In response to high salinity, algal endosymbionts of corals produce floridoside, an osmolyte with antioxidant potential.
Abstract
The endosymbiosis between Symbiodinium dinoflagellates and stony corals provides the foundation of coral reef ecosystems. The survival of these ecosystems is under threat at a global scale, and better knowledge is needed to conceive strategies for mitigating future reef loss. Environmental disturbance imposing temperature, salinity, and nutrient stress can lead to the loss of the Symbiodinium partner, causing so-called coral bleaching. Some of the most thermotolerant coral-Symbiodinium associations occur in the Persian/Arabian Gulf and the Red Sea, which also represent the most saline coral habitats. We studied whether Symbiodinium alter their metabolite content in response to high-salinity environments. We found that Symbiodinium cells exposed to high salinity produced high levels of the osmolyte 2-O-glycerol-α-d-galactopyranoside (floridoside), both in vitro and in their coral host animals, thereby increasing their capacity and, putatively, the capacity of the holobiont to cope with the effects of osmotic stress in extreme environments. Given that floridoside has been previously shown to also act as an antioxidant, this osmolyte may serve a dual function: first, to serve as a compatible organic osmolyte accumulated by Symbiodinium in response to elevated salinities and, second, to counter reactive oxygen species produced as a consequence of potential salinity and heat stress.
INTRODUCTION
Coral reefs are biodiversity hot spots of immense biological and economical value (1). The relationship between Symbiodinium—an autotrophic endosymbiotic dinoflagellate—and scleractinian corals form the basis of coral reef ecosystems (2). The coral host provides inorganic nutrients and carbon dioxide to the dinoflagellate in exchange for energy in the form of photosynthetically produced carbohydrates (3, 4). This symbiotic relationship is highly sensitive to environmental disturbance. For instance, increases in temperature, salinity, nutrients, and/or high solar irradiance can impair photosynthetic efficiency and enhance the formation of harmful reactive oxygen species (ROS) that must be detoxified by the organism via antioxidants and ROS scavengers (5–8). Otherwise, environmental stress can ultimately lead to coral bleaching, the visual whitening of corals due to loss of their endosymbionts (5, 8). As a consequence, increasing exposure to environmental stress, in particular to rising seawater temperatures, is threatening the existence of coral reefs at a global scale (9, 10).
In comparison to the detrimental effects of elevated seawater temperatures, much less is known about the effects of increased salinities on corals and their endosymbionts. Yet, reef corals from the Red Sea and the Persian/Arabian Gulf (PAG) are commonly found at salinities of up to 41 [Practical Salinity Scale 1978 (PSS-78)] in the Red Sea and up to 50 in the PAG, at summer temperatures exceeding 32° and 35°C in parts of the Red Sea and PAG, respectively (11–14). Both salinity and temperature in these regions are the highest globally to support reef growth (15). Although the osmotic response of Symbiodinium at the molecular level is virtually unknown (16–18), studies on free-living algae suggest that production and accumulation of compatible organic osmolytes (COOs), referred to as osmoadaptation (19), is the most widespread mechanism for adjusting intracellular osmotic pressure in response to elevated salinity; accumulation of inorganic ions and cell volume changes may occur as well. However, because the latter two processes are stressful and can disturb cellular function, they are not considered to represent viable long-term solutions for osmoadaptation (20–23). In contrast, the accumulation of COOs adjusts the osmotic pressure and protects proteins from increased ion concentrations (22).
To test whether Symbiodinium is capable of synthesizing COOs, we subjected Symbiodinium strains from different clades and origins to conditions of high salinity, both in vitro and in their coral hosts (hereafter referred to as in hospite), and screened for the presence of COOs using gas chromatography–mass spectrometry (GC-MS). We hypothesized that Symbiodinium would increase cellular concentrations of COOs in response to elevated salinities and that the response would be similar in vitro and in hospite. Among the COOs employed by Symbiodinium, we identified the carbohydrates floridoside, inositol, and mannitol in vitro and in hospite. These compounds can act as both osmolytes and antioxidants, thereby having the potential capacity to convey osmoadaptation to increased salinities and the ability to counter ROS produced as a consequence of salinity or other forms of stress, including heat stress (6, 8, 24–30).
RESULTS
High levels of floridoside in Symbiodinium exposed to high salinities
We screened Symbiodinium cultures exposed to different salinities (that is, 25 for low salinity, 38 for ambient salinity, and 55 for high salinity) for the presence of carbohydrate COOs. A markedly high abundance of a compound at 31.5-min retention time in the GC-MS trace was detected in all four tested Symbiodinium cultures under high-salinity conditions (that is, Symbiodinium microadriaticum type A1, Symbiodinium sp. type A1, Symbiodinium minutum type B1, and Symbiodinium psygmophilum type B2) (Fig. 1 and Table 1). This compound was identified as the derivative of floridoside [2-O-glycerol-α-d-galactopyranoside-(hexa-trimethylsilane)] by a search against the National Institute of Standards and Technology Mass Spectrometry (NIST MS) library with a reverse match factor of 971 of 1000 (table S1). Floridoside levels of Symbiodinium strains exposed to high salinity ranged from 50.3 ± 4.3 nmol (S. minutum) to 489.5 ± 31.9 nmol (S. psygmophilum) and were consistently represented among the most abundant carbohydrates quantified in this analysis (Table 1). In contrast, floridoside was nondetectable under low-salinity conditions for all Symbiodinium strains (Fig. 2A and Table 1). At an ambient salinity level of 38, floridoside was only detected in S. psygmophilum at a level of 51.0 ± 8.0 nmol (Fig. 2A and Table 1). This strain also accounted for the highest measured amounts of floridoside under a high-salinity level of 55 (489.5 ± 31.9 nmol). In comparison, inositol and mannitol were consistently present at low salinities and showed reduced levels at higher salinities for some strains (Table 1).
Fig. 1. GC analysis of carbohydrate osmolyte diversity in S. psygmophilum at three salinity levels.
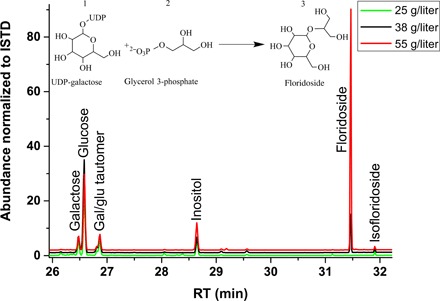
Green, low-salinity level of 25; black, ambient-salinity level of 38; red, high-salinity level of 55. The trace from 26- to 32-min retention time (RT) shows a floridoside peak at 31.5-min RT. The abundances are normalized to an internal standard (ISTD). The chemical reaction depicts synthesis of floridoside (3) as a product of uridine diphosphate (UDP)–galactose (1) and glycerol 3-phosphate (2). Notably, nearly identical traces were obtained for the other three Symbiodinium strains. Gal/glu, galactose/glucose.
Table 1. COO levels at three salinities of four Symbiodinium strains.
Carbohydrates (floridoside, inositol, mannitol, glycerol, glucose, galactose, ribose, and fructose) and amino acids (glycine, alanine, valine, and proline) were quantified by GC-MS. COO levels are provided in nanomoles and normalized to the GC-MS internal standard hydroxy benzylic acid (HBA) and 1 × 105 cells ml−1 [that is, concentration (Conc.) is in nanomoles per 105 cells or in nanomoles per milliliter culture]. S. microadriaticum, type A1; S. minutum, type B1; S. psygmophilum, type B2.
| Metabolite | Salinity | S. microadriaticum |
Symbiodinium sp. type A1 |
S. minutum | S. psygmophilum | ||||
| Conc. | SE | Conc. | SE | Conc. | SE | Conc. | SE | ||
| Floridoside | 25 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 38 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 51.0 | 8.0 | |
| 55 | 144.2 | 15.0 | 273.8 | 28.5 | 50.3 | 4.3 | 489.5 | 31.9 | |
| Inositol | 25 | 0.7 | 0.1 | 1.2 | 0.2 | 3.7 | 0.5 | 19.6 | 2.5 |
| 38 | 2.2 | 0.1 | 0.4 | 0.1 | 1.2 | 0.1 | 51.5 | 8.1 | |
| 55 | 14.3 | 1.5 | 2.6 | 0.3 | 35.2 | 3.0 | 111.1 | 7.2 | |
| Mannitol | 25 | 3.1 | 0.3 | 3.0 | 0.4 | 3.0 | 0.4 | 0.1 | 0.0 |
| 38 | 3.7 | 0.2 | 2.5 | 0.2 | 7.6 | 0.5 | 1.8 | 0.3 | |
| 55 | 2.8 | 0.3 | 2.0 | 0.2 | 4.6 | 0.4 | 1.6 | 0.1 | |
| Glycerol | 25 | 59.5 | 6.2 | 92.9 | 11.7 | 25.7 | 3.4 | 29.1 | 3.6 |
| 38 | 130.2 | 8.3 | 198.8 | 15.2 | 68.2 | 4.3 | 37.4 | 5.9 | |
| 55 | 107.0 | 11.1 | 37.2 | 3.9 | 6.9 | 0.6 | 10.9 | 0.7 | |
| Glucose | 25 | 85.3 | 8.9 | 181.0 | 22.7 | 27.3 | 3.6 | 50.8 | 6.4 |
| 38 | 111.9 | 7.2 | 69.9 | 5.4 | 32.7 | 2.0 | 193.9 | 30.5 | |
| 55 | 152.8 | 15.9 | 232.8 | 24.2 | 25.0 | 2.1 | 429.4 | 27.9 | |
| Galactose | 25 | 89.5 | 9.3 | 106.3 | 13.4 | 21.9 | 2.9 | 12.3 | 1.6 |
| 38 | 94.4 | 6.1 | 63.6 | 4.8 | 26.0 | 1.6 | 41.3 | 6.5 | |
| 55 | 127.2 | 13.2 | 166.2 | 17.3 | 27.2 | 1.4 | 74.3 | 4.8 | |
| Ribose | 25 | 4.6 | 0.5 | 8.8 | 1.1 | 2.8 | 0.3 | 2.9 | 0.3 |
| 38 | 8.3 | 0.5 | 0.2 | 0.0 | 1.0 | 0.1 | 5.9 | 0.9 | |
| 55 | 8.3 | 0.9 | 17.9 | 1.8 | 1.4 | 0.1 | 8.1 | 0.5 | |
| Fructose | 25 | 1.6 | 0.2 | 1.7 | 0.2 | 8.3 | 1.1 | 1.1 | 0.1 |
| 38 | 2.6 | 0.2 | 1.2 | 0.1 | 0.2 | 0.0 | 2.9 | 0.5 | |
| 55 | 2.4 | 0.2 | 7.8 | 0.8 | 1.3 | 0.1 | 3.7 | 0.2 | |
| Glycine | 25 | 18.6 | 2.0 | 6.2 | 0.8 | 4.1 | 0.5 | 0.0 | 0.0 |
| 38 | 31.5 | 2.0 | 1.3 | 0.1 | 5.1 | 0.5 | 13.3 | 2.1 | |
| 55 | 37.9 | 3.9 | 11.9 | 1.2 | 63.0 | 5.4 | 86.4 | 5.6 | |
| Alanine | 25 | 21.3 | 2.2 | 20.8 | 2.6 | 26.1 | 3.5 | 30.0 | 3.8 |
| 38 | 154.9 | 9.9 | 27.0 | 2.1 | 169.7 | 10.6 | 14.8 | 2.3 | |
| 55 | 53.5 | 5.6 | 108.3 | 11.3 | 30.0 | 2.5 | 108.2 | 7.0 | |
| Valine | 25 | 19.1 | 2.0 | 14.2 | 1.8 | 2.6 | 0.3 | 27.7 | 3.5 |
| 38 | 1.0 | 0.1 | 6.0 | 0.5 | 2.0 | 0.1 | 8.3 | 1.3 | |
| 55 | 7.7 | 0.8 | 9.8 | 1.0 | 3.6 | 0.3 | 10.7 | 0.7 | |
| Proline | 25 | 6.5 | 0.7 | 3.7 | 0.5 | 19.1 | 2.5 | 0.0 | 0.0 |
| 38 | 12.2 | 0.8 | 5.3 | 0.4 | 8.5 | 0.5 | 5.4 | 0.9 | |
| 55 | 9.8 | 1.0 | 3.4 | 0.3 | 3.5 | 0.3 | 19.1 | 1.3 | |
Fig. 2. Floridoside levels of Symbiodinium in vitro and in hospite at different salinities.
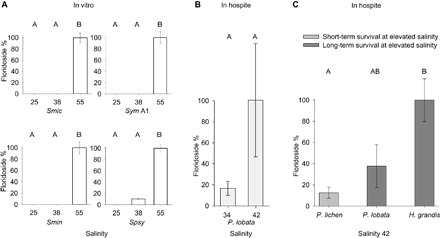
Floridoside levels represent measured amounts (in nanomoles) per 105 cells ml−1 for cultured Symbiodinium and per dry weight (in milligrams) of isolated Symbiodinium for coral samples. Data obtained for each experiment were normalized to the highest value (set to 100%). (A) Floridoside levels in four cultured Symbiodinium strains in vitro at low (25), ambient (38), and high salinities (55). Smic, S. microadriaticum (type A1); Sym A1, Symbiodinium sp. type A1; Smin, S. minutum (type B1); Spsy, S. psygmophilum (type B2). (B) Floridoside levels for S. thermophilum isolated from P. lobata cultured at salinities of 34 and 42. (C) Floridoside levels of Symbiodinium from corals with a different long-term survival capacity at high salinities after incubation at a salinity of 42. Floridoside levels were determined for Symbiodinium sp. type C96 (P. lichen), S. thermophilum (P. lobata), and Symbiodinium sp. type C40 (Hydnophora grandis). Error bars denote SE. Letters indicate Tukey’s HSD (honestly significant different) post hoc test differences based on pairwise comparisons of analysis of variance (ANOVA) results [groups with different letters are significantly different at P < 0.01 for (A) and P < 0.05 for (C)]. Osmolyte levels represent measured amounts (in nanomoles) per 105 cells ml-1.
We identified several other metabolites in the same GC-MS trace and could quantify a total of five additional carbohydrates (that is, glycerol, glucose, galactose, ribose, and fructose) and four amino acids (that is, glycine, alanine, valine, and proline) that serve as putative osmolytes for each of our samples. Metabolite levels were significantly different between different salinities (except proline) and Symbiodinium strains, as well as combinations thereof (all PANOVA < 0.01) (Table 2). This indicates that these metabolites are differentially regulated under changing salinities and in different Symbiodinium strains. Only the production of floridoside showed a substantial increase at high salinities, whereas the levels of all other metabolites (including mannitol and inositol) showed inconsistent patterns (Table 1).
Table 2. Statistical evaluation of metabolite changes of carbohydrates and amino acids at three salinities across four Symbiodinium strains.
Symbiodinium strain and salinity level are fixed factors, and strain*salinity serves as the interaction effect. Two-way ANOVA was used. Significance levels at P < 0.01 are in boldface.
| Measured metabolite | Symbiodinium strain | Salinity | Strain*salinity | |||
| F | P | F | P | F | P | |
| Floridoside | 84.9329 | <0.0001 | 408.1242 | <0.0001 | 61.0533 | <0.0001 |
| Inositol | 199.4686 | <0.0001 | 116.6508 | <0.0001 | 37.0999 | <0.0001 |
| Mannitol | 86.3291 | <0.0001 | 30.0316 | <0.0001 | 14.8469 | <0.0001 |
| Glycerol | 97.7188 | <0.0001 | 92.3146 | <0.0001 | 22.3281 | <0.0001 |
| Glucose | 74.5158 | <0.0001 | 66.2231 | <0.0001 | 32.1908 | <0.0001 |
| Galactose | 77.2113 | <0.0001 | 31.7549 | <0.0001 | 7.7703 | 0.0001 |
| Ribose | 45.7089 | <0.0001 | 47.2761 | <0.0001 | 33.2876 | <0.0001 |
| Fructose | 6.0851 | 0.0031 | 23.1370 | <0.0001 | 52.0966 | <0.0001 |
| Glycine | 55.2650 | <0.0001 | 282.0940 | <0.0001 | 56.3923 | <0.0001 |
| Alanine | 15.1440 | <0.0001 | 123.7890 | <0.0001 | 97.8385 | <0.0001 |
| Valine | 42.2583 | <0.0001 | 71.2012 | <0.0001 | 12.3394 | <0.0001 |
| Proline | 23.6038 | <0.0001 | 2.8518 | 0.0774 | 57.3734 | <0.0001 |
Because floridoside can be derived from glycerol and glucose/galactose (31), we investigated changes in the abundance of these molecules in detail (Fig. 3). In all Symbiodinium strains, a decrease in glycerol coincided with the accumulation of floridoside when comparing low- to high-salinity conditions (Fig. 3 and Table 1). Notably, glucose and galactose were enriched under high-salinity conditions in S. microadriaticum, Symbiodinium sp. type A1, and S. psygmophilum; it was only in S. minutum that these sugars remained at the same level or showed a slight decrease between low- and high-salinity conditions (Fig. 3 and Table 1).
Fig. 3. Osmolyte levels of floridoside and intermediates (glycerol, glucose, and galactose) at three salinities across four Symbiodinium strains.
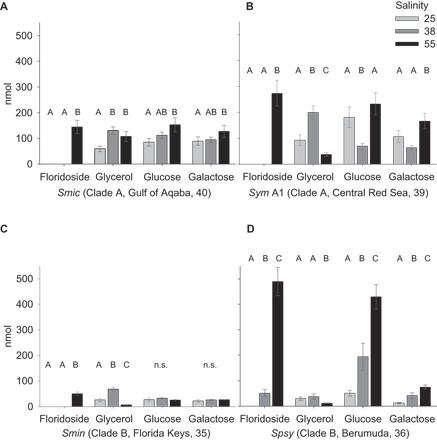
Glucose and galactose can be metabolized to glycerol (via the Calvin cycle) and cover UDP-galactose and glycerol 3-phosphate demands for floridoside synthesis. Bar graphs show floridoside, glycerol, glucose, and galactose levels for S. microadriaticum (type A1) (A), Symbiodinium sp. type A1 (B), S. minutum (type B1) (C), and S. psygmophilum (type B2) (D) cultures at low salinity (25; light gray), ambient salinity (38; gray), and high salinity (55; black) after 4 hours at 108 μmol photons m−a s−1. Clade designation, origin of strain, and respective average salinity are provided following strain abbreviations. Error bars denote SE. Letters indicate Tukey’s HSD post hoc differences based on pairwise comparisons of ANOVA results (groups with different letters are significantly different at P < 0.05). Tukey’s post hoc tests were not performed if ANOVAs yielded a nonsignificant F ratio, designated as n.s. (not significant). Osmolyte levels represent measured amounts (in nanomoles) per 105 cells ml−1.
High levels of floridoside in Symbiodinium from coral holobionts exposed to high salinities
To assess the importance of floridoside in coral holobionts, we exposed the coral Porites lobata associated with Symbiodinium thermophilum originating from the southern PAG (11) to different salinities and measured floridoside levels. All samples exposed to high salinity displayed a substantial increase of floridoside (~6-fold increase) (Fig. 2B and table S2).
We then compared floridoside levels in Symbiodinium from coral holobionts that show different capacities to survive at high salinities. We found that corals that were actively growing and surviving at a salinity of 42 for >24 months (that is, P. lobata and H. grandis) had higher floridoside levels than Porites lichen, which is only capable of surviving for a short period of time at this salinity (Fig. 2C and table S2) (11).
We also checked for homologs of the putative enzyme that converts glycerol 3-phosphate to floridoside in the available genomes of the corals Acropora digitifera and Stylophora pistillata to assess whether coral hosts are, in principle, able to synthesize floridoside. Following the study of Pade et al. (31), we used the gene sequence that encodes the enzymatically active floridoside phosphate synthase/phosphatase from the red alga Galdieria sulphuraria (Gasu_26940) to search for homologs in coral. We found no homologs for the corals A. digitifera and S. pistillata. Conversely, we found putative full-length homologs of this gene in all available Symbiodinium genomes, that is, S. microadriaticum (Smic14738, Smic32192, and Smic6078), S. minutum (symbB.v1.2.003359, symbB.v1.2.013114, and symbB.v1.2.013196), and Symbiodinium kawagutii (Skav203497). Hence, the coral genomes investigated do not harbor the floridoside phosphate synthase/phosphatase enzyme required to produce floridoside.
DISCUSSION
Here, we determined levels of the carbohydrates floridoside, inositol, and mannitol in response to high salinity in Symbiodinium in vitro and in hospite to assess the capacity of these COOs to fulfill a function in osmoadaptation to high salinities. Notably, other osmolytes that were not measured in our study, for example, taurines, betaines, and dimethylsulfoniopropionate (32–34), might also contribute to the osmoadaptation of Symbiodinium. Consequently, our data should not be considered a complete assessment of all osmolytes in Symbiodinium. However, we identified the osmolyte floridoside consistently and in increased amounts in Symbiodinium at high salinities. This shows that cultured Symbiodinium cells produce floridoside in response to salinity stress. We also found elevated floridoside levels in Symbiodinium of corals capable of long-term survival under high-salinity conditions (P. lobata and H. grandis). Hence, our work has uncovered a putative key COO that allows Symbiodinium to osmoadapt to extreme salinities in vitro and in hospite. The fact that our results show a consistent accumulation of floridoside across a range of Symbiodinium strains and experimental conditions provides strong support for the notion that increased floridoside levels constitute part of the osmoadaptive response to high salinities. Our findings also provide insight for our understanding of the role of osmoadaptation in the coral-Symbiodinium endosymbiosis, with implications for the coral stress response, as further discussed below.
Floridoside as a key osmolyte in Symbiodinium
Synthesis of the osmolyte floridoside has been identified as a conserved pathway in evolutionary distinct organisms, such as red algae, green algae, and Cryptophyceae (24, 31, 35). Floridoside is produced by UDP galactosyltransferases via condensation of glycerol 3-phosphate and UDP-galactose (31). UDP-galactose demands can be supplied via starch mobilization, resulting in increased glucose/galactose pools, as described for the green algae Dunaliella sp. under conditions of high salinity (36). A similar mechanism might explain the increased levels of glucose (in cultures of S. microadriaticum and Symbiodinium sp. type A1) and galactose (in all cultured strains) that we measured in Symbiodinium at high salinity (Fig. 3 and Table 1). The consistent increase of glucose and galactose in concert with up-regulation of floridoside suggests that these compounds fulfill a conserved osmotic adjustment function within the genus Symbiodinium (Fig. 3 and Table 1). Notably, the different Symbiodinium strains exhibited a differential response in regard to absolute floridoside production levels, but these differences did not follow a geographic (considering salinity levels at origin) or phylogenetic pattern (Fig. 3). Besides UDP-galactose, the second component required for floridoside synthesis, glycerol 3-phosphate, is likely supplied from photosynthesis or via the Calvin cycle (37). Glycerol 3-phosphate can be produced from glycerol, which is considered to be one of the main COOs in marine algae (38), although it has been shown to be released under osmotic pressure in Symbiodinium (16, 18, 39). Given that our analysis on available Symbiodinium genomes confirmed the presence of homologs for the enzyme that produces floridoside, it will be interesting to check for the presence and identity of the enzyme(s) required for floridoside synthesis in ecologically relevant Symbiodinium (for example, S. thermophilum) and to determine whether gene expression or duplication can be aligned with strain- or species-specific differences (40).
Osmoadaptation in the coral-Symbiodinium endosymbiosis, with implications for the coral stress response
Our results demonstrate that exposure to high salinities leads to higher endosymbiont floridoside levels in vitro and in hospite. This may point to the fact that elevated floridoside levels increase not only the capacity of Symbiodinium, but also that of the holobiont, to cope with the effect of high osmotic pressure in extreme environments. Furthermore, our results suggest that osmolarity changes within the coral tissue are noticed by endosymbiotic Symbiodinium; thus, both the coral and Symbiodinium respond to salinity changes, presumably by adjusting the inner osmolarity to the higher outside salinity. However, it remains to be determined whether Symbiodinium adjust their inner osmolarity the same way in hospite as in vitro (32). Since potentially any metabolite contributes to the osmolarity in hospite, the endosymbiotic environment in coral cells might differ from the in vitro seawater environment (38).
Beyond its function as an osmolyte, floridoside has been shown to act as an antioxidant with ROS-scavenging properties (25, 26). Hence, floridoside has the capability to convey osmoadaptation as well as to counter ROS produced in response to salinity or other forms of stress (6, 8, 24, 26–28). In particular, increased ROS is detrimental to photosystem II in photosynthetic organisms (5, 41–43). Therefore, the production of antioxidants at high salinities is potentially important for Symbiodinium, and floridoside represents an osmolyte that fulfills a ROS-scavenging function at the same time (26). Increasing levels of floridoside and oxidative stress in response to increased salinities were shown in the red algae Gracilaria sordida (44) and Gracilaria corticata (45), respectively. Future work should determine the exact role that floridoside plays in response to conditions of high salinity, as either an antioxidant, a COO, or both, for example, by comparing floridoside and ROS levels at ambient and increased salinities.
The notion that ROS-producing mechanisms for photosynthetic organisms are similar (if not identical) under salinity and heat stress (42) also has interesting implications for our understanding of the response to heat stress in Symbiodinium and, by extension, for the coral hosts. Similar to salt stress, heat stress results in malfunction of the photosynthetic machinery of Symbiodinium and in the production of ROS that may damage the algal cells and, in the case of the coral-algal endosymbiosis, may trigger bleaching (46). We find that Symbiodinium exposed to high salinity in vitro and in hospite accumulate high amounts of floridoside. Hence, elevated floridoside levels in high-salinity environments may increase the ability to tolerate heat stress in Symbiodinium and, by extension, their coral hosts through scavenging of increased ROS levels. Consequently, the thermal resilience of coral holobionts may potentially increase under conditions of high salinity because of the accumulation and inherent antioxidative capabilities of floridoside. Experimental data connecting increased floridoside levels to decreased ROS and bleaching levels at increased salinities are in demand to support this potential link.
MATERIALS AND METHODS
Symbiodinium cultures
S. microadriaticum CCMP2467 (type A1; originally isolated from S. pistillata, Red Sea, Gulf of Aqaba) (40), Symbiodinium sp. type A1 (originally isolated from Astreopora sp., Central Red Sea) (47), S. minutum Mf1.05b (type B1; isolated from Orbicella faveolata, Florida Keys, United States) (48, 49), and S. psygmophilum Mf10.14b.02 (type B2; isolated from Oculina diffusa, Bermuda, UK) (50, 51) were cultured in f/2 medium without silicium under a photon flux of 108 μmol m−2 s−1 at 26°C (52). The f/2 medium was prepared from sterile filtered Red Sea water [with a salinity of 38 and complemented with NaNO3, NaH2PO4, vitamins, and trace metals (53)]. For each strain, we used replicate culture flasks and prepared three salt-adjusted f/2 media [salinities of 25, 38, and 55; following the study of van der Merwe et al. (54)] either by adding appropriate amounts of NaCl or by diluting the media with double-distilled H2O (ddH2O). Triplicates of 5 ml of Symbiodinium cultures at exponential growth (105 to 106 cells ml−1) were transferred to 35 ml of salt-adjusted f/2 media for each salinity and incubated for 4 hours under culturing conditions. Symbiodinium cells were subsequently harvested by centrifugation (4500g, 10 min, 4°C). Cells were counted by fluorescence-activated cell sorting (FACS). To do this, 1 ml of each Symbiodinium culture was collected and fixed with formalin. After washing, samples were resuspended in 1 ml of ddH2O and labeled with SYBR Gold (Thermo Fisher Scientific), of which 150-μl aliquots were supplied for FACS (50-μl counting volume). FACS measurements were conducted in triplicate on a cell analyzer (LSRFortessa, BD Biosciences). Stained DNA or RNA was excited via a 488-nm blue laser and emission-detected for total nucleotide detection (Alexa Fluor 488 filters, Life Technologies). Detection of valid signals was a combined measure of forward and side scattering and of both fluorescence signals (that is, SYBR Gold and chlorophyll autofluorescence). FACS data were analyzed by FlowJo 10 flow cytometry analysis software (FlowJo LLC).
Coral cultures
Corals were kept in long-term culture (>24 months) in different compartments of the experimental coral mesocosm facility at the University of Southampton at salinity levels mimicking those of their habitats of origin [salinity of 42 for P. lobata from the PAG (11) and salinity of ~36.5 for P. lichen and H. grandis from the Indo-Pacific (11, 55, 56)]. P. lobata was additionally cultured under a reduced salinity condition of 34 for >24 months. Corals were kept at a temperature of 26°C with a 10-hour/14-hour light/dark cycle under a photon flux of 150 μmol m−2 s−1 (11). Light and temperature levels suitable for long-term culture of the corals were established during previous work (55, 56). These three species were studied owing to their different capacity for survival at elevated salinities: P. lichen associated with Symbiodinium sp. type C96 exhibits short-term survival (11), whereas P. lobata with S. thermophilum (11, 14) and H. grandis with Symbiodinium sp. type C40 both show long-term survival.
Survival capacity was determined before the experiment by incubating 10 replicate colonies for >24 months at a salinity of 42. In contrast to other Indo-Pacific species (including P. lichen) (11), P. lobata and H. grandis have not suffered any mortality and have been actively growing during this time (56). In the present experiments, replicate colonies of P. lichen and H. grandis previously cultured at lower salinities (34 and 36.5) were gradually adjusted to a high salinity of 42 over 2 days before being moved to the high-salinity compartment for 12 days before sampling. Replicate colonies were produced by earlier fragmentation.
Using an airbrush, coral tissue was blasted off the skeleton with ice-cold, sterile-filtered, freshly prepared artificial seawater with the same salinity as the culture-rearing water. Three coral colonies were used per species to extract zooxanthellae. Symbiodinium cells were precipitated from the homogenate by centrifugation at 2500g for 5 min at 4°C. The cells were washed twice (to remove the host tissue fraction and to prevent residual salt to infer with the downstream GC-MS analyses), first in precooled seawater and then in ddH2O, each step followed by centrifugation and resuspension. Exposure to ddH2O was limited to ~30 s to minimize potential effects of the hypo-osmotic environment. It should also be noted that because of the slow diffusion of water through the cell wall/membrane, the Symbiodinium cells experienced only a minimal influx of water and were protected from bursting by their rigid cell walls. Furthermore, any water that had entered the cells was removed in the process of freeze-drying and hence could not interfere with osmolyte downstream analysis. After the final centrifugation step, all liquid was removed, and the cell pellet was lyophilized for 14 hours. Although this procedure might still retain traces of host animal tissue/cells and osmolyte profiles may contain a host component, we consider the potential carry-over minor, in particular because corals do not seem to harbor the gene to produce floridoside.
Metabolite extraction and recovery
Cell pellets from Symbiodinium cultures and Symbiodinium extracted from coral tissues were resuspended and washed with 30 ml of sterile seawater on ice, pelleted, and washed for ~30 s, with further 5 ml of ddH2O to remove residual salt. After a further centrifugation step, pellets were resuspended in 5 ml of ddH2O, and cells were disrupted by tip ultrasonication for 4 min at 3-s pulsing and 6-s pause. Cell debris was removed by centrifugation at 20,000g for 20 min at 4°C. Proteins, DNA, or RNA was removed by ethanol precipitation by adding nine parts of −20°C ethanol to one part of supernatant. The precipitate was pelleted and removed by centrifugation, whereas the supernatant was frozen in liquid nitrogen and lyophilized. Dry samples were dissolved in 240 μl of ddH2O, spiked with 10 μl of internal standard [HBA (1 μg/μl) in ddH2O], transferred into GC vials, and dried under vacuum. For derivatization, 50 μl of MOX reagent (2% methoxamine HCl in pyridine) was added to each sample, and the solution was heated to 75°C for 1 hour. Afterward, 100 μl of MSTFA solution [N-methyl-N-(trimethylsilyl)trifluoroacetamide, 1% trimethylchlorosilane; Thermo Scientific] was added, and samples were heated for 1 hour at 75°C. Each sample vial was centrifuged at 2000g for 10 min, and 100 μl of the supernatant was transferred to glass inserts placed inside GC vials.
GC-MS analysis, quantification, and analysis
Derivatized carbohydrates, amino acids, and further intracellular compounds were characterized and quantified by GC-MS. For separation, an HP-5ms column (Agilent Technologies) and a temperature profile starting at 70°C were chosen. Temperature was increased in increments of 6°C min−1 up to 230°C, followed by increments of 60°C min−1 to a maximum of 280°C where it was held for 4 min. Metabolites were quantified by standard curves produced with pure glucose (99.5%; Sigma) and glycerol (≥99.5%, ACS Reagent–grade; Sigma), with 60, 30, 10, 1, and 0.1 μg of both compounds. The calibration standards were spiked with 1 μg of HBA, derivatized for GC-MS, and analyzed as described above. All samples were prepared and measured in triplicate. GC-MS data were processed (that is, background subtraction, peak picking, and integration; OpenChrom v. 0.901, Lablicate UG) and MS ionization spectra–identified (NIST MS Software 2.0, Agilent Technologies). Statistical testing was conducted on normalized quantities of metabolites (in nanomoles) using ANOVAs and Tukey’s HSD post hoc tests to assess differences between pairwise comparisons. In the case of cultured Symbiodinium, we normalized to 105 cells ml−1. For corals, Symbiodinium extracts were normalized over dry weights in milligrams.
Floridoside homologs in coral and Symbiodinium genomes
We searched for homologs of the putative enzyme that converts glycerol 3-phosphate to floridoside (31) in the available coral and Symbiodinium genomes via BLASTp on reefgenomics.org (57) using an e value cutoff of <10−5. Briefly, the amino acid sequence for the gene (Gasu_26940) coding for floridoside phosphate synthase/phosphatase from the red alga G. sulphuraria was queried against the genomes of A. digitifera (58) and S. pistillata (59), as well as against the genomes of S. microadriaticum (40), S. minutum (49), and S. kawagutii (60).
Acknowledgments
We thank N. M. Kharbatia and A. Ortega for assistance in method development, measurements, and analysis; M. Ziegler and A. Roik for support in statistical analysis; X. Gong for Symbiodinium extraction; and T. LaJeunesse for strain determination. Funding: This study was supported by the King Abdullah University of Science and Technology under baseline funds to C.R.V. and the Center Competitive Fund Program FCC/1/1973-22-01. Further funding was contributed by the Natural Environment Research Council (NE/K00641X/1 to J.W.) and the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/European Research Council (grant agreement no. 311179 to J.W.). Author contributions: M.A.O., T.R., and C.R.V. designed and conceived the experiments. M.A.O., T.R., C.R.V., C.D., and J.W. generated, analyzed, and interpreted data. C.R.V., C.D., and J.W. contributed cultures, reagents, and materials. C.R.V. wrote the manuscript, with contributions from T.R., C.D., J.W., and M.A.O. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Contact the corresponding author for animal specimens.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1602047/DC1
table S1. Metabolites in GC-MS traces and identification information for NIST MS 2.0 Library Search.
table S2. Overview of floridoside amounts from Symbiodinium of coral holobionts at different salinities.
REFERENCES AND NOTES
- 1.M. L. Reaka-Kudla, D. E. Wilson, E. O. Wilson, Biodiversity II: Understanding and Protecting Our Biological Resources (Joseph Henry Press, 1996). [Google Scholar]
- 2.Muscatine L., Porter J. W., Reef corals: Mutualistic symbioses adapted to nutrient-poor environments. BioScience 27, 454–460 (1977). [Google Scholar]
- 3.Falkowski P. G., Dubinsky Z., Muscatine L., Porter J. W., Light and the bioenergetics of a symbiotic coral. BioScience 34, 705–709 (1984). [Google Scholar]
- 4.Muscatine L., Porter J. W., Kaplan I. R., Resource partitioning by reef corals as determined from stable isotope composition. Mar. Biol. 100, 185–193 (1989). [Google Scholar]
- 5.M. P. Lesser, in Coral Reefs: An Ecosystem in Transition, Z. Dubinsky, N. Stambler, Eds. (Springer, 2011), pp. 405–419. [Google Scholar]
- 6.Wiedenmann J., D’Angelo C., Smith E. G., Hunt A. N., Legiret F.-E., Postle A. D., Achterberg E. P., Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 160–164 (2013). [Google Scholar]
- 7.Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C., Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 23, 490–497 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Brown B. E., Coral bleaching: Causes and consequences. Coral Reefs 16, S129–S138 (1997). [Google Scholar]
- 9.Ainsworth T. D., Heron S. F., Ortiz J. C., Mumby P. J., Grech A., Ogawa D., Eakin C. M., Leggat W., Climate change disables coral bleaching protection on the Great Barrier Reef. Science 352, 338–342 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Bruno J. F., Valdivia A., Coral reef degradation is not correlated with local human population density. Sci. Rep. 6, 29778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Angelo C., Hume B. C. C., Burt J., Smith E. G., Achterberg E. P., Wiedenmann J., Local adaptation constrains the distribution potential of heat-tolerant Symbiodinium from the Persian/Arabian Gulf. ISME J. 9, 2551–2560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röthig T., Ochsenkühn M. A., Roik A., van der Merwe R., Voolstra C. R., Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngugi D. K., Antunes A., Brune A., Stingl U., Biogeography of pelagic bacterioplankton across an antagonistic temperature–salinity gradient in the Red Sea. Mol. Ecol. 21, 388–405 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Hume B. C. C., Voolstra C. R., Arif C., D’Angelo C., Burt J. A., Eyal G., Loya Y., Wiedenmann J., Ancestral genetic diversity associated with the rapid spread of stress-tolerant coral symbionts in response to Holocene climate change. Proc. Natl. Acad. Sci. U.S.A. 113, 4416–4421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coles S. L., Coral species diversity and environmental factors in the Arabian Gulf and the Gulf of Oman: A comparison to the Indo-Pacific region. Atoll Res. Bull. 507, 497–508 (2003). [Google Scholar]
- 16.Suescún-Bolívar L. P., Iglesias-Prieto R., Thomé P. E., Induction of glycerol synthesis and release in cultured Symbiodinium. PLOS ONE 7, e47182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goiran C., Allemand D., Galgani I., Transient Na+ stress in symbiotic dinoflagellates after isolation from coral-host cells and subsequent immersion in seawater. Mar. Biol. 129, 581–589 (1997). [Google Scholar]
- 18.Suescún-Bolívar L. P., Traverse G. M. I., Thomé P. E., Glycerol outflow in Symbiodinium under osmotic and nitrogen stress. Mar. Biol. 163, 128 (2016). [Google Scholar]
- 19.Reed R. H., Use and abuse of osmo-terminology. Plant Cell Environ. 7, 165–170 (1984). [Google Scholar]
- 20.Ahmad I., Hellebust J. A., Osmoregulation in the extremely euryhaline marine micro-alga Chlorella autotrophica. Plant Physiol. 74, 1010–1015 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadka A., Lers A., Zamir A., Avron M., A critical examination of the role of de novo protein synthesis in the osmotic adaptation of the halotolerant alga Dunaliella. FEBS Lett. 244, 93–98 (1989). [Google Scholar]
- 22.Kirst G. O., Salinity tolerance of eukaryotic marine algae. Annu. Rev. Plant Biol. 41, 21–53 (1990). [Google Scholar]
- 23.Chen H., Jiang J. G., Osmotic responses of Dunaliella to the changes of salinity. J. Cell. Physiol. 219, 251–258 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Hoef-Emden K., Osmotolerance in the cryptophyceae: Jacks-of-all-trades in the Chroomonas clade. Protist 165, 123–143 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Kim M., Li Y.-X., Dewapriya P., Ryu B., Kim S.-K., Floridoside suppresses pro-inflammatory responses by blocking MAPK signaling in activated microglia. BMB Rep. 46, 398–403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y.-X., Lee S.-H., Qian Z.-J., Kim S.-K., Inhibitors of oxidation and matrix metalloproteinases, floridoside, and d-isofloridoside from marine red alga Laurencia undulata. J. Agric. Food Chem. 58, 578–586 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa P. M., Bressan R. A., Zhu J.-K., Bohnert H. J., Plant cellular and molecular responses to high salinity. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 51, 463–499 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Bose J., Rodrigo-Moreno A., Shabala S., ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Shen B., Jensen R. G., Bohnert H. J., Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 115, 527–532 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogoreutz C., Rädecker N., Cárdenas A., Gärdes A., Voolstra C. R., Wild C., Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob. Chang. Biol. 10.1111/gcb.13695 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Pade N., Linka N., Ruth W., Weber A. P. M., Hagemann M., Floridoside and isofloridoside are synthesized by trehalose 6-phosphate synthase-like enzymes in the red alga Galdieria sulphuraria. New Phytol. 205, 1227–1238 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Yancey P. H., Heppenstall M., Ly S., Andrell R. M., Gates R. D., Carter V. L., Hagedorn M., Betaines and dimethylsulfoniopropionate as major osmolytes in cnidaria with endosymbiotic dinoflagellates. Physiol. Biochem. Zool. 83, 167–173 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Pierce S. K., Invertebrate cell volume control mechanisms: A coordinated use of intracellular amino acids and inorganic ions as osmotic solute. Biol. Bull. 163, 405–419 (1982). [Google Scholar]
- 34.Eierman L. E., Hare M. P., Transcriptomic analysis of candidate osmoregulatory genes in the eastern oyster Crassostrea virginica. BMC Genomics 15, 503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Eggert, U. Karsten, in Red Algae in the Genomic Age, J. Seckbach, D. J. Chapman, Eds. (Springer, 2010), vol. 13, pp. 443–456. [Google Scholar]
- 36.Liska A. J., Shevchenko A., Pick U., Katz A., Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol. 136, 2806–2817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Amotz A., Grunwald T., Osmoregulation in the halotolerant alga Asteromonas gracilis. Plant Physiol. 67, 613–616 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayfield A. B., Gates R. D., Osmoregulation in anthozoan–dinoflagellate symbiosis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 147, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Burriesci M. S., Raab T. K., Pringle J. R., Evidence that glucose is the major transferred metabolite in dinoflagellate–cnidarian symbiosis. J. Exp. Biol. 215, 3467–3477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aranda M., Li Y., Liew Y. J., Baumgarten S., Simakov O., Wilson M. C., Piel J., Ashoor H., Bougouffa S., Bajic V. B., Ryu T., Ravasi T., Bayer T., Micklem G., Kim H., Bhak J., LaJeunesse T. C., Voolstra C. R., Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latifi A., Ruiz M., Zhang C.-C., Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S. I., Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lesser M. P., Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Ekman P., Yu S., Pedersen M., Effects of altered salinity, darkness and algal nutrient status on floridoside and starch content, α-galactosidase activity and agar yield of cultivated Gracilaria sordida. Brit. Phycol. J. 26, 123–131 (1991). [Google Scholar]
- 45.Kumar M., Kumari P., Gupta V., Reddy C. R. K., Jha B., Biochemical responses of red alga Gracilaria corticata (Gracilariales, Rhodophyta) to salinity induced oxidative stress. J. Exp. Mar. Biol. Ecol. 391, 27–34 (2010). [Google Scholar]
- 46.Lesser M. P., Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 16, 187–192 (1997). [Google Scholar]
- 47.X. Gong, “A survey into taxonomic and physiological differences of Symbiodinium sp., the photosynthetic symbiont of reef-building corals,” thesis, King Abdullah University of Science and Technology (2012). [Google Scholar]
- 48.Bayer T., Aranda M., Sunagawa S., Yum L. K., DeSalvo M. K., Lindquist E., Coffroth M. A., Voolstra C. R., Medina M., Symbiodinium transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLOS ONE 7, e35269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., Takeuchi T., Hisata K., Tanaka M., Fujiwara M., Hamada M., Seidi A., Fujie M., Usami T., Goto H., Yamasaki S., Arakaki N., Suzuki Y., Sugano S., Toyoda A., Kuroki Y., Fujiyama A., Medina M., Coffroth M. A., Bhattacharya D., Satoh N., Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408 (2013). [DOI] [PubMed] [Google Scholar]
- 50.LaJeunesse T. C., Parkinson J. E., Reimer J. D., A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J. Phycol. 48, 1380–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Parkinson J. E., Baumgarten S., Michell C. T., Baums I. B., LaJeunesse T. C., Voolstra C. R., Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 8, 665–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimann B. E. F., Lewin J. M. C., Guillard R. R. L., Cyclotella cryptica, a new brackish-water diatom species. Phys. Chem. Chem. Phys. 3, 75–84 (1963). [Google Scholar]
- 53.Guillard R. R. L., Ryther J. H., Studies of marine planktonic diatoms: I. Cyclotella Nana Hustedt, and Detonula Confervacea (Cleve) Gran. Can. J. Microbiol. 8, 229–239 (1962). [DOI] [PubMed] [Google Scholar]
- 54.van der Merwe R., Röthig T., Voolstra C. R., Ochsenkühn M. A., Lattemann S., Amy G. L., High salinity tolerance of the Red Sea coral Fungia granulosa under desalination concentrate discharge conditions: An in situ photophysiology experiment. Front. Mar. Sci. 1, 58 (2014). [Google Scholar]
- 55.D’Angelo C., Denzel A., Vogt A., Matz M. V., Oswald F., Salih A., Nienhaus G. U., Wiedenmann J., Blue light regulation of host pigment in reef-building corals. Mar. Ecol. Prog. Ser. 364, 97–106 (2008). [Google Scholar]
- 56.D’Angelo C., Wiedenmann J., An experimental mesocosm for long-term studies of reef corals. J. Mar. Biol. Assoc. U.K. 92, 769–775 (2012). [Google Scholar]
- 57.Liew Y. J., Aranda M., Voolstra C. R., Reefgenomics.Org—A repository for marine genomics data. Database 2016, baw152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., Fujie M., Fujiwara M., Koyanagi R., Ikuta T., Fujiyama A., Miller D. J., Satoh N., Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharya D., Agrawal S., Aranda M., Baumgarten S., Belcaid M., Drake J. L., Erwin D., Foret S., Gates R. D., Gruber D. F., Kamel B., Lesser M. P., Levy O., Liew Y. J., MacManes M., Mass T., Medina M., Mehr S., Meyer E., Price D. C., Putnam H. M., Qiu H., Shinzato C., Shoguchi E., Stokes A. J., Tambutté S., Tchernov D., Voolstra C. R., Wagner N., Walker C. W., Weber A. P., Weis V., Zelzion E., Zoccola D., Falkowski P. G., Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5, e13288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin S., Cheng S., Song B., Zhong X., Lin X., Li W., Li L., Zhang Y., Zhang H., Ji Z., Cai M., Zhuang Y., Shi X., Lin L., Wang L., Wang Z., Liu X., Yu S., Zeng P., Hao H., Zou Q., Chen C., Li Y., Wang Y., Xu C., Meng S., Xu X., Wang J., Yang H., Campbell D. A., Sturm N. R., Dagenais-Bellefeuille S., Morse D., The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1602047/DC1
table S1. Metabolites in GC-MS traces and identification information for NIST MS 2.0 Library Search.
table S2. Overview of floridoside amounts from Symbiodinium of coral holobionts at different salinities.


