Abstract
Mechanistic target of rapamycin (mTOR) is a serine/threonine kinase originally discovered as the molecular target of the immunosuppressant rapamycin. mTOR forms two compositionally and functionally distinct complexes, mTORC1 and mTORC2, which are crucial for coordinating nutrient, energy, oxygen, and growth factor availability with cellular growth, proliferation, and survival. Recent studies have identified critical, non-redundant roles for mTORC1 and mTORC2 in controlling B cell development, differentiation, and functions, and have highlighted emerging roles of the Folliculin-Fnip protein complex in regulating mTOR and B cell development. In this review, we summarize the basic mechanisms of mTOR signaling; describe what is known about the roles of mTORC1, mTORC2, and the Folliculin/Fnip1 pathway in B cell development and functions; and briefly outline current clinical approaches for targeting mTOR in B cell neoplasms. We conclude by highlighting a few salient questions and future perspectives regarding mTOR in B lineage cells.
Keywords: mTOR, B lymphocyte development, Folliculin, Fnip1
1. Introduction
Mechanistic Target of Rapamycin (mTOR) is a highly conserved serine-threonine kinase expressed in all eukaryotes. mTOR was originally discovered when an agent produced by the bacterium Streptomyces hygroscopicus isolated from Easter Island (also known as Rapa Nui) was found to arrest the growth of yeast in the G1-phase of the cell cycle [1,2]. The specificity of this new compound named rapamycin after the island from which it was isolated, was subsequently defined using classical genetics in yeast, which resulted in the identification of a rapamycin-resistant mutant called Tor (target of rapamycin) [3,4]. The mammalian ortholog of Tor was later cloned by multiple research groups [5–8], and although several names were initially proposed, Mammalian (now Mechanistic) Target of Rapamycin (mTOR) evolved as the name of choice.
Although rapamycin was initially developed as an anti-fungal agent, researchers recognized early on that it also blocked cell cycle progression in T lymphocytes, which led to its approval in 1999 by the Food and Drug Administration as an immunosuppressant to help prevent rejection in organ transplant recipients. Subsequent studies revealed that mTOR, similar to the yeast ortholog, is a central regulator of cellular growth and proliferation in response to diverse environmental cues including nutrients, oxygen, and energy levels (reviewed in [9–11]). Not surprisingly, mTOR was also found to be deregulated in a number of disease conditions including certain types of cancers, type-II diabetes, obesity, and several neurodegenerative disorders [9,11]. Intense efforts to develop pharmacological mTOR inhibitors in addition to the allosteric inhibitor rapamycin (also known as sirolimus) and its analogs, resulted in the development of ATP-competitive inhibitors such as Torin. In addition to its use in transplant recipients, mTOR inhibitors are now being utilized, or are proposed to be utilized, in treatment regimens for many diseases including cancers such as lymphoma and renal carcinomas [12]; autoimmune disease such as systemic lupus erythematosus [13]; neurodegenerative diseases including Alzheimer’s and Parkinson’s [14]; lysosomal storage diseases [15]; and for the extension of a healthy lifespan [16]. The increased and widespread use of rapamycin and other mTOR inhibitors highlights the need to more fully understand the molecular mechanisms of how mTOR functions, the potential toxicities of mTOR inhibitors, and the biological and molecular consequences of inhibiting mTOR in many different cell types.
Recent studies in immune cells have highlighted that mTOR not only couples nutrient availability to cell growth and proliferation, but also controls cell differentiation and activation-induced responses in B and T lymphocytes (reviewed in [17–19]), as well as natural killer cells, neutrophils, macrophages, and dendritic cells (reviewed in [20]). The biological complexity of mTOR signaling has been most elegantly demonstrated in T lymphocytes, in which multiple studies have demonstrated the evolution of mTOR from being primarily a “nutrient sensor” in yeast, to a highly complex orchestrator of mammalian cell growth and cell fate determination in response to a diverse array of inputs. In this review, we will highlight the basic cellular and molecular mechanisms of mTOR signaling derived from studies in mostly non-B cells, outline what is known about the importance of mTOR signaling in B lymphocyte development and functions, summarize current clinical approaches to targeting mTOR in B cell neoplasms, and conclude with a few salient questions and future perspectives regarding mTOR in B lineage cells.
2. Overview of mTOR Signaling Pathways
2.1. mTORC1 and mTORC2
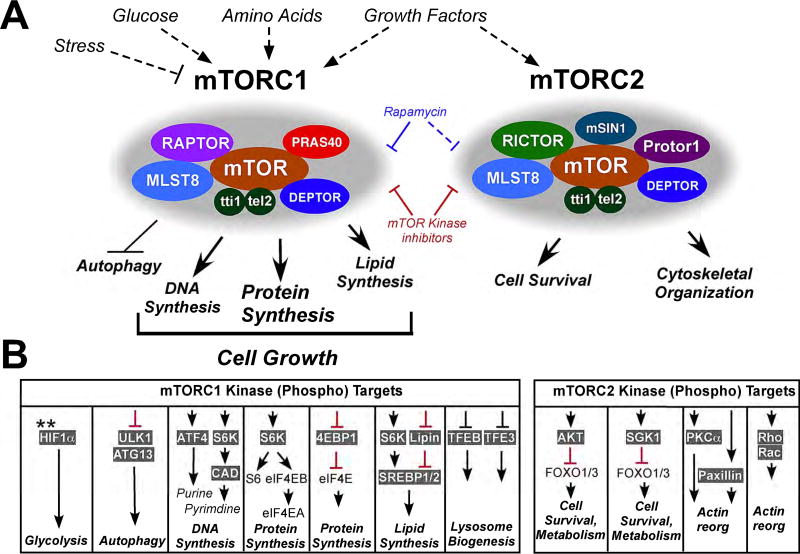
After the initial discovery of mTOR, follow-up studies in yeast and mammalian cells revealed that mTOR forms the catalytic core of two important but functionally distinct multi-protein complexes, mTORC1 and mTORC2, which are composed of both unique and shared components (Figure 1A) (reviewed in [9,11,21]). Specifically, mTORC1 is composed of mTOR in association with two unique regulatory protein subunits, Raptor (rapamycin-sensitive adapter protein of mTOR) and Pras40 (proline-rich AKT substrate 40 kDa), and the shared components mLST8 (mammalian lethal with Sec-13 protein 8), Tti1/Tel2 (Tel2 interacting protein 1/telomere maintenance 2), and Deptor (dep domain continingTOR-interacting protein). In contrast, mTORC2 contains the unique components Rictor (rapamycin-insensitive companion of mTOR), mSin1 (stress-activated protein kinase-interacting protein 1), and Protor 1/2 (protein binding rictor 1/2), in addition to the shared components mLST8, Tti1/Tel2, and Deptor. Although both mTORC1 and mTORC2 are central mediators of growth factor responses and cellular metabolism, mTORC1 is uniquely activated by environmental cues such as adequate nutrients (amino acids), energy (ATP/AMP), and oxygen availability, which results in activation of pathways leading to cellular growth (protein, DNA, and lipid synthesis) and inhibition of autophagy. Conversely, mTORC1-mediated cellular growth is inhibited by cellular stresses such as DNA damage, low energy states, and hypoxia. mTORC2, by comparison, is more specifically activated by growth factor signaling, and facilitates cell survival and cytoskeletal reorganization to promote cell migration and adhesion [9,11]. Importantly, mTORC1 and mTORC2 are differentially inhibited by rapamycin: mTORC1 is sensitive to acute rapamycin treatment, whereas mTORC2 assembly and activity is altered only after chronic treatment or after high doses of rapamycin [22,23] . In contrast, both mTORC1 and mTORC2 are inhibited by mTOR kinase inhibitors (mTOR-KIs) such as Torin [24–27]. Understanding how mTORC1 and mTORC2 differentially control the development and function of specific immune cells is essential to effectively design more effective therapeutic strategies using rapalogs and mTOR kinase inhibitors, with fewer undesirable side-effects.
Figure 1. mTORC1 and mTORC2 complexes and downstream signaling pathways.
(A) The mTOR protein forms two unique complexes called mTORC1 and mTORC2. mTORC1 is activated by growth factors, amino acids, sufficient energy and oxygen, whereas mTORC2 is primarily responsive to growth factors. mTORC1 consists of mTOR, Raptor, Pras40, Deptor, MLSt8, and Tti/tel2; mTORC2 consists of mTOR, Rictor, Protor1, as well as Deptor, Mlst8, and Tti/Tel2. mTORC1 is inhibited by rapamycin, whereas mTORC2 is relatively rapamycin resistant except at high doses. However, both mTORC1 and mTORC2 are sensitive to mTOR kinase inhibitors. (B) mTORC1 regulates numerous processes integral to cell growth and proliferation including protein synthesis, DNA synthesis, lipid synthesis, and glycolysis while inhibiting autophagy. In contrast, mTORC2 enhances cell survival and stimulates cytoskeletal reorganization. Important downstream phosphorylation targets of mTORC1 and mTORC2, and the specific processes regulated by each signaling event are shown.
**mTORC1 regulates HIF1a levels by translational control.
2.2. Regulation of mTOR complexes by antigen receptor, growth factor, and nutrient signals
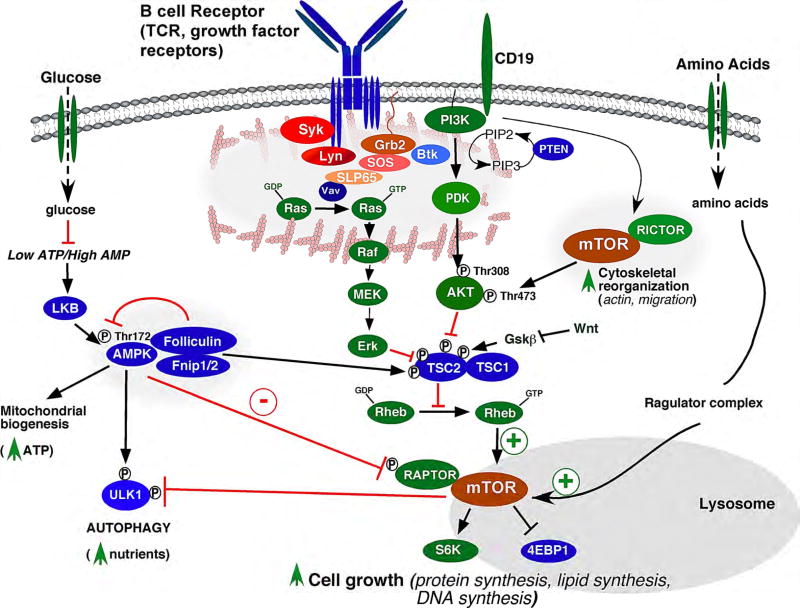
Depending on the organism and cell type, mTORC1 is regulated by specific growth factor signals, nutrients, oxygen levels, and cellular stress (see [9] for review). While details of the signaling networks that regulate mTOR complexes in B cells are not completely understood, signals derived from the B cell receptor (BCR), toll-like receptor 4 (TLR-4), and CD40 co-stimulatory receptor all require intact mTORC1 signaling [28,29]. In response to antigen, aggregation of the BCR results in phosphorylation of the transmembrane subunits of the BCR (Igα and Igβ) by Src-family protein tyrosine kinases (PTKs), including Lyn, Blk and Fyn, followed by recruitment and activation of the Syk PTK (Figure 2). Syk phosphorylates Blnk/SLP65 (B-cell linker protein), permitting recruitment of Grb2, Btk, Vav and other Src homology 2 (SH2) domain-containing proteins [30,31]. The guanine nucleotide exchange factor protein Vav activates the Ras GTPase by promoting GDP exchange for GTP. Active Ras in turn activates the Raf/Mek (MAP kinase kinase)/Erk1/2 (MAP kinase 1/2) signaling cascade, resulting in phosphorylation of ERK target proteins. In parallel, the CD19 co-receptor is phosphorylated by Lyn, resulting in recruitment of PI3K to the plasma membrane where it activates PDK and Akt/PKB. Activation of both ERK and Akt leads to dual phosphorylation of TSC2 (tuberous sclerosis complex protein 2, or tuberin) [32], which, together with TSC1, inactivate mTOR by forming a GTPase activating protein (GAP) complex for Rheb (Ras homolog enriched in brain). Thus, phosphorylation of TSC2 results in inactivation of the TSC1/TSC2 complex, which in turn promotes Rheb-GTP activation of mTORC1.
Figure 2. Activation of the mTOR signaling pathway modeled in B cells.
Antigen binding and aggregation of the B cell receptor (BCR) combined with co-receptor ligation leads to activation of the Ras/Raf/Mek/Erk and PI3 kinase/Akt protein kinase cascades, which have been shown to activate mTORC1 (mTOR/Raptor complex) activation through phosphorylation and inhibition of TSC2 (tuberous sclerosis complex protein 2). The TSC complex negatively regulates mTORC1 by suppressing activation of the Rheb GTPase, which necessary for activating mTORC1 catalytic activity. Although not demonstrated in B cells, amino acid sufficiency is also necessary to activate mTORC1 by association with active v-ATPase/Ragulator/Rag GTPase complexes on the surface of the lysosome (see Figure 3 for details). mTOR is also negatively regulated by low energy (low ATP/high AMP) levels through activation of AMP kinase (AMPK). Activated AMPK inhibits mTORC1 activity through phosphorylation and inhibition of Raptor (a positive regulator of mTOR) and phosphorylation and activation of the TSC complex. AMPK supports the generation of ATP and nutrients through upregulation of mitochondrial biogenesis and autophagy, while mTORC1 suppresses autophagy and promotes cell growth and proliferation. The PI3K pathway also activates mTORC2 (mTOR/Rictor complex), which also increases mTORC1 activity through direct phosphorylation and activation of Akt. Wnt signaling inhibits mTORC1 by inhibiting Gsk3b, which is a positive regulator of the TSC complex.
*Positive regulators of mTORC1 signaling are depicted in green, and negative regulators of mTORC1 are depicted in blue. Folliculin and Fnip1/2 have been described as both positive and negative regulators of mTORC1.
In addition to growth factor and antigen receptor signaling, sufficient amino acids have also been shown to be necessary for mTORC1 activation, although these studies have not been explored in B cells. In particular, sufficient cytosolic and lysosomal concentrations of amino acids (AAs) are required for mTORC1 recruitment and activation at the lysosomal surface (Figure 3) (reviewed in [33,34]). Intra-lysosomal AAs including leucine and arginine promote the loading of two heterodimers of the Rag GTPases RagA/B and RagC/D, with GTP and GDP respectively via “inside-out signaling”. The Rag GTPases are bound to the lysosomal surface in association with the vacuolar-ATPase (v-ATPase) and the scaffolding/activating Ragulator complex. Loading of RagA/B and RagC/D with their respective activating nucleotides enables the complex to interact with Raptor and recruit mTORC1 to the lysosomal surface where it can be activated by Rheb-GTP. In the absence of sufficient AAs, the mTORC1 complex dissociates from the lysosome and is inactivated. Thus, mTORC1 activation and cellular growth are tightly regulated by amino acid levels which modulate mTORC1 recruitment to the lysosome for subsequent activation by a “second” signal originating from growth factor and/or antigen receptors.
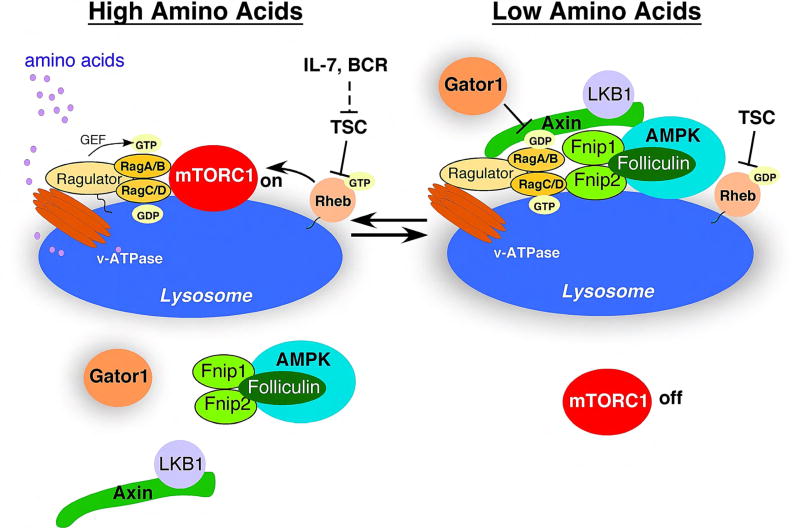
Figure 3. Model of mTORC1 regulation by amino acids and growth factors.
(i) When nutrients are abundant, the lysosomal v-ATPase is activated by amino acids (leucine and arginine in particular) transported into the lysosome, which stimulates the GEF activity of the Ragulator towards the RagA/B GTPases (“inside-out signaling”), resulting in mTORC1 recruitment to the lysosome. Concurrent growth factor and/or BCR stimulation results in phosphorylation and inhibition the TSC complex, permitting mTORC1 activation at the lysosome by Rheb-GTP.
(ii) During amino acid starvation, the AMPK/Flcn/Fnip1/2 complex is bound to inactive v-ATPases through Axin, which inhibits the GEF activity of the Ragulator and switches the Rag GTPases to an inactive state, leading to dissociation of mTORC1 from the lysosome. The Gator1 complex also prevents mTORC1 activation under conditions of amino acid insufficiency by acting as a GTPase activating protein (GAP) for Rag A/B.
(iii) When energy is also low, high AMP levels increase the affinity of AMPK for LKB1 bound to Axin, which increases recruitment of LKB1/Axin complexes to the lysosome, and subsequently AMPK activation by LKB1. Activated AMPK further inhibits mTORC1 by phosphorylation of Raptor and TSC2.
2.3. Regulation of mTOR signaling by AMPK and Fnip/Folliculin Complex
Whereas mTORC1 activity is positively regulated by sufficient amino acids, mTORC1 is also negatively regulated by low energy levels through the master energy sensor AMPK (AMP-activated protein kinase). Specifically, a low ATP/AMP or ATP/ADP ratios activate AMPK by activation of its upstream kinase, LKB1; inhibition of phosphatases that act to dephosphorylate AMPK; and induction of allosteric activation of AMPK (reviewed in [35]). Activated AMPK, in turn, inhibits mTORC1 by multiple mechanisms, including phosphorylation and activation of TSC2 [36,37]; phosphorylation and inhibition of Raptor [38]; and localization to the lysosome through interaction with the scaffolding protein Axin, where in association with the v-ATPase, AMPK stimulates GTPase-activating protein (GAP) activity for Rag A/B, thus antagonizing lysosomal recruitment of mTOR [39] (Figures 2 and 3).
Recent studies have identified intriguing new players in the regulation of AMPK and mTORC1 at the lysosome. The tumor suppressor Folliculin (Flcn) and Flcn-interacting proteins 1 and 2 (Fnip1 and Fnip2) have been shown to form a complex with AMPK (Figure 2) and can modulate AMPK and mTOR activity [40–42]. In humans, inactivating mutations in the BHD gene encoding FLCN result in Birt Hogg-Dubé Syndrome (BHDS), which is characterized by skin hamartomas, lung cysts, and renal cancers (see [43] for review). Although the exact mechanism of how Flcn/Fnip functions is still unclear and controversial, there is general agreement that the complex regulates, and is regulated by, the AMPK and mTORC1 signaling pathways. Using siRNA knockdown approaches in human cell lines, three groups have presented evidence showing that Flcn/Fnip regulates the recruitment of mTOR to the lysosome in response to amino acid stimulation [44–46]. Tsun et al. found that the Flcn/Fnip complex acts as a GAP for RagC/D [45], while Petit et al. found that Flcn may act as a GEF (guanine nucleotide exchange factor) for RagA/B [44]. Both types of interaction are predicted to facilitate mTOR activation. Interestingly, all three groups found Flcn localized to the lysosome and bound to inactive Rag GTPases only under conditions of amino acid starvation, when mTORC1 does not localize to the lysosome (Figure 3). The conclusion made from this data was that the Flcn/Fnip complex is necessary for GTP/GDP exchange in order to activate the Rag GTPases and to permit mTOR recruitment to the lysosome during recovery from starvation.
In contrast, various tissues and cells from patients with BHDS, or from mice with deletion of Flcn or deletion of both Fnip1 and Fnip2, exhibit hyperactive mTORC1 signaling [47–52]. In addition, pre-B cells and skeletal muscle from mice deficient in Fnip1, as well as cardiac tissue from Flcn knockout mice also exhibit increased mTOR activation [53–55]. These results are consistent with a model in which Flcn/Fnip inhibits mTORC1 activation, in contrast with the siRNA knockdown studies outlined previously. One potential mechanism for how Flcn/Fnip might alter mTORC1 activation was proposed by Nagashima et al., who found that ubiquitination and degradation of Fnip2 under fed conditions led to lysosomal dissociation of FLCN and subsequent constitutive lysosomal association of mTOR irrespective of nutrient status [56] (Figure 3). Overall, these studies highlight the complexity of the mTORC1 signaling pathway in response to Flcn/Fnip, and indicate that the control of mTORC1 by the Flcn/Fnip complex is highly dependent on the nutritional state and the type of cells studied.
Along with its role in lysosomal localization of mTOR, Flcn/Fnip has also been reported to modulate AMPK activity. Pre-B cells from mice deficient in Fnip exhibit increased basal AMPK activation and AMPK-target gene activation [53]. Similarly, skeletal muscle [54] and cardiac tissue [57] from Fnip1 null mice were also found to have increased AMPK activation, as did Flcn null mouse embryonic fibroblasts (MEFs) [58] and flcn-1 mutant C. elegans [59]. These results suggest that the Flcn/Fnip complex may directly inhibit AMPK (Figure 2), or alternatively, that loss of Flcn/Fnip could indirectly result in AMPK activation through negative feedback from hyperactive mTORC1-mediated energy consumption, or by induction of cellular stress. As mentioned above, recent studies have revealed that the scaffold protein Axin binds to LKB1 and tethers AMPK to the surface of endosomes and lysosomes where it is activated by the v-ATPase-Ragulator complex in response to glucose deprivation [39]. Based on the collective data, we propose a model whereby nutrient restriction results in recruitment of the AMPK/Flcn/Fnip complex to the lysosome where the complex associates with LKB1/Axin and AMPK is activated. Activated AMPK then inhibits mTORC1 either by phosphorylating TSC2 and Raptor (Figure 2), and/or by inhibiting further mTORC1 recruitment to the lysosome (Figure 3). Under fed conditions, AMPK/Flcn/Fnip would dissociate from the lysosome, thus allowing lysosomal association of mTORC1, which is then poised for activation by growth factors or BCR signals.
2.4. Downstream targets of mTORC1
mTORC1 and mTORC2 regulate unique biological responses by differentially phosphorylating specific molecular targets (reviewed in [9,11]). mTORC1 signaling results in increased protein synthesis through direct phosphorylation of two key substrates, S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E–BP1) (Figure 1B). 4E–BP1 phosphorylation increases 5’cap-dependent translation by relieving 4EBP1 sequestration of eIF4E [60], whereas S6K1 activation upregulates mRNA biogenesis, and translation initiation and elongation by phosphorylating a number of targets including eIF4B, a positive regulator of the 5’cap binding eIF4F complex [61,62], and PDCD4, an inhibitor of eIF4B [63]. To further facilitate cell growth and proliferation, mTORC1 stimulates lipid synthesis largely by promoting the expression and activity of transcription factors such as SREBPs (sterol regulatory element-binding proteins) [64] and PPAR-γ (peroxisome proliferator-activated receptor gamma) [65], which are important in fatty acid and cholesterol synthesis and adipogenesis. mTORC1 also promotes nucleic acid synthesis through regulation of ATF4 (activating transcription factor 4), which stimulates de novo purine biosynthesis [66], and S6K1, which phosphorylates and activates the multifunctional enzyme CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase), resulting in enhanced pyrimidine biosynthesis [67,68].
In addition to regulating biosynthetic functions, mTORC1 signaling regulates energy production and consumption by increasing glycolytic flux through the HIF1α (hypoxia inducible factor 1alpha) transcription factor [69], and by negatively regulating cellular degradation processes such as autophagy and lysosome biogenesis. mTORC1 directly phosphorylates and inhibits the Ulk1/Atg13 (unc-51-like kinase 1/mammalian autophagy-related gene 13) complex, which is required to initiate autophagy [70– 72]. mTORC1 also phosphorylates Dap1 (death-associated protein 1) which results in Dap1 suppression of autophagy [73]. mTORC1 also inhibits lysosomal biogenesis by phosphorylating TFEB (transcription factor EB), which blocks TFEB translocation to the nucleus thus preventing its transcription of lysosome-related genes [74–77]. In a similar manner, mTORC1 phosphorylates and inhibits the nuclear localization of Tfe3, another transcription factor responsible for lysosome biogenesis [46].
2.5. Downstream targets of mTORC2
While multiple diverse inputs feed into mTORC1 regulation, mTORC2 is predominantly activated by growth factor signaling through the PI3K pathway (Figures 1A and 2). Activated mTORC2 stimulates cell survival by phosphorylating AGC kinases including Akt and the serine/threonine protein kinase SGK1 (serum and glucocorticoid-regulated kinase 1) [78,79]. Activation of both Akt and SGK1 results in inhibition of the FOXO1/3 (forkhead box proteins O1 and O3) transcription factors by inducing translocation from the nucleus to the cytoplasm, thus preventing transcription of apoptosis-inducing genes such as Bim [80] (Figure 1B). mTORC2-mediated activation of Akt can, in turn, activate mTORC1 through phosphorylation and inhibition of the tuberous sclerosis 1/2 (TSC1/2) complex. Conversely, the mTORC1 substrate S6K1 modulates Sin1 to reduce mTORC2 activity through a negative feedback loop.
mTORC2 also regulates cytoskeletal organization by phosphorylating and inhibiting PKC (protein kinase C) family members [81–84], and altering cellular migration through activation of the plasma membrane-associated Rho and Rac GTPases, which induce actin polymerization [85].
3. Roles of mTOR in B cell development
3.1. Introduction to early B cell development
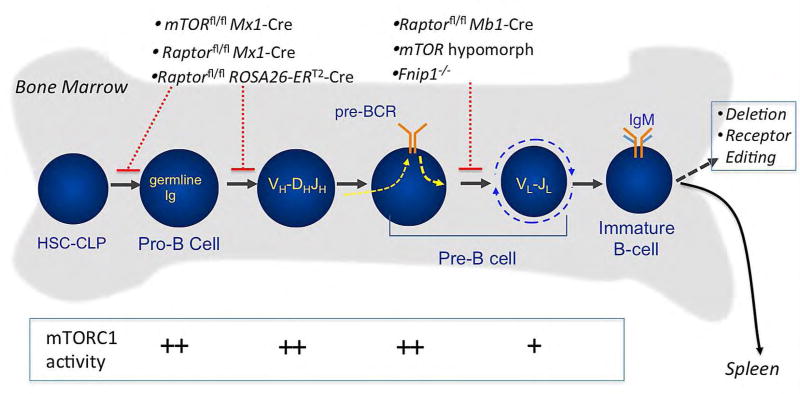
Prior to becoming a functional mature B cell, B cell progenitors must successfully proceed through a series of developmental checkpoints that ensure that mature B cells will express a diverse repertoire of functional immunoglobulin receptors capable of recognizing a wide-array of foreign proteins while exhibiting limited self-reactivity [86]. In the bone marrow, early committed B cell progenitors (pro-B cells), with their immunoglobulin heavy (IgH) and light chain (IgL) genes in the germline configuration, rely on stromal cell derived IL-7 for survival and proliferation [87,88]. During the pro-B cell stage, the protein products of the recombination activating genes Rag-1 and Rag-2 mediate VH(DH)JH rearrangement of the Ig heavy chain gene locus. Productive in-frame VH(DH)JH rearrangement results in expression of the pre-B cell receptor complex (pre-BCR), which is composed of the Ig µ-heavy chain protein in conjunction with the surrogate light chains lambda5 and VpreB and the signal transducing Igα and Igβ proteins, which marks the transition to the early (large) pre-B cell stage (Figure 4). Formation of the pre-BCR and associated signaling pathways are required to promote IgH allelic exclusion, clonal expansion of the pre-B cells, and activation of IgL chain gene rearrangements. Following in-frame VL-JL rearrangements and IgL expression, the IgL and IgH chains pair to form IgM on the surface of immature B cells. Immature B cells are then tested for reactivity against self-antigens in the bone marrow (central tolerance). B cells that react to self-antigens with high avidity are either deleted (negative selection) or undergo receptor editing resulting in expression of alternative IgL chains [86]. B cells with low-avidity interactions or no reactivity to self-antigens migrate to the spleen where development continues.
Figure 4. Regulation of B cell development in the bone marrow by mTORC1 and mTORC2 pathways.
B cell development in the bone marrow begins with the first committed B cell lineage progenitors (pro-B cells) undergoing Ig heavy chain gene rearrangement. Pre-B cells expressing a properly rearranged Ig heavy chain form the pre- B cell receptor (pre-BCR) complex along with surrogate light chains, which stimulates pre-B cell expansion and Ig light chain gene rearrangement. The Ig heavy and light chain proteins then form surface IgM, at which point immature B cells are then tested for self-reactivity. Immature B cells with high self-reactivity undergo deletion or receptor editing, while B cells with low or no self-reactivity migrate to the spleen, where development continues (see Figure 5). Genetic disruption of mTOR, Raptor, and Fnipl lead to blocks in B cell development, as depicted. Also shown is the relative activity of the mTORC1 signaling pathway during early B cell development.
3.2. Development and functions of mature B cells
Immature B cells that successfully escape central tolerance in the bone marrow continue developing through transitional B cell stages T1 and T2, where they are further tested for self-reactivity before becoming mature follicular (FO) or innate-like marginal zone (MZ) B cells (Figure 5). T1 or T2 cells that signal too strongly through the BCR either undergo apoptosis, or become anergic [86,89]. Appropriate signals derived from the B cell receptor (BCR), BAFF receptor, Notch2, and integrin or chemokine signaling pathways allow further survival and selection of T2 cells into the FO or MZ compartments [89], which are among the major functional B cell subsets responsible for humoral immunity.
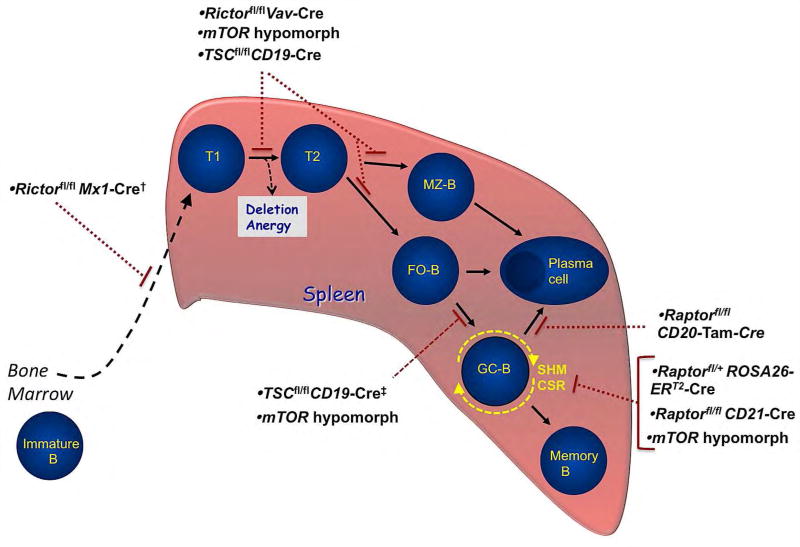
Figure 5. Model of peripheral B cell development in the spleen with associated blocks in mTORC1 and mTORC2 mutants.
Immature B cells migrate from the bone marrow and fetal liver to the spleen and proceed through transitional stages Tl and T2, where they continue to be tested for self-reactivity before differentiating into mature marginal zone (MZ-B) or follicular (FO) B cells. Upon encounter with specific antigen and with the requisite T-cell-derived co-stimulation, activated follicular B cells may differentiate into either short-lived antibody secreting plasma cells or into germinal center B cells (GC-B). GC-B cells undergo clonal expansion, somatic hypermutation, and class-switch recombination before terminally differentiating into long-lived antibody-secreting plasma cells or memory B cells. MZ-B cells are also capable of differentiating into plasma cells. Gene-targeted deletion of specific mTORC1 and/or mTORC2 signaling components in mice disrupt splenic B cell development at the stages shown.
†The specific developmental block in Rictorfl/flMx1-Cre mice has not been characterized. These mice develop a normal immature B cell populations in the bone marrow, but have reduced mature B cells.
‡One study showed TSC1fl/flCD19-Cre mice have a defect in GC-B cell formation, while the other study showed no impairment.
FO B cells represent the majority of mature B cells, and typically reside in primary and secondary lymphoid follicles in the spleen and lymph nodes. However, FO B cells also recirculate and can home to primary (such as bone marrow), secondary and tertiary lymphoid organs where they mount responses to T-dependent antigens through signals from the BCR, CD40, and TLRs. Activated FO B cells proliferate and differentiate into short-lived antibody-secreting cells, or can become germinal center (GC) B cells, where they interact with follicular T cells (TFH) and follicular dendritic cells (FDCs). This multi-cellular interaction allows GC B cells to undergo somatic hypermutation and class switch recombination (CSR), mediated by activation-induced cytidine deaminase (AID), before terminally differentiating into plasma cells or memory B cells [89,90].
In contrast to FO B cells, MZ B cells are innate-like B cells which are primed to respond rapidly to pathogens [91,92]. MZ B cells express polyreactive BCRs and share features of memory B cells, such as being long-lived and possessing the ability to self-renew. In response to antigen, MZ B cells quickly differentiate into plasmablasts and produce large amounts of IgM, IgG and some IgA via CSR. This rapid production of low affinity antibodies by the MZ B cells is thought to provide protection prior to the production of high-affinity antibodies by FO B cells [92].
In addition to the developmental pathways that generate conventional B-2 populations in mice (described above), an alternative pathway leads to the development of the B-1 B cell lineage. B-1 B cells are predominantly fetal-derived, reside in the pleural and peritoneal cavities, and, like MZ B cells, produce natural antibodies with polyreactive receptors [91,93]. B-1 B cells are produced during fetal development and for a limited time post-natally. However in adulthood, this population is maintained through self-renewal processes rather than by ongoing development [93].
3.3. Roles of mTORC1 in early B cell development
By assessing the total expression of mTORC1 signaling proteins Raptor and mTOR, as well as phosphorylation of mTORC1 downstream targets S6 ribosomal protein (S6R) and 4EBP1 (see Figure 2), we found that the mTORC1 pathway is predominantly activated during the pro-B, large pre-B, and small pre-B cells stages, and to a lesser extent in resting immature and recirculating mature B cells in the bone marrow (Figure 4 and [94]). These results are consistent with expression of the IL-7 receptor (IL-7R) during the pro-B and pre-B cell stages, and activation of mTORC1 downstream of the IL7R. Confirmation of the importance of mTORC1 in early B cell development has come from studies using genetic modifications in mice to alter mTORC1 activity. One of the first studies examined the consequences of constitutive or inducible deletion of mTOR signaling components throughout all tissues. Examination of a constitutive, total-body hypomorphic mutant of mTOR revealed a partial block in early B cell development, characterized by reduced pro-B, pre-B and immature B cell populations in the bone marrow, and reduced mature B cell populations in the spleen [95]. Inducible total-body disruption of mTOR was accomplished with mTORfl/fl mice crossed with Mx1-Cre and treated with the synthetic dsRNA analogue poly-IC (polyinosinic:polycytidylic acid). These mice exhibited pancytopenia with decreases in all hematopoietic lineages [96]. However, given that the mTOR protein was disrupted in both of these studies, the differential contributions of mTORC1 versus mTORC2 could not be determined. By contrast, analysis of mice with inducible deletion of Raptor identified mTORC1 specific effects. Raptorfl/fl mice crossed with either Mx1-Cre [97] or Rosa-CreERT2 mice [98] were treated with poly-IC or tamoxifen respectively to induce Cre-mediated deletion. Both mutants showed defects across multiple hematopoietic lineages including a block in early B cell development [97,98]. However, since both Raptorfl/flMx1-Cre and Raptorfl/flRosa-CreERT2 induce widespread deletion of mTORC1 in most tissues, conclusions about the cell-autonomous role of mTORC1 in B cell development could not be made. Recently, we generated a mouse model with B cell-specific deletion of Raptor by crossing Raptorfl/fl mice with B cell specific Mb1-Cre transgenic mice [94], which results in Cre expression and Raptor deletion at the pro-B cell stage and beyond. Raptorfl/flMb1-Cre mice exhibited a block in B cell development at the early pre-B cell stage, with a complete lack of immature and mature peripheral B cells. Raptor-null pre-B cells exhibited reduced proliferative capacity, reduced survival, and reduced oxidative and glycolytic metabolic capacity [94], consistent with the known role of mTORC1 in mitochondrial metabolism and glycolysis [11,99]. Rapamycin-treatment also recapitulated the early block in B cell development as evidenced by accumulation of pre-B cells and loss of immature B cells in mice receiving rapamycin daily for 3 weeks [94]. These results definitively established a requirement for mTORC1 signaling specifically in early B cell development.
3.4. mTORC1 in peripheral B cell development and function
A number of studies have also focused on mTORC1 signaling in peripheral B cells, in particular the functions of mTORC1 in mediating humoral immunity. Using the constitutive hypomorphic mTOR mutants described earlier, Zhang and colleagues found these mice produced fewer GCs following immunization, and had fewer antigen-specific antibodies. Furthermore, the antibodies produced were of lower affinity compared to wildtype controls [100]. Further analysis showed that the mTOR-hypomorphic B cells exhibited lower levels of activation-induced cytidine deaminase (AID), reduced somatic hypermutation, and reduced CSR, which could be rescued by transduction of a retroviral AID vector [100]. To further establish whether these phenotypes are B cell-autonomous, the authors analyzed mTORfl/flCD19-Cre mutant mice, which undergo B cell-specific deletion of mTOR beginning at the pre-B and immature stages of development. Similar to mTOR hypomorphs, mTORfl/flCD19-Cre mice exhibited fewer GCs after immunization, fewer high-affinity antibodies, and reduced CSR [100]. Again, because these studies utilized mTOR deficient mice, the contributions of mTORC1 and mTORC2 could not be differentiated. To examine mTORC1-specific contributions to humoral immunity, Keating and colleagues treated mice with rapamycin and analyzed their response to influenza infection [101]. Interestingly, rapamycin treatment resulted in reduced GC formation and CSR leading to an altered antibody repertoire with improved protection against a heterotypic influenza strain. The authors also showed that total-body deletion of Raptor using a floxed Raptor allele and tamoxifen-responsive Cre recombinase resulted in reduced levels of AID mRNA, consistent with the results seen with the hypomorphic mTOR mice [101]. However, both tamoxifen-responsive Cre and rapamycin inhibit mTORC1 in most tissues, and hence the B cell autonomous effects could not be discerned.
More recently, a mouse model with inducible deletion of Raptor in mature B cells (hCD20-Tam (ERT2)-Cre) was used to show that ablation of mTORC1 signaling before or after primary immunization impaired GC responses resulting in reduced plasma cell differentiation and antibody secretion. The authors found similar effects with rapamycin treatment [102]. Furthermore, while mature plasma cell numbers were not affected by rapamycin treatment, antibody production by these cells was decreased in a reversible manner [102]. In another study, GCs, particularly light zone regions, were found to be relatively hypoxic environments, and hypoxia was shown to reduce AID expression and CSR in a hypoxia-inducible factor (HIF)-dependent manner [103]. Hypoxia was also shown to inhibit mTORC1 activity, and similar to previous studies, deficiency in Raptor led to decreased AID expression, reduced CSR, and high-affinity antibody production [103].
Whereas rapamycin treatment, mTOR hypormorphism, and Raptor-deficiency all inhibited CSR, Limon and colleagues found that mTOR catalytic site inhibitors, which block both mTORC1 and mTORC2 function, paradoxically increased CSR and AID transcript levels in B cells. Furthermore, mice treated transiently with a mTOR kinase inhibitor exhibited greater production of high-affinity class-switched antibodies [104]. The effect of the mTOR kinase inhibitors on class-switching appeared to be a result of mTORC2 inhibition as Rictorfl/flCD19-Cre mice also exhibited increased CSR. In contrast, Raptorfl/flCD21-Cre mice, in which Raptor is deleted in transitional B cells, exhibited decreased CSR [104], which was consistent with the previously mentioned studies [100,101]. However, the results with the Rictorfl/flCD19-Cre mice and mTOR kinase inhibitors contrast with the previous findings indicating that Rictor-deficient cells as well as mTOR hypomorphic cells have impaired CSR activity [100,105]. The discrepancy was attributed to the incomplete inhibition of mTOR achieved with the low dose of kinase inhibitor used by Limon, et al., as well as the partial deletion of Rictor in the CD19-Cre model, compared to the more efficient Rictor deletion observed in the Vav-Cre model [104,105]. It is likely that relative impairment of the mTORC1 versus mTORC2 pathway, as well as their individual and combined effects on B cell development, cell proliferation, and cell survival alter the balance between increased and decreased CSR. Altogether, these studies show that mTORC1 activity is critical for proper AID expression, CSR and somatic hypermutation in response to T-dependent antigens, while mTORC2 has more nuanced effects on CSR depending on relative signaling activity. The phenotypes of mTORC1 and mTORC2 deletion models are further complicated by the fact that mTORC1 complexes and mTORC2 complexes regulate each other, and some phenotypic effects of constitutive deletion models could be due to regulatory feedback.
Effects of mTORC1 hyperactivation on B cell development have also been examined. Two groups independently generated and analyzed TSC1fl/flCD19-Cre mice, which exhibit constitutive B cell-specific mTORC1 hyperactivation [106,107]. Both groups reported a partial block in splenic B cell development with a relative accumulation of transitional T1 B cells and a loss of mature splenic B cells. While the FO mature population was markedly reduced in TSC1 knockout mice, the MZ B cell population was abolished [106,107]. Interestingly, both groups immunized these mice with the T-dependent antigen NP-CGG with varying results. While Benhamron and Tirosh observed impaired GC formation and variable NP-specific antibody production [106], Ci and colleagues observed normal GC formation and relatively normal antibody responses [107]. The cause of this discrepancy is not clear as background strain did not affect the phenotype observed by Ci and colleagues [107]. In addition to examining T-dependent immune responses, Benhamron and Tirosh also analyzed T-independent antibody production in response to NP-Ficoll. Consistent with a deficiency in MZ B cell development, TSC1 knockout mice failed to produce antibody in response to NP-Ficoll [106]. Collectively, these findings indicate that while mTORC1 signaling is required for early B cell development and for primary and secondary immune responses to T-dependent antigens, excessive mTORC1 activation is also detrimental to maturation of splenic B cells, and in particular, MZ B cells. This finding is interesting given that previous studies have also indicated relatively high levels of mTORC1 activation in MZ B cells [29]. MZ B cells are considered to exist in a pre-activated state, and produce natural antibodies under homeostatic conditions [92]. However, MZ B cells also require weak BCR signaling for differentiation as relatively strong BCR signals steer the differentiation of transitional B cells into FO B cells, and very strong signals lead to anergy or deletion [89]. Hyperactivation of mTORC1 may thus be an indication of high self-reactivity, and is especially detrimental to developing MZ B cells.
3.5. mTORC2 in B cell development
Several studies using gene targeting or chemical inhibition approaches have revealed the importance of the mTORC2 signaling pathway in B cell development and function. Two groups examined the consequences of Rictor deletion using Rictorfl/flMx1-Cre mice treated with polyIC. Both groups noted normal bone marrow B cell cellularity, but reduced splenic B cells in these mice [97,108]. However, only one group described the bone marrow B cell populations in detail and found an increased percentage of pro-B, pre-B and immature B cells with a marked reduction in mature B cells [108]. This defect in B cell development was shown to be B cell-autonomous based on bone marrow transplantation experiments [108]. Rictor-deficient B cells also exhibited increased expression of Rag1 and IL-7R, as well as decreased Foxo1 phosphorylation, which is indicative of increased nuclear Foxo1 transcriptional activity [108]. In another study, ablation of Sin1, a component of the mTORC2 complex, displayed a more robust early B cell development defect. Because constitutive Sin1 knockout mice are embryonic lethal, the authors utilized fetal liver hematopoietic stem cell transplantation to examine consequences of Sin1 loss on bone marrow B cell development. Analysis of reconstituted mice revealed that Sin1 deficiency leads to increased pro- and pre-B cells but decreased immature and mature bone marrow B cells [109]. The expansion of pro-B cells seen in recipients of Sin1 knockout HSCs is associated with increased IL-7R and Rag gene expression, and decreased Foxo1 phosphorylation [109], consistent with the phenotype observed in the Rictorfl/flMx1-Cre mice [108]. These studies collectively suggest that mTORC2 signaling regulates PI3K signaling in early B cells, and disruption of this pathway leads to a failure to properly downregulate IL-7R signaling and Rag gene expression, leading to disrupted B cell development.
More prominent phenotypes in peripheral B cell homeostasis and antibody production were observed in Rictor-deleted mice using Vav-Cre and ROSA26-Cre-ERT2 systems. Analysis of Rictorfl/flVav-Cre mice revealed impaired splenic B cell development as evidenced by increased transitional B cells and impaired maturation of MZ and FO B cells. In addition, the peritoneal B1a B cell population was also reduced [105]. The authors of this study also analyzed Rictorfl/flROSA26-Cre-ERT2 mice following tamoxifen-induced deletion of Rictor. Consistent with a role in maintenance of mature B cells, deletion of Rictor resulted in reduced peripheral B cells and MZ B cells, reduced B cell survival, and reduced B cell proliferation [105]. Furthermore, disruption of mTORC2 led to impaired expression of NF-κB target genes, which mediate cell survival downstream of BCR and BAFF-R signaling, and also resulted in compromised antibody production in Rictor-deficient mice. As mentioned earlier, the Rictorfl/flVav-Cre mice, as well as recipients of tamoxifen-deleted Rictorfl/flROSA26-Cre-ERT2 B cells, exhibited reduced antibody responses [105]. These findings are in contrast to results showing that moderate levels of mTORC2 inhibition (via chemical or genetic manipulation) results in increased class-switching [104]. As discussed, these B cell phenotypes appear to be highly sensitive to the degree of mTORC2 inhibition, as increasing the concentration of mTOR kinase inhibitors used results in decreased CSR as well as reduced cell proliferation [104]. Thus, mTORC2 signaling plays a major role in B cell-specific responses to signals originating from the BCR and BAFF-R to mediate survival, proliferation and antibody production.
3.6. Folliculin/Fnip1 in early B cell development
Recent studies on Folliculin and Fnip1 have highlighted essential roles for these adaptor proteins in cellular growth and differentiation, particularly in highly metabolic tissues such as heart, skeletal muscle, kidney, and immune cells. Utilizing an ENU chemical mutagenesis strategy in mice to identify novel immune regulating genes, we identified a mutant pedigree that lacks B lymphocytes due to a non-coding mutation in the Fnip1 gene [53]. Fnip1 null mice have a complete block in B cell development at the pre-B cell stage (Figure 4), and proliferate normally in response to IL-7. Whereas the pre-BCR and associated signaling molecules appeared to be expressed normally, Fnip1 null pre-B cells are larger in size, exhibit increased oxidative phosphorylation and glycolysis, and are more sensitive to nutrient deprivation-induced apoptosis relative to wildtype pre-B cells. These observations correlate with increased AMPK activation and inefficient termination of mTOR activity in response to the AMPK activator AICAR. Interestingly, Fnip1 null pre-B cells are also relatively resistant to transformation induced by the Myc oncogene, in part due to increased apoptosis of Myc-overexpressing pre-B cells. These results suggest that Fnip1, in conjunction with AMPK, may help maintain metabolic homeostasis in response to metabolic stresses such as nutrient deprivation or oncogene activation (Figure 3). Baba et al. also generated constitutive Fnip1 null or tamoxifen-inducible Bhd (Flcn) null pre-B cells using Bhdfl/− Cre-ERT2 mice [110] and also found that disruption of either gene led to a block in B cell development at the pro-B/pre-B cell stage due to increased apoptosis. Interestingly, humans with BHD syndrome have not been reported to have deficiencies in B cells, suggesting that perhaps Fnip1 or Fnip2 could compensate for the absence of FLCN in human B cells. Consistent with this notion, mice singly deficient in either Fnip1 or Fnip2 fail to develop significant renal disease or renal cancer, whereas conditional deletion of Flcn results in severe polycystic kidney disease that progresses to cancer [48]. However, deletion of both Fnip1 and Fnip2 in mice does recapitulate severe renal disease seen with disruption of Bhd, indicating that Fnip1 and Fnip2 can compensate for each other in renal tissue [51].
Although Fnip1 deficiency blocked B cell development, T cell development and functions were not found to be significantly impaired, perhaps due to compensation by Fnip2. However, disruption of Fnip1 does block invariant natural killer T (iNKT) cell development [111], similar to the phenotype observed with loss of c-Myc [112] and TSC1 [113] in these cells. These findings are consistent with the hypothesis that Fnip1 helps maintain metabolic homeostasis in response to cellular stress, such as occurs during oncogene activation and nutrient deprivation.
4. mTOR and B cell neoplasms
4.1 mTOR activation in B cell neoplasms
The PI3K/Akt signaling cascade and downstream regulator mTOR are deregulated in many human cancers (reviewed in [114]). The roles of PI3K and Akt signaling in B cell lymphomas have been extensively reviewed elsewhere [115,116], and hence we will focus more specifically on mTOR in B cell neoplasms in this review. Hyperactivation of the mTOR pathway in B cell lymphomas results in increased proliferation, cell survival, and invasion, all of which contribute to cancer progression and adaptive resistance to chemotherapies. For example, AKT and/or mTOR activation correlates with poor prognosis in B cell acute lymphoblastic leukemia (B-ALL) and non-Hodgkin’s lymphoma (NHL) patients [117–120]. Oncogenic drivers of B cell transformation such as BCR-ABL1 (Philadelphia chromosome [Ph+] positive ALL) translocations result in constitutive kinase activation with corresponding downstream activation of the PI3K/mTOR pathway. Similarly Philadelphia chromosome-like (Ph-like) BALL carries a poor prognosis and exhibits high rates of activating mutations in various kinases, although it lacks the BCR-ABL1 fusion protein. Kinases commonly activated in B-ALL include cytokine receptorlike factor 2 (CRLF2), Janus kinases (JAKs), Abelson murine leukemia viral oncogene homolog 1 (ABL1), colony-stimulating factor 1 receptor (CSF1R), platelet-derived growth factor receptor beta (PDGFRB), fms-related tyrosine kinase 3 (FLT3), and IL7R. The majority of these growth factors, kinases, and receptors activate the mTOR pathway. Loss of function mutations in tumor suppressors such as PTEN (Figure 2), or activating mutations in upstream kinases such as protein kinase C delta (PKCδ) or Syk, are the most common causes of mTOR hyperactivity, although various mutations in mTOR itself have also been associated with increased cancer risk [121]. For example, recent studies indicate that up to 17% of follicular lymphoma patients have activating mutations in the mTORC1 regulator RRAGC, enabling constitutive mTORC1 activation independent of amino acid levels [122,123]. Conversely, a polymorphism in the mTOR promoter that leads to decreased mTOR expression has been negatively associated with several different cancer types including acute lymphoblastic leukemia (ALL) [124]. These studies highlight the important contributions of mTORC1 signaling to lymphomagenesis and cancer progression, and reinforce the therapeutic potential of targeting mTOR in cancer.
4.2. Rapalogs as chemotherapy adjuvants
The partial mTORC inhibitors rapamycin and rapamycin analogs (rapalogs) were the first generation of mTOR inhibitors tested in preclinical studies for anti-leukemia activity (reviewed in [125– 129]). Rapamycin and rapalogs showed early promise as potent single chemotherapy agents in genetically modified mouse models as well as primary human tumor cells grown in vitro, and in tumor xenograft models [130–133]. Although rapalogs did not exhibit dose-limiting toxicities when used as monotherapies in clinical trials, they showed reduced efficacy compared to standard chemotherapeutic regimens except in the case of mantle cell lymphoma (MCL), which responds poorly to other chemotherapies. Indeed, temsirolimus has been approved as a single agent therapy for relapsed MCL in Europe [134], and is currently the only B cell cancer subtype for which rapalogs are clinically approved to treat.
While not generally effective as single chemotherapy agents, rapalog-induced cytostatic responses can synergize with cytotoxic chemotherapy to more effectively kill cancer cells and limit disease relapse (Table 2). For example, dual treatment of xenograft ALL models with sirolimus and methotrexate decreases levels of dihydrofolate reductase, a protein which confers methotrexate resistance by rescuing DNA biosynthesis [135]. Similarly, combinations of mTOR and histone deacetylase inhibitors (HDACs) have shown efficacy in preclinical models of ALL [136]. Dual inhibition of mTOR and the anti-apoptotic protein Bcl2 also increased death of ALL cells by repressing Mcl1 expression [137]. Although tyrosine kinase inhibitors such as imantinib are used clinically to treat Ph+ B-ALL, survival rates are still significantly lower than other types of ALL likely because the BCR-ABL1 fusion gene characteristic of this disease directly activates the mTOR pathway. Imatinib itself may also activate part of the mTOR pathway, leading to treatment resistance. Yang et al. showed that combining imatinib therapy with rapamycin overcame this effect and enabled induction of autophagy and cell cycle arrest in primary Ph+ B-ALL cells in vitro [138].
Table 2.
Summary of studies discussed using mTOR inhibitors as chemotherapy for B cell neoplasms.
| Compound | Drug type | Combination therapy |
Type of B cell neoplasm |
Efficacy versus control or single agent |
Type of study |
Reference |
|---|---|---|---|---|---|---|
| Sirolimus (rapamycin) | Rapalog | Imatinib | ALL | More effective | preclinical | [138] |
|
|
||||||
| Sirolimus | Rapalog | Methotrexate | ALL | More effective | preclinical | [135] |
|
|
||||||
| Temsirolimus | Rapalog | None | Relapsed MCL | More effective | clinical | [134] |
|
|
||||||
| Everolimus | Rapalog | Hyper-CVAD | Relapsed ALL | Similar efficacy (more effective for T-ALL) | clinical | [139] |
|
|
||||||
| Everolimus | Rapalog | Panobinostat | Relapsed HL, NHL | More effective | clinical | [140] |
|
|
||||||
| CCI-779 | mTOR KI | ABT737 | ALL | More effective in cells resistant to ABT737 | preclinical | [137] |
|
|
||||||
| AZD2014 | mTOR KI | None | ALL | More effective | preclinical | [145] |
|
|
||||||
| Gedatolisib | PI3K/mTOR KI | None | ALL | More effective | preclinical | [145] |
Abbreviations: Rapalog = rapamycin analog, KI = kinase inhibitor, ALL= acute lymphoblastic leukemia, MCL= mantle cell lymphoma, HL = Hodgkin’s lymphoma, NHL = non-Hodgkin’s lymphoma, Hyper-CVAD = multi-drug chemotherapy regimen involving cyclophosphamide, vincristine, doxorubicin, and dexamethasone.
The rapalogs sirolimus, temsirolimus, and everolimus are all being tested in various clinical trials for ALL and NHL in combination with multi-agent chemotherapy, with promising outcomes including complete remission of relapsed childhood ALL [125]. However, in some cases, rapalogs combined with multi-agent chemotherapy have shown remission rates no better than standard chemotherapeutic regimens [139], and therapeutic efficacy was sometimes accompanied by adverse toxicity, such as significant thrombocytopenia [140]. In addition, another disadvantage of rapalog therapy is upregulation of the pro-survival/proliferation PI3K pathway, which occurs through loss of mTORC1 negative feedback on PI3K and mTORC2 [141–143], and the incomplete suppression of some mTORC1 substrates such as 4E–BP1 [144]. Thus, investigations to identify rapalog/multi-drug chemotherapy combination regimens which are effective and well-tolerated continues, with many clinical trials currently underway.
4.3. mTOR kinase inhibitors as chemotherapy
In order to circumvent feedback activation of AKT and mTORC2, which can occur with rapalog therapy, there have been intense efforts to develop mTOR-KIs as chemotherapeutic agents. TOR-KIs compete with ATP for binding to the mTOR active site, and thus more effectively block mTORC1 substrate phosphorylation relative to rapalogs, while also inhibiting mTORC2 activity and attenuating the AKT pathway. Tasian et al found that treatment of patient-derived Ph-like ALL murine xenografts with the TOR-KI AZD2014 or the dual PI3K/mTOR inhibitor gedatolisib (PKI587) downregulated the mTOR pathway, and significantly decreased leukemia burden in 6/10 or 10/10 different tumors, respectively [145]. Interestingly, therapeutic responses to the TOR-KI AZD2014 were greatest in the two tested relapsed ALL samples. PI3Kα and PI3Kδ inhibitors alpelisib and idelalisib were also tested in this study, and the dual PI3K/mTOR inhibitor gedatolisib was the most effective treatment in 9/10 tumors. This finding confirmed earlier studies comparing dual PI3K/mTOR inhibitors with selective PI3K, mTORC1, and mTORC1/2 kinase inhibitors [146]. Examples of TOR-KIs being tested in clinical trials as single-agent treatments for B cell neoplasms (such as ALL and NHL) include MLN0128, CC-115, PQR-309 and CC-223. As discussed with rapalog therapy, combining mTOR kinase inhibitors with other drugs can also be utilized to target chemotherapy resistance mechanisms such as upregulation of the parallel Ras/MAPK/ERK pathway and receptor tyrosine kinases (RTKs), or to induce apoptosis by inhibiting pro-survival proteins such as BCL-2 and BCL-XL. In addition, mTOR inhibition can be combined with drugs targeting specific oncogenic drivers, such as various combinations of mTORC/PI3K inhibitors with the Btk inhibitor ibrutinib to combat the chronic BCR activation which drives activated B cell-like diffuse large B cell lymphoma [147].
One disadvantage noted in clinical trials is that, whereas TOR-KIs may be more efficacious in controlling tumor growth than rapalogs, they may also be associated with greater toxicities [125]. In addition, despite promising synergistic combinatorial therapies, mTOR inhibitors can also antagonize other chemotherapeutic agents by inducing autophagy and thus providing nutrients and cellular products to continue supporting tumor growth [148]. Therefore, dual inhibition of both the mTOR pathway and autophagy may prove useful, especially in certain cancers in which mTOR upregulation is paradoxically associated with increased autophagy, such as chronic lymphoid leukemia and acute myeloid leukemia [126]. The generation of more effective therapies will require thorough knowledge of the molecular defects observed in each cancer type combined with an understanding of how drugs modulate the interconnected signaling pathways.
5. Concluding remarks
The mTORC pathway is highly complex and regulates many essential parts of cell growth, proliferation, survival, and function. Although this pathway is clearly important for many aspects of B cell development and function, much of the regulatory circuitry modulating the activity of mTOR and the biological consequences of that activity remain unknown in B cells. Elegant biochemical studies in non-B cell lines are defining the molecular mechanisms of how nutrients such as amino acids and glucose, and growth factor signals converging on the TSC1/TSC2 complex, interplay to regulate the localization and activation of mTORC1 at the lysosome surface. This emerging mechanism is particularly intriguing in B cells given the high metabolic demands of activated B cells during clonal expansion, and the requirements for high protein synthetic capacity to fuel antibody production. However, whether amino acids specifically control B cell development and functions have not been defined, and how amino acid signaling might intersect with BCR, TLR, and co-receptor signaling are unclear. Similarly, although genetic studies in mice are defining the importance of mTOR signaling at specific stages of B cell development and for B cell functions, the specific receptors and upstream signaling pathways leading to mTOR activation during B cell development are unclear. Given that clinical trials in humans are revealing that mTOR kinase inhibitors can be very toxic, due in part to the widespread importance of mTOR signaling in many cell types, better understanding of the molecules and pathways activating mTOR specifically in B cells will assist in developing more targeted and less toxic approaches to inhibit mTOR in B cells and B cell neoplasms. Equally interesting questions remain regarding the signaling molecules and pathways downstream of mTOR that control B cell development and functions. For example, numerous molecules and transcription factors are being defined in non-B cells that mediate mTOR-driven protein, lipid, and nucleic acid synthesis, as well as autophagy inhibition and lysosome biogenesis. Determining which of these molecules and pathways are important in mediating mTOR activities during B cell development will assist in better defining the “metabolic checkpoint” pathways that govern maturation, differentiation, clonal expansion, and antibody production in B cells. Finally, improved systems for investigating mTOR signaling in B cells will enable clearer understanding of the molecules and pathways controlling mTOR in B cells. Whereas non-B cell lines such as human embryonic kidney (HEK) 293T and HeLa cervical cancer cells have been the workhorses for discovering important mTOR signaling molecules, the “wiring” between these cell lines and primary B cells could be significantly different, and some cell lines like HeLa, have evolved to lack expression of important signaling molecules such as LKB1, an upstream activator of AMPK. Similarly, constitutive and tissue specific disruption of important signaling molecules in mice can impose extensive selective pressures that can result in feedback signaling, which can be incorrectly interpreted as a primary consequence of a specific gene disruption. Hence, the development and utilization of better B cell-specific and inducible (acute) gene disruption models, along with systems to specifically mark deleted cells, will help clarify these issues.
Regardless of these challenges, the mTOR pathway is emerging as one of the most interesting and essential pathways controlling B cell development and functions. The widespread utilization of rapamycin and mTOR kinase inhibitors as potential therapies in a large array of diseases and for lifespan extension, further highlight the importance of completely understanding how this pathway modulates individual cell types such as B cells and other immune cells.
Table 1.
B cell phenotypes associated with aberrant mTORC1 and mTORC2 signaling.
| Gene | Mutation | Effect | B Cell Phenotypes | References |
|---|---|---|---|---|
| mTOR | mTORfl/flMx1-Cre | ↓mTORC1 & ↓mTORC2 | Pancytopenia (decrease in all hematopoietic lineages) | [96] |
| hypomorphic mTOR Kl allele | Partial block at large pre-B to small pre-B cell stage; decreased mature splenic B cells; decreased AID expression, reduced somatic hypermutation; reduced class switch recombination and reduced high-affinity antibody production. | [95][100] | ||
| mTORfl/flCD19-Cre | Reduced germinal center formation, reduced class switch recombination and reduced high-affinity class switched antibodies | [100] | ||
| Raptor | Raptorfl/flMb1-Cre | ↓mTORC1 | Block in B cell development at pro-B/pre-B cell stage; lack of peripheral B cells and fail to produce antigen-specific antibody | [94] |
| Raptorfl/flMx1-Cre | Pancytopenia including impaired B cell development with increased bone marrow pro-B and decreased mature peripheral B cells | [97] | ||
| Raptorfl/flROSA26-ERT2-Cre | Block in granulocyte and B cell development with reduced pro-B, pre-B and immature bone marrow B cells | [98] | ||
| Reduced AID expression | [101] | |||
| Raptorfl/flCD20-Tam-Cre | Decreased plasma cell differentiation with no change in mature plasma cell survival; reduced antibody synthesis | [102] | ||
| Raptorfl/flCD21-Cre | Normal B cell development; reduced class switch recombination and antibody secreting cells | [104] | ||
| Raptorfl/+ ROSA26-ERT2-Cre | Reduced AID expression; reduced class switch recombination; decreased high-affinity antigen-specific class-switched antibody production | [103] | ||
| Tsc1 | Tsc1fl/flCD19-Cre | ↑mTORC1 | Impaired mature B cell development; increased transitional B cells; loss of MZ/FO B cells; decreased Tl-antigen response; impaired GC formation | [106] |
| Tsc1fl/fl CD19-Cre | Block at transitional splenic B cell stage; decreased mature MZ and FO B cells; normal GC formation; mild reduction in class switch recombination | [107] | ||
| Tsc1fl/fl ERT2-Cre | ||||
| Rictor | Rictorfl/flVav-Cre | ↓mTORC2 | Impaired development of mature FO, MZ and peritoneal Bla B cells | [105] |
| Rictorfl/flROSA26-ERT2Cre | Reduced peripheral and MZ B cells | |||
| Rictorfl/flMx1-Cre | Block in B cell development with increased BM pro-B, pre-B, immature B cells; reduced mature B cells | [108] | ||
| Rictorfl/flCD19-Cre | Increased class switch recombination | [104] |
Acknowledgments
Funding sources: This work was partially supported by NIH grants R56 AI092093, R21 AI109020, RO1 AI092093 (to BMI), KO1 OD10554 (to JAR-K), and KO1 OD21421 (to TNI).
Biographies
Terri Iwata received her Ph.D. in Comparative Biomedical Sciences and her D.V.M. from Cornell University. She received post-doctoral training in Laboratory Animal Medicine and Pathology at the University of Washington. She is currently an Acting Assistant Professor in the Department of Comparative Medicine at the University of Washington where is investigating the role of the mTORC1 signaling pathway in B cell development and function.

Heon Park graduated from the University of Mississippi Medical Center in 2002 with PhD in Microbiology and Immunology. He then pursued Post-doctoral training at the University of Washington, Department of Immunology where his research focused on T cell biology and the roles of IL-17 family members in autoimmunity. He is now a Staff Scientist in the Department of Comparative Medicine, and his major research focus is on understanding the roles of specific metabolic regulating proteins in peripheral B cell development.

Julita Ramirez-Komo received her PhD in Biochemistry and Molecular Biology from Colorado State University in the laboratory of Jennifer Nyborg, where she studied transcriptional activation of the Human T cell Leukemia Virus-1 provirus. She received postdoctoral training in Immunology in the laboratory of James Hagman at National Jewish Health, where she investigated transcriptional control of early B cell development. She is currently an Acting Assistant Professor and Postdoctoral fellow in the laboratory of Brian Iritani at the University of Washington, where her work is focused on the metabolic control of B cell development and tumor growth by Folliculin interacting protein-1 (Fnip1).

Iritani Bio
Brian Iritani received his D.V.M. from Washington State University and Ph.D. in Immunology from the University of Washington. After Post-doctoral training in Molecular Oncology at Fred Hutchinson Cancer Research Center, Brian joined the Department of Comparative Medicine, University of Washington where is currently a Professor. Brian has had longstanding interests in understanding the molecular and cellular biology of oncogenes, and how they normally function to control the development of lymphocytes. More recently his laboratory has become interested in molecules that control the metabolism of lymphocytes, as well as other highly metabolic tissues including heart and skeletal muscle.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest: The authors have no conflicts of interest
References
- 1.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. (Tokyo) 1975;28:721–6. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal SN, Baker H, Vézina C. Rapamycin (AY-22,989), a new antifungal Fermentation antibiotic. II. isolation and characterization. J. Antibiot. (Tokyo) 1975;28:727–732. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- 3.Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 1993;13:6012–23. doi: 10.1128/MCB.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-F. [DOI] [PubMed] [Google Scholar]
- 5.Brown EJ, Albers MW, Bum Shin T, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 6.Chiu MI, Katz H, Berlin V. RAPT1 a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12574–8. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 8.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995;270:815–22. doi: 10.1074/JBC.270.2.815. [DOI] [PubMed] [Google Scholar]
- 9.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014;15:155–62. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 11.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim L, Cook R, Chen J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene. 2016:1–11. doi: 10.1038/onc.2016.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oaks Z, Winans T, Huang N, Banki K, Perl A. Activation of the Mechanistic Target of Rapamycin in SLE: Explosion of Evidence in the Last Five Years. Curr. Rheumatol. Rep. 2016;18:73. doi: 10.1007/s11926-016-0622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahrling JB, Laberge R-M. Age-Related Neurodegeneration Prevention Through mTOR Inhibition: Potential Mechanisms and Remaining Questions. Curr. Top. Med. Chem. 2015;15:2139–51. doi: 10.2174/1568026615666150610125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim J, Li L, Shirihai OS, Trudeau KM, Puertollano R, Raben N. Modulation of mTOR signaling as a strategy for the treatment of Pompe disease. EMBO Mol. Med. 2017;9:353–370. doi: 10.15252/emmm.201606547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamming DW, Ye L, Sabatini DM, Baur JA. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J. Clin. Invest. 2013;123:980–9. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat. Rev. Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Chapman NM, Karmaus PWF, Zeng H, Chi H. mTOR and metabolic regulation of conventional and regulatory T cells. J. Leukoc. Biol. 2015;97:837–847. doi: 10.1189/jlb.2RI0814-408R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman NM, Chi H. mTOR links environmental signals to T cell fate decisions. Front. Immunol. 2015;6:1–11. doi: 10.3389/fimmu.2014.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jhanwar-Uniyal M, Amin AG, Cooper JB, Das K, Schmidt MH, Murali R. Discrete signaling mechanisms of mTORC1 and mTORC2: Connected yet apart in cellular and molecular aspects. Adv. Biol. Regul. 2017 doi: 10.1016/j.jbior.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Sarbassov DD, Ali SM, Sengupta S, Sheen J-H, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 23.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, Guertin DA, Sabatini DM, Baur JA. Rapamycin-Induced Insulin Resistance Is Mediated by mTORC2 Loss and Uncoupled from Longevity. Science (80) 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Martínez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem. J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu K, Toral-Barza L, Shi C, Zhang W-G, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, Cole DC, Ellingboe J, Ayral-Kaloustian S, Mansour TS, Gibbons JJ, Abraham RT, Nowak P, Zask A. Biochemical, Cellular, and In vivo Activity of Novel ATP-Competitive and Selective Inhibitors of the Mammalian Target of Rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 28.Donahue AC, Fruman DA. Proliferation and survival of activated B cells requires sustained antigen receptor engagement and phosphoinositide 3-kinase activation. J. Immunol. 2003;170:5851–60. doi: 10.4049/jimmunol.170.12.5851. [DOI] [PubMed] [Google Scholar]
- 29.Donahue AC, Fruman DA. Distinct signaling mechanisms activate the target of rapamycin in response to different B-cell stimuli. Eur. J. Immunol. 2007;37:2923–2936. doi: 10.1002/eji.200737281. [DOI] [PubMed] [Google Scholar]
- 30.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol. Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat. Rev. Immunol. 2013;13:578–591. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- 32.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K–mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goberdhan DCI, Wilson C, Harris AL. Amino Acid Sensing by mTORC1: Intracellular Transporters Mark the Spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 37.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C-S, Jiang B, Li M, Zhu M, Peng Y, Zhang Y-L, Wu Y-Q, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu J-W, Yin H, Lin S-Y, Lin S-C. The Lysosomal v-ATPase-Ragulator Complex Is a Common Activator for AMPK and mTORC1, Acting as a Switch between Catabolism and Anabolism. Cell Metab. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 40.Baba M, Hong S-BS-B, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR, Linehan WM, Schmidt LS, Zbar B. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK and is involved in AMPK and mTOR signaling. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takagi Y, Kobayashi T, Shiono M, Wang L, Piao X, Sun G, Zhang D, Abe M, Hagiwara Y, Takahashi K, Hino O. Interaction of folliculin (Birt-Hogg-Dubé gene product) with a novel Fnip1-like (FnipL/Fnip2) protein. Oncogene. 2008;27:5339–47. doi: 10.1038/onc.2008.261. [DOI] [PubMed] [Google Scholar]
- 42.Hasumi H, Baba M, Hong S-B, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene. 2008;415:60–67. doi: 10.1016/j.gene.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt LS, Linehan WM. Molecular genetics and clinical features of Birt-Hogg-Dubé syndrome. Nat. Rev. Urol. 2015;12:558–569. doi: 10.1038/nrurol.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini D. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Sci. Signal. 2014;7:ra9–ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Futami K, Petillo D, Peng J, Wang P, Knol J, Li Y, Khoo S-K, Huang D, Qian C-N, Zhao P, Dykyma K, Zhang R, Cao B, Yang XJ, Furge K, Williams BO, Teh BT, Teh BT. Deficiency of FLCN in Mouse Kidney Led to Development of Polycystic Kidneys and Renal Neoplasia. PLoS One. 2008;3:e3581. doi: 10.1371/journal.pone.0003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baba M, Furihata M, Hong S-BS-B, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, Yao M, Bernardo M, Ileva L, Choyke P, Warren MB, Zbar B, Linehan WM, Schmidt LS. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 Akt-mTOR activation, cell hyperproliferation and polycystic kidneys. J. Natl. Cancer Inst. 2008;100:140–154. doi: 10.1093/jnci/djm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong S-B, Yamaguchi TP, Schmidt LS, Linehan WM. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18722–7. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuya M, Nakatani Y. Birt-Hogg-Dubé syndrome: clinicopathological features of the lung. J. Clin. Pathol. 2013;66:178–186. doi: 10.1136/jclinpath-2012-201200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Ito Y, Furuya M, Iribe Y, Kodama T, Southon E, Tessarollo L, Nagashima K, Haines DC, Linehan WM, Schmidt LS. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1624–31. doi: 10.1073/pnas.1419502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong M, Zhao X, Li J, Yuan W, Yan G, Tong M, Guo S, Zhu Y, Jiang Y, Liu Y, Jiang Y. Tumor Suppressor Folliculin Regulates mTORC1 through Primary Cilia. J. Biol. Chem. 2016;291:11689–11697. doi: 10.1074/jbc.M116.719997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park H, Staehling K, Tsang M, Appleby MW, Brunkow ME, Margineantu D, Hockenbery DM, Habib T, Liggitt HD, Carlson G, Iritani BM. Disruption of Fnip1 Reveals a Metabolic Checkpoint Controlling B Lymphocyte Development. Immunity. 2012;36:769–781. doi: 10.1016/j.immuni.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur-S DK, Hockenbery DM, Raftery D, Iritani BM. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2015;112:424–429. doi: 10.1073/pnas.1413021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasumi Y, Baba M, Hasumi H, Huang Y, Lang M, Reindorf R, Oh H-b, Sciarretta S, Nagashima K, Haines DC, Schneider MD, Adelstein RS, Schmidt LS, Sadoshima J, Marston Linehan W. Folliculin (Flcn) inactivation leads to murine cardiac hypertrophy through mTORC1 deregulation. Hum. Mol. Genet. 2014;23:5706–5719. doi: 10.1093/hmg/ddu286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagashima K, Fukushima H, Shimizu K, Yamada A, Hidaka M, Hasumi H, Ikebe T, Fukumoto S, Okabe K, Inuzuka H. Nutrient-induced FNIP degradation by SCF β-TRCP regulates FLCN complex localization and promotes renal cancer progression. Oncotarget. 2016;8:1–14. doi: 10.18632/oncotarget.14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siggs OM, Stockenhuber A, Deobagkar-Lele M, Bull KR, Crockford TL, Kingston BL, Crawford G, Anzilotti C, Steeples V, Ghaffari S, Czibik G, Bellahcene M, Watkins H, Ashrafian H, Davies B, Woods A, Carling D, Yavari A, Beutler B, Cornall RJ. Mutation of Fnip1 is associated with B-cell deficiency, cardiomyopathy, and elevated AMPK activity. Proc. Natl. Acad. Sci. 2016:201607592. doi: 10.1073/pnas.1607592113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan M, Gingras M-C, Dunlop EA, Nouët Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, Faubert B, Kamps M, Sabourin S, Preston RS, Davies DM, Roughead T, Chotard L, van Steensel MAM, Jones R, Tee AR, Pause A. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J. Clin. Invest. 2014;124:2640–2650. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Possik E, Jalali Z, Nouët Y, Yan M, Gingras M-C, Schmeisser K, Panaite L, Dupuy F, Kharitidi D, Chotard L, Jones RG, Hall DH, Pause A. Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival. PLoS Genet. 2014;10:e1004273. doi: 10.1371/journal.pgen.1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunn GJ, Hudson CC, Sekulić A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 61.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 62.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JWB, Blenis J, Pende M, Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and TRCP-Mediated Degradation of PDCD4 Promotes Protein Translation and Cell Growth. Science (80-) 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 64.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung Y-L, Schulze A. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim JE, Chen J. Regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–56. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 66.Ben-sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Brendan D. mTORC1 Induces Purine Synthesis Through Control of the Mitochondrial Tetrahydrofolate Cycle. Science. 2015;351:728–33. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de Novo Pyrimidine Synthesis by Growth Signaling Through mTOR and S6K1. Science (80-) 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative Phosphoproteomics Reveal mTORC1 Activates de Novo Pyrimidine Synthesis. Science (80-) 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 69.Land SC, Tee AR. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J. Biol. Chem. 2007;282:20534–43. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 70.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan J-L, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 Association with the ULK1 - Atg13 - FIP200 Complex Required for Autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koren I, Reem E, Kimchi A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 2010;20:1093–1098. doi: 10.1016/j.cub.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 74.Peña-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TAT, Zou L, Xie X-J, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–58. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci. Signal. 2012;5:ra42–ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Eur. Mol. Biol. Organ. J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 79.García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem. J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Zhou Y, Graves DT. FOXO transcription factors: Their clinical significance and regulation. Biomed Res. Int. 2014;2014:925350. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dos DS, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 82.Gan X, Wang J, Wang C, Sommer E, Kozasa T, Srinivasula S, Alessi D, Offermanns S, Simon MI, Wu D. PRR5L degradation promotes mTORC2-mediated PKC-δ phosphorylation and cell migration downstream of Gα12. Nat. Cell Biol. 2012;14:686–96. doi: 10.1038/ncb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Gao T. MTORC2 phosphorylates protein kinase Cζ to regulate its stability and activity. EMBO Rep. 2014;15:191–198. doi: 10.1002/embr.201338119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomanetz V, Angliker N, Cloëtta D, Lustenberger RM, Schweighauser M, Oliveri F, Suzuki N, Rüegg MA. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J. Cell Biol. 2013;201:293–308. doi: 10.1083/jcb.201205030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 86.Melchers F. Checkpoints that control B cell development. J. Clin. Invest. 2015;125:2203–2210. doi: 10.1172/JCI78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hardy RR, Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 88.Reth M, Nielsen P. Signaling circuits in early B-cell development. Adv. Immunol. 2014;122:129–75. doi: 10.1016/B978-0-12-800267-4.00004-3. [DOI] [PubMed] [Google Scholar]
- 89.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 2009;9:767–77. doi: 10.1038/nri2656. [DOI] [PubMed] [Google Scholar]
- 90.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 91.Allman D, Pillai S. Peripheral B cell subsets. Curr. Opin. Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 2013;13:118–32. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat. Rev. Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 94.Iwata TN, Ramírez JA, Tsang M, Park H, Margineantu DH, Hockenbery DM, Iritani BM. Conditional Disruption of Raptor Reveals an Essential Role for mTORC1 in B Cell Development, Survival and Metabolism. J. Immunol. 2016;197:2250–60. doi: 10.4049/jimmunol.1600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang S, Readinger Ja, DuBois W, Janka-Junttila M, Robinson R, Pruitt M, Bliskovsky V, Wu JZ, Sakakibara K, Patel J, Parent Ca, Tessarollo L, Schwartzberg PL, Mock Ba. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 2011;117:1228–38. doi: 10.1182/blood-2010-05-287821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo F, Zhang S, Grogg M, Cancelas JA, Varney ME, Starczynowski DT, Du W, Yang JQ, Liu W, Thomas G, Kozma S, Pang Q, Zheng Y. Mouse gene targeting reveals an essential role of mTOR in hematopoietic stem cell engraftment and hematopoiesis. Haematologica. 2013;98:1353–1358. doi: 10.3324/haematol.2012.080424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, Anastasiou D, Gilliland DG, Scadden DT, Guertin DA, Armstrong SA. MTOR complex 1 plays critical roles in hematopoiesis and pten-loss-evoked leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoshii T, Tadokoro Y, Naka K, Ooshio T, Muraguchi T, Sugiyama N, Soga T, Araki K, Yamamura K-I, Hirao A. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J. Clin. Invest. 2012;122:2114–29. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang S, Pruitt M, Tran D, Du Bois W, Zhang K, Patel R, Hoover S, Simpson RM, Simmons J, Gary J, Snapper CM, Casellas R, Mock Ba, Du Bois W, Zhang K, Patel R, Hoover S, Simpson RM, Simmons J, Gary J, Snapper CM, Mock Ba, Casellas R. B cell-specific deficiencies in mTOR limit humoral immune responses. J. Immunol. 2013;191:1692–703. doi: 10.4049/jimmunol.1201767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keating R, Hertz T, Wehenkel M, Harris TL, Edwards BA, McClaren JL, Brown SA, Surman S, Wilson ZS, Bradley P, Hurwitz J, Chi H, Doherty PC, Thomas PG, McGargill MA. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 2013;14:1266–76. doi: 10.1038/ni.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jones DD, Gaudette BT, Wilmore JR, Chernova I, Bortnick A, Weiss BM, Allman D. MTOR has distinct functions in generating versus sustaining humoral immunity. J. Clin. Invest. 2016;126:4250–4261. doi: 10.1172/JCI86504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, Thomas JW, Hiebert S, Haase VH, Boothby MR. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537:234–238. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Limon JJ, So L, Jellbauer S, Chiu H, Corado J, Sykes SM, Raffatellu M, Fruman DA. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5076–85. doi: 10.1073/pnas.1407104111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee K, Heffington L, Jellusova J, Nam KT, Raybuck A, Cho SH, Thomas JW, Rickert RC, Boothby M. Requirement for Rictor in homeostasis and function of mature B lymphoid cells. Blood. 2013;122:2369–2379. doi: 10.1182/blood-2013-01-477505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benhamron S, Tirosh B. Direct activation of mTOR in B lymphocytes confers impairment in B-cell maturation andloss of marginal zone B cells. Eur. J. Immunol. 2011;41:2390–6. doi: 10.1002/eji.201041336. [DOI] [PubMed] [Google Scholar]
- 107.Ci X, Kuraoka M, Wang H, Carico Z, Hopper K, Shin J, Deng X, Qiu Y, Unniraman S, Kelsoe G, Zhong XP. TSC1 promotes B cell maturation but is dispensable for germinal center formation. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0127527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y, Hu T, Hua C, Gu J, Zhang L, Hao S, Liang H, Wang X, Wang W, Xu J, Liu H, Liu B, Cheng T, Yuan W. Rictor is required for early B cell development in bone marrow. PLoS One. 2014;9:e103970. doi: 10.1371/journal.pone.0103970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lazorchak AS, Liu D, Facchinetti V, Di Lorenzo A, Sessa WC, Schatz DG, Su B. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol. Cell. 2010;39:433–443. doi: 10.1016/j.molcel.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Baba M, Keller JR, Sun HW, Resch W, Kuchen S, Suh HC, Hasumi H, Hasumi Y, Kieffer-Kwon KR, Gonzalez CG, Hughes RM, Klein ME, Oh HF, Bible P, Southon E, Tessarollo L, Schmidt LS, Linehan WM, Casellas R. The folliculin-FNIP1 pathway deleted in human Birt-Hogg-Dubé syndrome is required for murine B-cell development. Blood. 2012;120:1254–1261. doi: 10.1182/blood-2012-02-410407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park H, Tsang M, Iritani BM, Bevan MJ. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7066–71. doi: 10.1073/pnas.1406473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 2011;12:888–97. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lien EC, Lyssiotis CA, Cantley LC. Metabolic Reprogramming by the PI3K–Akt-mTOR Pathway in Cancer. Recent Results Cancer Res. 2016:39–72. doi: 10.1007/978-3-319-42118-6_3. [DOI] [PubMed] [Google Scholar]
- 115.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jellusova J, Rickert RC. The PI3K pathway in B cell metabolism. Crit. Rev. Biochem. Mol. Biol. 2016;51:359–378. doi: 10.1080/10409238.2016.1215288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Morishita N, Tsukahara H, Chayama K, Ishida T, Washio K, Miyamura T, Yamashita N, Oda M, Morishima T. Activation of Akt is associated with poor prognosis and chemotherapeutic resistance in pediatric B-precursor acute lymphoblastic leukemia. Pediatr. Blood Cancer. 2012;59:83–89. doi: 10.1002/pbc.24034. [DOI] [PubMed] [Google Scholar]
- 118.Nemes K, Sebestyén A, Márk Á, Hajdu M, Kenessey I, Sticz T, Nagy E, Barna G, Váradi Z, Kovács G, Kopper L, Csóka M. Mammalian Target of Rapamycin (mTOR) Activity Dependent Phospho-Protein Expression in Childhood Acute Lymphoblastic Leukemia (ALL) PLoS One. 2013;8:e59335. doi: 10.1371/journal.pone.0059335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peponi E, Drakos E, Reyes G, Leventaki V, Rassidakis GZ, Medeiros LJ. Activation of mammalian target of rapamycin signaling promotes cell cycle progression and protects cells from apoptosis in mantle cell lymphoma. Am. J. Pathol. 2006;169:2171–80. doi: 10.2353/ajpath.2006.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu B, Zhou X, Xiao X, Yan S, Qin T, Shi D. [Activation clinicopathologic significance of AKT/mTOR signaling pathway in diffuse large B-cell lymphoma] Zhonghua Bing Li Xue Za Zhi = Chinese J. Pathol. 2009;38:35–41. [PubMed] [Google Scholar]
- 121.Zining J, Lu X, Caiyun H, Yuan Y. Genetic polymorphisms of mTOR and cancer risk: a systematic review and updated meta-analysis. Oncotarget. 2016;7:57464–57480. doi: 10.18632/oncotarget.10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Okosun J, Wolfson RL, Wang J, Araf S, Wilkins L, Castellano BM, Escudero-Ibarz L, Al Seraihi AF, Richter J, Bernhart SH, Efeyan A, Iqbal S, Matthews J, Clear A, Guerra-Assunção JA, Bödör C, Quentmeier H, Mansbridge C, Johnson P, Davies A, Strefford JC, Packham G, Barrans S, Jack A, Du M-Q, Calaminici M, Lister TA, Auer R, Montoto S, Gribben JG, Siebert R, Chelala C, Zoncu R, Sabatini DM, Fitzgibbon J. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat. Genet. 2015;48:183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ying ZX, Jin M, Peterson LF, Bernard D, Saiya-Cork K, Yildiz M, Wang S, Kaminski MS, Chang AE, Klionsky DJ, Malek SN. Recurrent Mutations in the MTOR Regulator RRAGC in Follicular Lymphoma. Clin. Cancer Res. 2016;22:5383–5393. doi: 10.1158/1078-0432.CCR-16-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang L, Huang J, Wu P, Li Q, Rong L, Xue Y, Lu Q, Li J, Tong N, Wang M, Zhang Z, Fang Y. Association of genetic variations in mTOR with risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk. Lymphoma. 2012;53:947–951. doi: 10.3109/10428194.2011.628062. [DOI] [PubMed] [Google Scholar]
- 125.Lee J-HS, Vo T-T, Fruman DA. Targeting mTOR for the treatment of B cell malignancies. Br. J. Clin. Pharmacol. 2016;82:1213–1228. doi: 10.1111/bcp.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dinner S, Platanias LC. Targeting the mTOR Pathway in Leukemia. J. Cell. Biochem. 2016;117:1745–52. doi: 10.1002/jcb.25559. [DOI] [PubMed] [Google Scholar]
- 127.Calimeri T, Ferreri AJM. m-TOR inhibitors and their potential role in haematological malignancies. Br. J. Haematol. 2017 doi: 10.1111/bjh.14529. [DOI] [PubMed] [Google Scholar]
- 128.Tasian SK, Teachey DT, Rheingold SR. Targeting the PI3K/mTOR Pathway in Pediatric Hematologic Malignancies. Front. Oncol. 2014;4:108. doi: 10.3389/fonc.2014.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vu C, Fruman DA. Target of Rapamycin Signaling in Leukemia and Lymphoma. Clin. Cancer Res. 2010;16:5374–5380. doi: 10.1158/1078-0432.CCR-10-0480. [DOI] [PubMed] [Google Scholar]
- 130.Brown VI, Fang J, Alcorn K, Barr R, Kim JM, Wasserman R, Grupp SA. Rapamycin is active against B-precursor leukemia in vitro and in vivo an effect that is modulated by IL-7-mediated signaling. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15113–8. doi: 10.1073/pnas.2436348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen I-M, Collins JR, Mullighan CG, Hunger SP, Harvey RC, Willman CL, Fridman JS, Loh ML, Grupp SA, Teachey DT. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Avellino R, Romano S, Parasole R, Bisogni R, Lamberti A, Poggi V, Venuta S, Pik T. Rapamycin stimulates apoptosis of childhood acute lymphoblastic leukemia cells. Apoptosis. 2005;106:1400–1406. doi: 10.1182/blood-2005-03-0929.Supported. [DOI] [PubMed] [Google Scholar]
- 133.Teachey DT, Obzut DA, Cooperman J, Fang J, Carroll M, Choi JK, Houghton PJ, Brown VI, Grupp SA. The mTOR inhibitor CCI-779 induces apoptosis and inhibits growth in preclinical models of primary adult human ALL. Blood. 2006;107:1149–55. doi: 10.1182/blood-2005-05-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hess G, Herbrecht R, Romaguera J, Verhoef G, Crump M, Gisselbrecht C, Laurell A, Offner F, Strahs A, Berkenblit A, Hanushevsky O, Clancy J, Hewes B, Moore L, Coiffier B. Phase III study to evaluate temsirolimus compared with investigator’s choice therapy for the treatment of relapsed or refractory mantle cell lymphoma. J. Clin. Oncol. 2009;27:3822–9. doi: 10.1200/JCO.2008.20.7977. [DOI] [PubMed] [Google Scholar]
- 135.Teachey DT, Sheen C, Hall J, Ryan T, Brown VI, Fish J, Reid GSD, Seif AE, Norris R, Chang YJ, Carroll M, Grupp SA. mTOR inhibitors are synergistic with methotrexate: an effective combination to treat acute lymphoblastic leukemia. Blood. 2008;112:2020–2023. doi: 10.1182/blood-2008-02-137141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Beagle BR, Nguyen DM, Mallya S, Tang SS, Lu M, Zeng Z, Konopleva M, Vo T-T, Fruman DA. mTOR kinase inhibitors synergize with histone deacetylase inhibitors to kill B-cell acute lymphoblastic leukemia cells. Oncotarget. 2015;6:2088–2100. doi: 10.18632/oncotarget.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iacovelli S, Ricciardi MR, Allegretti M, Mirabilii S, Licchetta R, Bergamo P, Rinaldo C, Zeuner A, Foà R, Milella M, McCubrey JA, Martelli AM, Tafuri A. Co-targeting of Bcl-2 and mTOR pathway triggers synergistic apoptosis in BH3 mimetics resistant acute lymphoblastic leukemia. Oncotarget. 2015;6:32089–103. doi: 10.18632/oncotarget.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang X, He G, Gong Y, Zheng B, Shi F, Shi R, Yang X. Mammalian target of rapamycin inhibitor rapamycin enhances anti-leukemia effect of imatinib on Ph + acute lymphoblastic leukemia cells. Eur. J. Haematol. 2014;92:111–120. doi: 10.1111/ejh.12202. [DOI] [PubMed] [Google Scholar]
- 139.Daver N, Boumber Y, Kantarjian H, Ravandi F, Cortes J, Rytting ME, Kawedia JD, Basnett J, Culotta KS, Zeng Z, Lu H, Richie MA, Garris R, Xiao L, Liu W, Baggerly KA, Jabbour E, O’Brien S, Burger J, Bendall LJ, Thomas D, Konopleva M. A Phase I/II Study of the mTOR Inhibitor Everolimus in Combination with HyperCVAD Chemotherapy in Patients with Relapsed/Refractory Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2015;21:2704–14. doi: 10.1158/1078-0432.CCR-14-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oki Y, Buglio D, Fanale M, Fayad L, Copeland A, Romaguera J, Kwak LW, Pro B, de Castro Faria S, Neelapu S, Fowler N, Hagemeister F, Zhang J, Zhou S, Feng L, Younes A. Phase I study of panobinostat plus everolimus in patients with relapsed or refractory lymphoma. Clin. Cancer Res. 2013;19:6882–90. doi: 10.1158/1078-0432.CCR-13-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, Khuri FR. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 142.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K–dependent feedback loop in human cancer. J. Clin. Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Choo AY, Blenis J. Not all substrates are treated equally: Implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle. 2009;8:567–572. doi: 10.4161/cc.8.4.7659. [DOI] [PubMed] [Google Scholar]
- 145.Tasian SK, Teachey DT, Li Y, Shen F, Harvey RC, Chen I-M, Ryan T, Vincent TL, Willman CL, Perl AE, Hunger SP, Loh ML, Carroll M, Grupp SA. Potent efficacy of combined PI3K/mTOR and JAK or ABL inhibition in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2017;129:177–187. doi: 10.1182/blood-2016-05-707653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Badura S, Tesanovic T, Pfeifer H, Wystub S, Nijmeijer BA, Liebermann M, Falkenburg JHF, Ruthardt M, Ottmann OG. Differential effects of selective inhibitors targeting the PI3K/AKT/mTOR pathway in acute lymphoblastic leukemia. PLoS One. 2013;8:e80070. doi: 10.1371/journal.pone.0080070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, Duveau DY, Jiang J-K, Michael S, Mierzwa T, Huang W, Walsh MJ, Mott BT, Patel P, Leister W, Maloney DJ, Leclair CA, Rai G, Jadhav A, Peyser BD, Austin CP, Martin SE, Simeonov A, Ferrer M, Staudt LM, Thomas CJ. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc. Natl. Acad. Sci. 2014;111:2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang S, Yang ZJ, Yu C, Sinicrope FA. Inhibition of mTOR Kinase by AZD8055 Can Antagonize Chemotherapy-induced Cell Death through Autophagy Induction and Down-regulation of p62/Sequestosome 1. J. Biol. Chem. 2011;286:40002–40012. doi: 10.1074/jbc.M111.297432. [DOI] [PMC free article] [PubMed] [Google Scholar]