Abstract
Morphological plasticity such as the yeast-to-hyphae transition is a key virulence factor of the human fungal pathogen Candida albicans. Hyphal formation is controlled by a multilayer regulatory network composed of environmental sensing, signaling, transcriptional modulators as well as chromatin modifications. Here, we demonstrate a novel role for the replication-independent HIR histone chaperone complex in fungal morphogenesis. HIR operates as a crucial modulator of hyphal development, since genetic ablation of the HIR complex subunit Hir1 decreases sensitivity to morphogenetic stimuli. Strikingly, HIR1-deficient cells display altered transcriptional amplitudes upon hyphal initiation, suggesting that Hir1 affects transcription by establishing transcriptional thresholds required for driving morphogenetic cell-fate decisions. Furthermore, ectopic expression of the transcription factor Ume6, which facilitates hyphal maintenance, rescues filamentation defects of hir1Δ/Δ cells, suggesting that Hir1 impacts the early phase of hyphal initiation. Hence, chromatin chaperone-mediated fine-tuning of transcription is crucial for driving morphogenetic conversions in the fungal pathogen C. albicans.
Introduction
Chromatin plays fundamental roles in gene regulation during most cellular differentiation processes. Of note, chromatin architecture and functions have been conserved from human embryonic development to morphogenetic cell fate decisions in unicellular eukaryotes including the fungal pathogen Candida albicans 1–3. C. albicans, a normal commensal colonizer of most healthy individuals, is present on skin, gut and mucosal surfaces, and thus well-adapted to host niches with distinct immune surveillance4–6. However, immunosuppression can trigger C. albicans to switch into an invasive pathogenic mode, causing more than 400,000 life-threating invasive infections worldwide per year. Remarkably, together with other major pathogens, fungal infections claim about 1.5 million lives each year7.
The ability to undergo morphogenesis arguably constitutes one of the major virulence traits of C. albicans. The genomic plasticity is manifested by a transcriptional plasticity that enables a switch between a unicellular yeast-like morphology and the pseudohyphal/hyphal growth phases, which display characteristic septated filaments8–10. Hyphal formation is triggered by various environmental host signals. For instance, elevated temperatures, serum, nutrient availability, carbon dioxide, certain amino acids or N-acetyl glucosamine (GlcNAc) activate a complex signaling network, including the mitogen-activated protein kinase (MAPK) cascades and the cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA) signaling pathway, resulting in the activation of a panel of dedicated transcription factors such as Efg19, 11–15. Of note, besides the induction of transcriptional activators, hyphal triggers occlude transcriptional repressors of hyphal-specific genes (HSGs) such as Nrg1 from target promoters16. To sustain hyphal development, additional transcription factors such as Ume6 and Brg1 are recruited to HSGs, whereas prolonged binding of Nrg1 is inhibited. Thus, two distinct transcriptional phases modulate filamentation, involving both decoration and exclusion of transcriptional modulators to initiate and maintain hyphal growth3, 17.
Recent studies suggest a pivotal role for chromatin modification and organization in controlling fungal morphogenesis18–20. Moreover, Efg1 cooperates with the NuA4 histone acetyltransferase (HAT) complex and the ATP dependent chromatin remodeling complex SWI/SNF during hyphal initiation21. In addition, the Set3 histone deacetylase (HDAC) complex controls the threshold sensitivity of the cAMP/PKA pathway to hyphal stimuli22, while the Hda1 HDAC functions in prolonged hyphal growth17. Hence, the dynamic interplay of transcription factors and chromatin modification status integrates diverse triggers from upstream signaling cascades to set levels of coordinated transcriptional responses required for morphogenetic cell fate decisions.
The functional status of chromatin as well as its architecture can be altered by chromatin modifications including chromatin remodeling through the concomitant assembly and disassembly of nucleosomes. This process is guarded and facilitated by conserved histone chaperones acting in replication-dependent and –independent pathways23. The central hub for histone H3 and H4 turnover in the cytoplasm is the histone chaperone Asf124. Newly synthesized histone H3/H4 dimers are acetylated by the HAT Hat1 at H4K5 and K12 and delivered to Asf125. The dimer is further acetylated by the Rtt109 HAT at H3K56 in Saccharomyces cerevisiae 26. Asf1 shuttles modified H3/H4 dimers into the nucleus to a panel of other histone chaperones for chromatin assembly coupled to DNA replication, DNA damage repair, heterochromatin maintenance or transcription24, 27. For instance, the CAF-1 histone chaperone complex incorporates histones H3 and H4 into chromatin coupled to DNA-replication28. Notably, CAF-1 is involved in epigenetic white-opaque switching and in the oxidative stress response in C. albicans 29, 30. The HIR histone chaperone complex consists of four subunits in S. cerevisiae and assembles nucleosomes in a replication-independent manner31–33. The complex was originally discovered in S. cerevisiae as transcriptional repressor for 6 out of 8 histone genes34. Of note, functions of HIR related to non-histone gene regulation have been proposed, including cryptic promoter repression and suppression of Ty elements32, 35. Interestingly, loss of the HIR complex subunit Hir1 decreases nucleosome occupancy at various gene promoters36. Hence, Hir1 might affect the fine-tuning of transcriptional responses by regulating local chromatin architecture in target genes, thereby altering affinities of dedicated transcriptional regulators to cognate cis-acting sites, as well as the decoration by and displacement of transcriptional co-factors and modulators.
Initiation of C. albicans morphogenesis is accompanied by massive transcriptional changes affecting up to 15% of the genome37. Hence, we hypothesized that defects in replication-independent chromatin assembly may deregulate HSG expression and thus, influence the C. albicans yeast-to-hyphae transition. Indeed, here we show that chromatin assembly during filamentation is facilitated by HIR1, but not by the replication-dependent CAF-1 machinery. We find that HSG transcriptional amplitudes dramatically change upon genetic removal of HIR1. Furthermore, we provide compelling evidence that the HIR complex acts downstream of cAMP/PKA signaling, possibly in concert with other transcriptional activators such as Efg1, to regulate the initial onset of hyphal growth. Surprisingly, transcriptomics of HIR1-deficient cells revealed that the gene expression profile in cells lacking HIR1 was qualitatively very similar to the wild type (WT) following hyphal induction. However, the maximal transcriptional amplitudes are remarkably altered in the HIR1 mutant compared to the WT, which results in strongly decreased sensitivity of cells to filamentation signals. Strikingly, ectopic expression of Ume6 bypasses the requirement of Hir1-mediated transcriptional initiation for hyphal differentiation, which further substantiates our conclusion that Hir1 plays an important role during early steps of hyphal initiation. Our data are of general relevance, since they suggest a novel mechanism whereby chromatin chaperones like the HIR complex can impact fine-tuning of transcription to establish minimal threshold transcriptional levels required for triggering developmental changes or morphogenetic cell fate decisions independently of DNA replication or cell cycle control.
Results
The HIR complex facilitates hyphal initiation
Several previous studies suggest that chromatin alterations contribute to yeast-to-hyphae transitions16, 17, 19, 21, 37, 38. Moreover, recent work also uncovered a role of the HIR histone chaperone member Hir1 in white-opaque switching29 and azole tolerance30 in C. albicans. Hence, we reasoned that a functional connection may exist between hyphal formation and chromatin assembly mediated by the HIR histone chaperone complex. Thus, we genetically removed HIR1 30 and subjected the hir1Δ/Δ mutant to filament-inducing conditions. Strikingly, loss of Hir1 strongly decreased hyphal formation on solid YPD containing 10% serum at 37 °C as indicated by smooth colony morphology (Fig. 1A). Reintegration of HIR1 into its original genomic locus fully restored the WT phenotype. In addition, hir1Δ/Δ cells were defective for hyphal formation in response to other stimuli such as GlcNAc or Spider medium (Figure S1A). To investigate the filamentation defects of mutant cells in greater detail, we examined the hyphal initiation rates of HIR1-deficient cells in liquid YPD medium with 10% serum at 37 °C. The hir1Δ/Δ mutant showed severe hyphal formation defects at all time points, where only 30% of the hir1Δ/Δ cells initiated hyphal induction (Figs 1B and S1B).
Figure 1.
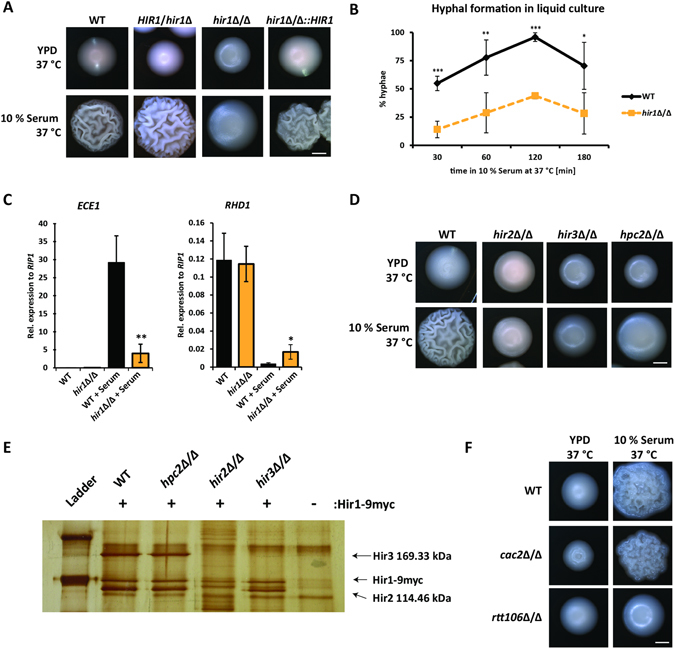
Regulation of hyphal formation by the HIR complex. (A) hir1Δ/Δ cells are defective in serum-induced hyphal formation on solid medium. Colony morphology was inspected after 3 days. Scale bar corresponds to 1 mm. (B) Loss of HIR1 decreases hyphal formation in liquid medium. Hyphal formation of WT and hir1Δ/Δ cells was evaluated in YPD supplemented with 10% FCS at 37 °C after the indicated time points. (C) Hir1 controls hyphal-specific gene (HSG) expression. ECE1 and RHD1 gene expression was measured after 30 min of hyphal induction via RT-qPCR. Transcript levels were normalized to the expression of the reference gene RIP1 22. (D) HIR complex mutants display decreased hyphal formation. Colony morphology of hpc2Δ/Δ, hir2Δ/Δ and hir3Δ/Δ was examined as in (A). Scale bar corresponds to 1 mm. (E) Hir1 associates with Hir2 and Hir3 in C. albicans. Hir1 was epitope-tagged with 9myc in either WT background or in strains lacking one HIR complex component and subjected to native immunoprecipitation. Precipitated proteins were separated through a 10% SDS-PAGE gel and visualized by silver staining. Detected protein bands were cut out and identified via mass spectrometry analysis. The indicated molecular weight for Hir3 and Hir2 are derived from mass spectrometry analysis. (F) Hyphal formation is Cac2-independent. Colony morphology of WT cells and cells lacking CAC2 and RTT106 was analyzed as above (A). Scale bar corresponds to 1 mm. (B,C) Data are presented as mean + SD of three independent experiments. For significance testing, hir1Δ/Δ cells were compared to WT cells. n.s. not significant, *P < 0.05, **P < 0.01, ***P < 0.001 with Student’s t-test.
Hyphal initiation is triggered by the so-called hyphal-specific transcriptional program37. Therefore, we examined the dynamics of HSG expression in hir1Δ/Δ cells using qRT-PCR. Induction of HSGs encoding the cell wall proteins Ece1 and Hwp1 or the G1-cyclin Hgc19 was remarkably reduced in hir1Δ/Δ cells after 30 min growth in YPD plus 10% serum at 37 °C (Figs 1C and S1D). This result suggested that Hir1 is required for the activation of hyphal-specific transcriptional programs. Since hyphal induction not only drives gene activation39, we also tested expression of the β-mannosyltransferase Rhd1 which is normally repressed upon the hyphal switch40. Indeed, RHD1 mRNA abundance was decreased during hyphal initiation in WT cells. A qualitatively similar response was observed in hir1Δ/Δ cells, although the amplitude of RHD1 transcriptional repression was decreased (Fig. 1C). In addition, Ywp1, a yeast phase cell wall protein41, was not downregulated in hir1Δ/Δ cells in response to hyphal induction. Interestingly, basal YWP1 expression was already substantially increased in hir1Δ/Δ cells during yeast phase growth (Figure S1D). These data strongly suggest that the transcriptional program during hyphal initiation cannot be fully activated in the absence of HIR1, thus impairing the yeast-to-hyphae transition. Since Hir1 might act globally during replication-independent chromatin assembly, we tested whether transcriptional amplitudes in response to other specific environmental cues are affected upon the loss of HIR1. Therefore, WT and hir1Δ/Δ cells were exposed to oxidative stress with H2O2 and the mRNA expression rate of the catalase CAT1 42 was quantified using RT-qPCR. The amplitude and kinetics of CAT1 induction were almost identical in the WT and the HIR1 mutant (Figure S2), showing that Hir1 function is dispensable for at least one stress response mechanism that requires rapid transcriptional reprogramming.
The HIR histone chaperone complex consists of the four subunits Hir1, Hir2, Hir3 and Hpc232 encoded in the C. albicans genome29. The removal of complex subunits such as Hir2, Hir3 and Hpc2 phenocopied the hir1Δ/Δ deletion (Figs 1D and S1C). This demonstrates that the HIR complex is conserved between S. cerevisiae and C. albicans. Additionally, mass spectrometry analysis of the immunopurified HIR complex from C. albicans showed that Hir3 and Hir2 co-purify with functionally myc-tagged Hir1 (Fig. 1E). Of note, we did not detect Hpc2 in our Co-IP experimental set-up. This might be due to a low silver staining efficiency of Hpc2 as reported earlier33. Since the loss of each individual complex subunit had the same effect on hyphal formation, we also tested how complex formation is affected by the loss of one member. Therefore, Hir1 was myc-tagged in hir2Δ/Δ, hir3Δ/Δ and hpc2Δ/Δ strains and subjected to co-immunoprecipitation. Upon loss of HPC2, Hir2 and Hir3 remained associated with Hir1-myc, while deletion of HIR2 abolishes binding of Hir3 to Hir1-myc. Furthermore, Hir1-myc and Hir2 interaction was still maintained in the absence of HIR3 (Fig. 1E). These data suggest that specific HIR complex subunits may execute distinct functions, ranging from assembly (Hir2) to the recruitment to target genes (Hpc2), as previously speculated in S. cerevisiae 35.
Since chromatin assembly uses two distinct pathways43, we analyzed the specificity of the HIR complex for regulating the yeast-to-hyphae transition. Cells lacking the CAF-1 subunit Cac2 implicated in the replication-coupled chromatin assembly44, showed no defects in hyphal formation (Fig. 1F). By contrast, deletion of RTT106, a histone chaperone involved in both pathways45, phenocopied Hir1 ablation on solid medium, albeit filamentation defects were less pronounced than in hir1Δ/Δ cells, since hyphal formation was not disrupted in liquid medium (data not shown). Hence, hyphal induction rather requires mainly HIR-mediated replication-independent chromatin assembly.
Loss of Hir1 mimics lack of Efg1
HSG induction depends on a complex signaling network. Two major signaling pathways, the cAMP/PKA and a MAPK pathway converge at the activation of downstream transcription factors such as Efg1 and Cph19, 46, and relief from Tup1 and Nrg1 repression16, 47 as shown in Fig. 2A. In S. cerevisiae, the Hir1-containing HIR complex acts as transcriptional repressor34, 36, 48. Hence, we reasoned that transcriptional repressors of HSGs may become deregulated and thus overexpressed in the absence of HIR1. A well-characterized transcription factor for hyphal formation is Nrg1, which represses filamentation. Consequently, its removal leads to constitutive filamentation9, 49. First, we tested whether deletion of HIR1 could alleviate the hyperfilamentation phenotype of nrg1Δ/Δ cells. Epistasis analysis of the hir1Δ/Δ nrg1Δ/Δ double mutant revealed that deletion of HIR1 in the nrg1Δ/Δ background failed to ameliorate the hyperfilamentation phenotype of nrg1Δ/Δ cells (Figure S3A). Of note, transcriptional downregulation of NRG1 was not impaired in hir1Δ/Δ cells upon hyphal triggers, whereas the activation of HSGs like ECE1 was strongly decreased (Figure S3C). Monitoring protein levels of functionally myc-tagged Nrg1 mirrored these data. Nrg1 was degraded independently of HIR1, as soon as cells activated the hyphal program (Figure S3D). These results suggest that hir1Δ/Δ cells are still able to sense hyphal-inducing conditions as NRG1 mRNA downregulation and Nrg1 degradation was fully functional in the mutant.
Figure 2.
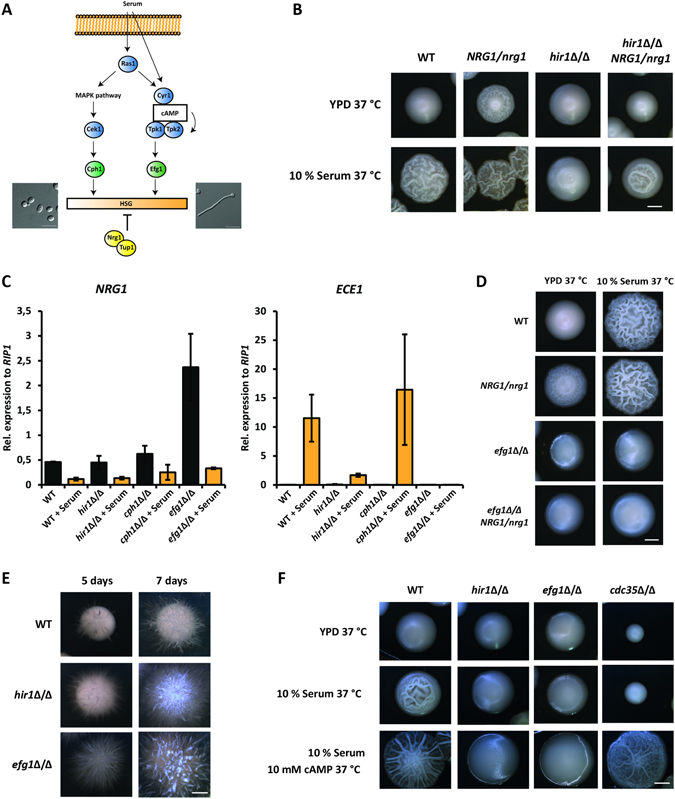
Loss of HIR1 phenocopies deletion of EFG1. (A) Simplified scheme of the major signaling cascades regulating the hyphal transcriptional program. The MAPK cascade and the cAMP/PKA pathway activate transcription factors such as Efg1 and Cph1 to initiate HSG expression. Additionally, transcriptional repressors such as Nrg1 and Tup1 control HSG expression and are themselves regulated by upstream signaling cascades such as cAMP/PKA. (B) Reduction in NRG1 gene dosage alleviates hyphal formation defects of hir1Δ/Δ cells on solid medium. Colony morphology was inspected after 3 days at 37 °C. Scale bar corresponds to 1 mm. (C) NRG1 downregulation is independent of EFG1, CPH1 and HIR1. Gene expression of NRG1 and ECE1 was measured after 30 min of hyphal induction in YPD with 10% FCS by RT-qPCR. Transcript levels were normalized to the expression of the reference gene RIP1. Data are presented as mean + SD of three independent experiments. (D) Reduction of NRG1 gene dosage does not affect efg1∆/∆ hyphal formation. Colony morphology was assessed after 3 days at 37 °C. Scale bar corresponds to 1 mm. (E) Loss of HIR1 partially phenocopies efg1Δ/Δ under embedded growth conditions. Colony pictures were taken after 5 and 7 days at 25 °C. Scale bar corresponds to 1 mm. (F) HIR1 mutant cells are irresponsive to exogenous cAMP supplementation. Colony morphology was assessed after 3 days at 37 °C. Scale bar corresponds to 1 mm.
As stated before, loss of NRG1 leads to a massive hyperfilamenting phenotype due to de-repression of HSGs. To uncover more subtle effects of possible interactions between Hir1 and Nrg1, we tested the effect of the deletion of one NRG1 allele on Hir1-mediated filamentation. In liquid medium, NRG1 heterozygosity in the hir1Δ/Δ background had no effect on hyphal formation after 60 minutes of serum-induced filamentation (Figure S3B). On solid YPD medium at 37 °C, unlike the smooth colony appearance of WT cells, NRG1/nrg1 cells formed wrinkled colonies, indicating filamentous growth. This phenotype was reverted upon genetic removal of HIR1 in the NRG1/nrg1 strain (Fig. 2B). In addition, the NRG1/nrg1 hir1Δ/Δ mutant showed reduced hyphal formation in response to serum when compared to NRG1/nrg1 cells (Fig. 2B). Thus, a subtle de-repression of HSGs by decreased NRG1 gene dosage is not sufficient to trigger full filamentous growth of HIR1-deficient cells.
As mentioned above, major positive regulators of hyphal formation are Cph1 and Efg1, as their deletion results in hyphal formation defects47, 50. We speculated that Hir1 could act in concert with Cph1 or Efg1 to adjust hyphal initiation. Therefore, we directly compared the behavior of cells lacking HIR1, EFG1 or CPH1 during growth in serum-containing YPD at elevated temperatures. Like hir1Δ/Δ cells, efg1Δ/Δ and cph1Δ/Δ cells downregulated NRG1 levels upon hyphal induction (Fig. 2C). However, only hir1Δ/Δ and efg1Δ/Δ knock-out cells lacked full activation of the HSG ECE1. Moreover, loss of CPH1 had no influence on hyphal formation on serum-containing YPD (data not shown) or ECE1 transcript abundance (Fig. 2C). Genetic removal of EFG1 altered ECE1 expression and hyphal formation more severely than a HIR1 deletion (Fig. 2C). Of note, efg1Δ/Δ cells are locked in the yeast phase during serum-induced filamentation15, 51. Furthermore, NRG1 heterozygosity failed to enhance hyphal growth in efg1Δ/Δ cells (Fig. 2D). Interestingly, hir1Δ/Δ cells in part phenocopied efg1Δ/Δ cells under embedded conditions, where Efg1 can act as a repressor of filamentation. Consequently, loss of EFG1 results in hyperfilamentation under these conditions52, 53. Likewise, HIR1-deficient cells showed enhanced hyphal formation during embedded growth (Fig. 2E). Of note, EFG1 transcription in yeast morphology growth was not affected by a HIR1 deletion (Figure S4).
Because cAMP/PKA signaling is essential for hyphal formation3, 54, we tested the integrity of this pathway in hir1Δ/Δ cells. If loss of HIR1 debilitates cAMP generation, exogenous cAMP should restore hyphal formation in hir1Δ/Δ cells. However, exogenous cAMP in hyphal-inducing medium failed to trigger the yeast-to-hyphae transition in HIR1-deficient cells or in cells lacking EFG1 (Fig. 2F). Cdc35, also known as Cyr1, is the adenylate cyclase synthesizing cAMP55. Unlike hir1Δ/Δ and efg1Δ/Δ cells, hyphal defects seen in a cdc35Δ/Δ mutant are fully restored upon the addition of exogenous cAMP (Fig. 2F). Notably, cAMP/PKA signaling is also involved in transcriptional downregulation of Nrg116, 56. Thus, NRG1 expression analysis (Figure S3C) suggests that the unresponsiveness of HIR1-deficient cells to cAMP signals (Fig. 2F) cannot be a consequence of defective cAMP/PKA signaling. These results suggest that Hir1 might affect hyphal formation downstream of cAMP/PKA signaling in the same pathway as Efg1.
HIR1-deficiency impacts the amplitude of hyphal-specific gene transcription
We37 and others40 demonstrated that the yeast-to-hyphae transition is accompanied by massive transcriptional changes. To determine how removal of HIR1 affects transcriptional responses, we performed RNA-seq of hir1Δ/Δ versus WT cells under yeast phase promoting conditions and after 30 minutes in YPD plus 10% serum at 37 °C. During yeast phase growth, some 189 genes were at least 1.5-fold upregulated, while 100 genes were downregulated to the same extent in hir1Δ/Δ cells when compared to WT cells. This indicates that Hir1 is acting not only as a transcriptional repressor, but is also implicated in transcriptional activation. The amplitude of the majority of differentially expressed genes covered a fold-change of 1.5 to 2.8 (Fig. 3A). Remarkably though, the number of differentially expressed genes between hir1Δ/Δ and WT almost doubled upon hyphal initiation when compared to the yeast phase. We found 284 genes at least 1.5-fold upregulated and 280 genes downregulated (Fig. 3A). As already observed for the yeast growth phase, the amplitude of transcriptional changes after removing HIR1 ranged from a fold-change of 1.5 and 2.8 (Fig. 3A). These data strongly suggest that Hir1 affects fine-tuning of transcriptional amplitudes upon hyphal formation.
Figure 3.

Hir1 affects the transcriptional amplitude during initiation of hyphal formation. (A) Absolute numbers of differentially expressed transcripts between WT and the hir1Δ/Δ mutant during yeast (y) or hyphal (h) growth. Differentially expressed genes were defined by FDR < 0.05. FC, fold change. (B) The transcriptional profile of hir1Δ/Δ cells showed high similarity to WT cells. log2-fold changes in RNA expression in WT cells during the yeast-to-hyphae transition (x-axis) were plotted against the log2-fold change in mRNA levels in hir1Δ/Δ cells during the yeast-to-hyphae transition (y-axis). (C) Loss of HIR1 causes transcriptional deregulation during hyphal initiation. Each dot corresponds to one ORF. The log2-fold changes in RNA expression in WT cells during the yeast-to-hyphae transition (x-axis) were plotted against the log2-fold changes between HIR1 mutant and WT cells in response to hyphal-inducing conditions (y-axis). (B,C) Significantly differentially expressed genes in the hir1Δ/Δ knock-out versus WT are depicted in green and were defined by an at least 1.5-fold change and FDR < 0.05.
Detailed inspection of transcriptomes from the WT and hir1Δ/Δ cells during the yeast-to-hyphae transition revealed qualitatively similar transcriptional responses concerning the nature and type of genes regulated in both strains (Fig. 3B). For instance, ECE1 and HWP1 showed the highest induction in WT cells following hyphal growth stimulation. The same was true for HIR1-deficient cells (Fig. 3B, top right corner). Similarly, FET3 and CFL2 were the most repressed genes in the WT and hir1Δ/Δ mutant (Fig. 3B, bottom left corner). Remarkably, the same genes (ECE1, HWP1, FET3, CFL2) were also among the most up- as well as downregulated genes in hir1Δ/Δ hyphae when compared to WT hyphae (Fig. 3C). However, it was obvious that genes with increased expression in WT during the yeast-to-hyphae transition showed much lower transcript levels in the absence of HIR1 and vice versa. In summary, these data demonstrate that transcriptional amplitudes of both activated and repressed HSG sets dramatically change during morphogenesis-associated transcriptional reprogramming upon loss of the Hir1 chaperone complex subunit (Fig. 3C). Hence, Hir1 can function both as transcriptional activator and repressor, thereby affecting the transcriptional fine-tuning in response to hyphal stimuli.
The loss of HIR1 affects different biological processes upon hyphal induction
The RNA-seq data of hir1Δ/Δ cells in the yeast growth phase revealed that major transcriptional changes already occurred during this growth phase (Figure S5A). Gene ontology (GO) analysis identified the gene sets affected by HIR. Differentially expressed genes in hir1Δ/Δ cells fall into fatty acid metabolism, cell surface remodeling and oxido-reduction processes, including genes involved in iron acquisition (FRP1 and CFL22) and anti-oxidative enzymes (SOD5 and GPX2). Furthermore, genes associated with DNA repeat maintenance and various transcription factors showed altered expression upon HIR1 deletion (Figure S5A,C). Of note, less than 30% of all upregulated genes in hir1Δ/Δ cells (either during yeast growth or hyphal induction) were mapped to a well-characterized GO category.
Interestingly, transcriptional changes owing to the loss of HIR1 were more severe during hyphal initiation than during the yeast phase (Fig. 3A). Further characterization of this gene set showed that upregulated transcripts encoded metabolism genes involved in glycolysis, fatty acid catabolism and the glyoxysome function (Figure S5B,D).
Downregulated genes are implicated in ER to Golgi vesicle trafficking, cell wall remodeling, filamentous growth and thus, fungal virulence (Figure S5B,D). The GO term analysis suggests that similar gene clusters are differentially regulated during hir1Δ/Δ yeast and hyphal growth. This was substantiated by the fact that many of the genes included in one GO group for hyphal hir1Δ/Δ cells were found differentially expressed in hir1Δ/Δ yeast growth (Figure S5A,B, gene*). However, the total number of overlapping genes with altered expression in hir1Δ/Δ yeast or hyphal phase is lower than suggested by the GO term analysis. Roughly one third of upregulated genes in HIR1-deficient hyphal cells were already elevated during yeast growth (106 out of 289). Moreover, only one fifth of all downregulated genes upon hyphal initiation in hir1Δ/Δ cells were also found in the yeast phase of the mutant (57 out of 280; Figure S5E,F). The majority of differentially expressed genes in hir1Δ/Δ cells were detected specifically upon hyphal induction. Hence, Hir1 function becomes more important for sustaining transcriptional responses to changing growth conditions, thereby affecting various biological processes, ranging from transcription control of metabolism, ER-Golgi trafficking to cell wall modulation.
In addition to common biological functions of Hir1-regulated genes, we analyzed common genomic sequence elements of gene sets. Of note, previous studies reported unusually long intergenic regions upstream of HSGs in C. albicans and upstream of developmentally regulated genes in S. cerevisiae 57–59. To evaluate whether Hir1-affected genes harbor unusually long upstream intergenic regions, we analyzed those sequences of genes differentially regulated in hir1∆/∆ cells during yeast phase growth (hir1-y) and in response to hyphal stimuli (hir1-h). We found that the median upstream intergenic regions of hir1-y- and hir1-h-affected genes are above-average in size when compared to all expressed genes in our RNA-seq dataset (Fig. 4A). Furthermore, almost 50% of differentially regulated transcripts in hir1-y and hir1-h cells belonged to the top 25% upstream intergenic length quartile (4th quartile) (Fig. 4B). These data provide compelling evidence that Hir1-regulated genes tend to have above-average upstream intergenic regions, which is implying complex transcriptional control by chromatin architecture as well as by a variety of transcriptional regulators57, 60.
Figure 4.

Hir1-affected genes harbor above-average intergenic upstream regions. (A) Intergenic upstream regions were extracted from the C. albicans genome assembly 22 and length distribution of those elements is displayed for all genes detected in the RNA-seq (all genes), for differentially expressed genes (DEGs) in hir1∆/∆ cells during yeast growth (hir1-y vs WT-y) and upon hyphal induction (hir1-h vs WT-h). Boxplot whiskers represent the 5–95 percentiles and the dotted line the median upstream intergenic length in “all genes”. Statistical analysis was carried out using Kruskal-Wallis with Dunn’s post hoc test. ***P < 0.001. (B) The “all genes” data set was split into quartiles based on the overall upstream intergenic length distribution. The relative gene number and the proportions (%) of DEGs in hir1-y vs WT-y and hir1-h vs WT-h in each quartile are displayed. (A,B) Differentially expressed genes were defined by an at least 1.5-fold change with an FDR value < 0.05.
HIR1 deficiency alters the histone densities at the HWP1 and UME6 upstream intergenic regions
Transcriptomic analysis of hir1Δ/Δ cells suggested that HIR1-deficiency strongly affects the transcriptional amplitudes of regulated genes during hyphal formation. Hir1 and homologues are thought to act in promoter regions of actively transcribed genes to aid deposition of histone H3/H4 dimers32. Therefore, we hypothesized that the loss of HIR1 might result in an altered chromatin state at gene promoters including HSGs. Since histone enrichment in the HWP1 promoter revealed chromatin dynamics during hyphal formation16, 17, and because HWP1 showed highest induction upon hyphal stimuli in WT and hir1Δ/Δ cells (Fig. 3B), we determined histone H3 occupancy in the HWP1 promoter. The HWP1 upstream intergenic region of about 2 kb contains several putative binding sites for transcription factors implicated in hyphal formation, including Efg1, Brg1 and Nrg117, 59, 61 (Fig. 5D). Chromatin immunoprecipitation analysis of histone H3 density at the HWP1 promoter in close proximity to the start codon and near a putative Nrg1 binding site showed a significantly increased histone density in hir1Δ/Δ yeast cells when compared to WT yeast cells (Fig. 5A, yeast and Fig. 5D). When cells were subjected to hyphal-inducing conditions, histone levels significantly decreased in the WT, reflecting the onset of HWP1 transcription. In contrast, histone density was only slightly affected in the mutant (Fig. 5A, hyphae). A similar behavior was observed upstream in the HWP1 promoter region about 1.25 kb upstream from the translational start site (Fig. 5B, yeast and hyphae). This HWP1 promoter element has been described as crucial region activating transcription61, since it harbors putative binding sites for hyphal activators, including Brg1 and Efg1, as well as for the Nrg1 repressor (Fig. 5D). Based on these results, we hypothesized that altered histone occupancy in the HWP1 promoter affects the induction rate of transcription of HSGs such as HWP1. Indeed, the maximal amplitude of HWP1 expression was altered in hir1Δ/Δ cells, while the overall pattern per se was qualitatively similar to WT cells (Fig. 5C).
Figure 5.

Hir1 affects the chromatin density in the HWP1 and UME6 promoter. (A,B) Loss of HIR1 increases the histone density in the HWP1 promoter. Histone density was measured using histone H3 ChIP and qPCR at different HWP1 promoter regions. (C) HWP1 expression is reduced in hir1Δ/Δ cells. WT and hir1Δ/Δ cells were grown in YPD with 10% FCS at 37 °C. Cultures were collected at the indicated time points followed by RNA extraction. Gene expression of HWP1 was measured via RT-qPCR and transcript levels were normalized to the reference gene RIP1. (D) Representative illustration of transcription factor binding sites in the HWP1 upstream intergenic sequence. Efg1 (yellow) and Nrg1 (red) binding motifs were taken from ref. 59 and Tec1 (green) and Brg1 (blue) from ref. 13. Note that no putative Tec1 binding site was identified in our in silico scan. (E) The same as in (C), but for UME6. (F,G) Genetic removal of HIR1 alters histone occupancy at distinct UME6 promoter regions. Histone density was measured using histone H3 ChIP and qPCR at different UME6 promoter regions. (H) Putative transcription factor binding sites for the upstream intergenic region of UME6 are represented as in (D). Transcription factor sites with asterisk (*) indicate multiple sites for a given regulator within less than 100 bp. (I) HIR1-deficient cells require stronger signal intensity to initiate hyphal formation. WT and hir1Δ/Δ cells were spotted on YPD supplemented with a continuous GlcNAc concentration gradient of 0–10 mM. Colony morphology was inspected after growth for 3 days at 37 °C. (A,B,F,G) The qPCR signals from Input and IP were normalized to an intergenic region on chromosome R. The ratio of normalized Input/IP values is shown on the y-axis labeled as “H3 density”. (A–C,E–G): Data are presented as mean + SD of three independent experiments. For significance testing, hir1Δ/Δ cells were compared to WT cells. *P < 0.05, **P < 0.01 with Student’s t-test.
The UME6 gene encodes a transcription factor that sustains hyphal induction60, 62. Ume6, unlike Hwp1, is essential for hyphal maintenance63, 64. Interestingly, the RNA-seq data showed that UME6 is significantly downregulated in hir1Δ/Δ hyphal cells when compared to the WT. This was confirmed by quantifying UME6 mRNA levels in the WT and hir1Δ/Δ cells upon hyphal induction for 2 hours (Fig. 5E). Like HWP1, UME6 expression followed the same kinetics in WT and hir1Δ/Δ cells. However, UME6 transcript induction rate remained lower in hir1Δ/Δ cells (Fig. 5E). We then analyzed the histone density of the UME6 promoter at around 5.3 kb upstream of the start codon. This region harbors a potential binding site for Brg160, Efg1 and Tec1 (Fig. 5H). As seen for the HWP1 promoter, loss of HIR1 increased histone occupancy at the UME6 promoter site in the yeast phase (Fig. 5F, yeast). Upon hyphal induction, WT and hir1Δ/Δ cells decreased histone enrichment, whereas hir1Δ/Δ hyphal cells retained a significantly higher histone density than the WT (Fig. 5F, hyphae). Inspection of the UME6 promoter around 4.6 kb upstream of the start codon showed a similar pattern albeit histone density was increased significantly only after hyphal induction in hir1Δ/Δ cells (Fig. 5G). This region contains putative binding sites for the Nrg1 repressor, as well as for Brg1 (Fig. 5H). Taken together, these results indicate that the HIR chaperone complex affects the local chromatin density in HSGs already during yeast phase growth, which might impact the recruitment of transcriptional regulators to their target promoters65. We identified several putative regulators involved in the control of genes differentially expressed only in hir1Δ/Δ yeast or hyphal cells and of genes deregulated in both growth phases (Figure S6A). Thereby, six regulators (Ndt80, Tec1, Sfl2, Fkh2, Mrr1 and Tye7) were associated with all three gene sets. Strikingly, four of these factors, Ndt80, Tec1, Sfl2 and Fkh2, are known regulators of the yeast-to-hyphae transition13, 66, 67. Of note, TEC1 and SFL2 are downregulated upon the loss of HIR1 (Figure S5A,B) both in the yeast and hyphal growth phase.
In addition to HIR1-affected genes such as UME6 and HWP1, we also assessed histone occupancy in NRG1 and CAT1 promoter elements. The transcriptional regulation of these genes did not require Hir1 (as shown in Figures S2 and S3C). We found slightly increased histone levels in the NRG1 and CAT1 promoter only upon hyphal induction in HIR1-deficient cells (Figure S6B,C), which could be a consequence of general chromatin alterations in the HIR1 mutant that might not necessarily affect transcription.
Due to increased basal histone densities at the HWP1, UME6 and possibly other gene promoters, the threshold sensitivity to signals required for switching to the hyphal growth phase could be increased in HIR1-deficient cells. To test this notion, we spotted WT and hir1Δ/Δ cells onto YPD plates containing a GlcNAc concentration gradient as the main filamentation trigger (Fig. 5I). Indeed, the WT was dramatically more sensitive to morphogenetic signals along the entire concentration range of GlcNAc. In contrast, hir1Δ/Δ cells required much higher GlcNAc concentrations to initiate hyphal growth. This result provides evidence that, HIR1 deficiency alters the responsiveness to hyphal growth signals.
Ectopic expression of UME6 restores hyphal formation in hir1Δ/Δ cells
Hyphal induction entails transcriptional activation of circuits acting at different layers. For example, Efg1 is one of the first transcription factors to integrate primary signals, which are transduced to another set of regulators required to maintain HSG expression and hence, filamentation3, 9, 46. One of the key downstream regulators is Ume6, which suppresses defective hyphal formation of efg1Δ/Δ cells68, 69. Our expression and ChIP analysis showed that UME6 expression was compromised upon genetic removal of HIR1. Given that Ume6 is indispensable for sustaining hyphal programs, we investigated whether ectopic overexpression of UME6 can rescue defective hir1Δ/Δ hyphal formation. Therefore, we used the conditional tet-ON system70, where UME6 expression is under the control of a tetracycline-inducible promoter, allowing for UME6 transcription by adding tetracycline. The tet-ON system had no effect on hyphal formation of WT or hir1Δ/Δ cells without tetracycline (Fig. 6A, upper panel). Tetracycline addition increased hyphal formation in WT cells when compared to the respective background control strain on serum-containing YPD at 37 °C (Fig. 6A, lower panel). Strikingly, hyphal induction of the tet-UME6 expressing hir1Δ/Δ cells was fully restored upon tetracycline supplementation. To determine how sensitive hir1Δ/Δ cells are to transcriptional induction of UME6, we cultivated WT and hir1Δ/Δ cells in the presence of different tetracycline concentrations to modulate UME6 levels. Remarkably, WT cells responded to 5 µg/ml tetracycline and hir1Δ/Δ cells required at least a four-fold higher concentration of tetracycline (20 µg/ml) for triggering hyphal induction (Fig. 6B).
Figure 6.

Ectopic expression of UME6 restores hyphal formation in the absence of HIR1. (A,B) The UME6 gene was placed under conditional expression control using the tet-ON system70. Expression was triggered by adding 50 µg/ml tetracycline (A) or various concentrations of tetracycline (B). Colony morphology was assessed after growth for 2 days at 37 °C. Scale bar corresponds to 1 mm.
In summary, ectopic overexpression of Ume6 rescued filamentation defects of hir1∆/∆ cells, suggesting that Hir1 acts at the primary transcriptional valve or molecular clutch, possibly acting in concert with Efg1 upstream of Ume6. These data provide compelling evidence for a tightly interlocked dual-layer regulatory mechanism, which involves both dedicated transcription factors and chromatin architecture to control developmental changes such as morphogenetic cell fate decisions in response to environmental stimuli.
Discussion
Here, we demonstrate a genetic link between replication-independent chromatin assembly and the transcriptional control of morphogenesis in the human fungal pathogen C. albicans. We show that Hir1, a subunit of the conserved HIR histone chaperone complex, affects transcriptional amplitudes during the yeast-to-hyphae transition, as activator and repressor of gene expression. The HIR complex is an evolutionary conserved complex initially discovered as a cell cycle-dependent repressor for 6 out of the 8 core histone genes in S. cerevisiae 34, 71. In multicellular organisms, HIR orthologues are implicated in embryonic as well as plant development72–74. In S. cerevisiae, bulk chromatin alterations that affect heterochromatin maintenance, chromosome segregation and DNA damage sensitivity have only been observed in combinatorial deletions of HIR1 and the CAF-1 complex subunit CAC2 75, 76. Recently, the conservation of these overlapping functions has been revealed in C. albicans. It was further demonstrated that Hir1 only affects the morphogenetic opaque to white cell switching upon deletion of the CAF-1 complex subunit CAC2 29. We previously demonstrated a CAF-1-independent role of Hir1 in antifungal azole tolerance30. Here, we explore further novel functions of Hir1 in fungal morphogenesis, which are independent of CAF-1. We reveal a plausible mechanism for HIR-mediated expression control of genes required for cell fate decisions during morphogenetic alterations in C. albicans.
The ability to undergo and regulate hyphal formation has been viewed as an important virulence factor of C. albicans 10, 77. The main pathway integrating environmental stimuli to drive HSG transcription is the cAMP/PKA pathway, which is, among many other functions, essential for hyphal growth54, 55, 78. To translate environmental stimuli into cell-fate changes, this pathway induces a hyphal-specific transcriptional program by activating Efg1 thereby relieving repression by Nrg19, 56, 79. Additionally, earlier reports demonstrated that sustained hyphal growth requires the concerted action of transcriptional activators such as Brg1 and Ume6, the full relief of Nrg1-mediated repression, but also chromatin-associated activities16, 17. Thus, at least two regulatory mechanisms execute the yeast-to-hyphae transition, namely hyphal initiation and its maintenance as long as the appropriate signals are present16. Our data suggest that Hir1 is involved in the first transcriptional wave, acting downstream of cAMP/PKA signaling. First, although defective in hyphal formation, hir1Δ/Δ cells maintain normal cAMP/PKA signaling, since exogenous supplementation of cAMP fails to restore hyphal growth of hir1Δ/Δ mutants. Moreover, the PKA subunit Tpk2 and the adenylate cyclase Cyr1 are required for transcriptional downregulation of Nrg156, which is not affected by the loss of HIR1 following hyphal stimuli. Second, ectopic expression of UME6, which is required for hyphal elongation and constitutes a key element of the second transcriptional phase to maintain the morphological stage, can rescue the filamentation defect of HIR1-deficient cells, as well as of the efg1Δ/Δ mutant68. Interestingly, hir1Δ/Δ cells hyperfilament under embedded conditions, which closely resembles the lack of EFG1 and CYR1 80. Thus, it is tempting to speculate that the action of Hir1 is linked to Efg1-mediated hyphal initiation.
Hir1 is part of a histone chaperone complex, which incorporates histones onto chromatin in a replication-independent manner32. Hence, the chromatin architecture at gene promoters could be specifically affected upon the loss of HIR1. Here, we show that hir1Δ/Δ cells have increased histone levels already during the yeast growth phase at distinct promoter elements of the HSGs HWP1 and UME6. Moreover, both genes displayed elevated histone densities at relevant promoter regions in the hir1Δ/Δ mutant in response to hyphal induction, which is reflected in decreased transcript abundance. At this point, we do not know whether increased histone occupancy at these promoters is the cause or the consequence of reduced transcriptional induction of UME6 or HWP1. Moreover, hir1Δ/Δ hyphal cells showed slightly increased histone densities at promoter regions of NRG1 and CAT1, for which transcriptional control is unaffected by HIR1-deletion. Elevated histone levels in those regions only occurred in response to hyphal induction. Since the HIR complex is a transcriptional repressor of histone genes, we cannot entirely exclude subtle effects of HIR1-deficiency on the global chromatin architecture. Given that these effects exist, they most likely do not influence bulk gene transcription, as otherwise expression of the entire genome would become deregulated in hir1Δ/Δ cells, which was clearly not the case. Strikingly, in contrast to constitutively expressed genes, highly regulated genes differ in their promoter constellation and often harbor multiple transcription factor binding sites. Hence, they are more likely to be influenced by subtle changes in nucleosome pattern in promoters rather than by the total nucleosome occupancy81, 82. Therefore it is plausible that the deregulation of chromatin architecture upon loss of HIR1 may primarily affect genes with complex regulation. Due to the limited number of loci for which we could inspect nucleosome occupancy, conclusions about a global impact of the HIR complex on chromatin organization are speculative at this point. Thus, further experiments are required to unravel a possible role for the HIR complex in chromatin homeostasis in C. albicans. However, in silico analysis of intergenic upstream regions from Hir1-dependent genes during yeast or hyphal growth showed that those regions are surpassingly long compared to the median length in the C. albicans genome. Genes implicated in developmental changes such as HSGs often harbor long 5 prime intergenic regions with multiple binding sites for transcriptional regulators, which imply complex transcriptional control, including chromatin remodeling57, 59. Hence, it is tempting to speculate that transcription of HSGs, which are regulated by long promoter regions, are more likely to be subject to transcriptional de-regulation when chromatin homeostasis is disturbed. This potentially impacts early transcriptional fine-tuning following hyphal induction stimuli. Defective transcriptional amplitudes can then be compensated by enhancing upstream signaling intensity for hyphal induction. Thus, the interplay of chromatin state and transcriptional control could provide a platform to efficiently adjust the sensitivity towards stimulating cues, and thus, modulate transcriptional threshold levels required for developmental changes or environmental responses.
Here we show that although hir1Δ/Δ cells induce the hyphal-specific transcriptional program in response to hyphal triggers, efficient hyphal formation is impaired due to altered amplitudes of this response. Indeed, we demonstrate that HIR1-deficient cells are less sensitive to hyphal-inducing conditions and that this defect can be compensated by stronger trigger for hyphal development or by ectopic overexpression of the UME6 transcription factor for hyphal maintenance. Therefore, it is tempting to speculate that Hir1 affects initial fine-tuning of transcription by assisting in homeostasis of the chromatin template and thereby safe-guarding the transition from the yeast to the hyphal growth phase. A similar mechanism has been shown for the mammalian homologue of Hir1, HIRA, which deposits the histone variant H3.3 preferentially at gene promoters83. In a mouse oogenesis model, HIRA is required for normal chromatin homeostasis and transcriptional fine-tuning. Similarly, binding of the polycomb repressive complex 2 (PRC2) to developmental genes upon differentiation stimuli depends on HIRA-mediated H3.3 deposition in mouse embryonic stem cells84.
Hir1/HIRA may be recruited via different components, including transcription factors84, 85, RNA Polymerase II and potentially even naked DNA86. We identified six transcription factors (Ndt80, Tec1, Sfl2, Fkh2, Mrr1 and Tye7) with target genes differentially regulated only in hir1Δ/Δ yeast or hyphal cells and also genes that are differentially expressed in both growth phases. Interestingly, Ndt80, Tec1, Sfl2 and Fkh2 are implicated in hyphal formation and TEC1 and SFL2 expression seems to depend on Hir1 action, since both genes are downregulated upon HIR1-deletion. Further experiments are still required in order to reveal to what extent HIR modulates the affinity of transcription factors or the general transcription machinery to HSGs. Since only half of all differentially expressed genes in the hir1Δ/Δ mutant are altered in a hyphal-specific context, it will be of great interest to investigate how Hir1 affects the chromatin architecture and to unravel the precise mechanism of Hir1 recruitment to target genes in C. albicans. Of note, we were unable to perform ChIP experiments with the Hir1-myc tagged strain to show a direct interaction of Hir1 with HSG promoters. However, this could be explained by transient nature of interactions, insufficient cross-linking of Hir1 and target DNA caused by a possible as yet unknown intermediate regulatory factor recruiting Hir1, as well as by high off rates of regulators.
Taken together, our study provides evidence for the necessity of the tight control of gene expression amplitudes upon encounter of changing environmental conditions exemplified here with the C. albicans yeast-to-hyphae transition. The HIR complex seems to contribute to the regulation of the initial phase of hyphal formation, where it might control signal sensitivity by acting as a valve or clutch that controls transcriptional loads via chromatin homeostasis. This way, the HIR complex could assist in the swift execution of responses to stimuli, and in the case of C. albicans, ascertains rapid adaptions to changing host environments. Importantly, the replication-independent nature of HIR function offers a major advantage for fungal cells, since it enables adaptive changes in gene expression without involving excessive and energy-intensive replication or cell cycle control. Hence, further exploring the general relevance of HIR-dependent transcriptional regulation will provide exciting new insights into C. albicans pathophysiology and host adaption as well as fungal virulence.
Materials and Methods
Media, chemicals and growth conditions, colony morphology and microscopy
Rich medium (YPD) was prepared as described previously87. Spider medium contained 1% Bacto yeast extract (BD Biosciences, Vienna, Austria), 1% mannitol (Merck, Vienna, Austria), 0.2% KH2PO4, 2% histidine, 2% leucine (all Sigma-Aldrich, Vienna, Austria) and 2% Bacto agar (BD Biosciences) for solid medium. C. albicans strains were routinely grown on YPD at 30 °C. For hyphal induction, a fresh saturated overnight culture was harvested, washed once with distilled water, resuspended in an equal volume corresponding to the starting overnight culture and diluted 1:10 in pre-warmed fresh YPD containing 10% fetal calf serum (FCS; serum)39. These cultures were then further grown at 37 °C for the indicated periods of time. Cultures were either used directly for further analysis or snap-frozen at −80 °C.
To investigate hyphal induction on solid medium, overnight cultures were diluted to an OD600 of 0.1 in fresh YPD and then propagated to the logarithmic growth phase. Cells were counted on a CASY® cell counter and approximately 50 (10 cm dishes) or 10 (6 cm dishes) cells were plated on YPD agar plates supplemented with FCS, N-acetyl glucosamine (GlcNAc), 3′,5′-Cyclic adenosine monophosphate (cAMP) (all Sigma-Aldrich) or Spider medium.
Colony morphology and microscopic inspection of single cells was performed as described previously22. For quantification of the yeast and hyphal fraction of a population, at least 200 cells were counted.
Plasmid and strain constructions
The C. albicans strains, primers and plasmids used in this study are listed in Table S1. All C. albicans strains were derived from the MTL a/α clinical isolate SC531488. The strains lacking HIR1, CDC35, CAC2, RTT106, CPH1 and EFG1 have been reported earlier22, 30, 89.
The HPC2 and HIR3-deficient mutants were generated using a modified SAT1-flipper technique90. Briefly, approximately 500 bp up- and downstream sequences of the target gene ORF were PCR amplified and cloned into the plasmid pSFS3b using ApaI, KpnI and BglII, NotI, respectively. Deletion of NRG1 was done in a similar way using the plasmid pSFS2a91. The plasmid for HIR2 deletion was constructed by fusing approximately 500 bp up- and downstream region of the target gene, the FLP-FRT-NAT1-FRT marker from pSFS3b and the pSFS3b backbone by an in vivo cloning approach in E. coli EL350 as described in ref. 92. Ectopic overexpression of UME6 was accomplished by the tet-ON system using the pNIM1 plasmid described earlier70. Gene-complemented strains were generated by cloning the HIR1 ORF with ApaI and KpnI upstream of the FLP recombinase and NAT1 into pSFS3b. 9myc epitope-tagging of HIR1 and NRG1 alleles was carried out by fusion PCR93 using plasmid pFA6a-9myc-NAT1 as a template. Correct genomic integration of the deletion cassette and re-integration constructs was verified with colony PCR, Southern Blot (for HIR1 knock-out, 9myc-tagged and complementation strain only) and/or immunoblotting (for epitope-tagged strains only).
Immunoblotting, immunoprecipitation and mass spectrometry analysis
Whole-cell-free extracts and immunoblotting was performed exactly as described earlier90. For detection of Nrg1-9myc, a mouse monoclonal anti-myc antibody (clone 4A6, MFPL monoclonal antibody facility, Vienna, Austria) was used. An anti-PSTAIRE antibody recognizing Cdc28 (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a protein loading control. Protein bands on the nitrocellulose membrane were visualized using an Odysee® CLx scanner (Li-Cor®, Lincoln, NE). Cells were prepared for immunoprecipitation exactly as described previously90. Whole-cell extracts corresponding to 50 OD600 units were incubated with 4 µl of the monoclonal anti-myc antibody. After an overnight incubation at 4 °C, 30 µl of Protein-G-coupled Dynabeads (Invitrogen, Vienna, Austria) were added for 2 hours at 4 °C. Subsequent washing steps were performed as described for the ChIP experiments, except that PBS instead of 1xTE was used for the final washing steps. The beads were then resuspended in Laemmli buffer for SDS-PAGE analysis. Silver staining of 10% SDS-PAGE gels was performed essentially as described elsewhere94. To identify resolved proteins, protein bands were cut out from the silver-stained gel, digested with trypsin and separated using an LC system for subsequent mass spectrometry analysis. Mass spectrometry analysis was carried out at the mass spectrometry facility at the Vienna Biocenter. The raw spectra were matched against the Candida albicans genome database (http://www.candidagenome.org/).
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described earlier37. For histone ChIP, 1 mg cell free protein extract was incubated with 1 µl of anti-Histone H3 antibody (#1791, Abcam, Cambridge, UK). After overnight incubation at 4 °C, 30 µl of Protein-G-coupled Dynabeads (Invitrogen) were added for 2 hours at 4 °C. Subsequent washing steps and DNA purification were performed as described previously using minor modifications95. Input and ChIP DNA were incubated with 5 µl of 10 mg/ml DNase-free RNase A at 65 °C for 16 hours and recovered using a MiniElute® PCR Purification Kit (Qiagen, Hilden, Germany). Input DNA was quantified on a NanoDrop-2000 spectrophotometer (Peqlab, Erlangen, Germany) and ChIP DNA concentration was measured using the Quantifluor® dsDNA System (Promega, Mannheim, Germany) in accordance to the manufacture’s protocol. Histone density at the HWP1 promoter was analyzed using primers amplifying the region ranging from −243 to −106 and from −1299 to −1195 with respect to the start codon. To analyze the UME6 promoter region, primers ranging from −4534 to −4676 and from −5210 to −5371 relative to the start codon were used. For the NRG1 and CAT1 promoter, a region ranging from −1702 to −150237 and −162 to −30630, respectively, was inspected. Input and IP qPCR signals were normalized to an intergenic region on chromosome R (RT5_tC_inter and RT3_tC_inter primers), before calculating the Input/IP ratio. The displayed “H3 density” in Figs 5A,B,F,G and S6B,C represents the enrichment over Input relative to the aforementioned intergenic region.
RNA isolation, RT-qPCR analysis, RNA-seq analysis and bioinformatics
RNA isolation, cDNA synthesis and qPCR analysis was performed exactly as described previously90. Relative quantification of mRNA levels was done by qPCR using the efficiency corrected ΔΔCt method96. RIP1 was used as a reference gene22. Statistical analysis was performed in Excel (Microsoft). DNase-treated RNA was subjected to rRNA depletion using the RiboMinusTM Eukaryote System v2 (Life Technologies, Vienna, Austria) adhering to the recommended protocol. The efficiency of rRNA depletion was checked using a Bioanalyzer with a RNA6000 Nano chip (Agilent, Vienna, Austria). Fragmentation of rRNA-depleted RNA was done using the NEBNext Magnesium RNA fragmentation module (New England BioLabs, Vienna, Austria) following the manufactures instructions. Samples were incubated at 94 °C for 4 minutes in a thermocycler. Fragmented samples were recovered using the RNeasy MiniElute Cleanup Kit (Qiagen) and subjected to 1st strand synthesis using SuperScript III Reverse Transcriptase (Life Technologies) and 3 µg random primers (Life Technologies, Vienna, Austria). Mini Quick Spin columns (Qiagen) were used to clean up the single-stranded cDNA, which was subsequently subjected to 2nd strand synthesis using the NEBNext mRNA Second Strand Synthesis Module (New England BioLabs) following the manual. Additionally, T4 DNA polymerase (New England BioLabs) was added for the last 5 minutes to polish overhanging ends. Double-stranded cDNA was cleaned up using a MiniElute PCR Purification Kit (Qiagen). DNA concentration was measured using the PicoGreen® dsDNA quantification reagent (ThermoFisher Scientific, Vienna, Austria).
Double-stranded cDNA was further processed and sequenced on a HiSeq. 2500 instrument (Illumina, San Diego, CA) by the Vienna Biocenter Campus Support Facility CSF (NGS unit, http://www.csf.ac.at). Two biological replicates for the WT and the hir1Δ/Δ cells during yeast growth or hyphal induction were subjected to RNA-seq analysis.
RNA-seq reads were mapped to the C. albicans genome Assembly 21 using TopHat, allowing for only uniquely mapped reads97. All reads mapping to rRNA loci were removed. Read counts were determined with HTSeq using the union mode98 and a reference annotation (C_albicans_SC5314_version_A21-s02-m07-r10; http://www.candidagenome.org). Differential gene expression was analyzed using edgeR99. P-values were adjusted to determine differentially regulated genes100. Venn diagrams were created by Venny 2.0.2 (http://bioinfogp.cnb.csic.es/tools/venny/index.html)101. GO analysis was carried out with FungiFun2 (https://elbe.hki-jena.de/fungifun/fungifun.php)102. The whole dataset of detected protein-coding genes and their expression can be found in Table S2.
In silico upstream intergenic sequence analysis
Sequence information for all C. albicans ORFs plus intergenic up- and downstream sequences were retrieved from the Candida genome database (CGD; http://www.candidagenome.org; C_albicans_SC5314_version_current_orf_plus_intergenic.fasta accessed on April 24th, 2017). Putative transcriptional regulators of differentially expressed genes in HIR1-deficient yeast and hyphal cells were identified using the PathoYeastract Database “Rank by TF” tool, allowing only for hits with verified target binding and expression data103.
Data availability
The authors confirm that all data underlying the findings are fully available without restrictions. All relevant data are within the paper and its Supporting Information files.
Electronic supplementary material
Acknowledgements
We thank all laboratory members for helpful discussions. We are grateful to Joachim Morschhäuser for providing tools for the tet-ON conditional expression system. This work was supported by a grant from the Austrian Science Found FWF (FWF-P25333-B22) to KK, and by NIH-R01 Grant R01AI124499 to NC and KK. MG and CJN were supported by the National Institutes of Health (NIH) grant R00AI100896 and by a Biomedical Scholar Award from the Pew Charitable Trusts. Sequence data for C. albicans were retrieved from the Stanford Genome Center (http://www.candidagenome.org).
Author Contributions
Wrote the manuscript: S.J. and K.K. Conceived and designed experiments: S.J., M.T., K.K. Performed experiments: S.J., M.G., C.J.N. and N.C. Analyzed data: S.J. and K.K. Contributed materials: N.C.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08239-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boskovic A, et al. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes & development. 2014;28:1042–1047. doi: 10.1101/gad.238881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y, Su C, Liu H. Candida albicans hyphal initiation and elongation. Trends in microbiology. 2014;22:707–714. doi: 10.1016/j.tim.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS pathogens. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan L, Yang C, Tang J. Disruption of the intestinal mucosal barrier in Candida albicans infections. Microbiological research. 2013;168:389–395. doi: 10.1016/j.micres.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Barelle CJ, et al. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cellular microbiology. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, G. D. et al. Hidden killers: human fungal infections. Science translational medicine4, 165rv113, doi:10.1126/scitranslmed.3004404 (2012). [DOI] [PubMed]
- 8.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nature reviews. Microbiology. 2012;10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudbery PE. Growth of Candida albicans hyphae. Nature reviews. Microbiology. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 10.Carlisle PL, et al. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:599–604. doi: 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown AJ, Gow NA. Regulatory networks controlling Candida albicans morphogenesis. Trends in microbiology. 1999;7:333–338. doi: 10.1016/S0966-842X(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 12.Enjalbert B, Whiteway M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryotic cell. 2005;4:1203–1210. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nobile CJ, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiology and molecular biology reviews: MMBR. 2011;75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoldt VR, Sonneborn A, Leuker CE, Ernst JF. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. The EMBO journal. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS biology. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Su C, Liu H. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS pathogens. 2012;8:e1002663. doi: 10.1371/journal.ppat.1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Chang P, Ding J, Chen J. Distinct and redundant roles of the two MYST histone acetyltransferases Esa1 and Sas2 in cell growth and morphogenesis of Candida albicans. Eukaryotic cell. 2013;12:438–449. doi: 10.1128/EC.00275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JE, et al. Ssn6 has dual roles in Candida albicans filament development through the interaction with Rpd31. FEBS letters. 2015;589:513–520. doi: 10.1016/j.febslet.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Wurtele H, et al. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nature medicine. 2010;16:774–780. doi: 10.1038/nm.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, et al. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Molecular biology of the cell. 2008;19:4260–4272. doi: 10.1091/mbc.E08-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hnisz D, Majer O, Frohner IE, Komnenovic V, Kuchler K. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS pathogens. 2010;6:e1000889. doi: 10.1371/journal.ppat.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. The EMBO journal. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nature reviews. Molecular cell biology. 2017 doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nature structural & molecular biology. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. The Journal of biological chemistry. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 27.Campos EI, Reinberg D. New chaps in the histone chaperone arena. Genes & development. 2010;24:1334–1338. doi: 10.1101/gad.1946810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson JS, Liu H. Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryotic cell. 2013;12:591–603. doi: 10.1128/EC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tscherner M, et al. The Candida albicans Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways. PLoS pathogens. 2015;11:e1005218. doi: 10.1371/journal.ppat.1005218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva AC, et al. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. The Journal of biological chemistry. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochimica et biophysica acta. 2013;1819:264–276. doi: 10.1016/j.bbagrm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Green EM, et al. Replication-independent histone deposition by the HIR complex and Asf1. Current biology: CB. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Molecular and cellular biology. 1997;17:545–552. doi: 10.1128/MCB.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vishnoi N, et al. Separation-of-function mutation in HPC2, a member of the HIR complex in S. cerevisiae, results in derepression of the histone genes but does not confer cryptic TATA phenotypes. Biochimica et biophysica acta. 2011;1809:557–566. doi: 10.1016/j.bbagrm.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fillingham J, et al. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Molecular cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 37.Hnisz D, et al. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS genetics. 2012;8:e1003118. doi: 10.1371/journal.pgen.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Lee JE, Lee JS. Histone deacetylase-mediated morphological transition in Candida albicans. Journal of microbiology. 2015;53:805–811. doi: 10.1007/s12275-015-5488-3. [DOI] [PubMed] [Google Scholar]
- 39.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Molecular biology of the cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nantel A, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Molecular biology of the cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. Yeast wall protein 1 of Candida albicans. Microbiology. 2005;151:1631–1644. doi: 10.1099/mic.0.27663-0. [DOI] [PubMed] [Google Scholar]
- 42.Wysong DR, Christin L, Sugar AM, Robbins PW, Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infection and immunity. 1998;66:1953–1961. doi: 10.1128/iai.66.5.1953-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. The EMBO journal. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira ME, Flaherty K, Prochasson P. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PloS one. 2011;6:e21113. doi: 10.1371/journal.pone.0021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiology and molecular biology reviews: MMBR. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun BR, Johnson AD. TUP1, CPH1 and EFG1 make independent contributions to filamentation in candida albicans. Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amin AD, et al. The mitotic Clb cyclins are required to alleviate HIR-mediated repression of the yeast histone genes at the G1/S transition. Biochimica et biophysica acta. 2012;1819:16–27. doi: 10.1016/j.bbagrm.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. The EMBO journal. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 51.Doedt T, et al. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Molecular biology of the cell. 2004;15:3167–3180. doi: 10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giusani AD, Vinces M. & Kumamoto, C. A. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics. 2002;160:1749–1753. doi: 10.1093/genetics/160.4.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langford ML, et al. Candida albicans Czf1 and Efg1 coordinate the response to farnesol during quorum sensing, white-opaque thermal dimorphism, and cell death. Eukaryotic cell. 2013;12:1281–1292. doi: 10.1128/EC.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocha CR, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Molecular biology of the cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Molecular microbiology. 2008;67:47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Y, Su C, Unoje O, Liu H. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1975–1980. doi: 10.1073/pnas.1318690111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Argimon S, et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryotic cell. 2007;6:682–692. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. The EMBO journal. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin R, et al. A core filamentation response network in Candida albicans is restricted to eight genes. PloS one. 2013;8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Childers DS, Kadosh D. Filament condition-specific response elements control the expression of NRG1 and UME6, key transcriptional regulators of morphology and virulence in Candida albicans. PloS one. 2015;10:e0122775. doi: 10.1371/journal.pone.0122775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S, Wolyniak MJ, Staab JF, Sundstrom P. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryotic cell. 2007;6:693–709. doi: 10.1128/EC.00341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Childers DS, Mundodi V, Banerjee M, Kadosh D. A 5′ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Molecular microbiology. 2014;92:570–585. doi: 10.1111/mmi.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee M, et al. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Molecular biology of the cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeidler U, et al. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS yeast research. 2009;9:126–142. doi: 10.1111/j.1567-1364.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- 65.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Developmental biology. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Znaidi S, Nesseir A, Chauvel M, Rossignol T, d’Enfert C. A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS pathogens. 2013;9:e1003519. doi: 10.1371/journal.ppat.1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greig JA, et al. Cell cycle-independent phospho-regulation of Fkh2 during hyphal growth regulates Candida albicans pathogenesis. PLoS pathogens. 2015;11:e1004630. doi: 10.1371/journal.ppat.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee M, et al. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryotic cell. 2013;12:224–232. doi: 10.1128/EC.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Y, Su C, Solis NV, Filler SG, Liu H. Synergistic regulation of hyphal elongation by hypoxia, CO(2), and nutrient conditions controls the virulence of Candida albicans. Cell host & microbe. 2013;14:499–509. doi: 10.1016/j.chom.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryotic cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zunder RM, Rine J. Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Molecular and cellular biology. 2012;32:4337–4349. doi: 10.1128/MCB.00871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadasivam DA, Huang DH. Maintenance of Tissue Pluripotency by Epigenetic Factors Acting at Multiple Levels. PLoS genetics. 2016;12:e1005897. doi: 10.1371/journal.pgen.1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duc C, et al. The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. The Plant journal: for cell and molecular biology. 2015;81:707–722. doi: 10.1111/tpj.12758. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, Z. et al. PHB Associates with the HIRA Complex to Control an Epigenetic-Metabolic Circuit in Human ESCs. Cell stem cell, doi:10.1016/j.stem.2016.11.002 (2016). [DOI] [PubMed]
- 75.Sharp JA, Rizki G, Kaufman PD. Regulation of histone deposition proteins Asf1/Hir1 by multiple DNA damage checkpoint kinases in Saccharomyces cerevisiae. Genetics. 2005;171:885–899. doi: 10.1534/genetics.105.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011;187:9–19. doi: 10.1534/genetics.110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bahn YS, Sundstrom P. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. Journal of bacteriology. 2001;183:3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadosh D. Shaping up for battle: morphological control mechanisms in human fungal pathogens. PLoS pathogens. 2013;9:e1003795. doi: 10.1371/journal.ppat.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao F, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Molecular biology of the cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome research. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 83.Pchelintsev NA, et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell reports. 2013;3:1012–1019. doi: 10.1016/j.celrep.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soni S, Pchelintsev N, Adams PD, Bieker JJ. Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for beta-globin gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13337–13342. doi: 10.1073/pnas.1405422111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ray-Gallet D, et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Molecular cell. 2002;9:1091–1100. doi: 10.1016/S1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 87.Kaiser, C., Michaelis, S. & Mitchell, A. Methods in Yeast Genetics. A Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press (1994).
- 88.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Molecular & general genetics: MGG. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 89.Hnisz D, Schwarzmuller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Molecular microbiology. 2009;74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tscherner M, Stappler E, Hnisz D, Kuchler K. The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans. Molecular microbiology. 2012;86:1197–1214. doi: 10.1111/mmi.12051. [DOI] [PubMed] [Google Scholar]
- 91.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 92.Lee EC, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 93.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nature genetics. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophor. 1987;8:93–99. doi: 10.1002/elps.1150080203. [DOI] [Google Scholar]
- 95.Hernday AD, Noble SM, Mitrovich QM, Johnson AD. Genetics and molecular biology in Candida albicans. Methods in enzymology. 2010;470:737–758. doi: 10.1016/S0076-6879(10)70031-8. [DOI] [PubMed] [Google Scholar]
- 96.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Benjamini, Y. & Hochber, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, 289–300 (1995).
- 101.Oliveros, J. C. Venny. An interactive tool for comparing lists with Venn’s diagrams. (2007–2015).
- 102.Priebe S, Kreisel C, Horn F, Guthke R, Linde J. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics. 2015;31:445–446. doi: 10.1093/bioinformatics/btu627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monteiro PT, et al. The PathoYeastract database: an information system for the analysis of gene and genomic transcription regulation in pathogenic yeasts. Nucleic acids research. 2017;45:D597–D603. doi: 10.1093/nar/gkw817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restrictions. All relevant data are within the paper and its Supporting Information files.


