Abstract
The human ventral visual stream consists of several areas considered processing stages essential for perception and recognition. A fundamental microanatomical feature differentiating areas is cytoarchitecture, which refers to the distribution, size, and density of cells across cortical layers. Because cytoarchitectonic structure is measured in 20-micron-thick histological slices of postmortem tissue, it is difficult to assess (a) how anatomically consistent these areas are across brains and (b) how they relate to brain parcellations obtained with prevalent neuroimaging methods, acquired at the millimeter and centimeter scale. Therefore, the goal of this study was to (a) generate a cross-validated cytoarchitectonic atlas of the human ventral visual stream on a whole brain template that is commonly used in neuroimaging studies and (b) to compare this atlas to a recently published retinotopic parcellation of visual cortex (Wang, 2014). To achieve this goal, we generated an atlas of eight cytoarchitectonic areas: four areas in the occipital lobe (hOc1-hOc4v) and four in the fusiform gyrus (FG1–FG4) and tested how alignment technique affects the accuracy of the atlas. Results show that both cortex-based alignment (CBA) and nonlinear volumetric alignment (NVA) generate an atlas with better cross-validation performance than affine volumetric alignment (AVA). Additionally, CBA outperformed NVA in 6/8 of the cytoarchitectonic areas. Finally, the comparison of the cytoarchitectonic atlas to a retinotopic atlas shows a clear correspondence between cytoarchitectonic and retinotopic areas in the ventral visual stream. The successful performance of CBA suggests a coupling between cytoarchitectonic areas and macroanatomical landmarks in the human ventral visual stream, and furthermore that this coupling can be utilized towards generating an accurate group atlas. In addition, the coupling between cytoarchitecture and retinotopy highlights the potential use of this atlas in understanding how anatomical features contribute to brain function. We make this cytoarchitectonic atlas freely available in both BrainVoyager and FreeSurfer formats. The availability of this atlas will enable future studies to link between the cytoarchitectonic organization and other parcellations of the human ventral visual stream with potential to advance the understanding of this pathway in typical and atypical populations.
Keywords: Visual Cortex, Brain Parcellation, Postmortem, Cortex-based alignment, Human Brain Atlas, Cytoarchitectonic-retinotopic correspondence
1. Introduction
The ventral visual pathway, a stretch of cortex including the ventral aspects of the occipital and temporal lobes, is a key processing stream involved in visual perception and recognition (Goodale et al., 1991; Mishkin et al., 1983; Grill-Spector & Weiner, 2014). A major neuroscientific goal is to understand the anatomical infrastructure composing this pathway. A classic microanatomical feature defining areas is cytoarchitecture – or the spatial arrangement of cell bodies and types in the six-layered cortical ribbon (Amunts and Zilles, 2015; Brodmann, 1909; Campbell, 1905; Smith, 1907; v.Economo and Koskinas, 1925), which is considered to be tightly linked to the functional properties of an area. To characterize the distribution and density across cortical layers, which is the main criterion to delineate boundaries between cytoarchitectonic brain areas, postmortem brains are stained to differentiate cell bodies of neurons and glial cells from other compartments of the cortical tissue (e.g. myelinated nerve fibers, axons, dendrites). Even though established more than a century ago, a large majority of present neuroimaging studies still relate their findings to Brodmann’s (1909) classic cytoarchitectonic parcellation.
While influential, there are several limitations to Brodmann’s and other classical approaches (Bailey and Bonin, 1951; Brodmann, 1909; Campbell, 1905; Smith, 1907; v.Economo and Koskinas, 1925) used to define brain areas based on cytoarchitectonics (Zilles and Amunts, 2010). First, classical approaches lack important information regarding the inter-subject variability of cytoarchitectonic structures. Second, the criteria used for histological distinctions were not clearly defined and researchers identified boundaries based on visual inspection of histological sections. Consequently, definitions of cytoarchitectonic boundaries depend on subjective judgments made by particular observers, leading to contention among researchers regarding the location of boundaries especially in higher sensory and association cortices. Third, the cytoarchitectonic areas that were identified with this approach were typically summarized by schematic drawings indicating their location on the brain. As such, they are often simplified. Further, as cytoarchitecture is one of several methods (Amunts et al., 2013; Glasser et al., 2016; Glasser and Van Essen, 2011; Yeo et al., 2011; Zilles and Amunts, 2009) to parcellate the brain into areas, an additional issue is that there is no precise method to project these schematics onto actual anatomical brain volumes obtained by modern magnetic resonance imaging (MRI) techniques. A common solution to project these areas onto anatomical MRIs has been to manually approximate locations of these cytoarchitectonic areas relative to cortical folding patterns (e.g. Talairach and Tournoux, 1988, Scholtens et al., 2015; van den Heuvel et al., 2015). This, however, results in considerable subjectivity and uncertainty in localizing cytoarchitectonic areas.
To overcome the problems of classical approaches, methodological advancements in the last 20 years have yielded new methods enabling more accurate delineation of cytoarchitectonic areas in the human brain (Schleicher et al., 2005, 1999, 1998). The main advantages of these modern techniques are that (1) the definitions of areal boundaries are observer-independent and statistically testable, and (2) properties of each cytoarchitectonic area are determined based on the analysis of histological structure of multiple brains. Specifically, an observer-independent algorithm identifies locations along the cortical ribbon in which the density and layering of cell bodies across the cortical depth (referred to as gray level index, GLI), show a significant change (Schleicher et al., 1999, 1998). For a cytoarchitectonic boundary to be accepted, it has to be consistently found in neighboring serial sections and across brains. Finally, cytoarchitectonic areas identified in histological sections are registered to the anatomical MRI volumes of each brain taken prior to histological processing. This not only identifies the precise location of a cytoarchitectonic area in each brain, but also corrects for shrinkage artifacts associated with histological processing (Amunts et al., 2000).
Using this approach, eight cytoarchitectonic areas have been identified in the human ventral visual stream (hOc1-hOc4v: Amunts et al., 2000; Rottschy et al., 2007, FG1–FG4: Caspers et al., 2013; Lorenz et al., 2015, see Figure 1). Additionally, a probabilistic atlas of these cytoarchitectonic areas has been incorporated into the SPM toolbox (Eickhoff et al., 2005). However, it is unknown how well these probabilistic maps predict the location of these cytoarchitectonic areas in new subjects, and whether cytoarchitectonic parcellations correspond to other parcellations of the human ventral visual stream (e.g., Glasser et al., 2016; Wang et al., 2014).
Figure 1. Eight cytoarchitectonic areas in human occipital and ventral temporal cortex.
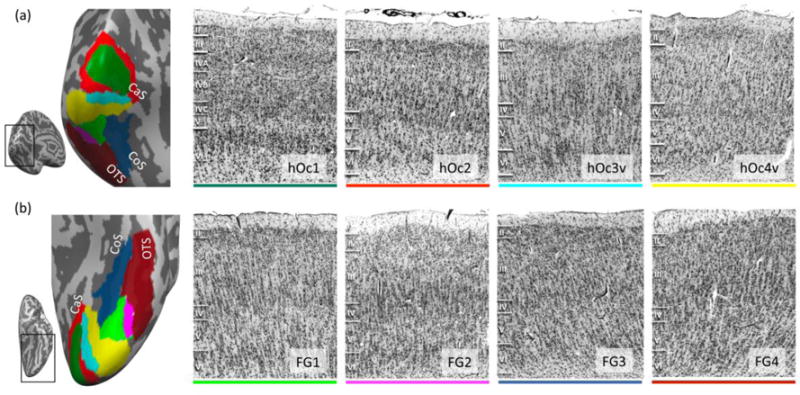
(a) Four occipital cytoarchitectonic regions of interest (cROIs). Left: occipital cROIs on an inflated cortical surface of an example postmortem brain. The surface is shown in a posterior-medial view (see inset). Color coding of cROIs corresponds to the colors on the right panels. Right 4 panels: Each panel depicts an example histological slice of a cROI. Cell bodies were stained using the Merker method (Merker, 1983) and borders were defined using a quantitative observer-independent algorithm. All sections are taken from the same brain. Note differences in the packing density of cells, the size and shape of cell bodies, and the width of cortical layers, which form the basis to distinguish cytoarchitectonic areas from one another. Roman numerals indicate cortical layer. (b) Four fusiform gyrus (FG) cROIs. Left: ventral cROIs on the same example inflated cortical surface. Right 4 panels: histological slices of FG cROIs. Acronyms: hOc1-2: human occipital cytoarchitectonic areas 1 and 2; hOc3v-4v: human occipital cytoarchitectonic ventral areas 3 and 4; FG1–4: fusiform gyrus areas 1–4; CaC: Calcarine sulcus; CoS: Collateral sulcus; OTS: Occipitotemporal sulcus.
To fill these gaps in knowledge we (1) generated a cross-validated probabilistic atlas of cytoarchitectonic areas in the human ventral visual stream extending from the occipital lobe through ventral temporal cortex (VTC) and (2) compared the cross-validated cytoarchitectonic atlas to a recently published retinotopic atlas of human visual cortex (Wang et al., 2014).
Importantly, we tested how different alignment approaches affect the accuracy of the generated atlas. Specifically, we generated atlases using three different methods: (1) an affine volumetric alignment (AVA) to the MNI305 brain, (2) a nonlinear volumetric alignment (NVA) to the Colin27 brain, and (3) cortex-based alignment (CBA), which align brains based on cortical folding patterns (Fischl et al., 1999; Frost and Goebel, 2012) to the FreeSurfer average brain, which is an average cortical surface of 39 independent living participants. We quantitatively assessed the performance of the different methods by (1) evaluating the consistency of cytoarchitectonic areas across brains and (2) quantifying how well each method predicts the location of cytoarchitectonic areas in new brains using an exhaustive leave-one-out cross-validation procedure. Recent reports find a strong coupling between cytoarchitectonic boundaries relative to macroanatomical landmarks in ventral occipital-temporal cortex (Lorenz et al., 2015; Rottschy et al., 2007; Weiner et al., 2014) and provide evidence that cortex-based alignment improves the inter-subject registration of two cytoarchitectonic areas within the human occipital lobe (Fischl et al., 2008). Thus, we hypothesized that registering brains using alignment methods that use macroanatomical features would increase the inter-subject consistency of cytoarchitectonic areas and produce a more precise group atlas of cytoarchitectonic areas of the human ventral visual stream compared to affine volumetric alignment that does not use macroanatomical landmarks.
2. Methods
2.1. Definition of cytoarchitectonic areas (cROIs)
Cytoarchitectonic regions of interest (cROIs) were defined previously from a sample of 11 postmortem (PM) adult brains (5 females; Amunts et al., 2000; Caspers et al., 2013; Lorenz et al., 2015; Rottschy et al., 2007). Each cROI was defined in 10 PM brains. hOc1-hoc4v, as well as FG1 and FG2, were defined on the same 10 brains. FG3 and FG4 were defined on 9 of these brains and in one additional PM brain, resulting in 11 brains in total. The brains were obtained from the body donor program of the Institute of Anatomy at the University of Düsseldorf. Donors had no neurological or psychiatric diseases, except one case with transitory motor disease (for details see Table 1 in Amunts et al., 2000; Caspers et al., 2013; Rottschy et al., 2007). Within 8–24 hours after death the brains were removed and fixated in 4% formalin or Bodians’s fixative for a time period of at least six months. Before the brains were histologically processed, each brain was scanned at a Siemens 1.5 T scanner (Erlangen, Germany). A high-resolution anatomical image of the whole brain was acquired, using a T1-weighted 3D-FLASH sequence (1 mm isotropic, TR = 40ms, TE = 5 ms, flip angle = 40°). Following that, the brains were embedded in paraffin, serially sectioned in oblique close to coronal sections of 20 μm and every 15th slice was stained for cell bodies (Merker, 1983). Detailed methods of histology and 3D reconstruction have been described previously (Amunts et al., 2005; Amunts et al., 1999; Zilles, Schleicher, Palomero-Gallagher, & Amunts, 2002).
2.2. Characteristics of ventral visual stream cROIs
There are four cROIs located in the occipital lobe:
Human occipital cytoarchitectonic area 1, hOc1 (Amunts, Malikovic, Mohlberg, Schormann, & Zilles, 2000) is located in the calcarine sulcus (Fig. 1a – dark green). This area closely matches functionally defined area V1 (Abdollahi et al., 2014; Hinds et al., 2009; Wohlschläger et al., 2005) and corresponds to Brodmann area 17 (Brodmann, 1909). hOc1 is characterized by a clear layered structure with prominent layer IV, which is further subdivided into sublayers, small cells in layer III and V, and a dense layer VI (Amunts et al., 2000).
Human occipital cytoarchitectonic area 2, hOc2 (Amunts et al., 2000) surrounds hOc1 both inferiorly and superiorly. It has a thinner layer IV compared to hOc1, a cell dense layer III, and shows a columnar organization (Fig. 1a - red); hOc2 matches area 18 of Brodmann (1909).
Human occipital cytoarchitectonic area 3 ventral, hOc3v (Rottschy et al., 2007) follows hOc2 ventrally and is located in the collateral sulcus, CoS. It does not have a clear border between layer II and III, it has a moderately dense and indistinctive layer IV, and shows a columnar organization (Fig. 1a - cyan).
Human occipital cytoarchitectonic area 4 ventral, hOc4v (Rottschy et al., 2007) is located on the ventral aspect of the occipital lobe, anterior to hOc3v. Compared to hOc3v, it is characterized by a dense layer II and a cell dense, prominent layer IV (Fig. 1b - yellow).
There are four cROIs in the fusiform gyrus (FG) and surrounding sulci:
FG1 is located on the posterior and medial fusiform gyrus and extends to the collateral sulcus (CoS, Caspers et al., 2013). It displays a columnar arrangement of small pyramidal cells and a thin and cell sparse layer IV (Caspers et al., 2013, Fig. 1b - light green).
FG2 is located on the posterior fusiform gyrus, lateral to the MFS, and extending to the occipito-temporal sulcus (OTS, Fig. 1b - pink). It shows large pyramidal cells in layer III, a prominent layer IV, but a less pronounced columnar organization. Additionally, FG2 is characterized by a higher cell density compared to FG1 (Caspers et al., 2013).
FG3 is located on the medial fusiform gyrus, anterior to FG1 and covers the medial aspect of the FG extending to the CoS (Lorenz et al., 2015). FG3 shows a compact and dense layer II, a prominent sub-layer IIIc with medium-sized pyramidal cells, and little clusters of granular cells in layer IV (Lorenz et al., 2015, Fig. 1b - dark blue).
FG4 is located on the lateral FG, anterior to FG2 and covering the lateral aspect of the FG, extending to the OTS (Fig. 1b - dark red). It has a less densely packed layer II, broad layer III, a thin, moderately dense layer IV, and a cell dense layer VI (Lorenz et al., 2015, Fig. 1b - dark red).
2.3. Analysis pipeline
Each of the postmortem brains was manually segmented to separate gray from white matter using ITK-SNAP (http://www.itksnap.org/pmwiki/pmwiki.php). Subsequently, each brain’s anatomical T1-weighted image, cortical segmentation, and cytoarchitectonic areas were further analyzed in BrainVoyager QX 2.8 (Brain Innovation, Maastricht, The Netherlands) if not stated otherwise. The anatomy of each postmortem brain was aligned via an affine transformation to the MNI305 template (Evans et al., 1993; Evans et al., 1992). The same transformation was then applied to each cytoarchitectonic area to bring it to the MNI305 template. The segmentations were then used to create a cortical surface reconstruction for each individual brain and each hemisphere, separately (Fig. 2). Subsequently, cytoarchitectonic areas were projected from each brain’s volume to their cortical surface reconstruction.
Figure 2. Generating a group cytoarchitectonic atlas.
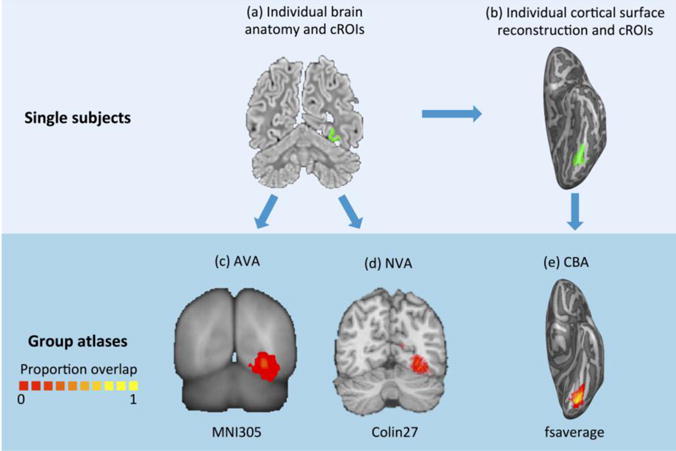
The pipeline involves the following steps: (a) At the individual subject level, cytoarchitectonic areas from histological slices are transformed to the MRI of that brain. (b) The individual cytoarchitectonic areas are projected to the cortical surface reconstruction of that brain. (a,b) show a coronal slice and cortical surface, respectively, of an individual postmortem (PM) brain and cytoarchitectonic area FG1 (green). Individual subject cROIs are transformed to a common brain template using one of 3 methods described in (c–e). In this common space, cROIs are averaged across subjects to yield a map of proportion overlapping subjects at each point in the brain (see colorbar). (c) Affine volume alignment (AVA) to the MNI 305 brain: Volume anatomies and cROIs were aligned to the MNI305 template brain. (d) Nonlinear volume alignment (NVA) to Colin 27: Volume anatomies and cROIs were warped to the Colin27 template using ANTS. (e) Cortex-based alignment (CBA) to the Freesurfer average brain (fsaverage): Cortical surfaces of PM brains and cROIs were aligned to the FreeSurfer average surface using CBA.
2.4. Comparison of different brain alignment methods and their effect on the between-subject consistency of cROIs
To investigate whether alignment of macroanatomical structures influences the correspondence of cytoarchitectonic areas across brains, we performed three different brain alignments to generate a group atlas: (1) affine volume-based alignment (AVA) to the MNI305 template (http://www.bic.mni.mcgill.ca/ServicesAtlases/MNI305), (2) nonlinear volumetric alignment (NVA) to the Colin27 brain (http://www.bic.mni.mcgill.ca/ServicesAtlases/Colin27) brain, and (3) cortex-based alignment (CBA) to the FreeSurfer average (fsaverage) brain.
2.4.1. Affine volume alignment to the MNI305 brain (AVA)
As each of the individual brain anatomies and cytoarchitectonic areas were normalized to the MNI305 template in the previous step, the cytoarchitectonic areas of all brains were already in the common coordinate system of the MNI305 anatomy.
2.4.2. Nonlinear volume alignment to the Colin27 brain (NVA)
Brains in native space were first aligned to Colin27 using an affine volume registration using AFNI tools to bring them to the same orientation and reference frame. Then, we utilized a nonlinear alignment to align each brain’s anatomy to the Colin27 brain using a nonlinear transformation implemented in ANTS (https://sourceforge.net/projects/advants/). Code used for the affine and nonlinear transformation can be found here: https://github.com/VPNL/cyto-functional. The same two-step transformation was applied to the cROIs to register them from the individual brain to the common volume of the Colin27 brain.
2.4.3. Cortex-based alignment to the FreeSurfer average brain (CBA)
Each brain’s left and right cortical surfaces were inflated into a sphere and curvature maps of gyri and sulci were created. First, each brain was rotated to best match the curvature pattern of the fsaverage (an average cortical surface of 39 independent living adult subjects) using an initial rigid alignment. The best match was established by the lowest variability in curvature between each respective brain and the target brain (Details in Frost & Goebel, 2012; Goebel, Esposito, & Formisano, 2006). Second, a non-rigid cortex-based alignment was initiated by iteratively aligning curvature maps of all PM brains to the target through vertex movements. This process is done iteratively, yielding a coarse to fine curvature alignment with four levels of anatomical detail. For each postmortem hemisphere, the sphere-to-sphere mapping file aligning the individual brain’s cortical surface to the fsaverage was saved during the alignment process. We applied this transformation to each cytoarchitectonic area to bring it into alignment with the common fsaverage surface.
To evaluate whether the template used for generating the atlas affects the accuracy of the atlas we compared CBA to the fsaverage to CBA (CBAfs) to the mean cortical surface of the postmortem brains (CBApm, see accompanying Data in Brief, DiB). Results show that performance is similar across the two templates with slight advantage to CBAfs over CBApm for hOc1 and FG3 (Fig. 1 in DiB). As the fsaverage is (1) more commonly used in fMRI analyses, (2) based on independent data (same as the other templates used for AVA and NVA), and (3) its performance is the same or superior to CBApm, all subsequent analyses in the main text use CBA to the fsaverage brain.
2.4.4. Creation of group cytoarchitectonic probability maps
We transformed each individual brain’s cytoarchitectonic areas to each of three common brain spaces: MNI305 (Fig. 2c), Colin27 (Fig. 2d), and fsaverage (Fig. 2e). A group map of each cytoarchitectonic area (cROI) was generated by averaging the binarized cROIs (1: cROI present; 0: cROI absent) of each brain in each of the common brain spaces. Thus, the value at each voxel (or vertex on the cortical surface for CBA) in the group map represents the proportion of subjects in which this voxel/vertex of the brain belongs to a given cytoarchitectonic area. For example, a value of 0 means that this voxel/vertex did not belong to that cytoarchitectonic area in any brain, a value of .5 means that it belonged to the area in half the brains, and a value of 1 indicates that it belonged to the cytoarchitectonic area in all brains.
2.4.5. Evaluating the cross-validation performance of the different alignment methods
We quantified how well the group cROI predicts cROIs in individual brains using an exhaustive leave-one-out cross-validation procedure. In each iteration, we created a group probabilistic map of each cROI based on all brains but one, and tested how well it predicted the location and extent of the cytoarchitectonic area in the left-out brain. This procedure was repeated for all combinations of left-out brains, and separately for each of the three alignment methods. We estimated the predictability of the group probabilistic cROI (G) and the left-out cROI (I) by calculating the dice coefficient (dc) between these cROIs:
The dice coefficient is a statistic used for comparing the similarity of two samples (Dice, 1945; Sørensen, 1948). A dice coefficient of zero indicates no predictability and a dice coefficient of 1 indicates perfect predictability. We applied different threshold levels to the group probabilistic cROI (G) to predict the location of the left-out-brain.
To perform statistical analyses comparing dice coefficients across alignment methods, we used two different thresholds, as thresholds are non-independent from each other: (1) unthresholded data and (2) the threshold level that generated the highest predictability across alignments and cROIs. To determine the latter, we averaged dice coefficient values across alignment methods, hemispheres and cROIs, resulting in one dice coefficient per threshold level. Comparison across thresholds revealed that the threshold of .33 produced the highest values. Therefore, this threshold was used for the comparison.
To evaluate whether there were statistically significant differences in cross-validation performance across alignment methods and hemispheres we used a nonparametric version of the analysis of variance, the Friedman test (Upton and Cook, 2008) as our data is not normally distributed. Once establishing significant differences across alignment methods, we performed pairwise comparisons between methods using paired permutation tests within each cROI. We tested all three possible pairwise comparisons: AVA vs. NVA, AVA vs. CBA, and NVA vs. CBA. To perform the paired permutation test, we generated a null distribution for which alignment labels were randomly shuffled 10,000 times. Using this null distribution, we estimated a p-value indicating the probability that the measured dice coefficients were derived from this null distribution. Statistical significance was Bonferroni corrected to adjust for the increased Type I error rate due to multiple comparisons.
2.4.5.1 Evaluating whether cROI size affects cross-validation performance
The probability that a cROI overlaps across subjects and its predictability may be related to the size of the cROI. To test the effect of area size on the dice coefficient we determined if there is a correlation between the cross-validated dice coefficient and average cROI size. We also estimated the chance level dice coefficient for each cROI size. To do so we generated synthetic data of cortical disks matched for each cROI size, randomly placed them within the anatomical mask of the ventral part of occipital cortex and ventral temporal cortex (Fig. 4, right inset), and estimated the chance level dice coefficient given this area size for each threshold level. For each iteration, we randomly placed 8 discs (as for the cross-validated data) to generate the group probability map, and one additional random disk that simulated the left-out subject. The dice coefficient was computed the same way as for the actual data. We ran 10000 iterations per simulated cROI size and calculated the chance dice coefficient for each threshold level.
Figure 4. Exhaustive leave-one-out cross-validation performance of three alignment methods.
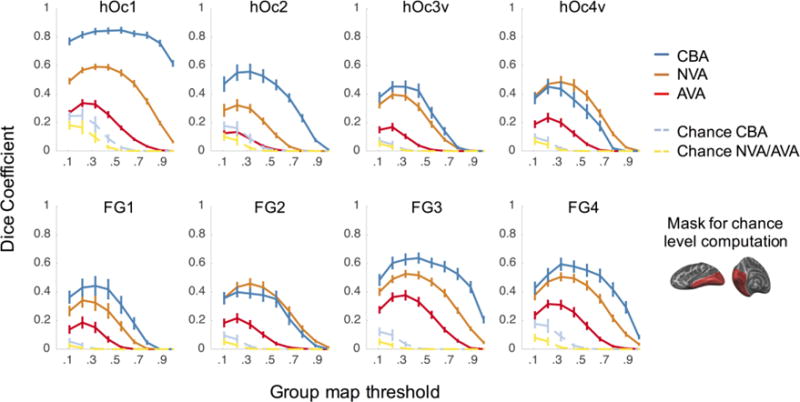
Each panel shows the cross-validated dice coefficient for each cROI as a function of the threshold applied to the group cROI map. The threshold determines the minimal proportion of overlapping brains at the voxel/vertex level included in the group cROI. A dice coefficient of 0 indicates no predictability and a dice coefficient of 1 indicates perfect predictability. Blue (CBA): cortex-based alignment (CBA) to the fsaverage brain. Orange (NVA): nonlinear volumetric alignment (NVA) to the Colin27 brain. Red (AVA): affine volumetric alignment (AVA) to the MNI305 template. Light blue: chance level for CBA. Yellow: chance level for volumetric alignments (AVA and NVA). Chance level was estimated by calculating the dice coefficient of randomly placed disk-shaped cROIs, matched in size to the average size of each cROI within the ventral part of occipital and ventral temporal cortex (right inset). Chance level was estimated for 10,000 iterations per cROI. Error bars: Standard error (SE) across the 10-fold cross-validation.
Chance level was estimated separately for cortex-based alignment (CBA) and volume based alignment (AVA and NVA) for several reasons: (1) the number of voxels in the volume is greater than on the cortical surface, as the volume, but not the surface, has a depth component, (2) the spatial variability of cROIs on the cortical surface is limited to the 2 dimensions of the cortical surface but in the volume they can vary in 3 dimensions, and (3) the resolution of the standard cortical surface in BrainVoyager is coarser than the resolution of the volume. Results of these simulations are shown in the dashed yellow (volume) and light blue (surface) lines in Fig. 4. To establish whether the measured dice coefficients were significantly higher than the empirical chance level we used permutation testing, comparing AVA and NVA to volume chance level (Fig. 4 – dashed yellow), and CBA to surface chance level (Fig. 4 – dashed light blue).
2.5. Creating a ventral visual stream cytoarchitectonic atlas
After comparing alignment methods, we used cortex-based alignment method to generate a human ventral visual stream cytoarchitectonic atlas.
2.5.1. Maximum-probability map (MPM) of cROIs
The probability maps generated above determine the probability that each voxel/vertex belongs to a given cROI. However, it is possible that a point on the brain may belong to more than one probabilistic cROI. This overlap is more likely to occur along boundaries of neighboring cytoarchitectonic areas. In order to assign a unique cytoarchitectonic label to each vertex in the atlas, we generated a maximum-probability map (MPM) of each area (Eickhoff et al., 2005). We used the probabilistic cROIs generated with CBA to the FreeSurfer average (fsaverage) brain, as this method yielded on average the highest dice coefficients (Fig. 4) and the fsaverage brain is widely used in the neuroimaging community (Benson et al., 2014, 2012; Glasser et al., 2016; Hinds et al., 2009; Wang et al., 2014).
Using probabilistic cROIs generated with CBA we determined which vertices were shared by more than one probabilistic cROI and assigned these vertices to a single cROI based on the group cROI that contained the highest probability at that vertex (Eickhoff et al., 2005). In cases where one vertex had the same probability across multiple cROIs, we used the probability of the neighboring vertices neighborhood to make the decision. The neighborhood was determined iteratively starting from the nearest neighbors. In brief, for vertices that were assigned to more than one cROI, we first calculated the average probability of each cROI across the vertex and its nearest neighbors. The vertex was then assigned to the cROI that yielded the highest probability. If the neighborhood probability was still identical across cROIs, the neighborhood was increased by the next set of nearest neighbors and the neighborhood probability was evaluated again. This process was repeated iteratively until the probability for one cROI was greater, or the neighborhood size exceeded the size of the cROI (a case that did not happen in this dataset). As a last we removed speckles from the MPM map, by searching for vertices that did not have any neighbors. Vertices that did not have at least one 3rd degree neighbor were either (a) deleted if they did not belong to any probability map before overlap removal, or (b) were assigned to the cROI with the 2nd highest probability if it was surrounded by neighbors of that cROI.
This process creates an MPM of each cytoarchitectonic area that eliminates vertices that are shared across cROIs at the group level, thus generating a unique, non-overlapping tiling of the ventral visual stream by the eight cytoarchitectonic areas (Fig. 6). MPM atlases were created in both BrainVoyager and FreeSurfer formats and can be downloaded here: http://vpnl.stanford.edu/vcAtlas.
Figure 6. Maximum probability map (MPM) of human ventral visual stream cytoarchitectonic areas.
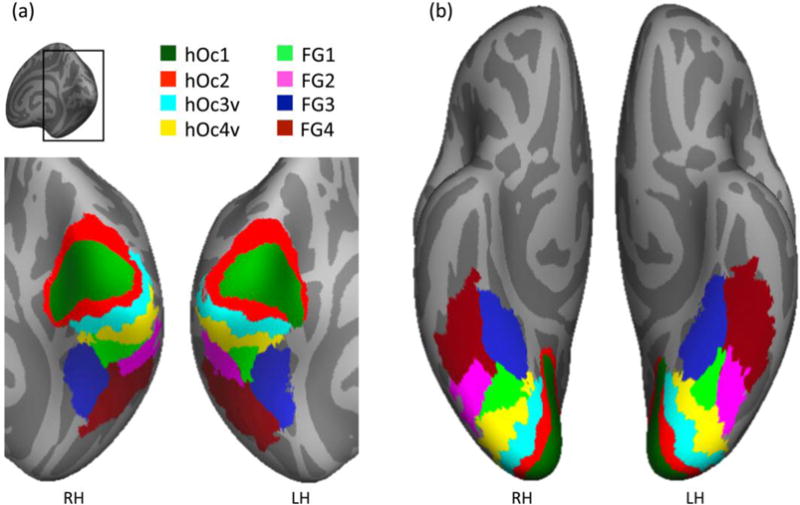
The MPM was generated after CBA to the fsaverage brain and is shown on the fsaverage cortical surface. (a) Posterior view zoomed on the occipital cortex. (b) Ventral view. RH: right hemisphere; LH: left hemisphere.
2.6. Determining the relationship between cytoarchitectonic and retinotopic parcellations of human ventral visual cortex
One utility of this cytoarchitectonic atlas is that it can be compared to other atlases and parcellations of human visual cortex. Here, we compared our atlas to a functional atlas of retinotopic visual areas (Wang et al., 2014). We compared our cROI atlas to the retinotopic atlas for the following reasons: (1) cytoarchitectonic and retinotopic atlases are based on independent methods, cytoarchitecture and visual field topography, (2) both of these atlases are based on key features scientists use to parcellate the brain (Felleman and Van Essen, 1991), (3) retinotopy is the single most commonly agreed method used to define areas in visual cortex (Wandell and Winawer, 2015, 2011) and (4) both atlases are aligned to the fsaverage brain using CBA, thus we compared different data that were aligned using the same algorithm as well as common brain template.
To test the correspondence between cROIs and retinotopic ROIs, we calculated the percentage overlap between each of the group fROIs and each of the individual PM cROIs aligned to the fsaverage brain. This resulted in 10 values per fROI-cROI combination. We performed this analysis for all retinotopic visual areas in the Wang et al. (2014) atlas that were in occipital and ventral temporal cortex: V1 (combination of their V1d and V1v), V2d, V2v, V3v, VO1, VO2, PHC1, PHC2.
Additionally, we computed the chance level overlap between each fROI-cROIs to test if the overlap between a retinotopic area and cROI is significantly different from chance. We estimated the chance level by calculating the average percentage overlap between a given retinotopic ROI and a randomly chosen cROI from a randomly chosen PM brain. This procedure was repeated 1000 times with replacements. We used a permutation test to evaluate whether the reported percentage overlap between retinotopic fROIs and cROIs were above chance. Those comparisons were Bonferroni corrected for each fROI by a factor of 7 (8 cROIs per fROI). We also used a Friedman’s test (Upton and Cook, 2008) to test for hemispheric differences.
3. Results
3.1. Cortex-based alignment improves the precision of group cROI maps compared to volumetric alignments
Using data from 10 postmortem brains, we generated a group map of 8 cytoarchitectonic ventral visual stream areas using three methods: (1) affine volumetric alignment (AVA) to the MNI305 template, (2) nonlinear alignment to the Colin27 template (NVA), and (3) cortex-based alignment to the fsaverage atlas brain (CBA, see Methods and Figure 2 for methodological details). Figure 3 illustrates resulting group maps derived by each of these methods for an example cytoarchitectonic area in occipital cortex (hOc2) and an example area in ventral temporal cortex (FG1).
Figure 3. Comparison between single brain and group cytoarchitectonic maps generated by the three alignment methods.
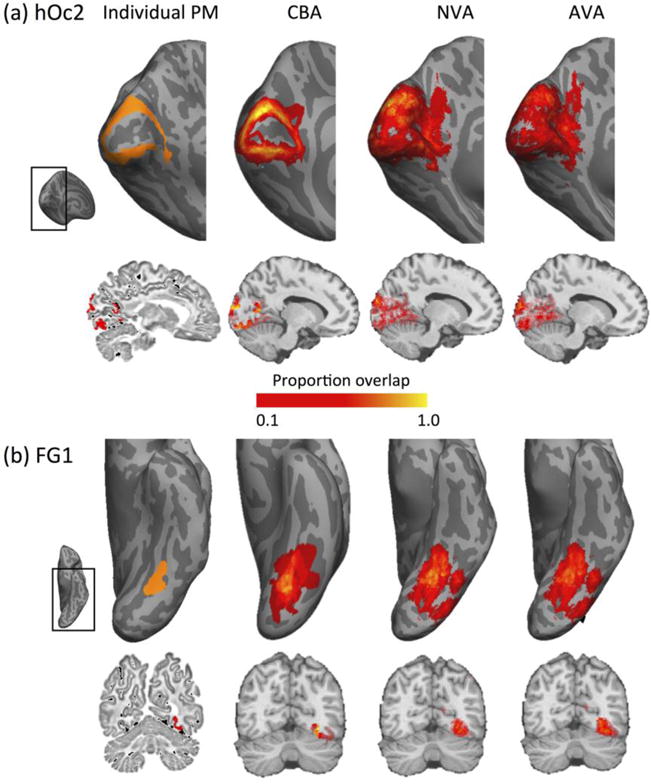
(a) Comparison of individual brain and group human occipital cytoarchitectonic area two, hOc2. Top row: inflated cortical surface view. Left inset: Anatomical region of the brain zoomed in the respective panels in each row. Left: Representative hOc2 shown in orange in an example inflated surface of a postmortem brain. Three panels on the right: Group cytoarchitectonic region of interest (cROI) of hOc2 for each of three alignment methods shown on an inflated cortical surface. CBA: Cortex-based alignment to the FreeSurfer average (fsaverage) cortical surface from 39 independent brains. NVA: Nonlinear alignment to the Colin27 template. AVA: Affine volumetric alignment to the MNI305 template, displayed on Colin27. Bottom row: same data displayed in a sagittal brain slice. The individual postmortem cROI is displayed in the native brain space, group maps are displayed on Colin27. (b) Same as (a) but for cytoarchitectonic area fusiform gyrus 1 (FG1). Color bar: proportion of brains containing the cROI.
Qualitative examination of these maps highlights two observations. First, the location and shape of the group cROI generated by CBA largely resembles the individual brain example for CBA, but the resemblance is poorer for the group cROIs generated by AVA and NVA (Fig. 3). For example, in the representative brain hoC2 illustrates a ringlike arrangement surrounding the calcarine sulcus (Fig. 3a - top left). Likewise, the group hOc2 generated by CBA illustrates a similar arrangement surrounding the calcarine in the respective common cortical surface. In contrast, AVA and NVA generate a group hOc2 that does not illustrate a ring shape around the calcarine sulcus, but instead, almost entirely encompass the calcarine sulcus. The latter alignments are clearly erroneous, as it is well established that the calcarine sulcus is the locus of the first visual occipital area hOc1, which has distinct anatomical (Amunts et al., 2000; Brodmann, 1909; Hinds et al., 2011; v.Economo and Koskinas, 1925) and functional characteristics (Engel et al., 1994; Holmes, 1945; Sereno et al., 1995).
Likewise, both NVA and AVA generate a group cROI of hOc2 that extends into white matter (Fig. 3b, brain slices). This is erroneous as cytoarchitecture, and consequently cROIs, are only measured and exist in the gray matter. Second, our results show that CBA produces a higher between-subject consistency of these two example areas as compared to AVA and NVA. This is evident by a larger number of vertices showing a higher proportion of overlap in the group cROI in the CBA alignment as compared to the volume alignments (Fig. 3, yellow vertices).
To quantitatively determine which method produces group cROIs with the highest predictability, we implemented a 10-fold leave-one-out cross-validation procedure for each of the three different alignment methods across a range of thresholds. Then we used the dice coefficient to evaluate how well the group cROI predicts the cROI in the left-out brain. Assessment of hemispheric difference revealed no difference (Chi-sq(1,23) = 2.67, P = .10). Therefore, we present data averaged across hemispheres. Across cROIs and thresholds, both CBA to the fsaverage brain and NVA to the Colin27 brain more accurately predicted the left-out cROI compared to AVA to the MNI305 brain (Fig. 4). Comparison between CBA and NVA revealed that except for two ROIs (hOc4v and FG2), CBA outperformed NVA (compare blue and orange curves in Fig. 4). Consistent with our qualitative observations, the dice coefficient is close to zero for group cROIs generated with AVA at thresholds > 0.5, due to the low number of voxels that pass these thresholds in the group cROI. In contrast, there is a substantial number of vertices that are consistent across more than half of the brains in group cROIs generated from either NVA or CBA.
We determined if differences in performance across alignment methods are statistically significant for two cases: (1) unthresholded maps and (2) the threshold level that yielded consistent successful performance across cROIs and alignment methods. The highest leave-one-out cross-validation performance across methods and cROIs was 0.40 which was observed for two thresholds: 0.22 and 0.33. We chose the threshold of 0.33 (excluding vertices shared by less than three brains) as it is a more conservative threshold which lets us compare methods across a range of criteria.
Results show that the dice coefficient significantly differed across alignment methods (significant main effect of alignment method for both unthresholded: Chisq(2,30) = 27.12, P<.001, and thresholded data: Chi-sq(2,30) = 27.12, P<.001). Posthoc pairwise permutation tests showed that differences between methods were significant within specific cROIs. In all cROIs, both CBA and NVA produced significantly better performance than AVA for both unthresholded and .33 thresholded data (Ps < .05), except for one case (FG3 for AVA vs. NVA and unthresholded data, P = .33). Permutation tests comparing CBA and NVA performance showed that for unthresholded maps CBA outperformed NVA in 6 out of 8 cROIs (except for FG2: P = .21, hOc4v: P = .09; others P < .001, Fig. 4). For data thresholded at 0.33, CBA outperformed NVA for 5 out of 8 cROIs (hOc1, hOc2, FG1, FG3, FG4: P < .05), while NVA outperformed CBA for 2 cROIs, hOc4v and FG2 (Ps < .05) and there were no significant differences across CBA and NVA for hOc3v (P = .38).
The dice coefficient analysis also shows differences across cytoarchitectonic areas. Specifically, across thresholds and methods, the dice coefficient was highest for hOc1 compared to other ventral visual stream cytoarchitectonic areas. As hOc1 is also the largest area (surface area sizes mean±SD: hOc1: 2,292±689 [mm2], hOc2: 2,115±953 [mm2], hOc3v: 1,172±490 [mm2], hOc4v: 898±322 [mm2], FG1: 529±201 [mm2], FG2: 620±238 [mm2], FG3: 954±321 [mm2], FG4: 1,222±448 [mm2]), we examined if the size of the area affects predictability in two complementary analyses. First, we calculated whether there is a significant correlation between the average area size and the dice coefficient. Correlating the dice coefficient obtained with CBA at a threshold of .33 revealed a significant and positive correlation between the size of the cytoarchitectonic area and the dice coefficient (r = .83, p<.05). That is, larger cROIs have larger dice coefficients than smaller cROIs. As the dice coefficient was correlated with cROI size, we estimated the chance level dice coefficient for each cROI size and tested if the dice coefficient obtained by the various methods was higher than chance (see Methods). We reasoned that if cROI size determines the dice coefficient, then the measured dice coefficients should be similar to chance. However, if cROI size does not solely determine the dice coefficient, the measured dice coefficient should be larger than chance. Results show that the chance level dice coefficient varied with both cROI size and thresholds (Fig. 4 – yellow and light blue dashed lines). Critically, however, dice coefficients obtained by both CBA and NVA were significantly higher than chance level (permutation tests: Ps < .005 for unthresholded; Ps < .005 thresholded at .33). Even the poorest alignment (AVA) performed consistently better than chance (unthresholded: hOc2: P < .05, rest Ps < .005, thresholded at .33: Ps < .005).
3.2. A cytoarchitectonic atlas of human ventral visual stream cROIs
We next created a group probability map for each of the eight ventral cytoarchitectonic areas based on CBA to the fsaverage atlas brain which allows visualization of the cROIs on a common brain space (Fig. 5) and comparisons to other atlases. We chose this approach because (1) In general, CBA outperformed both affine and nonlinear volume based alignments, and (2) the fsaverage brain is a common brain space that has been widely used to generate atlases (Benson et al., 2014, 2012; Glasser et al., 2016; Hinds et al., 2009; Wang et al., 2014), which allows comparison between this cytoarchitectonic atlases to other atlases in the field.
Figure 5. Probabilistic group maps for each of the ventral visual stream cytoarchitectonic areas.
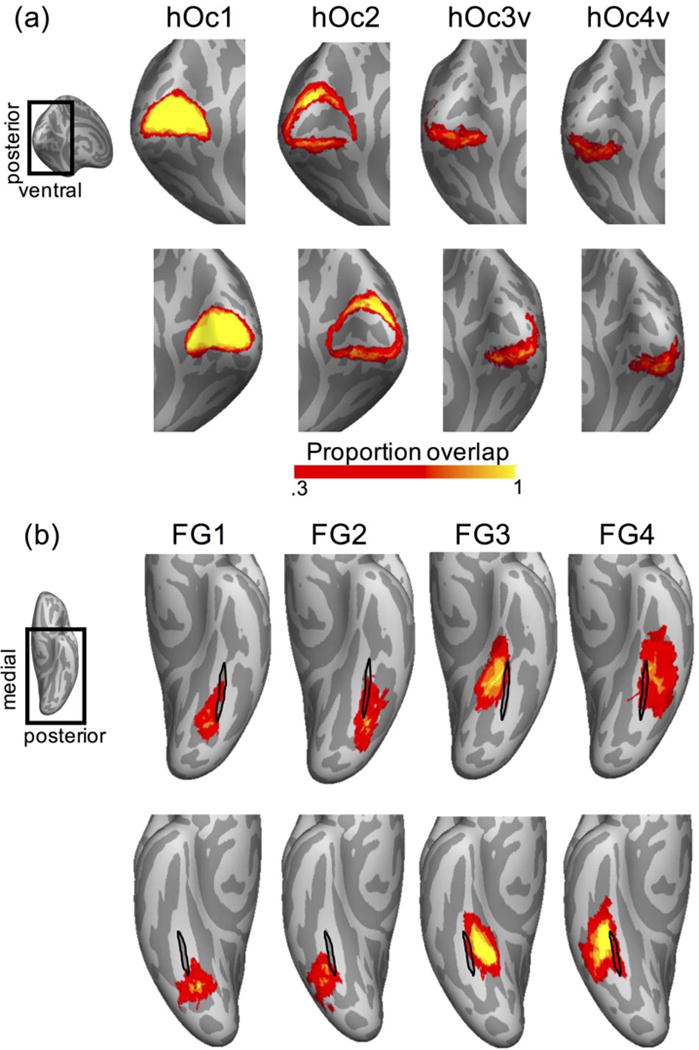
Maps are based on the CBA alignment to the fsaverage cortical surface thresholded at 0.33 and displayed on the inflated cortical surface of the fsaverage brain. Color bar: proportion of overlapping brains at each vertex. (a) Occipital group cROIs: hOc1-hOc4v. (b) Fusiform gyrus group cROIs: FG1-FG4. Black outline: mid-fusiform sulcus (MFS) of the fsaverage brain. Insets: the magnified brain area in the corresponding panels.
Consistent with the results of the dice coefficient analysis and prior results (Fischl et al., 2008), visualization of the cROI atlas shows that hOc1 shows the highest consistency across PM brains (Fig. 5). Between-subject consistency is somewhat lower in subsequent areas. However, consistency across brains does not further decline across the ventral visual steam processing hierarchy. In fact, between-subject consistency is as high in areas FG3 and FG4 on the fusiform gyrus, as it is in hOc2 (Fig. 5).
As a final step, we generated a maximum probability map (MPM) of human ventral visual stream cROIs. Different than the probabilistic maps in Fig. 5, which illustrate the probability of each vertex belonging to each cytoarchitectonic area, the MPM assigns each vertex to a unique cytoarchitectonic area. The vertex is assigned to a cytoarchitectonic area which shows the highest overlap that is greater than .33 (see Methods). Consequently, this generates a discrete tiling of the human ventral visual stream by cytoarchitectonic areas such that there is a unique labeling of each vertex on the cortical surface (Fig. 6). We make the MPM atlas publically available in both BrainVoyager and FreeSurfer file formats (http://vpnl.stanford.edu/vcAtlas).
It is interesting that the cytoarchitectonic organization of the MPM atlas of the ventral visual stream preserves the relationship between cytoarchitectonic areas and cortical folding reported in single brains. First, the spatial layout of the MPM based on group data mirrors the spatial layout of the cytoarchitectonic tiling in individual brains (compare Figures 6 and 1). Second, as in the individual brains (Lorenz et al., 2015; Weiner et al., 2014), the MFS of the fsaverage brain serves as a boundary between FG1 and FG2 as well as between FG3 and FG4.
3.3 The relationship between retinotopic regions and cytoarchitecture
The present cROI atlas of the human ventral visual stream generated on the cortical surface can be compared to other anatomical and functional parcellations of the ventral stream derived from other anatomical or functional metrics. As retinotopy is a key feature used to define visual areas (Wandell and Winawer, 2015, 2011) and there is a group atlas of retinotopic visual areas aligned to the fsaverage brain (Wang et al., 2014), we sought to compare the two atlases in order to assess how cytoarchitectonic and retinotopic areas align in the human ventral visual stream. To quantify their correspondence, we computed the percentage overlap between each retinotopic ROI and each cytoarchitectonic ROI (Fig. 7). As there were no significant differences across hemispheres (Friedman’s test: chi-sq(1,71) = 1.09, P = .30), we report data averaged across hemispheres.
Figure 7. Coupling between cytoarchitectonic and retinotopic parcellations of the human occipito-temporal and ventral temporal cortex.
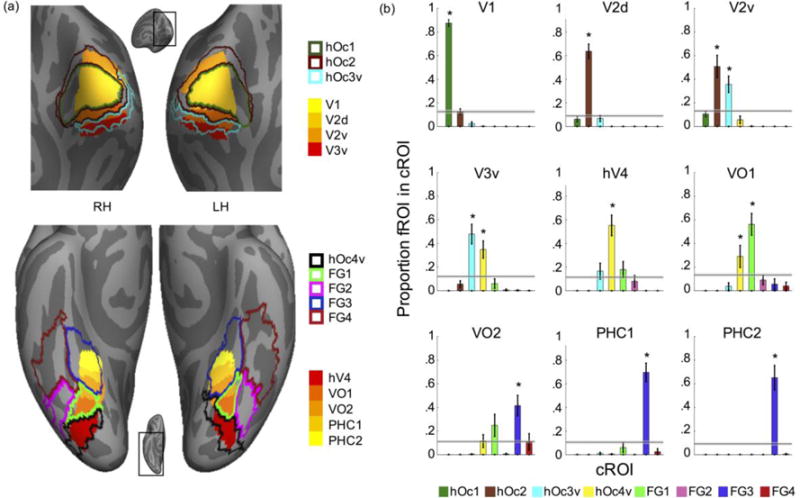
(a) Superposition of our cROI MPM atlas (Fig. 6) and the Wang (2014) retinotopic MPM atlas on the fsaverage brain. Top: Occipital view of the fsaverage brain, inset shows zoomed region of the brain displayed. Bottom: Ventral view of the fsaverage brain; Solid colors: retinotopic ROIs; Outlines: cROIs; (b) Quantification of correspondence between retinotopic ROIs and cROIs. Each graph shows the average overlap between an MPM of a retinotopic area and individual subject’s cROI. X-axis: cROIs, y-axis: proportion of retinotopic ROI contained in each cROI. Horizontal bars: Chance level with 95% confidence interval. Errorbars: Standard error across postmortem subjects. Asterisks indicate significant difference relative to chance (P<.01).
Consistent with prior results (Fischl et al., 2008), these analyses reveal that the strongest coupling between cytoarchitectonic and retinotopic areas occurs within striate compared to extrastriate cortex. Specifically, the cytoarchitectonic-retinotopic coupling within striate cortex (V1-hOc1 coupling=88±3%, significantly greater than chance, permutation testing, P<.001 Fig. 7b, top left) exceeds that among the second, third, and fourth visual areas in extrastriate cortex (V2d-hOc2 coupling=64±6%; V2v-hOc2 coupling=51±10%; V3v-hOc3V coupling=48±9%; hV4-hOc4v coupling=55±9%, all Ps<.001, permutation testing, Fig. 7b).
Crucially, cytoarchitectonic – retinotopic coupling does not further decline in the temporal lobe relative to the occipital lobe: VO1 largely aligns with FG1 (56±9%), while PHC1 and PHC2 are largely contained within FG3 (69±8%, 65±10% respectively). Nevertheless, these analyses also reveal that while occipital cROIs are largely functionally homogenous, cROIs in the ventral temporal cortex can be functionally heterogeneous. Specifically, retinotopically-defined VO2, PHC1, and PHC2 are all located within cytoarchitectonically-defined FG3 (Fig. 7a-bottom, this coupling is significantly different than chance Ps < .001). Together these analyses precisely quantify the complex correspondence between cytoarchitectonic and retinotopic parcellations of the human visual cortex.
4. Discussion
In the present study, we generated a cross-validated cytoarchitectonic atlas of the human ventral visual stream. We compared cortex-based alignment (CBA), nonlinear volumetric alignment (NVA) and affine volumetric alignment (AVA). We found that CBA and NVA generated better inter-subject consistency of group cROIs and a higher predictability as compared to an affine volumetric transformation to the MNI305 template. Furthermore, we found that CBA was superior to NVA for six of the eight cytoarchitectonic areas. Below we discuss the implications of these results for generating cytoarchitectonic atlases as well as the potential utility of this atlas in future research.
4.1. The relationship among cytoarchitectonic areas and cortical folding across the ventral visual processing hierarchy
The present study shows that there is improved between-subject correspondence and predictability of ventral visual stream cytoarchitectonic areas with cortex-based alignment and nonlinear volumetric alignment compared to an affine volumetric alignment that does not take macroanatomy into account. This improved alignment, notable also in higher-order visual cortices, may seem surprising since classic (Brodmann, 1909; Smith, 1907) and modern (Van Essen and Drury, 1997; Zilles et al., 2013) studies provide evidence for significant variability between cytoarchitectonic boundaries and cortical folding in several cortical areas. Specifically, these studies showed that there is often not a 1:1 mapping between either the crown of a gyrus and a cytoarchitectonic boundary or the fundus of a sulcus and a cytoarchitectonic boundary (Amunts et al., 2007). Although cytoarchitectonic boundaries do not perfectly follow gyral crowns or sulcal fundi, accumulating evidence suggests that there is a gross coupling between cytoarchitectonic areas and macroanatomical structures. For example, area hOc3v is likely to be found in the collateral sulcus, not the fusiform gyrus, whereas the reverse is true for area FG4 (Lorenz et al., 2015; Rottschy et al., 2007). Additionally, there is even more fine-grained coupling as some cytoarchitectonic boundaries tend to align with specific macroanatomical features with millimeter precision. For example, the cytoarchitectonic boundary between areas FG1 and FG2 occurs within the posterior mid-fusiform sulcus (MFS, Weiner et al., 2014; Weiner and Zilles, 2016) and the boundary between areas FG3 and FG4 occurs within more anterior portions of the MFS (Lorenz et al., 2015). We speculate that the combination of the general, gross, topological relationship between macroanatomical structures and cytoarchitectonic areas, as well as the more fine-grained (but infrequent) relationships, contribute to the improved between-subject correspondence and predictability of cytoarchitectonic areas with macroanatomical alignments compared to AVA, which ignores these features.
While our results illustrate that the highest cytoarchitectonic-macroanatomical coupling occurs for area hOc1, as reported previously (Fischl et al., 2008), it is interesting that after hOc2 the level of cytoarchitectonic-macroanatomical coupling does not further decrease across the ventral visual stream hierarchy. Rather, the consistency of the cROI group maps across brains for other intermediate and high-level cROIs – hOc3v, hOc4v, FG1 and FG2 – is similar to the hOc2 level and even slightly increases for FG3 and FG4, which are located in higher stages of the ventral processing stream.
Despite this consistency of the group cytoarchitectonic maps relative to cortical folding, there are also two main sources of variability. First, the precise boundaries of a given cytoarchitectonic area can vary across different brains (Amunts et al., 2007; Van Essen and Drury, 1997; Zilles et al., 2013). For example, the transition between FG1 and FG2 as described above, as well as the transition between FG3 and FG4 are located at the MFS. However, the boundary between FG3 and FG4 is better predictable by the MFS than the FG1 and FG2 boundary (Lorenz et al., 2015). Second, macroanatomical landmarks vary in their morphological features. For example, the MFS is variable in length, and shows higher variability in its posterior than anterior extent (Weiner et al., 2014). That results in higher consistency in cROIs tightly linked to the anterior tip of the MFS, as FG3 and FG4, compared to FG1 and FG2, which are located more posteriorly. Another important characteristic of the morphology of the brain is that macroanatomical landmarks vary in the extent they are fractionated. As an example, the occipito-temporal sulcus (OTS) can vary between having one continuous sulcal bed or having several fractionated components. This variability can occur even across hemispheres in the same brain. The greater the variability of a given landmark across brains, the more noise is introduced during CBA. Future research can examine anatomical and cytoarchitectonic variability, as well as the coupling between these features across the brain more generally, to test if these features are unique to the ventral visual stream or reflect general principles of brain architecture.
4.2. Comparison between the applied alignment methods: implications
Volume-based atlases integrate both brain areas along the cortical ribbon and areas within nuclei located deep in the brain. In generating volumetric atlases, our analyses suggest that nonlinear volumetric analysis to a template with macroanatomical details, such as the Colin27 brain, is superior to AVA to a group average brain template, such as the MNI305. Two methodological constraints of the AVA may have contributed to its decreased performance compared to NVA: (1) The rigid affine transformation, and (2) the usage of the MNI305 template that lacks macroanatomical information. While NVA to a specific brain is better than AVA to a group average, it is also important to be cautious about possible idiosyncrasies of that specific brain that may affect the outcome. We have used NVA to the Colin 27 brain as it has been used previously to align these cROIs (Evans et al., 2012).
While cortex-based alignment does not enable generating atlases of deep brain structures, it enables generating precise atlases of cortical regions. In the ventral visual system, cortex-based alignment of cROIs outperformed both types of volumetric alignments. This advantage is particular prominent for cortical areas that display a consistent coupling to macroanatomical landmarks (Fig. 3, 4, see also Fischl et al., 2008; Frost and Goebel, 2013). Additionally, CBA allows aligning brains to an average template that contains macroanatomical landmarks, which may be a more representative brain than any specific single brain such as the Colin27 brain.
For this atlas containing 8 cROIs spanning the occipito and temporal cortex, CBA performed similarly or better than NVA. However, whether CBA, in general, outperforms NVA for other parts of the brain is an open question for future research. For example, the dorsal visual stream also contains cytoarchitectonic (Kujovic et al., 2013; Malikovic et al., 2015; Scheperjans et al., 2008) and retinotopic regions (Silver and Kastner, 2009; Swisher et al., 2007; Wang et al., 2014). However, macroanatomical-cytoarchitectonic coupling has not yet been investigated in the dorsal visual stream with the same level of detail as in the ventral stream. Thus, a thorough investigation of this structural coupling and examination of how the alignment method affects an atlas of dorsal regions in occipital and parietal cortex is an important topic for future research.
The finding that CBA to the fsaverage brain vs. CBA to the postmortem average brain yielded atlases with similar accuracy (DiB, Fig. 1) has both theoretical and practical implications. On the theoretical side, the similar cROI consistency across alignments to different brain templates suggests that distortions introduced by the storage of PM brains outside of the skull prior to scanning (Amunts et al., 2000) and the possible idiosyncrasies of the anatomical features of these particular brains do not prevent generalization from these PM data to a typical set of living subjects. Furthermore, the finding that there were no significant differences in predictability or between-subject correspondence across the two types of alignments for 6 of the 8 cROIs (DiB, Fig. 1) suggests that the variability of the PM brains is not significantly different from the variability of an independent group of in-vivo brains. In turn, this suggests that having an average brain anatomy derived from CBA of many subjects (>=39, such as the fsaverage), produces a stable average anatomy that is likely to generalize to independent brains, either in-vivo or postmortem.
While our cytoarchitectonic atlas of the ventral visual stream is cross-validated and created as accurately as possible, we acknowledge that this atlas has one main limitation: It provides an estimated location of ventral visual stream cytoarchitectonic areas based on a group of postmortem brains, rather than a method to directly identify cytoarchitectonic areas in-vivo within individual subjects. Nevertheless, currently there are no methods that enable cytoarchitectonic analyses at microscopic resolution in-vivo in individual brains. It is important to consider that even with the most advanced brain alignments available, alignments will never result in 100% consistency across brains, due to individual differences in both cytoarchitectonic areas and macroanatomical landmarks. Therefore, a future goal would be to compare the predicted locus of cytoarchitectonic areas with anatomical delineations based on MRI scans in-vivo. Indeed, recently developed methods of quantitative MRI (qMRI), enable quantifying in-vivo anatomical tissue properties (Glasser and Van Essen, 2011; Mezer et al., 2013; Sereno et al., 2013). By comparing in-vivo quantitative MRI measurements with the predicted loci of cROIs (Gomez et al., 2016), future studies could potentially further validate these measurements by identifying what exactly the tissue properties are measuring with qMRI scans.
4.3. Utility of the cytoarchitectonic atlas of the ventral visual stream
The present cROI atlas of the human ventral visual stream generated on the cortical surface has many applications. First, cytoarchitecture can be used as a method for anatomical localization and labeling of brain areas. The introduction of this newly generated, cross-validated cytoarchitectonic atlas enables a more robust and accurate localization of cytoarchitectonic areas on MRI data than the widely used Brodmann parcellation, which is still commonly used to localize functional brain areas in the neuroimaging community. We make the cROI atlas publicly available on the fsaverage brain in both the FreeSurfer and BrainVoyager platforms to enable researchers to use and integrate the atlas in their studies (http://vpnl.stanford.edu/vcAtlas). Second, these cytoarchitectonic parcellations can be compared to other parcellations of the ventral stream such as those derived from other anatomical or functional metrics (Amunts et al., 2013; Glasser et al., 2016; Glasser and Van Essen, 2011; Wang et al., 2014; Yeo et al., 2011). For example, this cROI atlas can be compared to the recent atlas based on multimodal properties of the human brain (Glasser et al., 2016, DiB, Fig. 2). Third, cytoarchitectonic parcellations can be used to test differences in the ventral visual stream between typical and atypical populations. In particular, this cROI atlas can be useful for participant populations for which it is relatively easy to obtain anatomical measurements of the brain, but for which it is difficult to obtain functional scans of the visual system. For example, participants who are congenitally blind (Bedny et al., 2011; Mahon et al., 2009; Striem-Amit et al., 2012a, 2012b), autistic (Osterling and Dawson, 1994; Pierce et al., 2001; Schultz et al., 2000; Van Kooten et al., 2008), brain lesioned (Barton, 2008; Gilaie-Dotan et al., 2009; Schiltz and Rossion, 2006; Sorger et al., 2007; Steeves et al., 2006; Susilo et al., 2015), or very young (Cantlon et al., 2011; Golarai et al., 2007; Peelen et al., 2009; Scherf et al., 2007; Sowell et al., 2003).
4.4. Coupling between cytoarchitectonic and retinotopic parcellations of the ventral visual stream
Comparing this cytoarchitectonic atlas to functional parcellations of the ventral visual stream (Abdollahi et al., 2014; Amunts et al., 2007; Hinds et al., 2009; Malikovic et al., 2015) can address central questions regarding how cytoarchitectonic features may constrain functional features (Felleman and Van Essen, 1991; Grill-Spector and Weiner, 2014; Hubbel, 2013; Kravitz et al., 2013; Orban et al., 2004; Van Essen et al., 1992). As retinotopy is considered a fundamental organizational principle of the ventral visual stream (Arcaro et al., 2009; Brewer et al., 2005; Kolster et al., 2010; Orban et al., 2004; Roe et al., 2012; Sereno et al., 1995; Silson et al., 2016, 2015; Wang et al., 2014), we compared the present atlas to a recently published retinotopic atlas (Wang et al., 2014).
This comparison revealed a coupling between cytoarchitectonic and retinotopic areas in the ventral visual stream. The correspondence between cytoarchitectonic and retinotopic parcellations is particularly striking in striate cortex, which replicates prior findings (Abdollahi et al., 2014; Amunts et al., 2007; Hinds et al., 2009; Malikovic et al., 2015). While this coupling declines in extrastriate areas, most of the retinotopic areas in the ventral visual stream are confined to a single cytoarchitectonic area.
Discrepancies between cytoarchitectonic and retinotopic areas (e.g. hOc2-V2v or hOc3-V3v) may be due to imprecisions in comparisons across group atlases, or may indicate differences between cytoarchitectonic and retinotopic parcellations. We favor the former interpretation for two reasons. First, we believe that the accuracy of comparisons between cytoarchitecture and function may improve if these comparisons were done within the same participant. Future developments enabling measurements of cytoarchitecture in-vivo could test this hypothesis. Second, other evidence suggests that functional category-selective regions in the ventral stream are largely cytoarchitectonically homogenous (Weiner et al., 2016).
Interestingly, this comparison also reveals differences between structural-functional relations in the occipital and ventral temporal cortices. In occipital cortex there is largely a 1-to-1 mapping between cytoarchitecture and retinotopy, but in ventral temporal cortex, there is evidence for a 1-to-many mapping between cytoarchitecture and retinotopy. For example, FG3 contains three retinotopic areas: VO2, PHC1, PHC2, which is consistent with findings of a 1-to-many mapping between cytoarchitecture and category-selective regions in FG2 and FG4 (Weiner et al., 2016). Overall, these findings suggest that comparisons across different types of cortical parcellations (see also DiB) have the potential to advance understanding of how anatomical features contribute to brain function.
4.5. Conclusions
In sum, the present study shows that macroanatomical alignment of postmortem brains using cortex-based alignment results in a more accurate alignment of cytoarchitectonic areas compared to volumetric alignments. These results indicate that cytoarchitectonic areas of the ventral visual stream are largely coupled to macroanatomy. Additionally, we find a correspondence between cytoarchitectonic and retinotopic visual areas in the human ventral visual stream. Finally, we are hopeful that with the publication of this cROI atlas it will be broadly utilized to advance the understanding of the architecture and function of the human ventral visual stream. The atlas can be downloaded here: http://vpnl.stanford.edu/vcAtlas.
Highlights.
We provide a cross-validated cytoarchitectonic atlas of the human ventral stream
The atlas is publically available in BrainVoyager and FreeSurfer formats
Study compared how alignment methods affect atlas cross-validation performance
Cortex-based alignment outperforms both affine and nonlinear volume alignments
Coupling between cytoarchitectonic and retinotopic parcellations of ventral stream
Acknowledgments
Support for this research was provided by grants: 1RO1EY 02231801A1 and 1R01EY02391501A1 to KGS, the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 604102 (Human Brain Project) to KA and KZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdollahi RO, Kolster H, Glasser MF, Robinson EC, Coalson TS, Dierker D, Jenkinson M, Van Essen DC, Orban GA. Correspondences between retinotopic areas and myelin maps in human visual cortex. Neuroimage. 2014;99:509–524. doi: 10.1016/j.neuroimage.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau M-étienne, Bludau S, Bazin P, Lewis LB, Shah NJ, Lippert T, Zilles K, Evans AC. BigBrain: An Ultrahigh-Resolution 3D Human Brain Model. Science. 2013;340:1472–1475. doi: 10.1126/science.1235381. 80- [DOI] [PubMed] [Google Scholar]
- Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann’s areas 17 and 18 brought into stereotaxic pace - where and how variable? Neuroimage. 2000;11:66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Zilles K. Cytoarchitecture of the cerebral cortex–more than localization. Neuroimage. 2007;37:1061–1068. doi: 10.1016/j.neuroimage.2007.02.037. [DOI] [PubMed] [Google Scholar]
- Amunts K, Zilles K. Architectonic Mapping of the Human Brain beyond Brodmann. Neuron. 2015;88:1086–1107. doi: 10.1016/j.neuron.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Arcaro MJ, McMains SA, Singer BD, Kastner S. Retinotopic organization of human ventral visual cortex. J Neurosci. 2009;29:10638–10652. doi: 10.1523/JNEUROSCI.2807-09.2009. doi:29/34/10638 [pii]\r10.1523/JNEUROSCI.2807-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Bonin G. The isocortex of man. Univ Illinois Press Urbana 1951 [Google Scholar]
- Barton JJS. Structure and function in acquired prosopagnosia: lessons from a series of 10 patients with brain damage. J Neuropsychol. 2008;2:197. doi: 10.1348/174866407X214172. [DOI] [PubMed] [Google Scholar]
- Bedny M, Pascual-Leone A, Dodell-Feder D, Fedorenko E, Saxe R. Language processing in the occipital cortex of congenitally blind adults. Proc Natl Acad Sci U S A. 2011;108:4429–34. doi: 10.1073/pnas.1014818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson NC, Butt OH, Brainard DH, Aguirre GK. Correction of Distortion in Flattened Representations of the Cortical Surface Allows Prediction of V1–V3 Functional Organization from Anatomy. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson NC, Butt OH, Datta R, Radoeva PD, Brainard DH, Aguirre GK. The retinotopic organization of striate cortex is well predicted by surface topology. Curr Biol. 2012;22:2081–2085. doi: 10.1016/j.cub.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer AA, Liu J, Wade AR, Wandell BA. Visual field maps and stimulus selectivity in human ventral occipital cortex. 2005;8:1102–1109. doi: 10.1038/nn1507. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokal der Grosshirnrinde ihren Prinz dargestellt auf Grund des Zellenbaues. Barth; Leipzig: 1909. No Title. [Google Scholar]
- Campbell AW. Histological studies on the localisation of cerebral function. Cambridge Univ. Press; 1905. [Google Scholar]
- Cantlon JF, Pinel P, Dehaene S, Pelphrey KA. Cortical representations of symbols, objects, and faces are pruned back during early childhood. Cereb Cortex. 2011;21:191–199. doi: 10.1093/cercor/bhq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers J, Zilles K, Eickhoff SB, Schleicher A, Mohlberg H, Amunts K. Cytoarchitectonical analysis and probabilistic mapping of two extrastriate areas of the human posterior fusiform gyrus. Brain Struct Funct. 2013;218:511–526. doi: 10.1007/s00429-012-0411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR. Measures of the Amount of Ecologic Association Between Species Author (s): Lee R. Ecology. 1945;26:297–302. Dice Published by : Wiley Stable URL : http://www.jstor.org/stable/1932409 Accessed : 08-04-2016 13 : 33 UTC Your use of the JSTOR archive indicates your acceptance of th. [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Engel SA, Rumelhart DE, Wandell BA, Lee AT, Glover GH, Chichilnisky EJ, Shadlen MN. fMRI of human visual cortex. Nature. 1994 doi: 10.1038/369525a0. [DOI] [PubMed] [Google Scholar]
- Evans AC, Janke AL, Collins DL, Baillet S. Brain templates and atlases. Neuroimage. 2012;62:911–922. doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1. [DOI] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BT, Mohlberg H, Amunts K, Zilles K. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost MA, Goebel R. Functionally informed cortex based alignment: an integrated approach for whole-cortex macro-anatomical and ROI-based functional alignment. Neuroimage. 2013;83:1002–1010. doi: 10.1016/j.neuroimage.2013.07.056. [DOI] [PubMed] [Google Scholar]
- Frost MA, Goebel R. Measuring structural-functional correspondence: Spatial variability of specialised brain regions after macro-anatomical alignment. Neuroimage. 2012;59:1369–1381. doi: 10.1016/j.neuroimage.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Frost MA, Goebel R. Measuring structural-functional correspondence: spatial variability of specialised brain regions after macro-anatomical alignment. Neuroimage. 2012;59:1369–1381. doi: 10.1016/j.neuroimage.2011.08.035. [DOI] [PubMed] [Google Scholar]
- Gilaie-Dotan S, Perry A, Bonneh Y, Malach R, Bentin S. Seeing with profoundly deactivated mid-level visual areas: Non-hierarchical functioning in the human visual cortex. Cereb Cortex. 2009;19:1687–1703. doi: 10.1093/cercor/bhn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E. A multi-modal parcellation of human cerebral cortex. Nat Publ Gr. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel R, Esposito F, Formisano E. Analysis of functional image analysis contest (FIAC) data with brainvoyager QX: From single-subject to cortically aligned group general linear model analysis and self-organizing group independent component analysis. Hum Brain Mapp. 2006;27:392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golarai G, Ghahremani DG, Whitfield-Gabrieli S, Reiss A, Eberhardt JL, Gabrieli JDE, Grill-Spector K. Differential development of high-level visual cortex correlates with category-specific recognition memory. Nat Neurosci. 2007;10:512–22. doi: 10.1038/nn1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J, Barnett M, Natu V, Mezer A, Weiner K, Amunts K, Zilles K, Grill-Spector K. Macromolecular proliferation in human high-level visual cortex constrains development of function and behavior. Vision Science Society; 2016. [DOI] [Google Scholar]
- Goodale MA, Milner AD, Jakobson LS, Carey DP. A neurological dissociation between perceiving objects and grasping them. Nature. 1991 doi: 10.1038/349154a0. [DOI] [PubMed] [Google Scholar]
- Grill-Spector K, Weiner KS. The functional architecture of the ventral temporal cortex and its role in categorization. Nat Rev Neurosci. 2014;15:536–548. doi: 10.1038/nrn3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds O, Ghosh S, Thompson TW, Yoo JJ, Whitfield-Gabrieli S, Triantafyllou C, Gabrieli JDE. Computing moment-to-moment BOLD activation for real-time neurofeedback. Neuroimage. 2011;54:361–368. doi: 10.1016/j.neuroimage.2010.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds O, Polimeni JR, Rajendran N, Balasubramanian M, Amunts K, Zilles K, Schwartz EL, Fischl B, Triantafyllou C. Locating the functional and anatomical boundaries of human primary visual cortex. Neuroimage. 2009;46:915–922. doi: 10.1016/j.neuroimage.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G. Ferrier Lecture: The Organization of the Visual Cortex in Man. 1945;132:348–361. [Google Scholar]
- Hubbel Eye, Brain and Vision. J Chem Inf Model. 2013;53:1689–1699. doi: 10.1017/CBO9781107415324.004. [DOI] [PubMed] [Google Scholar]
- Kolster H, Peeters R, Orban Ga. The retinotopic organization of the human middle temporal area MT/V5 and its cortical neighbors. J Neurosci. 2010;30:9801–20. doi: 10.1523/JNEUROSCI.2069-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz DJ, Saleem KS, Baker CI, Ungerleider LG, Mishkin M. The ventral visual pathway: An expanded neural framework for the processing of object quality. Trends Cogn Sci. 2013;17:26–49. doi: 10.1016/j.tics.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujovic M, Zilles K, Malikovic A, Schleicher A, Mohlberg H, Rottschy C, Eickhoff SB, Amunts K. Cytoarchitectonic mapping of the human dorsal extrastriate cortex. Brain Struct Funct. 2013;218:157–172. doi: 10.1007/s00429-012-0390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz S, Weiner KS, Caspers J, Mohlberg H, Schleicher A, Bludau S, Eickhoff SB, Grill-Spector K, Zilles K, Amunts K. Two New Cytoarchitectonic Areas on the Human Mid-Fusiform Gyrus. Cereb Cortex. 2015:bhv225. doi: 10.1093/cercor/bhv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon BZ, Anzellotti S, Schwarzbach J, Zampini M, Caramazza A. Category-Specific Organization in the Human Brain Does Not Require Visual Experience. Neuron. 2009;63:397–405. doi: 10.1016/j.neuron.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikovic A, Amunts K, Schleicher A, Mohlberg H, Kujovic M, Palomero-Gallagher N, Eickhoff SB, Zilles K. Cytoarchitecture of the human lateral occipital cortex: mapping of two extrastriate areas hOc4la and hOc4lp. Brain Struct Funct. 2015 doi: 10.1007/s00429-015-1009-8. [DOI] [PubMed] [Google Scholar]
- Mezer A, Yeatman JD, Stikov N, Kay KN, Cho NJ, Dougherty RF, Perry ML, Parvizi J, Hua LH, Butts-Pauly K, Wandell BA. Quantifying the local tissue volume and composition in individual brains with magnetic resonance imaging. Nat Med. 2013;19:1667–72. doi: 10.1038/nm.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishkin M, Ungerleider LG, Macko Ka. Object vision and spatial vision: Two central pathways. Trends Neurosci. 1983;6:414–417. doi: 10.1016/0166-2236(83)90190-X. [DOI] [Google Scholar]
- Orban GA, Van Essen D, Vanduffel W. Comparative mapping of higher visual areas in monkeys and humans. Trends Cogn Sci. 2004;8:315–324. doi: 10.1016/j.tics.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Osterling J, Dawson G. Early Recognition of Children With Autism - a Study of 1St Birthday Home Videotapes. J Autism Dev Disord. 1994;24:247–257. doi: 10.1007/bf02172225. [DOI] [PubMed] [Google Scholar]
- Peelen MV, Glaser B, Vuilleumier P, Eliez S. Differential development of selectivity for faces and bodies in the fusiform gyrus. Dev Sci. 2009;12:16–25. doi: 10.1111/j.1467-7687.2009.00916.x. [DOI] [PubMed] [Google Scholar]
- Pierce K, Müller Ra, Ambrose J, Allen G, Courchesne E. Face processing occurs outside the fusiform “face area” in autism: evidence from functional MRI. Brain. 2001;124:2059–2073. doi: 10.1093/brain/124.10.2059. [DOI] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. MSM: A new flexible framework for multimodal surface matching. Neuroimage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AW, Chelazzi L, Connor CE, Conway BR, Fujita I, Gallant JL, Lu H, Vanduffel W. Toward a Unified Theory of Visual Area V4. Neuron. 2012;74:12–29. doi: 10.1016/j.neuron.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottschy C, Eickhoff SB, Schleicher A, Mohlberg H, Kujovic M, Zilles K, Amunts K. Ventral visual cortex in humans: Cytoarchitectonic mapping of two extrastriate areas. Hum Brain Mapp. 2007;28:1045–1059. doi: 10.1002/hbm.20348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheperjans F, Hermann K, Eickhoff SB, Amunts K, Schleicher A, Zilles K. Observer-independent cytoarchitectonic mapping of the human superior parietal cortex. Cereb Cortex. 2008;18:846–867. doi: 10.1093/cercor/bhm116. [DOI] [PubMed] [Google Scholar]
- Scherf KS, Behrmann M, Humphreys K, Luna B. Visual category-selectivity for faces, places and objects emerges along different developmental trajectories. Dev Sci. 2007;10 doi: 10.1111/j.1467-7687.2007.00595.x. [DOI] [PubMed] [Google Scholar]
- Schiltz C, Rossion B. Faces are represented holistically in the human occipito-temporal cortex. Neuroimage. 2006;32:1385–1394. doi: 10.1016/j.neuroimage.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Amunts K, Geyer S, Kowalski T, Zilles K. An Observer-Independent Cytoarchitectonic Mapping of the Human Cortex Using A Stereological Approach. Acta Stereol. 1998;17:75–82. [Google Scholar]
- Schleicher A, Amunts K, Geyer S, Morosan P, Zilles K. Observer-independent method for microstructural parcellation of cerebral cortex: A quantitative approach to cytoarchitectonics. Neuroimage. 1999;9:165–77. doi: 10.1006/nimg.1998.0385. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Morosan P, Amunts K, Zilles K. Quantitative architectural analysis: A new approach to cortical mapping. J Autism Dev Disord. 2005;39:1568–1581. doi: 10.1007/s10803-009-0790-8. [DOI] [PubMed] [Google Scholar]
- Scholtens LH, de Reus MA, van den Heuvel MP. Linking contemporary high resolution magnetic resonance imaging to the von economo legacy: A study on the comparison of MRI cortical thickness and histological measurements of cortical structure. Hum Brain Mapp. 2015;3046:3038–3046. doi: 10.1002/hbm.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RT, Gauthier I, Klin A, Fulbright RK, Anderson AW, Volkmar F, Skudlarski P, Lacadie C, Cohen D, Gore JC. Abnormal ventral temporal cortical activity during face discrimination among individuals with autism and Asperger syndrome. Arch Gen Psychiatry. 2000;57:331–40. doi: 10.1001/archpsyc.57.4.331. [DOI] [PubMed] [Google Scholar]
- Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, Rosen BR, Tootell RBH, Series N, May N. Borders of Multiple Visual Areas in Humans Revealed by Functional Magnetic Resonance Imaging Borders of Multiple Visual Areas in Humans Revealed by Functional Magnetic Resonance Imaging. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- Sereno MI, Lutti A, Weiskopf N, Dick F. Mapping the human cortical surface by combining quantitative T1 with retinotopy. Cereb Cortex. 2013;23:2261–2268. doi: 10.1093/cercor/bhs213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silson EH, Chan AW-Y, Reynolds RC, Kravitz DJ, Baker CI. A Retinotopic Basis for the Division of High-Level Scene Processing between Lateral and Ventral Human Occipitotemporal Cortex. J Neurosci. 2015;35:11921–11935. doi: 10.1523/JNEUROSCI.0137-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silson EH, Groen IIA, Kravitz DJ, Baker CI. Evaluating the correspondence between face-, scene-, and object-selectivity and retinotopic organization within lateral occipitotemporal cortex. J Vis. 2016;16:1–21. doi: 10.1167/16.6.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver MA, Kastner S. Topographic maps in human frontal and parietal cortex Michael. Trends Cogn Sci. 2009;13:488–495. doi: 10.1016/j.tics.2009.08.005.Topographic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GE. A new topographical survey of human cerebral cortex. J Anat. 1907;41:237–254. [PMC free article] [PubMed] [Google Scholar]
- Sørensen T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol Skr. 1948;5:1–34. [Google Scholar]
- Sorger B, Goebel R, Schiltz C, Rossion B. Understanding the functional neuroanatomy of acquired prosopagnosia. Neuroimage. 2007;35:836–852. doi: 10.1016/j.neuroimage.2006.09.051. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6:309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Steeves JKE, Culham JC, Duchaine BC, Pratesi CC, Valyear KF, Schindler I, Humphrey GK, Milner AD, Goodale MA. The fusiform face area is not sufficient for face recognition: Evidence from a patient with dense prosopagnosia and no occipital face area. Neuropsychologia. 2006;44:594–609. doi: 10.1016/j.neuropsychologia.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Striem-Amit E, Cohen L, Dehaene S, Amedi A. Reading with Sounds: Sensory Substitution Selectively Activates the Visual Word Form Area in the Blind. Neuron. 2012a;76:640–652. doi: 10.1016/j.neuron.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Striem-Amit E, Dakwar O, Reich L, Amedi A. The large-scale organization of “visual” streams emerges without visual experience. Cereb Cortex. 2012b;22:1698–1709. doi: 10.1093/cercor/bhr253. [DOI] [PubMed] [Google Scholar]
- Susilo T, Yang H, Potter Z, Robbins R, Duchaine B. Normal Body Perception despite the Loss of Right Fusiform Gyrus. J Cogn Neurosci. 2015;27:614–622. doi: 10.1162/jocn_a_00743. [DOI] [PubMed] [Google Scholar]
- Swisher JD, Halko MA, Merabet LB, McMains SA, Somers DC. Visual Topography of Human Intraparietal Sulcus. J Neurosci. 2007;27:5326–5337. doi: 10.1523/JNEUROSCI.0991-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain 1988 [Google Scholar]
- Upton G, Cook I. A dictionary of statistics. Oxford University Press; 2008. [Google Scholar]
- vEconomo C, Koskinas GN. No Title Die Cytoarchitektonik der Hirnrinde des Erwachsenen Menschen. Springer; Berlin: 1925. [Google Scholar]
- van den Heuvel MP, Scholtens LH, Feldman Barrett L, Hilgetag CC, de Reus MA. Bridging Cytoarchitectonics and Connectomics in Human Cerebral Cortex. J Neurosci. 2015;35:13943–13948. doi: 10.1523/JNEUROSCI.2630-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Anderson CH, Felleman DJ. Information Processing in the Primate Visual System: An Integrated Systems Perspective. Science. 1992;255:419–423. doi: 10.1126/science.1734518. 80- [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA. Structural and functional analyses of human cerebral cortex using a surface-based atlas. J Neurosci. 1997;17:7079–7102. doi: 10.1523/JNEUROSCI.17-18-07079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kooten IAJ, Palmen SJMC, Von Cappeln P, Steinbusch HWM, Korr H, Heinsen H, Hof PR, Van Engeland H, Schmitz C. Neurons in the fusiform gyrus are fewer and smaller in autism. Brain. 2008;131:987–999. doi: 10.1093/brain/awn033. [DOI] [PubMed] [Google Scholar]
- Wandell BA, Winawer J. Computational neuroimaging and population receptive fields. Trends Cogn Sci. 2015;19:349–357. doi: 10.1016/j.tics.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandell BA, Winawer J. Imaging retinotopic maps in the human brain. Vision Res. 2011;51:718–737. doi: 10.1016/j.visres.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mruczek REB, Arcaro MJ, Kastner S. Probabilistic Maps of Visual Topography in Human Cortex. Cereb Cortex. 2014:1–21. doi: 10.1093/cercor/bhu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner KS, Barnett M, Lorenz S, Caspers J, Stigliani A, Amunts K, Zilles K, Fischl B, Grill-Spector K. The Cytoarchitecture of Domain-specific Regions in Human High-level Visual Cortex. Cereb Cortex. 2016:1–16. doi: 10.1093/cercor/bhw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner KS, Golarai G, Caspers J, Chuapoco MR, Mohlberg H, Zilles K, Amunts K, Grill-Spector K. The mid-fusiform sulcus: a landmark identifying both cytoarchitectonic and functional divisions of human ventral temporal cortex. Neuroimage. 2014;84:453–465. doi: 10.1016/j.neuroimage.2013.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner KS, Zilles K. The anatomical and functional specialization of the fusiform gyrus. Neuropsychologia. 2016;83:48–62. doi: 10.1016/j.neuropsychologia.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschläger AM, Specht K, Lie C, Mohlberg H, Wohlschläger A, Bente K, Pietrzyk U, Stöcker T, Zilles K, Amunts K, Fink GR. Linking retinotopic fMRI mapping and anatomical probability maps of human occipital areas V1 and V2. Neuroimage. 2005;26:73–82. doi: 10.1016/j.neuroimage.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology. 2011 doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centerary of Brodmann’s map - Conception and Fate. Nat Rev Neurosci. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Receptor mapping: architecture of the human cerebral cortex. Curr Opin Neurol. 2009;22:331–339. doi: 10.1097/WCO.0b013e32832d95db. [DOI] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Amunts K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]


