Abstract
To better understand Simian betaretrovirus (SRV) seropositivity in virus negative macaques, we transfused blood from SRV infected or suspect donors into immunosuppressed naive recipients. Our results do not support typical SRV1-5 infection as the cause, but provide evidence for several possibilities including serological artifact, new/different SRV, or an endogenous virus.
Keywords: transfusion, serology, PCR
Introduction
Simian betaretrovirus (SRV) has been a model of immunodeficiency and a target of exclusion in colony management of macaques since the 1980’s [1–3]. With the implementation of improved diagnostic testing and management practices, the prevalence of naturally occurring SRV in captive colonies has declined dramatically [4, 5]. However, despite best practices, small but growing numbers of seropositive, virus-negative animals with no plausible history of exposure have been confirmed by multiple laboratories using various methods including antibody assays with viral lysate and recombinant protein targets on platforms including enzyme immunoassays, microbead arrays, immunofluorescence and Western Blot. Virus detection assays have included PCR and virus isolation in multiple cell lines [6]. While some results could perhaps be attributed to assay artifact, samples with reproducible antibody in the absence of any virus detection have been identified by all assays. These observations raise questions which our transfusion studies begin to address: Have current diagnostic methods become so exquisitely sensitive and the virus so rare that we are detecting noise? Is the host making an immune response to endogenous virus? Is this a new serotype? Have selection pressures changed the characteristics of the virus and/or its host?
Materials and Methods
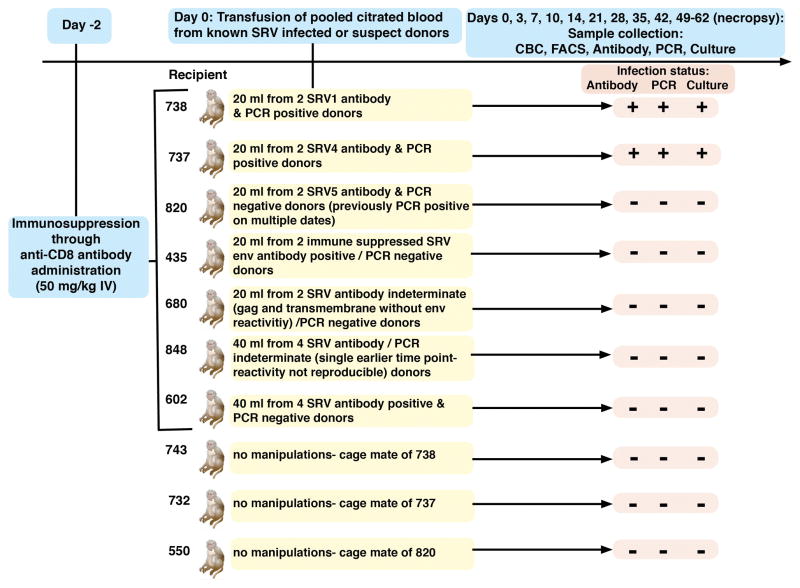
All animals were maintained in fully AAALAC-accredited facilities in accordance with the Animal Welfare Act, Regulations, and the Guide for the Care and Use of Laboratory Animals [7–9]. All procedures involving animals in this study were approved by each institution’s IACUC. As detailed in figure 1, citrated whole blood was collected from known SRV infected or SRV suspect (incomplete or full antibody reactivity patterns without PCR confirmed virus detection) pigtailed macaques and transfused into seven naive, CD8+ cell-depleted adult rhesus macaques [10]. Depletion was accomplished by the intravenous administration of monoclonal anti-CD8alpha rhesus recombinant M-T807R1 (NIH Nonhuman Primate Reagent Resource) at 50 mg/kg 48 hours prior to transfusion, to reduce CD8+ cell-mediated immune responses and increase the chances of inducing infection [11]. Animal health was monitored daily by trained animal care and veterinary staff. Blood samples for complete blood count (CBC), CD3/4/8/20 FACS analysis, SRV antibody, SRV PCR, and culture were collected at days 0, 3, 7, 10, 14, 21, 28, 35, 42 and necropsy (days 49–56). Each animal receiving known SRV positive blood was pair housed with a cage mate that was not subjected to any study manipulations. The four receiving SRV suspect blood were paired together. CBCs were performed on EDTA-anticoagulated blood using a Pentra 60C+ analyzer (ABX Diagnostics) with differentials determined manually. Lymphocyte subpopulations were analyzed by four-color flow cytometry using a FACSCalibur flow cytometer. Multiplex microbead (Charles River Laboratories, Wilmington, MA) and Western blot immunoassays were employed for antibody [13]. The microbead assays used recombinant and viral lysate antigens with biotinylated goat anti-human IgG and streptavidin peroxidase for detection on the Luminex platform. For immunoblots, SRV1 and SRV2 viral lysate were electrophoresed through a 4–12% gradient gel and transblotted onto PVDF membrane. Peroxidase conjugated goat anti-monkey IgG and 4-chloro-1-naphthol were used for detection. Real time PCR was performed using primers validated to detect SRV1-5 serotypes with a sensitivity of one to ten copies [14]. PBMCs were isolated, stimulated with SEA, and co-cultured on Raji cells in fetal calf sera supplemented RPMI media for six weeks [15, 16]. Cultures were observed twice weekly for CPE and supernatant was collected for PCR.
Figure 1.
Results
As shown in Table 1, all animals were efficiently CD8+ cell depleted for ≥ 3 weeks. SRV viral DNA was detected directly from PBMCs and in Raji cultures beginning two weeks post transfusion in the monkeys that received either SRV1 or SRV4 virus and antibody positive pooled blood from known infected donors. Rising antibody levels were detected in those same monkeys by four weeks post transfusion. No virus or seroconversion was detected in the monkey receiving antibody positive/virus negative blood from the known SRV5 infected donors. Virus or seroconversion was also not detected in the monkeys receiving blood from SRV suspect antibody indeterminate or positive but virus negative donors. No recipients exhibited clinical or hematological signs of disease during the course of this study. No pathology indicative of SRV infection was seen at necropsy. Furthermore, none of the newly SRV infected macaques transmitted infection to their naïve cage mates. However, subsequent transfusion from these known SRV recipients into another generation of naïve macaques did result in the expected new infections with SRV1 and 4 but not SRV5.
Table 1.
| Day −2 | Day 0 | Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Necropsy | |
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| MMU 738: received known SRV1 (antibody & PCR positive) transfusion) | |||||||||
|
|
|||||||||
| Antibody (WB bands) | neg | POS | POS | POS | POS | POS | POS | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | POS (37) | POS (31.6) | POS (26.5) | POS (27.5) | POS (25.5) | POS (24.8) | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | POS | POS | POS | POS | POS | POS | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 4.91/33.3/8.7/15 | 5.23/36.0/6.5/17 | 5.32/36.3/7.6/20 | 4.87/33.3/6.5/21 | 5.34/36.5/9.6/16 | 5.15/34.2/5.3/27 | 4.67/30.9/7.3/53 | 5.05/33.1/9.0/15 | |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 366/322/447/154 | 510/8/499/9 | 839/4/586/3 | 682/4/560/1 | 880/32/486/14 | 525/161/525/111 | 885/722/828/1471 | 342/194/276/575 | |
|
|
|||||||||
| MMU 737: received known SRV4 (antibody & PCR positive) transfusion | |||||||||
|
|
|||||||||
| Antibody (WB confirmed) | neg | none | POS | POS | POS | POS | POS | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | Ind (48) | POS (38.3) | POS (28.8) | POS (30.1) | POS (30.0) | POS (30.6) | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | POS | POS | POS | POS | POS | POS | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 4.63/34.9/3.6/46 | 5.22/39.8/5.3/19 | 5.12/38.5/4.7/29 | 4.46/33.3/3.2/41 | 4.85/36.3/2.8/44 | 5.12/37.9/3.2/53 | 4.73/34.7/4.3/44 | 4.53/32.9/3.8/50 | |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 582/417/346/291 | 521/4/371/2 | 941/7/315/1 | 795/2/351/1 | 819/2/280/1 | 1111/2/426/1 | 1075/8/560/2 | 828/204/522/369 | |
|
|
|||||||||
| MMU 820: received known SRV5 (antibody positive/PCR negative) transfusion | |||||||||
|
|
|||||||||
| Antibody (WB bands) | neg | neg | neg | neg | neg | neg | neg | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 5.03/35.2/4.09/35 | 5.46/38.5/8.9/14 | 5.25/36.7/6.0/24 | 5.08/35.5/3.7/44 | 5.53/38.6/4.6/35 | 5.25/36.7/3.4/38 | 5.30/37.0/4.0/31 | 5.32/37.2/4.8/58 | 4.90/34.0/4.2/57 |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 88/416/325/169 | 726/9/388/2 | 1075/5/260/1 | 1205/9/268/0 | 1252/5/208/0 | 880/6/259/2 | 819/3/262/1 | 1793/12/604/5 | |
|
|
|||||||||
| MMU 435: received Suspect SRV (env Ab positive/PCR negative) transfusio | |||||||||
|
|
|||||||||
| Antibody (WB confirmed) | neg | neg | neg | neg | neg | neg | neg | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 5.35/39.1/4.7/38 | 5.87/42.8/5.1/31 | 5.21/37.7/4.0/52 | 4.78/34.2/4.7/57 | 5.17/37.1/5.6/59 | 5.43/38.6/5.8/40 | 5.20/37.1/4.5/62 | 5.31/38.1/5.8/43 | |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 838/2389/392/213 | 986/9/465/3 | 1403/7/534/2 | 1705/8/639/0 | 1877/5/1143/2 | 1123/14/854/8 | 1556/13/934/3 | 1395/202/803/77 | |
|
|
|||||||||
| MMU 680: received Suspect (SRV gag and transmembrane Ab positive/PCR negative) transfusion | |||||||||
|
|
|||||||||
| Antibody (WB bands) | neg | neg | neg | neg | neg | neg | neg | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 4.70/34.9/5.9/19 | 5.36/40.0/5.9/17 | 4.93/36.5/6.4/28 | 4.53/33.5/5.8/33 | 4.81/35.6/5.2/43 | 5.02/37.2/7.4/25 | 4.83/35.5/6.4/48 | 4.59/34.1/7.8/38 | 4.56/33.5/6.4/36 |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 502/164/296/134 | 524/3/361/2 | 965/7/706/3 | 1054/6/604/0 | 1243/2/657/2 | 889/5/546/3 | 1591/365/500/462 | 1259/293/644/662 | |
|
|
|||||||||
| MMU 848: received (Suspect SRV Ab positive/PCR*) transfusion *single unconfirmed, non-reproducible PCR signal at an earlier date | |||||||||
|
|
|||||||||
| Antibody (WB bands) | neg | neg | neg | neg | neg | neg | neg | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 5.37/39.4/9.7/46 | 5.24/38.1/4.4/50 | 4.82/35.0/7.9/28 | 4.67/34.0/5.8/48 | 5.25/38.2/8.6/43 | 5.60/40.6/8.2/43 | 5.19/37.2/6.2/46 | 4.80/34.9/7.4/34 | 4.58/33.0/6.3/66 |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 1414/616/1759/602 | 861/10/1161/3 | 1054/6/1024/6 | 1160/6/1353/3 | 1737/9/1389/2 | 1610/8/1356/10 | 1390/155/975/212 | 1191/162/971/157 | |
|
|
|||||||||
| MMU 602: received Suspect (SRV Ab positive/PCR negative) transfusion | |||||||||
|
|
|||||||||
| Antibody (WB bands) | neg | neg | neg | neg | neg | neg | neg | ||
|
|
|||||||||
| PCR (Ct <55 = POS) | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Raji Cell Culture | neg | neg | neg | neg | neg | neg | neg | neg | |
|
|
|||||||||
| Health Report | normal | normal | normal | normal | normal | normal | normal | normal | |
|
|
|||||||||
| CBC: RBC#/Hct%/WBC#/Lymph % | 5.19/38.4/5.8/49 | 5.31/39.35/7.4/30 | 5.08/37.3/8.6/23 | 4.61/33.7/5.4/51 | 5.16/37.4/7.1/41 | 5.34/38.9/8.9/26 | 5.09/37.2/8.6/25 | 4.82/35.5/7.2/33 | |
|
|
|||||||||
| FACS: 3+4+/3+8+/3–20+/3–8+(NK) | 807/491/1339/140 | 609/11/1445/12 | 700/7/1159/3 | 846/7/1631/2 | 696/5/2018/5 | 535/7/1595/7 | 496/3/1483/6 | 676/3/1531/5 | |
|
|
|||||||||
Discussion
Although SRV1 and 4 infection were successfully transmitted from donor pools to recipient animals, the demonstrated lack of transmission to cage mates and lack of pathology (i.e. anemia, immunosuppression, wasting) contrasts with the historical descriptions of SRV [1, 2]. Could evolutionary pressures over time be selecting out a specific population of macaques or virus strains? The inability to confirm infection by either antibody or PCR in any recipients of SRV suspect blood supports the possibilities that these non-negative antibody/PCR negative profiles do not indicate conventional SRV1-5 infection but could perhaps represent a serological assay artifact; a different, low level, difficult to detect virus; or reactivity to an endogenous virus. With the current exquisitely sensitive diagnostic methods and low virus prevalence in many established colonies, statistical principles favor the greater probability of false as compared to true positives [6, 17, 18]. Recent publications have reported additional serotypes beyond the well-established SRV1-5 [19–23]. We (R. Grant, manuscript submitted) and others have demonstrated confounding detection of host immune responses to endogenous viruses in SRV serology [24, 25]. Although both SRV5 donors were antibody positive/PCR negative on the day of transfusion, they were infected as shown by not only antibody but also PCR positivity on five of ten and one of ten dates tested during the prior two months. Without a better understanding of the mechanisms and stresses triggering PCR positivity at any time point [26], transmission risks remain [26, 27]. Studies to further address and differentiate possible explanations to unravel the mystery of SRV seropositive/virus-negative and potential infection risk to other macaques are in progress.
Acknowledgments
We thank the animal care and laboratory staff at both the California and Washington National Primate Research Centers for providing excellent technical support for this study. This work was supported by NIH grants CNPRC P51 OD011107 and WANPRC P51OD010425.
References
- 1.Lerche NW, et al. Epidemiologic aspects of an outbreak of acquired immunodeficiency in rhesus monkeys (Macaca mulatta) Lab Anim Sci. 1984;34(2):146–50. [PubMed] [Google Scholar]
- 2.Lerche NW, et al. Natural history of endemic type D retrovirus infection and acquired immune deficiency syndrome in group-housed rhesus monkeys. J Natl Cancer Inst. 1987;79(4):847–54. [PubMed] [Google Scholar]
- 3.Lerche NW, Yee JL, Jennings MB. Establishing specific retrovirus-free breeding colonies of macaques: an approach to primary screening and surveillance. Lab Anim Sci. 1994;44(3):217–21. [PubMed] [Google Scholar]
- 4.Lerche NW, Marx PA, Gardner MB. Elimination of type D retrovirus infection from group-housed rhesus monkeys using serial testing and removal. Lab Anim Sci. 1991;41(2):123–7. [PubMed] [Google Scholar]
- 5.Schroder MA, Fisk SK, Lerche NW. Eradication of simian retrovirus type D from a colony of cynomolgus, rhesus, and stump-tailed macaques by using serial testing and removal. Contemp Top Lab Anim Sci. 2000;39(4):16–23. [PubMed] [Google Scholar]
- 6.Yee JL, et al. Specific pathogen free macaque colonies: a review of principles and recent advances for viral testing and colony management. J Med Primatol. 2016;45(2):55–78. doi: 10.1111/jmp.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. 8. Washington (DC): National Academies Press; 2011. [Google Scholar]
- 8.Animal Welfare Act as Amended. 2013. 7 USC §2131–2159.
- 9.Animal Welfare Regulations. 2013. 9 CFR § 3.129.
- 10.Jehuda-Cohen T, et al. Transmission of retroviral infection by transfusion of seronegative blood in nonhuman primates. J Infect Dis. 1991;163(6):1223–8. doi: 10.1093/infdis/163.6.1223. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz JE, et al. A nonhuman primate model for the selective elimination of CD8+ lymphocytes using a mouse-human chimeric monoclonal antibody. Am J Pathol. 1999;154(6):1923–32. doi: 10.1016/S0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serologic Methods Manual: Multiplexed Fluorometric ImmunoAssay (MFIA) Charles River Laboratories; Wilmington, MA: 2011. [Google Scholar]
- 13.Kuller L, et al. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn Microbiol Infect Dis. 2005;53(3):185–93. doi: 10.1016/j.diagmicrobio.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 14.White JA, et al. Development of a generic real-time PCR assay for simultaneous detection of proviral DNA of simian Betaretrovirus serotypes 1, 2, 3, 4 and 5 and secondary uniplex assays for specific serotype identification. J Virol Methods. 2009;162(1–2):148–54. doi: 10.1016/j.jviromet.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel MD, et al. A new type D retrovirus isolated from macaques with an immunodeficiency syndrome. Science. 1984;223(4636):602–5. doi: 10.1126/science.6695172. [DOI] [PubMed] [Google Scholar]
- 16.Lerche NW, et al. Evidence of infection with simian type D retrovirus in persons occupationally exposed to nonhuman primates. J Virol. 2001;75(4):1783–9. doi: 10.1128/JVI.75.4.1783-1789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons JH. Development, application, and quality control of serology assays used for diagnostic monitoring of laboratory nonhuman primates. ILAR J. 2008;49(2):157–69. doi: 10.1093/ilar.49.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstman BB. Basic biostatistics : statistics for public health practice. Sudbury, Mass: Jones and Bartlett Publishers; 2008. p. xvii.p. 557. [Google Scholar]
- 19.Fujiomto K, et al. Simian betaretrovirus infection in a colony of cynomolgus monkeys (Macaca fascicularis) Comp Med. 2010;60(1):51–3. [PMC free article] [PubMed] [Google Scholar]
- 20.Hara M, et al. Isolation and characterization of a new simian retrovirus type D subtype from monkeys at the Tsukuba Primate Center, Japan. Microbes Infect. 2005;7(1):126–31. doi: 10.1016/j.micinf.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Nandi JS, et al. New simian beta retroviruses from rhesus monkeys (Macaca mulatta) and langurs (Semnopithecus entellus) from Rajasthan, India. Virus Genes. 2006;33(1):107–16. doi: 10.1007/s11262-005-0032-x. [DOI] [PubMed] [Google Scholar]
- 22.Zao CL, et al. The complete genome and genetic characteristics of SRV-4 isolated from cynomolgus monkeys (Macaca fascicularis) Virology. 2010;405(2):390–6. doi: 10.1016/j.virol.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zao CL, et al. A novel simian retrovirus subtype discovered in cynomolgus monkeys (Macaca fascicularis) J Gen Virol. 2016;97(11):3017–3023. doi: 10.1099/jgv.0.000601. [DOI] [PubMed] [Google Scholar]
- 24.van der Kuyl AC, et al. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J Virol. 1997;71(5):3666–76. doi: 10.1128/jvi.71.5.3666-3676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HL, et al. Identification and spontaneous immune targeting of an endogenous retrovirus K envelope protein in the Indian rhesus macaque model of human disease. Retrovirology. 2016;13:6. doi: 10.1186/s12977-016-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zao CL, et al. Virological and serological characterization of SRV-4 infection in cynomolgus macaques. Arch Virol. 2011;156(11):2053–6. doi: 10.1007/s00705-011-1068-y. [DOI] [PubMed] [Google Scholar]
- 27.Lerche NW. Simian retroviruses: infection and disease--implications for immunotoxicology research in primates. J Immunotoxicol. 2010;7(2):93–101. doi: 10.3109/15476911003657406. [DOI] [PubMed] [Google Scholar]