Abstract
Phelan-McDermid Syndrome (PMDS) is a complex neurodevelopmental disorder characterized by global developmental delay, severely impaired speech, intellectual disability, and an increased risk of Autism Spectrum Disorders (ASDs)1. PMDS is caused by heterozygous deletions of chromosome 22q13.3. Among the genes in the deleted region is SHANK3, which encodes a protein in the postsynaptic density (PSD)2,3. Rare mutations in SHANK3 have been associated with idiopathic ASDs4–7, non-syndromic intellectual disability8, and schizophrenia9. Although SHANK3 is considered to be the most likely candidate gene for the neurological abnormalities in PMDS patients10, the cellular and molecular phenotypes associated with this syndrome in human neurons are unknown. We generated induced pluripotent stem cells (iPSCs) from individuals with PMDS and autism and used them to produce functional neurons. We show that PMDS neurons have reduced Shank3 expression and major defects in excitatory but not inhibitory synaptic transmission. Excitatory synaptic transmission in PMDS neurons can be corrected by restoring Shank3 expression or by treating neurons with insulin-like growth factor 1 (IGF1). IGF1 treatment promotes formation of excitatory synapses that lack Shank3 but contain PSD95 and NMDA receptors with fast deactivation kinetics. Our findings provide direct evidence for a disruption in the ratio of cellular excitation and inhibition in PMDS neurons, and point to a molecular pathway that can be recruited to restore it.
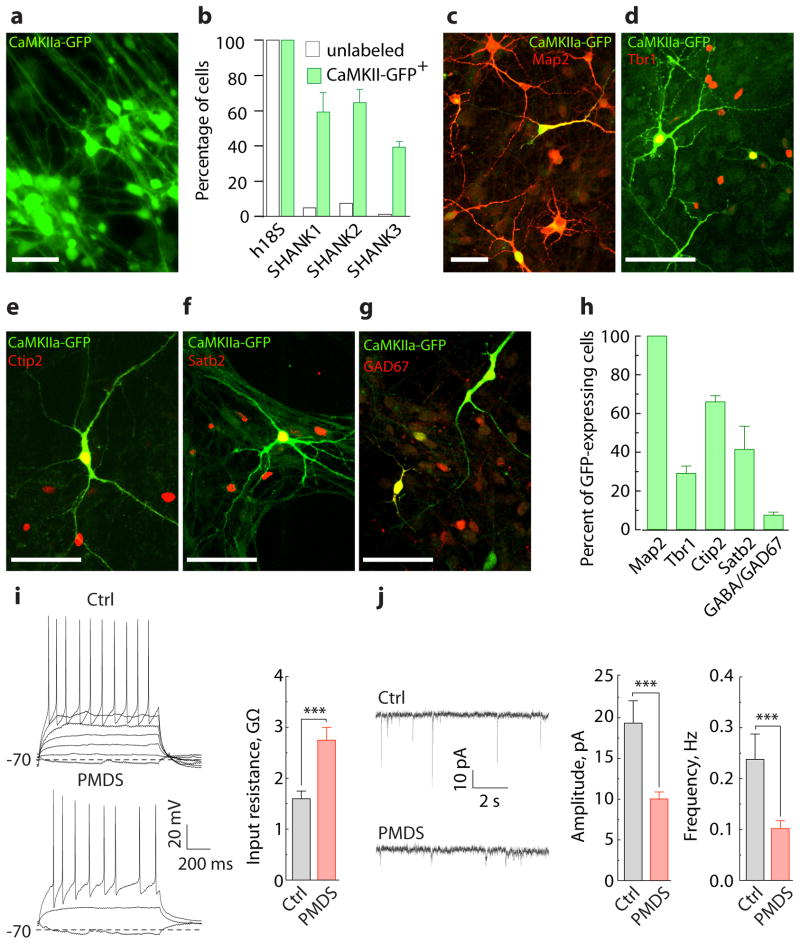
To study cellular and molecular phenotypes associated with PMDS in humans, we generated six iPSC lines from two patients with PMDS (Supplementary Table 1) that had deletions of approximately 1 Mb in chromosome 22 (Supplementary Fig. 1 and Supplementary Table 2) by using standard reprogramming techniques11,12. As controls, we used iPSC and human ES lines that were characterized in previous studies12,13. Control and PMDS iPSC lines had typical human ES cell-like morphologies, expressed endogenous pluripotency marker proteins but had suppressed the expression of the exogenous reprogramming factors, had normal karyotypes, and formed teratomas in-vivo (Supplementary Fig. 2 and Supplementary Table 3). We differentiated control and PMDS iPSCs into neurons using a modified monolayer differentiation protocol (Supplementary Fig. 3) and characterized the population of differentiated cells by quantitative real-time PCR (qRT-PCR) (Supplementary Fig. 4). Similar populations of neuronal cells were found in cultures of control and PMDS neurons. Analysis of gene expression with multiplex single cells qRT-PCR13,14 demonstarted that approximately 42% of cells expressed pan-neuronal markers, 15% expressed cortical layer markers, and only 2–5% of the cells expressed the synaptic markers SHANK1-3 (Supplementary Fig. 5), suggesting that only a small fraction of the cells were synaptically mature neurons. To enrich for mature forebrain neurons, we infected cells with EGFP under the control of the CaMKIIα promoter (CaMKIIα-GFP)15,16 (Fig. 1a). Single-cell profiling of GFP-expressing cells showed an approximate 12-fold enrichment in cells expressing SHANK1-3 (Fig. 1b). These cells also expressed synaptic markers and genes typically found in the cortex and striatum, including TBR1, CUX1, SATB2, SOX5, FOXP1, CTIP2, and FOXP2 (Supplementary Fig. 5) and were stained with antibodies recognizing Map2 (~99%), Tbr1 (~29%), Ctip2 (~66%), Satb2 (~42%), and GABA or GAD67 (~7.5%) (Fig. 1c–h and Supplementary Fig. 6).
Figure 1. CaMKIIα-GFP labels functional iPSC-derived forebrain neurons that express SHANK3.
a, Image of iPSC-derived neurons labeled with EGFP under the control of CaMKIIα promoter. b, Fraction of cells expressing SHANK1-3, selected either randomly (unlabeled, n = 1 line/90 cells) or based on CaMKIIα-GFP expression (CaMKIIα-GFP+, n = 8 lines/90 cells) and assessed by multiplex single cell qRT-PCR (Supplementary Fig. 6). c–g, Images of iPSC-derived neurons immunostained with antibodies against GFP and Map2 (c), Tbr1 (d), Ctip2 (e), Satb2 (f), and GAD67 (g). h, Fraction of cells expressing Map2 (n = 3 cover slips (682 cells)), Tbr1 (n = 4 (566)), Ctip2 (n = 8 (853)), Satb2 (n = 6 (646)), and GABA/GAD67 (n = 5 (493)) among GFP-expressing cells. i, Representative recordings of APs (left, induced by somatic current injections, ΔI = 5 pA) and input resistance measurements (right) obtained from GFP-expressing control (n = 22 cells/3 lines) and PMDS neurons (n = 36 cells/5 lines). j, Representative recordings (left) and quantification of amplitude and frequency (right) of spontaneous EPSCs obtained from GFP-expressing control (n = 19 cells/3 lines) and PMDS neurons (n = 26 cells/5 lines) (Supplementary Table 4). Data presented as means ± SEMs; *** p < 0.001, Mann-Whitney test. Scale bars = 50 μm.
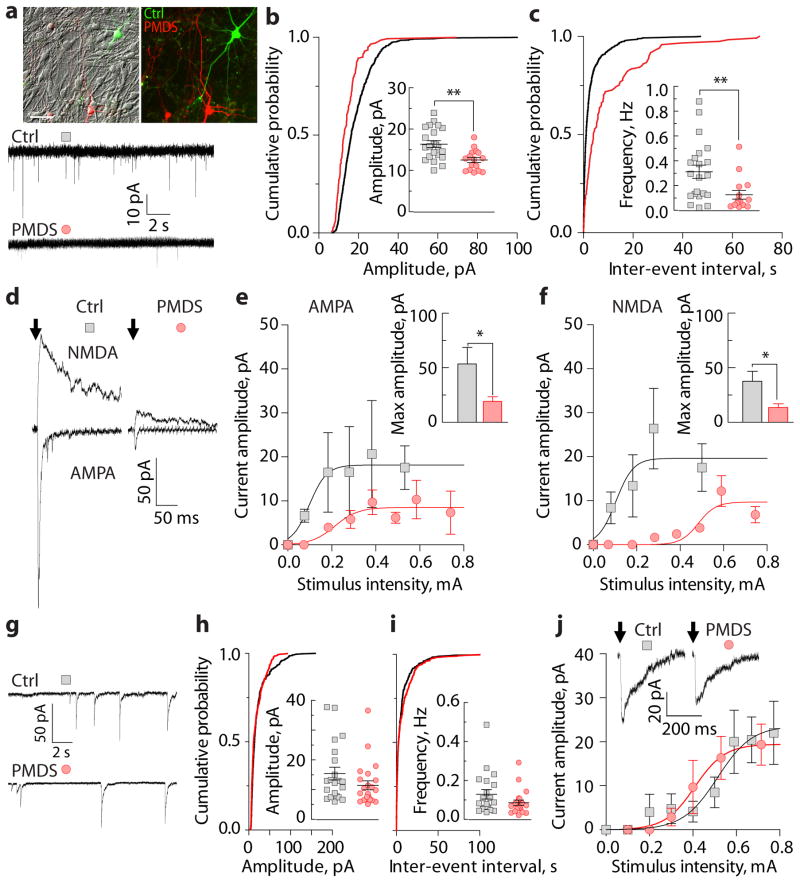
We next investigated the electrophysiological properties of control and PMDS neurons. Control and PMDS neurons expressing GFP had similar AP characteristics, resting membrane potential, and capacitance (Supplementary Table 4). PMDS neurons, however, had an increased input resistance and reduced amplitude and frequency of spontaneous synaptic events (Fig. 1i–j and Supplementary Table 4). A reduction in synaptic transmission in PMDS neurons could arise from differences in the number of neurons present in cultures of PMDS and control neurons. In fact, anti-Map2 antibody staining revealed that our PMDS iPSC lines produced slightly fewer neurons than controls (Supplementary Fig. 7). To control for this difference in neuronal density and to study cells of both genotypes under identical experimental conditions, we infected control and PMDS neurons with either CaMKIIα-GFP or -mKate2 and cultured them together in the same dish (Fig. 2a). We found that the amplitude and frequency of spontaneous excitatory postsynaptic currents (EPSCs) recorded from PMDS neurons were significantly reduced relative to controls (Fig. 2b–c), suggesting that synaptic defects in PMDS neurons are postsynaptic and cell autonomous. To determine if the changes in spontaneous EPSCs were also reflected in differences in evoked AMPA- and NMDA-mediated synaptic responses, we measured currents induced by local field stimulation at −70 mV (AMPA-EPSCs) and +60 mV (NMDA-EPSCs). Smaller AMPA- and NMDA-EPSCs were measured in PMDS neurons in response to different stimulus intensities (Fig. 2d–f), indicating that both AMPA- and NMDA-mediated synaptic transmissions are significantly impaired.
Figure 2. PMDS neurons display impaired excitatory synaptic transmission.
a, Images of control (green) and PMDS (red) neurons co-cultured on a bed of rat cortical astrocytes (scale bar = 50 μm), top. Traces of spontaneous EPSCs recorded at −70 mV, bottom. b–c, Cumulative distribution and quantification of amplitude (b) and frequency (c) of spontaneous EPSCs (n = 22 control and 16 PMDS cells). d, Traces of evoked AMPA- (Vh = −70 mV) and NMDA-EPSCs (Vh = +60 mV). Arrows indicate the time of stimulation. Stimulation artifacts were blanked. e-f, Input-output curves of evoked AMPA- (measured at the peak; n = 15 control and 23 PMDS cells) and NMDA-EPSCs (measured 50 ms post-stimulus; n = 14 control and 20 PMDS cells) recorded in response to different stimulus intensities. Bar graphs show quantification of maximum current amplitude. g, Traces of spontaneous IPSCs recorded at −70 mV. h–i, Cumulative distribution and quantification of amplitude (h) and frequency (i) of spontaneous IPSCs (n = 21 control and 23 PMDS cells). j, Representative traces and input-output curves of evoked IPSCs (n = 17 control and 16 PMDS cells). Data presented as means ± SEMs; * p < 0.05, ** p < 0.01, Mann-Whitney test.
To determine if inhibitory synaptic transmission was also altered in PMDS neurons, we recorded spontaneous and evoked inhibitory postsynaptic currents (IPSCs) (Fig. 2g–j). There were no differences in IPSCs measured from co-cultured control and PMDS neurons. To determine the direction of the voltage shifts induced by IPSCs at this developmental stage, we measured voltage deflections induced by focal GABA application (Supplementary Fig. 8). Most control and PMDS neurons responded with hyperpolarizing shifts in membrane potential. These results indicate that the defects in PMDS neurons are limited to excitatory synaptic transmission and suggest that there is an alteration in the balance of cellular excitation and inhibition in PMDS neurons.
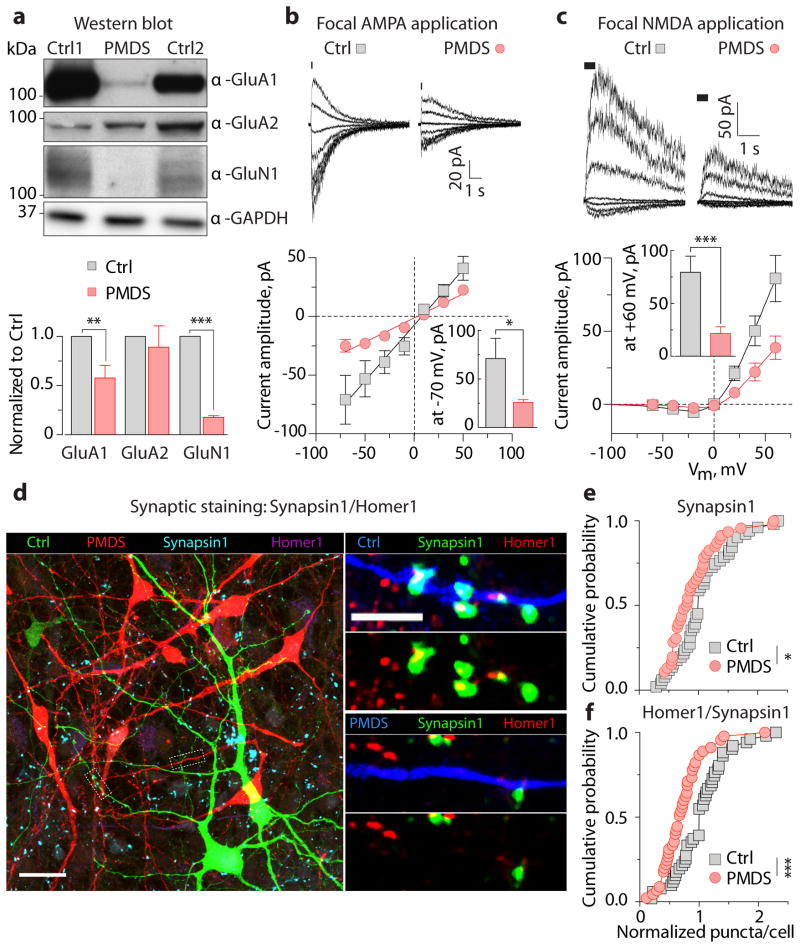
To gain insight into the mechanism underlying the impairment of excitatory synaptic transmission in PMDS neurons, we measured the expression levels of AMPA and NMDA receptors. PMDS neurons expressed significantly less GluA1 and GluN1 proteins (Fig. 3a) and generated significantly smaller currents in response to the focal application of either AMPA (Fig. 3b) or NMDA (Fig. 3c). In contrast, the amplitude of GABA-evoked currents was similar for both control and PMDS neurons (Supplementary Fig. 9), suggesting that PMDS neurons express a decreased number of excitatory neurotransmitter receptors and normal levels of inhibitory receptors.
Figure 3. PMDS neurons show reduced expression of glutamate receptors and decreased number of synapses.
a, Quantification of expression of AMPA and NMDA receptors in control and PMDS neurons using Western blot (D 45 – 50; n = 3 – 11 pairs of samples; ** p < 0.01, *** p < 0.001, one-sample t-test). GAPDH was used as a loading control. b–c, Representative currents (top) and peak current IV-curves (bottom) recorded in control and PMDS neurons in response to focal application of 200 μM AMPA (−70 – +50, Δ20 mV, n = 7 control and 11 PMDS cells) and 100 μM NMDA/10 μM Gly (−60 – +60, Δ20 mV, n = 21 control and 30 PMDS cells). Data presented as means ± SEMs; * p < 0.05, *** p < 0.001, Mann Whitney test. d, Images of co-cultured control (green) and PMDS (red) neurons immunostained for mKate2, GFP, Synapsin1, and Homer1. Scale bars = 20 (left) and 5 μm (right). e–f, Cumulative distributions of the number of Synapsin1+ (e) and Homer1+/Synapsin1+ (f) puncta on co-cultured control and PMDS neurons; * p < 0.05, *** p < 0.001, Kolmogorov-Smirnov test.
To determine whether the impairment in excitatory synaptic transmission in PMDS neurons is due to a reduced number of synapses, we immunostained co-cultured control and PMDS neurons with antibodies that recognize the presynaptic protein Synapsin1 and postsynaptic protein Homer1 (Fig. 3d). We observed a significant decrease in the number of puncta that were Synapsin1+ (Fig. 3e) and Homer1+/Synapsin1+ (Fig. 3f) in PMDS neurons as compared to controls, indicating that PMDS neurons have significantly fewer excitatory synapses than controls.
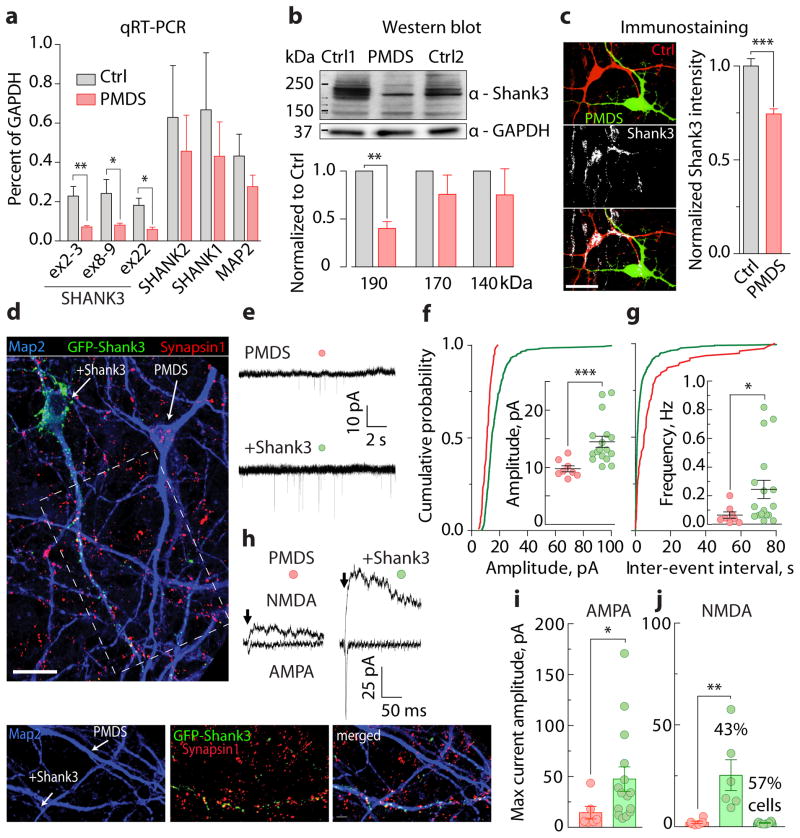
While SHANK3 has been proposed to be primarily responsible for the neurological phenotype in PMDS, the vast majority of PMDS patients have 10–100 additional genes deleted. To determine whether the synaptic phenotypes in PMDS neurons are due to reduced Shank3 expression, we measured the levels of SHANK3 mRNA and protein in PMDS neurons. While MAP2, SHANK1, and SHANK2 were expressed at similar levels in control and PMDS neurons (Fig. 4a), SHANK3 mRNA was significantly reduced in PMDS (Fig. 4a). Western blot analysis showed a significant reduction in the expression of a 190 kDa Shank3 protein isoform (Fig. 4b), which was expressed primarily in neurons (Supplementary Fig. 10). Immunostaining with anti-Shank3 antibodies on co-cultured control and PMDS neurons also revealed a significantly reduced level of Shank3 protein expression in PMDS neurons (Fig. 4c), suggesting that loss of a copy of SHANK3 in PMDS significantly downregulates the expression of the protein.
Figure 4. Reduced Shank3 expression contributs to synaptic defects in PMDS neurons.
a, Relative expression of SHANK1, SHANK2, SHANK3, and MAP2 mRNAs in iPSC-derived neurons (D30 – D35; n = 5 control and 6 PMDS biological replicates; * p < 0.05, ** p < 0.01, Student’s t-test). b, Quantification of Shank3 expression in control and PMDS neurons as assessed by Western blot (D45 – 50; n = 4 pairs of samples; ** p < 0.01, one-sample t-test). GAPDH was used as a loading control. c, Images (left) and quantification (right) of Shank3 expression in co-cultured control (red) and PMDS (green) neurons (scale bar = 20 μm). d, Images of PMDS neurons infected with GFP-Shank3 lentiviruses and immunostained with antibodies against Map2, GFP, and Synapsin1 (scale bars = 20 (top) and 5 (bottom) μm). e, Traces of spontaneous EPSCs recorded from uninfected and Shank3-infected PMDS neurons at −70 mV. f–g, Cumulative distribution and quantification of amplitude (f) and frequency (g) of spontaneous EPSCs (n = 8 uninfected and 17 Shank3-infected cells). h, Traces of evoked AMPA- (Vh = −70 mV) and NMDA-EPSCs (Vh = +60 mV). i–j, Quantification of maximum amplitudes of evoked AMPA- (i, measured at peak) and NMDA-EPSCs (j, measured 50 ms post stimulus) recorded from uninfected (n = 6) and Shank3-infected (n = 14) PMDS neurons. Data presented as means ± SEMs; * p < 0.05, *** p < 0.001, Mann-Whitney test.
We next investigated whether we could reverse the synaptic phenotype in PMDS neurons by increasing Shank3 expression. We infected PMDS neurons with a lentivirus containing the longest isoform of Shank3 fused to EGFP (Fig. 4d and Supplementary Fig. 11). Shank3 expression in PMDS neurons restored both the amplitude and frequency of spontaneous EPSCs (Fig. 4e–g). It also completely rescued evoked AMPA- and NMDA-EPSCs in 43% of recorded cells, and in the remaining cells, it rescued only AMPA-EPSCs (Fig. 4h–j). This suggests that loss of SHANK3 substantially contributes to the impaired excitatory synaptic transmission detected in PMDS neurons; however, it also suggests that Shank3 acts differently on AMPA and NMDA receptors.
We next tested if we could restore synaptic defecits in PMDS neurons pharmacologically, using agents that have been previously reported to increase the level of Shank3 expression or to increase synaptic transmission, including Trichostatin A (TSA)17,18, valproic acid (VPA)17,19, Nifedipine (Nif)20, IGF121–23, and IGF224. We treated control and PMDS neurons over the course of two to twenty days and measured the number of Homer1+/Synapsin1+ puncta. Only IGF1 increased the number of Synapsin1+/Homer1+ puncta to the levels observed in control neurons in the same culture (Fig. 5a–b). We found that IGF1 increased the average amplitude and frequency of spontaneous EPSCs in PMDS neurons to control levels (Fig. 5c), but had no effect on spontaneous IPSCs (Supplementary Fig. 12). IGF1 also restored the amplitude of evoked AMPA- and NMDA-EPSCs (Fig. 5d) and reduced the input resistance of PMDS neurons to that observed in control neurons (Supplementary Fig. 13). Finally, IGF1 completely restored the whole cell NMDA receptor currents evoked by focal application of NMDA, but failed to fully restore whole cell AMPA receptor currents elicited by application of AMPA (Supplementary Fig. 14). Together, these results suggest that treatment with IGF1 restores synaptic transmission in PMDS neurons by increasing the number of synaptic NMDA and AMPA receptors.
Figure 5. IGF1 treatment restores excitatory synaptic transmission in PMDS neurons.
a, Quantification of the number of Homer1+/Synapsin1+ puncta in co-cultured neurons after treatment with different agents (untreated: n = 51 control, 43 PMDS cells; TSA: n = 11/12, VPA: n = 25/29, Nif: n = 9/8, IGF2: n = 31/40, IGF1: n = 35/32). b, Images of co-cultured control (green) and PMDS (red) neurons after IGF1 treatment. Scale bars = 20 (top) and 5 (bottom) μm. c, Recordings (top) and quantification of amplitude and frequency (bottom) of spontaneous EPSCs recorded in IGF1-treated cultures at −70 mV (n = 18 control and 25 PMDS cells). d, Recordings (top) and quantification of amplitudes (bottom) of evoked AMPA- (measured at the peak, Vh = −70 mV) and NMDA-EPSCs (measured 50 ms post-stimulus, Vh = +60 mV) acquired in IGF1-treated cultures (n = 14 control and 21 PMDS cells). Dashed lines represent data originally presented in Fig. 2. e, Images of co-cultured control (green) and PMDS (red) neurons in untreated (top) and IGF1-treated (bottom) cultures, immunostained with antibodies against GFP, mKate2, Shank3, and PSD95. Scale bars = 20 (left) and 5 (right) μm. f, Quantification of the number of Shank3+, PSD95+, and Shank3/PSD95+ puncta in co-cultured control (w/o, n = 25 cells; IGF1, 21 cells) and PMDS (w/o, n = 33 cells; IGF1, 25 cells) neurons. Values were normalized to the mean values acquired from control neurons in untreated cultures. g, Mean normalized traces (left) and quantification of the decay kinetics (right) of NMDA-EPSCs recorded from control (w/o, n = 9; IGF1, 12) and PMDS (w/o, n = 18; IGF1, 18, GFP-Shank3, 6) neurons. Data presented as means ± SEMs; *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with Bonferroni’s test. h, Cartoon depicting possible mechanisms for the recovery of synaptic transmission in PMDS neurons.
To get insight into the mechanism of IGF1 action, we measured Shank3 expression in control and PMDS neurons treated with IGF1. Surprisingly, we found that IGF1 caused a dramatic decrease of Shank3 protein expression in the cell body and processes of both control and PMDS neurons (Supplementary Fig. 15). We next examined Shank3 expression at synapses by infecting cells with GFP-tagged Shank3 and staining neurons with the postsynaptic antibody anti-PSD95 (Supplementary Fig. 16a). We found only 50% overlap between Shank3- and PSD95-containing puncta in human iPSC-derived neurons. A similar pattern of Shank3 and PSD95 expression was detected in the human fetal brain tissue immunostained with anti-Shank3 and anti-PSD95 antibodies (Supplementary Fig. 16b). This suggests that in developing neurons there are three populations of excitatory synapses: those that contain Shank3 alone, those that contain Shank3 and PSD95, and those that contain PSD95 alone. We investigated the effect of IGF1 on these three populations of synapses by staining neurons with anti-Shank3 and anti-PSD95 antibodies (Fig. 5e). Treatment with IGF1 caused a decrease in the number of puncta expressing Shank3 alone in control neurons but had little effect in PMDS neurons, which already had reduced levels of Shank3 (Fig. 5f). In contrast, IGF1 caused a 45% and 340% increase in the fraction of puncta expressing PSD95 in control and PMDS neurons, respectively, while decreasing the number of puncta containing both markers (Fig. 5f). To determine if synapses that lack Shank3 are functionally different, we analyzed the physiological properties of synapses in PMDS and control neurons. We found that PMDS neurons have a significantly faster rate of decay of NMDA-EPSCs relative to control cells and treatment with IGF1 increased the decay rate of NMDA-EPSCs in control neurons to match the rate of decay in PMDS neurons (Fig. 5g). In contrast, Shank3 expression in PMDS neurons caused a decrease in the rate of decay of NMDA-EPSCs to match that in untreated control neurons (Fig. 5g). These results are consistent with the idea that IGF1 treatment rescues synaptic transmission in PMDS neurons by causing loss of Shank3-containing synapses and gain of PSD95-containing synapses, which are characterized by rapidly decaying NMDA-EPSCs (Fig. 5h).
Here, we show that neurons derived from iPSCs from PMDS patients have major defects in excitatory synaptic transmission arising from both a failure to form the correct number of excitatory synapses and a reduction in the expression of glutamate receptors. As no significant impairments in inhibitory synaptic transmission were found, this resulted in an imbalance between excitation and inhibition at a cellular level. Only some aspects of the phenotype detected in human cells are similar to those found in mouse models of PMDS25–27. Why there is a discrepancy is unclear. One potentially important issue is that there are multiple SHANK3 isoforms. PMDS patients generally lack the entire gene on one chromosome, whereas mice typically lack only some of the isoforms and have generally been studied as homozygote knockouts. We also demonstrated that Shank3 and IGF1 largely restore synaptic deficits in PMDS neurons. Interestingly, IGF1 decreases Shank3 expression and promotes the formation of a class of synapses containing PSD95 but lacking Shank3. One possibility is that Shank3 is required at an early stage of synapse formation, and that IGF1 promotes maturation of synapses by triggering the loss of Shank3 and recruitment of PSD95. Consistent with this view, Shank3-containing synapses have slowly decaying NMDA receptor currents that resemble those of GluN2B receptors that occur early in development, whereas synapses that contain PSD95 but lack Shank3 have rapidly decaying NMDA receptor currents that resemble the GluN2A-containing synapses that appear later in development28. In summary, we have identified synaptic defects in iPSC-derived neurons from patients with PMDS and we show that these defects depend on loss of Shank3 and can be reversed by IGF1.
Methods
Recruitment of patients with PMDS and skin biopsy procedure
Participants diagnosed with 22q13 deletion syndrome were recruited from ongoing research studies, the medical genetics service at Lucile Packard Children’s Hospital, and interest groups for patients and families with genetic disorders associated with autistic spectrum manifestations. Consenting participants were evaluated with the Autism Diagnostic Observation Schedule (ADOS), and, when possible, the Stanford-Binet Intelligence Scales, Fifth Edition (SB5). In addition, parents of participants completed the Autism Diagnostic Interview-Revised (ADI-R) and questionnaires about the participant, including a medical history questionnaire and the Social Responsiveness Scale (SRS). All assessors were trained nationally and certified as reliable in the administration of ADOS and ADI-R assessments. Skin punch biopsies were performed as follows: after local anesthesia with topical and injected xylocaine, a shallow, 3mm punch biopsy sample was obtained from either the forearm or thigh.
This study was approved by both the Stanford University Institutional Review Board (IRB) and the Stem Cell Research Oversight Committee (SCRO).
Generation and characterization of iPSC lines
Detailed procedures used for the generation, maintenance, and characterization of iPSCs were previously described12. Briefly, primary fibroblasts from patients with PMDS were acquired by skin-punch biopsy following approved SCRO and IRB protocols. Cells were propagated for 2–3 passages, plated at 105 cells/well in 6-well plates, and infected with retroviruses carrying SOX2, OCT3/4, MYC, and KLF4 transcription factors the next day. iPSCs colonies, normally detected 21–30 days after the first infection, were routinely maintained on irradiated CF1 feeder cells in iPSC medium (DMEM/F12 (11330-032, Gibco), containing 20% Knockout™ SR (10828-028, Gibco), 1% MEM NEAA (11140, Gibco), 0.5% L-glutamine (25030, Gibco), 1% penicllin/streptomycin (15140, Gibco), 0.1 mM ″-mercaptoethanol (Sigma), and 10 ng/ml bFGF (R&D Systems)).
iPSCs were immunostained with antibodies against Nanog (AF1997, R&D Systems) and TRA-2-49/6E (developed by P. W. Andrews and obtained from the Development Studies Hybridoma Bank developed under the auspices of the NICHD and maintained in the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242).
SKY analysis of all iPSC lines was performed according to a standard procedure. Briefly, metaphases were extracted after O/N incubation in 0.03 μg/ml KaryoMAX Colcemid solution in iPSC media. Chromosomes were isolated and stained with the SKYPaint™ probe mixture and CAD kit (Applied Spectral Imaging), following the manufacturer’s protocol. Spreads were imaged on a Leica microscope (DM400B), equipped with SpectraCube™SD300 and a 63x objective, using Case Data Manager 6.0 and HiSKY 6.0 software (Applied Spectral Imaging, Inc., Vista, CA, USA). Twenty spreads from each iPSC line with well-separated chromosomes were imaged and analyzed for visible chromosome abnormalities.
For the teratoma formation assay, iPSCs from a confluent 10 cm dish were enzymatically harvested, washed, and resuspended in 1.5 ml of PBS. Cells were then injected into the kidney capsules of SCID mice (Charles River Laboratories International, Inc.). Visible tumors were dissected 3–8 weeks post-transplantation and fixed in 4% PFA. Fixed samples were sent to the Cureline Biopathology Laboratory (San Francisco, Bay Area) for paraffin embedding, sectioning, and staining with hematoxylin and eosin. Sections were then examined for the presence of tissue representatives of all three germ layers.
Genomic DNA from fibroblasts and iPSCs was isolated using the DNeasy Blood & Tissue kit (Qiagen). Haploinsufficiency of SHANK3 in fibroblasts and all iPSCs of patients, was verified using the SALSA MLPA kit (P188, MRC-Holland), following the manufacturer’s protocol.
Neural differentiation
For neural differentiation, a combination of two protocols was used29,30. The rationale and detailed procedures are presented in Supplementary Fig. 3.
qRT-PCR
qRT-PCR was performed in a real-time thermal cycler (Mastercycler ep realplex, Eppendorf) using either FastStart Universal SYBR Green (Rox) (Roche) or RT qPCR master mixes (SABiosciences). Total RNA was isolated using the RNeasy Mini kit (Qiagen). cDNA was synthesized with either the SuperScript III First-Strand Synthesis kit (Invitrogen) or the RT First Strand kit (SABiosciences). All primers are listed in Supplementary Table 5.
Multiplex single cell qRT-PCR
Single cells were harvested either by aspiration with a patch glass pipette filled with 4 μl of sterile intracellular solution or by FACS sorting using a BD influx sorter (BD Biosciences) into 10μl of a pre-amplification mix containing 40nM of all primers for the genes of interest and the following components of the CellsDirect One-Step qRT–PCR kit (Invitrogen): 2× Reaction Mix, SuperScript III RT/Platinum Taq Mix. After collection, samples were reverse transcribed and pre-amplified for 18 cycles. Pre-amplified samples were diluted (3×) with TE buffer and stored at −20°C. Sample and assay (primer pairs) preparation for 96.96 Fluidigm Dynamic arrays was done according to the manufacturer’s recommendation. Briefly, each sample was mixed with 20× DNA binding dye sample loading reagent (Fluidigm), 20× EvaGreen (Biotium) and TaqMan Gene Expression master mix (Applied Biosystems). Assays were mixed with 2× assay loading reagent (Fluidigm) and TE to a final concentration of 5μM. The 96.96 Fluidigm Dynamic Arrays (Fluidigm) were primed and loaded on an IFC Controller HX (Fluidigm) and qPCR experiments were run on a Biomark System for Genetic Analysis (Fluidigm). Melting curves were used to determine the specificity of each reaction. In addition to single-cell material, every experiment contained four standard dilutions of a mixed human cDNA library. Data were collected and analyzed using Fluidigm Real-Time PCR Analysis software (v.2.1.3 and v.3.0.2). Threshold cycle (CT) as a measure for original template amount was used for quantification. To compare the data acquired from different chips, the CT values were normalized to the median intensity of the entire chip. Because single cell gene expression in mammals occurs in bursts and follows a binary distribution, we established 32 cycles (at least five standard deviations from the mean of the lowest expressed genes) as a general threshold for whether or not a gene is expressed.
Electrophysiology
All recordings were performed at room temperature (22–25°C) on EGFP- or mKate2-expressing iPSC-derived neurons on Day 31–45 (separate cultures without astrocytes) or Day 45–52 (co-cultured on rat cortical astrocytes). Briefly, neurons grown on cover slips or 35 mm plastic dishes were visualized with a 40X air objective on an inverted Nikon microscope (Ellipse TE2000) and recorded using an EPC10 amplifier (HEKA). The following solutions were used (in mM): extracellular, 140 NaCl, 2.5 KCl, 2.5 CaCl2, 2 MgCl2, 1 NaH2PO4, 20 Glucose, 10 HEPES, pH 7.4; intracellular, 135 CsMeS, 5 CsCl, 10 HEPES, 0.5 EGTA, 1 MgCl2, 4 Mg2ATP, 0.4 NaGTP, 5 QX-314, pH 7.4 with CsOH (for EPSCs, AMPA-, NMDA-, and GABA-current measurements) and 125 CsCl, 10 EGTA, 1 MgCl2, 0.1 CaCl2, 4 MgATP, 0.3 NaGTP, 10 HEPES, 5 QX-314 pH 7.2 with CsOH (for IPSCs measurements). Recording pipettes made of borosilicate glass (BF150-110-10, Sutter Instruments) had a resistance of 3–6 MΩ when filled with intracellular solution. The following combinations of antagonists were present in the extracellular solutions to isolate different postsynaptic currents (in μM): 20 bicuculline or 50 picrotoxin (spontaneous and evoked EPSCs), 10 NBQX and 50 APV (spontaneous and evoked IPSCs), 10 NBQX and 20 bicuculline or 50 picrotoxin (NMDA-current, focal application), 50 APV and 50 picrotoxin (AMPA-current, focal application), 10 NBQX and 50 APV (GABA-current, focal application). Spontaneous EPSCs and IPSCs were recorded for 3 minutes (1 min after break-in to block sodium current by QX-314) at holding potential −70 mV. Recordings were filtered at 2 kHz and digitized at 4 kHz. Evoked postsynaptic currents were measured after spontaneous currents and induced by brief (1 ms) unipolar current pulses of various amplitudes (0.1–0.9 mA, Δ = 0.05–0.1 mA), delivered with a stimulating electrode (CBAEC75, FHC) positioned 100–150 μm from the cell soma and connected to an external stimulator (A365, WPI). Recordings were filtered at 2 kHz and digitized at 10 kHz. NMDA-, AMPA-, and GABA-currents were induced by a focal transient application of the following agonists (in μM): 100 NMDA and 10 Gly, 200 AMPA, or 100 GABA. An injection pipette (patch pipette with resistance of 1–2 MΩ when filled with extracellular solution) was positioned about 20 μm from the cell soma and connected to a pico-spritzer (PDES-02DX, npi Electronics) with 10 psi in-line pressure. In all experiments, access resistance was monitored throughout the experiment (~15 MΩ), but not compensated. Cells were included for analysis only if the change of resistance was < 25% throughout the experiment.
Data were collected and initially analyzed with Patchmaster software (HEKA). Further analysis was performed using Clampfit 10 (Molecular Devices), IgorPRO (Wavemetrics), Excel (Microsoft), and Prism (GraphPad). The baseline noise amplitude in our experiments was about 2.5 pA. Spontaneous EPSCs were detected using a template search procedure in Clampfit 10. A template for automatic EPSC detection was created by averaging 50 manually picked synaptic events. Spontaneous IPSCs were detected using MiniAnalysis software. The decay kinetics of NMDA-EPSCs was estimated using a weighted decay time constant (τdecay)31, calculated from the area under the normalized current on an interval from 16.4 ms (3XτAMPA decay) to 1.1 s after the peak of AMPA-EPSC.
Western blot analysis
Cells grown in 2 cm dishes were lysed in a standard RIPA buffer containing a protease inhibitor cocktail (Roche Diagnostics). Protein concentrations were measured using a BCA protein assay kit (Thermo). Proteins were denatured with Laemmli buffer at 70°C for 10 min, separated on a NuPAGE 4–12% Bis-Tris gel (Novex), and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked in 5% Milk (1h at RT) and blotted with the following primary antibodies (O/N at 4°C): guinea pig anti-Shank3 (1:700, gift from Carlo Sala), mouse anti-GluN1 (1:1000, SYSY), rabbit anti-GluA1 (1:1000, Upstate), mouse anti-GluA2 (1:1000, NeuroMab), and mouse anti-GAPDH (MS, 1:1000, Ambion). To control loading protein amounts, we first quantified the relative concentrations of GAPDH in all lysates. Afterwards, 15–25 μg of proteins were loaded into a gel and intensities of appropriate bands were measured and normalized to the intensity of the GAPDH band in the same lane. Bands were visualized and quantified using the FluorChemQ System (Alpha Innotech).
Immunocytochemistry, imaging, and synaptic assays
Cells were fixed in 4% paraformaldehyde (15 min. at RT), washed (3x) with PBS glycine, permeabilized with 0.5% Nonidet P40 (7 min. at RT), washed (3x) with PBS, and blocked in 3% BSA (O/N at 4°C). Primary antibodies were diluted in 3% BSA and applied O/N at 4°C. The following primary antibodies were used: chicken anti-GFP (Abcam, 1:1000), rabbit anti-tRFP (Axxora, 1:1000), guinea pig anti-Shank3 (produced by TM Boeckers, 1:800), mouse anti-Synapsin1 (SYSY, 1:500), guinea pig anti-Homer1 (SYSY, 1:200), mouse anti-PSD95 (Abcam, 1:200), rat anti-Ctip2 (Abcam, 1:200), mouse anti-Satb2 (Abcam, 1:10), mouse anti-GAD67, and anti-Nuclei (Millipore, 1:100). Secondary antibodies were diluted (1:500–1000) in 3% BSA and applied for 1 hour at RT. The following secondary antibodies were used: Alexa-405, -488, -594, and -647 (Invitrogen). Neurons were visualized using either a Zeiss spinning disc confocal microscope, equipped with 405/488/594/640 lasers (Perkin Elmer) or a laser-scanning confocal microscope equipped with 405/488/594/633 lasers (Stanford Neuroscience Microscopy Services), using either Volocity (Improvision) or Zenn (Zeiss) software. Z-stack images of fluorescent cells were taken with a 0.2–0.7 μm interval using 100x or 63x objectives that cover an 80×80 or 150×150 μm2 area, correspondingly. Images within an experiment were acquired with identical acquisition settings. Approximately 20 images (10 GFP+ and 10 mKate2+ neurons) were collected from each coverslip. Visualization and analysis of all images were performed using Volocity 5.1 and ImageJ. For quantification of total Shank3 expression, a 3D mask of a neuron was created based on GFP or mKate2 fluorescence and the mean Shank3 intensity within the masked area was calculated. For quantification of the number of synapses, Z-stacks were merged using maximum intensity readings for each of the channels and a 2D mask of a neuron was created based on the GFP or mKate2 fluorescence reading. Presynaptic (Synapsin1) and postsynaptic (Shank3, Homer1, or PSD-95) puncta within a mask were found either manually using a cell counter plug-in for ImageJ or automatically using a custom measurement protocol created in Volocity. During automatic quantification, once settings for puncta detection were chosen, they were uniformly applied for all images within an experiment. Results of automatic puncta quantification were always validated by visual inspection and manual counting for a subset of the images. All cover slips were coded and processed in a blinded manner. In order to combine data acquired from different coverslips in different experiments, we normalized all puncta counts to the median number of puncta detected on control neurons for a particular cover slip.
Construction of Shank3 lentiviral vector
The translated region of the rat Shank3 cDNA (NM_021676.1, generously provided by Dr. Ramnik Xavier) was isolated in pieces by PCR and flanked with NdeI (5′) and ClaI (3′) restriction enzyme sites, and then subcloned into the pSC-B-amp/kan vector using the StrataClone Blunt PCR Cloning kit (Stratagene). Similarly, a DNA fragment of EGFP flanked with NotI (3′ UTR), NdeI (5′ TR before stop codon), and ClaI (3′ UTR) restriction enzyme sites was obtained by PCR and subcloned into a pSC-B-amp/kan vector. Afterwards, NotI-EGFP-ClaI (0.7 kb) and NdeI-Shank3-ClaI (5.2 kb) fragments were isolated, gel-purified, and sequentially inserted into the NotI and ClaI sites and then into an NdeI- and ClaI-digested pHAGE-EF1a lentiviral vector backbone32 to generate the pHAGE-EF1a-EGFP-Shank3 (13 kb) vector. The sequence was confirmed by sequencing.
Lentivirus production, titering, and infection
Lentiviruses were generated with a five-plasmid transfection system using Fugene 6 transfection reagent (Roche) in 293T cells as previously described32. 48 hours after transfection, supernatants were collected at 12-hour intervals, 4–5 times in total. Viral particles were concentrated using Lenti-X concentrator (Clontech), aliquoted (200 μl), and stored at −80°C.
Different amounts of virus (5–100 μl) were placed on ~80% confluent HEK293T cells for 3–4 days. Approximately 80% of the cells in the 35 mm plate were fluorescent upon addition of 100 μl of virus. We used 100 μl of GFP-Shank3 virus to transduce human iPSC-derived neurons (Day 25–30) growing in 35-mm culture plates. The medium was replaced 10–20 hours later. Neurons were transferred onto rat primary astrocytes after 5–10 days.
Drugs and treatment procedure
The following agents were tested: 0.1 μM Trichostatin A (TSA, Tocris), 1 mM valproic acid (VPA, Calbiochem), 3 μM nifedipine (Nif, Tocris), 20 ng/ml insulin growth factor 1 (IGF1, R&D Systems), and 20 ng/ml insulin growth factor 2 (IGF2, R&D Systems). Concentrations were chosen based on previous reports of their use available in the literature. Effects of TSA, VPA, and Nif were assessed after 24–36 hours of treatment and IGF1 and IGF2 after 2–3 weeks, with approximately 25% of the media changed every other day.
Data collection and statistics
All experiments were replicated at least three times using iPSC lines indicated in Supplementary Table 3. The sample size and description of the sample collection are reported in each figure legend. The number of samples was selected based on the variability of the particular assay. Samples were excluded from analysis only when they were clear outliers (identified visually and confirmed with the Grubbs’ test) or were erroneously generated as a result of a technical problem. Wells with neurons were randomly selected for an assay/drug treatment, coded, and processed in a blinded manner. Statistical tests used for comparison are reported in each figure legends. For data sets where parametric tests were used for analysis, the normal distribution of data was confirmed using the Kolmogorov-Smirnov test.
Supplementary Material
Acknowledgments
We are grateful to participants and their families for their support; M. Adam for assistance with recruitment; to X. Jia, A. Cherry, C. Bangs, P. Jones, and J. Williams for assistance with tissue culture; P. Liao for help with MLPA; M. Fabian for astrocyte preparations; H.N. Nguyen for consultations on the neural differentiation protocol and SKY; V. Vu, and G. Lin for help with data analysis; T. Sudhof, T. Boeckers, A. Grabruker, C. Garner, and C. Sala for antibodies; R. Xavier for Shank3 cDNA; R. Reijo-Pera and members of the Dolmetsch lab for commenting on the manuscript; E. Nigh for editing the manuscript. We also thank the Stanford Neuroscience Microscopy Service (supported by NIH NS069375). Support for this study came from CIRM, Autism Science Foundation and Phelan-McDermid Syndrome Foundation (AS), Swiss National Science Foundation (TP), Japan Society for the Promotion of Research Abroad and American Heart Association (MY), NIMH grant R33MH087898 (JFH), NIH Pioneer Award (5DP1OD3889), CIRM (RT2-01906), and Simons Foundation (RED). We are also grateful for funding from JDH research fund, N. Juaw, B. and F. Horowitz, M. McCafferey, B. and J. Packard, P. Kwan and K. Wang, and the Flora foundation.
Footnotes
Contributions: AS and RED designed experiments and wrote the manuscript; AS performed iPSC maintenance, neural differentiation, electrophysiology, cloning, and immunocytochemistry; OS maintained and characterized iPSCs, performed WB and qRT-PCR; MY generated and characterized iPSCs; TP performed multiplex single cell qRT-PCR; VS performed teratoma assay; RS and AK performed qRT-PCR and data analysis; WF, JAB, and JFH recruited and characterized patients and performed the MLPA assay.
References
- 1.Phelan K, McDermid HE. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. 000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- 3.Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 4.Boccuto L, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gauthier J, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 7.Moessner R, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamdan FF, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier J, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Yazawa M, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nature medicine. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittgen T, et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc Natl Acad Sci U S A. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akhtar MW, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29:8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinaldi T, Kulangara K, Antoniello K, Markram H. Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proc Natl Acad Sci U S A. 2007;104:13501–13506. doi: 10.1073/pnas.0704391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Kusky JR, Ye P, D’Ercole AJ. Insulin-like growth factor-I promotes neurogenesis and synaptogenesis in the hippocampal dentate gyrus during postnatal development. J Neurosci. 2000;20:8435–8442. doi: 10.1523/JNEUROSCI.20-22-08435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tropea D, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen DY, et al. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozdagi O, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 29.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaspard N, et al. Generation of cortical neurons from mouse embryonic stem cells. Nat Protoc. 2009;4:1454–1463. doi: 10.1038/nprot.2009.157. [DOI] [PubMed] [Google Scholar]
- 31.Bellone C, Nicoll RA. Rapid bidirectional switching of synaptic NMDA receptors. Neuron. 2007;55:779–785. doi: 10.1016/j.neuron.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 32.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.