Abstract
Human echovirus 18 (E-18) is a member of the enterovirus B species. To date, sixteen full-length genome sequences of E-18 are available in the GenBank database. In this study, we describe the complete genomic characterization of two E-18 strains isolated in Yunnan, China. Pairwise comparisons of the nucleotide sequences and the deduced amino acid sequences revealed that the two Yunnan E-18 strains had 87.5% nucleotide identity and 96.3–96.5% amino acid identity with the Chinese strain. Phylogenetic and bootscanning analyses revealed the two E-18 strains had the highest identity with other several EV-B serotypes than the other E-18 strains in the P3 coding region, especially, 3B region of the Swine Vesicular disease virus (SVDV) strain HK70, indicated that frequent intertypic recombination might have occurred in the two Yunnan strains. This study contributes the complete genome sequences of E-18 to the GenBank database and provides valuable information on the molecular epidemiology of E-18 in China.
Introduction
The genus Enterovirus, within family Picornaviridae, order Picornavirales, can be divided into 13 species: enterovirus A–J, and rhinovirus A–C (www.picornaviridae.com), comprising more than 100 serotypes. Enterovirus B (EV-B) containing 63 serotypes consists coxsackieviruses B (CV-B1-6), CV-A9, echoviruses (E-1–7, 9, 11–21, 24–27, 29–33) and enteroviruses named with digital serial numbers (EV-B69, 73-75, 77-88, 93, 97-101, 106-107,110-113) and the simian enterovirus SA5 (www.picornaviridae.com/enterovirus/ev-b/ev-b.htm). EVs are small, single-stranded, non-enveloped and positive-sense RNA viruses. The EV genome consisting of approximately 7,400 nucleotides (nt), contains a long open reading frame (ORF) flanked by a 5′-untranslated region (UTR) and a 3′-UTR1, 2. The 5′-UTR of EVs has an internal ribosomal entry site that drives translation initiation. The 3′-UTR, containing a long poly (A) stretch, is very important in RNA replication3, 4. The ORF encodes a polyprotein that can be processed into three polyprotein precursors: P1, P2, and P3. P1 encodes VP1–4 structural proteins. P2 and P3 encode 2A–C and 3A–D non-structural proteins, respectively5.
EVs infect about a billion people annually worldwide. Though the majority of clinical symptoms are asymptomatic or mild, such as fever, irritation, agitation, sore throat, headache, myalgia, vomiting, diarrhoea, hand-foot-and-mouth disease (HFMD) and others, a few patients may develop severe neurological syndromes as aseptic meningitis, acute flaccid paralysis (AFP), hypertensive heart failure (HHF), dilated cardiomyopathy and death5. Of these symptoms, the viruses of species EV-B are the most common viral cause of aseptic meningitis6–8.
Echovirus (E-18) belongs to species EV-B. The prototype strain (Metcalf) of E-18 was isolated in 1995 from a patient with diarrhoea9. Outbreaks of aseptic meningitis caused by E-18 have been frequently reported10–16. To date, except for the prototype strain, there are fifteen E-18 full-length genome sequences in the GenBank database, isolated from the patients with meningitis or contact persons, respectively. In this study, we determined the full-length genome sequences of two E-18 strains (strain A83/YN/CHN/2016, and strain A86/YN/CHN/2016), isolated in the Yunnan province of China in 2016 from two children with HFMD aged 5 and 4.2 years, respectively. This is the first report of E-18 and its probable association with HFMD.
Results
Molecular serotyping and primary characterization
The isolates A83/YN/CHN/2016 and A86/YN/CHN/2016 (abbreviated as A83 and A86) were recovered from human rhabdomyosarcoma (RD) cells. They cannot produce a cytopathic effect in KMB17 human embryonic lung diploid fibroblasts (KMB17) and human lung cancer (A549) cell lines. Partial VP1 sequencing and molecular typing by the online genotyping tool17 showed that the serotype of the two isolates is E-18. In addition, complete VP1-coding region alignment revealed the two strains had 79.0–79.2% nt and 92.0–92.7% amino acid identities with E-18 prototype strain Metcalf (AF317694).
Complete genome analysis
The complete genomes of the two strains consisted of 7,404 and 7,399 nt, respectively. They encoded a polypeptide of 2,192 amino acids. The coding sequences were flanked by a non-coding 5′-UTR of 745 nt and a non-coding 3′-UTR of 94 or 100 nt, respectively. The two Yunnan E-18 strains shared 80.4% similarities in the complete genome and 95.4–95.7% similarities in the deduced amino acid sequence with the prototype strain Metcalf, there were 11-nt and 16-nt deletions at the 3′-end of their genomes, and 5 nt insertions and one nt deletion at the 5′-UTR, respectively. The overall base compositions of the strains A83 and A86 genomes were 28.3% A, 22.8% G, 24.9% C and 23.9% U, and 28.2% A, 22.9% G, 24.9% C and 24.0% U, respectively. Compared with other E18 strains, the nt sequences of non-structural regions were more variable (74.2–91.5%), while, the sequences of structural regions were highly conserved (91.3–98.3%). Strain A83 shared 99.7% nt and 99.5% amino acid identities with A86 strain. A comprehensive comparison of different genomic regions of the nt sequences and deduced amino acid sequences of strains A83 and A86 with the E-18 prototype strain and other E-18 strains are shown in Table 1.
Table 1.
Nucleotide and amino acid identities between the two Yunnan E-18 strains, Metcalf and other E-18 in all sequenced genomic regions.
| Nucleotide identity (%) [Amino acid identity (%)] | ||||
|---|---|---|---|---|
| Region | A83/YN/CHN/2016 | A86/YN/CHN/2016 | ||
| Metcalf | Other E-18 | Metcalf | Other E-18 | |
| 5′UTR | 81.7 | 85.1–97.7 | 81.7 | 84.8–97.0 |
| VP4 | 80.7(92.8) | 85.0–96.6 (95.7–98.6) | 78.7(89.9) | 83.6–94.7(95.7–100) |
| VP2 | 80.5(96.5) | 86.0–98.3 (98.5–100) | 80.5(96.5) | 86.0–98.3(98.5–100) |
| VP3 | 79.6(96.2) | 87.2–97.6 (96.7–97.9) | 79.9(97.1) | 87.4–97.8(97.5–98.7) |
| VP1 | 79.0(92.0) | 86.9–97.2 (94.8–97.9) | 79.2(92.7) | 87.5–97.7(95.8–99.3) |
| 2A | 77.8(93.3) | 78.4–90.7 (90.7–96.0) | 77.8(93.3) | 78.7–90.7(90.7–96.0) |
| 2B | 79.1(97.0) | 75.1–90.9 (90.9–100) | 79.1(97.0) | 75.1–90.9(90.9–100) |
| 2C | 82.4(97.0) | 79.4–91.5 (94.8–97.3) | 82.3(96.7) | 79.3–91.4(94.5–97.0) |
| 3A | 77.2(92.1) | 75.7–81.3 (93.3–100) | 77.2(92.1) | 75.7–81.3(93.3–95.5) |
| 3B | 78.7(95.5) | 74.2–81.8 (95.5–100) | 78.8(95.5) | 77.3–81.8(95.5–100) |
| 3C | 82.3(97.3) | 76.3–81.1 (95.6–97.3) | 82.3(97.3) | 76.3–81.1(95.6–97.3) |
| 3D | 80.2(96.6) | 77.5–80.9 (94.3–95.7) | 80.2(96.1) | 77.5–80.9(94.3–95.7) |
| 3′UTR | 85.9 | 81.0–84.2 | 85.1 | 80.0–85.1 |
| Genome | 80.4(95.4) | 81.8–88.5 (95.8–97.2) | 80.4(95.5) | 81.8–88.5(95.9–97.3) |
Phylogenetic analysis
The phylogenetic tree was conducted with 80 strains available in GenBank based on the complete VP1 sequences (Fig. 1). The 82 strains (including the two Yunnan strains) could be divided into five different genogroups (E-18:1-5). Except for 05430/SD/CHN/2005 (GQ329813), the other Chinese strains clustered together and belonged to genogroup E-18:5, which was also prominent epidemic strains worldwide.
Figure 1.
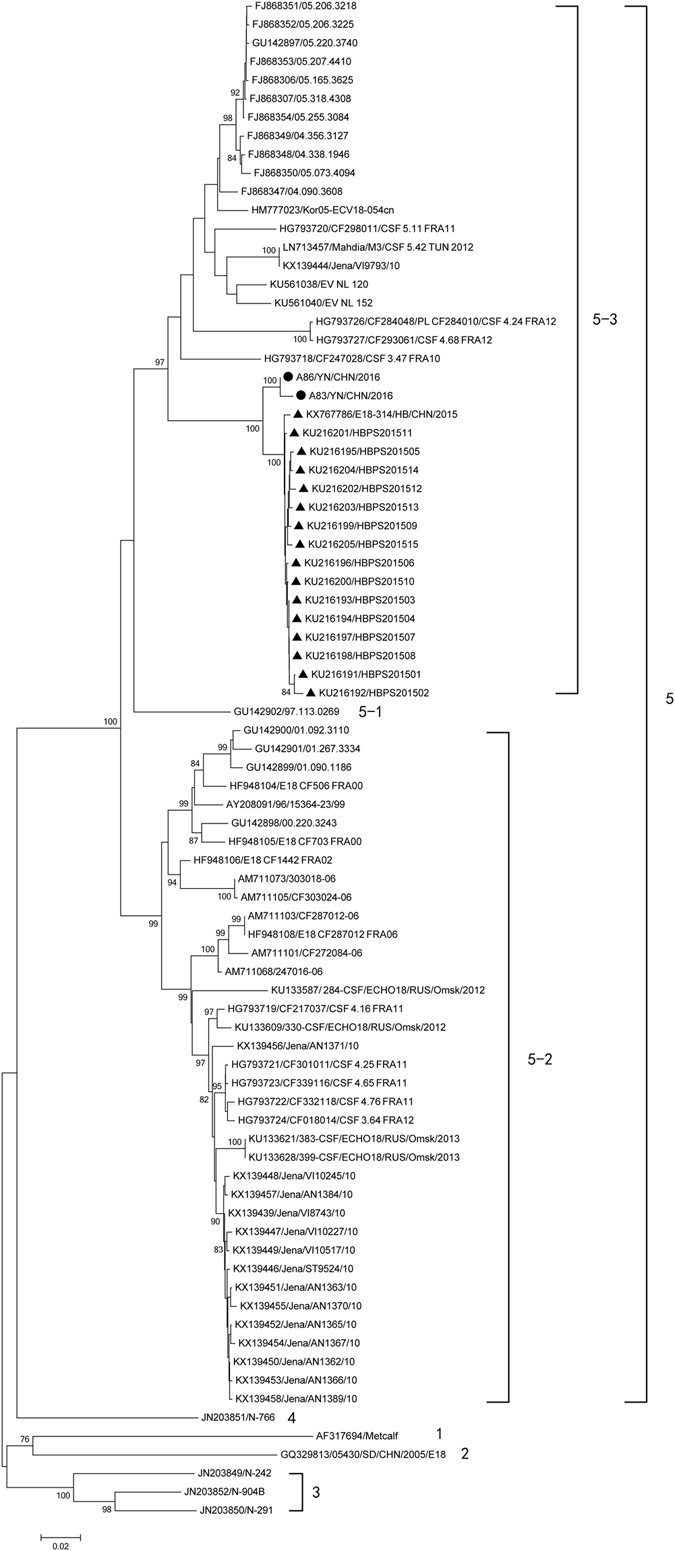
Phylogenetic relationships based on the complete VP1 sequences of 82 E-18 strains. Two Yunnan E-18 strains and the other 80 E-18 strains available in GenBank database were analysed by nucleotide sequence alignment using the neighbor joining algorithms implemented in the MEGA 6.06 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. • Strains isolated in this investigation. ▲ Strains isolated from other provinces of China.
Phylogenetic analysis of strains A83 and A86 with 377 EV-B strains available in GenBank database were performed based on the other genomic regions respectively (Figs 2–5). In P1 capsid coding region, the Yunnan E-18 strains only clustered together with other E-18 strains, consistent with the preliminary molecular typing results. However, in the P3 (or 3A, 3B, 3C and 3D) regions (Figs 3 and 5), the two Yunnan strains did not form a lineage with E-18 strains, and grouped together with other EV-B serotypes such as Swine Vesicular disease virus (SVDV) strain HK70. For 3′UTR and P2 (or 2A , 2B and 2C ) regions (Figs 2, 3 and 4), the two isolates clustered together with one E-18 strain and other EV-B serotypes. Among these, the nt sequence of the 3B genomic region was the most homologous (93.5%) to the corresponding sequence of SVDV strain HK70. Thus, these results suggest the occurrence of one or more putative recombination events between the two E-18 Yunnan strains and other EV-B serotypes.
Figure 2.
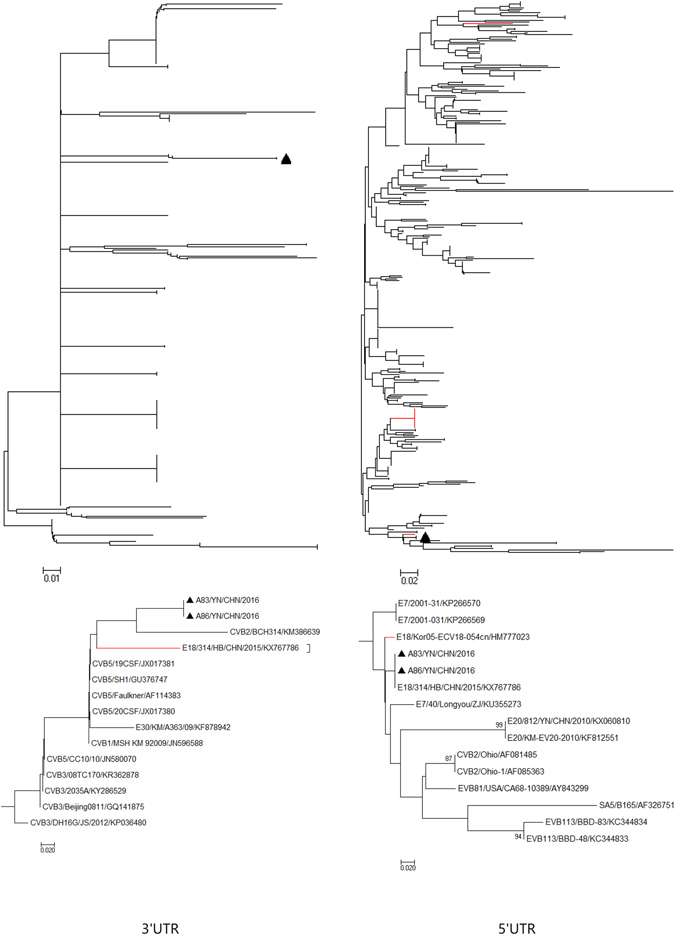
Phylogenetic relationships based on the 5′UTR and 3′UTR sequences of EV-B strains. Two E-18 strains and 343 EV-B strains for 5′UTR, and 349 EV-B strains for 3′UTR available in GenBank database were analysed by nucleotide sequence alignment using the neighbor joining algorithms implemented in the MEGA 6.06 program, respectively. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. Only high bootstrap values (>75%) are shown. ▲ Strains isolated in this investigation. Red denote the other E-18 strains.
Figure 5.
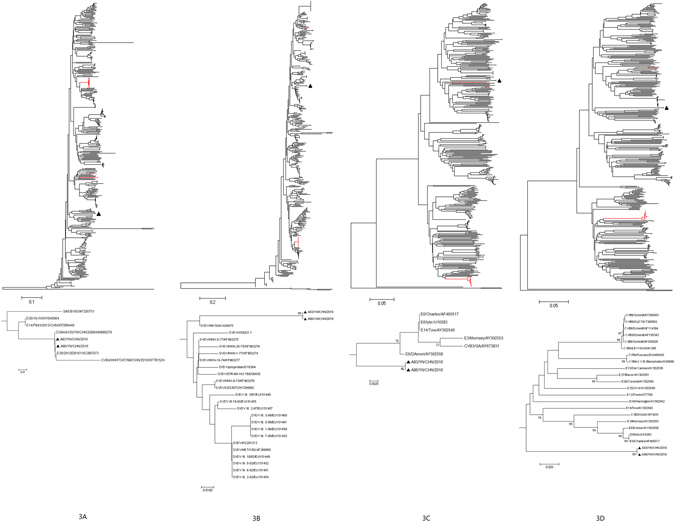
Phylogenetic relationships based on the 3 A, 3 B, 3C, and 3D coding sequences of 379 EV-B strains. Two E-18 strains and 377 EV-B strains available in GenBank database were analysed by nucleotide sequence alignment using the neighbor joining algorithms implemented in the MEGA 6.06 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. Only high bootstrap values (>75%) are shown. ▲ Strains isolated in this investigation. Red denote the other E-18 strains.
Figure 3.
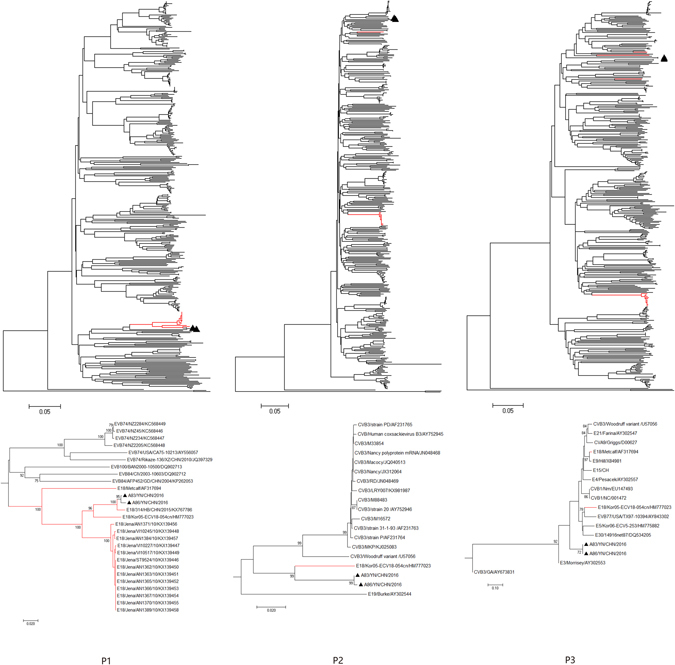
Phylogenetic relationships based on the P1, P2, and P3 coding sequences of 379 EV-B strains. Two E-18 strains and 377 EV-B strains available in GenBank database were analysed by nucleotide sequence alignment using the neighbor joining algorithms implemented in the MEGA 6.06 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. Only high bootstrap values (>75%) are shown. ▲ Strains isolated in this investigation. Red denote the other E-18 strains.
Figure 4.
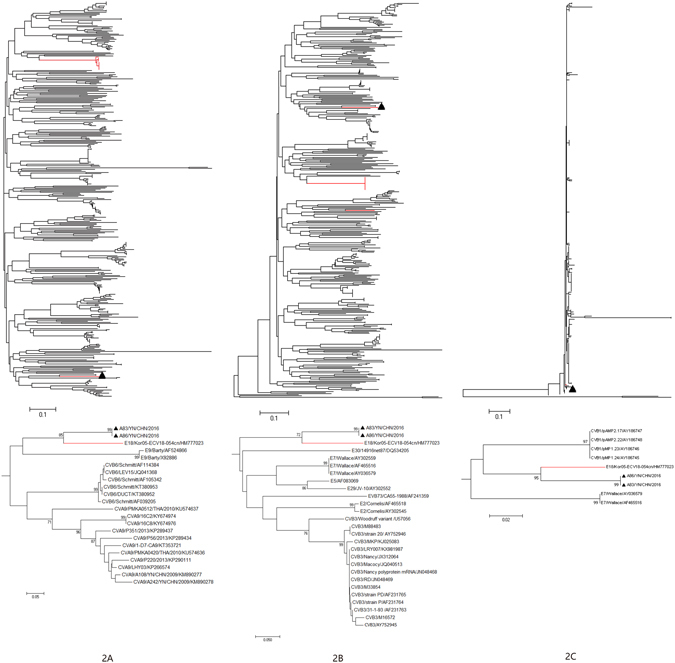
Phylogenetic relationships based on the 2 A, 2 B, and 2C coding sequences of 379 EV-B strains. Two E-18 strains and 377 EV-B strains available in GenBank database were analysed by nucleotide sequence alignment using the neighbor joining algorithms implemented in the MEGA 6.06 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The scale bars represent the genetic distance. Only high bootstrap values (>75%) are shown. Only high bootstrap values (>75%) are shown. ▲ Strains isolated in this investigation. Red denote the other E-18 strains.
In addition, the VP1 nt and amino acid sequences were compared between the E-18 genogroups (Table 2). The average VP1 nt sequence divergence between the E-18 genogroups E-18-1, 2, 3, 4, and 5 was 18.6% (range: 15.2–22.1%). However, the divergence was higher than the 14.95% divergence value assigned for EV-A71 subgenotyping18. This further confirmed that the E-18 sequences segregated into five distinct genogroups.
Table 2.
The complete VP1 nucleotide and amino acid sequences comparisons between the E-18 gene clusters.
| genotype | A | B | C | D | E |
|---|---|---|---|---|---|
| A | 79.4 | 79.6–80.6 | 78.7 | 77.9–78.7 | |
| B | 95.5 | 82.0–83.0 | 81.4 | 79.1–79.8 | |
| C | 92.3–94.4 | 94.8–97.2 | 83.9–84.8 | 82.7–83.0 | |
| D | 94.1 | 96.9 | 95.6–98.6 | 83.3–83.6 | |
| E | 91.3–93.0 | 92.0–95.5 | 90.9–96.5 | 94.1–95.8 |
Note: the data in the upper right corner was for nucleotide homology analysis and the data in the lower left corner was for amino acid homology analysis.
Recombination analysis
To confirm the recombination events between the Yunnan E-18 strains and other EV-B serotypes, bootscanning analyses were performed (Fig. 6). The results also revealed multiple recombination events between the genomic sequence of strains A83 and A86 with other EV-B serotypes, especially, the 3B region of SVDV strain HK70.
Figure 6.
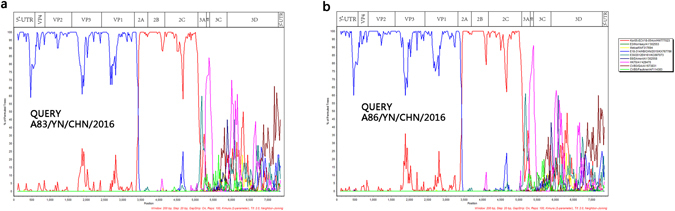
Bootscanning analyses of the complete genome of the two Yunnan E-18 strains. Bootscanning analysis of complete EV-B genomes using a sliding window of 200 nt moving in 20-nt steps. The two Yunnan strains were used as query sequences independently.
Discussion
Since 2008, millions of children have developed HFMD each year in mainland China and HFMD has become a serious public health issue. EVs are the causative agents of HFMD. Of these, CV-A16 and EV-A71 are the predominant etiologic agents. Other EV serotypes including CV-A2–6, CV-A8–10, CV-A12, CV-A14, CV-B1–6 and E-4–7, E-9, E-11, E-18, E-25, and E-30 were occasionally detected in sporadic cases or outbreaks of HFMD19, 20. Thus, persistent surveillance of HFMD pathogens might help to predict potential predominant serotypes and related disease outbreaks. Recently CV-A6 and CV-A10 have been the prevalent etiologic agents in some provinces of China and in a few countries19. Although a vaccine for EV-A71 has been developed, it is important to develop multivalent vaccines against other main serotypes to protect children.
E-18 infection is associated with a variety of clinical presentations from asymptomatic to aseptic meningitis and death21. Outbreaks of aseptic meningitis caused by E-18 have been reported in China and worldwide10–15. Currently, the E-18 strains available in GenBank were all isolated from aseptic meningitis patients and contact persons. The serotypes of causative agents for aseptic meningitis may overlap with that of HFMD22. Thus, the two Yunnan E-18 strains were isolated from HFMD patients, indicating possible connections between E-18 infections and HFMD.
There are currently 80 E-18 complete VP1 sequences available in the GenBank database, and among these, fifteen strains are full-length genome sequences. The 80 E-18 strains were isolated from different countries, China, India, South Korea, Australia, Netherlands, Germany, Sweden, Russia and France, indicating that E-18 infection is a global distribution. The large divergences between the complete VP1 coding sequences result in the formation of five clusters in phylogenetic analysis, which indicates that the circulation of distinct E-18 lineages exist in the same and different countries. And the large divergences between Yunnan isolates (A83 and A86) and Hebei strain E18-314/HB/CHN/2015 in non-capsid sequences showed the existence of Chinese E-18 strain apparently evolved along two lineages independently of the other E-18 strains. Additionally, the complete genome sequences of the two strains were highly homologous, indicating that E-18 had been circulating in the environment for an extended period of time.
Recombination is a universal phenomenon for EVs23, 24. According to the phylogenetic analyses in different genomic regions, proposed recombination events were observed between the two Yunnan strains and E-30 strain 2012EM161, SVDV strain HK70, E-6 strain D’Amori, and CV-B3 strain GA in corresponding regions, respectively. And bootscanning analyses for the two strains also showed intertypic recombination between E-18, E-30, CV-B3, SVDV and E-6 with the P3 non-capsid region. Interestingly, the nt sequences of the 3B genomic region of the two Yunnan strains were the most homologous (93.5%) to the corresponding sequence of the SVDV strain HK70. Whereas, other E-18 strains were no more closely related to HK70 than it was to other CV-B serotypes.
SVD is a highly contagious disease of pigs and is classified as a list A disease by the International Office of Epizootics25, 26. Clinically, SVD cannot be distinguished from foot-and-mouth disease (FMD), vesicular stomatitis (VS) and vesicular exanthema of swine (VES), and, therefore, diagnosis depends on laboratory tests. Due to the persistence of the virus in the environment, once the disease spreads, it is very difficult to control. Thus, SVD is notifiable in the world and is strictly controlled by livestock movement. SVDV is the causative agent of SVD25, 26. SVDV share a high degree of sequence similarity with CV-B5 in P1 region and the cross-neutralization was observed between the two viruses27. But other genetic regions of SVDV are less homologous to CV-B5. The P1 protein contains neutralization sites and the (partial) P1 region-based molecular typing result corresponds well to enterovirus serotypes28. At present, the enterovirus typing is generally defined by analysis of the P1 region. So SVDV is currently classified as a porcine variant of CV-B526. However, it is unclear whether the recombination of SVDV occurred in humans or pigs29. In addition, the non-structural protein and un-translated regions, which have diverse functions in the viral replication are characterized by species-specific11. Recombination events usually occur in non-structural regions of EV-B23, 30–32, which is consistent with our report. E-18 prototype Metcalf strain also is a recombinant with E-9 Hill, which share highly homologous in the P3 region33, 34, and SVDV also is a recombinant with CV-A9 Net/1/63 in the 3D region. Thus, recombination plays a key role on the evolution and adaptation of the members of EV-B species.
Duo to the number of complete sequences of circulating strains is rather limited, this could result a bias of intra-and inter-typic recombination. In addition, silent transmissions could be the reason of underestimation of recombination events35. Thus, although our isolates A83 and A86 share high degree of sequence similarity with SVDV in 3B region, it is difficult to determine whether the SVDV strain was the donor of Yunnan E-18 strains and more complete genome sequences are required for analysis. In addition, the two Yunnan strains in this study were isolated from patients with HFMD, but we could also not conclude that E-18 was the causative agent of HFMD, for samples not taken from the throat swab, serum and other sites of lesions in the two patients. But after all the two strains were isolated from feces of patients with HFMD. So, further research is needed to elucidate the correlation.
In conclusion, we have reported the full-length genome sequences of two E-18 strains during HFMD surveillance in Yunnan, China. Sequence analysis suggested that the two strains have high genetic diversity compared with the other E-18 strains, intertypic recombination in the non-structural protein region. The high divergence among the E-18 strains reflects that this virus has circulated in the environment for many years and recombination drives the evolution of E-18. This study expands the number of complete genome sequences of E-18 in the GenBank database and provides the molecular epidemiology of E-18 in China, which can help to evaluate the association between E-18 and E-18-related diseases.
Material and Methods
Ethics statement
All participants gave written informed consent. The protocol was in accordance with the Helsinki Declaration, and was approved by the Institutional Ethics Boards of the Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College. The human material used were stool samples collected from two patients aged 5 and 4.2 years with HFMD. Stool samples were collected and processed according to standard procedures. Three cell lines, RD, KMB17, and A549 were used to isolate viruses from the stool specimens according to standard procedures36. All positive isolates were stored at −80 °C.
Molecular typing
For molecular serotyping, the viral RNAs were extracted from cell culture supernatants with a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA). Reverse transcription-polymerase chain reaction (RT-PCR) was carried out using a PrimeScriptTM One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) with primer pairs 222 and 22437. The PCR-positive products were sequenced using an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA, USA) at BGI Sequencing Company (Beijing, China). The partial VP1 sequences were compared with sequences from GenBank using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Full-length genome amplification
Two long-distance PCR amplifications were performed using a PrimeScriptTM One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China). The primers used for RT-PCR and sequencing of the full-length genome were designed by a “primer-walking” strategy38 and are listed in Table 3. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germany), and sequenced in both directions at least two from each strand using ABI 3130 Genetic Analyser (Applied Biosystems, USA).
Table 3.
Primers used for complete genome amplification and sequencing.
| primer | Sequence (5′-3′) | Nucleotide position* | Orientation |
|---|---|---|---|
| 224 | GCIATGYTIGGIACICAYRT | 1977–1996 | Forward |
| 222 | CICCIGGIGGIAYRWACAT | 2969–2951 | Reverse |
| E181F | TTAAAACAGCCTGTGGGTTG | 1–20 | Forward |
| E181R | TGTGCCATGAAGGGTGTA | 2431–2414 | Reverse |
| E181r | GGAACGCGGTGACTCATC | 340–323 | Reverse |
| E181f | CTATAAGGATGCGGCATC | 839–856 | Forward |
| E182F | CATAAACGTTAGGGAGA | 2714–2730 | Forward |
| E182f | GTACTTCTCGCAGCTGGG | 3600–3617 | Forward |
| E184f | CAGAGTGATCAAGAGCA | 4263–4269 | Forward |
| E185r | CCCAACTGGGATGTACAT | 5749–5732 | Reverse |
| E186r | CCTGGGTTTTGGTGAAAG | 6520–6503 | Reverse |
| E187f | GACAAGGGAGAGTGTTT | 7018–7034 | Forward |
| E188R | ACCGAATGCGGAGAATTTAC | 7410–7391 | Reverse |
*Numbering according to the genome of E-18 strain E18-314/HB/CHN/2015.
Sequence analysis and recombination analysis
Nucleotide and amino acid sequence alignment were performed by using Geneious (5.6.5)16. Phylogenetic analysis was conducted via MEGA version 6.06 using the neighbour-joining method with 1000 duplicates and bootstrap values greater than 75% were considered statistically significant. Bootscanning analysis were performed using the Simplot 3.5.1 program with a 200-nt window moving in 20 nt steps38.
Nucleotide sequence accession number
The complete genome sequences of the E-18 strains A83/YN/CHN/2016 and A86/YN/CHN/2016 described in this study were deposited in the GenBank database under the accession number: KY828851-KY828852.
Acknowledgements
This work was supported by the Basic Research Projects of Yunnan Province, China (2013FZ136 and 2017FA006) and Chinese Academy of Medical Sciences Initiative for Innovative Medicine (2016-I2M-3-026).
Author Contributions
H.Z., Y.Z., Z.Y., and S.M. conceived the study and drafted the paper, H.L., and X.H. performed the experiments. H.S. helped to interpret results and contributed to the writing. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Haihao Zhang and Yilin Zhao contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhaoqing Yang, Email: zyang@imbcams.com.cn.
Shaohui Ma, Email: shaohuima70@hotmail.com.
References
- 1.Minor, P. Picornaviridae. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses (Elsevier, San Diego, 2011).
- 2.Zhang Y, et al. Molecular typing and characterization of a new serotype of human enterovirus (EV-B111) identified in China. Virus research. 2014;183:75–80. doi: 10.1016/j.virusres.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA (New York, N.Y.) 2000;6:1781–1790. doi: 10.1017/S1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoll J, Heus HA, van Kuppeveld FJ, Melchers WJ. The structure-function relationship of the enterovirus 3′-UTR. Virus research. 2009;139:209–216. doi: 10.1016/j.virusres.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Pallansch, M. A., Oberste, M. S. & Whitton, J. L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. Fields virology, 6th edn, Vol. 1 (eds Knipe, D. M. & Howley, P. M.) 490–530 (Lippincott Williams & Wilkins, Philadelphia, 2013).
- 6.Holmes CW, et al. Predominance of enterovirus B and echovirus 30 as cause of viral meningitis in a UK population. Journal of Clinical Virology. 2016;81:90–93. doi: 10.1016/j.jcv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, et al. Molecular identification of human enteroviruses associated with aseptic meningitis in Yunnan province, Southwest China. Springerplus. 2016;5 doi: 10.1186/s40064-016-3194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mladenova Z, et al. Aseptic meningitis outbreak caused by echovirus 30 in two regions in Bulgaria, May–August 2012. Epidemiology & Infection. 2014;142:2159–2165. doi: 10.1017/S0950268813003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichenwald HF, Ababio A, Arky AM, Hartman AP. Epidemic diarrhea in premature and older infants caused by ECHO virus type 18. Journal of the American Medical Association. 1958;166:1563–1566. doi: 10.1001/jama.1958.02990130023005. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X. P., Sun, S. Z., Guo, J. Y., Li, J. J. & Xie, Z. D. Complete Genome Sequence Analysis of Echovirus 18 Associated with Aseptic Meningitis in Hebei Province, China, in 2015. Genome Announcements4 (2016). [DOI] [PMC free article] [PubMed]
- 11.Krumbholz A, et al. Analysis of an echovirus 18 outbreak in Thuringia, Germany: insights into the molecular epidemiology and evolution of several enterovirus species B members. Medical Microbiology & Immunology. 2016;205:1–13. doi: 10.1007/s00430-016-0464-z. [DOI] [PubMed] [Google Scholar]
- 12.Tsai HP, et al. An echovirus 18-associated outbreak of aseptic meningitis in Taiwan: epidemiology and diagnostic and genetic aspects. Journal of Medical Microbiology. 2011;60 doi: 10.1099/jmm.0.027698-0. [DOI] [PubMed] [Google Scholar]
- 13.Mclaughlin JB, Gessner BD, Lynn TV, Funk EA, Middaugh JP. Association of regulatory issues with an echovirus 18 meningitis outbreak at a children’s summer camp in Alaska. Pediatric Infectious Disease Journal. 2004;23:875–877. doi: 10.1097/01.inf.0000136867.18026.22. [DOI] [PubMed] [Google Scholar]
- 14.Turabelidze G, Lin M, Butler C, Fick F, Russo T. Outbreak of echovirus 18 meningitis in a rural Missouri community. Missouri Medicine. 2009;106 [PubMed] [Google Scholar]
- 15.Wang SM, Ho TS, Shen CF, Wang JR, Liu CC. Echovirus 18 meningitis in southern Taiwan. Pediatric Infectious Disease Journal. 2011;30:259–260. doi: 10.1097/INF.0b013e3181f7cb69. [DOI] [PubMed] [Google Scholar]
- 16.X C, et al. An outbreak of echovirus 18 encephalitis/meningitis in children in Hebei Province, China, 2015. Emerging microbes & infections. 2017;6 doi: 10.1038/emi.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroneman A, et al. An automated genotyping tool for enteroviruses and noroviruses. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Chan YF, Sam IC, AbuBakar S. Phylogenetic designation of enterovirus 71 genotypes and subgenotypes using complete genome sequences. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2010;10:404–412. doi: 10.1016/j.meegid.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Klein M, Chong P. Is a multivalent hand, foot, and mouth disease vaccine feasible? Human vaccines & immunotherapeutics. 2015;11:2688–2704. doi: 10.1080/21645515.2015.1049780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma S, et al. Dynamic Constitution of the Pathogens Inducing Encephalitis in Hand, Foot and Mouth Disease in Kunming, 2009–2011. Japanese journal of infectious diseases. 2015;68:504–510. doi: 10.7883/yoken.JJID.2014.428. [DOI] [PubMed] [Google Scholar]
- 21.Brunel D, Jacques J, Motte J, Andreoletti L. Fatal echovirus 18 leukoencephalitis in a child. Journal of clinical microbiology. 2007;45:2068–2071. doi: 10.1128/JCM.00320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao Z, et al. Molecular epidemiology of human enterovirus associated with aseptic meningitis in Shandong Province, China, 2006–2012. PloS one. 2014;9 doi: 10.1371/journal.pone.0089766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukashev AN, et al. Recombination in circulating Human enterovirus B: independent evolution of structural and non-structural genome regions. Journal of General Virology. 2005;86:3281–3290. doi: 10.1099/vir.0.81264-0. [DOI] [PubMed] [Google Scholar]
- 24.Lukashev AN, et al. Recombination strategies and evolutionary dynamics of the Human enterovirus A global gene pool. Journal of General Virology. 2014;95:868–873. doi: 10.1099/vir.0.060004-0. [DOI] [PubMed] [Google Scholar]
- 25.Bellini S, Santucci U, Zanardi G, Brocchi E, Marabelli R. Swine vesicular disease surveillance and eradication activities in Italy. Revue Scientifique Et Technique. 2007;26:585–593. doi: 10.20506/rst.26.3.1766. [DOI] [PubMed] [Google Scholar]
- 26.Lin F, Kitching RP. Swine Vesicular Disease: An Overview. Veterinary Journal. 2000;160 doi: 10.1053/tvjl.2000.0505. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G, Haydon DT, Knowles NJ, Mccauley JW. Molecular evolution of swine vesicular disease virus. Journal of General Virology. 1999;80(Pt 3) doi: 10.1099/0022-1317-80-3-639. [DOI] [PubMed] [Google Scholar]
- 28.Xiang Z, et al. Coxsackievirus A21, Enterovirus 68, and Acute Respiratory Tract Infection, China. Emerging Infectious Diseases. 2012;18:821–824. doi: 10.3201/eid1805.111376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruhn CAW, et al. Viral meningitis epidemics and a single, recent, recombinant and anthroponotic origin of swine vesicular disease virus. Evolution, Medicine, and Public Health,2015,1(2015-10-27) 2015;2015:289–303. doi: 10.1093/emph/eov026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, et al. Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China. Sci Rep. 2017;7 doi: 10.1038/srep43080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jie, Z. et al. Molecular characterization of a new human coxsackievirus B2 associated with severe hand-foot-mouth disease in Yunnan Province of China in 2012. Archives of Virology, 1–5 (2016). [DOI] [PubMed]
- 32.Zheng H, et al. Isolation and Characterization of a Highly Mutated Chinese Isolate of Enterovirus B84 from a Patient with Acute Flaccid Paralysis. Scientific Reports. 2016;6 doi: 10.1038/srep31059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson P, Edman K, Lindberg AM. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Research. 2002;85:71–83. doi: 10.1016/S0168-1702(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 34.Oberste MS, Maher K, Pallansch MA. Evidence for Frequent Recombination within Species Human Enterovirus B Based on Complete Genomic Sequences of All Thirty-Seven Serotypes. Journal of Virology. 2004;78:855–867. doi: 10.1128/JVI.78.2.855-867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kyriakopoulou Z, Pliaka V, Amoutzias GD, Markoulatos P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes. 2015;50:177–188. doi: 10.1007/s11262-014-1152-y. [DOI] [PubMed] [Google Scholar]
- 36.Prim N, et al. Combining cell lines to optimize isolation of human enterovirus from clinical specimens: report of 25 years of experience. J Med Virol. 2013;85:116–120. doi: 10.1002/jmv.23426. [DOI] [PubMed] [Google Scholar]
- 37.Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. Journal of clinical microbiology. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. Complete genome sequence analysis of two human coxsackievirus A9 strains isolated in Yunnan, China, in 2009. Virus genes. 2015;50:358–364. doi: 10.1007/s11262-015-1180-2. [DOI] [PubMed] [Google Scholar]


