Abstract
Intraoperative identification of carcinoma at lumpectomy margins would enable reduced re-excision rates, which are currently as high as 20–50%. While imaging of disease-associated biomarkers can identify malignancies with high specificity, multiplexed imaging of such biomarkers is necessary to detect molecularly heterogeneous carcinomas with high sensitivity. We have developed a Raman-encoded molecular imaging (REMI) technique in which targeted nanoparticles are topically applied on excised tissues to enable rapid visualization of a multiplexed panel of cell surface biomarkers at surgical margin surfaces. A first-ever clinical study was performed in which 57 fresh specimens were imaged with REMI to simultaneously quantify the expression of four biomarkers HER2, ER, EGFR and CD44. Combined detection of these biomarkers enabled REMI to achieve 89.3% sensitivity and 92.1% specificity for the detection of breast carcinoma. These results highlight the sensitivity and specificity of REMI to detect biomarkers in freshly resected tissue, which has the potential to reduce the rate of re-excision procedures in cancer patients.
Keywords: breast cancer, surgery, techniques and strategies, multiplexed molecular imaging, intraoperative imaging, Raman spectroscopy
Introduction
Approximately 200,000 patients are diagnosed with early-stage breast carcinoma each year in the United States, for which breast-conserving surgery (a.k.a. partial mastectomy or lumpectomy) is a standard intervention (1). Unfortunately, amongst various institutions, between 20% and 50% of these patients require additional surgery if post-operative pathology reveals that the resection margins are positive for carcinoma (2). Recent studies and consensus guidelines recommend that lumpectomy procedures be considered complete when a negative margin is observed for invasive carcinoma (3) – otherwise referred to as “no tumor on ink” (i.e. no carcinoma at the surgical margin surface). Whether invasive or non-invasive (in situ), there is no debate that the presence of carcinoma at the surgical margin surface would warrant re-excision at all institutions and that intraoperative identification of residual carcinoma at these surfaces would thereby greatly reduce the rates of re-excision surgeries.
Frozen section analysis has been explored for the intraoperative assessment of surgical margins. However, frozen sectioning is particularly difficult to perform for fatty breast tissues, requires additional surgical time, introduces freezing artifacts that compromise postoperative histological diagnosis, and can yield significant false-negative rates since only a small number of thin tissue sections can be rapidly prepared and evaluated during surgery (e.g. low sensitivities of <70% have been reported (4,5)). A number of other ex vivo imaging techniques have been proposed to assess breast margins, such as confocal microscopy (6), nonlinear microscopy (95.4% sensitivity, 93.3% specificity) (7,8), optical coherence tomography (90–100% sensitivity, 75–82% specificity) (9,10), optical coherence microelastography (11), light reflectance spectroscopy (74% sensitivity, 86% specificity) (12,13), autofluorescence lifetime measurement (12) and intrinsic Raman spectroscopy (94–100% sensitivity, 96–100% specificity) (14–16). Optical-sectioning microscopy techniques, such as confocal microscopy and nonlinear microscopy, can obtain high-resolution and high-contrast images of freshly resected tissues. However, applying these techniques for intraoperative imaging of large tissue surfaces is challenging due to the relatively slow speed of these point-by-point imaging techniques. In addition, these techniques suffer from an extremely limited depth of focus, necessitating elaborate tissue-flattening or volumetric imaging strategies to visualize a large tissue surface with topological irregularities (6,7). Spectroscopic methods (e.g. reflectance, Raman and autofluorescence lifetime) reveal the relative concentrations of chemical constituents such as hydrocarbons, lipids, nucleic acids, and/or the size distribution of photon-scattering objects in tissues, and have yielded promising results for tumor detection (12–16). However, the ability to image the expression of molecular biomarkers, which play a prominent role in recent advances in precision medicine (17,18), would offer complementary information that could further improve the sensitivity and specificity of detection.
Over the past few decades, there has been increasing confidence in the presumption that molecular-imaging approaches – namely, the imaging of protein biomarkers of cancer – can identify tumors with a high degree of sensitivity and specificity (19). High specificity would be valuable for intraoperative cancer detection to minimize over-excision and to optimize patient cosmesis (a major goal of breast-conserving surgeries). However, in order to achieve high detection sensitivity, multiple biomarkers should be evaluated since the molecular profiles of most cancers, including breast carcinoma, vary greatly between patients as well as spatially and temporally within a single tumor mass (20,21). While quantum dots (QDs) have been used for multiplexed imaging of 3–5 biomarkers in breast cancer cells and formalin-fixed tissue sections, (22), no clinical studies have been reported to evaluate the efficacy of QD-based imaging technologies for guiding lumpectomy. Recently, surface-enhanced Raman-scattering (SERS) nanoparticles (NPs) have attracted wide interest due to their excellent multiplexing capabilities over quantum dots and fluorescent dyes (23,24). These SERS NPs are available in many “flavors,” each of which emits a characteristic Raman fingerprint spectrum when illuminated at a common wavelength (25–27). By functionalizing various flavors of SERS NPs with different targeting molecules (e.g. antibodies (28), affibodies (29), etc.), the NPs can be multiplexed to simultaneously target and image a large panel of protein biomarkers.
We have recently demonstrated that a Raman-encoded molecular imaging (REMI) technique, which utilizes the topical application of multiplexed SERS NPs, enables the rapid visualization of multiple cell-surface biomarkers at the surfaces of fresh tissues (28,30–32). In our REMI approach, toxicity and sterility concerns are circumvented by staining and imaging fresh surgical specimens ex vivo, with the potential to allow for expedited regulatory approval and rapid clinical translation.
A critical component of REMI is a ratiometric imaging method that enables accurate quantification of biomarker expression levels by utilizing one untargeted NP flavor to normalize for the nonspecific accumulation exhibited by all of the NPs (both the targeted and untargeted NPs), as for example due to off-target binding, uneven NP delivery and washout, and variations in tissue permeability and retention (33–39). The SERS NPs used for REMI are particularly well-suited for accurate ratiometric quantification due to the fact that they are all excited at a single wavelength (785 nm) and emit Raman spectra within the same narrow spectral range (approximately 850 – 900 nm), which ensures that all NPs are irradiated identically and are affected by the same tissue optical properties. This is in contrast to fluorescent agents, in which a large range of laser-illumination wavelengths and collection bands are typically needed for multiplexed imaging. As a result, each fluorescent agent is affected by different tissue-scattering and absorption properties (wavelength-dependent), which requires calibration with wavelength-dependent light propagation models and thus complicates ratiometric quantification (40). Our previous studies have verified that REMI provides a linear measurement of NP concentrations/ratios when applied on tissues (Supplementary Fig. S1A and B)(28,30,31,33,41,42) and, more importantly, provides a quantitative measure of biomarker expression levels that agrees with both flow cytometry and gold-standard immunohistochemistry (Supplementary Fig. S2A–D)(28,30,32).
Here we assess the diagnostic accuracy of REMI for identifying carcinoma at the surfaces of freshly excised breast specimens through simultaneous quantification of four cell-surface biomarkers - human epidermal growth factor receptor 2 (HER2), membrane estrogen receptor (mER), epidermal growth factor receptor (EGFR) and CD44 (a brief description of these biomarkers is provided in the Supplementary Information). By imaging 57 fresh specimens from 29 patients, we validated and optimized the sensitivity and specificity of REMI to identify the elevated expression of the four biomarkers mentioned above, in comparison to IHC, which is the clinical gold-standard method for assessing protein expression in tissues. In addition, by combining all four biomarkers, we assessed the overall sensitivity and specificity of REMI for identifying breast carcinoma at surgical margin surfaces, in comparison to the diagnostic gold standard of H&E histopathology.
Materials and Methods
REMI system
A customized spectral-imaging system has been developed to measure the concentration and concentration ratio of SERS NPs that are topically applied on tissue specimens (30). The NP-stained surface (surgical margin) of the tissue specimen was raster scanned with a two-axis stage (Newmark systems Inc., ET-50-11) and imaged using a fixed spectral-imaging probe (FiberTech Optica Inc.). The imaging probe utilizes a multimode fiber (100-μm core, 0.10 NA) at the center of the probe for illumination, and 27 surrounding multimode fibers (200-μm core, 0.22 NA) for light collection (Raman, autofluorescence and back-scattered laser light). A 785-nm diode laser (18 mW at the tissue) is used to illuminate the tissue, creating a laser spot with a diameter of 0.5 mm (imaging resolution). Light collected by the 27 multimode fibers are transmitted to a customized spectrometer (Andor Holospec), where they are filtered (to remove autofluorescence and back-scattered laser light) and then dispersed onto a cooled deep-depletion spectroscopic CCD (Andor, Newton DU920P-BR-DD). The detector integration time (i.e. the spectral acquisition rate, which equals the pixel rate) utilized in this study was 50 ms. A direct-classical-least-squares (DCLS) algorithm was employed to calculate the concentrations and ratios of various SERS NP flavors as described previously (30,31) and as summarized in Supplementary Information. Figure 1 provides an illustration of the REMI process.
Figure 1. REMI for intraoperative guidance of lumpectomy.
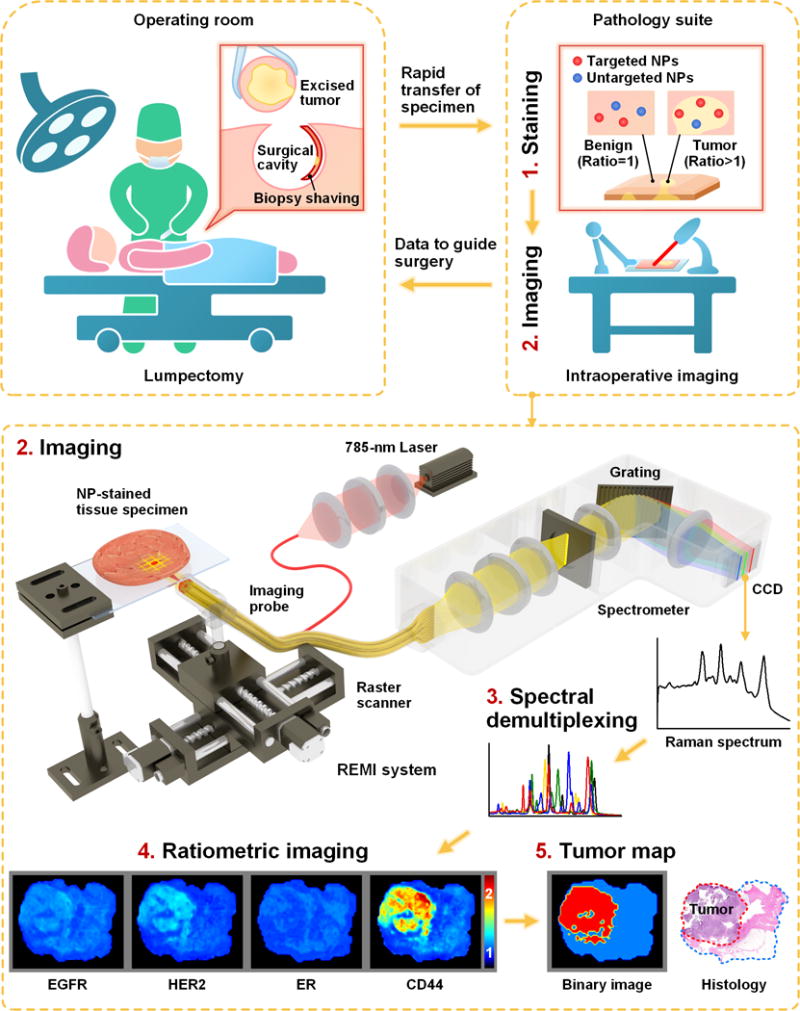
In a clinical implementation of REMI, freshly resected human breast tissues from lumpectomy procedures are immediately transferred to a pathology suite for intraoperative consultation. Each specimen is topically stained with a mixture of SERS NPs (multiple biomarker-targeted NPs and at least one untargeted control NP, step 1), followed by spectroscopic imaging of the surgical margin surface (step 2). The acquired SERS spectra are demultiplexed to determine the ratio of the targeted vs. untargeted NPs (step 3), which enables the quantification of various biomarker targets (e.g. EGFR, HER2, mER and CD44 in this study, step 4). REMI images of the individual biomarkers are combined to detect the presence of residual tumors at the surgical margin surfaces of the specimens (step 5). The entire REMI procedure (staining, rinsing, imaging, spectral demultiplexing) was performed within 10–15 min depending upon the size of the specimen.
Tissue staining and imaging
Unstained specimens were first imaged to acquire a set of background spectra for the calculation of their principal components (to account for the tissue background during least-squares demultiplexing). The specimen surfaces (surgical margins) were then topically stained with the NP staining solution using a convection-enhanced staining method (28). Based on a previously optimized staining protocol (30), the staining solution consisted of a mixture of 5 flavors of SERS NPs (150 pM per flavor) supplemented with 1% BSA to minimize nonspecific binding. After 5 min of staining, the tissue sample was rinsed in 50-mL PBS with gentle agitation for 10 s, followed by raster-scanned imaging of the entire stained tissue surface (>3 cm2/min), followed by spectral demultiplexing (~1000 spectra/s) and ratiometric mapping (<1 min). The entire REMI procedure (including image processing) can be performed within 15 min.
SERS NPs and functionalization
The SERS NPs were purchased from BD (Becton, Dickinson and Company). These NPs consist of a 60-nm-diameter gold core, a layer of Raman reporters adsorbed onto the surface of the gold cores, and a 60-nm-thick silica coating (Supplementary Fig. S3A). The thick silica shell makes the SERS NP signals insensitive to the environment and immune from signal changes induced by aggregation of the NPs. Previous studies have demonstrated excellent linearity of NP measurements in a variety of animal and human tissues over a wide range of concentrations (25). Five “flavors” of NPs were used in this study, identified as S420, S421, S440, S481 and S493, each of which emits a characteristic Raman spectrum due to chemical differences in the Raman reporter layer (Supplementary Fig. S3B).
Using a previously described conjugation protocol (33), unique SERS NP flavors were functionalized with different monoclonal antibodies (mAb) targeting either EGFR, HER2, ER or CD44. In addition, negative-control NPs were prepared by conjugating one NP flavor with an isotype control antibody (mouse IgG1). The NPs, which were functionalized with active thiols at their surface, were first reacted with a fluorophore, DyLight 650 Maleimide (Thermo Scientific, 62295), for the purposes of flow-cytometry characterization. The NPs were then conjugated with either an isotype control (Thermo Scientific, MA110407), an anti-EGFR mAb (Thermo Scientific, MS378PABX), an anti-HER2 mAb (Thermo Scientific, MS229PABX), an anti-ER mAb (Thermo Scientific, RM9101S0) or an anti-CD44 mAb (Abcam plc., ab6124) at 500 molar equivalents per NP. To ensure reproducible tissue imaging throughout the duration of this study, mAb-conjugated NPs were used for tissue imaging experiments only when they yielded flow-cytometry results that were similar (±20% error) to the ones shown in Supplementary Fig. S4A–D. The flow-cytometry methods are described in the Supplementary Information.
Acquisition and handling of human breast tissues
This study was approved by the University of Washington institutional review board, and was conducted in accordance with the provisions of the Declaration of Helsinki. De-identified human breast tissue specimens were obtained from consenting patients and imaged within 1 hour after lumpectomy or mastectomy at the University of Washington Medical Center (informed consent was obtained from all patients). Tissue collection was managed by the Northwest BioTrust (NWBT) under an IRB exemption for these de-identified tissues. After imaging, the tissues were fixed with 10% formalin and submitted for histopathology (IHC and H&E staining). To facilitate the correlative analysis between REMI and histopathology images, the fixed tissues were flattened against the bottom of the tissue mold, and sectioned as close as possible to the imaged surface (within ~200 μm of the surface). According to a pathological modeling study (43), a tissue section at a depth of <250 μm is predicted to exhibit minimal variation in important pathologic parameters compared with the surface. The histopathology slides were scanned using a whole slide scanner (Hamamatsu Nanozoomer).
Pathology for validation
Histology slides from the breast tissue specimens were independently interpreted by two pathologists who were blinded to the REMI results. For each specimen, a slide-scanned composite H&E image was first analyzed to annotate the carcinomatous regions. The corresponding four IHC images (HER2, EGFR, ER and CD44) were then scored according to standard-of-care scoring criteria (44–47). For IHC of HER2, a score of 0 or 1+ is considered negative, a score of 2+ is considered equivocal, and 3+ is considered positive (for a full discussion of HER2 scoring criteria, see (45)). For IHC of ER, an Allred score of 0–2 is considered negative, and an Allred score of 3–8 is considered positive. For IHC of CD44 and EGFR, positive expression requires that >1% of carcinoma cells exhibit membranous staining.
Statistical analysis
Statistical analysis was performed in Matlab or Origin. ROC curves were plotted in MedCalc. Statistical significance was calculated by a student’s t-test (two-sample, unpaired), and the level of significance was set at P < 0.001. For all of the box plots, the bottom and top of the box represent the 1st and 3rd quartiles of the dataset, respectively, and the band inside the box represents the median (2nd quartile) of the data.
Results
REMI workflow
We performed REMI of de-identified fresh tissue specimens obtained from lumpectomy and mastectomy procedures (57 specimens from 29 patients). Each specimen was topically stained with a mixture of 5 flavors of SERS NPs (4 targeted and 1 untargeted control, 150 pM/flavor) using a convection-enhanced staining method (28), followed by raster-scanned imaging and spectral demultiplexing to simultaneously quantify the expression levels of 4 biomarkers - HER2, mER, EGFR and CD44. The entire REMI procedure (staining, rinsing, imaging, spectral demultiplexing) was performed within 10–15 min depending upon the size of the specimen (see Fig. 1 for an illustration of the REMI process). This time frame is comparable to that of current intraoperative guidance techniques such as frozen-section analysis, which typically requires 15–30 min and suffers from sampling errors and freezing artifacts due to the high lipid content in breast tissues. After imaging, specimens were submitted for standard formalin-fixed paraffin-embedded (FFPE) histology with hematoxylin and eosin (H&E) staining, as well as immunohistochemistry (IHC) of the relevant protein targets. Additional details regarding the study design and biostatistics, the technical robustness of REMI, and the biomarkers assessed in this study are provided in the Supplementary Information.
REMI enables detection of various molecular subtypes of carcinoma
Representative REMI images and corresponding histology images (H&E and IHC) are shown in Fig. 2A–C from a specimen containing HER2-positive DCIS. Previously, we had demonstrated the ability to perform REMI of the HER2 receptor in fresh breast tissues by multiplexing two NP flavors (HER2-NPs and isotype-NPs) (30). By improving the tissue staining protocol and spectral resolution of our system (28), we are now able to image four biomarkers (with five multiplexed NP flavors) in the same amount of time as the previous single-biomarker imaging procedure (< 15 min). REMI of all four biomarkers exhibit good agreement with H&E and IHC data (Fig. 2B and C). REMI also enables the detection of double-positive (ER+ and HER2+) breast tumors (Supplementary Fig. S5A–C).
Figure 2. REMI enables multiplexed detection of disease-associated biomarkers.
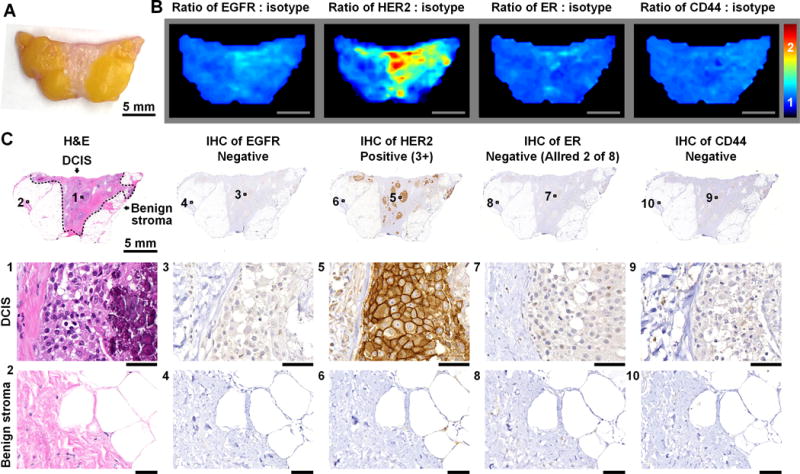
Here, an example is shown of a patient with a HER2-positive neoplasm in which REMI successfully identifies the over expression of this cell-surface biomarker. A, Photograph of a human breast specimen with DCIS. B, REMI results. Unlabeled scale bars represent 5 mm. The color bar indicates NP ratios. C, Validation data: H&E histology and immunohistochemistry (IHC). H&E histology is the clinical gold-standard method for the detection of carcinoma, and IHC is a clinical gold-standard method for the assessment of protein expression. In this example, the specimen is positive for HER2 and negative for ER, EGFR and CD44. Unlabeled scale bars represent 50 μm.
Triple-negative breast carcinomas, which express negligible levels of ER, PR and HER2, but which frequently overexpress EGFR and/or Ki-67 (47), are an aggressive breast carcinoma subtype with a high rate of recurrence. Supplementary Fig. S6A–C provides an example of REMI for a specimen with triple-negative invasive ductal carcinoma (IDC). The images in Supplementary Fig. S6B reveal elevated EGFR expression and low HER2 and ER expression, in agreement with IHC (Supplementary Fig. S6C). These results demonstrate the value of including EGFR in the REMI biomarker panel for guiding the complete removal of triple-negative breast tumors.
REMI enables discrimination between malignant and benign lesions
Intraoperative evaluation of margins is especially challenging when malignant lesions are mixed with benign lesions such as usual ductal hyperplasia (UDH). Fig. 3A–C shows two specimens from a single patient, in which there is a benign lesion (UDH) in one specimen and a malignant lesion (IDC) in the other. Postoperative IHC validation data agree with the REMI results (Fig. 3B and C). REMI shows that the expression of ER and CD44 are slightly elevated in the benign lobule and UDH regions (where moderate nuclear or cytoplasmic staining is shown in Fig. 3C), but are significantly elevated in the IDC regions (where strong nuclear or membranous staining is shown in Fig. 3C). These results show that the IDC regions and benign regions (UDH and benign lobules and stroma) can be well distinguished by setting an appropriate REMI threshold, as will be discussed later. A total of 7 patient specimens were found to contain benign lesions, including 2 cases of UDH, 1 case of fibroadenoma (Supplementary Fig. S7A–C) and 4 cases of fibrocystic changes (see Supplementary Table S1 for a summary of these benign lesions).
Figure 3. REMI enables discrimination between benign and malignant lesions.
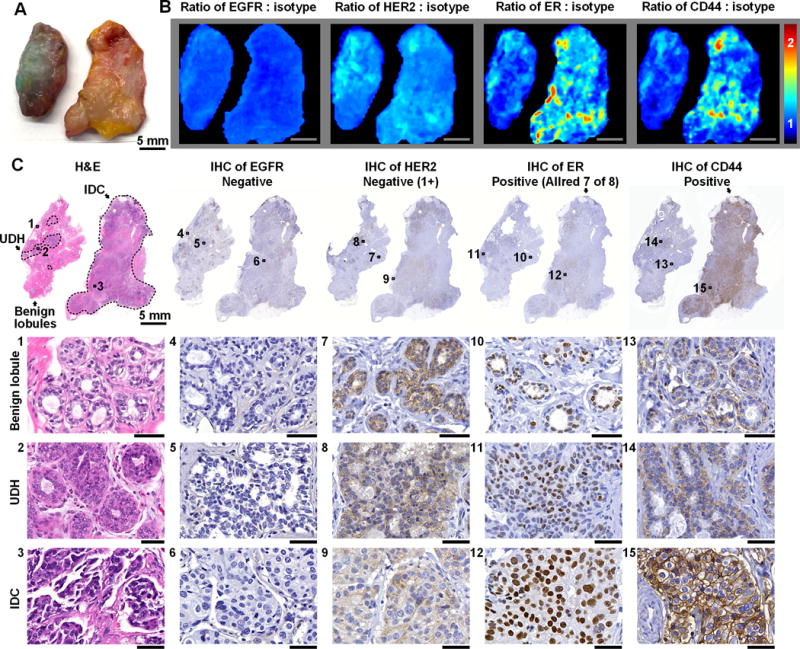
A, Photograph of two tissue specimens from a single patient. The specimen on the left presents regions of usual ductal hyperplasia (UDH, benign lesion), and the specimen on the right presents invasive ductal carcinoma (IDC, malignant lesion). B, REMI reveals that the overexpression of ER and CD44 is associated with IDC (specimen on right) but not UDH (specimen on left). Unlabeled scale bars represent 5 mm. The color bar indicates NP ratios. C, Validation data: H&E and IHC. The specimen with IDC is positive for ER and CD44, which is concordant with the REMI results. The specimen with UDH is negative for all four biomarkers. See text for details on how the IHC results are scored based on standard-of-care methods. Unlabeled scale bars represent 50 μm.
REMI enables detection of carcinoma in patients whose molecular phenotype varies over time
The molecular profile of breast carcinoma in a single patient often varies spatially and over time, such that a single targeted imaging agent may not detect the tumor at all locations within the tumor mass, or at all times (20). For example, several of the patients who were diagnosed with ER-positive carcinoma (by core needle biopsy) underwent endocrine therapy before lumpectomy. Resected specimens from these patients were imaged using REMI and submitted for histology, which showed that three of these patient’s tumors no longer expressed ER (as confirmed through repeated rounds of IHC, Supplementary Table S2). The ability to detect alternative tumor biomarkers (CD44, in the case shown in Fig. 4A–C) is essential to detect carcinoma in these situations where a previously expressed biomarker is no longer present, either due to neoadjuvant treatment or natural disease progression.
Figure 4. The multiplexed imaging of multiple biomarkers, enabled by REMI, improves detection sensitivity for malignancies with molecular phenotypes that are spatially and/or temporally varying.
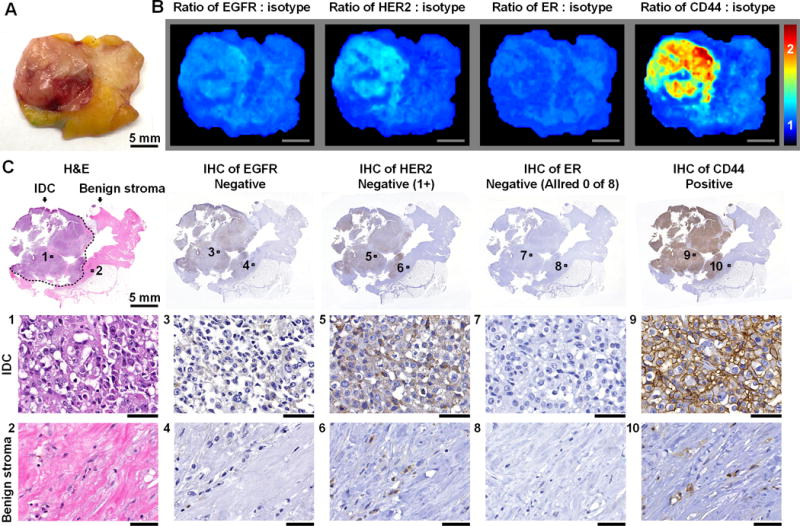
A, Photograph of a human breast specimen with IDC that was ER-positive on biopsy but ER-negative on lumpectomy, presumably as a result of neoadjuvant endocrine therapy. B, REMI results. Unlabeled scale bars represent 5 mm. The color bar indicates NP ratios. C, Validation data: H&E and IHC. The specimen is positive for CD44 and negative for HER2 (cytoplasmic staining only), ER and EGFR. See text for details on how the IHC results are scored based on standard-of-care methods. Unlabeled scale bars represent 50 μm.
Sensitivity and specificity of REMI for the detection of individual biomarkers and carcinoma
Prior to assessing the overall sensitivity and specificity of REMI for carcinoma detection, a receiver operating characteristic (ROC) analysis was first performed for the detection of biomarker overexpression (for each of the four biomarkers), with IHC as the gold standard. REMI images and corresponding IHC images were uniformly divided into larger regions of interest (ROI) of 2 mm × 2 mm (a 4 × 4-pixel bin) to improve the correlative analysis between REMI and IHC (Fig. 5A). This was done because it is technically challenging to cut an en face histology section that approximates a large tissue surface, which results in imperfect spatial co-registration between REMI images and corresponding IHC images on a pixel-to-pixel basis. Using ROIs rather than pixels, specimens, or patients, is also a standard method for quantitative analysis of digitized H&E and IHC slides (48,49). Imperfect co-registration was only found in 8 specimens that contained mixed tumor and benign regions (Fig. 5A). To ensure accurate ROC analyses, a total of 74 ROIs that contained a mixture of both tumor and benign tissues were excluded to minimize co-registration issues (i.e. only pure tumor or pure benign ROIs were used for ROC analyses). Each ROI was assigned a “gold-standard” diagnostic result of 0 (negative) or 1 (positive) for the overexpression of each cell-surface biomarker based upon the consensus interpretation of IHC slides by two pathologists, N.P.R and S.M.D (Fig. 5A). The average NP ratio from each ROI in a REMI image was calculated, and its value was compared with a threshold value to determine if the NP ratio was indicative of overexpression of a biomarker (ratio > threshold). By varying this threshold value, it was possible to construct an ROC plot that displays the tradeoff between sensitivity and specificity for detecting biomarker overexpression. Figure 5B shows the ROC curves for each biomarker (based on 2106 ROIs). REMI achieves a sensitivity of between 90.0 and 93.7% to detect the elevated expression of HER2, EGFR or CD44, when a high specificity (>90%) is enforced. A post-hoc power analysis of our ROC curve results showed that we had a power of 0.99 for detecting a significant difference from the null for HER2, EGFR, and CD44, and a power of 0.36 for ER at an alpha value of 0.05. The sensitivity for detecting mER is low (if >90% specificity is enforced) due to the limited expression of ER at cell surfaces, a topic that will be discussed in greater depth later.
Figure 5. ROC analysis for the identification of biomarker overexpression by REMI.
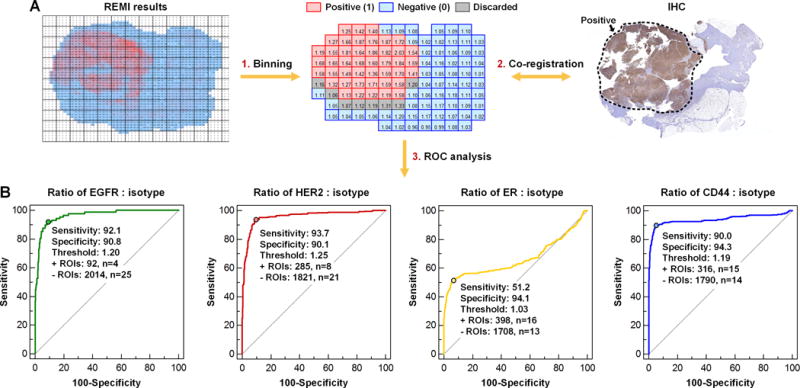
A, Co-registration of IHC and REMI data. B, ROC curves for the biomarkers EGFR, HER2, ER and CD44. The ratio of targeted NPs vs. isotype-NPs from each of 2106 ROIs were used to generate the ROC curves, in which IHC was used as the gold standard for detection of overexpressed biomarkers. For the detection of EGFR overexpression, the area under the curve (AUC) = 0.96 with a 95% confidence interval (CI) of 0.96–0.97. For the detection of HER2 overexpression, the AUC = 0.96 with a 95% CI of 0.95–0.97. For the detection of mER overexpression, the AUC = 0.67 with a 95% CI of 0.64–0.69. For the detection of CD44 overexpression, the AUC = 0.94 with a 95% CI of 0.93–0.95.
In this early-stage feasibility study, the overall sensitivity and specificity for the detection of breast carcinoma was calculated by assuming that a tissue region would be considered malignant if any one (or more) of the four candidate biomarkers was positively expressed in that region (Fig. 1). For tumor detection, H&E histology was used as a gold standard. Based on these assumptions, the overall sensitivity and specificity of REMI for breast carcinoma detection (based on ROIs) in this study was 89.3% and 92.1%, respectively (Supplementary Table S2). On a per-patient basis, REMI achieved 92.9% sensitivity (two patients had ER+ tumors misdiagnosed as benign) and 89.7% specificity (three patients had benign specimens misdiagnosed as HER2+ tumors). In the future, it may be possible to utilize machine-learning methods to more-accurately identify tumor regions based on unique biomarker-expression signatures, rather than our simple algorithm that assumes that a tissue region is malignant if any of the biomarker targets is overexpressed.
REMI is less sensitive than histology to intracellular background staining
For IHC evaluation of cell-surface receptors such as HER2 and EGFR, only membranous staining is classified as positive, whereas cytoplasmic staining is ignored (50). This is a potential source of error in IHC, where pathologists must carefully differentiate between membranous and cytoplasmic expression. Interestingly, REMI of fresh tissue is intrinsically insensitive to cytoplasmic targets (Fig. 6A) since these fresh tissue surfaces are predominantly composed of intact cells in which cell membranes are preferentially exposed to the SERS NPs (Supplementary Fig. S8). Note that the large SERS NPs (120 nm) used in our REMI approach exhibit negligible internalization through cell membranes after 5–10 minutes of topical application (28,31), and therefore do not have efficient access to intracellular proteins. This differs from thin tissue sections on a pathology slide, in which intracellular contents are preferentially exposed (and stained) but cell membranes are minimally sampled (Supplementary Fig. S8). The insensitivity of REMI to intracellular protein also explains the low sensitivity to detect ER expression (Fig. 5B), which is primarily a nuclear biomarker with weak cell-surface expression. Despite the low sensitivity of REMI to detect mER, the overall sensitivity for tumor detection is aided by the ability to image a multiplexed panel of 4 biomarkers.
Figure 6. REMI can quantify the expression of cell-surface biomarkers but is intrinsically insensitive to intracellular (e.g. cytoplasmic and nuclear) expression.
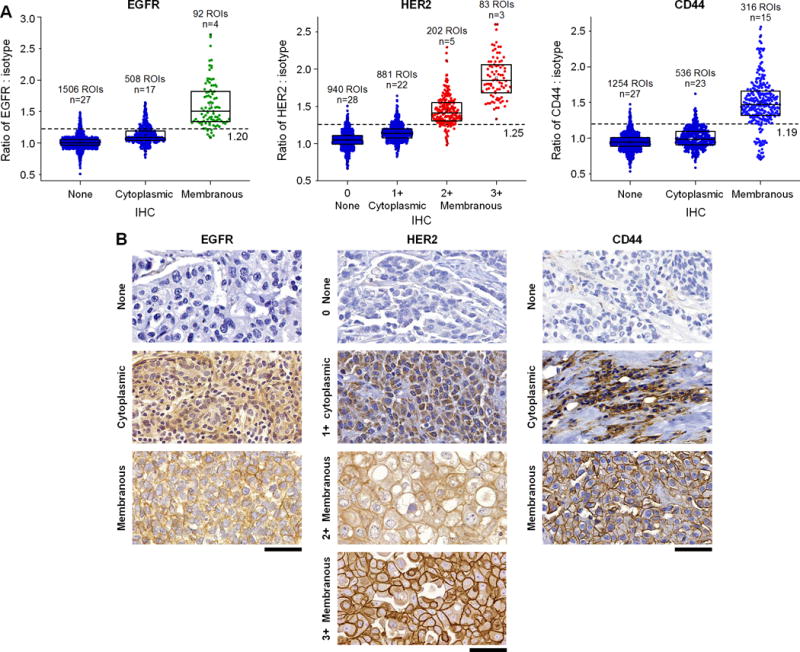
A challenging issue for pathologists, when assessing the expression of cell-surface biomarkers with IHC, is differentiating between intracellular and membranous staining and only using the latter as a basis for scoring the expression levels of cell-surface proteins. REMI stains and images fresh tissue surfaces that are composed of mostly intact cells, in which the large SERS NPs are not easily internalized. Therefore, REMI is intrinsically less sensitive to intracellular targets and provides reliable quantification of cell-surface biomarker expression, with good correlation to IHC scores (as determined by expert pathologists). A, Correlation between REMI and IHC results for cytoplasmic vs. cell-surface membrane targets (2106 ROIs in total). B, Examples of IHC images showing nonexistent or weak cytoplasmic staining (row 1), intermediate or strong cytoplasmic staining (row 2) and cell-membrane staining (rows 3 and 4). Unlabeled scale bars represent 50 μm. The box plots in panel A are all significantly different from each other (p < 0.001).
Discussion
Through imaging experiments with fresh human breast tissues, we have demonstrated that REMI allows for the rapid (<15 min) quantification of multiple biomarkers for the purposes of detecting positive margins during lumpectomy procedures (Fig. 2–4, Supplementary Fig. S5–S7). Since the molecular profiles of breast carcinoma vary greatly between patients (e.g. Fig. 2–4, Supplementary Fig. S5, S6) as well as within a single patient over time (e.g. Fig. 4), the ability to image various carcinoma biomarkers, enabled by REMI, is necessary to detect malignancies with high sensitivity (89.3%). In addition, REMI can achieve high detection specificity (92.1%), thereby allowing for the discrimination between benign and malignant lesions (e.g. UDH and IDC in Fig. 3). Note that the presence of hemoglobin (Fig. 3,4, Supplementary Fig. S6), surgical inks (Fig. 3,4, Supplementary Fig. S7) and/or fluorescent dyes do not interfere with the accurate measurement of SERS NPs due to the well-defined fingerprint spectra of SERS NPs and a robust demultiplexing algorithm (30).
As mentioned previously, a critical component of REMI is a ratiometric imaging strategy (33–35,37,38) that enables accurate and sensitive identification of biomarker overexpression without the confounding effects of nonspecific background signals (Supplementary Fig. S9A–E and S10A and B). As shown in Supplementary Fig. S9B and S10B, the imaging of the raw concentration (signal) of targeted NPs fails to differentiate between malignant and benign regions due to the misleading nonspecific accumulation of the NPs, a phenomenon that, to some degree, affects all molecular imaging approaches in which exogenous contrast agents are delivered either systemically or topically. For example, the nonspecific accumulation of NPs is heavily influenced by the mechanical properties of a tissue, such as porosity and interstitial pressure, which are often higher in benign tissues compared with dense tumors (Supplementary Fig. S9B) (31,33,51). In addition, the imaging of raw NP concentrations (signals) suffers from other confounding effects such as variations in illumination power and detector working distance (for example due to tissue-surface irregularities), while ratiometric imaging is insensitive to those effects (33). Several features make our REMI approach particularly well-suited for a ratiometric or “paired-agent” imaging strategy (35,39). (i) All of the SERS NP flavors are identical in terms of geometry and surface properties (same silica coating), which allow them to exhibit identical nonspecific behavior. (ii) Multiplexed SERS NPs are excited at a single illumination wavelength (785 nm), thereby obviating wavelength-dependent effects and ensuring that all NP flavors are illuminated identically in terms of intensity and spot size (unlike multiplexed fluorescent agents, which are often excited at disparate wavelengths).
REMI possesses a number of advantages over conventional frozen-section and FFPE histopathology, but can also complement these methods because it is nondestructive (no sectioning is necessary) and has not been shown to interfere with downstream pathology (Fig. 2–4, Supplementary Fig. S5–S7) (30,31). By comprehensively imaging an entire surgical margin surface, REMI can overcome the sampling limitations associated with slide-based histology, in which tissues are sectioned in the vertical direction at periodic intervals with minimal sampling of the tissue surfaces (i.e. the inked margin). An interesting finding is that REMI is quite insensitive to intracellular (e.g. cytoplasmic and nuclear) targets (Fig. 6A), which can often confound the quantification of cell-surface receptors via microscopic visualization of IHC slides (Fig. 6B). This is due to the fact that intracellular proteins are minimally exposed at fresh (unsectioned) tissue surfaces as compared to thin tissue sections on histology slides (Supplementary Fig. S8), as well as the relatively large size of the SERS NPs (~120 nm, Supplementary Fig. S3A), which are largely confined to the tissue surface and are not internalized by cells during the brief staining durations utilized in REMI.
There are several clear opportunities to further improve the REMI technique. Increasing the number of biomarker targets can potentially allow for improved sensitivity of REMI-based detection of molecularly heterogeneous tumors. For example, a cell-surface biomarker that could be considered in future REMI panels is MUC1, which has been reported to be expressed in up to 90% of breast tumors (52). Although a few studies have shown the feasibility of demultiplexing up to 10 flavors of untargeted SERS NPs (25,53), the ability to accurately quantify larger panels of biomarkers (>5) with targeted NPs remains to be demonstrated. As an intraoperative imaging technique, the current implementation of REMI allows for accurate detection of positive margins with sub-millimeter spatial resolution under time-constrained intraoperative conditions (imaging requires <5 min). In the future, the imaging speed and resolution of REMI can be further improved by using brighter SERS NPs (e.g. resonant Raman nanoparticles (54)) and/or advanced detection schemes such as a spectral compression technique that our group has recently described (41). Finally, REMI may potentially be combined with wide-area fresh-tissue microscopy techniques, such as structured-illumination microscopy or light-sheet microscopy (55), to allow surgeons and pathologists to visualize biomarker expression patterns in the context of their tissue microenvironment (i.e. intraoperative wide-area immunofluorescence microscopy).
Translational impact and summary
At most medical institutions, at the final stages of lumpectomy, resected tissues or biopsy shavings at the edges of the resection cavity are marked (“inked”) at the surgical margin surfaces, and are submitted for histopathology, a process that takes days. In pathology, these tissue specimens are physically sectioned at approximately 0.5-cm intervals such that each histology slide displays a cross-section of the specimen from the inked surface down towards the direction of the original tumor core. Consequently, less than 1% of the total inked margin surface is sampled using this approach. A surface-imaging approach like REMI is ideally suited for guiding lumpectomy procedures, especially because it enables comprehensive nondestructive imaging of an entire surgical margin surface without the undersampling of conventional post-operative histology. For in situ breast carcinomas, close margins (e.g. within 1- to 2-mm of the inked margin) continue to be of concern for most institutions, and REMI should be supplemented with standard-of-care post-operative pathology to identify such cases. However, it should be noted that the historical preference to re-excise patients with close margins (especially for DCIS), may have arisen because of the sampling limitations of conventional pathology, and that this safety margin may prove to be unnecessary in future outcomes-based clinical studies in which a comprehensive surface-imaging approach like REMI is utilized.
In summary, multiplexed molecular imaging, as enabled by REMI, has value for the detection of tumors that exhibit significant inter-patient and intra-patient heterogeneity. Through comprehensive molecular imaging of the surfaces of resected breast tissues, in which four biomarkers are simultaneously visualized and quantified, we have demonstrated that REMI has the feasibility to provide rapid (<15 min) detection of positive surgical margins with 89.3% sensitivity and 92.1% specificity. There is potential for REMI to guide a variety of other tumor-resection procedures in which large surface areas should be inspected for the presence of residual tumor, such as radical prostatectomies and the resection of head and neck carcinomas.
Supplementary Material
Acknowledgments
The authors acknowledge the technical assistance of the Histology and Imaging Core Facility at the University of Washington.
Financial support: The authors acknowledge support from the NIH/NIBIB R21 EB015016 (J.T.C. Liu), the NIH/NCI R21 CA215561 (J.T.C. Liu), the Department of Mechanical Engineering at the University of Washington, and the department of education GAANN fellowship program (S. Kang). The Northwest BioTrust (NWBT) is supported, in part, by the NIH (P30-CA015704).
Footnotes
Conflicts of Interest: Jonathan T.C. Liu has an issued patent: “Raman imaging devices and methods of molecular imaging.” The other authors declare no competing financial interests.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs L. Positive margins: the challenge continues for breast surgeons. Ann Surg Oncol. 2008;15(5):1271–2. doi: 10.1245/s10434-007-9766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran MS, Schnitt SJ, Giuliano AE, Harris JR, Khan SA, Horton J, et al. Society of Surgical Oncology–American Society for Radiation Oncology Consensus Guideline on Margins for Breast-Conserving Surgery With Whole-Breast Irradiation in Stages I and II Invasive Breast Cancer. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.53.3935. [DOI] [PubMed] [Google Scholar]
- 4.Jorns JM, Daignault S, Sabel MS, Wu AJ. Is intraoperative frozen section analysis of reexcision specimens of value in preventing reoperation in breast-conserving therapy? Am J Clin Pathol. 2014;142(5):601–8. doi: 10.1309/AJCPRSOA2G8RLEXY. [DOI] [PubMed] [Google Scholar]
- 5.Cendan JC, Coco D, Copeland EM., 3rd Accuracy of intraoperative frozen-section analysis of breast cancer lumpectomy-bed margins. J Am Coll Surg. 2005;201(2):194–8. doi: 10.1016/j.jamcollsurg.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Abeytunge S, Larson B, Peterson G, Morrow M, Rajadhyaksha M, Murray MP. Evaluation of breast tissue with confocal strip-mosaicking microscopy: a test approach emulating pathology-like examination. J Biomed Opt. 2017;22(3):034002–02. doi: 10.1117/1.JBO.22.3.034002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao YK, Shen D, Sheikine Y, Ahsen OO, Wang HH, Schmolze DB, et al. Assessment of breast pathologies using nonlinear microscopy. Proc Natl Acad Sci U S A. 2014;111(43):15304–09. doi: 10.1073/pnas.1416955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu H, Liu Y, Turchinovich D, Marjanovic M, Lyngsø JK, Lægsgaard J, et al. Stain-free histopathology by programmable supercontinuum pulses. Nat Photon. 2016;10(8):534–40. doi: 10.1038/nphoton.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen FT, Zysk AM, Chaney EJ, Kotynek JG, Oliphant UJ, Bellafiore FJ, et al. Intraoperative Evaluation of Breast Tumor Margins with Optical Coherence Tomography. Cancer Res. 2009;69(22):8790–96. doi: 10.1158/0008-5472.CAN-08-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assayag O, Antoine M, Sigal-Zafrani B, Riben M, Harms F, Burcheri A, et al. Large field, high resolution full-field optical coherence tomography: a pre-clinical study of human breast tissue and cancer assessment. Technol Cancer Res Treat. 2014;13(5):455–68. doi: 10.7785/tcrtexpress.2013.600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy BF, McLaughlin RA, Kennedy KM, Chin L, Wijesinghe P, Curatolo A, et al. Investigation of Optical Coherence Microelastography as a Method to Visualize Cancers in Human Breast Tissue. Cancer Res. 2015;75(16):3236–45. doi: 10.1158/0008-5472.CAN-14-3694. [DOI] [PubMed] [Google Scholar]
- 12.Sharma V, Shivalingaiah S, Peng Y, Euhus D, Gryczynski Z, Liu H. Auto-fluorescence lifetime and light reflectance spectroscopy for breast cancer diagnosis: potential tools for intraoperative margin detection. Biomed Opt Express. 2012;3(8):1825–40. doi: 10.1364/BOE.3.001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JQ, Bydlon TM, Kennedy SA, Caldwell ML, Gallagher JE, Junker M, et al. Optical spectral surveillance of breast tissue landscapes for detection of residual disease in breast tumor margins. PLoS One. 2013;8(7):e69906. doi: 10.1371/journal.pone.0069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller MD, Vargis E, de Matos Granja N, Wilson RH, Mycek MA, Kelley MC, et al. Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation. J Biomed Opt. 2011;16(7):077006. doi: 10.1117/1.3600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu CH, Zhou Y, Sun Y, Li JY, Zhou LX, Boydston-White S, et al. Resonance Raman and Raman spectroscopy for breast cancer detection. Technol Cancer Res Treat. 2013;12(4):371–82. doi: 10.7785/tcrt.2012.500325. [DOI] [PubMed] [Google Scholar]
- 16.Kong K, Rowlands CJ, Varma S, Perkins W, Leach IH, Koloydenko AA, et al. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc Natl Acad Sci U S A. 2013;110(38):15189–94. doi: 10.1073/pnas.1311289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadeghipour N, Davis SC, Tichauer KM. Generalized paired-agent kinetic model for in vivo quantification of cancer cell-surface receptors under receptor saturation conditions. Phys Med Biol. 2016;62(2):394–414. doi: 10.1088/1361-6560/62/2/394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissleder R, Ross BD, Rehemtulla A, Gambhir SS. Molecular imaging: principles and practice. People’s Medical Publishing House. 2010 [Google Scholar]
- 20.Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12(7):381–94. doi: 10.1038/nrclinonc.2015.73. [DOI] [PubMed] [Google Scholar]
- 21.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–17. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M, et al. In Situ Molecular Profiling of Breast Cancer Biomarkers with Multicolor Quantum Dots. Advanced Materials. 2007;19(20):3146–51. [Google Scholar]
- 23.Wang Y, Yan B, Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113(3):1391–428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 24.McClatchy DM, 3rd, Krishnaswamy V, Kanick SC, Elliott JT, Wells WA, Barth RJ, Jr, et al. Molecular dyes used for surgical specimen margin orientation allow for intraoperative optical assessment during breast conserving surgery. J Biomed Opt. 2015;20(4):040504. doi: 10.1117/1.JBO.20.4.040504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavaleta CL, Garai E, Liu JT, Sensarn S, Mandella MJ, Van de Sompel D, et al. A Raman-based endoscopic strategy for multiplexed molecular imaging. Proc Natl Acad Sci U S A. 2013;110(25):E2288–97. doi: 10.1073/pnas.1211309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jokerst JV, Cole AJ, Van de Sompel D, Gambhir SS. Gold nanorods for ovarian cancer detection with photoacoustic imaging and resection guidance via Raman imaging in living mice. ACS Nano. 2012;6(11):10366–77. doi: 10.1021/nn304347g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohndiek SE, Wagadarikar A, Zavaleta CL, Van de Sompel D, Garai E, Jokerst JV, et al. A small animal Raman instrument for rapid, wide-area, spectroscopic imaging. Proc Natl Acad Sci U S A. 2013;110(30):12408–13. doi: 10.1073/pnas.1301379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YW, Doerksen JD, Kang S, Walsh D, Yang Q, Hong D, et al. Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles. Small (Weinheim an der Bergstrasse, Germany) 2016;12(40):5612–21. doi: 10.1002/smll.201601829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jokerst JV, Miao Z, Zavaleta C, Cheng Z, Gambhir SS. Affibody-functionalized gold-silica nanoparticles for Raman molecular imaging of the epidermal growth factor receptor. Small (Weinheim an der Bergstrasse, Germany) 2011;7(5):625–33. doi: 10.1002/smll.201002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Kang S, Khan A, Ruttner G, Leigh SY, Murray M, et al. Quantitative molecular phenotyping with topically applied SERS nanoparticles for intraoperative guidance of breast cancer lumpectomy. Sci Rep. 2016;6:21242. doi: 10.1038/srep21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Kang S, Doerksen J, Glaser A, Liu J. Surgical Guidance via Multiplexed Molecular Imaging of Fresh Tissues Labeled with SERS-Coded Nanoparticles. IEEE J Sel Top Quantum Electron. 2016;22(4):154–64. doi: 10.1109/JSTQE.2015.2507358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YW, Kang S, Khan A, Bao PQ, Liu JTC. In vivo multiplexed molecular imaging of esophageal cancer via spectral endoscopy of topically applied SERS nanoparticles. Biomed Opt Express. 2015;6(10):3714–23. doi: 10.1364/BOE.6.003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YW, Khan A, Som M, Wang D, Chen Y, Leigh SY, et al. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology (Singap World Sci) 2014;2(2):118–32. doi: 10.1142/S2339547814500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JTC, Helms MW, Mandella MJ, Crawford JM, Kino GS, Contag CH. Quantifying Cell-Surface Biomarker Expression in Thick Tissues with Ratiometric Three-Dimensional Microscopy. Biophys J. 2009;96(6):2405–14. doi: 10.1016/j.bpj.2008.12.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tichauer KM, Samkoe KS, Gunn JR, Kanick SC, Hoopes PJ, Barth RJ, et al. Microscopic lymph node tumor burden quantified by macroscopic dual-tracer molecular imaging. Nat Med. 2014;20(11):1348–53. doi: 10.1038/nm.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YW, Khan A, Leigh SY, Wang D, Chen Y, Meza D, et al. Comprehensive spectral endoscopy of topically applied SERS nanoparticles in the rat esophagus. Biomed Opt Express. 2014;5(9):2883–95. doi: 10.1364/BOE.5.002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallia RJ, McVeigh PZ, Fisher CJ, Veilleux I, Wilson BC. Wide-field multiplexed imaging of EGFR-targeted cancers using topical application of NIR SERS nanoprobes. Nanomedicine (Lond) 2015;10(1):89–101. doi: 10.2217/nnm.14.80. [DOI] [PubMed] [Google Scholar]
- 38.Oseledchyk A, Andreou C, Wall MA, Kircher MF. Folate-Targeted Surface-Enhanced Resonance Raman Scattering Nanoprobe Ratiometry for Detection of Microscopic Ovarian Cancer. ACS Nano. 2016 doi: 10.1021/acsnano.6b06796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tichauer KM, Wang Y, Pogue BW, Liu JTC. Quantitative in vivo cell-surface receptor imaging in oncology: kinetic modeling and paired-agent principles from nuclear medicine and optical imaging. Phys Med Biol. 2015;60(14):R239. doi: 10.1088/0031-9155/60/14/R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat Meth. 2010;7(8):603–14. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 41.Kang S, Wang Y, Reder NP, Liu JTC. Multiplexed Molecular Imaging of Biomarker-Targeted SERS Nanoparticles on Fresh Tissue Specimens with Channel-Compressed Spectrometry. PLoS One. 2016;11(9):e0163473. doi: 10.1371/journal.pone.0163473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leigh SY, Som M, Liu JTC. Method for Assessing the Reliability of Molecular Diagnostics Based on Multiplexed SERS-Coded Nanoparticles. PLoS One. 2013;8(4):e62084. doi: 10.1371/journal.pone.0062084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cserni G. A model for determining the optimum histology of sentinel lymph nodes in breast cancer. J Clin Pathol. 2004;57(5):467–71. doi: 10.1136/jcp.2003.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J Clin Oncol. 2010;28(16):2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 46.Honeth G, Bendahl P-O, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, et al. The CD44+/CD24−phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):1–12. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 48.Kothari S, Phan JH, Stokes TH, Wang MD. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc. 2013;20(6):1099–108. doi: 10.1136/amiajnl-2012-001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzger GJ, Dankbar SC, Henriksen J, Rizzardi AE, Rosener NK, Schmechel SC. Development of multigene expression signature maps at the protein level from digitized immunohistochemistry slides. PLoS One. 2012;7(3):e33520. doi: 10.1371/journal.pone.0033520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor SL, Platt-Higgins A, Rudland PS, Winstanley JH, Barraclough R. Cytoplasmic staining of c-erbB-2 is not associated with the presence of detectable c-erbB-2 mRNA in breast cancer specimens. Int J Cancer. 1998;76(4):459–63. doi: 10.1002/(sici)1097-0215(19980518)76:4<459::aid-ijc2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 51.Tichauer KM, Samkoe KS, Sexton KJ, Gunn JR, Hasan T, Pogue BW. Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging. J Biomed Opt. 2012;17(6):0660011–06600110. doi: 10.1117/1.JBO.17.6.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Regimbald LH, Pilarski LM, Longenecker BM, Reddish MA, Zimmermann G, Hugh JC. The Breast Mucin MUC1 as a Novel Adhesion Ligand for Endothelial Intercellular Adhesion Molecule 1 in Breast Cancer. Cancer Res. 1996;56(18):4244–49. [PubMed] [Google Scholar]
- 53.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci U S A. 2009;106(32):13511–16. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harmsen S, Huang R, Wall MA, Karabeber H, Samii JM, Spaliviero M, et al. Surface-Enhanced Resonance Raman Scattering Nanostars for High Precision Cancer Imaging. Sci Transl Med. 2015;7(271):271ra7–71ra7. doi: 10.1126/scitranslmed.3010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Kimbrell HZ, Sholl AB, Tulman DB, Elfer KN, Schlichenmeyer TC, et al. High-Resolution Rapid Diagnostic Imaging of Whole Prostate Biopsies Using Video-Rate Fluorescence Structured Illumination Microscopy. Cancer Res. 2015;75(19):4032–41. doi: 10.1158/0008-5472.CAN-14-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


