Abstract
The relevance of the transcription factor p53 in cancer is inarguable, and numerous lncRNAs are involved in the p53 regulatory network as either regulators or effectors, triggering a transcriptional response that causes either cell arrest or apoptosis following DNA damage in a p53-dependent manner. Despite the fact that the therapeutic response is improved in NPC, heterogeneity among people remains with regard to the susceptibility of adverse effects and the efficacy of treatments. Therefore, we analysed eight potentially functional SNPs of five genes in the lncRNA-p53 regulatory network in a discovery cohort of 505 NPC patients. By performing multivariate logistic regression, the impact of genetic variations on the efficacy and risk of CRT-induced toxicities was investigated. The most dramatic finding was that the MEG3 rs10132552 CC genotype had a greater than three-fold increased risk of developing grade 3–4 anaemia (OR = 3.001, 95%CI = 1.355–6.646, P = 0.007). Furthermore, the rs10132552 CT genotype had a better response to treatment (OR = 0.261, 95%CI = 0.089–0.770, P = 0.015). Individuals carrying LINC-ROR rs2027701 with one or two variant alleles had significant associations with a reduced risk of neutropaenia (OR = 0.503, 95%CI = 0.303–0.835, P = 0.008). In conclusion, our results suggested that genetic polymorphisms of the lncRNA-p53 regulatory network could play a potential role in reducing treatment-related toxicities and improving outcomes for NPC patients.
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy with extremely skewed ethnic and geographic distributions and a particularly high prevalence in southern China1. Concurrent chemoradiotherapy (CRT), an important therapeutic milestone, is the standard treatment for locally advanced NPC. Although overall survival has been dramatically improved by the advancement of radiotherapy technology as well as the broader application of chemotherapy, CRT-induced acute toxicity remains a challenge, as it is multifactorial and difficult to predict2. Chemoradiotherapy is invariably associated with higher incidences of haematological and non-haematological acute toxic effects compared with radiotherapy alone3–5. There is significant variation in prognosis and the risk of toxicities among patients, even if they are exposed to the same therapeutic regimens, suggesting that genetic polymorphisms play a crucial role in individual susceptibility to toxicities and sensitivity to treatments6,7.
In support of this notion, many studies have illustrated that single nucleotide polymorphisms (SNPs) may be useful as independent factors for predicting the toxicities and curative efficacy of chemoradiotherapy in human cancers including NPC. For example, the rs1982073 polymorphism of TGFB1 seems to trend with a higher risk of mucositis in head and neck squamous cell carcinoma when patients underwent platinum-based CRT8. As indicated by Ming Jia9, the GADD45B rs2024144T variant allele correlated with a major event in response to severe haematologic toxicities in individuals with non-small cell lung cancer. Similarly, SNPs in DNA repair pathway genes were correlated with sensitivity to radiotherapy and chemotherapy10.
LncRNA is a type of long non-coding RNA with transcripts >200 nt in length that functioning as a master regulator controlling protein-coding and non-coding genes at multiple levels; lncRNAs could drive important cancer phenotypes and serve as a biomarker in diverse cancers such as NPC11–14. Undoubtedly, p53, a master human tumour suppressor, participates in all steps of tumour initiation and development by regulating the expression of many downstream genes, whose dysfunction is closely related to the occurrence and progression of NPC15. LncRNA has now been added to the p53 regulatory pathway (Fig. 1 ) to form a sophisticated regulatory network16,17 that has generated increased attention for its potential to contribute to disease.
Figure 1.
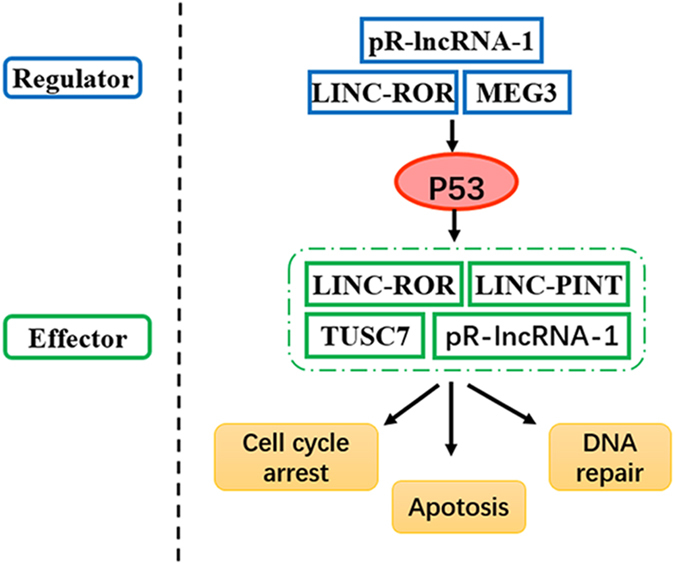
LncRNAs serve as p53 regulators and effectors, participating in the p53 regulatory pathway. On the one hand, lncRNAs are able to regulate p53 directly or indirectly at the transcriptional or posttranscriptional levels such as MEG3. On the other hand, several lncRNAs could be induced or suppressed by p53 such as LINC-ROR, pR-lncRNA-1, LINC-PINT and TUSC7.
On one hand, lncRNAs act as p53 effectors that are regulated by p53 directly and modulate gene expression downstream of p53. For example, LINC-ROR, pR-lncRNA-1, LINC-PINT and TUSC7 alter the interaction between p53 and potential p53 response elements (p53REs) when confronted with cellular stresses such as the DNA damage induced by radiation and/or chemotherapeutics by regulating cell proliferation, cell cycle and cell apoptosis18–21. On the other hand, lncRNAs such as MEG3 can serve as p53 regulators by controlling p53 stability17. Based on these examples, we selected eight potential SNPs in five genes MEG3 (rs10132552T > C), LINC-ROR (rs2027701A > G), pR-lncRNA-1 (rs73594404G > A and rs3743773G > A), LINC-PINT (rs1059698A > C and rs2293750T > A) and TUSC7 (rs1829346C > A and rs36080650T > C) to determine whether genetic polymorphisms of lncRNA-p53 regulatory network genes are associated with toxicities or the therapeutic efficacy of concurrent chemoradiotherapy in NPC in hopes of discovering valuable new biomarkers for personalized CRT among NPC patients.
Results
Patient Clinical Characteristics and Genotype Distribution
The studied cohort included 374 males and 131 females, with a mean age of 47.41 (ranging from 15 to 73) years old. Among these patients, 455 individuals (90.1%) were diagnosed at late stages (III and IV), and the other patients (9.9%) were at early stages (I and II). All of the patients were treated with IMRT, and the median total radiation dose was 71.34 Gy. Regarding chemotherapy, regimes of platinum-based inducing and concurrent chemotherapy were given to participants. The demographic characteristics of NPC patients are described in Table 1. Although none of the patients in this study suffered death caused by toxicities, 51 (10.1%), 129 (25.5%), 121 (24.0%), 73 (14.5%) and 94 (18.6%) experienced grade 3–4 dermatitis, mucositis, myelosuppression, leukopaenia and neutropaenia, respectively. Furthermore, 25 (6.0%) and 62 (15.0%) experienced worse treatment efficacy of CRT at the primary tumour and lymph node, respectively, months after treatments.
Table 1.
Patient demographics and clinical characteristics.
| Patient characteristics | N = 505(%) |
|---|---|
| Gender | |
| Male | 374 (74.1) |
| Female | 131 (25.9) |
| Age, years | |
| Mean ± SD | 47.41 ± 9.15 |
| <47 | 229 (45.3) |
| ≥47 | 276 (54.7) |
| BMI | |
| <18.5 | 30 (5.9) |
| 18.5 ~ 24 | 274 (54.3) |
| ≥24 | 201 (39.8) |
| Smoking status | |
| Smoker | 247 (48.9) |
| Nonsmoker | 258 (51.1) |
| Drinking status | |
| Drinker | 90 (17.8) |
| Nondrinker | 415 (82.2) |
| Histological type | |
| WHO type II | 214 (42.4) |
| WHO type III | 291 (57.6) |
| Clinical stage a | |
| I–II | 50 (9.9) |
| III–IV | 455 (90.1) |
| T-staging | |
| T1–T2 | 246 (48.7) |
| T3–T4 | 259 (51.3) |
| N-staging | |
| N0–N1 | 93 (18.4) |
| N2–N3 | 412 (81.6) |
| Grade 3–4 toxic reactions | |
| Dermatitis | 51 (10.1%) |
| Mucositis | 129 (25.5%) |
| Leukopenia | 73 (14.5%) |
| Neutropenia | 94 (18.6%) |
| Myelosuppression | 121 (24.0%) |
| IC regimen | |
| DP | 200 (39.6) |
| FP | 92 (18.2) |
| TP | 203 (40.2) |
| GP | 10 (2) |
| CRT regimen | |
| FP | 85 (16.8) |
| TP | 108 (21.4) |
| DDP | 83 (16.4) |
| NDP | 172 (34.1) |
| DP | 57 (11.3) |
| pGTVnx | |
| Mean ± SD | 71.34 ± 2.79 |
| <71.00 Gy | 261 (51.7) |
| ≥71.00 Gy | 234 (48.3) |
Abbreviations: BMI, Body Mass Index; IC regimen, Induction chemotherapy regimens; CRT regimen, concurrent chemoradiotherapy regimen; pGTVnx, irradiation dose.
The characteristics of the 8 SNPs are shown in Table 2. The allelice frequencies of the enrolled SNPs all fit Hardy-Weinberg equilibrium (P > 0.05).
Table 2.
Genotype distribution and MAF of 8 SNPs in LINC-ROR, pR-lncRNA-1, LINC-PINT, MEG3 and TUSC7.
| Gene | SNP | SNP Location | Alleles | Genotype Distributiona | HWE | MAF | Detectable Rate (%) |
|---|---|---|---|---|---|---|---|
| LINC-ROR | rs2027701 | chr18:54724945-54725445 | A/G | 174/247/81 | 0.670 | 0.3810 | 99.4 |
| pR-lncRNA-1 | rs73594404 | chr16:53079364-53079864 | G/A | 453/50/2 | 0.625 | 0.0714 | 100 |
| rs3743773 | chr16:53077757-53078257 | G/A | 313/167/25 | 0.656 | 0.2476 | 100 | |
| LINC-PINT | rs1059698 | chr7:130628844-130629344 | A/C | 223/222/60 | 0.677 | 0.3238 | 100 |
| rs2293750 | chr7:130629522-130630022 | T/A | 155/252/96 | 0.720 | 0.4143 | 99.6 | |
| MEG3 | rs10132552 | chr14:101300762-101301262 | T/C | 251/208/37 | 0.496 | 0.2952 | 98.2 |
| TUSC7 | rs1829346 | chr3:116428657-116429157 | C/A | 240/219/46 | 0.694 | 0.3476 | 100 |
| rs36080650 | chr3:116431337-116431837 | T/C | 241/219/45 | 0.635 | 0.3571 | 100 |
aIn the order of wild homozygote/heterozygote/mutant homozygote.
Abbreviations: HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency (Southern Han Chinese).
TUSC7 SNP and the Risk of CRT-induced Dermatitis
We demonstrated that TUSC7 rs1829346 and rs36080650 were significantly associated with dermatitis (Table 3 ). Patients carrying the AA genotype of rs1829346 were less resistant to grade 3–4 CRT-induced dermatitis (OR = 2.641, 95%CI = 1.118–6.243, P = 0.027). Similarly, the rs36080650 CC genotype was also associated with a prominently higher risk of dermatitis than the CT/TT genotypes (OR = 2.700, 95%CI = 1.141–6.386, P = 0.024), which was supported by the recessive model (CC vs CT + TT, OR = 2.544, 95%CI = 1.143–5.662, P = 0.022). However, no significant corrections between the risk of oral mucositis and SNPs were found.
Table 3.
Associations between genotypes and concurrent chemoradiotherapy-induced toxicities
| Toxic reactions | SNP | Genotypes | Toxicity grade | OR (95% CI) | P a | |
|---|---|---|---|---|---|---|
| Dermatitis | Grade ≤ 2 N (%) | Grade > 2 N (%) | ||||
| rs1829346 | CC | 219 (48.2) | 21 (41.2) | 1.00 (reference) | ||
| CA | 198 (43.6) | 21 (41.2) | 1.127 (0.597-2.130) | 0.712 | ||
| AA | 37 (8.1) | 9 (17.6) | 2.641 (1.118–6.243) | 0.027 | ||
| AA + CA vs CC | 1.407 (0.767–2.584) | 0.270 | ||||
| AA vs CA + CC | 2.492 (1.121–5.540) | 0.025 | ||||
| rs36080650 | TT | 220 (48.5) | 21 (41.2) | 1.00 (reference) | ||
| CT | 198 (43.6) | 21 (41.2) | 1.130 (0.598–2.135) | 0.706 | ||
| CC | 36 (7.9) | 9 (17.6) | 2.700 (1.141–6.386) | 0.024 | ||
| CC + CT vs TT | 1.414 (0.770–2.595) | 0.264 | ||||
| CC vs CT + TT | 2.544 (1.143–5.662) | 0.022 | ||||
| Neutropenia | Grade ≤ 2 N (%) | Grade > 2 N (%) | ||||
| rs2027701 | AA | 134 (32.6) | 40 (42.6) | 1.00 (reference) | ||
| GA | 205 (49.9) | 42 (44.7) | 0.545 (0.319–0.930) | 0.026 | ||
| GG | 70 (17.0) | 11 (11.7) | 0.378 (0.171–0.836) | 0.016 | ||
| GG + GA vs AA | 0.503 (0.303–0.835) | 0.008 | ||||
| GG vs GA + AA | 0.550 (0.264–1.146) | 0.110 | ||||
| rs73594404 | GG | 374 (91.0) | 79 (84.0) | 1.00 (reference) | ||
| GA | 36 (8.8) | 14 (14.9) | 2.118 (1.011–4.440) | 0.047 | ||
| AA | 1 (0.2) | 1 (1.1) | 3.484 (0.128–95.079) | 0.459 | ||
| AA + GA vs GG | 2.164 (1.049–4.464) | 0.037 | ||||
| AA vs GA + GG | 3.260 (0.121–87.981) | 0.482 | ||||
| rs1059698 | AA | 173 (42.1) | 50 (53.2) | 1.00 (reference) | ||
| CA | 185 (45.0) | 37 (39.4) | 0.780 (0.469–1.295) | 0.336 | ||
| CC | 53 (12.9) | 7 (7.4) | 0.395 (0.161–0.971) | 0.043 | ||
| CC + CA vs AA | 0.660 (0.408–1.070) | 0.092 | ||||
| CC vs CA + AA | 0.443 (0.185–1.058) | 0.067 | ||||
| Anemia | Grade ≤ 0 N (%) | Grade > 0 N (%) | ||||
| rs10132552 | TT | 141 (49.0) | 110 (50.7) | 1.00 (reference) | ||
| CT | 129 (44.8) | 79 (36.4) | 0.764 (0.500–1.169) | 0.215 | ||
| CC | 14 (4.9) | 23 (10.6) | 2.653 (1.172–6.008) | 0.019 | ||
| CC + CT vs TT | 0.929 (0.618–1.398) | 0.725 | ||||
| CC vs CT + TT | 3.001 (1.355–6.646) | 0.007 | ||||
| rs73594404 | GG | 263 (91.3) | 190 (87.6) | 1.00 (reference) | ||
| GA | 25 (8.7) | 25 (11.5) | 2.109 (1.062–4.188) | 0.033 | ||
| AA | 0 (0) | 2 (0.9) | NA | NA | ||
| AA + GA vs GG | 2.239 (1.138–4.405) | 0.020 | ||||
| AA vs GA + GG | NA | NA | ||||
| Myelosuppression | Grade ≤ 2 N (%) | Grade > 2 N (%) | ||||
| rs1059698 | AA | 161 (41.9) | 62 (51.2) | 1.00 (reference) | ||
| CA | 172 (44.8) | 50 (41.3) | 0.810 (0.510–1.287) | 0.373 | ||
| CC | 51 (13.3) | 9 (7.4) | 0.407 (0.180–0.920) | 0.031 | ||
| CC + CA vs AA | 0.708 (0.456–1.098) | 0.123 | ||||
| CC vs CA + AA | 0.449 (0.204–0.987) | 0.046 | ||||
Abbreviations: CI, confidence interval; OR, odds ratio.
P a values were calculated with adjustment for age, sex, BMI, smoking status, drinking status, histological type, clinical stage, T-staging, N-staging, Induction chemotherapy regimens, concurrent chemoradiotherapy regimen, irradiation dose.
P value < 0.05 is shown in bold.
Multivariate Analysis of Selected SNPs as Prognostic Factors of Haematological Toxicities
Neutropaenia
Three SNPs were significantly associated with neutropaenia: rs2027701, rs73594404 and rs1059698. LINC-ROR rs2027701 showed an obvious trend towards a superior reaction with toxic effects in patients with one or two variant alleles compared with those with the wild-type genotype (OR = 0.503, 95%CI = 0.303–0.835, P = 0.008). As demonstrated, the LINC-PINT rs1059698 CC genotype was a protective factor (OR = 0.395, 95%CI = 0.161–0.971, P = 0.043), whereas LINC-PINT rs2293750 had no association with the risk of neutropaenia. In contrast to rs2027701 and rs1059698, rs73594404 pR-lncRNA-1 had a weak correlation with increased risk of adverse reactions when patients possessed the GA genotype (OR = 2.118, 95%CI = 1.011–4.440, P = 0.047).
Anaemia
Patients with a minor A allele of pR-lncRNA-1 rs73594404 had an increased risk of anaemia (OR = 2.109, 95%CI = 1.062–4.188, P = 0.033). The MEG3 rs10132552 CC genotype correlated with a significantly inferior toxic reaction (OR = 2.653, 95%CI = 1.172–6.008, P = 0.019), which is an independent predictor for prognosis (CC vs CT + TT, OR = 3.001, 95%CI = 1.355–6.646, P = 0.007).
Myelosuppression
Statistical results indicated that only LINC-PINT rs1059698 polymorphisms had a correlation with CRT-induced myelosuppression. Although this SNP was not correlated with dermatitis and anaemia, we found that it had a significant association with not only the risk of neutropaenia (OR = 0.395, 95%CI = 0.161–0.971, P = 0.043), but also myelosuppression (OR = 0.407, 95%CI = 0.180–0.920, P = 0.031).
According to the multivariate logistic regression analyses, selected 8 SNPs did not interact with leukopaenia and thrombocytopenia in this study.
Stratification Analysis of the Association between SNPs in lncRNA-p53 Regulatory Network Genes and Toxic Reactions
Supplementary Table S1 lists the relevancies between patient-related, tumour-related and therapy-related characteristics and the risk of toxicities generated by CRT. There was a significant dependency between gender, IC regimen, CRT regimen and multifarious toxicities. Female gender was an adverse factor for toxic reactions. Patients treated with TP, DP or DDP during CRT had a higher risk of anaemia compared with those treated with FP. Moreover, both age and BMI impacted individual risks. Patients with advanced T stages had a more than 1.5-fold greater myelosuppression risk compared with those in the early stages.
According to the above evidence, stratified analysis by adjusting for sex, IR regimen and CRT regimen was conducted to estimate the associations between the enrolled SNP polymorphisms and adverse reactions (Table 4 ). The rs10132552 CT genotype had an increased risk of leukopaenia, neutropaenia and myelosuppression in subgroups of the DP induction protocol. pR-lncRNA-1 rs73594404 was another vital SNP that showed a strong relationships with toxicities in the IC regimen subgroup (OR = 3.394 and P = 0.015 for leukopaenia; OR = 3.540 and P = 0.036 for thrombocytopenia; OR = 3.054 and P = 0.022 for myelosuppression). Of significant interest, two SNPs, rs2027701 and rs1059698, had a weak correlation with oral mucositis in the subgroups of female (OR = 3.375 and P = 0.045) and IC regimen-DP (OR = 0.527 and P = 0.049), respectively.
Table 4.
Stratification analysis of association between SNPs in lncRNA-p53 regulatory network genes and the toxic reactions in NPC patients.
| Stratified factors | n | SNP | Toxic reactions | OR (95% CI) | P |
|---|---|---|---|---|---|
| IC regimen-TP | 203 | rs2027701 | Dermatitis | 4.721 (1.141–19.536) | 0.032 |
| rs73594404 | Leukopenia | 3.394 (1.263–9.123) | 0.015 | ||
| Myelosuppression | 3.054 (1.178–7.922) | 0.022 | |||
| IC regimen-DP | 200 | rs1059698 | Mucositis | 0.527 (0.279–0.997) | 0.049 |
| rs10132552 | Leukopenia | 4.300 (1.345–13.748) | 0.014 | ||
| Neutropenia | 5.462 (1.836–16.251) | 0.002 | |||
| rs73594404 | Thrombocytopenia | 3.540 (1.089–11.500) | 0.036 | ||
| Sex-Female | 131 | rs2027701 | Mucositis | 3.375 (1.025–11.108) | 0.045 |
| rs73594404 | Thrombocytopenia | 10.237 (2.530–41.429) | 0.001 | ||
| rs10132552 | Myelosuppression | 4.135 (1.303–13.119) | 0.016 | ||
| CCRT regimen-TP | 108 | rs73594404 | Leukopenia | 3.784 (1.086–13.184) | 0.037 |
Abbreviations: OR, odds ratio; CI, confidence interval;
P value < 0.05 is shown in bold.
Hence, to further obtain the predictive power of lncRNAs after adjusting for clinical variables, a risk score model was built in accordance with the regression coefficients of variables to predict each patient’s risk of developing toxicities including neutropaenia, anaemia and myelosuppression (factors involved in P < 0.05). Using receiver operating characteristic (ROC) curve analysis, the prognostic power was evaluated (Fig. 2 ). The area under the curve (AUC) of neutropaenia, anaemia and myelosuppression was 0.731, 0.750 and 0.648, respectively, indicating good performance of the lncRNAs combined with clinical information for predicting toxicities in NPC patients.
Figure 2.
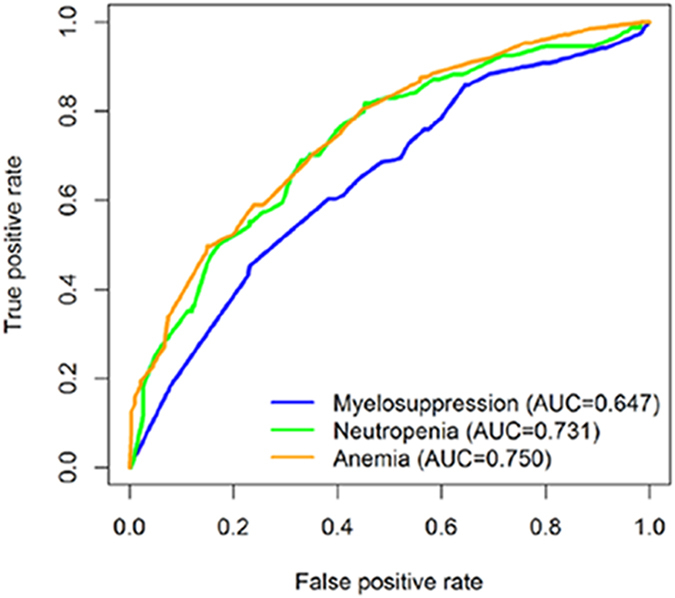
Analysis of receiver-operating characteristic curve to predict toxicities. ROC analysis showed the AUC of myelosuppression, neutropaenia and anaemia was 0.647, 0.731 and 0.750, respectively.
Interaction between Selected SNPs and the Efficacy of CRT 3 Months after Treatment
All positive results concerning the relationship between selected SNPs and the efficacy of CRT on the primary tumour and lymph nodes are listed in Table 5. Our results provide a plausible link between LINC-ROR rs2027701 polymorphisms and efficacy at the lymph node 3 months after CRT (OR = 2.266, 95%CI = 1.020–5.033, P = 0.045), while no significant difference was found on the primary tumour. In contrast, the MEG3 rs10132552 CT genotype had a better response to treatments for the primary tumour compared with the TT genotype (OR = 0.261, 95%CI = 0.089–0.770, P = 0.015). With respect to the lymph node, rs10132552 polymorphisms are nonfunctional. We also analysed the connections between clinical factors and treatment efficacy; positive results are displayed in Supplementary Tables S2.
Table 5.
Association between rs2027701 and rs10132552 and the efficacy of CRT at the primary tumor and lymph node 3 months after treatment in NPC patients
| SNP | Primary tumor 3 months after treatment | P c | Lymph node 3 months after treatment | P c | |||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype distribution (%) | OR (95% CI) | Genotype distribution | OR (95% CI) | ||||||
| Non-sensitivea | Sensitiveb | Non-sensitivea | Sensitiveb | ||||||
| rs2027701 | AA | 8 (32.0) | 125 (32.1) | 1.00 (reference) | 15 (24.2) | 118 (33.5) | 1.00 (reference) | ||
| GA | 14 (56.0) | 193 (49.6) | 1.289 (0.467–3.558) | 0.624 | 30 (48.4) | 177 (50.3) | 1.280 (0.643–2.549) | 0.483 | |
| GG | 3 (12.0) | 68 (17.5) | 0.564 (0.131–2.426) | 0.442 | 17 (27.4) | 54 (15.3) | 2.266 (1.020–5.033) | 0.045 | |
| rs10132552 | TT | 18 (72.0) | 187 (48.1) | 1.00 (reference) | 35 (56.5) | 170 (48.3) | 1.00 (reference) | ||
| CT | 6 (24.0) | 169 (43.4) | 0.261 (0.089–0.770) | 0.015 | 23 (37.1) | 152 (43.2) | 0.735 (0.398–1.358) | 0.326 | |
| CC | 1 (4.0) | 28 (7.2) | 0.219 (0.024–1.990) | 0.177 | 2(3.2) | 27 (7.7) | 0.331 (0.069–1.588) | 0.167 | |
aComplete response (CR) and partial response (PR).
bStable disease (SD) and progressive disease (PD).
cAdjusted for age, sex, BMI, smoking status, drinking status, histological type, clinical stage, T-staging, N-staging, induction chemotherapy regimens, concurrent chemoradiotherapy regimen, irradiation dose for the association between SNPs and the efficacy of CRT at the primary tumor and lymph node 3 months after treatment.
Abbreviations: OR, odds ratio; CI, confidence interval.
P value < 0.05 is shown in bold.
Discussion
In this study, we estimated the association of 8 SNPs in the lncRNA-p53 regulatory network of genes and the efficacy and toxic reactions in 505 NPC patients. To the best of our knowledge, this is the first study to demonstrate that 2 SNPs (TUSC7 rs1829346 and rs36080650) were correlated with dermatitis, 3 SNPs (LINC-ROR rs2027701, pR-lncRNA-1 rs73594404 and LINC-PINT rs1059698) were related to neutropaenia, 2 SNPs (MEG3 rs10132552, pR-lncRNA-1 rs73594404) and 1 SNP (LINC-PINT rs1059698) were associated with anaemia and myelosuppression in NPC patients, respectively. Of these, rs2027701 and rs10132552 were related to curative efficacy 3 months after treatment.
The regulation of the p53 tumour-suppressor pathway by lncRNAs, directly or indirectly, has been a hot topic of particularly intense interest. Non-coding mutations contributing to small changes in gene expression can have a large phenotypic impact on carcinoma, perhaps to an even greater degree than currently appreciated. As regulators, lncRNAs could affect p53 expression by influencing p53 mRNA stability or reducing its ability to recognize some of its binding sites, ultimately inhibiting p53 transcription22,23. lncRNAs also act as regulators that activate p53 directly by interacting with p53REs, regulating the gene expression of the p53 pathway at multiple levels and even establishing a regulatory feedback loop with p53. It is tempting to speculate that the interaction between p53 and lncRNAs has an impact on cell proliferation, cell cycle and cell apoptosis upon DNA damage, resulting in personalized differences in the toxic reactions and efficacy in NPC.
TUSC7 was significantly induced in cells expressing wild-type p53, serving as a putative tumour suppressor by inhibiting cell growth, which plays a critical role in cancer prognosis, including oesophageal squamous cell carcinoma (ESCC)24, colorectal cancer25 and osteosarcoma26. Low expression of TUSC7 was dramatically negatively correlated with the pathologic response to CRT and resulted in a poorer prognosis in cancers than the higher expression group16. Similarly, we found that rs1829346 and rs36080650 were significantly correlated with the risk of dermatitis under CRT. Patients carrying a homozygous mutation of these two SNPs were less resistant to grade 3–4 dermatitis. Unfortunately, the combination of the two SNPs did not increase risk. Accumulating studies have revealed that lncRNAs impact cellular functions through various mechanisms, most notably as a ‘sponge’ to titrate miRNAs27,28. The variation in the rs1829346 sequence may create a new binding site for miR-1304 (Supplementary Table S3 ), inhibiting the expression of TUSC7, regulating cell proliferation proteins and ultimately leading to a higher risk of CRT-induced dermatitis. For rs36080650, although there is no direct evidence to illustrate its function, the strong linkage disequilibrium (LD) with rs1829346 suggested that the regulation of gene expression is not mediated by this SNP but by a variant in rs1829346.
LINC-ROR, was first discovered in induced pluripotent stem cells (iPSCs); since it was discovered, the number of studies in this area have increased dramatically. LINC-ROR is not only a p53 effector but also a p53 regulator; its depletion would lead to the upregulation of genes involved in the p53 response to DNA damage-inducing agents and other stresses responses17,29. In our study, LINC-ROR rs2027701 showed an obvious trend towards a superior reaction with toxic effects in patients with one or two variant alleles, which may be explained by Zhang21, who found that LINC-ROR can significantly suppress p53 during DNA damage. Moreover, LINC-ROR suppression of the p53 pathway may account for patients who resist chemotherapy, thus playing a critical role in NPC. Our study found that rs2027701 polymorphisms were correlated with worse CRT efficacy at the lymph node.
pR-lncRNA-1, in a similar manner as LINC-ROR, is induced by p53 and modulates p53 transcriptional activity by forming an autoregulatory feedback loop with p53, enhancing p53 tumour suppressor activity and ultimately modulating the gene expression response to DNA damage20. pR-lncRNA-1 rs73594404 has a strong connection to both neutropaenia and anaemia. Patients with the GA genotype, but not the AA genotype, have increased risk of CRT-induced toxicities. Although rs73594404 was identified in the intron region, it had enhancer activity and had its maximum signal strength in the region of transcription factor binding sites, suggesting that the rs73594404 mutant may influence gene expression by destroying the site or increasing the affinity of the transcription factor, leading to inactivation of tumour suppressor genes30,31. This links to poorer prognosis and additional studies will be required to confirm these observations.
LINC-PINT is a bona fide p53 transcriptional target that regulates cell proliferation by inducing cell apoptosis19,32. As shown in Table 2, the LINC-PINT rs1059698 CC genotype was a protective factor in neutropaenia and myelosuppression. Although studies on LINC-PINT are limited, using a database, we found that rs1059698 is located in the predicted active promoter flanking regions. The contribution of rare variants to asthma susceptibility is principally due to noncoding variants in sequences flanking the exons33. Similarly, Johnson34 revealed that genetic variation in noncoding sequences flanking the CYP3A locus was associated with the risk of breast cancer. Therefore, we speculated that the genetic polymorphism of rs1059698 would control promoter activity, inhibit LINC-PINT expression, and regulate a multitude of signalling pathways including the p53 network. Furthermore, rs1059698 is also located in the DNase I Hypersensitivity (DHS) cluster. Mutation of rs1059698 was strongly associated with transcription initiation activity, highlighting the role of rs1059698. However, further investigation is needed to delineate the precise mechanism.
MEG3 is a p53 regulator that is downregulated by MDM2, a well-known negative p53 regulator, thus controlling p53 stability and regulating downstream genes. MEG3, a tumour suppressor, has a great capacity for prognosis in many cancers. The MEG3 rs7158663 AA genotype has significantly increased colorectal cancer risk, as revealed by Cao35. Analogously, we found that the MEG3 rs10132552 CC genotype correlates with a significantly inferior toxic reaction; however, individuals with the CT genotype had a better response to treatments, suggesting that rs10132552 polymorphisms may impact the expression of MEG3, thus influencing p53 and subsequently suppressing cell proliferation or promoting cell apoptosis. Furthermore, functional genomic analyses were designed to provide a potential biological basis for the observed associations. Rs10132552 polymorphisms could create miRNA (mir-564, mir-650 and mir-602) binding sites on MEG3, disturbing the lncRNA-miRNA interaction, acting as competing endogenous RNAs (ceRNA) and thereby negatively regulating miRNA expression36,37. Interestingly, it is clear that mir-650, mir-602 and mir-564 played roles in cancers such as breast cancer and lung cancer38–40. Performing an in silico analysis, we found that when the T allele was substituted by the C allele, the structure of the transcript was changed (Fig. 3 ), leading to a minimum free energy (MFE) change from −150.60 kcal/mol to −153.30 kcal/mol, thus elucidating an important role for SNPs on RNA structure and supporting the idea that even SNPs can alter local RNA folding structure41. The altered gene expression and secondary structure may eventually result in different sensitivities of individuals to anaemia.
Figure 3.
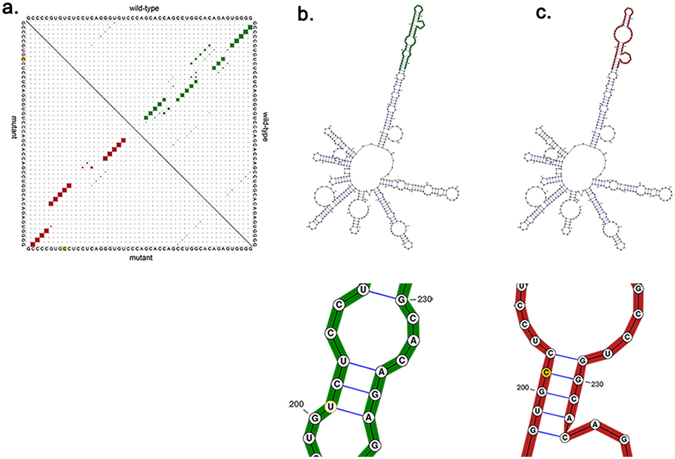
The predicting rs10132552 on MEG3 secondary structure. (a) Base pair probabilities of the local region rs10132552. (b) The optimal secondary structure of global wild-type sequence. Minimum free energy −150.60 kcal/mol. (c) The optimal secondary structure of global mutant sequence. Minimum free energy −153.30 kcal/mol.
However, genetic polymorphisms are not the only signature of neutropaenia in NPC patients, and gemcitabine plus cisplatin was associated with increased risk of grade 3–4 haematological adverse events such as neutropaenia compared with fluorouracil plus cisplatin42, which could be supported by our data (40% had grade 3–4 neutropaenia in the GP group vs 7.6% in the PF group). Furthermore, we combined SNPs (rs2027701, rs73594404 and rs1059698) and clinical information to conduct ROC curves; the AUC of neutropaenia was 0.731, indicating good performance for predicting an adverse effect.
Some strengths of this study should be noted. First, this is the first study to explore the impact of SNPs on genes of the lncRNA-p53 regulatory network and the efficacy and toxic reactions in NPC patients. Second, a well-defined cohort of pathological diagnosed cases and strict inclusion criteria were used to avoid possible confounding factors that could hinder analysis. Finally, we obtained all of the essential clinical data from the included individuals. However, several limitations should not be ignored. First, our study is limited as a retrospective study at a single centre, thus selection bias could not be avoided. Second, the sample size seems to be too small for stratification analysis; therefore, statistical power may be limited. Finally, this pathway is complex-the five genes included in this analysis were insufficient and further study is needed.
In conclusion, we found six potential SNPs in five genes in the lncRNA-p53 regulatory network that are significantly associated with the toxicities and efficacy of CRT in 505 patients with NPC, thus providing new biomarkers that can predict therapeutic effect and acute toxic reactions. This study represents a significant step forward toward a better understanding of the importance of lncRNA-p53 in NPC.
Methods
Patient selection
This study consisted of 505 newly diagnosed NPC cases from the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University between 2014 and 2016. Peripheral blood specimens for genetic analysis were collected from each patient at the time of diagnosis. Patients were enrolled if they met the following criteria: (a) pathologically confirmed NPC; (b) Karnofsky score ≥70; (c) received intensity modulated radiation-therapy (IMRT) and concurrent chemoradiotherapy; and (d) patients without recurrence, metastasis and other malignancy. Patient demographics and clinical characteristics are shown in Table 1. This study was performed with the approval of the Independent Ethical Committee of the Institute of Clinical Pharmacology, Central South University (CTXY-140007–2). At recruitment, written informed consent was obtained from all participants involved in this study. All experiments methods were performed in accordance with the relevant guidelines and regulations.
Efficacy regimen
All patients were treated with IMRT with the median total radiation dose of 71.34 Gy. The induction chemotherapy and concurrent chemotherapy were all performed with platinum-based chemotherapy regimens, including DP, docetaxel with cisplatin/ nedaplatin; FP, fluorouracil with cisplatin/nedaplatin; TP, paclitaxel with cisplatin/nedaplatin; GP, gemcitabine with cisplatin/ nedaplatin; DDP, cisplatin alone; NDP, nedaplatin alone.
SNP selection and genotyping assays
We selected the SNPs for TUSC7, LINC-ROR, pR-lncRNA-1, LINC-PINT and MEG3 by using databases including ENCODE, lncRNASNP, Hapmap and Ensembl to analyse their potential function. The SNPs were selected according to the following criteria: (1) with a minor allele frequency (MAF) ≥0.05 in a Southern Han Chinese population; (2) located in the promoter, miRNA binding site and other functional region; (3) SNPs have not been study before, not only NPC, but also other carcinomas. Genomic DNA was extracted from lymphocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions, and stored at -80 °C until use. The DNA purity and concentration were determined by spectrophotometric measurement of absorbance at 260 nm and 280 nm. The candidate SNPs were genotyped using the Sequenom MassARRAY iPLEX platform (Sequenom, Inc., San Diego, CA, USA). The detection rate of all SNPs was greater than 98%.
Evaluations of toxic reactions
Acute CRT-induced toxic reactions including dermatitis, mucositis, leukopaenia, myelosuppression, neutropaenia, anaemia and thrombocytopenia were recorded and evaluated according to the Common Terminology Criteria for Adverse Events (CTCAE 3.0). We defined grade 0–2 as mild toxic reactions and grade 3–4 as severe side effects, except for anaemia and thrombocytopenia, which have a lower incidence rate, and we chose 0 as the cutoff target. All acute toxic reactions were observed once a week from the first day to the end of treatment. RECIST (Response Evaluation Criteria in Solid Tumours) was used to evaluate efficacy three months after treatment.
Statistical analysis
Deviations from Hardy-Weinberg equilibrium were calculated using χ2 analysis. By computing the odds ratio (OR) and the corresponding 95% confidence interval (CI), and continuous variables such as age and BMI were switched to binary variables, multivariate logistic regression was performed to determine the association of each SNP of TUSC7, LINC-ROR, pR-lncRNA-1, LINC-PINT and MEG3 with the CRT efficacy and toxic reactions, with adjustments for age, sex, clinical stage, treatment modality and other clinical factors. In addition, stratification analyses were performed to characterize the associations between SNPs and toxic reactions in some subgroups when the corresponding clinical factors had an impact on toxic effects. We used receiver operating characteristic (ROC) curve analysis to evaluate the prognostic power of SNPs by comparing area under the curve (AUC) for each ROC when the P value < 0.05 was considered statistically significant. All statistical analyses were performed with SPSS19.0 and the R package. We also used lncRNASNP (http://bioinfo.life.hust.edu.cn/lncRNASNP/, accessed 30 January 2017) and RNASNP (http://rth.dk/resources/rnasnp/, accessed 30 January 2017) to predict the folding structure variants of genes due to SNP genotypes and the gain and loss of function of miRNA-lncRNA interactions through SNP polymorphisms, respectively.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). But the raw data used for the analysis and genotyping data for each patient is private.
Electronic supplementary material
Acknowledgements
This research was supported by grants from the National Key Research and Development Program (No. 2016YFC0905000), National High Technology Research and Development Program of China, “863” Project (No. 2012AA02A518), National Scientific Foundation of China (No. 81522048, 81573511, 81472802, 81273595) and Innovation Driven Project of Central South University (No. 2016CX024).
Author Contributions
Youhong Wang and Zhen Guo conceived and designed the study, performed mutation screening and the data analysis. Youhong Wang wrote the manuscript. Yu Zhao, Yi Jin, Liang An and Bin Wu collected and evaluated the clinical date. Zhaoqian Liu, Xiaoping Chen, Xiang Chen, Honghao Zhou, Hui Wang and Wei Zhang led and coordinated the study. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-017-16511-1.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08890-2
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hui Wang, Email: wanghui710327@163.com.
Wei Zhang, Email: yjsd2003@163.com.
References
- 1.Bruce JP, Yip K, Bratman SV, Ito E, Liu FF. Nasopharyngeal Cancer: Molecular Landscape. J Clin Oncol. 2015;33:3346–3355. doi: 10.1200/JCO.2015.60.7846. [DOI] [PubMed] [Google Scholar]
- 2.Lee AW, Ma BB, Ng WT, Chan AT. Management of Nasopharyngeal Carcinoma: Current Practice and Future Perspective. J Clin Oncol. 2015;33:3356–3364. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 3.Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 4.Sun X, et al. Long-term outcomes of intensity-modulated radiotherapy for 868 patients with nasopharyngeal carcinoma: an analysis of survival and treatment toxicities. Radiother Oncol. 2014;110:398–403. doi: 10.1016/j.radonc.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 5.Zhang AM, et al. Increased treatment-related mortality with additional cisplatin-based chemotherapy in patients with nasopharyngeal carcinoma treated with standard radiotherapy. Radiother Oncol. 2012;104:279–285. doi: 10.1016/j.radonc.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Guo Z, Shu Y, Zhou H, Zhang W, Wang H. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis. 2015;36:307–317. doi: 10.1093/carcin/bgv007. [DOI] [PubMed] [Google Scholar]
- 7.Feng XP, et al. Identification of biomarkers for predicting nasopharyngeal carcinoma response to radiotherapy by proteomics. Cancer Res. 2010;70:3450–3462. doi: 10.1158/0008-5472.CAN-09-4099. [DOI] [PubMed] [Google Scholar]
- 8.Lundberg M, Saarilahti K, Mäkitie AA, Mattila PS. TGFbeta1 genetic polymorphism is associated with survival in head and neck squamous cell carcinoma independent of the severity of chemoradiotherapy induced mucositis. Oral Oncol. 2010;46:369–372. doi: 10.1016/j.oraloncology.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Jia M, et al. Genetic variants of GADD45A, GADD45B and MAPK14 predict platinum-based chemotherapy-induced toxicities in Chinese patients with non-small cell lung cancer. Oncotarget. 2016;7:25291–25303. doi: 10.18632/oncotarget.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, et al. The study of the relation of DNA repair pathway genes SNPs and the sensitivity to radiotherapy and chemotherapy of NSCLC. Sci Rep. 2016;6:26526. doi: 10.1038/srep26526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 12.Bo H, et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404–20418. doi: 10.18632/oncotarget.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou J, et al. Nasopharyngeal carcinoma–review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30:946–963. doi: 10.1002/hed.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol. 2014;6:181–191. doi: 10.1093/jmcb/mju013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marín-Béjar O, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez Y, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang A, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Lu H. Noncoding RNAs: ‘our turn’ to join the p53 network. J Mol Cell Biol. 2014;6:179–180. doi: 10.1093/jmcb/mju022. [DOI] [PubMed] [Google Scholar]
- 23.Huarte M. p53 partners with RNA in the DNA damage response. Nat Genet. 2016;48:1298–1299. doi: 10.1038/ng.3702. [DOI] [PubMed] [Google Scholar]
- 24.Tong YS, et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi P, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasic I, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 27.Hua WF, et al. RBM24 suppresses cancer progression by upregulating miR-25 to target MALAT1 in nasopharyngeal carcinoma. Cell Death Dis. 2016;7:e2352. doi: 10.1038/cddis.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, et al. Linc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol Oncol. 2014;133:333–339. doi: 10.1016/j.ygyno.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 29.Pan Y, et al. The Emerging Roles of Long Noncoding RNA ROR (lincRNA-ROR) and its Possible Mechanisms in Human Cancers. Cell Physiol Biochem. 2016;40:219–229. doi: 10.1159/000452539. [DOI] [PubMed] [Google Scholar]
- 30.Khurana E, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17:93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 31.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiekhattar R. PINTing for p53. Genome Biol. 2013;14:132. doi: 10.1186/gb-2013-14-12-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torgerson DG, et al. Resequencing candidate genes implicates rare variants in asthma susceptibility. Am J Hum Genet. 2012;90:273–281. doi: 10.1016/j.ajhg.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson N, et al. CYP3A variation, premenopausal estrone levels, and breast cancer risk. J Natl Cancer Inst. 2012;104:657–669. doi: 10.1093/jnci/djs156. [DOI] [PubMed] [Google Scholar]
- 35.Cao X, et al. Associations between polymorphisms of long non-coding RNA MEG3 and risk of colorectal cancer in Chinese. Oncotarget. 2016;7:19054–19059. doi: 10.18632/oncotarget.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao S, et al. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget. 2016;7:1608–1618. doi: 10.18632/oncotarget.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, et al. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7:57228–57238. doi: 10.18632/oncotarget.11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang C, et al. MicroRNA-564 is downregulated in glioblastoma and inhibited proliferation and invasion of glioblastoma cells by targeting TGF-beta1. Oncotarget. 2016;7:56200–56208. doi: 10.18632/oncotarget.8987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Mraz M, et al. MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood. 2012;119:2110–2113. doi: 10.1182/blood-2011-11-394874. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, et al. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803–808. doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- 41.Li S, et al. Association of genetic variants in lncRNA H19 with risk of colorectal cancer in a Chinese population. Oncotarget. 2016;7:25470–25477. doi: 10.18632/oncotarget.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 2016;388:1883–1892. doi: 10.1016/S0140-6736(16)31388-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files). But the raw data used for the analysis and genotyping data for each patient is private.


