Abstract
Protein tagging with a wide variety of epitopes and/or fusion partners is used routinely to dissect protein function molecularly. Frequently, the required DNA subcloning is inefficient, especially in cases where multiple constructs are desired for a given protein with unique tags. Additionally, the generated clones have unwanted junction sequences introduced. To add versatile tags into the extracellular domain of the transmembrane protein THSD1, we developed a protein tagging technique that utilizes non-classical type IIS restriction enzymes that recognize non-palindromic DNA sequences and cleave outside of their recognition sites. Our results demonstrate that this method is highly efficient and can precisely fuse any tag into any position of a protein in a scarless manner. Moreover, this method is cost-efficient and adaptable because it uses commercially available type IIS restriction enzymes and is compatible with the traditional cloning system used by many labs. Therefore, precision tagging technology will benefit a number of researchers by providing an alternate method to integrate an array of tags into protein expression constructs.
Keywords: Recombinant DNA, Precision tagging, Type IIS restriction enzyme, non-palindromic, THSD1
Introduction
Protein tagging is a powerful technique that is routinely used in many research labs to investigate the molecular function of proteins. One of the best examples is use of fluorescent protein reporter tags from far-red to UV-excitable green that enable biological imaging of their fusion partners [1]. In addition, epitope tag-based affinity purification is widely used to identify and characterize protein-protein interactions [2].
Protein tagging becomes invaluable in immunostaining or immunoprecipitation experiments where antibodies against the protein of interest are not available, thus presenting a barrier to the characterization of its subcellular localization or its physical interactions. Even in cases where a high quality antibody exists, several tags provide useful information due to their unique properties. For example, Dentra2 is a photoconvertible (green to red) fluorescent protein for tracking protein dynamics in real time [3]. Also, Strep-tag II binds to Strep-Tactin with high affinity that helps purify protein complexes in physiological concentrations [4]. Another tag, miniSOG, permits both fluorescent and electron microscopy studies [5].
Currently, several labs use traditional subcloning for tagging of proteins. This entails the use of multiple type II restriction enzymes (e.g., BamHI, EcoRI, EcoRV, HindIII, among a multitude of other enzymes) that recognize and then cleave internally its palindromic DNA recognition sites. Typically, an expression vector is chosen that already contains a specific tag where a multiple cloning site is provided to add the open reading frame of interest. Alternatively, an expression construct of interest is generated containing the gene of interest that also has unique restriction sites introduced to allow for the subsequent addition of N-terminal and/or C-terminal tags. However, several shortcomings are largely unavoidable. First, traditional tagging introduces unwanted junction sequences due to the presence of the flanking restriction enzyme recognition site. Secondly, it becomes time-intensive procedure when a protein needs a variety of tags that often require different flanking restriction enzyme sequences. Next, traditional ligation is less efficient when multiple DNA fragments are joined in one reaction. In particular, when the extracellular domain of a transmembrane protein is tagged, four DNA fragments (signal peptide, tag, gene of interest, linearized vector) are commonly joined according to a traditional tagging protocol. Although recombination-based systems (e.g., Gateway cloning) have been developed to improve the cloning/tagging efficiency, these systems introduce even longer junction sequences due to the remaining recombination site, which may interfere with protein folding and/or function [6].
We have developed a precision tagging strategy that is low-cost and avoids the aforementioned issues caused by traditional tagging methods such as unwanted junction sequences and low efficiency. This technique combines the use of both type II and type IIS restriction enzymes. Type IIS restriction enzymes recognize non-palindromic DNA sequences and cleave the DNA outside of their recognition site. For example, BsaI recognizes non-palindromic DNA sequences (5′-GGTCTC) and cleaves the DNA downstream of its recognition site, resulting in a 4 base 5′ overhang [7]. Utilizing this characteristic of type IIS enzymes, the proposed precision tagging strategy generates a scarless clone by completely eliminating the junction sequences. Importantly, it is easy to fit in with the type II restriction enzyme-based traditional cloning system used by many labs.
As a proof-of-concept, we applied this new cloning strategy to thrombospondin type I domain containing protein 1 (THSD1), a transmembrane protein whose function remains poorly characterized [8]. We had recently identified THSD1 as the first gene that appears to cause both familial and sporadic intracranial aneurysms and subarachnoid hemorrhage in humans and provided supporting in vivo data from loss-of-function studies in both zebrafish and mice [9]. Antibodies qualified for immunostaining and immunoprecipitation against THSD1 are suboptimal and little is known about its protein interaction partners. To systematically study THSD1 protein-protein interaction, subcellular localization, and protein dynamics, we picked 11 different tags and using precision tagging, we obtained all the THSD1 tagged variants with a 100% success rate. Our data reveal that the precision tagging approach will become beneficial for many biological researchers who need to make scarless clones in a highly-efficient and low-cost way.
Results
Precision tagging is scarless by eliminating junction sequences
Using traditional subcloning, two different unique restriction enzyme sites on either end of a tag are frequently introduced. As a result, the flanking sequences of normally six nucleotides long will encode two unwanted amino acids on either end of tag (see junction sequences in green in the left panel of Figure 1). In contrast, our newly developed precision tagging strategy completely eliminates such unwanted junction sequences and produces a scarless clone (see the right panel of Figure 1). For instance, when tagging a trans-membrane protein at its N-terminus, we normally add a tag right after its signal peptide (Figure 1).
Figure 1. A scarless clone generated by precision tagging.
Type I transmembrane protein is shown as an example. N-terminal half of the protein is located outside the cytoplasmic membrane (extracellular domain) after signal peptide (SP) is cleaved, and C-terminal half resides inside (intracellular domain). TM is short for transmembrane and highlighted in yellow box. By traditional tagging, tag (highlighted in red box) is inserted between SP and extracellular domain with two unwanted junction sequences (JC in green). In contrast, precision tagging eliminates the unwanted junction sequences and generates a scarless clone.
As shown in Supplementary Figure 1, the traditional tagging mainly uses type II restriction enzymes that cleave nucleotides within their recognition sequences (these enzymes are abbreviated as TII-es). For example, EcoRI recognizes 5′-GAATTC and creates 5′-AATT overhang (red letters in Supplementary Figure 1 and Figure 2a). As expected, to tag the extracellular domain of a trans-membrane protein, four different TII-es (TII-e1, e2, e3, and e4 as labeled in Supplementary Figure 1) are needed to create different overhangs (see colored sticky ends in Supplementary Figure 1). After ligation, the destination clone contains two separate unwanted junction sequences encoded by restriction enzyme recognition sites on either side of the tag (green and purple box in Supplementary Figure 1).
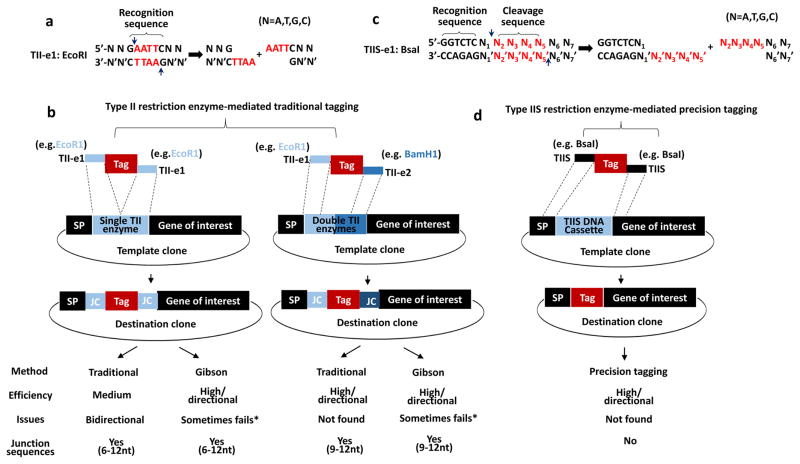
Figure 2. Comparison of type II and IIS restriction enzyme-mediated protein tagging strategy.
(a) Type II restriction enzymes (TII-es) recognize palindromic DNA sequences. For example, EcoRI recognizes 5′-GAATTC-3′ (marked by top curly bracket) and creates 4 base pairs overhangs highlighted in red.
(b) Single or double type II restriction enzymes cassette (highlighted in blue box) for traditional protein tagging. Note that in all destination clones, varying junction sequences exist adjacent to both sides of the tag.
(c) Type IIS restriction enzymes (TIIS-es) recognize non-palindromic, asymmetrical DNA sequences. For example, BsaI recognizes 5′-GGTCTC-3′ (marked by top curly bracket) and cleaves DNA one bp away (indicated by the two arrows), producing N2′N3′N4′N5′ custom sticky end (highlighted in red). N indicates four bases of DNA, including A, T, G and C. Apostrophe (’) indicates the complementary base of the DNA.
(d) Type IIS restriction enzyme DNA cassette (TIIS DNA cassette highlighted in blue box) for precision tagging. Note that on both ends of a tag, the flanking sequences (such as BsaI-released 5′-N2′N3′N4′N5′ and 5′-N2N3N4N5 belong to gene-specific sequences including SP or gene of interest indicated by two closely dotted lines. After Tag replaces type IIS DNA cassette, a scarless tagging clone can be generated. Comparison between traditional and precision tagging were summarized in the bottom table. * Gibson assembly sometimes fails due to certain DNA sequences such as repetitive region or creating one or two nucleotides deletion.
Multiple fragment ligation generally leads to lower frequency of correct clones. To reduce the number of fragments to ligate, we can turn to a two-step cloning. First, we perform overlapping PCR to build a template clone containing a cleavable cassette where the tag is to insert, and then obtain the tag clone/destination clone by two-fragment ligation. As shown in Figure 2b, we introduce a single (labeled as single TII enzyme and highlighted in light blue) or double restriction sites (labeled as double TII enzymes and left half in light blue and right half in dark blue) in between SP and the gene of interest. We can follow traditional method to fuse a tag in between, or alternatively use a Gibson assembly protocol, a restriction enzyme-independent method, to get the same result [10]. However, as summarized in the table below, all the destination clones using both methods have varying junction sequences. In addition, Gibson assembly sometimes fails due to redundant sequences and sequence-specific issues that lead to error in assembly or recurrent and invariant deletion of one or two nucleotides that introduce frame-shift mutations.
In precision tagging, a different cassette (labeled as TIIS DNA Cassette) is created between SP and the gene of interest. This cassette can be recognized by non-classical type IIS restriction enzymes. They recognize non-palindromic DNA sequences and cleave outside their recognition sites. For example, BsaI recognizes 5′-GGTCTC, cuts one nucleotide (indicated by N1) away downstream, and creates a 5′-N5′N4′N3′N2′ sticky end (red letters in Figure 2c). These custom overhangs make it possible for our destination clones to be scarless if we match them in tag flanking sequences with the gene-specific sequences (see the matched area indicated by two closely dotted lines in Figure 2d).
Procedure to make a template clone
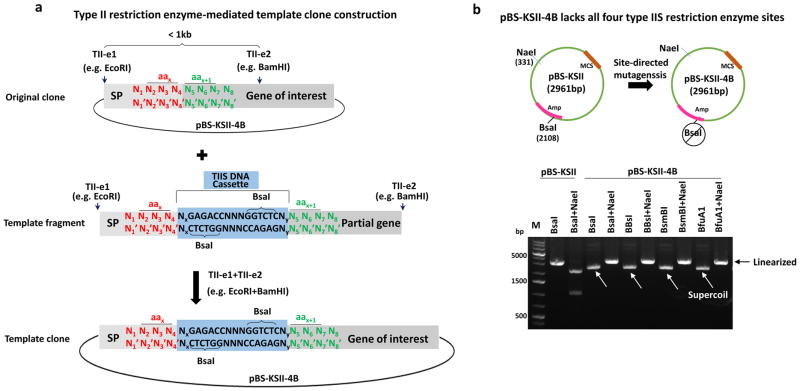
As shown in Figure 2d, a key step in precision tagging is building a template clone where TIIS DNA cassette is inserted in the gene of interest (see blue box in both Figure 2d and Figure 3a). First, we need to determine the position in the gene of interest for the tag to insert. For example, we highlight the number x amino acid (aa) in red (encoded by N2N3N4 and labeled as aax in Figure 3a) and the next number x+1 amino acid in green (encoded by N5N6N7 and labeled as aax+1 in Figure 3a), where the tag will be added in between. Second, we obtain a template fragment containing a TIIS DNA cassette that is composed of two inverted end-oriented type IIS restriction enzyme recognition sequences such as 5′-GGTCTC by BsaI as shown in the middle lane of Figure 3a. Such a fragment can be routinely obtained by following a standard overlapping PCR protocol that joins two fragments with overlapping sequences, or purchasing from gene synthesis companies (we acquired this DNA fragment as a gBlock from IDT DNA). Using traditional restriction enzymes (as labeled by TII-e1 and e2 in both the original clone and a template fragment in Figure 3a), the template fragment can replace the counterpart in the original clone, which generates a template clone we need for the next step.
Figure 3. Procedure to make a template clone.
(a) In the original clone, two unique sites recognized by two different type II restriction enzymes (labeled as TII-e1 and TII-e2) are selected in the gene of interest. The number x amino acid encoded by N2N3N4 (in red) is indicated by aax, or next amino acid by aax+1 (in green). A template fragment contains a TIIS DNA cassette (highlighted in blue box) in between aax and aax+1. All the cutting positions are indicated by arrows and complementary bases by apostrophe (’).
(b) pBS-KSII contains a multiple cloning site (MCS) and a BsaI in the ampicillin (Amp) coding gene, which was destroyed by site-directed mutagenesis to create pBS-KSII-4B. Wild-type pBS-KSII was linearized by BsaI and two overlapping fragments were released by BsaI and NaeI double digestion. pBS-KSII-4B does not have BsaI, BbsI, BsmBI, and BfuAI/BspMI and can be linearized by NaeI. Three brighter bands from GeneRuler 1kb plus DNA marker are labeled.
We chose the commonly used pBluescript KS II (pBS-KSII) vector for making a template clone (Figure 3a). The type IIS enzymes BbsI, BsmBI, and BfuAI/BspMI do not cut this plasmid and each generates a 4 base 5′ overhang upon DNA cleavage. However, another type IIS restriction enzyme BsaI that also creates a 4 base 5′ overhang is present in the coding region of ampicillin resistance (the upper panel and the first two lanes in lower panel in Figure 3b). Site-directed mutagenesis was performed to destroy it synonymously. We confirmed that this modified pBS-KSII lacks BsaI, BbsI, BsmBI and BfuAI/BspMI (lanes 3–10 in lower panel in Figure 3b) and named it pBS-KSII-4B, which is available immediately upon request and will be deposited at Addgene.
Precision tagging is a one-step procedure
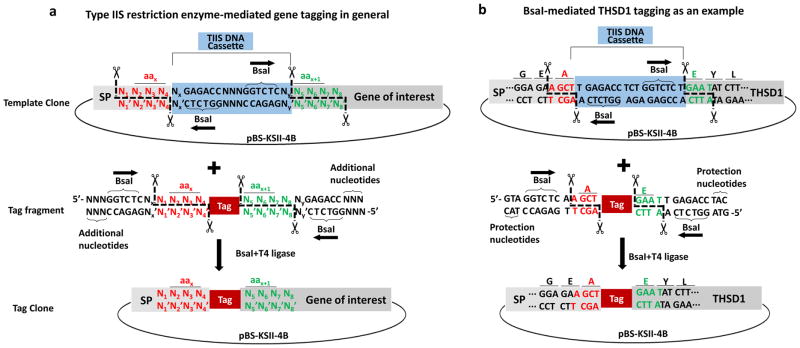
As shown in Figure 4a, a template clone after digestion generates two cohesive ends such as 5′-N4′N3′N2′N1′ (see red letters and the cutting positions indicated by cartoon scissors) and 5′-N5N6N7N8 (see green letters and the cutting positions indicated by cartoon scissors). They are complementary with the sequence flanking the tag (red and green letters in the middle panel). All tag fragments can be easily acquired by either standard PCR, gene synthesis, or annealed oligonucleotides. Once the tag fragment joins with the linearized template clone, the tag clone (see bottom panel) no longer contains any type II restriction enzyme recognition site. Therefore, both type IIS enzymes such as BsaI and T4 DNA ligase can be used simultaneously in the same tube, and a tag clone can be generated by this one-step digestion/ligation reaction, which cannot be achieved by traditional cloning. Notably, since the cohesive ends (labeled as “N” in either red or green) belong to the gene-specific sequence and are preselected in frame (N2N3N4 encodes aax and N5N6N7 encodes aax+1 in Figure 4), the tag clone will express the tag right between aax and aax+1 of the protein of interest (see tag in red box inserted between red letters and green letters in Figure 4a). In particular, we applied this method to THSD1, a transmembrane protein implicated in intracranial aneurysm formation and subarachnoid hemorrhage and of which little is known about its biochemical functions. As shown in Figure 4b, the TIIS DNA cassette will be removed after BsaI digestion and no longer exist in the final tag clone. All the primers used in this work are listed in Table 1.
Figure 4. Precision tagging is one-step.
(a) In a template clone, TIIS DNA cassette (shaded in blue) contains two reverted outside-oriented TIIS enzyme recognition sequences (indicated by horizontal arrows). For example, BsaI recognizes 5′-GGTCTC and produces two custom sticky ends that belong to SP or gene of interest sequence in an in-frame pattern (N2N3N4 encodes the number x amino acid of the protein and N5N6N7 the number x+1 amino acid). Tag flanked by complementary stick ends (indicated by the same colored letters) is ligated into a template clone in the presence of both BsaI and T4 DNA ligase.
(b) The last three amino acids of SP are labeled as G, E, A, while the first three amino acids of THSD1 trunk region (full-length THSD1 protein lacking SP) are labeled as E, Y, L. Precision tagging adds a tag in between SP and the trunk region of THSD1 without junction sequences.
Table 1. Primers of precision tagging for THSD1.
All the primers for THSD1 precision tagging were listed. Type IIS restriction enzyme recognition sequences are marked by red letters. Custom sticky ends belonging to the THSD1-specific sequences are highlighted in green letters. Tag-specific sequence were marked by square bracket. Note that different tags may require different Type IIS restriction enzymes (BsaI, BbsI and BsmBI) to avoid internal cleavage in the tag.
| Primer name | Sequence | Type IIS enzyme |
|---|---|---|
| HA-sense |
|
N/A |
| HA-antisense |
|
N/A |
| Myc-sense |
|
N/A |
| Myc-antisense |
|
N/A |
| FLAG-sense |
|
N/A |
| FLAG-antisense |
|
N/A |
| V5-sense |
|
N/A |
| V5-antisense |
|
N/A |
| AviTag-sense |
|
N/A |
| AviTag-antisense |
|
N/A |
| Strep-Tag II-sense |
|
N/A |
| Strep-Tag II-antisense |
|
N/A |
| EGFP-F |
|
BbsI |
| EGFP-R |
|
BbsI |
| mCherry-F |
|
BsaI |
| mCherry-R |
|
BsaI |
| Dendra2-F |
|
BsmBI |
| Dendra2-R |
|
BsmBI |
| SNAPf-F |
|
BbsI |
| SNAPf-R |
|
BbsI |
| miniSOG-F |
|
BbsI |
| miniSOG-R |
|
BbsI |
Precision tagging can be used for universal as well as multiple tandem tagging
Unlike traditional cloning, the overhangs for different tags by precision tagging can be identical gene-specific cohesive ends (e.g. GFP, Strep-tag II and SNAPf all have the same overhangs indicated by black color in Figure. 5a). Therefore, the template clone is universal for versatile tags, simplifying the tedious procedure when different tags require distinct flanking sequences (Supplementary Figure 1). For instance, the template clone can be linearized by BsaI to produce 5′-N4′N3′N2′N1′ and 5′-N5N6N7N8 two overhangs at its two ends (Figure 4), which can join with the complementary ones (labeled as TIIS on each end) in all different tags such as GFP, Strep-tag II and SNAPf. It is worth noting that precision tagging is often a two fragment ligation, which has much higher efficiency of getting positive clones in comparison with multiple fragments ligation by traditional cloning.
Figure 5. Universal and multiple tandem precision tagging strategy.
(a) A template clone contains a TIIS DNA cassette (shaded in blue). Different tags such as GFP (green), Strep-tag II (red), and SNAPf (blue) contain the same two custom sticky ends that belong to SP and gene of interest (indicated by overhangs in black). TIIS restriction enzymes and T4 DNA ligase join them together in a universal manner.
(b) One sticky end on the right side of GFP is complementary to another one on the left side of Strep-tag II (indicated by green color bar). Similarly, Strep-tag II has a complementary sticky end to the one in SNAPf (indicated by red color bar). TIIS restriction enzyme and T4 ligase can fuse all those tags together and generate a multiple tandem tag (indicated by sequentially arranged GFP, Strep-tag II and SNAPf).
Interestingly, if one end of a tag was made complementary to the other end of the second tag (e.g. green overhang in GFP is complementary to the same colored one in Strep-tag II in Figure 5b), multiple tandem tagging is practical and convenient. Such type of tagging has been widely used in protein functional studies. For example, the fate of autophagosome can be traced by its marker protein LC3 labeled with a tandem tag GFP-mCherry, since GFP is quenched in matured and acidic autophagosome while mCherry is stable [11]. Other tandem tags such as FLAG-HA or ProtA-CBP are routinely used for protein complexes purification [12]. Thus, precision tagging offers a convenient way to combine any tags in a scarless manner.
Precision tagging is a cost-efficient strategy
As shown in Figure 6, precision tagging was summarized as a schematic model. In step 1, we typically use type II restriction enzymes to swap a template fragment with its counterpart in the gene of interest so as to make a template clone that contains a TIIS DNA cassette. Next, non-classical type IIS restriction enzymes are employed to introduce a scarless tag by a one-step reaction. Finally, we again use type II restriction enzymes to move freely the tagged insert to various functional vectors that any lab has been already using for its own research needs. As described, the precision tagging system only requires a few extra type IIS enzymes (highlighted in red frame in Figure 6). In contrast, another tagging system based on gateway technology requires both BP and LR clonase that specifically catalyze recombination sequence exchange (e.g., attB and attP sequence can be changed to attL and attR sequence under BP clonase, or vice versa by LR clonase) [13], which is patented technology. Furthermore, the recombination sequences such as attB left in the final destination clone by the gateway system will introduce even longer unwanted junction sequence (about 8–13 amino acids) between the tag and the gene of interest.
Figure 6. Summary of precision tagging strategy.
Step 1: Type II restriction enzymes are used for swapping a template fragment containing TIIS DNA cassette with its counterpart in the original clone. Step 2: A template clone becomes a tag clone with the help of type IIS restriction enzymes by one-step reaction. Step 3: Type II restriction enzymes are used to freely move the insert in the tag clone to any designated destination clones. Type IIS restriction enzymes in Step 2 are highlighted in red frame.
Highly efficient precision tagging of THSD1, a trans-membrane protein, with versatile tags
Recently, using clinical samples, our lab identified thrombospondin type I domain containing 1 (THSD1), a transmembrane protein, whose cellular function remains poorly understood [9]. In addition, antibodies good for both immunostaining and immunoprecipitation of THSD1 are not available. Therefore, we decided to apply the newly developed precision tagging strategy to THSD1 as a proof-of-concept experiment.
A range of tags was selected to cover ones used frequently in labs for different purposes. As shown in Table 2, small epitope tags including HA, Myc, Flag, V5, AviTag, and Strep-tag II were chosen for studying protein-protein interactions in the future. It is worth noting that AviTag can strongly binds to streptavidin after biotinylation, while Strep-tag II binds to Strep-Tactin with high specificity [4,14]. Live-imaging can be performed by GFP or mCherry, while the protein lifetime can be traced by a photoconvertible fluorescent protein Dendra2 [green to red converted by UV-violet (360–420 nm) or blue region (460–500 nm)] [3]. We also selected two newly-developed tags including SNAPf and miniSOG. SNAPf is a chemical tag that can be labeled with synthetic probes whenever the chemicals with fluorophores are added [15], while miniSOG permits high quality preservation under electron microscopic processing [5].
Table 2. Versatile tags and their applications.
All the tags used in this work were listed together with their potential applications.
| Tag | Application |
|---|---|
| HA | Protein interaction/purification |
| Myc | Protein interaction/purification |
| FLAG | Protein interaction/purification |
| V5 | Protein interaction/purification |
| AviTag | Protein interaction/purification |
| Strep-Tag II | Protein interaction/purification |
| GFP | Live-imaging/green |
| mCherry | Live-imaging/red |
| Dendra2 | Live-imaging/photoconvertable |
| SNAPf | Pulse-chase imaging |
| miniSOG | Electron microscope |
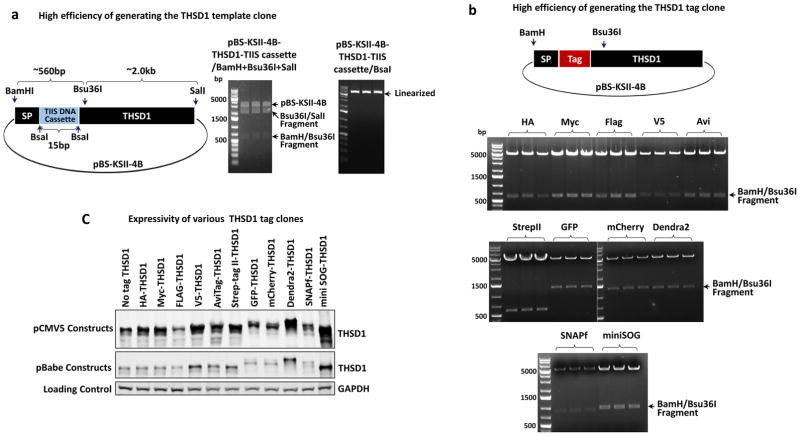
First, we constructed a template clone for THSD1 N-terminal tagging, a key component for precision tagging (Figure 7a). To do that, a template fragment was synthesized by overlapping PCR and used to replace the counterpart in the N-terminal region of THSD1 from BamHI to Bsu36I (about 560 bp long indicated by first top curly bracket in Figure 7a). As mentioned previously in Figure 3, this step only requires type II enzymes such as BamHI and Bsu36I and to perform a two-fragment ligation. Therefore, we expectedly obtained all the positive clones from three randomly-picked colonies after bacteria transformation. The template clones (pBS-KSII-4B-THSD1-TIIS cassette) were confirmed by three-enzyme digestion, as two expected fragments were released (see the first right panel in Figure 7a), as well as its ability to be linearized by BsaI (the second right panel in Figure 7a). Next, we used type IIS restriction enzymes to insert versatile tags into the template clone according to previous strategy (Figure 4). BamHI and Bsu36I were used for screening out the positive clones from three randomly picked colonies after bacteria transformation. As shown in the middle and bottom panels in Figure 7b, all the tag clones were positive and verified correct by later DNA sequencing. We further subcloned these tagged inserts into pCMV5 or pBabe vector for transiently or stably expressions in mammalian cells respectively. All of them are expressed well in HEK293T cells judged by western blotting analyses (Figure 7c).
Figure 7. Highly efficient precision tagging for THSD1.
(a) A template clone for THSD1. Three type II enzymes including BamHI, Bau36I, and SalI were selected. Three randomly picked colonies were characterized by these three enzymes. All are positive based on two released fragments (indicated by arrows) and single linearized fragment by BsaI.
(b) Various tag clones for THSD1. Different tags including HA, Myc, Flag, V5, AviTag, Strep-tag II, GFP, mCherry, Dendra2, SNAPf and miniSOG were inserted into the templet clone acquired from (a). Three random colonies from each transformation were picked up and BamHI/Bau36I double digestion was used for characterization. Note the different sizes of released fragments caused by the different tags.
(c) Expression of tagged THSD1 in two different destination vectors. 2 ug of each plasmid was transfected into HEK293T cells for 48 hours before harvesting in SDS sample buffer. THSD1 antibody was used for revealing the protein expression by immunoblotting. GAPDH was used as loading control. Note that pCMV5 plasmids express much higher levels of protein than pBabe plasmids as expected.
Discussion
High efficiency of precision tagging
Tagging using a traditional cloning strategy requires different type II restriction enzyme sites at two ends and amplification of the same set of DNA fragments repeatedly in order to add different flanking sequences (Supplementary Figure 1). In contrast, due to the universality of custom sticky end provided by precision tagging (Figure 5a), such a tedious process can be avoided. Furthermore, if tagging is needed within a gene such as labeling the extracellular domain of some transmembrane proteins (e.g. THSD1 tagging), traditional method normally joins four different fragments while precision tagging often builds a template clone by ligating two fragments, which greatly increases the cloning efficiency and success rate (Figure 7a). Importantly, precision tagging utilizes non-classical type IIS restriction enzymes (e.g. BsaI, BbsI, BsmBI, etc...) that generate custom sticky ends regardless of the enzyme selections as long as they are absent to both the insert and the vector. Once these custom sticky ends join in the new vector (such as the tag clone in pBSII-KS-4B in Figure 4), the type IIS restriction enzymes can no longer separate them away, which allows digestion and ligation occur in the same tube and simplifies cloning procedures. This unique property has been exploited in plant synthetic biology for large DNA assembly or effective library generation [16,17]. In our trial on the transmembrane protein THSD1 (Figure 7), we had a satisfying cloning result (100% success rate for getting the template clone and each tag clone).
Ease of use of precision tagging
In terms of cloning efficiency, Gateway technology provides an alternative way for gene tagging. However, BP and LR clonase as well as a set of destination vectors are patented technology. In contrast, precision tagging only requires a few extra type IIS restriction enzymes as shown in Figure 6. And though the gateway technology provides a variety of destination vectors that try to cover as many research purposes, such as expression, purification and imaging, as possible, it would be exhaustive to incorporate every new tag that any individual lab favors for its own research advantage. In comparison, precision tagging does not require predesigned destination vectors, which adds great flexibility for researchers to make their personalized tagging choices.
Our strategy is similar to traditional cloning with type II restriction enzymes and benefits from the array of current and future plasmids that are available through commercial and nonprofit sources such as Addgene. As a result, precision tagging strategy makes it easy to put a variety of tags in the gene of interest. It is worth of mentioning that the fragment by overlapping PCR can also be obtained via gene synthesis companies, which further simplifies the workflow of precision tagging. Besides, a modified cloning vector pBSII-KS-4B lacking four commonly used type IIS restriction enzymes is becoming an open resource and facilitate other researcher’s cloning work in the future.
Both traditional and gateway tagging will inevitably introduce unwanted junction sequences at either end of a tag, due to the presence of the type II restriction enzyme recognition site or of recombinase-mediated recombination sequences, respectively. In contrast, because of the unique feature of the type IIS restriction enzyme that cleaves DNA sequence outside of its recognition site, precision tagging completely eliminates such unwanted junction sequences, or if needed, can deliberately introduce custom junction sequence just by treating it as a part of a tag.
In summary, precision tagging provides a new strategy as cheap as a traditional cloning method, and as efficient as a gateway technology, but meanwhile makes a scarless clone that both the aforementioned approaches cannot achieve.
Materials and Methods
Plasmids
pBabe-GFP (#10668), mCherry-Talin-H-18 (#62749), Dendra2-Vinculin-N-21 (#57749), pSNAPf-C1 (#58186), miniSOG-Zyxin-6 (#57781) from Addgene provide template for these tags: GFP, mCherry, Dendra2, SNAPf and miniSOG, respectively. THSD1 in pBSII-KS-4B were made by subcloning previous plasmid pCR3.1-THSD1 [9,18]. pCMV5 and pBabe were used for transient and stable expression as previously described [19].
Chemicals
All the enzymes including type II restriction enzymes (EcoRI, BamHI, SalI and Bau36I), type IIS restriction enzymes (BsaI, BbsI and BsmBI), T4 DNA ligase, Phusion enzyme, and T5 exonuclease were purchased from New England BioLabs. All other chemicals such as agarose or protein gels were obtained from Sigma or BioRad.
Cell culture and transfection
HEK293T cells were maintained in DMEM medium (Mediatech) containing 10% fetal bovine serum (FBS, Invitrogen), 100 IU penicillin, and 100ug/ml streptomycin. Transfections of plasmid DNA were performed using lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions.
Western blotting and Antibodies
Transiently transfected 293T cells in 60-mm dishes were lysed in a Triton X-100 lysis buffer as previously described [20] and sonicated briefly before centrifuged at 13,000 rpm for 30 min at 4 °C. Total cell lysates were added by 2X SDS sample buffer and then subjected to discontinuous SDS-PAGE analysis. Proteins were transferred to nitro-cellulose membranes using Bio-Rad mini transfer apparatus followed by blocking with 5% non-fat milk. Primary antibodies and secondary antibodies were used usually at 1:1000 and 1:10000 dilutions respectively before using Odyssey system to detect fluorescence signal. THSD1 antibody was from Novus Biologicals (NBP1-86930).
Oligos and site-directed mutagenesis
All the oligonucleotides listed in Table 2 and the gBlock fragment containing BsaI-recognized TIIS cassette were synthesized by Integrated DNA Technology. For small epitope tags including HA, Myc, Flag, V5, AviTag and Strep-tag II, complementary oligonucleotides were annealed in PCR machine and phosphorylated by T7 polynucleotide kinase (NEB) before ligated into the linearized vector as previously described [21]. To destroy BsaI synonymously, QuikChange Lightning kit (Agilent Technologies) was applied by using two overlapping oligos “5-gctgcaatgataccgcgTgaTccacgctcaccggctc” and “5-gagccggtgagcgtggAtcAcgcggtatcattgcagc”. All the newly constructed clones were further verified by DNA sequencing (Lone Star Labs).
Supplementary Material
Type II restriction enzymes (TII-es) such as EcoRI recognizes palindromic DNA sequence “5′-GAATTC” and cleave it into two 5′-overhangs in red letters. SP, Tag, and gene of interest flanked with different TII-es (TII-e1, 2, 3 and 4) are added by polynucleotide chain reaction (PCR) followed by ligation with a linearized destination vector due to their complementary overhangs (marked by the same color). Note that two junction sequences are produced in the destination clone (highlighted in green and purple box).
Acknowledgments
We appreciate the critical reading of this manuscript by Dr. Bo Xiong. The project was supported by an R03NS087416 from the US National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 2.Morris JH, Knudsen GM, Verschueren E, Johnson JR, Cimermancic P, Greninger AL, Pico AR. Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat Protoc. 2014;9:2539–2554. doi: 10.1038/nprot.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker SM, Buckheit RW, 3rd, Falk MM. Green-to-red photoconvertible fluorescent proteins: tracking cell and protein dynamics on standard wide-field mercury arc-based microscopes. BMC Cell Biol. 2010;11:15. doi: 10.1186/1471-2121-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 5.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esposito D, Garvey LA, Chakiath CS. Gateway cloning for protein expression. Methods Mol Biol. 2009;498:31–54. doi: 10.1007/978-1-59745-196-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Lundin S, Jemt A, Terje-Hegge F, Foam N, Pettersson E, Kaller M, Wirta V, Lexow P, Lundeberg J. Endonuclease specificity and sequence dependence of type IIS restriction enzymes. PLoS One. 2015;10:e0117059. doi: 10.1371/journal.pone.0117059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayanagi S, Hiroyama T, Yamazaki S, Nakajima T, Morita Y, Usui J, Eto K, Motohashi T, Shiomi K, Keino-Masu K, Masu M, Oike Y, Mori S, Yoshida N, Iwama A, Nakauchi H. Genetic marking of hematopoietic stem and endothelial cells: identification of the Tmtsp gene encoding a novel cell surface protein with the thrombospondin-1 domain. Blood. 2006;107:4317–4325. doi: 10.1182/blood-2005-09-3747. [DOI] [PubMed] [Google Scholar]
- 9.Santiago-Sim T, Fang X, Hennessy ML, Nalbach SV, DePalma SR, Lee MS, Greenway SC, McDonough B, Hergenroeder GW, Patek KJ, Colosimo SM, Qualmann KJ, Hagan JP, Milewicz DM, MacRae CA, Dymecki SM, Seidman CE, Seidman JG, Kim DH. THSD1 (Thrombospondin Type 1 Domain Containing Protein 1) Mutation in the Pathogenesis of Intracranial Aneurysm and Subarachnoid Hemorrhage. Stroke. 2016;47:3005–3013. doi: 10.1161/STROKEAHA.116.014161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 11.Hansen TE, Johansen T. Following autophagy step by step. BMC Biol. 2011;9:39. doi: 10.1186/1741-7007-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y. Commonly used tag combinations for tandem affinity purification. Biotechnol Appl Biochem. 2010;55:73–83. doi: 10.1042/BA20090273. [DOI] [PubMed] [Google Scholar]
- 13.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairhead M, Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodor DL, Rodriguez MG, Moreno N, Jansen LE. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr Protoc Cell Biol. 2012;Chapter 8(Unit8):8. doi: 10.1002/0471143030.cb0808s55. [DOI] [PubMed] [Google Scholar]
- 16.Sarrion-Perdigones A, Vazquez-Vilar M, Palaci J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D. GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013;162:1618–1631. doi: 10.1104/pp.113.217661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quaglia D, Ebert MC, Mugford PF, Pelletier JN. Enzyme engineering: A synthetic biology approach for more effective library generation and automated high-throughput screening. PLoS One. 2017;12:e0171741. doi: 10.1371/journal.pone.0171741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko JM, Chan PL, Yau WL, Chan HK, Chan KC, Yu ZY, Kwong FM, Miller LD, Liu ET, Yang LC, Lo PH, Stanbridge EJ, Tang JC, Srivastava G, Tsao SW, Law S, Lung ML. Monochromosome transfer and microarray analysis identify a critical tumor-suppressive region mapping to chromosome 13q14 and THSD1 in esophageal carcinoma. Mol Cancer Res. 2008;6:592–603. doi: 10.1158/1541-7786.MCR-07-0154. [DOI] [PubMed] [Google Scholar]
- 19.Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, Zhang S. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui Y, Xu Z, Lin S, Li Q, Rui H, Luo W, Zhou HM, Cheung PY, Wu Z, Ye Z, Li P, Han J, Lin SC. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 2004;23:4583–4594. doi: 10.1038/sj.emboj.7600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rui Y, Xu Z, Xiong B, Cao Y, Lin S, Zhang M, Chan SC, Luo W, Han Y, Lu Z, Ye Z, Zhou HM, Han J, Meng A, Lin SC. A beta-catenin-independent dorsalization pathway activated by Axin/JNK signaling and antagonized by aida. Dev Cell. 2007;13:268–282. doi: 10.1016/j.devcel.2007.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Type II restriction enzymes (TII-es) such as EcoRI recognizes palindromic DNA sequence “5′-GAATTC” and cleave it into two 5′-overhangs in red letters. SP, Tag, and gene of interest flanked with different TII-es (TII-e1, 2, 3 and 4) are added by polynucleotide chain reaction (PCR) followed by ligation with a linearized destination vector due to their complementary overhangs (marked by the same color). Note that two junction sequences are produced in the destination clone (highlighted in green and purple box).