Abstract
A polyphasic study was undertaken to establish the taxonomic status of Streptomyces strains isolated from hyper-arid Atacama Desert soils. Analysis of the 16S rRNA gene sequences of the isolates showed that they formed a well-defined lineage that was loosely associated with the type strains of several Streptomyces species. Multi-locus sequence analysis based on five housekeeping gene alleles showed that the strains form a homogeneous taxon that is closely related to the type strains of Streptomyces ghanaensis and Streptomyces viridosporus. Representative isolates were shown to have chemotaxonomic and morphological properties consistent with their classification in the genus Streptomyces. The isolates have many phenotypic features in common, some of which distinguish them from S. ghanaensis NRRL B-12104T, their near phylogenetic neighbour. On the basis of these genotypic and phenotypic data it is proposed that the isolates be recognised as a new species within the genus Streptomyces, named Streptomyces asenjonii sp. nov. The type strain of the species is KNN35.1bT (NCIMB 15082T = NRRL B-65050T). Some of the isolates, including the type strain, showed antibacterial activity in standard plug assays. In addition, MLSA, average nucleotide identity and phenotypic data show that the type strains of S. ghanaensis and S. viridosporus belong to the same species. Consequently, it is proposed that the former be recognised as a heterotypic synonym of the latter and an emended description is given for S. viridosporus.
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-017-0886-7) contains supplementary material, which is available to authorized users.
Keywords: Streptomyces, Polyphasic taxonomy, Hyper-arid, Atacama Desert
Introduction
The prospect of isolating novel filamentous actinobacteria that synthesise new specialised metabolites is enhanced when bioprospecting strategies are focused on neglected and unexplored habitats (Hong et al. 2009; Tiwari and Gupta 2012; Guo et al. 2015), including desert soils (Meklat et al. 2011; Boubetra et al. 2013). The most extensive surveys of culturable actinobacterial diversity in desert biomes have been concentrated on sites in the Atacama Desert in northern Chile, the driest non-polar desert on the planet (Bull and Asenjo 2013; Bull et al. 2016). The application of a taxonomic approach to drug discovery (Goodfellow and Fiedler 2010) has been effective in the isolation of putatively novel filamentous actinobacteria from Atacama Desert habitats, some of which produce novel natural products (Bull et al. 2016; Wichner et al. 2016). Indeed, polyphasic taxonomic studies on dereplicated actinobacteria isolated from hyper-arid and extreme hyper-arid Atacama Desert soils have led to the description of novel species of Lechevalieria (Okoro et al. 2010), Lentzea (Idris et al. 2017a) and Modestobacter (Busarakam et al. 2016a) and to the detection of rare thermophilic Amycolatopsis species (Busarakam et al. 2016b). In addition, several new Streptomyces species have been described (Santhanam et al. 2012a, b, 2013; Idris et al. 2017b), one of which, Streptomyces leeuwenhoekii (Busarakam et al. 2014), encompasses strains that synthesise novel antibiotics (Nachtigall et al. 2011; Rateb et al. 2011a, b) and chaxapeptin, a new lasso peptide (Elsayed et al. 2015).
The present study was designed to establish the taxonomic position of several closely related Atacama Desert streptomycetes. These strains were the subject of a polyphasic taxonomic study which showed that they belong to a new species, Streptomyces asenjonii sp. nov.
Materials and methods
Isolation, maintenance and cultivation of strains
Isolates KNN6.11a, KNN35.1bT, KNN35.2b, KNN48.3e and KNN83.e were recovered from a hyper-arid soil collected in 2012 by one of us (ATB) from the Chaxa de Laguna, Salar de Atacama near Tocanão (23°17′33″S, 68°10′99″W at 2219 m above sea level), using the dilution plate procedure described by Okoro et al. (2009). The strains were isolated on Gauze’s No.1 agar (KNN6.11a) (Zakharova et al. 2003), humic acid-vitamin agar (KNN35.1bT, KNN35.2b) (Hayakawa and Nonomura 1987) and SM1 agar (KNN48.3e, KNN83.e) (Tan et al. 2006) following incubation for 14 days at 28 °C. Similarly, the final strain, KNN42.f, was isolated from a starch-casein agar plate (Küster and Williams 1964) following inoculation with a suspension of an extreme hyper-arid soil collected by ATB in 2010 from the Yungay core region of the Atacama Desert (24°06′18.6″S, 70°01′55.6″W at 1016 m asl). These strains, together with Streptomyces ghanaensis NRRL B12104T (Wallhäuser et al. 1965), were maintained on yeast extract—malt extract agar (International Streptomyces Project [ISP2] medium., Shirling and Gottlieb 1966) and as suspensions of spores and hyphal fragments in 20%, v/v glycerol at −20 and −80 °C. Biomass samples for most of the chemotaxonomic analyses and for the 16S rRNA gene sequencing studies were prepared in shake flasks (180 revolutions per minute) of ISP 2 broth after incubation at 28 °C for 14 days and washed twice in distilled water. Cells for the chemotaxonomic analyses were freeze-dried and those for the sequencing studies stored at room temperature. Biomass preparations for the fatty acid analyses were harvested from shake flasks of Tryptic Soy broth (Difco) following incubation at 28 °C for 7 days.
Phylogenetic analysis
16S rRNA gene sequencing. Genomic DNA extraction, PCR-mediated amplification of 16S rRNA genes and purification of the resultant products were carried out on all of the isolates using the procedures described by Kim and Goodfellow (2002). Identification of phylogenetic neighbours and calculation of pairwise 16S rRNA gene sequence similarities were achieved using the EzTaxon-e server (http://www.ezbiocloud.net/taxonomy; Yoon et al. 2017) and the resultant sequences aligned using the CLUSTAL W algorithm from the MEGA 6 software package (Tamura et al. 2013). Phylogenetic analyses using the maximum-likelihood (ML) (Felsenstein 1981) and maximum-parsimony (MP) algorithms (Fitch 1971) were also realised using the GGDC web server (Meier-Kolthoff et al. 2013a) of the DSMZ phylogenomics pipeline (Meier-Kolthoff et al. 2014) adapted to single genes available at http://ggdc.dsmz.de/. ML and MP trees were inferred from the alignment with RAxML (Stamatakis 2014) and TNT (Goloboff et al. 2008), respectively. The topologies of the resultant trees were evaluated by bootstrap analyses (Felsenstein 1985) based on 1000 replicates used in conjunction with tree-bisection-and-reconnection branch swapping and ten additional random sequence replicates for MP and rapid bootstrapping in conjunction with the auto MRE bootstopping criterion (Pattengale et al. 2010) for ML. The trees were rooted using the 16S rRNA gene sequence of Streptomyces albus subspecies albus DSM 40317T (GenBank accession number AJ621602). The Χ2 test implemented in PAUP* (Swofford 2002) was used to check for compositional bias. Pairwise sequence similarities were calculated using the method recommended by Meier-Kolthoff et al. (2013b) for 16S rRNA genes and a multiple sequence alignment was created with MUSCLE (Edgar 2004).
Multi-locus sequence analysis. Genomic DNA extracted from each of the isolates following growth in ISP2 broth at 28 °C was purified, as described by Idris et al. (2017a). The housekeeping genes used in previous analyses on streptomycetes (Busarakam et al. 2014; Labeda et al. 2017; Idris et al. 2017b; Labeda 2016), namely atpD (ATP synthase F1, beta subunit), gyrB (DNA gyrB subunit), rpoB (RNA polymerase beta subunit), recA (recombinase A) and trpB (tryptophane B, beta subunit), were amplified, sequenced, purified, deposited in the GenBank database and organised using the Bacterial Isolate Genome Sequence Database BIGSdb version 1.15.4 on the ARS Microbial Genome Sequence Database server (http://199.133.98.43). The sequences of the protein loci of the strains were aligned with one another and with those of their close neighbours and phylogenetic relationships established using the ML algorithm after Idris et al. (2017a). Pairwise distances between the sequences of each locus were established using the Kimura two-parameter model (Kimura 1980). Strain pairs having MLSA evolutionary distances ≤0.007 were considered conspecific based on the cut-off point empirically determined by Rong and Huang (2012, 2014), a value that corresponds to the 70% DNA:DNA threshold recommended for the delineation of prokaryotic species by Wayne et al. (1987).
Draft genome preparation and ANI calculations
The draft genome sequence of Streptomyces viridosporus NRRL 2414T was prepared following the protocol outlined in Labeda et al. (2016) with the exception that CLCbio Genomic Workbench Version 9.5.3 (CLCbio; Boston, MA) was used for contig trimming and de novo assembly. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession MSGP00000000.
The draft genome sequence of NRRL 2414T was compared with the draft genomes sequences of S. viridosporus T7A (Genbank accession number AJFD00000000), S. ghanaensis ATCC 14672T (GenBank accession number ABYA00000000), Streptomyces hirsutus NRRL B-3713T (GenBank accession number LIQT00000000), and Streptomyces cyanoalbus NRRL B-3040T (GenBank accession number LIPS00000000) obtained from Genbank utilising the calculate_ani.py script (https://github.com/widdowquinn/sripts/blob/master/bioinformatics/calculate_ani.py) which implements the methods described by Goris et al. (2007) and Richter and Rosselló-Móra (2009), with results shown in Supplemental Table S1.
Chemotaxonomy and morphology
Isolates KNN35.1bT and KNN35.2b were examined for spore chain arrangement and spore-surface ornamentation following growth on oatmeal agar (ISP 3 medium; Shirling and Gottlieb 1966) for 14 days at 28 °C, by scanning electron microscopy (Cambridge 240 instrument), using the protocol described by O’Donnell et al. (1993). Key chemotaxonomic markers were sought using standard chromatographic procedures. All of the isolates were examined for isomers of diaminopimelic acid (A2pm) after Staneck and Roberts (1974). Strains KNN35.1bT and KNN35.2b were analysed for menaquinone, whole cell sugar and polar lipid composition using the procedures described by Collins et al. (1985), Lechevalier and Lechevalier (1970) and Minnikin et al. (1984), respectively. S. ghanaensis NRRL B-12104T, the close phylogenetic neighbour of the isolates, was included in the sugar and polar lipid analyses. Fatty acids of representative isolates, namely strains KNN35.1bT, KNN35.2b and KNN42.f, and the S. ghanaensis type strain, were extracted, methylated and analysed using the established Sherlock Microbial Identification (MIDI) system and the ACTIN version 6 database (Sasser 1990).
Cultural characteristics
The cultural properties of the isolates were recorded on tryptone-yeast extract, yeast extract-malt extract, inorganic salts-starch, glycerol-asparagine, peptone-yeast extract-iron and tyrosine agar plates (ISP media 1-7, Shirling and Gottlieb 1966) after 14 days at 28 °C. Aerial and substrate mycelial colours and those of diffusible pigments were determined by comparison against chips from the Inter-Society Colour Council-National Bureau of Standard Colour charts (Kelly 1964).
Phenotypic tests
The isolates and S. ghanaensis NRRL B-12104T were examined for standard biochemical, degradative and physiological characteristics after Williams et al. (1983) and enzyme profiles determined using API-ZYM kits (BioMerieux) employing a standardised inoculum corresponding to 5 on the McFarland scale (Murray et al. 1999) and the protocol provided by the manufacturer. The oxidation of carbon sources and resistance to inhibitory compounds were determined using GENIII microplates in an Omnilog device (Biolog Inc., Haywood, USA). The microplates were inoculated with cell suspensions made in a ‘gelling’ inoculating fluid at a cell density of 98% transmittance with a run time of 7 days in phenotypic microarray mode at 28 °C. The exported data were analysed using the opm package for R version 1.0.6. (Vaas et al. 2012, 2013). The Biolog tests were carried out in duplicate.
Antibacterial sensitivity assays
Four of the isolates, strains KNN6.11a, KNN35.1bT, KNN35.2b and KNN83.e, were examined for their ability to inhibit the growth of wild type strains of Bacillus subtilis, Escherichia coli, Pseudomonas fluorescens and Staphylococcus aureus using a standard plug assay (Fiedler 2004). The isolates were grown on yeast extract-malt extract sloppy agar (0.8%, w/v agar) for 14 days at 30 °C and then plugs were transferred to nutrient agar plates which had been inoculated with 100 µl of the wild type strains grown overnight in lysogeny broth. The inoculated plates were incubated overnight and then examined for the presence of inhibition zones around the agar plugs.
Results and discussion
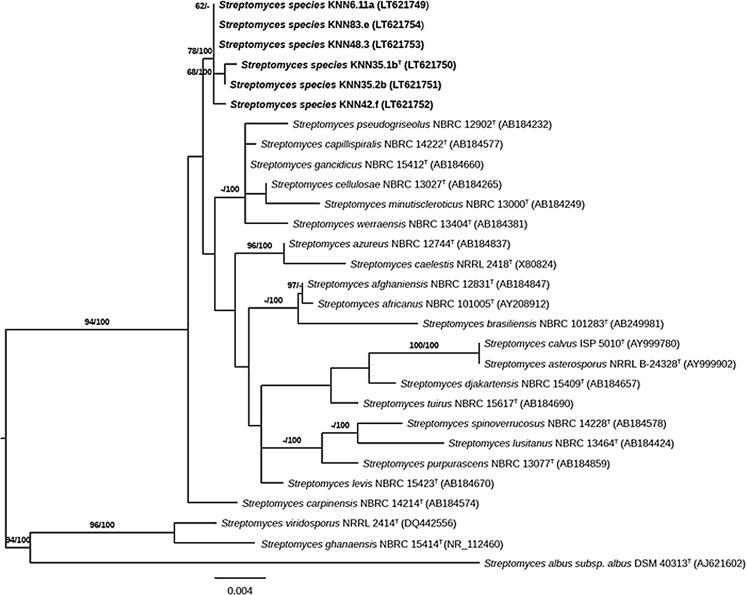
The six strains isolated from the hyper-arid and extreme hyper-arid Atacama Desert soils were shown to form a well delineated subclade in the Streptomyces 16S rRNA gene tree, a relationship that was supported by all of the tree-making algorithms and by a 78% bootstrap value (Fig. 1). The isolates were found to share 16S rRNA gene sequence similarities within the range 99.85–100%, which corresponds to up to 3 nucleotide (nt) differences at 1373 locations. The strains were seen to be closely related to the type strains of Streptomyces gancidicus DSM 40935 (99.57–99.64% similarity), Streptomyces pseudogriseolus DSM 40026T (99.49–99.58% similarity), Streptomyces capillispiralis DSM 41695T (99.49–99.57% similarity), Streptomyces werraensis DSM 40486T (99.34–99.51% similarity), Streptomyces minutiscleroticus DSM 40301T (99.05–99.15% similarity) and Streptomyces cellulosae DSM 40362T (99.35–99.50% similarity). These data suggest that the Atacama Desert isolates are not particularly closely related to any of their near phylogenetic neighbours in the Streptomyces 16S rRNA gene tree.
Fig. 1.
Maximum-likelihood phylogenetic tree based on 16S rRNA sequences showing relationships between isolates KNN6.11a, KNN 35.1b, KNN 35.2b, KNN 42.f, KNN 48.3, and KNN83.e and between them and the type strains of the most closely related Streptomyces species, the tree was inferred using the GTR+GAMMA model. The branches are scaled in terms of the expected number of substitutions per site. The numbers above the branches are support values when larger than 60% from ML (left) and MP (right) bootstrapping
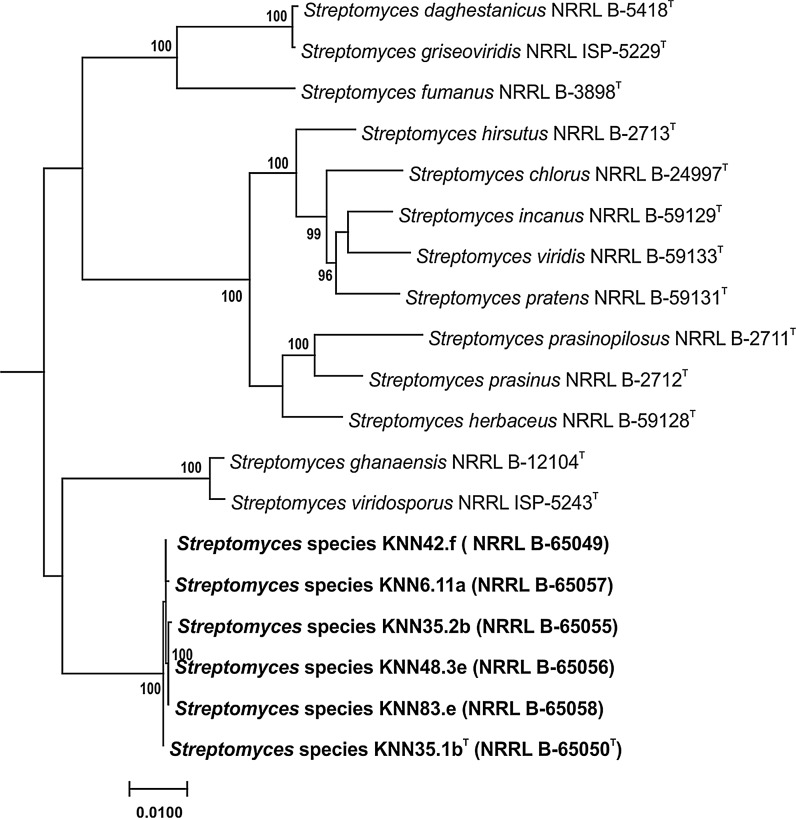
The isolates were found to belong to a distinct and homogeneous lineage in the Streptomyces MLSA gene tree based on concatenated partial sequences of the five housekeeping genes, a result supported by a 100% bootstrap value (Fig. 2). The MLSA evolutionary distances between the isolates ranged from <0.000 to 0.001 (Table 1), that is, well within the species level threshold of ≤0.007 proposed by Rong and Huang (2012, 2014). Members of this well delineated taxon were found to be closely related to the type strains of S. ghanaensis DSM 40746T and S. viridosporus DSM 40243T (Pridham et al. 1958), albeit with MLSA distances well above the species cut-off point (Table 1). These results provide further evidence of the value of MLSA sequence analyses in clarifying the subgeneric relationships of Streptomyces (Guo et al. 2008; Rong and Huang 2010, 2012, 2014; Busarakam et al. 2014; Idris et al. 2017b; Labeda et al. 2014, 2017; Labeda 2016). The S. ghanaensis and S. viridosporus strains formed a well-supported subclade in the 16S rRNA gene tree (Fig. 1) but were not particularly closely related (99.5% sequence similarity, 19 nt differences), results that are clearly more apparent than real given the corresponding MLSA data.
Fig. 2.
Subtree from the Streptomyces phylogenetic tree inferred from concatenated partial sequences of the house-keeping genes atpD, gyrB, recA, rpoB and trpB in IQ-Tree version 1.4.2 (Nguyen et al. 2015) as described by Labeda et al. (2017). Bootstrap values less than 95% were omitted as suggested by the IQ-Tree developers. Bar scale reflects number of substitutions per site
Table 1.
MLSA distances calculated for species phylogenetically near to the proposed new species, Streptomyces asenjonii
| MLSA (Kimura 2-parameter) distance | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
S. daghestanicus
NRRL B-5418T |
– | ||||||||
|
S. griseoviridis
NRRL ISP-5229T |
0.001 | – | |||||||
|
S. fumanus
NRRL B-3898T |
0.034 | 0.034 | – | ||||||
|
S. hirsutus
NRRL B-2713T |
0.060 | 0.060 | 0.062 | – | |||||
|
S. chlorus
NRRL B-24997T |
0.064 | 0.063 | 0.064 | 0.024 | – | ||||
|
S. incanus
NRRL B-59129T |
0.063 | 0.063 | 0.060 | 0.021 | 0.019 | – | |||
|
S. viridis
NRRL B-59133T |
0.063 | 0.063 | 0.062 | 0.016 | 0.021 | 0.016 | – | ||
|
S. pratens
NRRL B-59131T |
0.063 | 0.063 | 0.065 | 0.023 | 0.021 | 0.016 | 0.021 | – | |
|
S. prasinopilosus
NRRL B-2711T |
0.058 | 0.058 | 0.058 | 0.039 | 0.038 | 0.040 | 0.043 | 0.041 | – |
|
S. herbaceous
NRRL B-59128T |
0.060 | 0.060 | 0.058 | 0.029 | 0.032 | 0.026 | 0.031 | 0.031 | 0.027 |
|
S. prasinus
NRRL B-2712T |
0.058 | 0.058 | 0.056 | 0.030 | 0.035 | 0.036 | 0.035 | 0.036 | 0.023 |
|
S. ghanaensis
NRRL B-12104T |
0.052 | 0.051 | 0.052 | 0.052 | 0.056 | 0.057 | 0.054 | 0.060 | 0.059 |
|
S. viridosporus
NRRL ISP-5243T |
0.051 | 0.051 | 0.053 | 0.050 | 0.055 | 0.056 | 0.054 | 0.058 | 0.057 |
|
S. species KNN35.1bT
(NRRL B-65050T) |
0.043 | 0.043 | 0.043 | 0.051 | 0.055 | 0.054 | 0.051 | 0.058 | 0.055 |
|
S. species KNN42.f (NRRL B-65049) |
0.044 | 0.043 | 0.043 | 0.051 | 0.055 | 0.055 | 0.052 | 0.059 | 0.055 |
|
S. species KNN35.2b (NRRL B-65055) |
0.045 | 0.044 | 0.044 | 0.052 | 0.056 | 0.056 | 0.053 | 0.060 | 0.056 |
|
S. species KNN48.3e (NRRL B-65056) |
0.044 | 0.044 | 0.043 | 0.051 | 0.056 | 0.055 | 0.052 | 0.059 | 0.056 |
|
S. species KNN6.11a (NRRL B-65057) |
0.044 | 0.044 | 0.043 | 0.051 | 0.056 | 0.055 | 0.052 | 0.059 | 0.056 |
|
S. species KNN83.e (NRRL B-65058) |
0.044 | 0.044 | 0.043 | 0.051 | 0.056 | 0.055 | 0.052 | 0.059 | 0.056 |
| MLSA (Kimura 2-parameter) distance | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
S. daghestanicus
NRRL B-5418T |
|||||||||
|
S. griseoviridis
NRRL ISP-5229T |
|||||||||
|
S. fumanus
NRRL B-3898T |
|||||||||
|
S. hirsutus
NRRL B-2713T |
|||||||||
|
S. chlorus
NRRL B-24997T |
|||||||||
|
S. incanus
NRRL B-59129T |
|||||||||
|
S. viridis
NRRL B-59133T |
|||||||||
|
S. pratens
NRRL B-59131T |
|||||||||
|
S. prasinopilosus
NRRL B-2711T |
|||||||||
|
S. herbaceous
NRRL B-59128T |
– | ||||||||
|
S. prasinus
NRRL B-2712T |
0.020 | – | |||||||
|
S. ghanaensis
NRRL B-12104T |
0.056 | 0.053 | – | ||||||
|
S. viridosporus
NRRL ISP-5243T |
0.054 | 0.052 | 0.004 | – | |||||
|
S. species KNN35.1bT
(NRRL B-65050T) |
0.054 | 0.051 | 0.036 | 0.037 | – | ||||
|
S. species KNN42.f (NRRL B-65049) |
0.054 | 0.052 | 0.036 | 0.037 | 0.000 | – | |||
|
S. species KNN35.2b (NRRL B-65055) |
0.056 | 0.053 | 0.037 | 0.038 | 0.001 | 0.001 | – | ||
|
S. species KNN48.3e (NRRL B-65056) |
0.056 | 0.052 | 0.036 | 0.038 | 0.000 | 0.001 | 0.000 | – | |
|
S. species KNN6.11a (NRRL B-65057) |
0.056 | 0.052 | 0.036 | 0.038 | 0.000 | 0.001 | 0.001 | 0.001 | – |
|
S. species KNN83.e (NRRL B-65058) |
0.056 | 0.052 | 0.036 | 0.038 | 0.000 | 0.001 | 0.000 | 0.000 | 0.001 |
The isolates were shown to form extensively branched substrate mycelia bearing aerial hyphae, to contain LL-A2pm as the wall diamino acid and exhibited good growth on all of the ISP media, notably on oatmeal and yeast extract-malt extract agar (Table 2). In general, the substrate mycelia were observed to be grey to yellowish white and the aerial spore mass greyish yellow or bright orange yellow, as were the diffusible pigments. Isolates KNN35.1bT and KNN35.2b were seen to form open spirals of hairy ornamented surfaced spores, as shown in Fig. 3. These isolates and S. ghanaensis NRRL B-12104T, their close phylogenetic neighbour, were found to have glucose, mannose, ribose and xylose in whole organism hydrolysates, whilst the S. ghanaensis strain was also found to contain galactose. The polar lipid patterns of these strains showed the presence of diphosphatidylglycerol, glycophospholipid, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol and a number of unidentified components, as shown in Figure S1. The predominant isoprenologs seen in isolates KNN35.1b and KNN35.2bT were identified as MK9 (H6) (~35%), MK9 (H8) (~30%) and MK9 (H4) (~10%). All of these properties are consistent with the classification of the isolates in the genus Streptomyces (Kämpfer 2012; Idris et al. 2017b). Complex mixtures of saturated and branched chain fatty acids were found in the representative isolates and in the type strain of S. ghanaensis (Table 3). The predominant components in all of these organisms were found to be anteiso–C15:0 (11.5–17.8%) and iso–C16:0 (21.3–32.6%); quantitative differences were seen in these and other components while some of the minor fatty acids were discontinuously distributed, as exemplified by the presence of anteiso-C17:1 and C17:1 cis9 in the S. ghanaensis type strain and anteiso-C18:0 amongst the isolates.
Table 2.
Growth and cultural characteristics of all of the isolates on ISP media after incubation for 14 days at 28 °C
| Media | Growth | Substrate mycelium colour | Aerial spore mass colour | Diffusible pigment |
|---|---|---|---|---|
| Glycerol-asparagine agar (ISP 5) | +++ | Dark grey | Dark grey | None |
| Inorganic salts-starch agar (ISP 4) | +++ | Yellowish white | Light yellowish orange | Light yellowish orange |
| Oatmeal agar (ISP 3) | ++++ | Yellowish white | Light yellowish orange | Light yellowish orange |
| Peptone-yeast extract-iron agar (ISP 6) | +++ | Yellowish grey | Olivaceous grey green | Yellowish grey |
| Tryptone-yeast extract agar (ISP1a) | +++ | Yellowish white | Light yellowish orange | Light yellowish orange |
| Tyrosine agar (ISP 7) | +++ | Yellowish white | Light yellowish orange | Light yellowish orange |
| Yeast extract-malt extract agar (ISP 2) | ++++ | White | Dark yellowish orange | Yellowish grey |
++++ abundant growth; +++ very good growth
aISP1 agar medium
Fig. 3.

Scanning electron micrograph of isolate KNN35-1bT showing hairy ornamented spores in open spirals following growth on oatmeal agar at 28 °C for 14 days. Bar 1 µm
Table 3.
Fatty acid profiles (%) of representatives Streptomyces isolates and the type strain of S. ghanaensis
| Fatty acids | Isolate KNN 35.1bT | Isolate KNN 35.2b | Isolate KNN 42.f | S. ghanaensis NRRL B-12104T |
|---|---|---|---|---|
| Iso-C14:0 | 5.3 | 7.6 | 8.1 | 3.7 |
| Anteiso- C15:0 | 17.5 | 15.1 | 17.8 | 11.5 |
| Iso-C15:0 | 11.3 | 7.2 | 8.4 | 5.2 |
| C16:0 | 4.0 | – | 6.7 | 3.0 |
| Iso-C16:0 | 24.2 | 21.3 | 28.6 | 32.6 |
| Iso- H C16:0 | 3.0 | 8.3 | – | – |
| C16:0 | – | 8.8 | – | 3.0 |
| Iso-H C16:1 | – | – | 4.4 | 8.8 |
| Iso-C17:0 | 6.9 | 3.4 | 3.7 | 2.7 |
| Anteiso- C17:0 | 9.9 | 6.4 | 7.8 | 12.3 |
| C17:1 CIS 9 | – | – | – | 1.2 |
| C17:1 ω 8c | 0.9 | – | 0.7 | 1.2 |
| Anteiso- C17:1 ω 9c | 3.4 | 3.3 | 3.7 | 6.6 |
| Anteiso –C17:1 | – | – | – | 6.6 |
| C17:0 | 0.8 | – | 0.9 | 0.4 |
| C17:0 10-methyl | – | – | – | 0.2 |
| C18:0 | 0.3 | 4.2 | 1.3 | – |
| C18:0 ω 9c | – | – | – | – |
| Iso- H C18:1 | 0.9 | – | – | 1.4 |
| Summed feature 3 | 2.0 | 3.3 | 2.8 | 4.0 |
| Summed feature 5 | 6.1 | 1.7 | – | |
| Summed feature 9 | 6.6 | 5.1 | 3.0 | 3.8 |
Trace proportions (<0.9%) are only cited for strains where other fatty acids were found at levels beyond this cut-off point
Summed feature 3, C16:1 ω7cand/orC16:1 ω6c; summed feature 5, iso-C17:1 ω9c and/or C18:2 ω6,9c; summed feature 9, iso-C19 ω8a and/or iso-C17:1 ω9c
Identical results were obtained for nearly all of the duplicated strains included in the phenotypic tests, whilst the exceptions were a few of the carbon source features recorded from the GENIII microplates (Table 4). It can also be seen from Table 4 that the isolates can be distinguished from one another showing that they are not clones. In addition, several properties distinguished all of the isolates from the type strain of S. ghanaensis (Table 4). Thus, only the Atacama isolates produced N-acetyl-β-glucosaminidase, oxidised l-arginine, butyric acid, l-keto-butryric acid, citric acid, d-and l-fucose and d-sorbitol and grew in the presence of 4%, w/v sodium chloride, potassium tellurite and rapamycin SV and at 10 °C. In contrast, only S. ghanaensis NRRL B-12104T oxidised N-acetyl-β-d-mannosamine, N-acetyl-neuraminic acid and d-glucuronic acid. It is also apparent from Table 4 that all of the strains have many phenotypic properties in common.
Table 4.
Phenotypic tests that distinguish the isolates from one another and from Streptomyces ghanaensis NRRL B-12104T
| Characteristic | Isolate KNN 6.11a | Isolate KNN 35.1bT | Isolate KNN 35.2b | Isolate KNN 42.f | Isolate KNN 43.e | Isolate KNN 83.e | S. ghanaensis NRRL B-12104T |
|---|---|---|---|---|---|---|---|
| API ZYM tests | |||||||
| N-Acetyl-β-glucosaminidase | + | + | + | + | + | + | – |
| Esterase (C4) | + | – | – | – | – | + | – |
| α-Glucuronidase | + | – | + | – | – | + | + |
| α-Mannosidase | – | + | – | + | – | – | + |
| GEN III BIOLOG microplate tests | |||||||
| (a) Oxidation of sugars | |||||||
| N-acetyl-D-galactosamine | + | – | – | – | – | + | + |
| N-acetyl-β-d-mannosamine | – | – | – | – | – | – | + |
| N-acetyl-neuraminic acid | – | – | – | – | – | – | + |
| d-Fucose | + | + | + | + | + | + | – |
| l-Fucose | + | + | + | + | + | + | – |
| d-Glucose-6-phosphate | – | + | – | + | – | – | + |
| α-d-Lactose | + | + | + | – | + | – | + |
| d-Mannitol | + | + | + | – | + | – | + |
| β-methyl-d-Glucoside | + | + | – | – | – | – | – |
| d-Salicin | + | – | – | – | – | + | + |
| d-Sorbitol | + | + | + | + | + | + | – |
| (b) Oxidation of amino acids | |||||||
| l-Arginine | + | + | + | + | + | + | – |
| l-Serine | + | + | – | + | – | – | + |
| (c) Oxidation of organic acids | |||||||
| Bromo-succinic acid | + | + | – | – | – | – | – |
| Butyric acid | + | + | + | + | + | + | – |
| α-keto-Butyric acid | + | + | + | + | + | + | – |
| Citric acid | + | + | + | + | + | + | – |
| α-keto-Glutaric acid | + | – | – | + | + | + | + |
| d-Glucuronic acid | – | – | – | – | – | – | + |
| α-hydroxy-Butyric acid | + | – | – | + | – | – | – |
| l-Lactic acid | + | – | – | + | – | – | – |
| l-Malic acid | + | – | + | + | + | + | – |
| Methyl pyruvate | – | – | – | + | – | – | – |
| l-Pyroglutamic acid | + | + | + | + | + | – | – |
| (d) Resistance to inhibitory compounds | |||||||
| Lincomycin | + | – | – | – | – | + | – |
| Potassium tellurite | + | + | + | + | + | + | – |
| Rifamycin SV | + | + | + | + | + | + | – |
| Sodium chloride (4%, w/v) | + | + | + | + | + | + | – |
| Sodium lactate (1%) | + | + | – | + | + | + | – |
| Tetrazolium blue | + | – | – | – | – | + | – |
| Tetrazolium violet | + | – | – | – | – | + | – |
| Troleandomycin | + | – | – | – | – | + | – |
| (e) Growth at | |||||||
| pH 5 | + | + | – | – | + | + | – |
| Degradation test | |||||||
| Casein | + | + | – | – | – | – | – |
| Growth at | |||||||
| 10 °C | + | + | + | + | + | + | – |
| 45 °C | – | – | – | + | – | + | – |
+ positive result; − negative result
Positive results recorded for all of the isolates and the S. ghanaensis type strain:API ZYM tests: acid and alkaline phosphatases, cysteine arylamidase, esterase lipase (C8), β-galactosidase, leucine and valine arylamidasesGEN III BIOLOG microplate tests: utilization of d-alanine, l-glutamic acid, l-histidine, inosine (amino acids), N-acetyl-d-glucosamine (amino-monosaccharide), glycyl-l-proline (dipeptide), acetic acid, acetoacetic acid, γ-amino-l-butyric acid, p-hydroxy-phenylacetic acid, d-malic acid, propionic acid (organic acids), gelatin (polymer), d-cellobiose, dextrin, d-fructose, d-galactose, β-gentiobiose, d-glucose, 3-O-methyl-d-glucose, d-maltose, d-mannose, d-melibiose, sucrose, d-trehalose, d-turanose (sugars), d-galacturonic acid, l-galacturonic acid-Ý-lactone, d-gluconic acid, β-hydroxy-butyric acid (sugar acids), d-arabitol, glycerol, myo-inositol (sugar alcohols), growth at pH6, resistance to aztreonam, guanidine hydrochloride, lincomycin, nalidixic acid, niaproof and growth in the presence of, sodium bromate, and sodium formate (1%, w/v)Other phenotypic tests: aesculin and arbutin hydrolysis, degradation of adenine, elastin, hypoxanthine, starch, l-tyrosine, Tweens 40, 60 and 80 and growth at 20, 30 and 40 °C Negative results recorded for all of the isolates and for the S. ghanaensis type strain:API ZYM tests: α-chymotrypsin, α-fucosidase, α-galactosidase, β-glucosidase, β-glucuronidase, lipase (C14) and naphthol-AS-BI-phosphohydrolaseGEN III BIOLOG microplate tests: utilization of d-aspartic acid, d-serine #1, d-serine #2 (amino acids), d-fructose-6-phosphate, stachyose (sugars), glucuronamide (amine hexose), d-lactic acid methyl ester, mucic acid, quinic acid, d-saccharic acid (organic acids), pectin (polymer) and resistance to fusidic acid and minocyclineOther phenotypic tests: allantoin and urea hydrolysis, nitrate reduction, H2S production, degradation of cellulose, chitin, guanine, tributyrin, uric acid, xanthine, xylan and growth in the presence of sodium chloride (8%, w/v) and at 4 and 50 °C Non-reproducible results recorded for all of the strains:GEN III BIOLOG microplate tests: utilisation of d- raffinose (trisaccharide), l-rhamnose (monosaccharide), l-alanine, l-aspartic acid (amino acids), citric acid, formic acid, α-keto-glutaric acid (organic acids), Tween 40 (surfactant); resistance to vancomycin (antibiotic), lithium chloride (heavy metal) and growth in presence of sodium butyrate (salt)
Isolates KNN35.1bT and KNN35.2b were found to inhibit the growth of the wild type strains of B. subtilis, E. coli, P. fluorescens and S. aureus, whilst isolates KNN6.11a and KNN83.b only inhibited the growth of the B. subtilis strains; in all cases inhibition zones were extensive ranging from 13 to 24 mm.
It can be concluded that the six isolates from the hyper-arid Atacama Desert soils have identical or almost identical 16S rRNA and MLSA gene sequences and share many phenotypic features in common, some of which distinguish them from the type strain of S. ghanaensis, their close phylogenetic neighbour in the Streptomyces MLSA gene tree generated from the five housekeeping genes. It is, therefore, proposed that the isolates be recognised as a new species within the genus Streptomyces, named Streptomyces asenjonii sp. nov. It seems likely that S. asenjonii strains are common in hyper-arid Atacama Desert soils as additional isolates from the Salar de Atacama sampling site show the same aerial/substrate mycelial and diffusible pigment colours as the current isolates when grown on oatmeal agar. Colour groups such as these have been shown to be reliable indicators of Streptomyces species identity (Antony-Babu et al. 2010; Goodfellow and Fiedler 2010).
It is evident from Table 1 that the type strains of S. ghanaensis and S. viridosporus have a low MLSA distance consistent with their assignment to a single genomic species. Indeed, in their extensive MLSA study of type strains of the family Streptomycetaceae, Labeda et al. (2017) noted that S. ghanaensis NRRL B-12104T (also ATCC 14672T) is a later synonym of S. viridosporus NRRL ISP-5243T. This observation was confirmed by determination of the ANIm and ANIb average-nucleotide identity values between draft genome sequences of the type strains of these species using the calculate_ani.py script (https://github.com/widdowquinn/scripts/blog/master/bioinformatics/calculate_ani.py), as shown in Table S1. Note that the ANIm percentages between the genome sequences of S. viridosporus NRRL 2414T, S. viridosporus T7A and S. ghanaensis ATCC 14672T are >96% and the ANIb percentages between these genomes are >97% which is indicative of species level relatedness (Richter and Rosselló-Móra 2009). Thus, according to Rule 38 of the Bacteriological Code of Nomenclature of Bacteria (Lapage et al. 1992; Parker et al. 2015), S. viridosporus Pridham et al. 1958 has priority over S. ghanaensis Wallhäuser et al. 1965. The type strains of these taxa form spiral chains of spiny to hairy spores (Kämpfer 2012), properties known to be predictive in Streptomyces systematics (Labeda et al. 2012) and have many physiological features in common (Kämpfer et al. 1991). Consequently, on the basis of these observations an emended description is given of Streptomyces viridosporus Pridham et al. (1958).
Description of Streptomyces asenjonii sp. nov.
Streptomyces asenjonii (a.sen.jo’ni.i. N.L. gen. n., asenjonii, named after Juan A. Asenjo in recognition of his promotion of work on Atacama Desert actinobacteria).
Aerobic, Gram-positive actinobacteria which form an extensively branched substrate mycelium which carry aerial hyphae that differentiate into open spirals of hairy ornamented spores. Grows from 10 to 50 °C, optimally 37 °C; from pH 5 to 11, optimally 7.5; and in the presence of up to 5% w/v sodium chloride. Produces acid and alkaline phosphatase, cysteine arylamidase, esterase lipase (C8), β-galactosidase, N-acetyl-β-glucosaminidase and leucine and valine arylamidases (API ZYM tests), hydrolyses aesculin and arbutin, degrades adenine, elastin, hypoxanthine, starch, l–tyrosine and Tweens 20, 40, 60 and 80 and is resistant to aztreonam. Additional phenotypic properties are given in Table 4. The cell wall peptidoglycan contains LL-diaminopimelic acid and whole cell hydrolysates contain glucose, mannose, ribose and xylose. The major fatty acid is iso-hexadecanoic acid (iso-C16:0) and the predominant menaquinones are MK9(H6) and MK9(H8). The polar lipid profile contains diphosphatidylglycerol, glycophospholipid, phosphatidylethanolamine, phosphatidylglycerol and phosphatidylinositol.
The type strain KNN 35.1bT (NCIMB 15082T = NRRL B-65050T) and strains KNN 6.11a, KNN 35.2b, KNN 42.f, KNN 48.3e and KNN 83.e were isolated from hyper-arid Atacama Desert soils. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain KNN 35.1bT is LT621750. The Digital Protologue database TaxoNumber for strain KNN 35.1bT is TA00093.
Emended description of Streptomyces viridosporus Pridham, Hesseltine and Benedict 1958, 67AL
Heterotypic synonym: Streptomyces ghanaensis Wallhäusser, Nesemann, Präve and Steigler 1966, 734AL
Most of the data are taken from Kämpfer et al. (1991) and Kämpfer (2012).
Aerobic, Gram-stain positive actinobacteria that form substrate mycelia which bear a green aerial spore mass on glycerol-asparagine, salts-starch, oatmeal and yeast extract-malt extract agars. Short chains of over 10 spores are formed on these media. Spore surfaces are spiny to hairy. Melanoid pigments are not formed on peptone-yeast extract-iron or tyrosine agar or in tryptone-yeast broth. l-arabinose, d-cellobiose, d-fructose, d-galactose, d-glucose, d- glucosamine, glycogen, d-maltose, d-mannitol, d-mannose, starch, d-trehalose, d-xylose, acetate, fumarate, and pyruvate are used as sole carbon sources, but not myo-inositol, d-tagatose or d-turanose.
The source of the type strain NRRL ISP-5243T = NRRL 2414T is not known; ‘S. ghanaensis’ NRRL B-12104 was isolated from soil from Ghana.
The type strain is ATCC 27479 = CBS 654.72 = BCRC (formerly CCRC) 11870 = CCUG 37512 = DSM 40243 = NBRC 13353 = IMET 43514 = JCM 4859 = KCTC 9145 = NCIMB 9824 = NRRL 2414 = NRRL-ISP 5243 = RIA 1314 = VKM Ac-1769 = VKM Ac-618.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
KB and HI are grateful to the Thai and Malaysian Governments, respectively for PhD scholarships, IN for a postdoctoral fellowship from Newcastle University and ATB and MG for Emeritus Fellowships from the Leverhulme Trust. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer. DPL and the ARS Culture Collection CRIS project was supported by ARS National Program 301.
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical statement
This article does not contain any studies with human participants and/or animals performed any of the authors. Formal consent is not required in this study.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-017-0886-7) contains supplementary material, which is available to authorized users.
References
- Antony-Babu S, Stach JEM, Goodfellow M. Computer-assisted numerical analysis of colour-group data for dereplication of streptomycetes for bioprospecting and ecological purposes. Antonie Van Leeuwenhoek. 2010;9(7):231–239. doi: 10.1007/s10482-009-9404-x. [DOI] [PubMed] [Google Scholar]
- Boubetra D, Sabaoua N, Zitouni A, Bijani C, Lebrihi A, Mathieu F. Taxonomy and chemical characterisation of new antibiotics produced by Saccharothrix SA 198 isolated from a Saharan soil. Microbiol Res. 2013;168:223–230. doi: 10.1016/j.micres.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Bull AT, Asenjo JA. Microbiology of hyper-arid environments: recent thoughts from the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 2013;103:1173–1179. doi: 10.1007/s10482-013-9911-7. [DOI] [PubMed] [Google Scholar]
- Bull AT, Asenjo JA, Goodfellow M, Gómez-Silva B. The Atacama Desert: technical resources and the growing importance of novel microbial diversity. Ann Rev Microbiol. 2016;70:215–234. doi: 10.1146/annurev-micro-102215-095236. [DOI] [PubMed] [Google Scholar]
- Busarakam K, Bull AT, Girard G, Labeda DP, van Wezel GP, Goodfellow M. Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie van Leeuwenhoek. 2014;105:849–861. doi: 10.1007/s10482-014-0139-y. [DOI] [PubMed] [Google Scholar]
- Busarakam K, Bull AT, Trujillo ME, Riescu R, Sangal V, van Wezel GP, Goodfellow M. Modestobacter caceserii sp.nov., novel actinobacteria with an insight into their adaptive mechanisms for survival in extreme hyper-arid Atacama Desert soils. Appl Microbiol. 2016;39:243–251. doi: 10.1016/j.syapm.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Busarakam K, Brown R, Bull AT, Tan GYA, Zucchi TD, da Silva LJ, de Souza WR, Goodfellow M. Classification of thermophilic actinobacteria isolated from arid desert soils, including the description of Amycolatopsis deserti sp.nov. Antonie van Leewuenhoek. 2016;109:319–334. doi: 10.1007/s10482-015-0635-8. [DOI] [PubMed] [Google Scholar]
- Collins MD, Goodfellow M, Minnikin DE, Alderson G. Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J Appl Bacteriol. 1985;58:77–86. doi: 10.1111/j.1365-2672.1985.tb01431.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high through put. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed SS, Trusch F, Deng H, Raab A, Prokes I, Busarakam K, Asenjo JA, Andrews BA, van West P, Bull AT, Goodfellow M, Yi Y, Ebel R, Jaspars M, Rateb M. Chaxapeptin, a lasso peptide from extremotolerant Streptomyces leeuwenhoekii strain C58 from the hyperarid Atacama Desert. J Org Chem. 2015;80:10252–10260. doi: 10.1021/acs.joc.5b01878. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fiedler H-P. Screening for bioactivity. In: Bull AT, editor. Microbial diversity and bioprospecting. Washington, DC: American Society of Microbiology; 2004. pp. 324–335. [Google Scholar]
- Fitch WM. Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:400–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. doi: 10.1111/j.1096-0031.2008.00217.x. [DOI] [Google Scholar]
- Goodfellow M, Fiedler HP. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie van Leeuwenhoek. 2010;98:119–142. doi: 10.1007/s10482-010-9460-2. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Teidje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zheng W, Rong X, Huang Y. A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus gene analyses for streptomyces systematics. Int J Syst Evol Microbiol. 2008;58:149–159. doi: 10.1099/ijs.0.65224-0. [DOI] [PubMed] [Google Scholar]
- Guo X, Liu N, Li X, Ding Y, Shang F, Gao Y, Ruan J, Huang Y. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl Environ Microbiol. 2015;81:3086–3103. doi: 10.1128/AEM.03859-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M, Nonomura H. Humic acid vitamin agar: a new medium for the selective isolation of soil actinomycetes. J Ferm Technol. 1987;65:501–509. doi: 10.1016/0385-6380(87)90108-7. [DOI] [Google Scholar]
- Hong K, Gao A, Xie Q, Gao H, Zhuang L, Lin H, Yu H, Li J, Yao X, Goodfellow M, Ruan J. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar Drugs. 2009;7:24–44. doi: 10.3390/md7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris H, Nouioui I, Asenjo JA, Bull AT, Goodfellow M (2017a) Lentzia chajnantorensis sp.nov., a very high altitude actinobacterium isolated from Cerro Chajnantor gravel soil in northern Chile. Antonie van Leeuwenhoek. doi:10.1007/s10482-017-0851-5 [DOI] [PubMed]
- Idris H, Labeda DP, Nouioui I, Castro JF, Montero-Calasanz MC, Bull AT, Asenjo JA, Goodfellow M (2017b) Streptomyces aridus sp.nov., isolated from a high altitude Atacama Desert soil and emended description of Streptomyces noboritoensis Isono et al. 1957. Antonie van Leeuwenhoek 110(5):705–717. doi:10.1007/s10482-017-0838-2 [DOI] [PMC free article] [PubMed]
- Kämpfer P. Genus Streptomyces. In: Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2012. pp. 1455–1767. [Google Scholar]
- Kämpfer P, Kroppenstedt RM, Dott W. A numerical classification of the general Streptomyces and Streptoverticillium using miniaturized physiological tests. J Gen Microbiol. 1991;137:1331–1891. [Google Scholar]
- Kelly KL. Centroid notations for revised ISCC-NBS colour name blocks. J Res Nat Bur Stand USA. 1964;61:472. [Google Scholar]
- Kim SB, Goodfellow M. Streptomyces thermospinisporus sp. nov., a moderately thermophilic carboxydotrophic streptomycete isolated from soil. Int J Syst Evol Microbiol. 2002;52:1225–1228. doi: 10.1099/00207713-52-4-1225. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Küster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- Labeda DP. Taxonomic evaluation of putative Streptomyces scabiei strains held in the ARS Culture Collection (NRRL) using multi-locus sequence analysis. Antonie van Leeuwenhoek. 2016;109:349–356. doi: 10.1007/s10482-015-0637-6. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K. Phylogenetic study of the species within the family Streptomycetaceae. Antonie van Leeuwenhoek. 2012;101:73–104. doi: 10.1007/s10482-011-9656-0. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Doroghazi JR, Ju KS, Metcalf WW. Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int J Syst Evol Microbiol. 2014;64:894–900. doi: 10.1099/ijs.0.058107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Rong X, Huang Y, Doroghazi JR, Ju KS, Metcalf WW. Taxonomic evaluation of species in the Streptomyces hirsutus clade using multi-locus sequence analysis and proposals to reclassify several species in this clade. Int J Syst Evol Microbiol. 2016;66:2444–2450. doi: 10.1099/ijsem.0.001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Dunlap CA, Rong X, Huang Y, Duroghazi JR, Ju KS, Metcaff WW (2017) Phylogenetic relationships in the family Streptomycetaceae using multilocus sequence analysis. Antonie van Leeuwenhoek 10(4):563–583. doi:10.1007/s10482-016-0824-0 [DOI] [PMC free article] [PubMed]
- Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA. International code of nomenclature of bacteria (1990 revision) Washington, DC: ASM Press; 1992. [PubMed] [Google Scholar]
- Lechevalier MP, Lechevalier HA. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol. 1970;20:435–443. [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, Rohde C, Rohde M, Fartmann B, Goodwin LA, Chertkov O, Reddy T, Pati A, Ivanova N, Markowitz V, Kyrpides NC, Woyke T, Göker M. Klenk H-P (2014) Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;10:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meklat A, Sabaou N, Zitouni A, Mathieu F, Lebrihi F. Isolation, taxonomy and antagonistic properies of halophilic actinomycetes in Saharan soils of Algeria. Appl Environ Microbiol. 2011;77:6710–6714. doi: 10.1128/AEM.00326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- Murray PR, Boron EJ, Pfaller MA, Tenover FC, Yolken RH. Manual of clinical microbiology. 7. Washington, DC: ASM Press; 1999. [Google Scholar]
- Nachtigall J, Kulik A, Helaly S, Bull AT, Goodfellow M, Asenjo JA, Maier A, Wiese J, Imhoff JF, Süssmuth RD, Fiedler HP. Atacamycins A-C, 22 membered antitumor macrolide derivatives produced by Streptomyces sp. C38. J Antibiot (Tokyo) 2011;64:775–780. doi: 10.1038/ja.2011.96. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E. Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek. 1993;64:325–340. doi: 10.1007/BF00873091. [DOI] [PubMed] [Google Scholar]
- Okoro CK, Brown R, Jones AL, Andrews BA, Asenjo JA, Goodfellow M, Bull AT. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie van Leeuwenhoek. 2009;95:121–133. doi: 10.1007/s10482-008-9295-2. [DOI] [PubMed] [Google Scholar]
- Okoro CK, Bull AT, Mutreja A, Rong X, Huang Y, Goodfellow M. Lechevalieria atacamensis sp. nov., Lechevalieria deserti sp. nov. and Lechevalieria roselyniae sp. nov., isolated from hyperarid soils. Int J Syst Evol Microbiol. 2010;60:296–300. doi: 10.1099/ijs.0.009985-0. [DOI] [PubMed] [Google Scholar]
- Parker CT, Tindall BJ, Garrity GM (2015) International code of nomenclature of prokaryotes. Prokaryotic code (2008 revision). Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.000778 [DOI] [PubMed]
- Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Pridham TG, Hesseltine CW, Benedict RG. A guide for the classification of streptomycetes according to selected groups, placement of strains in morphological groups. Appl Microbiol. 1958;6:52–79. doi: 10.1128/am.6.1.52-79.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rateb MF, Houssen WE, Arnold M, Abdelrahman MH, Deng H, Harrison WTA, Okoro CY, Asenjo JA, Andrews BA, Ferguson G, Bull AT, Goodfellow M, Ebel R, Jaspars M. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J Nat Prod. 2011;74:1491–1499. doi: 10.1021/np200320u. [DOI] [PubMed] [Google Scholar]
- Rateb MF, Houssen WE, Harrison WTA, Deng H, Okoro CY, Asenjo JA, Andrews BA, Bull AT, Goodfellow M, Ebel R, Jaspars M. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod. 2011;74:1965–1971. doi: 10.1021/np200470u. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131 [DOI] [PMC free article] [PubMed]
- Rong X, Huang Y. Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA:DNA hybridisation with proposal to combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol. 2010;60:696–703. doi: 10.1099/ijs.0.012419-0. [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol. 2012;35:7–18. doi: 10.1016/j.syapm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y. Multi-locus sequence analysis: taking prokaryotic systematics to the next level. Methods Microbiol. 2014;41:221–251. doi: 10.1016/bs.mim.2014.10.001. [DOI] [Google Scholar]
- Santhanam R, Okoro CK, Huang Y, Bull AT, Andrews BA, Asenjo JA, Weon HY, Goodfellow M. Streptomyces deserti sp.nov., isolated from hyper-arid Atacama Desert soil. Antonie van Leeuwenhoek. 2012;101:575–581. doi: 10.1007/s10482-011-9672-0. [DOI] [PubMed] [Google Scholar]
- Santhanam R, Okoro CK, Rong X, Huang Y, Bull AT, Wen HY, Andrews BA, Asenjo JA, Goodfellow M. Streptomyces atacamensis sp. nov., isolated from an extreme hyper-arid soil of the Atacama Desert. Chile. Int J Syst Evol Microbiol. 2012;62:2680–2684. doi: 10.1099/ijs.0.038463-0. [DOI] [PubMed] [Google Scholar]
- Santhanam R, Rong X, Huang Y, Andrews BA, Asenjo JA, Goodfellow (2013) Streptomyces bullii sp.nov., isolated from a hyper-arid Atacama Desert soil. Antonie van Leeuwenhoek 103: 367–373 [DOI] [PubMed]
- Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, MIDI Inc., Newark, DE
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol. 1966;16:313–340. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0 b10. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GYA, Ward AC, Goodfellow M. Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst Appl Microbiol. 2006;29:557–569. doi: 10.1016/j.syapm.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Tiwari K, Gupta RK. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotech. 2012;32:108–132. doi: 10.3109/07388551.2011.562482. [DOI] [PubMed] [Google Scholar]
- Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE. 2012;7:e34846. doi: 10.1371/journal.pone.0034846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaas LAI, Sikorski J, Hofner B, Fiebig A, Buddruhs N, Klenk HP, Göker M. opm: an R package for analysing OmniLog® phenotype microarray data. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt291. [DOI] [PubMed] [Google Scholar]
- Wallhäuser KH, Nesemann G, Prave P, Steigler A. Moenomycin, a new antibiotic 1. Fermentation and isolation. J Antimicrob Agents Chemother. 1965;1996:734–736. [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- Wichner D, Idris H, Houssen WE, McEwan AR, Bull AT, Asenjo JA, Goodfellow M, Jaspars M, Ebel R, Rateb ME (2016) Isolation and anti-HIV-1 integrase activity of lentzeosides A–F from extremotolerant lentzea sp. H45, a strain isolated from a high-altitude Atacama Desert soil. J Antibiot doi:10.1038/ja.2016.78 [DOI] [PubMed]
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed]
- Zakharova OS, Zenova GM, Zvyagintsey DG. Some approaches to the selective isolation of actinomycetes of the genus Actinomadura from soil. Microbiology. 2003;72:110–113. doi: 10.1023/A:1022294526830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.