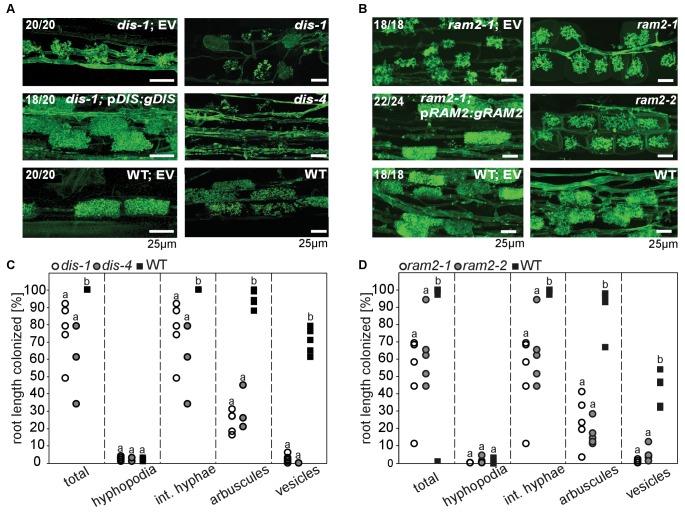
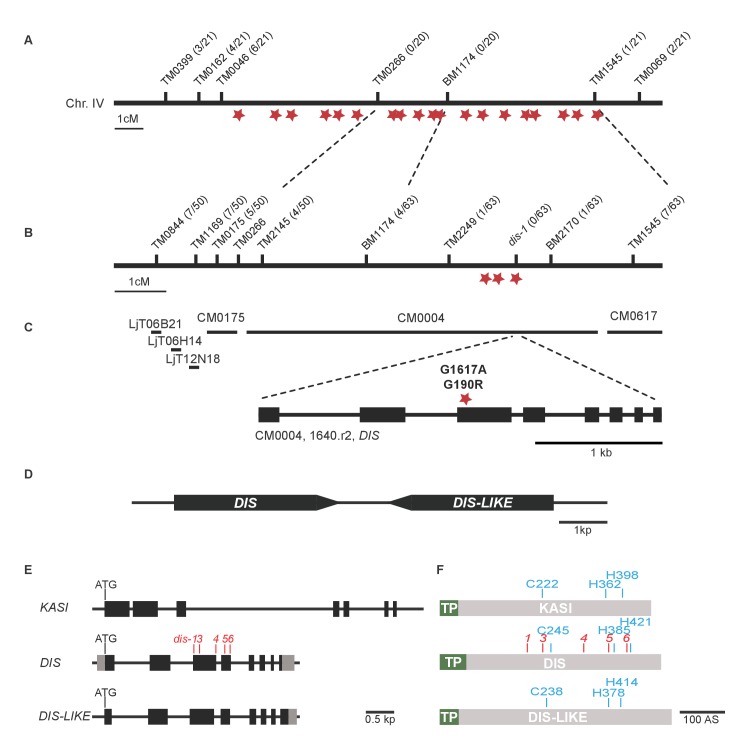
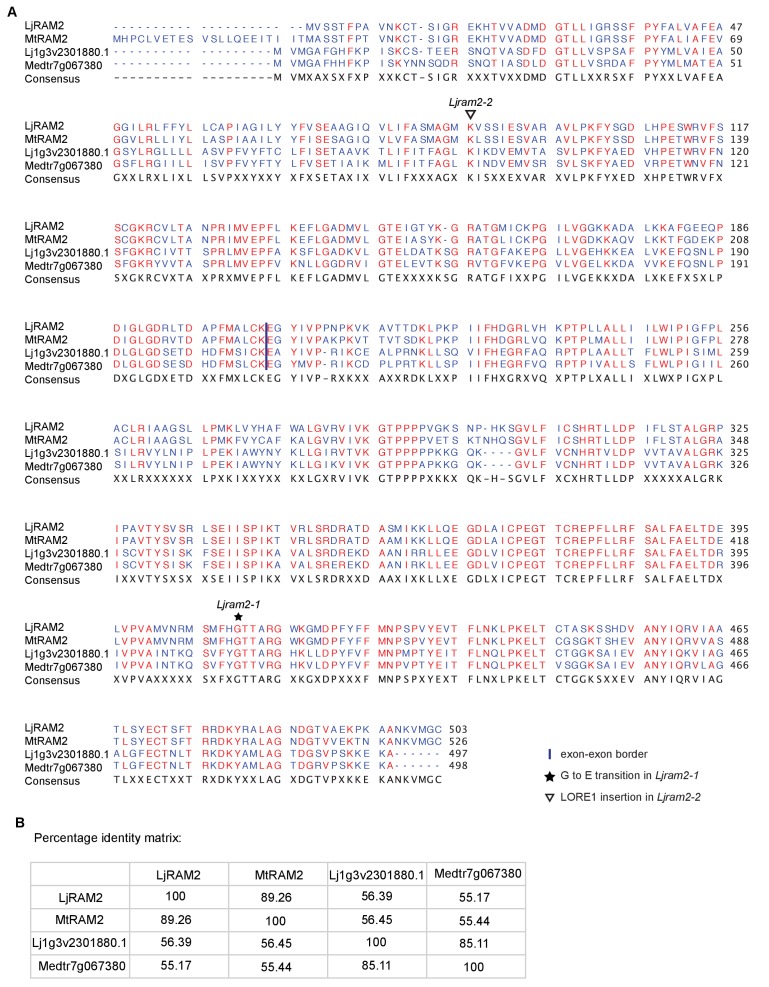
(
A–B) Genetic map of the
DIS locus on chromosome 4. Numbers next to marker positions refer to the proportion of recombinant individuals among the number of analyzed F2 mutant plants. Rough mapping had previously identified the position of the
dis mutation on the south arm of chromosome 4 (
Groth et al., 2013). (
A) In the first fine-mapping round, the interval narrowed down by recombinants comprised 19 EMS-induced SNPs (red stars), that could be confirmed by re-sequencing the mutant genome using next generation sequencing. (
B) Further fine mapping resulted in an interval with 3 of these confirmed SNPs. (
C) Physical map of the
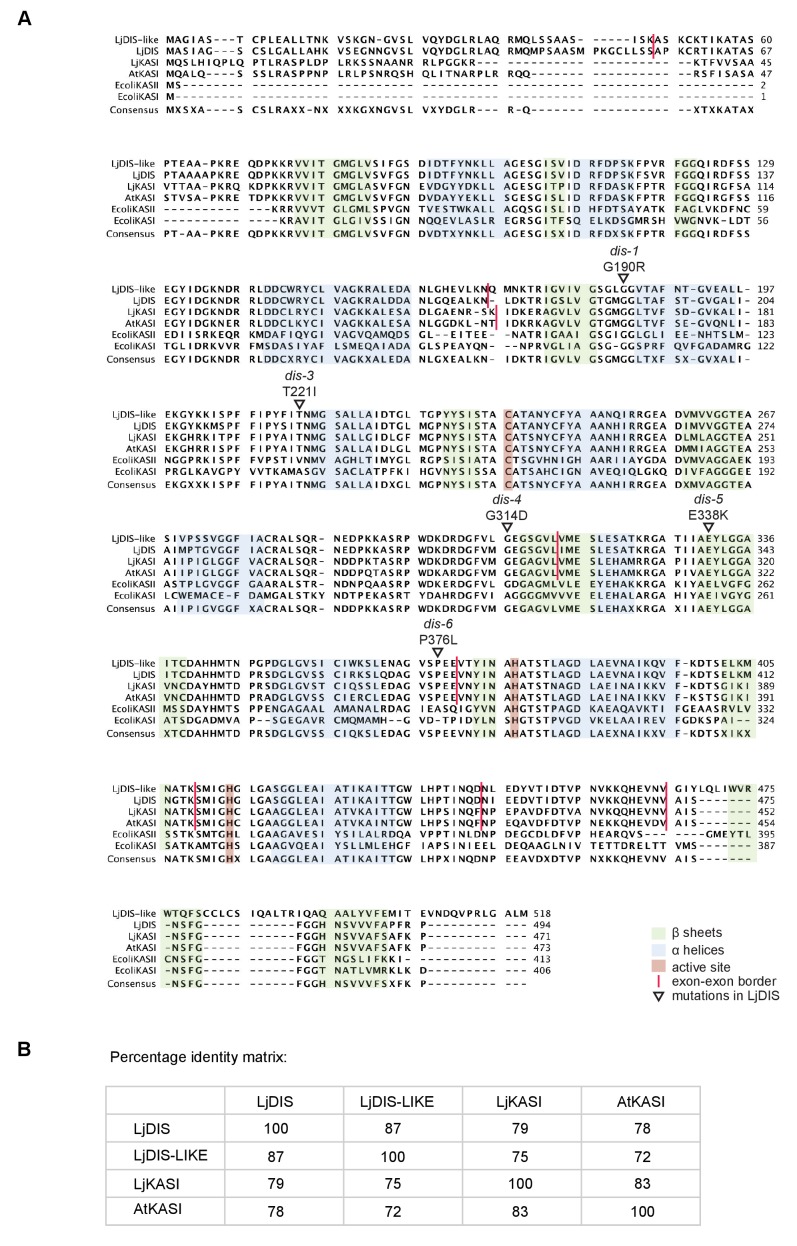
DIS locus. LjT followed by a number refers to TAC clones. CM followed by a number refers to contigs. One of the three SNPs causes a G to A transition in exon 3 of chr.4.CM004.1640.r2.a resulting in an amino acid change from glycine to arginine at position 190 of the protein product, which shares 79% sequence identity with a β-keto-acyl ACP synthase I (KASI) from
Arabidopsis thaliana. Black boxes indicate exons separated by introns. (
D) The
DIS gene is duplicated in tandem. (
E) Gene structure of
DIS,
DIS-LIKE and
KASI. Black boxes display exons separated by introns (black lines). Grey boxes indicate determined un-translated regions. (
F) DIS, DIS-LIKE and KASI are predicted to contain a plastid transit peptide (green). The catalytic triad is shown in blue and the location of mutations identified by TILLING in the
DIS gene are shown in red. We chose the
dis-4 mutant for further analysis because the mutation resulted in a glycine replacement, which likely affects the functionality of the protein.