Abstract
Soil salinity is a common abiotic stress affecting crop productivity. To identify favorable alleles from wild rice (Oryza rufipogon Griff.) that enhance salinity tolerance of rice (O. sativa L.), a set of introgression lines (ILs) were developed. The ILs were derived from an O. rufipogon accession collected from Chaling (Hunan Province, China) as the donor, and a widely grown O. sativa indica cultivar 93-11 as the recipient. Through evaluating the salt tolerance of 285 ILs at the seedling stage, a total of 10 quantitative trait loci (QTLs) related to salt tolerance were identified on chromosomes 1, 5, 7 and 9–12, with individual QTLs explaining 2–8% of phenotypic variance. The O. rufipogon-derived alleles at four QTLs improved salt tolerance in the 93-11 background. At the same time, a salt-tolerant IL, 9L136, was identified and characterized. Compared with the recipient parent 93-11, a total of 1,391 differentially expressed genes (DEGs) were detected specifically in 9L136 between salt stress and normal condition through genome-wide expression analysis. Of these, four DEGs located in the QTL regions carried by 9L136, suggesting that the four genes might be candidates associated with salt tolerance. Both the highly salt-tolerant ILs and the favorable O. rufipogon-derived QTLs identified in the present study will provide new genetic resources for improving the resistance of cultivated rice against salinity stress using molecular breeding strategies in the future.
Keywords: common wild rice, introgression line, seedling stage, salt tolerance, QTL analysis, RNA-seq
Introduction
Soil salinity is one of the most common abiotic stresses affecting crop growth and productivity worldwide (Hoang et al., 2016). It is estimated that 20% of the total cultivated and 33% of irrigated agricultural land are affected by salinity (Epstein et al., 1980). Many adaptation and mitigation strategies are required for crops to cope with the impacts of salinity. However, these strategies are cost intensive and time-consuming. Compared with improving saline soil, enhancing resistance of crop plants against salinity stress by breeding and genetic manipulation is an effective and low-cost approach. Rice (Oryza sativa L.), one of the most important food crops in the world, has medium sensitivity to salinity stress, and shows symptoms of damage and a dramatic reduction in productivity when the soil soluble salt reaches 0.3% (Akbar et al., 1972). Especially at the seedling stage, salt stress is more likely to cause growth retardation or death of rice, resulting in significant yield reductions (Zeng et al., 2001; Zhao et al., 2014). Therefore, further enhancing salt tolerance at the seedling stage would be helpful to improve environmental adaptation and increase yield in rice.
In the past decades, many QTLs/genes associated with adaptation to salt stress have been identified in rice (Lin et al., 1998, 2004; Koyama et al., 2001; Ren et al., 2005; Huang et al., 2009; Kumar et al., 2015; Liang et al., 2015). One QTL, related to survival time under salt stress at the seedling stage, was detected near the marker locus RG3 on chromosome 5 using a recombinant inbred population, and explained 11.6% of phenotypic variation (Lin et al., 1998). Through evaluating five phenotypic traits associated with salt tolerance, including sodium (Na+) and potassium (K+) content, a total of 11 QTLs were located on chromosomes 1, 4, 6, and 9, which explained 6.4–19.6% of phenotypic variation (Koyama et al., 2001). In addition, two major QTLs for shoot Na+ and K+ concentrations, qSNC-7 and qSKC-1, were found on chromosomes 7 and 1, and explained 48.5 and 40.1% of phenotypic variation, respectively (Lin et al., 2004). Using a genome-wide association study for 220 rice accessions, 20 single nucleotide polymorphisms (SNPs) significantly associated with Na+/K+ ratio, and 44 SNPs with other traits observed under stress conditions were identified at the reproductive stage (Kumar et al., 2015). Additionally, 11 loci containing 22 significant salt tolerance-associated SNPs were identified at the seed germination stage using a similar method (Shi et al., 2017). Through map-based cloning, a salt-tolerance related QTL, SKC1, was isolated, which encoded a member of the HKT-type transporters, regulating K+/Na+ homeostasis under salt stress (Ren et al., 2005). The DST gene (drought and salt tolerance), encoding a C2H2-type zinc finger transcription factor, was cloned and had a negative effect on drought and salt tolerances through regulating stomatal closure in rice (Huang et al., 2009). In conclusion, salt tolerance is controlled by complex genetic mechanisms in rice, which remain unclear.
Common wild rice (O. rufipogon Griff.), the wild progenitor of Asian cultivated rice (O. sativa L.), is an important primary gene pool for improving rice. Many favorable genes have been identified and characterized from wild rice (Tian et al., 2006, 2011; Zhang et al., 2006; Tan et al., 2007; Garcia-Oliveira et al., 2009), which can increase rice yield, improve grain quality, and enhance resistance to both biotic and abiotic stresses. Here, to exploit beneficial alleles for salt tolerance from wild rice, we evaluated the salt tolerance of a set of introgression lines (ILs) derived from a cross between an elite indica cultivar 93-11 and an O. rufipogon accession (Chaling common wild rice) from China, and identified QTLs for salt tolerance. Four candidate genes in QTL regions associated with salt tolerance were identified through integrating RNA sequencing and QTL mapping. The results will aid in utilizing alien alleles to improve tolerance to salt stress in modern rice breeding.
Materials and Methods
Plant Materials
A set of ILs was derived from a cross between elite indica cultivar, 93-11, and an O. rufipogon accession (Chaling common wild rice) from Chaling (Hunan Province, China) based on an advanced backcross (BC3) and consecutive selfing strategy (Supplementary Figure S1) as described previously (Ma et al., 2016). A total of 285 ILs were evaluated for salt tolerance and genotype using 142 simple sequence repeat (SSR) markers.
Phenotypic Evaluation
Seeds of both the recurrent parent 93-11 and 285 ILs were surface-sterilized in 5% sodium hypochlorite for 30 min and washed three times with distilled water, and then germinated at 30°C for 2 days. Finally, uniformly germinated seeds were sown in holes in thin Styrofoam board (with 96 holes and a nylon net bottom), floated on water for 4 days, and then transferred to float on Yoshida’s culture solution (Yoshida et al., 1976; Tian et al., 2011). About 14 days after germination, 10 normal seedlings of each line and the recurrent parent 93-11 with two and a half leaves were transferred to culture solution containing 125 mM NaCl and full-strength Yoshida’s solution. Both the survival rate and salt tolerance score (STS) of each line were measured after 9 days of salt treatment and 7 days of recovery. The survival rate was the percentage of living seedlings in the total number of seedlings. The STS was divided into five grades as described by Gregorio and Senadhira (1993) and Tian et al. (2011) with some modification (Supplementary Figure S2): Grade 0 included seedlings that died or were close to death; Grade 1 had significantly inhibited seedling growth and a survival rate of leaves of less than 20%; Grade 2 had some inhibition of seedling growth, plants had three leaves, and a survival rate of leaves of 20–40%; Grade 3 included seedlings with normal growth, plants that reached the four-leaf stage, and a survival rate of leaves of 40–60%; Grade 4 was seedling growth close to normal and a survival rate of leaves of 60–80%; and Grade 5 was normal seedling growth, with only the leaf tips of leaves being dry, and a survival rate of leaves of 80–100%.
DNA Extraction and SSR Analysis
Using the cetyltrimethyl ammonium bromide method (Rogers and Bendich, 1988), DNA of fresh leaves was extracted after grinding in liquid nitrogen. Primers to amplify SSR sequences were synthesized based on sequence information (McCouch et al., 2002). PCR amplification, polyacrylamide denaturing gel electrophoresis, and silver-staining were performed as described previously (Panaud et al., 1996).
Genotyping
Based on the genotypic analysis of 285 ILs using 142 polymorphic SSR markers, QTLs for salt tolerance were determined using single-point analysis in Map Manager QTXb17 (Manly et al., 2001). The statistical threshold for single-point analysis was P < 0.01 and LOD ≥ 2. The proportion of observed phenotypic variance explained by a QTL was estimated by the coefficient of determination (R2) from the corresponding linear model fitting.
Measurement of Physiological Characteristics
Six physiological characteristics associated with abiotic stress were measured as described previously (Yemm and Willis, 1954; Beauchamp and Fridovich, 1971; Dhindsa et al., 1981; Nakno and Asada, 1981; Ie-sung et al., 2003): contents of proline and soluble sugars, and activities of malondialdehyde, superoxide dismutase, catalase, and peroxidase. Both 93-11 and a highly salt-tolerant IL 9L136 were grown to the two and a half leaf-stage, and then transferred to culture solution containing 125 mM NaCl for the salt treatment. The seedlings were harvested quickly at different time points (0, 1, 2, 3, and 4 days after salt treatment) for measurement of physiological characteristics. Each experiment was replicated three times.
RNA-seq Analysis
Total RNA was isolated from roots of the recurrent parent 93-11 and IL 9L136 under salt treatment (1 h after treatment, ST) and control (0 h, CK) using TRizol (Invitrogen, Carlsbad, CA, United States) and purified using a Qiagen RNeasy kit (Qiagen, Germany). Each sample had three biological replicates. The purified RNA was used to construct the cDNA library using a NEBNext Ultra RNA Library Prep kit. The 12 libraries (including root samples from 9L136 and 93-11 before and 1 h after application of salt stress, and each sample had three biological repeats, n = 3) were subsequently sequenced using an Illumina HiSeq2500 Sequencer. After removing adaptor sequences and low-quality reads, the high-quality paired-end reads were mapped to the Nipponbare reference genome (MSU Rice Genome Annotation Project Release 7) using the spliced read mapper TopHat version 2.0.12 (Kim et al., 2013). Principal component analysis (PCA) was performed using the prcomp function in R software with default settings to interpret the relatedness among all replicas in each genotype (R Development Core Team, 2012). Significant differentially expressed genes (DEGs) were determined based on a threshold of twofold expression change and false discovery rate (FDR) < 0.05 using Cuffdiff, one module of Cufflinks (Trapnell et al., 2012).
Validation of RNA-Seq Data by Quantitative Real-Time RT-PCR (qRT-PCR)
RNA samples were reverse transcribed into cDNA using a SuperScript III RT kit (Invitrogen, Carlsbad, CA, United States; Cat. No. 18080-044). The cDNA samples were subjected to qRT-PCR quantification with three biological replicates and performed with SYBR Green Master Mix (Applied Biosystems, Foster City, CA, United States; PN 4309155) according to the product manual on a CFX96 Real Time System (Bio-Rad, Hercules, CA, United States). Reference genes and specific gene primers are listed in Supplementary Table S1. The relative quantification method was used to evaluate the quantitative variation between the replicates examined.
Results
Evaluation of Salt Tolerance of Wild Rice Introgression Lines at the Seedling Stage
To identify O. rufipogon-derived favorable alleles associated with salt tolerance, we investigated both STS and survival rate of 285 ILs and the recurrent parent 93-11 under 125 mM NaCl treatment. Phenotypic measurement showed that the frequency distributions of the two traits in the IL population accorded with the normal distribution, and both STS and the survival rate of the recurrent parent 93-11 (1.07 and 43%) were lower than the means for ILs (1.81 and 54%) at the seedling stage (Figure 1 and Table 1). The results indicated that the recurrent parent 93-11 was more susceptible to salt stress, and that the O. rufipogon genome might harbor beneficial alleles associated with enhanced salt tolerance in the cultivated rice background.
FIGURE 1.
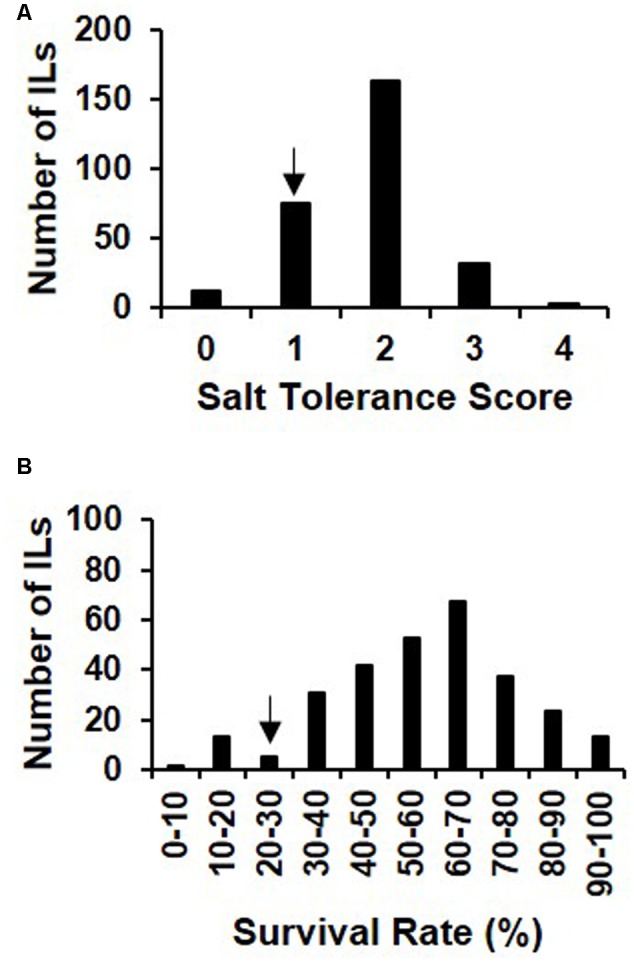
Frequency distribution of salt tolerance score (A) of seedlings and survival rate (B) in introgression lines under 125 mM NaCl treatment for 9 days and recovery for 7 days. The phenotype of the recurrent parent, 93-11, is indicated by arrows.
Table 1.
Observations of salt tolerance in the recipient parent 93-11 and introgression lines at the seedling stage.
| Traits | 93-11 | Introgression lines |
||||
|---|---|---|---|---|---|---|
| Average | Range | Coefficient of variation | Skewness | Kurtosis | ||
| STS | 1.07 | 1.81 ± 0.67 | 0.07–3.82 | 0.37 | –0.07 | 0.21 |
| Survival rate | 43% | 54 ± 16% | 7–100% | 0.3 | –0.43 | 0.33 |
QTL Analysis for Salt Tolerance at the Seedling Stage
Based on the genotypic analysis of 285 ILs using 142 polymorphic SSR markers, a total of 1358 chromosomal segments from O. rufipogon were detected, including 1082 homozygous and 276 heterozygous segments (Supplementary Table S2). Using single-point analysis, a total of 10 QTLs associated with salt response were identified (Figure 2 and Table 2). They were located on chromosomes 1, 5, 7 and 9–12, and each QTL explained 2–8% of the phenotypic variation (Figure 2 and Table 2). Additionally, the O. rufipogon-derived alleles at four loci (40% of QTLs) improved salt tolerance. Among these QTLs, six located on chromosomes 1 and 9–12 were detected using STS and survival rate as indexes. For example, qST10 was located near the SSR marker RM258 on chromosome 10, and qST12 near SSR marker RM7102 on chromosome 12. Moreover, alleles from O. rufipogon at the two QTLs could improve salt tolerance at the seedling stage in the 93-11 background. However, the other four QTLs (qST5-1, qST5-2, qST1-2 and qST7) were only detected by one index.
FIGURE 2.
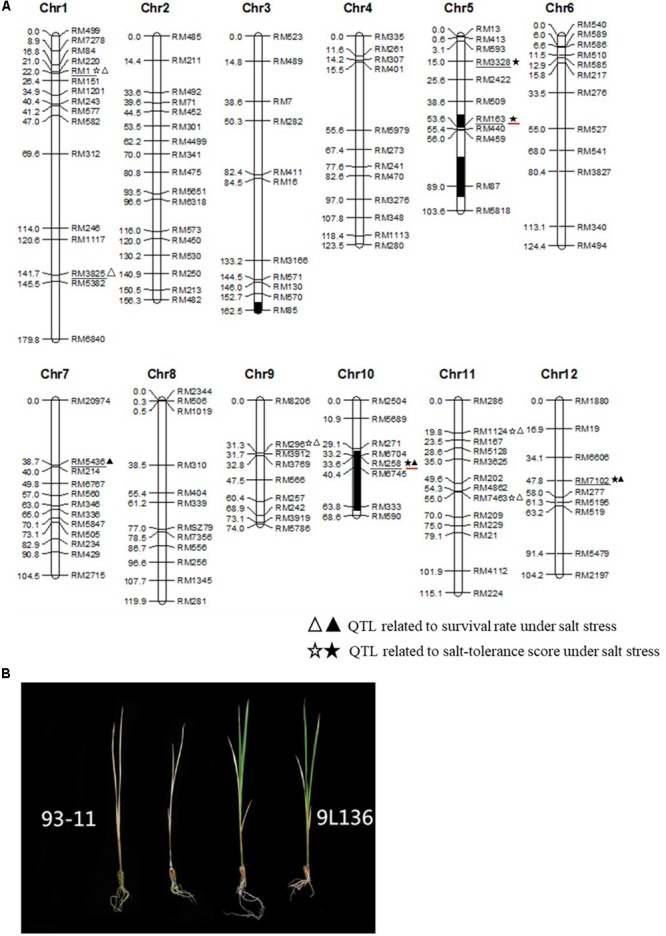
Chromosomal location of salt tolerance-related QTLs (A) and the 9L136 and 93-11 phenotypes treated with 125 mM NaCl for 9 days (B). Symbols on the right of chromosomes indicate the positions of putative QTLs for salt tolerance-related traits. The closed symbols indicate that the O. rufipogon-derived alleles conferred a positive effect on salt tolerance-related traits, and the open symbols indicate a negative effect. The markers related to QTLs for salt tolerance are underlined with black. In the graphical genotypes, the black regions indicate homozygous regions from Chaling common wild rice in 9L136 and the white regions indicate homozygous regions for recurrent parent 93-11. The QTLs enhancing salinity tolerance and located on the introgressed region in 9L136 are underlined with red.
Table 2.
QTLs for salt tolerance at the seedling stage identified using O. rufipogon introgression lines.
| Traits | Chr. | QTL | Marker | LOD | PV (%) | P-value | Add. |
|---|---|---|---|---|---|---|---|
| STS | 1 | qST1-1 | RM1 | 3 | 4 | 0.001 | –0.3 |
| 5 | qST5-1 | RM3328 | 2 | 8 | 0.003 | 0.64 | |
| 5 | qST5-2 | RM163 | 2 | 8 | 0.006 | 0.62 | |
| 9 | qST9 | RM296 | 3 | 4 | 0 | –0.26 | |
| 10 | qST10 | RM258 | 2 | 4 | 0.001 | 0.42 | |
| 11 | qST11-1 | RM1124 | 3 | 5 | 0.001 | –0.69 | |
| 11 | qST11-2 | RM7463 | 3 | 5 | 0.001 | –0.45 | |
| 12 | qST12 | RM7102 | 2 | 4 | 0.001 | 0.43 | |
| Survival rate | 1 | qST1-1 | RM1 | 3 | 5 | 0 | –0.08 |
| 1 | qST1-2 | RM3825 | 2 | 2 | 0.009 | –0.04 | |
| 7 | qST7 | RM5436 | 2 | 4 | 0.007 | –0.11 | |
| 9 | qST9 | RM296 | 3 | 5 | 0 | –0.06 | |
| 10 | qST10 | RM258 | 2 | 4 | 0.002 | 0.09 | |
| 11 | qST11-1 | RM1124 | 2 | 5 | 0.001 | –0.12 | |
| 11 | qST11-2 | RM7463 | 2 | 4 | 0.001 | –0.07 | |
| 12 | qST12 | RM7102 | 2 | 4 | 0.007 | 0.12 |
Chr., chromosome; PV, phenotypic variation; Add., additive effect.
Analysis of Salt-Tolerant Introgression Lines
To select the genetic materials for further breeding salt-tolerant rice varieties and cloning salt tolerance-related genes, we performed six independent-experiment evaluations and identified four ILs (9L19, 9L46, 9L136, and 9L201), which showed stable salt tolerance in multiple experiments, and had significantly higher STS and survival rates than those of the recurrent parent 93-11 (Table 3). Comparison of introgression segments with chromosomal location of QTLs carried by ILs showed that the four lines harbored two positive-effect QTLs for salt tolerance from O. rufipogon (Table 3 and Supplementary Figure S3). Notably, the highly salt-tolerant IL 9L136 carrying two salt tolerance-related QTLs (qST5-2 and qST10) (Figure 2), had the highest STS and survival rate among the four ILs, and its average survival rate was 84%, which increased by 95.3% compared with 93-11 (Table 3). Therefore, 9L136 was chosen for further characterization, as a highly salt-tolerant IL.
Table 3.
QTLs included in the introgressed segments carried by four selected salt-tolerant introgression lines.
| Number | Salt-tolerance | Survival | QTLs |
|---|---|---|---|
| of ILs | score | rate | included |
| 9L19 | 2.80 ± 1.83 | 63 ± 40% | qST5-1, qST5-2 |
| 9L46 | 3.11 ± 1.23 | 76 ± 25% | qST5-1, qST5-2 |
| 9L136 | 3.67 ± 0.69 | 84 ± 12% | qST5-2, qST10 |
| 9L201 | 2.48 ± 1.31 | 60 ± 25% | qST5-1, qST1-2 |
| 93-11 | 1.07 ± 0.53 | 43 ± 31% |
Physiological Characteristics of the Salt-Tolerant Line 9L136
Generally, salt stress is accompanied by osmotic stress, leading to reactive oxygen species damage and osmolyte accumulation (Abraham et al., 2003; Mittler et al., 2004; Miller et al., 2010; Farmer and Mueller, 2013). To investigate the physiological basis for the improved stress tolerance of 9L136, we measured the contents of proline and soluble sugar and activities of malondialdehyde, superoxide dismutase, catalase, and peroxidase in 9L136 and 93-11 under normal and salt-stress conditions. The activities of superoxide dismutase, catalase, and peroxidase were all significantly higher in 9L136 than 93-11 under salt stress (P < 0.05, Figures 3A–C), and 9L136 had a distinctly lower malondialdehyde content in response to 4 days of salt treatment compared with 93-11 (P < 0.01, Figure 3D). We also investigated the contents of soluble sugars, a class of compatible osmolytes, and observed that this was significantly higher in 9L136 than in 93-11 under salt treatment for 2, 3, and 4 days (Figure 3E). Additionally, the proline content in 9L136 was significantly higher than that in 93-11 under salt stress (Figure 3F). Overall, the results implied that 9L136 did not suffer from oxidative stress as much as the recurrent parent 93-11 during salt stress, suggesting that favorable alleles from wild rice carried by 9L136 might improve the salt tolerance of rice in the 93-11 background.
FIGURE 3.
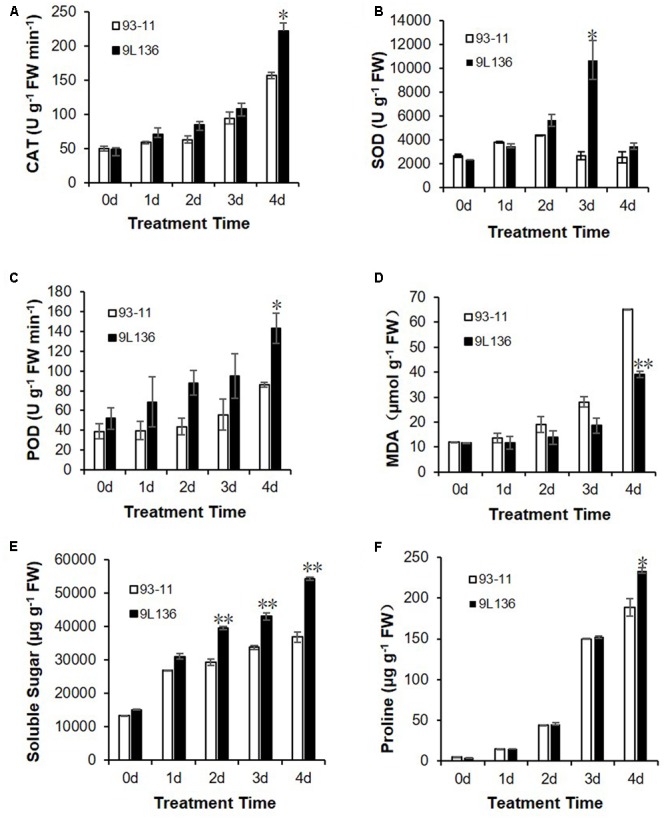
Comparison of physiological characteristics between salt-tolerant line 9L136 and recurrent parent 93-11 under salt stress conditions (125 mM NaCl). Physiological characteristics included the activities of catalase (CAT) (A), superoxide dismutase (SOD) (B), peroxidase (POD) (C), and malondialdehyde (MDA) (D), and the contents of soluble sugars (E) and proline (F). Data are means ± standard deviation with three replicates. Asterisks represent significant differences between 93-11 and 9L136 using Student’s t-test: ∗P < 0.05 and ∗∗P < 0.01.
Identification of Salt Tolerance-Related Candidate Genes in Wild Rice
To explore candidate genes related to salt tolerance, RNA-seq analysis was performed using RNA extracted from roots of 93-11 and 9L136 after 0 (CK) and 1 h (ST) treatment with 125 mM NaCl. A total of 692 million clean reads (FPKM of all replicas cufflinks showed in Supplementary Table S3) were obtained from 12 libraries, and about 80% of the reads could be mapped to predict gene regions. Sequence read information is summarized in Supplementary Table S4. Both Pearson’s correlation and PCA showed a high correlation among the replicas in each genotype (Supplementary Figure S4, S5 and Table S5). We also validated the expression levels of 15 randomly selected genes using qRT-PCR, which gave similar results to those of RNA-seq analysis – suggesting that RNA-seq data were reliable (Supplementary Figure S6).
Global expression analysis showed that a total of 4,650 DEGs, including 2,504 up-regulated and 2,146 down-regulated genes, were detected in IL 9L136 between salt stress (9L136-ST) and normal condition (9L136-CK); and a total of 4,291 DEGs, including 2,173 up-regulated and 2,118 down-regulated genes, were detected in the recipient parent 93-11 between salt stress (93-11-ST) and normal condition (93-11-CK) (Figure 4A). Further comparison of DEGs between 9L136-ST/9L136-CK and 93-11-ST/93-11-CK found that 1,391 genes showed differentially expressed only in 9L136-ST/9L136-CK but not in 93-11-ST/93-11-CK (Figure 4A), suggesting that these genes should be specifically induced in IL 9L136 under salt stress.
FIGURE 4.
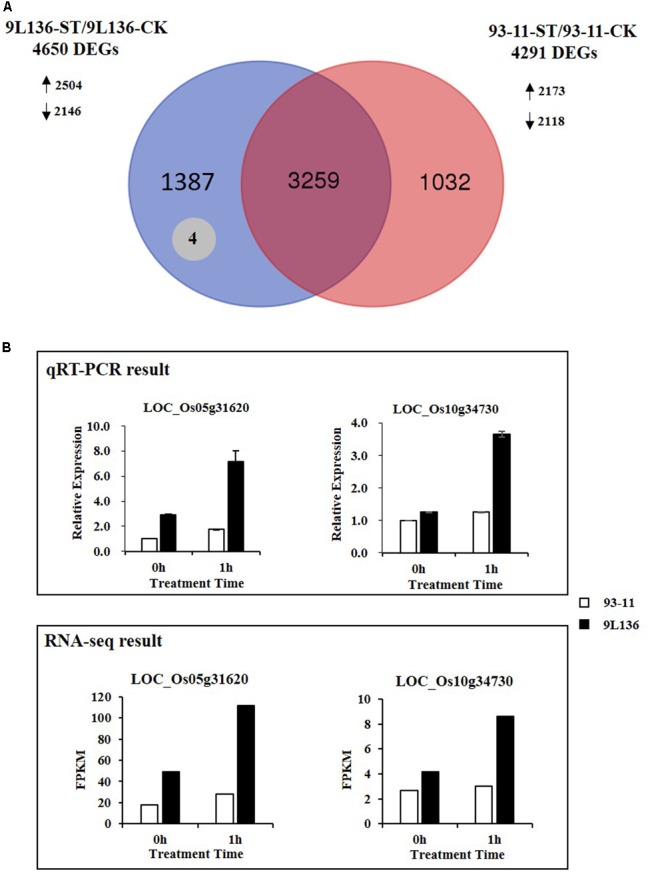
Identification of candidate genes related to salt tolerance from wild rice. (A) Venn diagram analysis for RNA-seq data showing differentially expressed genes between 9L136-ST/9L136-CK and 93-11-ST/93-11-CK. There were 1,391 genes showed differentially expressed only in 9L136-ST/9L136-CK but not in 93-11-ST/93-11-CK (A), suggesting these genes should be specifically induced in introgression line 9L136. Among the 1,391 genes, four were co-localized with QTLs for salt tolerance (shown in gray circle). Arrows indicated the up-regulated (↑) and down-regulated (↓) genes. (B) Expression profiles of LOC_Os05g31620 and LOC_Os10g34730 detected by qRT-PCR and RNA-seq, respectively.
Additionally, based on the gene annotation information of the rice reference genome (the TIGR Rice Genome Annotation Database), chromosomal regions of two QTLs (qST5-2 and qST10) carried by 9L136 harbored a total of 209 genes. To narrow down the candidate genes for salt tolerance, we further compared the chromosomal location between the 1,391 DEGs and QTL regions, and found that four genes, including acetyltransferase (LOC_Os05g31254), calmodulin-related calcium sensor protein (OsCML15, LOC_Os05g31620), LRP1 (LOC_Os05g32070), and GRAM domain containing protein (GEM, LOC_Os10g34730), were co-localized with the QTLs (qST5-2 and qST10) (Supplementary Table S6). Of the four genes, two ones (LOC_Os05g31620 and LOC_Os10g34730) were not only specifically induced by salt stress in 9L136 but also showed higher expression level in 9L136-ST compared with 93-11-ST (Figure 4B). Taken together, four candidate genes associated with salt tolerance at the seedling stage were identified through the strategy of combining QTL mapping and genome-wide expression analysis.
Discussion
Comparison of Salt Tolerance-Related QTLs in Different Populations
In previous studies, many QTLs related to salt tolerance were identified and located on 12 rice chromosomes, with most of them identified in cultivated rice. There are few reports of QTLs for salt tolerance in common wild rice. A total of 15 QTLs for salt tolerance were identified on chromosomes 1, 2, 3, 6, 7, 9, and 10 using a set of ILs derived from Yuanjiang common wild rice, and the wild rice derived alleles at 13 QTLs (86.7%) could enhance salt tolerance in the Teqing background (Tian et al., 2011). In our study, we used 285 ILs derived from a cross between elite indica variety 93-11 and an accession of Chaling common wild rice to identify 10 QTLs for salt tolerance on chromosomes 1, 5, 7, and 9–12. Comparison of salt tolerance-related QTLs in this study with those reported by Tian et al. (2011) showed that QTL qST7 mapped near RM5436 on chromosome 7 shared a similar chromosomal region with three QTLs (qRRW7, qRSW7, and qRTW7); and qST10, detected on chromosome 10 near RM258, was located on a similar region with a QTL cluster (qRRW10, qRSW10, and qRTW10) reported by Tian et al. (2011). Interestingly, the O. rufipogon-derived alleles at these QTLs/QTL cluster improved salt tolerance in the cultivated rice background.
We also compared the QTLs in this study with those detected in O. sativa populations. The results showed that qST5-1 near RM3328 on chromosome 5 was in a similar location to qDLRa5-2 and QSst5a (Qian et al., 2009; Liang et al., 2015), and qST5-2 was located on a similar location to qDSRs5-1, QDss5, QSf5, and QGW5 (Chen et al., 2008; Liang et al., 2015), further confirming the reliability of the salt stress-related QTLs detected in the present study. Moreover, the results indicated that the stable QTLs might be useful for rice breeding using marker-assisted selection and so accelerate the development of salt-tolerant rice varieties.
Possible Physiological and Molecular Mechanisms Underlying the Salt-Stress Response
Through both cellular and physiological changes, crop plants can respond and adapt to abiotic stresses, thus enabling them to survive (Huang et al., 2012; Kumar et al., 2013; Hoang et al., 2016). Under salt stress, the cell membrane is damaged due to salt damage, resulting in malondialdehyde production. Malondialdehyde is an indicator of reactive oxygen species level and the extent of damage to the membrane, and many free radicals are generated by this damage (Farmer and Mueller, 2013). Peroxidase and catalase are protective enzymes activated after superoxide dismutase, and further remove peroxide and hydrogen peroxide (Apel and Hirt, 2004; Jaleel et al., 2009). In our study, activity of protective enzymes catalase, superoxide dismutase, and peroxidase in 9L136 under salt-stress conditions were significantly higher than in the recurrent parent 93-11. Moreover, the superoxide dismutase increased significantly at 3 days of salt treatment and other substances (peroxidase, catalase, and malondialdehyde) had significant levels at 4 days of salt treatment. This indicated that the superoxide dismutase may remove the initially produced O2– and H2O2, and then peroxidase and catalase further removed or decomposed H2O2 (Pan et al., 2006), leading to lower reactive oxygen species levels in 9L136 than in 93-11 during salt stress. In addition, the contents of proline and soluble sugars in 9L136 under salt-stress conditions were significantly increased compared with 93-11. The results implied that the cell membrane in 9L136 was protected through increased activities of enzymes that scavenge reactive oxygen species, and more proline and soluble sugars under salt stress conditions, leading to improved salt tolerance.
Implication of Genes for Salt-Stress Tolerance
Through the platform combining QTL mapping and RNA-seq analysis, four candidate genes were co-localized with QTLs for salt tolerance, including acetyltransferase (LOC_Os05g31254), calmodulin-related calcium sensor protein (OsCML15, LOC_Os05g31620), LRP1 (LOC_Os05g32070), and GRAM domain containing protein (GEM, LOC_Os10g34730). Notably, both LOC_Os05g31620 (calmodulin-related calcium sensor protein, OsCML15) and LOC_Os10g34730 (GRAM domain containing protein, GEM) had higher expression level in 9L136-ST than in 93-11-ST. There are many GRAM domain-containing protein genes in rice, but only one GRAM domain-containing protein has been reported in rice and may function in plant development (Shu et al., 2005). In Arabidopsis, VAD1, a novel GRAM domain-containing protein was first characterized and its expression was in response to a pathogen-infection-dependent salicylic acid pathway (Lorrain et al., 2004). The gene LOC_Os10g34730 may function in stress defense and result in salt tolerance of rice by control of their expression.
We are most interested in OsCML15, the calmodulin-related calcium sensor protein gene located in the QTL region. In Arabidopsis, several CMLs have dramatic expression changes under various biotic and abiotic stimuli. CML10 can modulate stress responses in Arabidopsis by regulating ascorbic acid production (Cho et al., 2016). CML24 may enable responses to abscisic acid, daylength, and various salt stresses (Delk et al., 2005). In young Arabidopsis seedlings, AtCML9 expression is rapidly induced by abiotic stress and abscisic acid, and mutation analysis showed that it plays essential roles in modulating responses to salt stress and abscisic acid (Magnan et al., 2008). Additionally, in rice, a novel rice calmodulin-like gene, OsMSR2, was identified, which can enhance drought and salt tolerance and increase abscisic acid sensitivity in Arabidopsis (Xu et al., 2011). Consequently, further study of OsCML15 will be valuable for determining salt-stress mechanisms and in saline-resistance breeding.
Availability Of Supporting Data
The original RNA-seq data has been uploaded to NCBI (https://www.ncbi.nlm.nih.gov/geo/), and the GEO accession number is GSE101734.
Author Contributions
Manuscript draft: SW, FL, XM, LT, and CS. Experiments: SW, MC, JZ, and WC. Analyzing data: XM. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MJ and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 31371585).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01427/full#supplementary-material
References
- Abraham E., Rigo G., Szekely G., Nagy R., Koncz C., Szabados L. (2003). Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol. Biol. 51 363–372. 10.1023/A:1022043000516 [DOI] [PubMed] [Google Scholar]
- Akbar M., Yabuno T., Nakao S. (1972). Breeding for saline-resistant varieties of rice variability for salt tolerance among some rice varieties. Jpn. J. Breed. 22 277–284. 10.1270/jsbbs1951.22.277 [DOI] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Beauchamp C., Fridovich J. (1971). Superoxide dismutase: improved assays and assay applicable to polyacrylamide gels. Anal. Biochem. 44 276–287. 10.1016/0003-2697(71)90370-8 [DOI] [PubMed] [Google Scholar]
- Chen H. T., Jiang H., Yan M. X., Dong G. J., Qian Q., Guo L. B. (2008). QTL-mapping comparison of tolerance to alkali at germination period and early seedling stage between two different double haploid populations in rice. Mol. Plant Breed. 6 439–450. [Google Scholar]
- Cho K. M., Nguyen H. T., Kim S. Y., Shin J. S., Cho D. H., Hong S. B., et al. (2016). CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol. 209 664–678. 10.1111/nph.13612 [DOI] [PubMed] [Google Scholar]
- Delk N. A., Johnson K. A., Chowdhury N. I., Braam J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in response to abscisic acid, daylength, and ion stress. Plant Physiol. 139 240–253. 10.1104/pp.105.062612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa R. S., Plumb-Dhindsa P., Thorpe T. A. (1981). Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation and decreased level of superoxide dismutase and catalase. J. Exp. Bot. 32 93–101. 10.1093/jxb/32.1.93 [DOI] [Google Scholar]
- Epstein E., Norlyn J. D., Rush D. W., Kingsbury R. W., Kelley D. B., Cunningham G. A., et al. (1980). Saline culture of crops: a general approach. Science 210 399–404. 10.1126/science.210.4468.399 [DOI] [PubMed] [Google Scholar]
- Farmer E. E., Mueller M. J. (2013). ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 64 429–450. 10.1146/annurev-arplant-050312-120132 [DOI] [PubMed] [Google Scholar]
- Garcia-Oliveira A. L., Tan L. B., Fu Y. C., Sun C. Q. (2009). Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 51 84–92. 10.1111/j.1744-7909.2008.00730.x [DOI] [PubMed] [Google Scholar]
- Gregorio G. B., Senadhira D. (1993). Genetic analysis of salinity tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 86 333–338. 10.1007/BF00222098 [DOI] [PubMed] [Google Scholar]
- Hoang T. M. L., Tran T. N., Nguyen T. K. T., Williams B., Wurm P., Bellair S., et al. (2016). Improvement of salinity stress tolerance in rice: challenges and opportunities. Agronomy 6:54 10.3390/agronomy6040054 [DOI] [Google Scholar]
- Huang G. T., Ma S. L., Bai L. P., Zhang L., Ma H., Jia P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39 969–987. 10.1007/s11033-011-0823-1 [DOI] [PubMed] [Google Scholar]
- Huang X. Y., Chao D. Y., Gao J. P., Zhu M. Z., Shi M., Lin H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23 1805–1817. 10.1101/gad.1812409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ie-sung S., Yukie M., Akihiro Y., Dea-Wook K., Kenji U. (2003). Inhibition of catalase activity by oxidative stress and its relationship to salicylic acid accumulation in plants. Plant Growth Regul. 39 285–295. 10.1023/A:1022861312375 [DOI] [Google Scholar]
- Jaleel C. A., Riadh K., Gopi R., Manivannan P., Ines J., Al-Juburi H. J., et al. (2009). Antioxidant defense responses: physiological plasticity in higher plants under abiotic constraints. Acta Physiol. Plant. 31 427–436. 10.1007/s11738-009-0275-6 [DOI] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama M. L., Levesley A., Koebner R. M. D., Mikiko L., Flowers T. J., Yeo A. R. (2001). Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 125 406–422. 10.1104/pp.125.1.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Kumar M., Kim S. R., Ryu H., Cho Y. G. (2013). Insights into genomics of salt stress response in rice. Rice 6:27 10.1186/1939-8433-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Singh A., Mithra A. S. V., Krishnamurthy S. L., Parida S. K., Jain S., et al. (2015). Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). Genome Res. 22 133–145. 10.1093/dnares/dsu046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. L., Qu Y. P., Yang C. G., Ma X. D., Cao G. L., Zhao Z. W., et al. (2015). Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 201 441–452. 10.1007/s10681-014-1236-8 [DOI] [Google Scholar]
- Lin H. X., Yanagihara S., Zhuang J. Y., Senboku T., Zheng K. L., Yashima S. (1998). Identification of QTL for salt tolerance in rice via molecular markers. Chin. J. Rice Sci. 12 72–78. [Google Scholar]
- Lin H. X., Zhu M. Z., Yano M., Gao J. P., Liang Z. W., Su W. A., et al. (2004). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108 253–260. 10.1007/s00122-003-1421-y [DOI] [PubMed] [Google Scholar]
- Lorrain S., Lin B., Auriac M. C., Kroj T., Saindrenan P., Nicole M., et al. (2004). VASCULAR ASSOCIATED DEATH1 a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16 2217–2232. 10.1105/tpc.104.022038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Fu Y. C., Zhao X. H., Jiang L. Y., Zhu Z. F., Gu P., et al. (2016). Genomic structure analysis of a set of Oryza nivara introgression lines and identification of yield-associated QTLs using whole-genome resequencing. Sci. Rep. 6:27425 10.1038/srep27425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan F., Ranty B., Charpenteau M., Sotta B., Galaud J. P., Aldon D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56 575–589. 10.1111/j.1365-313X.2008.03622.x [DOI] [PubMed] [Google Scholar]
- Manly K. F., Cudmore J. R. H., Meer J. M. (2001). Map Manager QTX, cross platform software for genetic mapping. Mamm. Genome 12 930–932. 10.1007/s00335-001-1016-3 [DOI] [PubMed] [Google Scholar]
- McCouch S. R., Teytelman L., Xu Y. B., Lobos K. B., Clare K., Walton M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9 199–207. 10.1093/dnares/9.6.199 [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 33 453–457. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Breusegem F. V. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Nakno Y., Asada K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxide in spinach chloroplasts. Plant Cell Physiol. 22 867–880. [Google Scholar]
- Pan Y., Wu L. J., Yu Z. L. (2006). Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 49 157–165. 10.1007/s10725-006-9101-y [DOI] [Google Scholar]
- Panaud O., Chen X., McCouch S. R. (1996). Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Genet. Genomics 252 597–607. 10.1007/BF02172406 [DOI] [PubMed] [Google Scholar]
- Qian Y. L., Wang H., Chen M. Y., Zhang L. K., Chen B. X., Cui J. T., et al. (2009). Detection of salt-tolerance QTL using BC2F3 yield selected introgression lines of rice (Oryza sativa L.). Mol. Plant Breed. 7 224–232. [Google Scholar]
- R Development Core Team (2012). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ren Z. H., Gao J. P., Li L. G., Cai X. L., Huang W., Chao D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 1141–1146. 10.1038/ng1643 [DOI] [PubMed] [Google Scholar]
- Rogers O. S., Bendich A. J. (1988). “Extraction of DNA from plant tissue,” in Plant Molecular Biology Manual eds Glevin S. B., Schilperoort R. A. (Dordrecht: Kluwer Academic Publishers; ) 1–10. 10.1007/978-94-017-5294-7_6 [DOI] [Google Scholar]
- Shi Y. Y., Gao L. L., Wu Z. C., Zhang X. J., Wang M. M., Zhang C. S., et al. (2017). Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 17:92 10.1186/s12870-017-1044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y. J., Minnie C., Srinivasan R. (2005). The Oryza sativa no pollen (Osnop) gene plays a role in male gametophyte development and most likely encodes a C2-GRAM domain-containing protein. Plant Mol. Biol. 57 835–853. 10.1007/s11103-005-2859-x [DOI] [PubMed] [Google Scholar]
- Tan L. B., Liu F. X., Xue W., Wang G. J., Ye S., Zhu Z. F., et al. (2007). Development of Oryza rufipogon and Oryza sativa introgression lines and assessment for yield-related quantitative trait loci. J. Integr. Plant Biol. 49 871–884. 10.1111/j.1744-7909.2007.00497.x [DOI] [Google Scholar]
- Tian F., Li D. J., Fu Q., Zhu Z. F., Fu Y. C., Wang X. K., et al. (2006). Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112 570–580. 10.1007/s00122-005-0165-2 [DOI] [PubMed] [Google Scholar]
- Tian L., Tan L. B., Liu F. X., Cai H. W., Sun C. Q. (2011). Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J. Genet. Genomics 38 593–601. 10.1016/j.jgg.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. Y., Rocha P. S. C. F., Wang M. L., Xu M. L., Cui Y. C., Li L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234 47–59. 10.1007/s00425-011-1386-z [DOI] [PubMed] [Google Scholar]
- Yemm E. W., Willis A. J. (1954). The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 57 508–514. 10.1042/bj0570508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Forno D. A., Cock J. H., Gomez K. A. (1976). Laboratory Manual for Physiological Studies of Rice 3rd Edn. Manila: International Rice Research Institute. [Google Scholar]
- Zeng L., Shannon M. C., Lesch S. M. (2001). Timing of salinity stress affects rice growth and yield components. Agric. Water Manag. 48 191–206. 10.1016/S0378-3774(00)00146-3 [DOI] [Google Scholar]
- Zhang X., Zhou S. X., Fu Y. C., Su Z., Wang X. K., Sun C. Q. (2006). Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 62 247–259. 10.1007/s11103-006-9018-x [DOI] [PubMed] [Google Scholar]
- Zhao X. Q., Wang W. S., Zhang F., Deng J. L., Li Z. K., Fu B. Y. (2014). Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at seedling stage. PLoS ONE 9:e108020 10.1371/journal.pone.0108020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


