Abstract
Different pyruvate kinase isoforms are expressed in a tissue-specific manner, with pyruvate kinase M2 (PKM2) suggested to be the predominant isoform in proliferating cells and cancer cells. Due to differential regulation of enzymatic activities, PKM2 but not PKM1 has been thought to favor cell proliferation. However, the role of PKM2 in tumorigenesis has been recently challenged. Here we report that increased glucose catabolism through glycolysis and increased pyruvate kinase activity in c-MYC-driven liver tumors are associated with increased expression of both PKM1 and PKM2 isoforms and decreased expression of the liver-specific isoform of pyruvate kinase, PKL. Depletion of PKM2 at the time of c-MYC over-expression in murine livers did not affect c-MYC induced tumorigenesis and resulted in liver tumor formation with decreased pyruvate kinase activity and decreased catabolism of glucose into alanine and the Krebs cycle. An increased PKM1/PKM2 ratio by ectopic PKM1 expression further decreased glucose flux into serine biosynthesis and increased flux into lactate and the Krebs cycle, resulting in reduced total levels of serine. However, these changes also did not affect c-MYC-induced liver tumor development. These results suggest that increased expression of PKM2 is not required to support c-MYC-induced tumorigenesis in the liver and that various PKM1/PKM2 ratios and pyruvate kinase activities can sustain glucose catabolism required for this process.
Keywords: Pyruvate kinases, PKM1, PKM2, cancer metabolism, hepatocellular carcinoma, glycolysis, Krebs cycle, stable isotopes
Introduction
Aberrant cellular metabolism is one of the hallmarks of cancer (1). Increased glucose transport and metabolism are among the most prominent metabolic abnormalities observed in multiple tumors and cancer models (2, 3). Metabolic changes, in part triggered by expression changes of genes encoding metabolic enzymes, are thought to ensure survival and proliferation of cancer cells by providing enough ATP and macromolecule precursors. Several enzymes or enzyme isoforms over-expressed in different tumor types have been shown to be required for tumorigenesis or are even able to initiate tumorigenesis (2, 3).
Pyruvate kinase catalyzes the final step in glycolysis by transferring the phosphate from phosphoenolpyruvate (PEP) to ADP, thereby generating pyruvate and ATP (4). Mammalian pyruvate kinases are encoded by two genes (PKLR and PKM), and each can generate two isoforms, respectively (PKL and PKR; PKM1 and PKM2). These isoforms have distinct kinetic properties and tissue distribution. Different tissue promoters drive the expression of PKL or PKR isoforms from the PKLR locus, thus generating two tissue specific mRNA species (5, 6). PKR is exclusively expressed in erythrocytes, whereas PKL is primarily expressed in liver, kidney and small intestine (5, 6). PKM1 and PKM2 are alternative splicing products of the PKM gene and expressed in various tissues (4). PKM1 is found in some differentiated adult tissues, such as heart, muscle and brain (7, 8). PKM2 is widely expressed and a predominant isoform in many adult cell types, including kidney tubular cells, intestinal epithelial cells, and lung epithelial cells (8, 9); and it is also considered to be the dominant isoform in proliferating cells, including embryonic and adult stem cells as well as cancer cells (7). In contrast to PKM1, which has constitutively high pyruvate kinase activity, PKM2 was shown to exist in either a less active or a more active form (10-12). The transition between these two PKM2 forms can be regulated allosterically by the upstream intermediate of glycolysis, fructose-1,6-bisphosphate (FBP) and by serine, as well as by interaction with tyrosine phosphorylated proteins and by oxidation (11, 13-16). The differential characteristics of the two PKM isoforms have been shown to have a direct impact on glycolysis and cell proliferation. Christofk and colleagues showed that in cancer cells whose endogenous PKM2 expression was suppressed by shRNA, the ectopic expression of PKM2 resulted in higher ability to generate xenograft tumors than ectopic expression of PKM1 (15). Moreover, ectopic expression of PKM1 without silencing PKM2 expression resulted in increased PK activity and decreased ability to form xenograft tumors (12). Consistent with this, PKM2 activators, which increase its pyruvate kinase activity by promoting the formation of the tetrameric form, have been shown to reduce the ability of cells to form xenograft tumors (12, 17). Finally, knockout of PKM2 in mouse embryonic fibroblasts resulted in compensatory expression of PKM1, diversion of glucose flux from nucleotide biosynthesis into lactate and the Krebs cycle and, eventually, proliferation arrest (18). All these observations led to the hypothesis that expression of PKM2 rather than PKM1 would provide proliferating cells with a differential control over the glycolytic flux. Decreased PKM2 activity as the result of allosteric regulation could permit the accumulation of intermediates upstream of phosphoenolpyruvate, increasing the flux into biosynthetic and antioxidative pathways while activation of PKM2 or high pyruvate kinase activity as the result of PKM1 expression can inhibit tumorigenesis (19). A non-enzymatic pro-tumorigenic role of PKM2 has also been suggested (18, 20-22). However, recent precise absolute quantifications of the PKM1 and PKM2 protein content challenged the exclusive association of proliferative state with PKM2 expression. Bluemlein and co-authors demonstrated the presence of both PKM1 and PKM2 in most of the tumor and normal adjacent tissue samples analyzed, including kidney, bladder, colon, lung and thyroid (9). Although expression of both PKM1 and PKM2 were increased in tumors in comparison with their normal tissue counterparts, PKM2 was a predominant form in both normal and tumor tissues in most of the cases (9). Dayton T.L. et al. also demonstrated expression of PKM2 in kidney tubular cells, intestinal and lung epithelial cell, pancreatic islet cells (8). Strikingly, tissue-specific knockout of PKM2 did not affect the formation of mammary gland tumors in BRCA1-/-;p53+/- mice, which have high expression of PKM2 and low expression of PKM1 (23). The resulting PKM2-null tumors had heterogenous PKM1 expression, which led the authors to conclude that PKM1 was either present in non-proliferating or was low in proliferating tumor cells. The authors suggested that PKM1 expression was kept low to maintain the low pyruvate kinase activity supporting cell proliferation (23). In another study, inducible knock-down of either PKM2 alone or both PKM1 and PKM2 in cancer cells with predominant expression of PKM2 inhibited proliferation of cells in culture, but neither affected xenograft tumor progression (24). The proposed glycolysis-independent role of PKM2 in tumorigenesis has recently also been challenged (25). Finally, germline loss of PKM2 can be compensated by PKM1 expression in embryonic tissues and PKM2-expressing adult tissues and does not affect normal development (8). Altogether, these results suggest that increased PKM2 expression may not be essential for cell proliferation and tumorigenesis in vivo and the precise role of different pyruvate kinase isoforms in supporting these processes in different in vivo settings remains to be elucidated.
Amplification of the c-Myc protooncogene has been found in 40%-60% of early hepatocellular carcinoma (HCC) samples (26), where it acts as a master regulator of cellular proliferation, apoptosis, and metabolism (27). Our previous studies demonstrated that liver tumors induced by liver-specific over-expression of c-MYC have increased catabolism of glucose through glycolysis in comparison with normal liver tissue (28). Using the same mouse model, it was shown that c-MYC-induced liver tumors exhibit increased expression of the Pkm gene (29). Here we demonstrate that c-MYC induced liver tumors have significantly increased expression of both, PKM1 and PKM2, compared with normal liver tissue. Interestingly, significantly decreasing PKM2 expression in tumors results in the diminished pyruvate kinase activity to the level comparable to normal liver. However, this decrease does not reduce glucose flux proportionally and does not affect tumor formation and progression. Moreover, ectopic PKM1 overexpression in the context of PKM2 inhibition, which diverts glucose flux from serine biosynthesis into lactate and the Krebs cycle, does not affect tumorigenesis either. Our results suggest that increased PKM2 expression is not required for c-MYC induced tumorigenesis in the liver and that this process can be sustained by various pyruvate kinase activities and PKM1/PKM2 ratios.
Materials and Methods
Constructs and reagents
The constructs for mouse injection, including pT3-EF1α-c-MYC and pCMV/SB (which encodes for sleeping beauty transposase or SB), were previously described (30). Mcl1 and PKM1 plasmids were obtained from Addgene (Addgene Catalog number 25714 for Mcl1 (with HA tag) and 34607 for PKM1 (with V5 tag), and cloned into pT3-EF1α vector via the Gateway cloning strategy (Invitrogen, Carlsbad, CA). miR30 based shRNA targeting for PKM1/2 3’UTR as well as a previously published shRNA against Renilla Luciferase (31) were cloned into pT3-EF1α-c-MYC via the Gateway polymerase chain reaction (PCR) cloning strategy to generate pT3-EF1α-c-Myc-shPKM and pT3-EF1α-c-Myc-shLuc plasmids, respectively (Figure S1). Plasmids were purified using the Endotoxin-free Maxiprep kit (Sigma-Aldrich, St. Louis, MO). 13C6-glucose was purchased from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA).
Hydrodynamic injection and mouse monitoring
Wild-type FVB/N mice were obtained from Jackson Laboratory (Bar Harbor, ME). Hydrodynamic injection was performed as described (32). In brief, 10μg of pT3-EF1α-c-MYC-shPKM or pT3-EF1α-c-MYC-shLuc with 5μg of pT3-EF1α-Mcl1 along with sleeping beauty transposase in a ratio of 25:1 were diluted in 2 mL of saline (0.9% NaCl), filtered through 0.22 μm filter, and injected into the lateral tail vein of 6 to 8-week-old FVB/N mice in 5 to 7 seconds. Mice were housed, fed, and monitored in accordance with protocols approved by the Committee for Animal Research at the University of California, San Francisco.
Western blotting
Mouse liver tissues were processed as previously reported (33). Protein lysates were fractionated by SDS PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% non-fat dry milk for 1 h and incubated with primary antibodies against PKM1 (1:1000; Cell Signaling Technology), PKM2 (1:1000; Cell Signaling Technology), PKM1/2 (1:1000; Cell Signaling Technology), PKLR (1:1000, Abcam), c-MYC (1:1000; Cell Signaling Technology), PCNA (1:1000; Cell Signaling Technology), PLK1 (1:1000; Cell Signaling Technology), HK1 (1:1000; Cell Signaling Technology), HK2 (1:1000; Cell Signaling Technology), HK4 (1:1000; Santa Cruz Biotechnology, Inc.), PFKM (1:1000; GeneTex, Inc.), PFKL (1:1000; Proteintech Group, Inc.), PFKP (1:1000; Gene Tex, Inc.), GLS1 (1:1000; Abcam), β-Actin (1:4000; Sigma-Aldrich), overnight, and then incubated with secondary antibodies for 1 h. Bands were visualized using an enhanced chemiluminescence system (ECL, Amersham), according to the manufacturer’s instructions. β-Actin and/or GAPDH were used as loading control.
Immunohistochemical staining
Liver specimens were fixed in 4% paraformaldehyde and embedded in paraffin. Immunohistochemistry (IHC) was performed as previously described (34). The primary antibodies include: anti-c-MYC (1:200; Abcam), anti-PKM2 (1:800; Cell Signaling Technology), anti-V5 (1:5000; Invitrogen), anti-Ki67 (1:150; ThermoFisher), and anti-PKLR (1:500; Abcam).
Pyruvate Kinase assay
PK activity of liver and tumor tissues was measured using the Pyruvate Kinase Activity Assay Kit (Sigma-Aldrich), according to the manufacturer’s protocol. Protein concentration was quantified using BAC protein assay kit (Thermo Scientific). Pyruvate Kinase activity for each sample was normalized to the final protein concentration.
Assessment of proliferation
Proliferation index was calculated by counting malignant hepatocytes stained positive for Ki67 (Bethyl Laboratories) in at least 3000 cells per each tumor investigated.
13C-glucose experiment
Evaluation of glucose catabolism using 13C-glucose bolus injection has been described previously (28, 35). Briefly, 20mg of 13C6-glucose dissolved in 100μl saline was injected into the tail vein of each mouse. Fifteen minutes after the injection, mice were euthanized; the plasma and liver tissues were collected immediately and snap frozen in liquid nitrogen for subsequent metabolic analysis.
Stable isotope labeling and metabolite extraction from tissues
Freeze clamped tissues were ground in liquid nitrogen with mortar and pestle and subsequently lyophilized on a FreeZone Freeze Dry System (Labconco). Metabolite extraction in mice tissues and derivatization of polar samples were performed essentially as previously described (36). Briefly, 5 mg of dry weight per sample were extracted with 1.8 ml of chloroform:methanol (2:1 v/v) containing internal standards (10 nmol scylloinositol, 10 nmol L-Norleucine and 160 nmols of DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid)) for 1 hour at 4°C with intermittent sonication. After centrifugation (18,000g for 10 minutes at 4°C), the supernatant (SN1) was vacuum dried in rotational-vacuum-concentrator RVC 2-33 CD (Christ). The pellet was re-extracted with methanol:water (2:1 v/v) as described above. After centrifugation, supernatant (SN2) was vacuum dried in the SN1 tube. Phase partitioning (chloroform:methanol:water, 1:3:3 v/v) was used to separate polar and apolar metabolites. Fractions were vacuum dried as described above and the fraction containing the polar metabolites fraction were washed twice with methanol, derivatized by methoximation (Sigma, 20 μl, 20 mg/ml in pyridine) and trimethylsilylation (20 μl of N,O-bis(trimethylsilyl)trifluoroacetamide reagent (BSTFA) containing 1% trimethylchlorosilane (TMCS), Supelco), and analysed on an Agilent 7890A-5975C GC-MS system (36, 37). Splitless injection (injection temperature 270 °C) onto a 30 m + 10 m × 0.25 mm DB-5MS+DG column (Agilent J&W) was used, using helium as the carrier gas, in electron ionization (EI) mode. The initial oven temperature was 70 °C (2 min), followed by temperature gradients to 295 °C at 12.5 °C/min and then to 320 °C 25 °C/min (held for 3 min). Metabolite quantification and isotopomer distributions were corrected for the occurrence of natural isotopes in both the metabolite and the derivatization reagent. Data analysis and peak quantifications were performed using MassHunter Quantitative Analysis software (B.06.00 SP01, Agilent Technologies). The level of labeling of individual metabolites was corrected for natural abundance of isotopes in both the metabolite and the derivatization reagent (30). Abundance was calculated by comparison to responses of known amounts of authentic standards.
NMR spectra were acquired at 25°C with a Bruker Avance III HD instrument with a nominal 1H frequency of 800 MHz using 3 mm tubes in a 5 mm CPTCI cryoprobe. For 1H 1D profiling spectra the Bruker pulse program noesygppr1d was used with a 1 s presaturation pulse (50 Hz bandwidth) centred on the water resonance, 0.1 ms mixing time, and 4 s acquisition time at 25 °C. Typically 128 transients were acquired.
Statistical analysis
Student’s t and one-way ANOVA with Tukey’s post-hoc tests were used to evaluate statistical significance as indicated in figure legends; and Kaplan-Meier method was for survival analysis. Value of p <0.05 were considered significant. Data are expressed as mean ± SD.
Results
Increased glycolysis in c-Myc-induced liver tumors is associated with increased PKM1 and PKM2 expression
In our previous study, we demonstrated that liver tumors from CEBPb-tTA-TRE-c-MYC transgenic mice exhibit increased levels of glycolysis compared to normal liver tissue (28). In order to generate c-MYC driven tumors in adult murine livers we delivered a Sleeping-Beauty Transposon vector encoding the c-MYC oncogene into mice by hydrodynamic injection (30). We found that enforced c-MYC expression consistently led to tumor formation in FVB/N mice obtained from Jackson Laboratory, whereas c-MYC injection failed to induce liver tumor formation or induced liver tumors with significant delay and decreased penetrance in many other strains tested. This inconsistency could be explained by previous observations that overexpression of c-MYC induces apoptosis that can lead to elimination of c-MYC-transformed cells in vivo (38). The tissue environment may be more or less supportive of the survival of c-MYC-transformed hepatocytes in different genetic backgrounds (39), which would result in tumor formation upon c-MYC overexpression in some mouse strains but not others. To generate consistently highly penetrant liver tumors in all mouse strains, we co-injected plasmids expressing c-MYC and MCL1, an anti-apoptotic member of BCL2 protein family and found that this combination (c-MYC/MCL1) induces liver tumor formation in 5 to 8 weeks in all mouse strains tested, including C57BL/6, FVB/N and Balb/C mice (Figure S2).
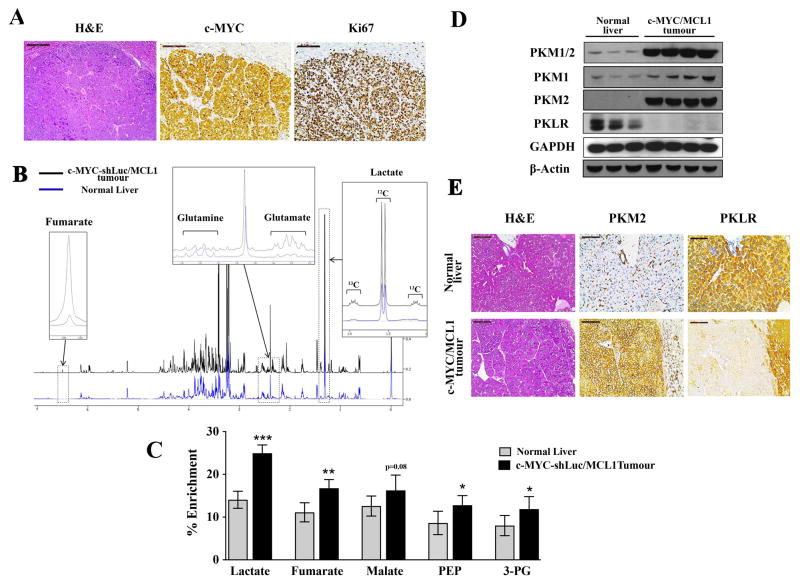
In the further experiments, pT3-EF1α-c-MYC vector also carried a small hairpin against Renilla luciferase (shLuc), which did not affect c-MYC/MCL1 tumor latency. c-MYC-shLuc/MCL1 liver tumors showed equivalent histopathological features to those of CEBPb-tTA-TRE-c-MYC liver tumors as well as c-MYC/MCL1. All tumor cells expressed ectopic human c-MYC oncogene and tumors were highly proliferative (Figure 1A). Similar to CEBPb-tTA-TRE-c-MYC tumors, c-MYC-shLuc/MCL1 tumors had increased glucose catabolism (28, 40), as suggested by increased contribution of glucose to the pools of lactate, glycolytic intermediates, phosphoenolpyruvate (PEP) and 3-phosphoglycerate (3-PG), and the Krebs cycle intermediates (Figure 1B and 1C).Importantly, increased glucose catabolism in c-MYC/MCL1 and c-MYC-shLuc/MCL1 tumors was associated with increased expression of both PKM1 and PKM2 (Figure 1C and 2A). c-MYC/MCL1 and c-MYC-shLuc/MCL1 tumors also had liver-specific isoform of pyruvate kinase, PKL, expressed as detected with the antibodies against the product of PKLR gene. However, its expression was very low (Figure 1D and 2A) and was not detected in all of the tumor cells (Figure 1E). These changes in the expression of pyruvate kinase isoforms in c-MYC-shLuc/MCL1 tumors were associated with increased pyruvate kinase activity in comparison with the normal liver (Figure 2B).
Figure 1. c-MYC-induced liver tumorigenesis is associated with increased glycolysis and a switch from PKL to PKM.
A) Immunohistochemical patterns of c-MYC and Ki67 in c-MYC/MCL1 tumors. Scale bars, 100μm. B)Representative 1H-NMR spectra from extracts of normal livers (blue) and c-MYC/MCL1 liver tumors (black) of mice injected with a bolus of 13C-Glucose. Regions of interest are magnified to show increased pools of glutamate, fumarate and lactate and decreased pool of glutamine in tumors in comparison with normal tissue. Increased 13C incorporation into lactate is denoted by the 13C satellites of the H3-lactate. C) % Enrichment from 13C6-glucose in glycolytic and Krebs cycle intermediates measured by GC-MS. Data are presented as mean ± SD. Student’s t-test was used; *p <0.05, **p <0.01, ***p <0.001. D)PKM1/2 and PKLR protein levels measured in whole-tissue lysates from normal and tumor samples by immunoblotting using β-Actin and GAPDH as a loading control. E) Expression of PKM2 and PKLR examined by immunochemistry in normal livers and c-MYC/MCL1 liver tumors. Scale bars, 100μm.
Figure 2. PKM1/2 knock-down does not affect c-MYC-induced liver tumorigenesis.
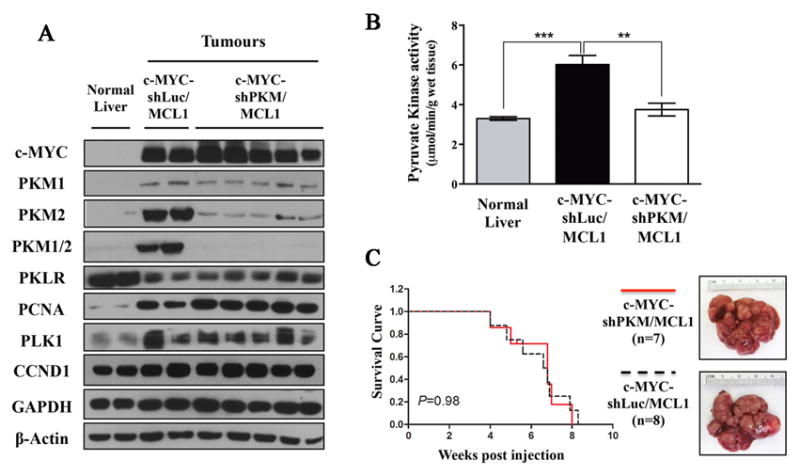
A) Total pyruvate kinase activity measured in normal livers (n=3), c-MYC-shLuc/MCL1 (n=3) and c-MYC-shPKM/MCL1 (n=3) tumors. Data are presented as mean ± SD. Student’s t-test was used; *p <0.05, **p <0.01, ***p <0.001. B) Western blot analysis of glycolytic enzymes and proliferation associated proteins in normal livers, c-MYC-shLuc/MCL1 and c-MYC-shPKM/MCL1 tumors. C) Survival analysis of mice bearing c-MYC-shLuc/MCL1 (n=8) and c-MYC-shPKM/MCL1 (n=7) tumors using Kaplan-Meier survival method. Gross images of both tumors are shown in the right panel.
Decreased PKM2 expression results in decreased pyruvate kinase activity but does not affect c-MYC-inducedliver tumorigenesis
Given the contradictory results about the role of PKM isoforms in tumorigenesis, we thought to evaluate if differential expression of the isoforms and pyruvate kinase activity would have any effect on c-MYC-driven liver tumorigenesis. Two miR-30 based shRNAs against mouse PKM 3’-UTR which are common for both PKM1 and PKM2 (shPKM.1 and shPKM.2, Figure S1) were first tested in vitro in a cell line derived from CEBPb-tTA-TRE-c-MYC tumors (41) (Figure S3). Both shPKMs significantly decreased the expression of PKM1 and PKM2 compared to a control shRNA (shLuc), but did not affect cell proliferation in culture (Figure S3).
To investigate whether silencing of PKM1 and PKM2 expression affects c-MYC-induced tumorigenesis in the liver in vivo, we cloned shPKM and shLuc into pT3-EF1α-c-MYC plasmid (Figure S4). The plasmids were hydrodynamically injected into mice together with the vector encoding MCL1 (c-MYC-shPKM/MCL1 and c-MYC-shLuc/MCL1, respectively, Figure S4). In contrast to its effect in vitro, shPKM strongly decreased PKM2 expression while having only minor effect on PKM1 expression (Figure 2A, S5, and S6). Pyruvate kinase activity was decreased in c-MYC-shPKM/MCL1 tumors to the level observed in the normal liver (Figure 2B), suggesting that increased PKM2 expression is mostly responsible for the increased pyruvate kinase activity in c-MYC liver tumors. c-MYC-shPKM/MCL1 tumors had PKL levels comparable to those in c-MYC-shLuc/MCL1 tumors (Figure 2A and S5), which together with PKM1 may be supporting the pyruvate kinase activity observed. Expression of other key enzymes involved in glycolysis including hexokinases (HK1, HK2 and HK4) and phosphofructokinases (PFKP, PFKM and PFKL) as well as the first enzyme of glutamine catabolism, GLS1 also did not change in c-MYC-shPKM/MCL1 tumors in comparison with c-MYC-shLuc/MCL1 tumors (Figure S6). Overall shPKM specifically decreased PKM2 expression leading to lower pyruvate kinase activity, but it did not affect the expression of other genes in either glucose or glutamine catabolism.
In both groups of mice tumors developed between 5 to 8 weeks post injection. There was no difference in survival rate, liver weight, and liver/body ratio between c-Myc-shLuc/Mcl1 and c-Myc-shPKM/Mcl1 mice (Figure 2C and Figures S7). Histologically, both c-MYC-shPKM/MCL1 and c-MYC-shLuc/MCL1 tumors were composed of small tumor cells with large nuclei, consistent with the features of c-MYC-induced liver tumors as described by others (Figure S7) (42). Furthermore, we did not observe significant differences in proliferation in c-MYC-shPKM/MCL1 and c-MYC-shLuc/MCL1 tumors with the proliferation index 65.7± 11.1 in c-MYC-shPKM/MCL1 and 68.8 ± 9.2 in c-MYC-shLuc/MCL1 (P>0.05; n=9 for each group). Consistent with this, no difference was observed in the expression of proliferation associated proteins, including cyclin D1, PCNA and Polo-like kinase (PLK1) between c-MYC-shLuc/MCL1 and c-MYC-shPKM/MCL1 tumors (Figure 2A).
Silencing of PKM2 expression results in altered glucose catabolism
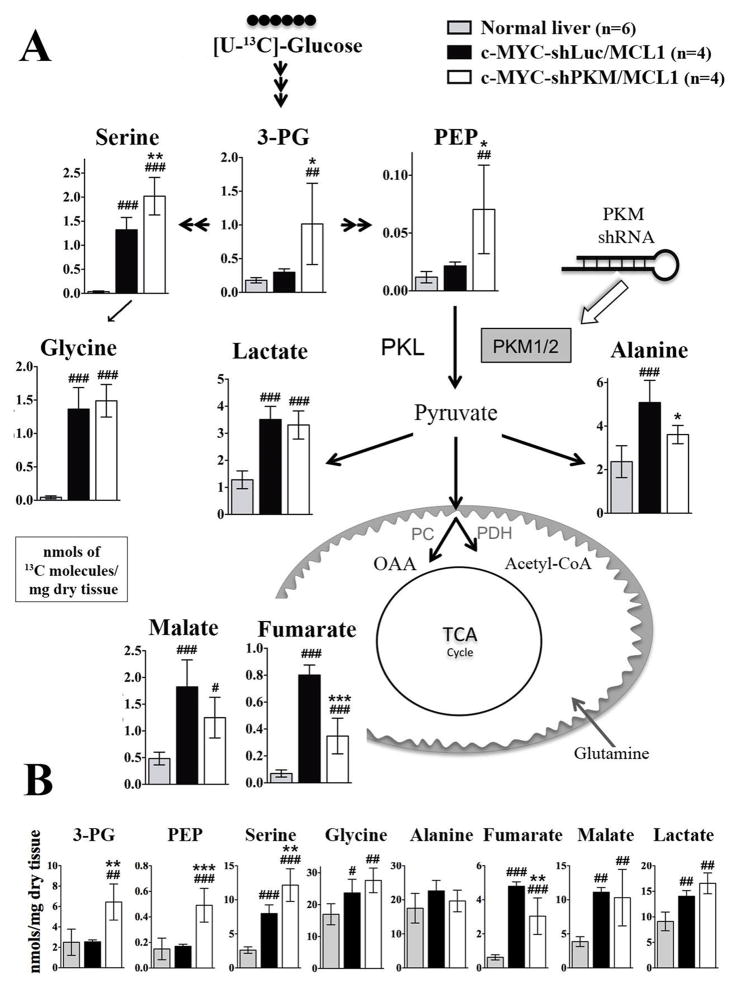
To evaluate how decreased expression of PKM isoforms and resultant decrease in pyruvate kinase activity affect glucose catabolism, we injected tumor-bearing animals with 13C6-glucose and evaluated the total levels and the levels of 13C-labeled metabolites by GC-MS and NMR. Consistent with decreased pyruvate kinase activity c-MYC-shPKM/MCL1 tumors had significantly higher levels of total and 13C-labeled PEP, 3-phosphoglycerate (3PG) and serine than c-MYC-shLuc/MCL1 tumors (Figure 3A and 3B). Increased levels of glycolytic intermediates upstream of pyruvate kinase in c-MYC-shPKM/MCL1 tumors were associated with decreased levels of 13C-labeled alanine and the Krebs cycle intermediates (Figure 3A), indicating that decreased pyruvate kinase activity in c-MYC-shPKM/MCL1 tumors resulted in decreased glucose catabolism below the pyruvate kinase step. These changes were associated with significant decrease in total fumarate levels, but not those of alanine or malate (Figure 3B). Interestingly, the levels of total and 13C-lactate were not affected in c-MYC-shPKM/MCL1 tumors in comparison with c-MYC-shLuc/MCL1 tumors (Figure 3A and 3B). These results demonstrate that increased pyruvate kinase activity in c-MYC-induced liver tumors supports increased glycolytic flux. However, pyruvate kinase activity at the levels observed in the normal liver tissue can still support the glycolytic flux higher than the one observed in the normal liver and presumably created by glycolytic steps upstream of pyruvate kinase. Significantly decreasing PKM2 expression in tumor cells (Figure 2A) does not affect the initiation of proliferation and survival of c-MYC transformed liver cells.
Figure 3. PKM1/2 knock-down reduces PK flux in vivo.
Livers and tumors from mice injected with 13C-glucose boluses were analysed by GCMS. A) The fate of 13C from 13C6-glucose through glycolysis, into the Krebs cycle, and amino acid synthesis is depicted; the levels of 13C-labeled metabolites are expressed as nmols of 13C molecules (sum of all isotopomers except M+0) per mg of dry tissue. B) Total concentration of metabolites is presented. In both panels data are presented as mean ± SD; #p <0.05; ##p <0.01; ###p <0.001 relative to Normal Liver and *p <0.05; **p <0.01; ***p <0.001 relative to the c-Myc-shLuc/Mcl1group. One-way ANOVA, with a Turkeys post hoc test was used.
Increasing PKM1/PKM2 ratio does not affect c-Myc-induced liver tumorgenesis
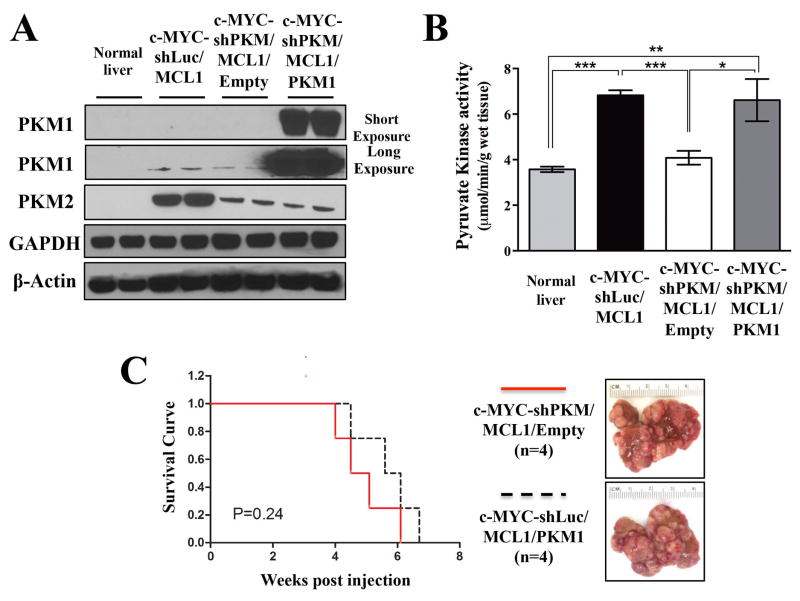
Increasing pyruvate kinase activity by ectopic overexpression of PKM1 or using PKM2 activators was previously shown to impair the ability of cancer cells to form xenograft tumors (12, 43). Moreover, mammary gland tumor-derived cells with deleted PKM2 and ectopic overexpression of PKM1 were not able to develop tumors, whereas cells with just deleted PKM2 were (23). These works suggested that increasing PKM1 levels or constitutively activating pyruvate kinase can be detrimental for cancer cell proliferation in vivo by, for example, diverting the glucose flux from the synthesis of anabolic precursors. In c-MYC-shPKM/MCL1 tumors, the decrease in PKM2 levels was more significant than the decrease in PKM1 levels in comparison with c-MYC-shLuc/MCL1 tumors, suggesting that PKM1/PKM2 ratio in c-MYC-shPKM/MCL1 was higher than in c-MYC-shLuc/MCL1 tumors. However, to ensure that PKM1/PKM2 ratio is significantly increased and to recapitulate the conditions used in other systems, we ectopically overexpressed PKM1 cDNA with C-terminal V5 tag together with decreasing PKM2 expression by expressing shPKM (Figure S8). In brief, we injected mice with pT3 vector expressing c-MYC-shPKM, pT3 vector expressing MCL1 and with either empty pT3 vector (c-MYC-shPKM/MCL1/Empty) or pT3 expressing PKM1 (c-MYC-shPKM/MCL1/PKM1). It is important to note that the co-injected PKM1 plasmid did not contain 3’-UTR and therefore could not be targeted by shPKM. PKM1 was confirmed to be uniformly expressed in c-MYC-shPKM/MCL1/PKM1 tumor cell via anti-V5 immunostaining (Figure S8).
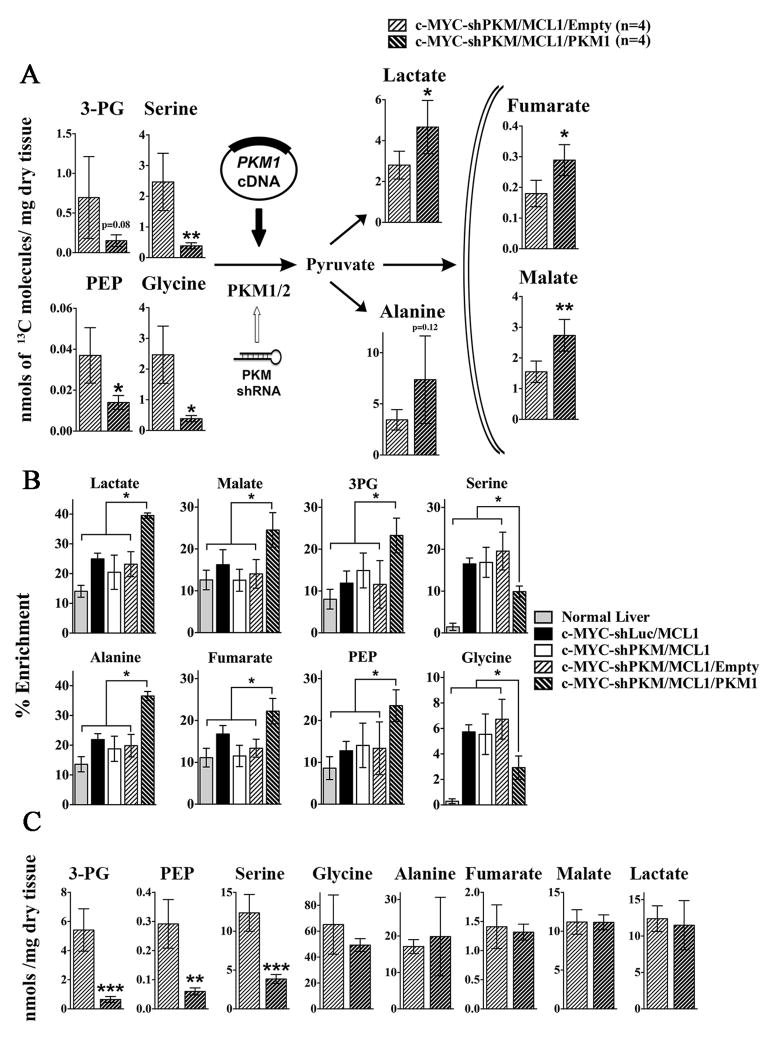
c-MYC-shPKM/MCL1/PKM1tumors demonstrated significantly higher PKM1/PKM2 ratio than c-MYC-shLuc/MCL1 and c-MYC-shPKM/MCL1/Empty tumors (Figure 4A). Pyruvate kinase activity measured in tissue lysates from c-MYC-shPKM/MCL1/PKM1 tumors was higher than in c-MYC-shPKM/MCL1/Empty tumors and comparable with c-MYC-shLuc/MCL1 tumors (Figure 4B). Evaluating glucose catabolism in c-MYC-shPKM/MCL1/PKM1 tumors demonstrated significant decrease in the levels 13C-labelled PEP, 3PG, serine but increased levels of 13C-labelled lactate and the Krebs cycle intermediates, malate and fumarate, in comparison with c-MYC-shPKM/MCL1/Empty tumors (Figure 5A). As demonstrated by total carbon enrichment from 13C-glucose in all experimental groups the flux of glucose into lactate and the Krebs cycle intermediates in c-MYC-shPKM/MCL1/PKM1 tumors was higher even in comparison with the initial c-MYC-shLuc/MCL1 tumors (Figure 5B). The redistribution of the glucose-derived carbons in c-MYC-shPKM/MCL1/PKM1 tumors resulted in the decreased total serine levels (Figure 5C), which were lower than in c-MYC-shPKM/MCL1/Empty (Figure 5C) and the initial cMYC-shLuc/MCL1 tumors (Figure 3B). Regardless of this diverted flux, however, the latency and burden of c-MYC-shPKM/MCL1/PKM1 tumors was not statistically significantly different from c-MYC-shPKM/MCL1 tumors (Figure 4C).
Figure 4. Increasing PKM1/PKM2 ratio does not suppress c-MYC-induced liver tumorigenesis.
A) Western blot analysis of the expression of PKM isoforms in normal livers, c-MYC-shLuc/MCL, c-MYC-shPKM/MCL1/Empty and MYC-shPKM/MCL1/PKM1 tumors B) Total pyruvate kinase activity measured in tissues from groups in (A) (n=3 in each group). Data are presented as mean ± SD. Student’s t-test was used; *p <0.05, **p <0.01, ***p <0.001. C) Survival analysis of of mice bearing c-MYC-shPKM/MCL1/Empty (n=4) and c-MYC-shPKM/MCL1/PKM1 (n=4) tumors using Kaplan-Meier survival method. Gross images of both tumors are shown in the right panel.
Figure 5. PKM1 overexpression increases PK flux in vivo.
Livers and tumors from mice injected with 13C-glucose boluses were analysed by GCMS. A) The levels of 13C-lableled metabolites (sum of all isotopomers except M+0) are presented. Data are presented as mean ± SD. *p <0.05; **p <0.01, unpaired two-sided Student’s t test. B) Total concentration of the metabolites shown in A. In A and B data are presented as mean ± SD. *p <0.05; **p <0.01, ***p <0.001, unpaired two-sided Student’s t test. C) 13C enrichment of metabolites is presented (experimental groups introduced in Figure 1 and 3 are included for comparison). Data are presented as mean ± SD; *p <0.05. One-way ANOVA, with a Turkeys post hoc test.
Overall, our data demonstrate that presence of a specific pyruvate kinase isoform is not required to support the initiation and progression of MYC-induced tumorigenesis in the liver. Moreover, partial suppression or activation of the pyruvate kinase flux is not sufficient to affect oncogenic transformation or tumor progression.
Discussion
The predominant expression of PKM2 in many embryonic tissues, its increased expression in cancer tissues and preferential expression in multiple cancer cell lines have led to a long-standing belief about its requirement for cell proliferation and tumorigenesis. However, several recent reports using PKM2 knockout demonstrating that PKM2 is dispensable for normal embryonic development and either mammary gland tumorigenesis induced by HER2 or spontaneous liver tumorigenesis (8, 24), have challenged this idea.
Since c-MYC has been demonstrated to increase PKM2/PKM1 ratio by regulating the expression of HnRNP proteins (44), we tested the expression of PKM isoforms and their role in c-MYC-induced tumorigenesis in the liver. In contrast to the results by David and co-authors, c-MYC-induced transformation resulted in significantly increased expression of both PKM1 and PKM2. However the fact that decreasing PKM2 expression resulted in decreasing tumor pyruvate kinase activity to the normal liver levels indicated that PKM2 is still mostly responsible for the increase in pyruvate kinase activity observed in c-MYC-induced liver tumors.
Inhibiting the expression preferentially of PKM2 together with ectopically expressing c-MYC in hepatocytes resulted in tumors with decreased pyruvate kinase activity and increased flux of glucose into serine biosynthesis on the expense of its catabolism through the Krebs cycle. However, consistent with the results obtained in PKM2 knockout mouse models (24), these changes in PKM2 expression and glycolytic flux did not affect either tumor initiation or progression. Pyruvate kinase activity at the level of the normal liver, which is maintained possibly by the expression of PKM1 and PKLR isoforms, was sufficient to support glycolytic flux and flux of glucose through the Krebs cycle required to sustain the level of vital metabolites and metabolic activities. Interestingly, one of the early studies evaluating the expression of PK isoforms in the liver of rats treated with choline/DL-methionine-deficient diet demonstrated that, although being decreased in liver parenchyma, the expression of liver isoform PKL was increased in some proliferating cells during early stages of the treatment and in glycogenolytic cells within dysplastic lesions at later stages (45). The expression of both PKL and PKM2 was also observed in renal cell carcinomas and their metastasis (46). Consistent with these earlier observations high expression of PKLR is reported in the liver and renal cancers by The Human Protein Atlas (47). Intriguingly, germline loss of PKM2 is associated with the formation of HCCs, in which most of the cells were PKM1-negative (8). These results may additionally suggest the role of PKL in supporting proliferation of tumor cells in the absence of PKM isoforms or the existence of pyruvate kinase-independent mechanism of PEP conversion into pyruvate (48).
Similar to the previously demonstrated effect of PKM2 activators (49) and selective overexpression of PKM1 (13), ectopically overexpressing PKM1 while decreasing the expression of PKM2 resulted in significantly decreased glucose flux into serine and increased flux into the Krebs cycle intermediates. As a result, tumors with significantly increased PKM1/PKM2 ratio had significantly decreased levels of serine. Interestingly pyruvate kinase activity measured in lysates from c-MYC-shPKM/MCL1/PKM1 tumors was comparable to the pyruvate kinase activity in the lysates from original tumors confirming the role of allosteric regulation of PKM2 in vivo in regulating glucose flux into the synthesis of biosynthetic precursors. Nevertheless, the significantly decreased levels of serine in c-MYC-shPKM/MCL/PKM1 tumors are still sufficient to support the pools of serine-derived metabolites and sustain cellular transformation and proliferation of transformed cells in the liver. These results are in contrast to the results demonstrating the deleterious effect of increased PKM1 expression in in vitro systems with ectopic overexpression of PKM1 (12) and in mammary gland tumors and normal fibroblasts, where increased PKM1 expression was the result of PKM2 knock-out (18, 24). The difference in the sensitivity of different systems to increasing pyruvate kinase activity and diverting the glucose flux can be determined by the differential requirements for certain levels of biosynthetic precursors and the ability to utilize the compensatory pathways. Based on the results demonstrating the serine auxotrophy in response to PKM2 activators (49), c-MYC-shPKM/MCL1/PKM1 tumors can be suggested to be sensitive to depleting serine from the diet.
It is also noteworthy that our present results do not confirm the previously proposed non-enzymatic role of PKM2 in tumorigenesis (20, 50).
In summary, our findings demonstrate that the control of the flux at the pyruvate kinase step is not a requirement for c-MYC-driven liver tumorigenesis, and suggest that different pyruvate kinase isoforms and various pyruvate kinase activities can equally sustain tumor progression. Our findings further denote the difference between previously demonstrated differential role of PKM1 and PKM2 isoforms in vitro and their requirement in tumors developing in their natural environment in vivo.
Supplementary Material
Acknowledgments
We would like to thank Dr. James I. MacRae for his support with metabolomic analysis. The NMR data were acquired at the MRC Biomedical NMR Centre at the Francis Crick Institute.
Financial support
This work was supported by grant from NIH R01CA136606 and R21CA198490 to X. Chen; grant P30DK026743 for UCSF Liver Center; National Natural Science Foundation of China grant No. 81602424 to J. Hu. This work was also supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001223), the UK Medical Research Council (FC001223), and the Wellcome Trust (FC001223) to M. Yuneva.
Footnotes
Conflict of interests: The authors declare no potential conflict of interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell metabolism. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirschey MD, DeBerardinis RJ, Diehl AM, Drew JE, Frezza C, Green MF, et al. Dysregulated metabolism contributes to oncogenesis. Seminars in cancer biology. 2015;35(Suppl):S129–50. doi: 10.1016/j.semcancer.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Seminars in cell & developmental biology. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262:14366–71. [PubMed] [Google Scholar]
- 6.Marie J, Simon MP, Dreyfus JC, Kahn A. One gene, but two messenger RNAs encode liver L and red cell L’ pyruvate kinase subunits. Nature. 1981;292:70–2. doi: 10.1038/292070a0. [DOI] [PubMed] [Google Scholar]
- 7.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–80. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Dayton TL, Gocheva V, Miller KM, Israelsen WJ, Bhutkar A, Clish CB, et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 2016;30:1020–33. doi: 10.1101/gad.278549.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bluemlein K, Gruning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda Y, Tanaka T, Noguchi T. Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution. J Biol Chem. 1997;272:20495–501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 11.Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991;266:16842–6. [PubMed] [Google Scholar]
- 12.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6904–9. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–62. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 16.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh MJ, Brimacombe KR, Anastasiou D, Yu Y, Israelsen WJ, Hong BS, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. [PubMed] [Google Scholar]
- 18.Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Molecular cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- 20.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Wang Y, Wang T, Hawke DH, Zheng Y, Li X, et al. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat Commun. 2014;5:5566. doi: 10.1038/ncomms6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Wang H, Yang JJ, Chen J, Jie J, Li L, et al. Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals. J Biol Chem. 2013;288:15971–9. doi: 10.1074/jbc.M112.448753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:489–94. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of Evidence for PKM2 Protein Kinase Activity. Molecular cell. 2015;59:850–7. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer research. 2009;69:2775–82. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism. 2012;15:157–70. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Morze C, Larson PE, Hu S, Yoshihara HA, Bok RA, Goga A, et al. Investigating tumor perfusion and metabolism using multiple hyperpolarized (13)C compounds: HP001, pyruvate and urea. Magnetic resonance imaging. 2012;30:305–11. doi: 10.1016/j.mri.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow EK, Fan LL, Chen X, Bishop JM. Oncogene-specific formation of chemoresistant murine hepatic cancer stem cells. Hepatology. 2012;56:1331–41. doi: 10.1002/hep.25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschaharganeh DF, Xue W, Calvisi DF, Evert M, Michurina TV, Dow LE, et al. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell. 2014;158:579–92. doi: 10.1016/j.cell.2014.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao J, Ji J, Li X, Ding N, Wu H, Liu Y, et al. Distinct anti-oncogenic effect of various microRNAs in different mouse models of liver cancer. Oncotarget. 2015;6:6977–88. doi: 10.18632/oncotarget.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SA, Ladu S, Evert M, Dombrowski F, De Murtas V, Chen X, et al. Synergistic role of Sprouty2 inactivation and c-Met up-regulation in mouse and human hepatocarcinogenesis. Hepatology. 2010;52:506–17. doi: 10.1002/hep.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–83. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan TW, Lane AN, Higashi RM, Yan J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–69. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, Kenny S, et al. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC biology. 2013;11:67. doi: 10.1186/1741-7007-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamboni N, Fendt SM, Ruhl M, Sauer U. (13)C-based metabolic flux analysis. Nature protocols. 2009;4:878–92. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- 38.Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS biology. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juric V, Ruffell B, Evason KJ, Hu J, Che L, Wang L, et al. Monocytes promote liver carcinogenesis in an oncogene-specific manner. Journal of hepatology. 2016;64:881–90. doi: 10.1016/j.jhep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell metabolism. 2011;14:131–42. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Z, Fan-Minogue H, Bellovin DI, Yevtodiyenko A, Arzeno J, Yang Q, et al. MYC phosphorylation, activation, and tumorigenic potential in hepatocellular carcinoma are regulated by HMG-CoA reductase. Cancer research. 2011;71:2286–97. doi: 10.1158/0008-5472.CAN-10-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–7. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Zhang X, Li BS, Zhai X, Yang Z, Ding LX, et al. Simultaneous targeting of PI3Kdelta and a PI3Kdelta-dependent MEK1/2-Erk1/2 pathway for therapy in pediatric B-cell acute lymphoblastic leukemia. Oncotarget. 2014;5:10732–44. doi: 10.18632/oncotarget.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hacker HJ, Steinberg P, Bannasch P. Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet. Carcinogenesis. 1998;19:99–107. doi: 10.1093/carcin/19.1.99. [DOI] [PubMed] [Google Scholar]
- 46.Brinck U, Eigenbrodt E, Oehmke M, Mazurek S, Fischer G. L- and M2-pyruvate kinase expression in renal cell carcinomas and their metastases. Virchows Archiv : an international journal of pathology. 1994;424:177–85. doi: 10.1007/BF00193498. [DOI] [PubMed] [Google Scholar]
- 47.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chemistry & biology. 2012;19:1187–98. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.