhances mitochondrial membrane potential, reduces reactive oxygen species within the cell, and increases replicative life span. These effects appear to be mediated in part by an interaction between p62/SQSTM1 and Keap1. This interaction allows nuclear accumulation of the nuclear factor erythroid 2-like 2 (NFE2L2, also known as nuclear factor related factor 2 or NRF2), increased expression of the nuclear respiratory factor 1 (NRF1), and increased expression of nuclear-encoded mitochondrial genes, such as the mitochondrial transcription factor A, and mitochondrial-encoded genes involved in oxidative phosphorylation. These findings reveal a portion of the intracellular signaling network that couples mitochondrial turnover with mitochondrial renewal to maintain homeostasis within the cell and suggest mechanisms whereby a reduction in mTOR activity may enhance longevity.
Keywords: mammalian target of rapamycin, mitochondria, senescence, rapamycin
Introduction
Impaired mitochondrial function is associated with aging and age-related disease, and decreased mitochondrial function can have a significant impact on ATP production, maintenance of NAD/NADH ratios, and reactive oxygen species (ROS) production, processes that are critical to cellular function. To preserve mitochondrial function and ensure that metabolic activity is adequately maintained, a decline or loss of mitochondrial membrane potential acts as a trigger to promote fission of damaged mitochondrial regions. These regions are subsequently recycled through fusion events or removed via autophagy (Twig et al., 2008a,b; Westermann, 2010). However, the clearance of mitochondrial components via autophagy must be balanced by mitochondrial gene expression, which maintains mitochondrial mass, and it would be expected that these two processes would be linked.
Mitochondrial biogenesis involves the expression of genes in both the nuclear and mitochondrial genomes. In the yeast Saccharomyces cerevisiae, a retrograde signaling response involving the transcription factors Rtg1p and Rtg3p is induced following mitochondrial damage (Parikh et al., 1987). Impaired mitochondria activate the transcription factors Rtg1p and Rtg3p, whose activity is modulated by the nutrient-sensing target of rapamycin (TOR) complex (Liu & Butow, 2006). Because TOR activity also modulates autophagy, this process links autophagy to mitochondrial biogenesis in yeast.
In mammalian cells, the mitochondrial retrograde response is not well defined. However, critical factors that regulate mitochondrial components have been identified. For example, the transcription activators nuclear respiratory factor 1 and GA-binding protein transcription factor (NRF1 and GABPA, also known as NRF2) regulate expression of the nuclear-encoded mitochondrial genes as well as the mitochondrial transcription factor (TFAm), which is required for both mitochondrial DNA replication and mitochondrial gene expression (Scarpulla, 2008). Additional transcription factors regulating mitochondria-related gene expression include the estrogen receptor-related receptor (ERR), yin-yang 1 (YY1), and the cyclic AMP response element binding protein (CREB; Hock & Kralli, 2009). In addition, the structurally related transcription coactivators PGC-1α, PGC-1β, and PRC can increase the expression of nuclear-encoded mitochondrial genes (Wu et al., 1999). However, the mechanisms linking these nuclear factors to mitochondrial turnover are not well defined.
In yeast, the TOR intracellular signaling cascade has been implicated in the control of mitochondrial function and autophagy. The TOR pathway (mTOR in mammalian systems) is an evolutionarily conserved intracellular network that serves to couple nutrient availability with cellular metabolism and cell division (Ma & Blenis, 2009). Two distinct complexes containing mTOR (mTORC1 and mTORC2) exist in mammalian cells and are distinguished by differential sensitivity to inhibition by rapamycin. The mTORC1 complex is primarily involved in the regulation of protein translation and growth (Ma & Blenis, 2009). It has been reported that mTOR activity is required to maintain oxidative phosphorylation (Schieke et al., 2006; Cunningham et al., 2007) and a chronic suppression of the mTOR complex over longer periods may also improve mitochondrial function (Bitto et al., 2010). In the current study, we examined the effects of reduced mTOR activity in both quiescent and proliferating nonimmortalized human fibroblasts to identify a novel mechanism linking mitochondrial clearance with mitochondrial gene expression. This link involves an interaction between p62/SQSTM1 and Keap1, which is driven by an increased rate of autophagy. This association promotes the accumulation of nuclear factor erythroid 2-like 2 (NFE2L2, also known as Nrf2) in the nucleus and increases mitochondrial gene expression, potentially through expression of NRF1 and the mitochondrial transcription factor TFAm. This process provides physiologic benefit to the cell as demonstrated by enhanced mitochondrial membrane potential, resistance to oxidative stress, and a delay in the onset of replicative senescence.
Results
Long-term exposure to rapamycin increases mitochondrial gene expression and reduces cellular ROS in quiescent human fibroblasts
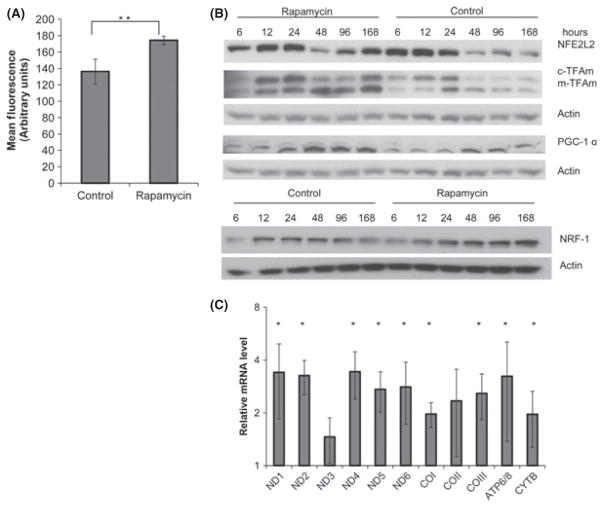
We have previously reported that inhibition of mTOR with rapamycin provides relief from stress induced by unopposed IGF-1 signaling (Bitto et al., 2010) and considered the possibility that this reduced cellular stress may be due to changes in mitochondrial homeostasis. To explore this possibility, we exposed quiescent human WI-38 fibroblasts to rapamycin over a period of 7 days and examined both mitochondrial mass and the levels of proteins involved in the regulation of mitochondrial gene expression. Mitochondrial mass was increased by 28% in cells exposed to 10 nM rapamycin relative to controls (Fig. 1A). We next examined the steady-state mRNA and protein levels of genes involved in mitochondrial biogenesis including NRF1, PGC-1α, TFAm, and NFE2L2. Steady-state protein levels were stabilized over a period of 72–96 h following the addition of rapamycin and were consistently elevated or maintained in rapamycin-treated cultures relative to control cells after this initial period (Fig. 1B, densitometry measurements presented in Fig. S2A). Additionally, mRNA levels of PGC-1, TFAm, and NFE2L2 were also elevated in cultures exposed to rapamycin (Fig. S2B). Furthermore, the mRNA levels of several mitochondrial DNA-encoded genes that are known targets of TFAm are increased in response to rapamycin treatment (Fig. 1C). Together, these data suggest that prolonged exposure to rapamycin increases the steady-state levels of several proteins involved with mitochondrial biogenesis and at least a subset of mitochondrial-encoded genes in human fibroblasts.
Fig. 1.
Inhibition of mammalian target of rapamycin (mTOR) promotes the expression of mitochondrial genes. WI-38 fibroblasts were placed into serum-free medium (MCDB-105) in the presence or absence of rapamycin (10 nM). (A) Mitochondrial mass in WI-38 fibroblasts. MitoTracker Green-associated fluorescence as measured by flow cytometry is presented in cells grown in MCDB-105 with or without 10 mM rapamycin for 2 weeks. Bars are average ± standard deviation of three independent samples. (B) Representative immunoblot examining steady-state protein levels of NFE2L2, NRF1 and TFAm in quiescent cells maintained in MCDB-105 with or without 10 mM rapamycin. Total protein lysates were analyzed at the indicated time point. (C) QRT-PCR of mitochondrial-encoded genes after 6 days’ treatment with 10 mM rapamycin. Bars are average ± standard deviation of three independent experiments. In all cases, measurements that differ significantly (P < 0.05) are marked with an asterisk. Differences marked with two asterisks are significant at the P < 0.01 level. Densitometry for the Western blot is provided in Fig. S2 (Supporting information).
Long-term exposure to rapamycin increases life span of human fibroblast cells
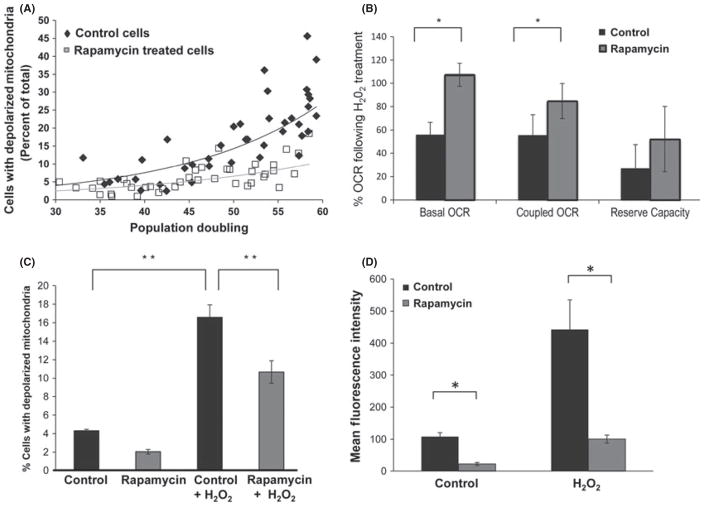
We next examined the influence of rapamycin on mitochondrial membrane potential during replicative life span of human fibroblasts. The concentration of rapamycin was reduced in this set of experiments (1 vs. 10 nM in the experiments outlined in Fig. 1) to allow cell proliferation while maintaining an inhibition of mTOR activity as judged by S6 phosphorylation (demonstrated in Fig. 3A,B). Cell cultures grown under standard conditions showed an accumulation of cells with depolarized mitochondria over time (Fig. 2A, Spearman’s ρ = 0.855, P = 0.002). In contrast, cultures grown in medium supplemented with rapamycin preserved mitochondrial membrane potential (Fig. 2A, Spearman’s ρ = 0.224, P = 0.533). Although there was an increase in the number of cells which had lost mitochondrial membrane potential with increased population doublings, the percentage of the total population remained far below the control cultures. Live cell imaging of JC-1-stained cells revealed that in cultures treated with rapamycin, cells maintained a more polarized mitochondrial network compared with control cultures (see Fig. S3A for examples of mitochondrial staining by flow cytometry and Fig. S3B for examples of live cell fluorescent microscopy). Similar results were obtained by flow cytometry using either JC-1 or TMRE as indicators of mitochondrial membrane potential (TMRE data not shown). We next examined oxygen consumption rates and extracellular acidification, as indirect measures of mitochondrial activity in control and rapamycin-treated cultures. Mitochondrial respiration rate was reduced by rapamycin treatment (Fig. S3C) and extracellular acidification was increased, consistent with enhanced glycolysis (not shown). However, cells grown in rapamycin-supplemented medium retained higher levels of both basal and ATP-coupled mitochondrial respiration when challenged with H2O2 (Fig. 2B). In addition, rapamycin-treated cultures were resistant to mitochondrial depolarization from exogenous H2O2 exposure (Fig. 2C). Consistent with a reduction in intracellular stress, ROS levels were reduced in rapamycin-treated cells (Fig. 2D).
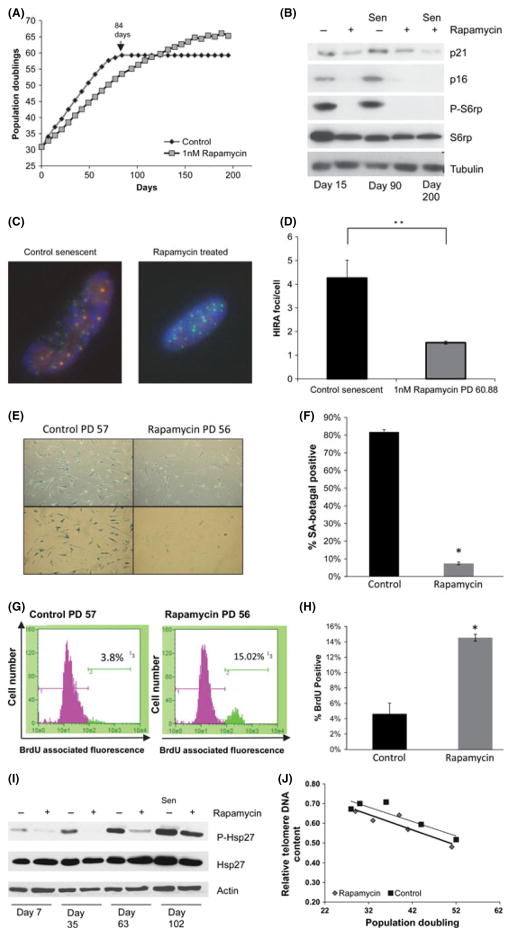
Fig. 3.
Rapamycin delays the onset of senescence and reduces the presence of senescence-associated markers. (A) Cumulative population doubling (CPD) curve of control and 1 nM rapamycin-supplemented WI-38 fibroblasts. Arrow indicates onset of senescence in control cells after 84 days in culture. The curve is representative of three independent experiments (see Fig. S5 for additional life span curves). Cells were cultured in control medium or medium supplemented with 1 nM rapamycin for life span analysis as described in Methods. Rapamycin was continuously present in culture medium during these analyses. (B) Levels of p21Cip1, p16INK4A, phospho-S6, and total S6 assayed by Western blot on proliferating cells and senescent cells. ‘Sen’: Senescent cells. (C) Representative fluorescence micrograph of HIRA/PML foci in control and CPD-matched rapamycin-supplemented cells. Blue: DAPI, Green: PML, Red: HIRA. (D) Number of HIRA/PML foci per cell in senescent and CPD-matched rapamycin-supplemented cells. Bars are average ± standard deviation of three independent samples (**P < 0.01). (E) Representative SA beta-gal staining in control and rapamycin-treated cultures at CPD 57 and CPD 56, respectively. Control cultures have entered senescence at this point. (F) Quantification of SA beta-gal-positive cells in control and rapamycin-treated cultures. (G) Representative flow cytometry analysis of control and rapamycin-treated cultures at CPD 57 and CPD 56, respectively, following BRDU labeling to measure percentage of cells in S phase. (H) Quantification of BRDU staining in control and rapamycin-treated cultures at CPD 57 and CPD 56, respectively. (I) Levels of phospho-Hsp27 and total Hsp27 assayed by Western blot on proliferating and senescent cells. (J) Relative telomere repeat content in control and rapamycin-treated cultures as a function of life span. Telomere DNA content quantified relative to a single gene locus is plotted against population doublings.
Fig. 2.
Improved mitochondrial profile upon inhibition of mammalian target of rapamycin (mTOR) with rapamycin. (A) Mitochondrial membrane potential was assessed by JC-1 staining and flow cytometry at the indicated time points during replicative life span of cells grown with or without 1 nM rapamycin (Life span curve presented in Fig. 3). The percentage of cells with depolarized mitochondria was calculated at each population doubling as described in Methods and in the legend for Fig. S3 (Supporting information). Vehicle-treated cells show a significant increase in the number of cells with reduced mitochondrial membrane potential with increasing population doubling (Spearman’s ρ = 0.855, P < 0.01), while the correlation between population doublings and percent of cells with depolarized mitochondria was not significant in cells maintained in the presence of 1 nM rapamycin (P = 0.533). WI-38 fibroblast cells were maintained in complete growth media for life span analysis as described in Methods. Cells were maintained in either normal growth media or in media containing rapamycin (1 nM). Rapamycin was present in culture media at all times during propagation. (B) Relative oxygen-coupled respiration (OCR) of cells growing in control and 1 nM rapamycin-supplemented medium upon exposure to H2O2 (200 μM for 30 min). Basal, ATP-coupled, and reserve OCR were measured using a Seahorse XF Analyzer. An average of three independent measurements is presented. C) Mitochondrial membrane potential of vehicle and rapamycin-treated cells 24 hours after exposure to H2O2 (100μM for 60 minutes) as measured by JC-1 staining. Mitochondrial membrane potential was determined as described in Methods and in the legend for Figure S3. The percentage of cells with depolarized mitochondria within the population are presented. Bars are average +/− standard deviation of 3 independent samples. (D) Accumulation of ROS as measured by DCFDA staining and fluorescence spectroscopy. Bars are average +/− standard deviation of 3 independent samples normalized for protein content. In all cases, measurements that differ significantly (P < 0.05) are marked with an asterisk. Differences marked with two asterisks are significant at the P < 0.01 level.
We also examined the life span of cells grown in the presence of rapamycin compared to control (Fig. 3). Addition of rapamycin increased the maximum life span of WI-38 fibroblasts by 14%, a result which was consistent in three independent trials (Fig. 3A, S5A). To more clearly define the effect of rapamycin on the senescent arrest, several cellular events associated with senescence were examined. The protein levels of both p21Cip1/Waf1 and p16INK4a were reduced in rapamycin-treated cultures at high cumulative population doubling levels relative to controls (Fig. 3B). Furthermore, chromatin remodeling events associated with senescence (Adams, 2007) were reduced, as evidenced by a decreased localization of the histone chaperone HIRA to PML bodies (Fig. 3C,D). The number of cells positive for senescence-associated beta-galactosidase (SA beta-gal) activity at high population doublings was decreased by rapamycin (Fig. 3E, F). Additionally, the percentage of cells that continued to incorporate BrdU in the rapamycin-treated cultures was increased relative to control cultures as they approached senescence (Fig. 3G,H). These findings are consistent with increased proliferation at late passages in the rapamycin-treated cultures relative to the control cultures.
We postulated that the alleviation of mitochondrial dysfunction and endogenous ROS production through rapamycin treatment might reduce the endogenous stress response and that this might underlie the increased life span observed in rapamycin-treated cultures. The p38 MAPK is a marker of endogenous stress associated with senescence arrest in response to telomere attrition, and genotoxic and oxidative stress (Wang et al., 2002; Iwasa et al., 2003). To determine the activity of the p38 MAPK pathway, we measured the phosphorylation status of the downstream target HSP27 (Beyaert et al., 1996) at early, middle, and late passage. As expected, human fibroblasts exhibited increased phosphorylation of HSP27 during life span, consistent with an activation of the p38 MAPK. Conversely, cells treated with rapamycin showed a reduced level of HSP27 phosphorylation relative to controls until the end of their life span (Fig. 3I), suggesting a reduced activity of p38 MAPK. Consistent with reduced p38 MAPK activity, the production of IL-6, part of the SASP known to be dependent upon p38 (Freund et al., 2011), was significantly reduced in rapamycin-treated cultures relative to controls (1 ng mL−1 vs. > 2.5 ng mL−1 respectively, P < 0.05).
If rapamycin attenuates endogenous stress associated with mitochondrial function, one might predict that telomere attrition would proceed in a manner similar to control cultures, in cells exposed to rapamycin. To test this possibility, telomere length was examined using quantitative PCR (Cawthon, 2002) during life span in cultures grown in control medium or medium supplemented with 1 nM rapamycin (Fig. 3J). Telomere DNA content decreased with increasing population doublings in both control and rapamycin-treated cultures. This suggests that the observed increase in replicative life span with rapamycin treatment was not due to an attenuation of telomere attrition.
Autophagy and p62/SQSTM1 mediate the mitochondrial response induced by rapamycin
Levels of LC3 and the accumulation of LC3 puncta were examined in cultures grown in the presence of rapamycin to determine whether the rate of autophagy was altered under these conditions. Exposure of these cultures to lysosomal inhibitors revealed that LC3 accumulated in both control and rapamycin-treated cultures consistent with an ongoing autophagy process. However, the relative increase in LC3 was greater in the rapamycin-treated cultures, suggesting increased autophagic flux (Fig. S4A). We then examined the presence of LC3 puncta in WI-38 cells transduced with a viral expression vector that produces an LC3-GFP fusion protein. The number of LC3 puncta was significantly greater in rapamycin-treated cells (Fig. S4B, S4C), further supporting an increase in autophagy in cells exposed to rapamycin.
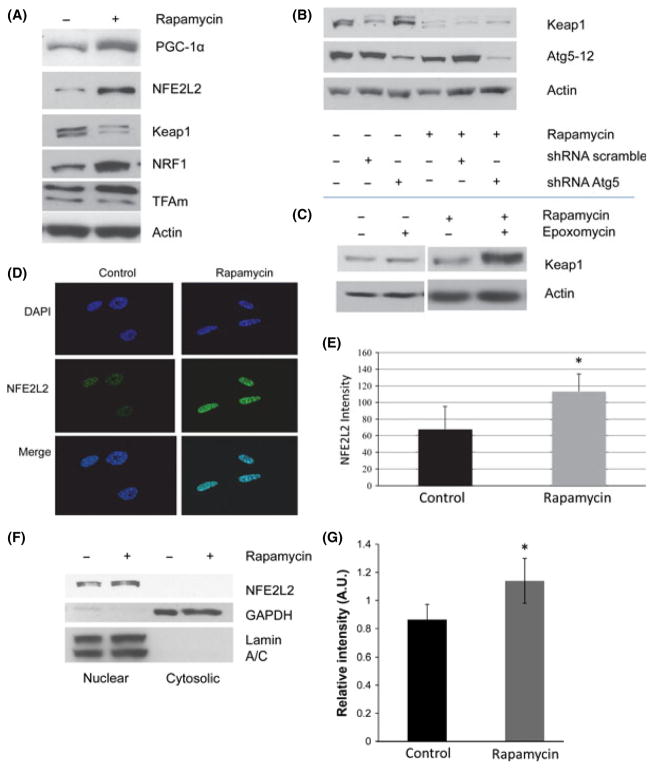
Next, we examined PGC-1α, Keap1, NFE2L2, NRF1, and TFAm expression in the human fibroblasts throughout their replicative life span. Similar to the quiescent cells examined in Fig. 1, we found increased steady-state levels of NFE2L2, NRF1, PGC-1α, and TFAm proteins in cells exposed to rapamycin while Keap1 levels were reduced (Fig. 4A). The steady-state mRNA levels of NFE2L2, NRF1, and PGC-1α were also increased while Keap1 mRNA levels were unaffected (Fig. S3D–S3G). To determine why Keap1 protein levels were decreased despite no change in mRNA level, we examined the role of both the proteasome and the autophagy pathway in modulating Keap1 levels in our system. Interestingly, Keap1 levels increased following the inhibition of either autophagy (Fig. 4B) or proteasome activity (Fig. 4C), suggesting that either pathway may degrade Keap1. However, in cultures exposed to rapamycin, proteasome inhibition had a more pronounced effect on Keap1 levels, while in control cultures targeting autophagy by ATG5 inhibition had a stronger effect. Based on the known interaction between Keap1 and NFE2L2 (Motohashi & Yamamoto, 2004), we examined the nuclear localization of NFE2L2 in rapamycin-treated cultures using both indirect immunofluorescence (Fig. 4D,E) and biochemical fractionation of nuclear proteins (Fig. 4F,G). Both methods indicated that nuclear localization of NFE2L2 increased in cultures treated with rapamycin. Interestingly, both the biochemical fractionation and the immunofluorescence analysis revealed that the majority of NFE2L2 is localized to the nucleus in WI-38 cells, suggesting that the differences in NFE2L2 abundance we observed in total cell lysates reflects differences in active NFE2L2.
Fig. 4.
Inhibition of mammalian target of rapamycin (mTOR) increases the levels of proteins associated with mitochondrial biogenesis and activation of NFE2L2. (A) Representative Western blots of the steady-state levels of PGC-1α, TFAm, NFE2L2, NRF1, and Keap1 in cultures grown in the presence of 1 nM rapamycin during the life span experiments outlined in Fig. 2. Cells were grown in the presence of 1 nM rapamycin for 10–14 days prior to analysis and were then continued for life span analysis as in Fig. 3. Similar increases in these proteins have been obtained in at least three independent measurements at different points during life span. (B) Representative levels of Keap1 following knockdown of Atg5. Analysis was performed 10 days following transduction with lentiviral vectors expressing shRNA targeting ATG5. (C) Representative levels of Keap1 following 24-h treatment with 10 nM epoxomicin. Results shown are representative of two independent experiments. (D) Representative fluorescence micrographs of NFE2L2 localization in vehicle and rapamycin-treated cells. (E) Quantification of Panel C. Bars are average ± standard deviation of three independent samples. At least 200 cells per sample were analyzed (**P < 0.01). (F) Steady-state levels of NFE2L2 in nuclear and cytoplasmic extracts of WI-38 fibroblasts grown in the presence or absence of rapamycin. Equal protein loading was verified using GAPDH for cytosolic extracts and lamin A/C for nuclear extracts. (G) Densitometric analysis of Western blot analysis from Panel F. Results are representative of three independent experiments. Differences marked with an asterisk are significantly different from control at P < 0.05.
Rapamycin increases p62 turnover through the autophagy machinery
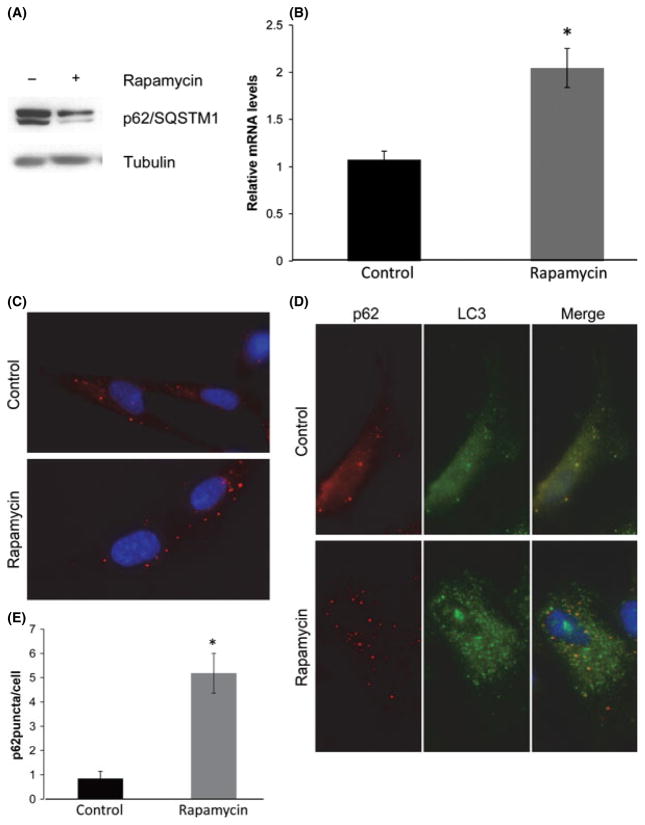
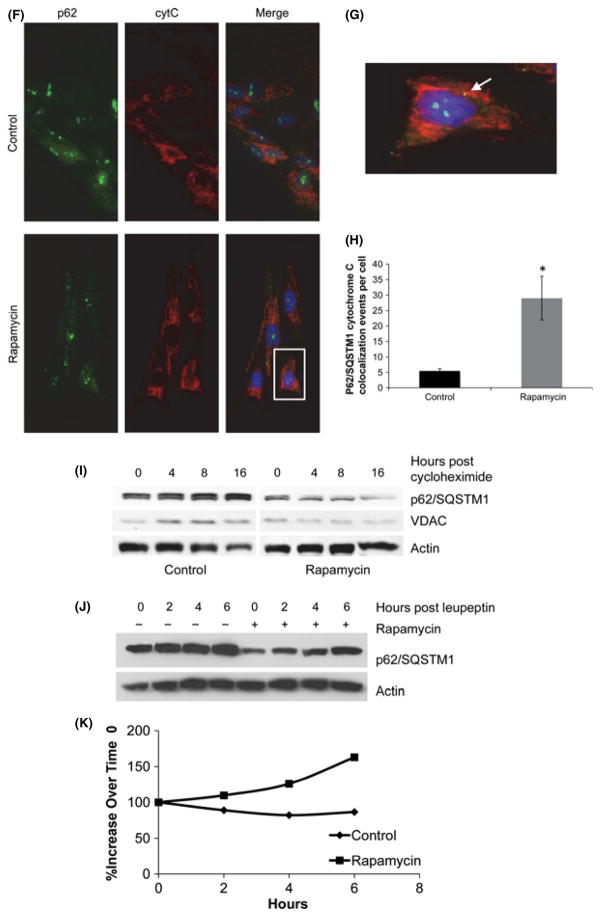
The autophagy adaptor p62/SQSTM1 has been involved in the clearance of depolarized mitochondria (Geisler et al., 2010) and in the activation of NFE2L2 (Komatsu et al., 2010). To determine whether p62/SQSTM1 is required for the changes in mitochondrial membrane potential and gene expression that occur in response to rapamycin, we first examined the steady-state levels of p62/SQSTM1 in cultures treated with rapamycin and controls. Protein levels of p62/SQSTM1 were reduced in rapamycin-treated cultures while mRNA levels were increased (Fig. 5A,B). Despite the reduced protein levels of p62/SQSTM1, rapamycin-treated cultures contained a number of prominent p62/SQSTM1-positive foci that were less pronounced and fewer in number in control cultures, and which colocalized with LC3 in all cases (Fig. 5C–E). In addition, we examined the colocalization of these p62/SQSTM1 puncta with cytochrome C. Rapamycin- treated cells contained a significantly greater number of cytochrome C-positive p62/SQSTM1 puncta compared to controls (Fig. 5F–H).
Fig. 5.
Rapamycin improves mitochondrial homeostasis by altering p62/SQSTM1 turnover. (A) Steady-state protein levels of p62/SQSTM1 in WI-38 fibroblasts grown in the presence or absence of rapamycin. (B) Relative mRNA levels of p62/SQSTM1 in vehicle and rapamycin-treated cells, measured by QRT-PCR. (C) Immunofluorescent staining for p62/SQSTM1 puncta in cells grown in the presence or absence of rapamycin. (D) Colocalization of p62/SQSTM1 and LC3 in WI-38 fibroblasts grown in the presence or absence of rapamycin. (E) Quantification of p62 puncta presented in Panel C. Immunostaining for p62SQSTM1 and LC3 was performed in three independent experiments and a minimum of 100 cells were examined for quantification of puncta. Differences that are significant (P < 0.05) are marked with an asterisk. (F) Colocalization of p62/SQSTM1 puncta and cytochrome C. (G) Enlarged area from Panel F demonstrating colocalization of p62/SQSTM1 and cytochrome C. (H) Quantification of the number of p62/SQSTM1/cytochrome C colocalization events in cells grown in growth medium without or with the addition of ramamycin (1 nM). (I) Representative Western blot of p62/SQSTM1 and VDAC upon exposure to cycloheximide in cultures maintained in standard culture medium with or without rapamycin (1 nM). (J) Representative Western blot of p62/SQSTM1 levels upon exposure to rapamycin and/or 0.1 mM leupeptin/20 mM NH4Cl. (K) Graphical representation of densitometry data depicting the rate of accumulation of p62/SQSTM1 in rapamycin-treated or control cultures following addition of leupeptin/20 mM NH4Cl.
We hypothesized that increased degradation of p62/SQSTM1 might account for reduced protein levels of p62/SQSTM1 in the cells treated with rapamycin and examined the half-life of the protein. The half-life of p62/SQSTM1 was dramatically reduced in cells that were exposed to rapamycin (Fig. 5I). The calculated half-life in cells exposed to rapamycin was 11.5 h, while in control cultures, the half-life was > 24 h. The half-life of VDAC, a resident mitochondrial protein, was decreased to 13 h in cultures exposed to rapamycin consistent with an increased turnover of mitochondrial components through mitophagy (see Table 1). In addition, treatment of cells with lysosomal inhibitors known to block autophagosome degradation resulted in a relatively greater accumulation of p62/SQSTM1 in cultures treated with rapamycin relative to cultures maintained under control conditions (Fig. 5J,K).
Table 1.
Half-life of p62/SQSTM1 and VDAC. Protein half-life was calculated from immunoblot analysis presented in Fig. 5I of cell grown in the presence or absence of rapamycin. Cultures were early passage, at population doubling 35 during these experiments.
| Protein half-life
|
||
|---|---|---|
| p62/SQSTM1 (h) | VDAC (h) | |
| Control | >24 | > 24 |
| Rapamycin | 11.5 | 13 |
p62/SQSTM1 protein complexes contain K63-linked ubiquitin conjugates
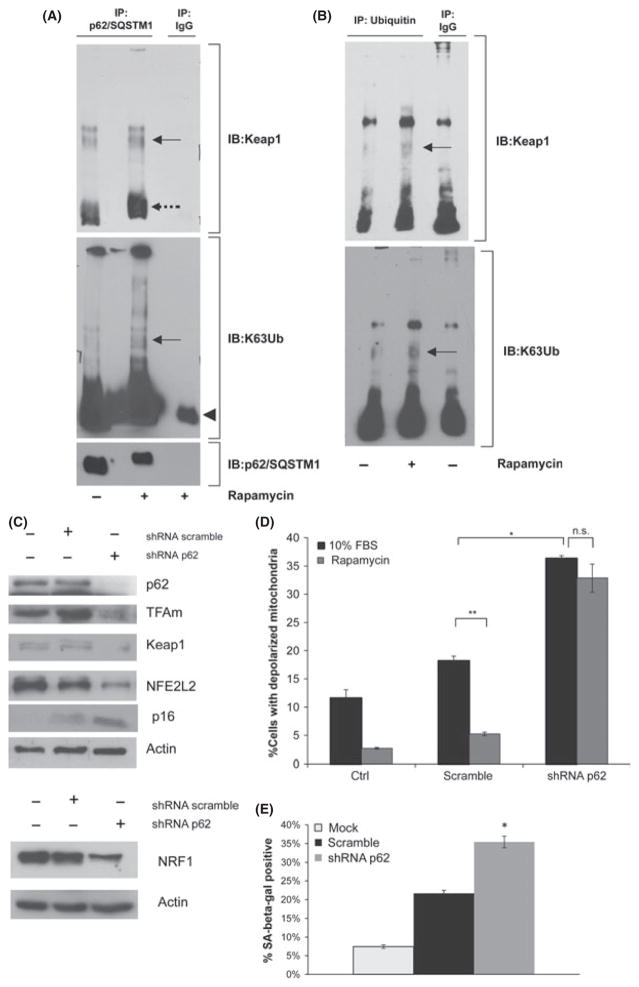
Given that rapamycin increases expression and turnover of p62/SQSTM1, we hypothesized that p62/SQSTM1 associates with Keap1, potentially allowing its degradation and activating NFE2L2. To determine whether p62/SQSTM1 and Keap1 associate when mTOR activity is reduced, we concentrated p62/SQSTM1 protein complexes by immunoprecipitation in cells treated with rapamycin and looked for the levels of Keap1 in the complex via Western blot (Fig. 6). A stronger Keap1 signal was detected associated with p62/SQSTM1 in cell lysates from cultures treated with rapamycin, despite the fact that the steady-state levels of p62/SQSTM1 are reduced in these cells relative to control cultures (Fig. 6A). Interestingly, the major band for Keap1 in the immunoprecipitate appeared to have a slightly higher molecular weight in the rapamycin-treated cultures which suggests that the protein might be modified in these cells (see Fig. 6A, dashed arrow). Furthermore, in rapamycin-treated cells, we also found enrichment in a Keap1-positive band associated with p62/SQSTM1 that migrated at a higher molecular weight than the major Keap1 protein detected in whole cell lysates (~80 kD; Fig. 6A, full arrow). This high molecular weight form of Keap1 can be detected with at least two commercial antibodies, suggesting it is not due to nonspecific interaction or antibody cross-reactivity. To determine whether this larger form of Keap1 might be the result of ubiquitin conjugation, we probed the immunoprecipitates with a set of linkage-specific anti-polyubiquitin antibodies. An 80-kD band appeared positive for K63-linked polyubiquitin in the p62/SQSTM1 complex in the rapamycin-treated cells, suggesting that this 80-kD Keap1 reactive band represented Keap1 that had been modified by ubiquitin conjugation with this specific linkage. Consistent with this interpretation, there was no reactivity with an antibody specific to the K-48 ubiquitin linkage (not shown). We next determined whether Keap1 could be detected when ubiquitin-conjugated proteins were concentrated. Proteins collected by immunoprecipitation using anti-polyubiquitin antibodies were probed by immunoblotting for the presence of Keap1. We were able to detect a higher level of Keap1 among the polyubiquitin-conjugated proteins isolated from rapamycin-treated cultures but not from the control cultures (Fig. 6B). Additionally, antibodies specific for the K-63 ubiquitin linkage detected a band of molecular weight similar to Keap1 among the polyubiquitin-conjugated proteins isolated from rapamycin-treated cultures but not from control cultures (Fig. 6B).
Fig. 6.
p62/SQSTM1 associates with K63- ubiquitin-conjugated Keap1 and is required for increased levels of mitochondrial biogenesis factors in response to rapamycin. (A). Representative Western blot of Keap1, K63-Ubiquitin, and p62/SQSTM1 in p62/SQSTM1 immunoprecipitates from control and rapamycin-treated cells. Full arrow: K63-Ubiquitin-associated Keap1 (~80 kD). Dashed arrow: Keap1 (~64 kD). Full arrowhead: IgG (~50 kD). (B) Representative Western blot of Keap1, K63-Ubiquitin, and p62/SQSTM1 in antiubiquitin immunoprecipitates from control and rapamycin-treated cells. Full arrow: K63-Ubiquitin-associated Keap1 (~80 kD). (C) Representative Western blot showing the steady-state levels of p62/SQSTM1, TFAm, Keap1, NFE2L2, and NRF1 in cells treated with 1 nM rapamycin for 2 weeks and infected with either a scramble sequence or an shRNA targeting p62/SQSTM1. (D) The percentage of cells with depolarized mitochondria was examined in cultures following targeted knockdown of p62/SQSTM1. Cells were infected with a lentiviral-targeting vector containing an shRNA that targets p62/SQSTM1 or an shRNA containing a scrambled sequence. Following selection with puromycin and 10 days’ acclimatization to rapamycin, cells were loaded with JC-1 stain, as described in material and methods, and analyzed using flow cytometry. Results shown represent an average of three independent samples (*P < 0.05, **P < 0.01, n.s. = not significant). (E) SA beta-gal activity in cells expressing an shRNA targeting p62/SQSTM1. Results are presented as the percentage of the population staining positive for SA beta-gal and are representative of two independent experiments. Cells harboring the p62/SQSTM1 shRNA were distinctly more positive than cells harboring the scramble shRNA. Differences that are significant at P < 0.05 are marked by an asterisk.
To test the role of p62/SQSTM1 in the mitochondrial changes induced by rapamycin, we stably reduced p62/SQSTM1 levels through the introduction of an shRNA targeting vector (Fig. 6C). Following p62/SQSTM1 reduction, the accumulation of cells with depolarized mitochondria and the levels of NFE2L2, NRF1, and TFAm were examined. Knockdown of p62/SQSTM1 greatly increased the subpopulation of cells with depolarized mitochondria (Fig. 6D), consistent with previously reported observations on the role of p62/SQSTM1 in mitochondrial clearance (Geisler et al., 2010). In addition, reduction of p62/SQSTM1 abrogated the beneficial effects of rapamycin on the mitochondrial profile (Fig. 6D) and reduced the protein levels of TFAm, NRF1, and NFE2L2 (Fig. 6C). In contrast, knockdown of NFE2L2 reduced the levels of TFAm and NRF1 without reducing the accumulation of cells with depolarized mitochondria (Fig. S6A, S6B). It was not possible to serially passage the shRNAp62-expressing cells for life span analysis. The growth of these cultures was initially suppressed, but after a period of 10–20 days, proliferation resumed and immunoblot analysis revealed a loss of p62/SQSTM1 silencing (not shown). However, we examined markers of senescence in the cells prior to 20 days and found an increase in p16 levels and a higher level of SA beta-gal staining (Fig. 6C,E).
Discussion
Our results suggest that a reduction in mTOR activity leads to increased turnover of p62/SQSTM1, allowing displacement of Keap1 from NFE2L2 and increased expression of NFE2L2 target genes. These results are consistent with the concept that the p62/SQSTM1 protein serves as a bridge between autophagosomes and cellular components destined for lysosomal degradation (Bjorkoy et al., 2005; Pankiv et al., 2007). However, p62/SQSTM1 also serves as a docking protein for signaling complexes that have a role in NFkB and mTOR signaling as well as apoptosis (Moscat & Diaz-Meco, 2009; Duran et al., 2011). Our results bridge these two functions of p62/SQSTM1 providing a physiologic feedback from mitochondrial clearance to mitochondrial biogenesis. The results are supported by previous studies indicating that NFE2L2 activation can occur through impairment of autophagy, due to an accumulation of p62/SQSTM1 (Komatsu et al., 2010), and that deletion of the Sqstm1 gene leads to an accumulation of Keap1 and inhibition of NFE2L2 in mice (Kwon et al., 2012). In addition, p62−/− mice show a striking loss of mitochondrial integrity and decreased life span (Kwon et al., 2012), while in Caenorhabditis elegans, SKN1/NFE2L2 has been implicated as a common regulator of life span extension in response to reduced mTOR signaling, starvation, and reduced INS/IGF signaling (Tullet et al., 2008; Paek et al., 2012; Robida-Stubbs et al., 2012). These observations are fully consistent with our findings, providing strong evidence that p62/SQSTM1 is required for mitochondrial integrity. Our results extend these observations demonstrating that reduced mTOR creates the proper conditions for Keap1 displacement from NFE2L2, potentially through clustering of p62SQSTM1 at sites of mitochondrial clearance. Interestingly, the route of Keap1 clearance appears to shift in the presence or absence of rapamycin and Keap1 levels decrease following targeting of p62 through shRNA (see Fig. 4B,C). These results suggest that the interaction between p62 and Keap1 serves to modulate Keap1 levels in a manner that is dependent upon multiple inputs.
Consistent with the association between p62/SQSTM1 and Keap1, we detect increased levels of K-63-linked ubiquitin conjugation of Keap1 in cells exposed to rapamycin. Using two separate antibodies to Keap1, we were able to visualize a high molecular weight form of Keap1 in association with p62/SQSTM1 that also reacts with an antibody specific for the K-63 ubiquitin linkage. Given the relatively greater affinity of p62/SQSTM1 for K-63-linked ubiquitin conjugates (Seibenhener et al., 2004), the results suggest that this modification of Keap1 provides an additional binding site for p62/SQSTM1 on Keap1, effectively disrupting the Keap1/NFE2L2 association. A model for the influence of reduced mTOR1 activity on mitochondria and life span in relation to proliferation rate is presented in Fig. S1 (Supporting information).
The connection between mitochondrial integrity and senescence appears to be a bidirectional relationship. Telomerase activity and the nontelomeric function of telomerase influence mitochondrial biogenesis and function potentially via a p53-mediated pathway (Sahin & DePinho, 2012), while dysfunctional mitochondrial have been shown to contribute to senescence in vitro and generate telomere attrition in vivo (Liu et al., 2002; Passos et al., 2007). Consistent with these observations, modeling theories predict that both mitochondrial and nuclear damage is likely to contribute to senescence (Sozou & Kirkwood, 2001), potentially involving p38 activity, which has been implicated as a common element in the senescence program (Iwasa et al., 2003). Thus, the increase in dysfunctional mitochondria that accumulate with senescence may be related to the gradual erosion of telomeres. Interestingly, both mTOR activity and autophagy have also been linked to the senescence arrest (Young et al., 2009; Kang et al., 2011; Leontieva et al., 2011) and the clearance of dysfunctional mitochondria may be one aspect of the connection between senescence and these processes.
A delay in the onset of senescence may have important implications at the organism level, given the long-lived phenotype of mice engineered to clear senescent cells as they arise (Baker et al., 2011). Senescent cells accumulate in the body with age and contribute to the aging phenotype. It is an intriguing hypothesis that a reduced accumulation of senescent cells, similar to our in vitro results, may contribute to the long-lived phenotype seen in mice treated with rapamycin (Harrison et al., 2009) or with reduced activity in signaling pathways upstream of mTOR, such as the GH/IGF-I axis (Bartke, 2011). Studies are beginning to emerge suggesting that rapamycin feeding may reduce senescence in mice (Hinojosa et al., 2012), although further work is required in this area. Future studies may reveal whether a reduction in the burden of cellular senescence contributes to the long life of these experimental models and may provide a target to ameliorate the aging process.
Methods
Cell culture
All culture reagents were from Cellgro (Manassas, VA, USA), unless otherwise stated. WI-38 human diploid fibroblasts were grown in MEM supplemented with 10% fetal bovine serum, 1% L-Glutamine, MEM nonessential amino acids, and MEM vitamins according to a standard culture protocol for life span analysis of human diploid fibroblasts (Cristofalo & Charpentier, 1980). For growth studies, rapamycin was added at 1 nM, a concentration that effectively reduces S6 phosphorylation but does not prevent cell growth (see Fig. 3). Studies using quiescent cells in MCDB105 in Fig. 1 were performed as described previously (Bitto et al., 2010). Briefly, cells were seeded at 1×104 cm−2 in complete growth medium that consisted of MEM containing 10% FBS (single lot MediaTech premium USA derived serum; endotoxin tested) and allowed to attach for 24–48 h. At this time, cells were placed into MCDB105 medium with no additions or with 10 nM rapamycin. Culture medium was replaced every 72 h for the duration of each experiment. Cell cycle analysis on cultures maintained in MCDB105 demonstrated that these cultures were quiescent (data not shown). For rapamycin studies on proliferating cells, 1 nM rapamycin (Enzo Biologicals, Plymouth Meeting, PA, USA) was added to the culture medium prior to feeding and passaging cultures.
Mitochondrial potential, mitochondrial function, ROS production, and mitochondrial mass evaluation
For mitochondrial potential studies, cells were incubated with 5 μg mL−1 JC-1 or 25 nM TMRE (Molecular Probes, Carlsbad, CA, USA) at 37 °C in 5% CO2 for 30 min and harvested in 2.5% trypsin–EDTA. Following the addition of four volumes of serum-containing medium, cells were analyzed immediately with a Guava EasyCyte Mini using the Guava Express Plus program (Guava Technologies, Millipore, Hayward, CA, USA). Gating was performed on the cell population by forward scatter and emission at 525 nm. JC-1 aggregate accumulation in the polarized environment of the mitochondria display maximal emission at 580 nm and were detected on the 583 (yellow channel) of the Guava EasyCyte to distinguish them from the JC-1 monomers that have maximal emission at 500 nm. The main population of cells is visualized as indicated in Fig. S3A (Supporting information), and cells that display JC-1 staining that is below the main population of cells are taken as those with loss of mitochondrial membrane potential. The number of cells in this population as a percentage of the total cell population is presented in Figs 2A,C, 6D, and S6B. Live cell imaging was performed using an EVOS FL microscope (AMG, Bothell, WA, USA) immediately following the 30-min incubation period. For mitochondrial mass evaluation, cells were incubated for 30 min in 100 nM Mitotracker Green FM or nonyl Acridine Orange (Molecular Probes) at 37 °C, harvested in 2.5% trypsin-EDTA, resuspended in 200 μL complete growth medium, and analyzed immediately with a Guava EasyCyte Mini. ROS accumulation was measured as previously described (Torres & Perez, 2008). Oxygen consumption rates were measured with a Seahorse XF analyzer (Seahorse Bioscience, North Billerica, MA, USA) as previously described (Diers et al., 2010). H2O2 treatments were performed using 200 μM H2O2 for 30 min. Culture medium was removed; the cells were washed three times with fresh medium and allowed to recover for 24 h prior to mitochondrial analysis. ROS accumulation was measured immediately after H2O2 treatment.
Western blotting and immunoprecipitation
For Western blot analyses, 30 μg of protein extracts was run on SDS-PAGE and transferred onto Immobilon P PVDF (EMD Millipore, Billerica, MA, USA) or nitrocellulose (Biorad, Hercules, CA, USA) membranes. Blots were incubated with antibodies specific for LC3B, beta-tubulin, Keap1, Phospho-S6, ribosomal protein S6 (Cell Signaling, Danvers, MA, USA), p62/SQSTM1 (Biomol, Plymouth Meeting, PA), beta-actin (Sigma, St Louis, MO, USA), Keap1, TFAm, p16, p21, p62/SQSTM1, NRF1 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA), or NFE2L2 (Epitomics, Burlingame, CA, USA). For immunoprecipitation, 0.5 mgof protein extracts was incubated with anti-p62/SQSTM1 rabbit polyclonal antibody (Biomol International) at a 1:75 weight per volume ratio or an anti-ubiquitin antibody (Santa Cruz Biotechnologies) at a 1:50 weight per volume ratio, overnight at 4 °C, and the immunocomplexes were precipitated with 100 μL of protein A/G agarose beads (Santa Cruz Biotechnologies).
RNA interference
Mission Short hairpin RNA lentiviral plasmids against Atg5 (cat. TRCN0000150645) and p62 (cat. TRCN0000007234 and TRCN0 000007236) were obtained from Sigma-Aldrich. The scramble shRNA plasmid (cat. 1864) was purchased from Addgene (Cambridge, MA, USA; Bitto et al., 2010). Lentiviral particles were produced in HEK 293T cells. All experiments were performed following a selection period of 72 h using 2 μg mL−1 puromycin (Sigma-Aldrich).
Quantitative PCR
Total RNA was isolated following standard guanidinium isothiocyanate extraction. For quantitative real-time PCR (QRT-PCR), the following primers were used: mitochondrial gene mRNA array (ND1-6, COI-III, ATP6/8, CytB) previously described (Nagao et al., 2008). SYBR Green-based QRT-PCR was performed on Stratagene mx3000p with Verso 1-Step RT-qPCR reagents (ThermoFisher Scientific, Pittsburgh, PA, USA). Single products for each reaction were verified by dissociation curve analysis and by gel electrophoresis. Telomere PCR was performed by the University of Washington genomic core facility on total DNA isolated from control or rapamycin-treated cultures at the population doublings indicated (Cawthon, 2002). Telomere content was normalized to signal derived from the beta-globin gene locus.
Fluorescence microscopy
Cells seeded on coverslips at 1–2 × 104 cells cm−2 were fixed in 4% paraformaldehyde after 48 h. Cells were permeabilized in 0.25% Triton X-100 prior to staining, incubated overnight with rabbit anti-LC3B, mouse anti-p62, goat anti-PML (Santa Cruz Biotechnologies), rabbit antip62 (Biomol), mouse anti-cytochrome C (Millipore, Billerica MA, USA), or mouse anti-HIRA (kindly provided by Dr. Peter Adams, University of Glasgow, Scotland), stained with goat anti-rabbit Alexa Fluor 488, donkey anti-mouse Alexa Fluor 555, and/or donkey anti-goat Alexa Fluor 488 (Invitrogen, Carlsbad, CA, USA), counterstained with DAPI (Sigma-Aldrich). Images were acquired using an Olympus BX61 fluorescence deconvolution microscope via a Hamamatsu CCD camera and Slidebook 4 (version 4.0.1.44) software (Olympus America, Center Valley, PA, USA). Confocal images of nuclear localization of NFE2L2 were obtained using a Leica laser scanning spectral confocal microscope TCS SP2 AOBS. Images were quantified using Image J software and were normalized to DAPI intensity. Live cell imaging was performed using an EVOS FL microscope (AMG).
Statistical analysis
Unless otherwise stated, results are representative of three independent experiments and statistical significance was assessed with unpaired, two-tailed, Student’s t-test.
Supplementary Material
The influence of mTOR1 activity on proliferation rate, autophagy, and mitophagy impact mitochondrial biogenesis.
Densitometric analysis of immunoblot data presented in Figure 1.
Evaluation of mitochondrial membrane potential and basal oxygen consumption rates in cells treated with rapamycin.
Analysis of LC3 in the presence or absence of rapamycin.
Lifespan analysis of cultures maintained in the presence of rapamycin.
Expression of mitochondrial biogenesis factors and mitochondrial membrane potential following NFE2L2 targeting.
Acknowledgments
M. Konigsberg was the recipient of a Fulbright and CONACYT Scholarship. This work was supported by grant AG022334 to C. Sell. A. Bitto is the recipient of a fellowship from the Drexel Aging Initiative.
Footnotes
Conflict of interest
The authors have no conflict of interest related to the work described in this manuscript.
Author contributions
AB, CL, DP, TN, MK, and CS performed experiments; AB, CL, CT, HR, and CS contributed experimental design; AB, CL, DP, MK, HR, CT, and CS involved in manuscript preparation.
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site.
References
- Adams PD. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397:84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Wijshake T, Tchkonia T, Lebrasseur N, Childs B, van de Sluis B, Kirkland J, van Deursen J. Clearance of p16(Ink4a)-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Pleiotropic effects of growth hormone signaling in aging. Trends Endocrinol Metab. 2011;22:437–442. doi: 10.1016/j.tem.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaert R, Cuenda A, Vanden Berghe W, Plaisance S, Lee JC, Haegeman G, Cohen P, Fiers W. The p38/RK mitogen-activated protein kinase pathway regulates interleukin-6 synthesis response to tumor necrosis factor. EMBO J. 1996;15:1914–1923. [PMC free article] [PubMed] [Google Scholar]
- Bitto A, Lerner C, Torres C, Roell M, Malaguti M, Perez V, Lorenzini A, Hrelia S, Ikeno Y, Matzko ME, McCarter R, Sell C. Long-term igf-I exposure decreases autophagy and cell viability. PLoS One. 2010;5:e12592. doi: 10.1371/journal.pone.0012592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofalo VJ, Charpentier R. A standard procedure for cultivating human diploid fibroblast like cells to study cellular aging. J Tissue Cult Methods. 1980;6:117–121. [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Diers AR, Higdon AN, Ricart KC, Johnson MS, Agarwal A, Kalyanaraman B, Landar A, Darley-Usmar VM. Mitochondrial targeting of the electrophilic lipid 15-deoxy-Delta12,14-prostaglandin J2 increases apoptotic efficacy via redox cell signalling mechanisms. Biochem J. 2010;426:31–41. doi: 10.1042/BJ20091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30:1536–1548. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinojosa CA, Mgbemena V, Van Roekel S, Austad SN, Miller RA, Bose S, Orihuela CJ. Enteric-delivered rapamycin enhances resistance of aged mice to pneumococcal pneumonia through reduced cellular senescence. Exp Gerontol. 2012;47:958–965. doi: 10.1016/j.exger.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Iwasa H, Han J, Ishikawa F. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 2003;8:131–144. doi: 10.1046/j.1365-2443.2003.00620.x. [DOI] [PubMed] [Google Scholar]
- Kang HT, Lee KB, Kim SY, Choi HR, Park SC. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, Choi YB, Lee AH, Lee KH, Park C, Obin MS, Park SK, Seo YJ, Oh GT, Lee HW, Shin J. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13:150–156. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontieva OV, Demidenko ZN, Gudkov AV, Blagosklonny MV. Elimination of proliferating cells unmasks the shift from senescence to quiescence caused by rapamycin. PLoS One. 2011;6:e26126. doi: 10.1371/journal.pone.0026126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Liu L, Trimarchi JR, Smith PJ, Keefe DL. Mitochondrial dysfunction leads to telomere attrition and genomic instability. Aging Cell. 2002;1:40–46. doi: 10.1046/j.1474-9728.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nagao A, Hino-Shigi N, Suzuki T. Measuring mRNA decay in human mitochondria. Methods Enzymol. 2008;447:489–499. doi: 10.1016/S0076-6879(08)02223-4. [DOI] [PubMed] [Google Scholar]
- Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, Suzuki T, Yamamoto M, Blackwell TK, Curran SP. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly toAtg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol. 2012;13:397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozou PD, Kirkwood TB. A stochastic model of cell replicative senescence based on telomere shortening, oxidative stress, and somatic mutations in nuclear and mitochondrial DNA. J Theor Biol. 2001;213:573–586. doi: 10.1006/jtbi.2001.2432. [DOI] [PubMed] [Google Scholar]
- Torres CA, Perez VI. Proteasome modulates mitochondrial function during cellular senescence. Free Radic Biol Med. 2008;44:403–414. doi: 10.1016/j.freeradbiomed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008a;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008b;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, Sun P. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–3403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The influence of mTOR1 activity on proliferation rate, autophagy, and mitophagy impact mitochondrial biogenesis.
Densitometric analysis of immunoblot data presented in Figure 1.
Evaluation of mitochondrial membrane potential and basal oxygen consumption rates in cells treated with rapamycin.
Analysis of LC3 in the presence or absence of rapamycin.
Lifespan analysis of cultures maintained in the presence of rapamycin.
Expression of mitochondrial biogenesis factors and mitochondrial membrane potential following NFE2L2 targeting.