Abstract
Wild birds are reservoirs for Chlamydia spp. Of the total 225 samples from wild birds during January to September 2016 in Korea, 4 (1.8%) and 2 (0.9%) showed positive for Chlamydia psittaci and Chlamydia gallinacea, respectively. Phylogenetic analyses and comparisons of sequence identities for outer-membrane protein A (ompA) revealed that Korean C. psittaci fall into three previously known genotypes; genotype E, 1V and 6N, whereas the Korean C. gallinacea were classified as new variants of C. gallinacea. Our study demonstrates that wild birds in South Korea carry at least two Chlamydia species: C. psittaci and C. gallinacea, and provides new information on the epidemiology of avian chlamydiosis in wild birds.
Keywords: Chlamydia spp., genetic diversity, South Korea, wild bird
Chlamydia spp. are the etiological agents of chlamydiosis in animals and humans [16, 22]. The family Chlamydiaceae encompasses the single genus Chlamydia, which comprises 11 species: Chlamydia psittaci, C. muridarum, C. suis, C. trachomatis, C. abortus, C. caviae, C. felis, C. pecorum, C. pneumoniae, C. avium and C. gallinacea [22, 24, 26, 27].
Of these species, C. psittaci is the most well-known zoonotic agent [13]. Isolates of C. psittaci have been reported to exist in approximately 460 species of birds [15]. C. psittaci infection in birds can persist for months to years, often without causing obvious illness [16]. Until recently, there were nine outer-membrane protein A (ompA) genotypes described in C. psittaci, designated A–F, E/B, M56 and WC [25], as well a number of provisional genotypes (1V, YP84, R54, 6N, CPX0308, I and J), representing strains that have thus far been un-typeable [20, 25]. The importance of genotyping lies in the fact that certain genotypes tend to be associated with certain hosts and differ in virulence [13]. Genotypes A and B are usually associated with psittacine birds and pigeons, respectively. Genotype C is primarily isolated from ducks and geese, and genotype D from turkeys. Genotype E is the most divergent among these types, which has been isolated from pigeons, ratites, ducks and turkeys, and occasionally from humans [13]. Regarding to C. psittaci in South Korea, association with ocular adnexal mucosa-associated lymphoid tissues (MALT) lymphoma in human has been reported [31]. Interestingly, this association between C. psittaci and the MALT lymphoma seems to show geographic variation. For example, C. psittaci was detected in the MALT patients in Italy and South Korea, but not in the patients in the Northern China and Japan [3].
To date, C. gallinacea has been mainly isolated from chickens, ducks, guinea fowl, turkeys and other domestic poultry in China and European countries [12, 32]. This new emerging agent was predominantly found in asymptomatic poultry; however, a decrease in the rate of weight gain was reported in C. gallinacea-infected chickens [12]. So far, at least 13 genotypes of ompA protein of C. gallinacea have been identified. Because it has been suggested that C. gallinacea may cause a potential zoonotic infection [18], the pathogenicity and characterization of each genotype require systemic investigation [12].
Although studies involving the genetic characterization of Chlamydia spp. are prevalent on a global scale [8, 17, 23, 30], there is little information on the occurrence or epidemiology of Chlamydia spp. infection in wild birds of South Korea. In the present study, polymerase chain reaction (PCR) was performed to obtain baseline data on the prevalence of Chlamydia spp. in various species of wild birds. We also performed sequence analysis of 16S rRNA and ompA genes to evaluate the genetic diversity and epidemiology of Chlamydia spp. in wild birds in South Korea.
Bird carcasses were obtained from wildlife rescue and conservation centers in Busan, Chungbuk, Daejeon, Gangwon, Gyeongnam, Jeonbuk, Jeonnam and Ulsan during January to September 2016. Tracheal swabs of bird carcass were collected individually from 225 birds, from 43 species belonging to 14 orders (Supplemental Table 1). Necropsies were performed on all dead birds. Tissue samples including lungs, spleens and livers were aseptically collected and examined for macroscopic lesions, including multifocal hepatic necrosis and spleen and liver enlargement. All swabs were immediately placed in BD Universal Viral Transport (UVT) tubes (BD Biosciences, Baltimore, MD, U.S.A.). Swabs were stored at −80°C until DNA extractions were performed.
DNA was extracted from tracheal swabs, using the automated Maxwell RSC Viral Total Nucleic Acid Purification Kit with the Maxwell 16 instrument (Promega Corporation, Madison, WI, U.S.A.). All steps were automated according to the pre-programmed viral nucleic acid protocol. An initial screening was performed with whole genomic DNAs by 23S rRNA-based, Chlamydia spp. -specific quantitative real time PCR with QuantStudio 6 flex (Thermo Scientific, Rockford, IL, U.S.A.), as described in a previous report [5, 23]. For further characterization of chlamydial ompA and 16S rRNA, fragments were amplified using previously described representative primer pairs and conditions: CTU/CTL (ompA sequencing) [4] and 16S1/rp2 (16S rRNA sequencing) [21]. PCR-amplified segments of ompA and 16S rRNA genes were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and directly sequenced (Macrogen, Seoul, Korea). Nucleotide sequences have been deposited under GenBank nos. KX603684-KX603697.
Nucleotide sequences were BLASTed against the NCBI database to identify related sequences and aligned using CLUSTALW2 [6]. The multiple alignments were used to infer the phylogenies with the maximum-likelihood (ML) method implemented in MEGA 5 [28]. To obtain the ML tree topologies, 1,000 bootstrap replicates were performed for each dataset. The inferred tree topologies were inspected visually using FigTree version 1.3.1.
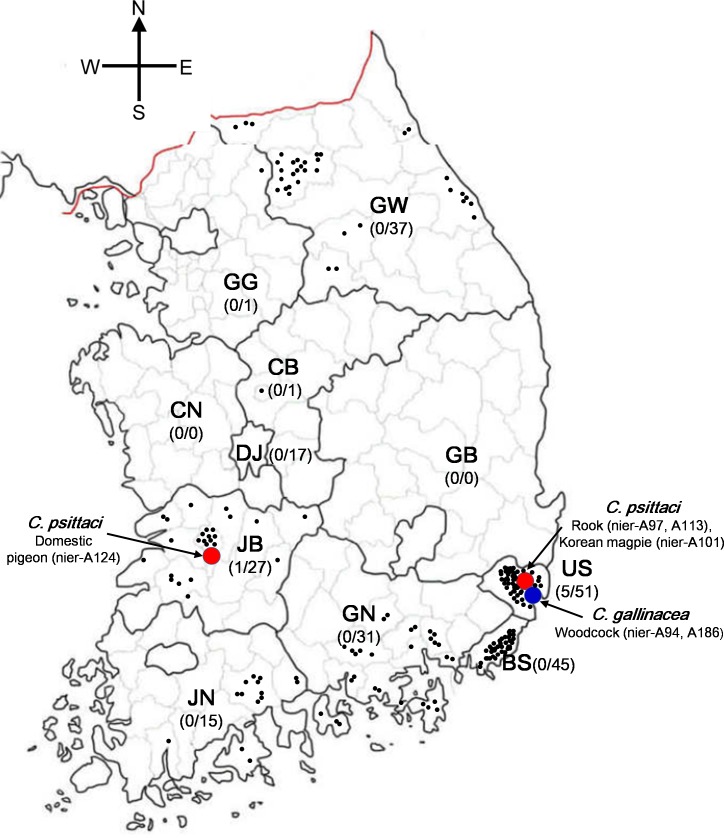
Chlamydia prevalence was defined as the number of PCR-positive samples described as a percentage of the total sample number. No macroscopic lesions were detected during the necropsy of 225 birds from wildlife rescue and conservation centers. However, six of 225 tracheal swabs tested positive for chlamydial DNA by a diagnostic real-time PCR assay for 23S rRNA, corresponding to 2.7% (6/225) of the examined population. Birds that tested positive for Chlamydia spp. belonged to four species in three orders (Charadriiformes, Columbiformes and Passeriformes), including woodcock (Scolopax rusticola), domestic pigeon (Columba livia var. domestica), the Korean magpie (Pica pica sericea) and rook (Corvus frugilegus) (Supplemental Table 1). Geographically, the Chlamydia spp. were detected in Jeonbuk province (one case in domestic pigeon) and Ulsan metropolitan city (two cases in woodcock, one case in Korean magpie and two cases in rook) (Fig. 1).
Fig. 1.
Chlamydia spp. prevalence in wild birds in South Korea. The red and blue filled circles with arrows indicate C. psittaci and C. gallinacea, respectively. The black filled circles indicate Chlamydia negative samples. GG, Gyeonggi province; GW, Gangwon province; CN, Chungnam province; DJ, Daejeon metropolitan city; CB, Chungbuk province; JB, Jeonbuk province; JN, Jeonnam province; GB, Gyeongbuk province; GN, Gyeongnam province; US, Ulsan metropolitan city; BS, Busan metropolitan city.
The confirmatory PCR targeting the variable domains of 16S rRNA genes enabled us to identify the presence of two Chlamydia spp. in the birds examined: C. psittaci (1.8%; 4/225) and C. gallinacea (0.9%; 2/225) (Table 1).
Table 1. Chlamydia spp. identification in South Korea from January to September 2016.
| Sample ID | Species | Region | Chlamydia spp. identification by 16s rRNA | Genotyping by ompA gene |
|---|---|---|---|---|
| nier-A94 | Woodcock | US | C. gallinacea | Un-typeable |
| nier-A97 | Rook | US | C. psittaci | Genotype 1V |
| nier-A101 | Korean Magpie | US | C. psittaci | Genotype 1V |
| nier-A113 | Rook | US | C. psittaci | Genotype 6N |
| nier-A124 | Domestic pigeon | JB | C. psittaci | Genotype E |
| nier-A186 | Woodcock | US | C. gallinacea | Un-typeable |
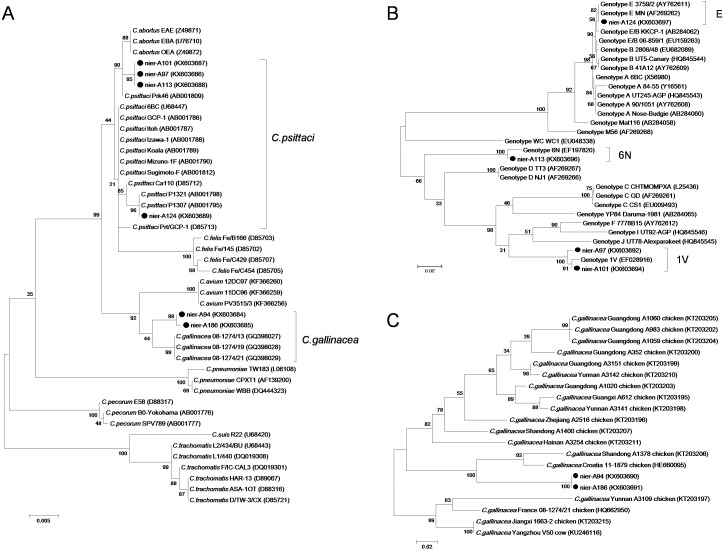
The Chlamydia 16S rRNA sequences from nier-A97, -A101 and -A113 were identical and displayed a high degree of conservation with that of C. psittaci strain Prk46 (AB001809, 99.8% identity), which represents an intermediate variant between C. psittaci and C. abortus. The sequence of nier-A124 is 100% identical to that of previously reported human and pigeon C. psittaci isolates. The nucleotide sequences of C. gallinacea (nier-A94 and -A186) shared 99.9% identity and were closely related to those of C. gallinacea 08-1274/13 (GQ398027, 99.0% identity). A comparative phylogenetic analysis based on the 16S rRNA sequence demonstrated that the C. psittaci and C. gallinacea strains detected in this study are all closely related to their representative C. psittaci and C. gallinacea (Fig. 2A).
Fig. 2.
Phylogeny of Chlamydia spp. A. The 16S rRNA gene (1,370 bp) of Chlamydia spp. B. The ompA gene (>886 bp) of C. psittaci. C. The ompA gene (>379 bp) of C. gallinacea. The phylogenetic distances were calculated using MEGA 5 with the maximum-likelihood algorithm, and the trees were visualized using FigTree 1.3.1. Bootstrap values are shown as percentages of 1,000 replicates. Bootstrap value less than 50% is shown in the phylogenetic trees. The scale bars represent the number of substitutions per site. Nucleotide sequences in the present study are indicated by black dots.
The ompA sequences of C. psittaci and C. gallinacea were amplified by PCR to analyze genetic diversity. The ompA sequences of nier-A97 and -A101 displayed a high level of nucleotide identity (99.2%) and conservation with ompA nucleotide sequences from genotype 1V (EF028916) isolates from Russian hooded crows (98.5% and 99.3% identity, respectively). In addition, nier-A113 showed a high degree of identity (98.9%) to genotype 6N (EF197820) isolates from Russian rooks [30]. The nier-A124 nucleotide sequence was 100% identical to that of genotype E isolate MN (AF269262), which has been isolated from humans and a variety of birds worldwide [1, 11]. A comparative phylogenetic analysis based on the ompA sequence revealed that the four C. psittaci sequences identified in this study are all closely related to their respective C. psittaci genotypes (Fig. 2B).
The two C. gallinacea sequences (nier-A94 and -A186) were 100% identical and showed 85.5% nucleotide identity to the sequence of C. gallinacea isolate 11-1879 from Croatia, which was the most closely related to the sequence of the previously characterized C. gallinacea isolates. A phylogenetic comparison showed that the ompA genes identified from two instances of C. gallinacea in this study clearly diverged to the previously reported C. gallinacea isolates from France and China (Fig. 2C).
To the best of our knowledge, our study is the first to characterize avian chlamydiosis in wild birds from South Korea. Diagnostic real-time PCR-based approaches for the detection of Chlamydia spp. from tracheal swab samples have been previously employed for 23S rRNA [5, 23]. In the present study, two Chlamydia species [C. psittaci (in 4/225) and C. gallinacea (in 2/225)] were detected, and their genetic variants were identified by analyzing the sequences of ompA and 16S rRNA.
In the present study, C. psittaci was detected in domestic pigeons, but did not appear to be associated with any signs of disease in these birds on necropsy. Thus, these birds could be asymptomatic carriers of C. psittaci. Studies describing the C. psittaci carrier status of the pigeon population have been published worldwide, thereby suggesting their potential zoonotic aspects [14, 29]. For example, the prevalence of C. psittaci was 7.9% (26/331) and 22.2% (103/463) in feral pigeons in Netherland and Japan, respectively. Sequence analyses of ompA and 16S rRNA revealed that C. psittaci is identical to genotype E, which was previously detected in both humans and pigeons worldwide [1]. Although our results showed a relatively low prevalence of C. psittaci in pigeons compared to that observed in other countries, this finding indicates that the pigeon could also be a natural vector for C. psittaci genotype E in South Korea and a zoonotic threat in the region.
C. psittaci genotypes 1V and 6N were first discovered in corvid species (crow and rook) in Russia in 2006 [30]. The discovery of C. psittaci genotypes 1V and 6N in the rooks and Korean magpies in the present study provides evidence of circulation of C. psittaci in the corvid species of our region, where the rook may be an asymptomatic carrier. The rook is characterized as a migratory bird, which breeds in central Europe or Siberia and migrates to South Korea or Japan during the winter season [2]. In particular, Taehwa River located in Ulsan metropolitan city is one of the most important resting sites for migratory rooks, which overwinter in South Korea from Siberia. The ompA sequences of C. psittaci genotypes 1V and 6N from rooks in our study showed a close association with those of isolates from Russian corvid species, although the 16S rRNA sequence of our isolates showed a close genetic similarity to C. psittaci prk/46, which is known to cause systemic infections in parakeets in Japan [9] (Fig. 2A and 2B). These results indicate that the migratory rook between Russia and South Korea plays an important role in the epidemiology of C. psittaci genotypes 1V and 6N.
This study presents the first instance of detection of C. gallinacea in South Korea. C. gallinacea was reported in chickens during an outbreak of psittacosis in Germany [10] and was subsequently isolated from chickens in France (C. gallinacea strain 08-1743/3) and Croatia (C. gallinacea strain 11-1879). In France, a slaughterhouse worker presenting with atypical pneumonia was reportedly exposed to C. gallinacea-carrying chickens [18]. The C. gallinacea in our study appeared to be segregated genetic variants in comparison to known isolates from China and Europe, based on a phylogenetic analysis of the ompA sequences (Fig. 2C). Our findings are in agreement with those in previous reports, which showed that geographically separated isolates displayed genetic diversity (85.7% similarity between isolates in China and Europe) [12, 18]. Furthermore, the partial sequencing of the 16S rRNA and ompA gene revealed that although the 16S rRNA sequences in our study were similar to those of previous C. gallinacea isolates (99% nucleotide identity), the ompA genes were highly distinguished (85.5% nucleotide identity). This is in agreement with a previous report, in which rRNA genes were subjected to evolutionary pressure to a far lesser extent than genes encoding outer membrane proteins [7].
Although the prevalence of C. gallinacea in woodcocks was very high (2/3, 66.7%), the sample size was too low for statistical analysis. Because our method of sample collection from wildlife health centers could have contributed to a bias, additional studies are required to assess the prevalence of C. gallinacea in woodcocks. Furthermore, because it was reported that bovine C. gallinacea had ompA sequences identical to those of avian species in China [19], additional studies should be conducted to understand the potential transmission of C. gallinacea between wild birds and domestic animals, including poultry and mammals, in Korea.
In conclusion, this study demonstrates the prevalence of Chlamydia spp. in wild birds, as well as the genetic diversity of C. psittaci and C. gallinacea, in South Korea. Chlamydia spp. in wild birds could be a potential source of infection in domestic animals and humans in South Korea, especially in people associated with handling of wild birds; hence, it is necessary to implement biosecurity measures to minimize the possibility of infection. The results obtained from this study contribute to improving the present understanding of the epidemiology of avian chlamydiosis in wild bird populations.
Supplementary
Acknowledgments
We thank the regional wildlife rescue and conservation centers for their efforts in the collection of wild bird carcasses. This research was supported by a grant from the NIER (No. 2016-01-01-033) of the Republic of Korea. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Andersen A. A.1997. Two new serovars of Chlamydia psittaci from North American birds. J. Vet. Diagn. Invest. 9: 159–164. doi: 10.1177/104063879700900209 [DOI] [PubMed] [Google Scholar]
- 2.Avibirds. Profile Rook. http://www.avibirds.com [accessed May 15, 2017].
- 3.Collina F., De Chiara A., De Renzo A., De Rosa G., Botti G., Franco R.2012. Chlamydia psittaci in ocular adnexa MALT lymphoma: a possible role in lymphomagenesis and a different geographical distribution. Infect. Agent. Cancer 7: 8. doi: 10.1186/1750-9378-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denamur E., Sayada C., Souriau A., Orfila J., Rodolakis A., Elion J.1991. Restriction pattern of the major outer-membrane protein gene provides evidence for a homogeneous invasive group among ruminant isolates of Chlamydia psittaci. J. Gen. Microbiol. 137: 2525–2530. doi: 10.1099/00221287-137-11-2525 [DOI] [PubMed] [Google Scholar]
- 5.Ehricht R., Slickers P., Goellner S., Hotzel H., Sachse K.2006. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell. Probes 20: 60–63. doi: 10.1016/j.mcp.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 6.European Bioinformatics Institute. http://www.ebi.ac.uk/Tools/msa/clustalo/ [accessed May 15, 2017].
- 7.Everett K. D., Bush R. M., Andersen A. A.1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49: 415–440. doi: 10.1099/00207713-49-2-415 [DOI] [PubMed] [Google Scholar]
- 8.Frutos M. C., Monetti M. S., Vaulet L. G., Cadario M. E., Fermepin M. R., Ré V. E., Cuffini C. G.2015. Genetic diversity of Chlamydia among captive birds from central Argentina. Avian Pathol. 44: 50–56. doi: 10.1080/03079457.2014.993593 [DOI] [PubMed] [Google Scholar]
- 9.Fukushi H., Nojiri K., Hirai K.1987. Monoclonal antibody typing of Chlamydia psittaci strains derived from avian and mammalian species. J. Clin. Microbiol. 25: 1978–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaede W., Reckling K. F., Dresenkamp B., Kenklies S., Schubert E., Noack U., Irmscher H. M., Ludwig C., Hotzel H., Sachse K.2008. Chlamydophila psittaci infections in humans during an outbreak of psittacosis from poultry in Germany. Zoonoses Public Health 55: 184–188. doi: 10.1111/j.1863-2378.2008.01108.x [DOI] [PubMed] [Google Scholar]
- 11.Geens T., Desplanques A., Van Loock M., Bönner B. M., Kaleta E. F., Magnino S., Andersen A. A., Everett K. D., Vanrompay D.2005. Sequencing of the Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J. Clin. Microbiol. 43: 2456–2461. doi: 10.1128/JCM.43.5.2456-2461.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W., Li J., Kaltenboeck B., Gong J., Fan W., Wang C.2016. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus). Sci. Rep. 6: 19638. doi: 10.1038/srep19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkinezhad T., Geens T., Vanrompay D.2009. Chlamydophila psittaci infections in birds: a review with emphasis on zoonotic consequences. Vet. Microbiol. 135: 68–77. doi: 10.1016/j.vetmic.2008.09.046 [DOI] [PubMed] [Google Scholar]
- 14.Heddema E. R., Ter Sluis S., Buys J. A., Vandenbroucke-Grauls C. M. E., van Wijnen J. H., Visser C. E.2006. Prevalence of Chlamydophila psittaci in fecal droppings from feral pigeons in Amsterdam, The Netherlands. Appl. Environ. Microbiol. 72: 4423–4425. doi: 10.1128/AEM.02662-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaleta E. F., Taday E. M.2003. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 32: 435–461. doi: 10.1080/03079450310001593613 [DOI] [PubMed] [Google Scholar]
- 16.Knittler M. R., Sachse K.2015. Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog. Dis. 73: 1–15. doi: 10.1093/femspd/ftu007 [DOI] [PubMed] [Google Scholar]
- 17.Krawiec M., Piasecki T., Wieliczko A.2015. Prevalence of Chlamydia psittaci and Other Chlamydia Species in Wild Birds in Poland. Vector Borne Zoonotic Dis. 15: 652–655. doi: 10.1089/vbz.2015.1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laroucau K., Vorimore F., Aaziz R., Berndt A., Schubert E., Sachse K.2009. Isolation of a new chlamydial agent from infected domestic poultry coincided with cases of atypical pneumonia among slaughterhouse workers in France. Infect. Genet. Evol. 9: 1240–1247. doi: 10.1016/j.meegid.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 19.Li J., Guo W., Kaltenboeck B., Sachse K., Yang Y., Lu G., Zhang J., Luan L., You J., Huang K., Qiu H., Wang Y., Li M., Yang Z., Wang C.2016. Chlamydia pecorum is the endemic intestinal species in cattle while C. gallinacea, C. psittaci and C. pneumoniae associate with sporadic systemic infection. Vet. Microbiol. 193: 93–99. doi: 10.1016/j.vetmic.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 20.Madani S. A., Peighambari S. M.2013. PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathol. 42: 38–44. doi: 10.1080/03079457.2012.757288 [DOI] [PubMed] [Google Scholar]
- 21.Pudjiatmoko F., Fukushi H., Ochiai Y., Yamaguchi T., Hirai K.1997. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 47: 425–431. doi: 10.1099/00207713-47-2-425 [DOI] [PubMed] [Google Scholar]
- 22.Rodolakis A., Yousef Mohamad K.2010. Zoonotic potential of Chlamydophila. Vet. Microbiol. 140: 382–391. doi: 10.1016/j.vetmic.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 23.Sachse K., Kuehlewind S., Ruettger A., Schubert E., Rohde G.2012. More than classical Chlamydia psittaci in urban pigeons. Vet. Microbiol. 157: 476–480. doi: 10.1016/j.vetmic.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 24.Sachse K., Laroucau K., Hotzel H., Schubert E., Ehricht R., Slickers P.2008. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 8: 63. doi: 10.1186/1471-2180-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachse K., Bavoil P. M., Kaltenboeck B., Stephens R. S., Kuo C. C., Rosselló-Móra R., Horn M.2015. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst. Appl. Microbiol. 38: 99–103. doi: 10.1016/j.syapm.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 26.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Creasy H. H., Weidmann M., Myers G., Vorimore F., Vicari N., Magnino S., Liebler-Tenorio E., Ruettger A., Bavoil P. M., Hufert F. T., Rosselló-Móra R., Marz M.2014. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst. Appl. Microbiol. 37: 79–88. doi: 10.1016/j.syapm.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 27.Stephens R. S., Myers G., Eppinger M., Bavoil P. M.2009. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol. Med. Microbiol. 55: 115–119. doi: 10.1111/j.1574-695X.2008.00516.x [DOI] [PubMed] [Google Scholar]
- 28.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka C., Miyazawa T., Watarai M., Ishiguro N.2005. Bacteriological survey of feces from feral pigeons in Japan. J. Vet. Med. Sci. 67: 951–953. doi: 10.1292/jvms.67.951 [DOI] [PubMed] [Google Scholar]
- 30.Yatsentyuk S. P., Obukhov I. L.2007. Molecular genetic characterization of avian Chlamydophila psittaci isolates. Russ. J. Genet. 43: 1215–1220. doi: 10.1134/S1022795407110026 [DOI] [PubMed] [Google Scholar]
- 31.Yoo C., Ryu M. H., Huh J., Park J. H., Kang H. J., Ahn H. S., Lee Y., Kim M. J., Lee H., Kim T. W., Chang H. M., Lee J. L., Kang Y. K.2007. Chlamydia psittaci infection and clinicopathologic analysis of ocular adnexal lymphomas in Korea. Am. J. Hematol. 82: 821–823. doi: 10.1002/ajh.20962 [DOI] [PubMed] [Google Scholar]
- 32.Zocevic A., Vorimore F., Marhold C., Horvatek D., Wang D., Slavec B., Prentza Z., Stavianis G., Prukner-Radovcic E., Dovc A., Siarkou V. I., Laroucau K.2012. Molecular characterization of atypical Chlamydia and evidence of their dissemination in different European and Asian chicken flocks by specific real-time PCR. Environ. Microbiol. 14: 2212–2222. doi: 10.1111/j.1462-2920.2012.02800.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.