Abstract
Potential O-acetylation of the sialic acid residues of Escherichia coli K1, groups W-135, Y, and C meningococci, and group B Streptococcus capsular polysaccharides modifies their immunogenicity and susceptibility to glycosidases. Despite the biological importance of O-acetylation, no sialic or polysialic acid O-acetyltransferase has been identified in any system. Here we show that the E. coli K1 O-acetyltransferase encoded by neuO is genetically linked to the endo-neuraminidase tail protein gene of a chromosomal accretion element, designated CUS-3, with homology to lambdoid bacteriophage. Molecular epidemiological analysis established concordance between O-acetyltransferase and CUS-3 in a set of E. coli K1 strains. Deleting neuO eliminated enzymatic activity, which was restored by complementation in trans, and confirmed by 13C-NMR analysis of the acetylated product. Analysis of mutants that accumulate intracellular polysialic acid because of export defects (kpsM and kpsS) or an inability to synthesize the sialic acid precursor, N-acetylmannosamine (neuC), indicated that NeuO does not require constant association with its substrate for activity. DNA sequencing and PCR analysis of neuO from strains that had undergone random capsule form variation showed that slip strand DNA mispairing or unequal recombination resulted in gain or loss of (5′-AAGACTC-3′)n heptanucleotide repeats (where n ≈ 14–39) located in the neuO 5′ region. These repeats code for a previously undescribed structure designated the poly(Ψ) motif. The unexpected discovery of the neuO contingency locus (hypervariable gene controlling expression of a surface epitope) in E. coli, and of a potential phage for redistributing variant neuO alleles, provides a robust system for investigating the functions of localized hypermutability in pathogen evolution.
Keywords: Escherichia coli K1 form variation, polysialic acid capsule acetylation, N-acetylneuraminic acid, lysogenic bacteriophage
With the advent of routine prophylaxis against Gram-positive bacterial infection in pregnant women, Escherichia coli K1 has replaced group B Streptococcus as the leading cause of neonatal sepsis and meningitis.§ In addition to being a predominant neonatal pathogen, E. coli K1 remains the most common cause of female urinary tract infections, a leading cause of childhood and adult bacteremia, and a major etiologic agent of mortality and carcass condemnation in the poultry industry resulting from respiratory infections and septicemia (1). In each of these diseases, the capsular polysialic acid, or K1 antigen, a linear homopolymer of α2,8-linked N-acetylneuraminic acid (the most common sialic acid) residues, plays an essential role in pathogenesis by protecting the invasive bacteria from host innate immunity (2). Molecular mimicry of poly(α2,8) linkages in vertebrates has impeded development of safe and effective anticapsule vaccines. Better understanding of microbial sialic acid metabolism is thus essential for the development of new treatment or prevention strategies.
E. coli K1 strains may also modify their capsules by O-acetylating sialyl units at the carbon-7 or -9 hydroxyl, thereby altering polysialic acid immunogenicity and susceptibility to glycosidases (3). Acetylation randomly form varies resulting in loss or gain of modified polysialic acid after each cell division (3). Although the O-acetyltransferase (O-AcTase) has been shown to act on polysialic acids with >10 sialyl residues and uses acetyl-CoA (Ac-CoA) as the two-carbon donor (4), the genetic basis for acetylation is unknown (3). Indeed, there has been no definitive evidence reported to date linking any putative sialic acid acetylase gene with defined acceptor or donor substrates (4–7). Here we demonstrate that K1-acetylation is catalyzed by an Ac-CoA-dependent O-AcTase, encoded by neuO, which functions as a receptor modifying enzyme as part of a previously undescribed K1-specific lysogenic bacteriophage-like element designated CUS-3. We also demonstrate that form variation involves slipped-strand DNA mispairing or unequal recombination of (5′-AAGACTC-3′)n heptanucleotides in the neuO 5′ region, with loss or gain of other than multiples of n = 3 resulting in translational frame shift and an inactive NeuO. Each set of three heptanucleotides encodes a protein structure designated the poly(Ψ) motif, which could account for the previously observed physical association between O-AcTase and its polysialic acid substrate (4). Our combined results provide discovery of both a contingency locus (8–12) in E. coli and a mobile delivery vehicle for redistributing variant neuO alleles among K1-encapsulated bacteria.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions. The bacterial strains and plasmids used in this study are given in Table 1. Bacteria were propagated from cryopreserved cultures in LB medium (13) with aeration at 37°C, or on plates containing 1.5% agar. Plasmids were maintained by addition of 100 μg/ml ampicillin. For mutant identification or selection, nalidixic acid (Nalr) or kanamycin (Kmr) resistance was selected in the presence of 20 or 50 μg/ml antibiotic, respectively. Plasmid pSX785 and pSX786 neuO inserts were sequenced on both strands by the Core Sequencing Facility of the University of Illinois W. M. Keck Center for Comparative and Functional Genomics by using an ABI 3730XL capillary sequencer and assigned GenBank accession numbers AY779018 and AY779019, respectively. Signature-tagged mutagenesis was carried out by a modification of our published methods (refs. 14 and 15 and unpublished data). Oligonucleotide primers were purchased gel-purified from IDT (Coralville, IA).
Table 1. E. coli strains and plasmids used in this study.
| Strain | Relevant property | Source (ref.) |
|---|---|---|
| Bacterial strains | ||
| BW30270 | Wild type K-12 | CGSC* |
| BOS-12 | K92 | W. Vann (46) |
| DH5α | RecA- K-12 | Laboratory collection |
| EV36 | K-12/K1 Hybrid | Vimr et al. (19) |
| EV291 | Nalr RS218 | Gonzalez et al. (14) |
| EV708 | EV291 ΔneuO1 (Kmr) | This study |
| EV709 | EV291 ΔneuO2 (Kmr) | This study |
| EV710 | EV291 ΔneuO3 (Kmr) | This study |
| EV711 | EV291 kpsM::kan | This study |
| EV712 | EV291 kpsS::kan | This study |
| EV713 | EV291 neuC::kan | This study |
| RS1085 | K1+ parent of EV36 | R. Silver (30) |
| Other “RS” strains | Clinical K1+ isolates | R. Silver (25) |
| 10809 | [O18A]† | DSMZ‡ |
| 10811 | RS164 | DSMZ |
| 10837 | [O18B] | DSMZ |
| 10890 | [O18A1] | DSMZ |
| 10892 | [O18B1] | DSMZ |
| Plasmids | ||
| pKD46 | Red functions | Datsenko and Wanner (17) |
| pKD4 | kan (Km) template | Datsenko and Wanner (17) |
| pGEM-T Easy | PCR cloning vector | Promega |
| pSX785 | 10811-on | This study |
| pSX786 | 10811-off | This study |
| pSX788 | 10811 ΔneuO | This study |
| pSX789 | 10811-off | This study |
CGSC, E. coli Genetic Stock Center (Yale University, New Haven, CT).
Brackets indicate O18 lipopolysaccharide chemotypes A, B, A1, or B1 as defined by Jann et al. (47).
DSMZ, German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany).
Acetylase Assay. Cell extracts were assayed by a modification of the procedure described by Higa and Varki (4). Cells (25 ml) were grown overnight and harvested by centrifugation, resuspended in 0.5 ml Triton X-100 lysis buffer, and stored overnight at 4°C (4). Cell lysis was completed by sonication for 5 s with a Branson Sonifier microtip. The cleared lysate containing the acetylase was prepared by centrifugation for 5 min at 8,000 × g and 4°C. Assay components included 1.25 mM 14C-labeled (4 mCi/mmol) acetyl-CoA purchased from American Radiochemical Corporation (St. Louis), 117 mM cacodylate buffer, pH 7.5, and protein extract in a total volume of 20 μl. Unless indicated otherwise, assays also contained 20 μg of colominic acid (Sigma) as exogenous acceptor. After incubation at 37°C for 30–60 min, assays were stopped by spotting 10-μl samples directly to What-man 3 MM paper for development by descending chromatography in a 95% ethanol/1 M ammonium acetate, pH 7.5 solvent as described (16). Radioactivity remaining at the origin representing acetylated colominic acid or endogenously radiolabeled polysialic acid was quantified by liquid scintillation spectrometry (16). Activities were expressed in units, where 1 unit transferred 1 nmol of O-acetyl groups per mg of protein in 1 h. Protein concentration was estimated by the one-tube dye-binding method (Geno Technology, St. Louis), and verified by using BSA as standard. Control experiments indicated that the assay was proportional to time and protein concentration, and that the product was sensitive to 0.1 M sodium hydroxide treatment at 37°C for 30 min, indicating the synthesis of base-labile O-acetyl esters. Sensitivity of the product to endo-N-acetylneuraminidase (endo-N) digestion demonstrated that O-acetyl groups were transferred to α2,8-linked sialyl residues. Natural abundance 13C-NMR was carried out as described in Supporting Text, which is published as supporting information on the PNAS web site.
Mutagenesis. Deletions of neuO were generated in strain EV291 by Red swap (17) essentially as described (18). This method relies on the overproduction of λ-derived recombination proteins encoded by the temperature-sensitive plasmid pKD46, and PCR amplification of a kan cassette in pKD4 flanked by 5′ and 3′ neuO sequences defined by forward and reverse oligonucleotide primers 5′-GACTGCATGATAGCAAGAGATGTTATT TTGCGTGCATCAcatatgaatatcctccttagttcc-3′ and 5′-GATGT T T TATAT T TAT TGCGTGAGCT TCGCATGATAGCtgtgtaggctggagctgcttc-3′, respectively, where lowercase letters indicate mutagenesis cassette-specific nucleotides. After electroporation and kan selection, the expected neuO deletions in the pKD46-cured EV291 derivatives were verified by diagnostic PCR (17).
Detection and Cloning of neuO. Form variants were identified by stabbing bacteria into LB agar plates containing 10% (vol/vol) horse-46 (H.46) antiserum. Unacetylated polysialic acid reacts strongly to produce antigen-antibody precipitin halos (halo test) surrounding colonies after overnight growth at 37°C and incubation for 2 days at 4°C, whereas acetylation results in a weak or undetectable halo response (3, 19). Selected colonies with undetectable (acetylation-on) or visible (acetylation-off) halos were picked and boiled in water. The extracted DNA was used as template for PCR amplification with neuO forward and reverse primers, 5′-AGCACTAAATGTTTCGTTGGCGTC-3′ and 5′-AATATTGGTAATATGTCTGCATGATG-3′, respectively. Amplicons were cloned directly into pGEM-T Easy PCR cloning vector (Promega), generating plasmid pSX785 (acetylation-on) and the acetylation-off plasmids pSX786 and pSX789. The putative catalytic NeuO domain was generated with neuO forward (5′-ATGATAGCAAGAGATGTTATTTTGCGTG-3′) and reverse (5′-TTGCGTGAGCTTCGCATGATAGC-3′) primers to generate plasmid pSX788, which included nucleotides 8737–9072 of contig 7505 in the unannotated E. coli K1 strain RS218 genomic DNA database (www.genome.wisc.edu/sequencing/rs218.htm). Flanking front (FF) and flanking back (FB) primers for amplification of the neuO heptanucleotide repeat region were 5′-GGTAAAATAACGTAGGATACTAATATG-3′ and 5′-GACCCATTATCATCAACGGAAAAC-3′, respectively, using Z-Taq (Takara Mirus Bio, Madison, WI) and the manufacturer's recommended reaction conditions.
Confirmation that NeuO is a fusion of a catalytic domain and a noncatalytic domain encoded by a tandem heptanucleotide repeat region was obtained by RT-PCR (Promega Access Quick System) using 5′ forward primers F-26 (5′-GTGGTAAAATAACGTAGGATACTAATATG-3′), where the underlined nucleotides represent the Shine–Dalgarno and translational start codon, and F132 (5′-AAGACTCGTTTTCCGTTGATAATAATGG-3′), where underlined nucleotides represent the 3′ most neuO heptanucleotide tandem repeat. A common reverse primer, R630 (5′-CTCCCATACCCAATGACAGATCCG-3′) complementary to the 5′ end of the hexapeptide repeat region (HRR) of the catalytic domain was used to demonstrate domain fusion in the message by agarose gel electrophoresis and ethidium bromide staining of the expected amplicons. Total RNA template was isolated by using RNeasy (Qiagen, Valencia, CA). Reverse transcriptase-negative control reactions were carried out to confirm specific message amplification.
Results and Discussion
Identification of neuO. During ongoing investigation of E. coli K1 systemic disease factors by a signature-tagged mutagenesis approach (14), we noted two ORFs potentially coding for endo-N and carbohydrate O-AcTase in the unannotated genomic DNA sequence database of strain RS218 (www.genome.wisc.edu/sequencing/rs218.htm). All previously identified endo-N structural genes encode the tail proteins of K1-specific lytic bacteriophage (phage) that depolymerize polysialic acid as an obligate step in the infectious process (20). K1 phage depolymerases cleave the internal α2,8-linkages of the K1 antigen or related polysialic acids composed of at least seven sialyl units (20–22), and the strain RS218 endo-N was found to be homologous with all known polysialic acid depolymerases (Fig. 4, which is published as supporting information on the PNAS web site). The weak similarity to the prototypic P22 tail spike endorhamnosidase is consistent with previous evidence identifying endo-N as a phage tail spike protein (20). Many of the ORFs flanking the endo-N gene could encode orthologues of gene products of the lambdoid family that includes HK620 (23), defining a 40,212-bp accretion domain integrated between dsdC and argW that we designated CUS-3 (Table 3 and Fig. 5, which are published as supporting information on the PNAS web site). The genetic organization of CUS-3 suggests it evolved, like other K1-specific phage, to use the polysialic acid capsule as receptor. We conclude that CUS-3, so designated because of its geographic site of discovery (24), is a K1-specific lambdoid phage that lysogenized RS218 before this strain's isolation in 1974 (25).
Genes encoding tail spike proteins with specificity for carbohydrate receptors are often linked to genes encoding receptor modifying enzymes (26). Inspection of the DNA sequences flanking endo-N indicated that the ORF immediately downstream of it could encode a polypeptide with the canonical HRR in O-AcTases of the NodL-LacA-CysE family (27). This family is part of a superfamily of enzymes designated hexapeptide acyltransferases (28) characterized by tandem repeats of the [LIV]-[GAED]-X2-[STAV]-X hexapeptide, where X can be any amino acid and the other residues represented by their standard one-letter abbreviations (27, 29). However, no putative sialic acid O-AcTase, including NeuD that also contains the HRR, has been definitively identified in any system (6). We concluded from its linkage to CUS-3 and homology with other known or putative O-AcTases that neuO may encode the polysialic acid acetylase. This conclusion implied that only strains harboring CUS-3 should express O-AcTase.
Molecular Epidemiology of neuO. If neuO encodes polysialic acid O-AcTase, we expected it to covary with enzymatic activity in a collection of E. coli K1 strains. Assuming that the endo-N gene is a marker of lysogenization by CUS-3, we used specific forward and reverse primers to investigate the presence of endo-N and neuO genes in the strain collection by PCR (Table 4, which is published as supporting information on the PNAS web site). Strains lacking CUS-3, as verified in most cases by PCR amplification of two additional CUS-3-specific markers, invariably lacked acetylase activity. The concordance between these results provided evidence that neuO encodes polysialic acid O-AcTase. Note that although the kps/neu gene cluster in EV36 was derived from an acetylase-positive K1 derivative, RS1085 (30), loss of CUS-3 in the construction of EV36 produces an acetylase-negative phenotype that does not form vary. A similar observation was made by NMR analysis when the K1 gene cluster was cloned into a non-K1 E. coli host (31), formally excluding neuD as the K1 acetylase gene. Therefore, the recent suggestion that sialic acid acetylation in both group B Streptococcus (GBS) and E. coli K1 is carried out by NeuD or its GBS orthologue is unlikely (6). This conclusion is supported by recent studies showing that the phenotype of a neuD mutant is loss of sialic acid synthesis (32, 33).
Characterization of NeuO. To directly demonstrate that neuO is necessary for polysialic acid acetylation, we constructed three independent deletions by Red swap. Table 2 shows undetectable O-AcTase compared to the EV291 parental strain. The loss of acetylase in these mutants suggested that neuO is essential for polysialic acid acetylation, a conclusion consistent with it being the only polysialic acid O-AcTase in E. coli K1. To confirm that the loss of acetylase in these mutants resulted from deletion of neuO, we amplified neuO+ from strain 10811 by PCR and cloned the fragment into pGEM, generating plasmid pSX785. Transformation of pSX785 into the neuO1 mutant restored enzyme activity (Table 2), demonstrating that neuO+ encodes the expected O-AcTase. 13C-NMR analysis confirmed that cloned NeuO produces an O-acetylated polysialic acid product (see Supporting Text and Fig. 6, which is published as supporting information on the PNAS web site). Expression of pSX785 in the naturally kps/neu-negative laboratory E. coli K-12 strain DH5α also resulted in an acetylase-positive extract (Table 2), indicating activity of recombinant NeuO. As expected from the results of Higa and Varki (4), activity in the DH5α transformant was only detected when exogenous acceptor was added to the extract (Table 2). As described below, pSX786 encodes an acetylation-off variant and, as expected, lacked acetylase activity (Table 2). The combination of genetic, biochemical and product characterization results confirms the epidemiological inference that neuO encodes E. coli K1 O-AcTase.
Table 2. O-AcTase specific activities in wild-type and neuO mutant E. coli extracts.
| Relevant properties
|
NeuO activity, units
|
||||
|---|---|---|---|---|---|
| Strain (plasmid) | CUS-3 | neuO | K1 | Without CA | With CA |
| BW30270 | - | - | - | 0.1 | 0.1 |
| EV36 | - | - | + | <0.1 | <0.1 |
| EV291 | + | + | + | 3.2 | 7.5 |
| RS189 | + | + | + | ND | 5.5 |
| RS354 | - | - | + | <0.1 | <0.1 |
| EV708 | + | - | + | 0.1 | 0.1 |
| EV709 | + | - | + | ND | 0.1 |
| EV710 | + | - | + | ND | 0.1 |
| 10811 | + | + | + | 18.9 | 36.4 |
| EV711 | + | + | (+)* | 7.8 | 12.0 |
| EV712 | + | + | (+) | 8.7 | 11.2 |
| EV713 | + | + | - | <0.1 | 7.6 |
| EV708 (pSX785) | + | - | + | ND | 32.5 |
| EV708 (pSX786) | + | - | + | ND | <0.1 |
| DH5α (pSX785) | - | - | - | <0.1 | 48.5 |
| EV36 (pSX788) | - | - | + | ND | <0.1 |
ND, not determined.
Parentheses indicate the production of intracellular K1 antigen.
Previous experiments indicated that O-AcTase binds polysialic acid and may require constant association with it for activity (4). To determine whether this finding was true in vivo, we assayed capsule-negative derivatives of EV291. Mutants that do not export polysialic acid because of kpsM (EV711) or kpsS (EV712) disruptions had high endogenous and exogenous O-AcTase, indicating that the intracellular K1 antigen that accumulates in these strains (34) functions as acceptor (Table 2). However, if NeuO requires constant association with polysialic acid for activity (4), we would expect a neuC mutant (EV713), which does not produce polysialic acid because of the failure to synthesize ManNAc (18), not to acetylate either endogenous or exogenous acceptors. In contrast, an extract of EV713 acetylated exogenous acceptor and, as expected, had no endogenous activity, indicating that NeuO is sufficiently stable in the absence of polysialic acid to detect its activity after cell disruption (Table 2). To demonstrate that NeuO modifies polysialic acid in vivo, pSX785 was transformed into EV36 and halo-tested on H.46 agar, demonstrating that this plasmid conferred an acetylation-on phenotype (Fig. 7, which is published as supporting information on the PNAS web site).
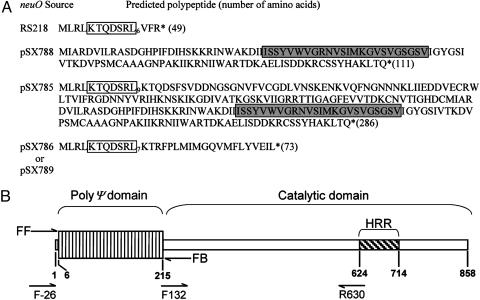
Mechanism of K1 Form Variation. The ATG codon beginning at nucleotide 8289 of the RS218 genomic contig-7505 is preceded by a correctly spaced (7-bp) ribosome-binding site (AGGA). The sequence ATGTT, where underlined nucleotides indicate the start codon, is followed by 19 tandem repeats of the AAGACTC heptanucleotide before a stop codon at nucleotides 8436–8438. This neuO allele is predicted to encode an inactive 49-aa polypeptide lacking the signature HRR (Fig. 1A). When a neuO amplicon including the putative catalytic domain was cloned from an alternative ATG (nucleotides 8737–9072) to generate plasmid pSX788, the predicted 111-aa polypeptide (Fig. 1 A) produced no detectable acetylase activity (Table 2). Therefore, because repetitive DNA is associated with DNA mispairing (35), we hypothesized that the sequence in the RS218 database (nucleotides 8289–9072) is an acetylation-off variant that resulted from deletions or insertions of the repetitive heptanucleotide sequence. This hypothesis was supported by sequencing the active neuO insert in pSX785, demonstrating that the ORF beginning at nucleotide 8289 in RS218 could code for a polypeptide of 286 aa, including the complete catalytic domain and nine repeats of the KTQDSRL heptad encoded by 30 heptanucleotide repeats (Fig. 1B). We propose the designation poly(Ψ) for this heptad motif, and that neuO is a fusion between poly(Ψ) and catalytic domains confirmed by RT-PCR using the primers shown in Fig. 1B (results not shown).
Fig. 1.
Variants of neuO gene products in E. coli K1 strains RS218 and 10811. (A) The complete double-stranded DNA sequences of the neuO inserts in all plasmids were determined and the derived O-acetyltransferases compared to that derived from strain RS218 (www.genome.wisc.edu/sequencing/rs218.htm). Subscripts (open boxes) indicate the number of repetitive poly(Ψ) motifs in each predicted NeuO derivative. Shaded boxes indicate the HRR. Asterisks indicate stop codons. (B) Schematic representation of the neuO allele in pSX785. The neuO ORF is numbered from the beginning of the ATG start codon to the 3′ end of the gene. The poly(Ψ) domain is indicated by the large, vertically sectored box where each sector represents one heptanucleotide tandem repeat. FF and FB refer to flanking forward and flanking back primers used by PCR analysis of poly(Ψ) domain length in natural E. coli K1 isolates. Primers F-26, F132, and R630 were used for RT-PCR analysis of neuO transcripts isolated from EV291-derived colonies 3 or 5 as described in the text.
The occurrence of multiple heptanucleotide repeats also explains the frequency of neuO form variation observed in strain 10811 (Fig. 8, which is published as supporting information on the PNAS web site), as indicated by sequencing two acetylation-off variants (pSX786 and pSX789 derived from colonies 1 and 2, respectively). Each clone was found to contain identical deletions resulting in 23 heptanucleotide repeats. As expected, the predicted 73-aa polypeptide encoded by these plasmids lacked acetylase activity (Table 2). Thus, loss or gain of heptanucleotide repeats is predicted to cause frame shifts resulting in an inactive, truncated acetylase.
To demonstrate that form variation involves frame shifting of chromosomal neuO, we passaged colony 1 from strain 10811 nonselectively on LB agar. Single colonies were halo-tested on H-46 agar resulting in an acetylation-on variant with 24 repeats. When this variant was repassaged and halo-tested, an acetylation-off variant with 23 repeats was reisolated. Therefore, after the original isolation of an inactive variant with 23 repeats (colony 1), form variation in the on and off directions resulted in strains with the addition or loss of one repeat that either restored or disrupted the neuO reading frame, respectively. This stereotyped genotypic progression has been observed in at least one Haemophilus influenzae contingency locus (36), leading us to investigate the extent of neuO variation among different CUS-3-infected E. coli K1 strains.
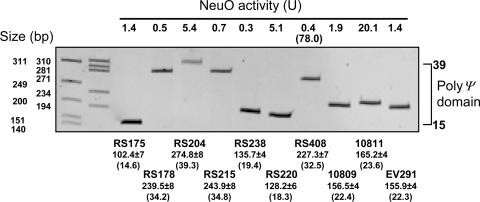
Allelic Variation of E. coli K1 neuO. Variation in neuO heptanucleotide repeat length was determined by using FF and FB PCR primers (Fig. 1B). As shown in Fig. 2, repeat length varied from ≈14 to 39 tandem heptanucleotides. The same colonies that were used for PCR were simultaneously halo-tested and inoculated into LB for enzyme assay. Although in most cases it was only possible to assign a repeat length of ± 1 heptanucleotide (Fig. 2), neuO in bacteria with activities between 0.3 and 1.9 units was likely to be predominantly in the off-form, consistent with halos detected in all such isolates. This hypothesis was confirmed by isolating a halo-negative (on) form variant of RS408 that differed from the off-form shown in Fig. 2 by the addition of one repeat, and by quantifying its O-AcTase, which exceeded the threshold for detection of an on-form variant by halo test (Fig. 2).
Fig. 2.
Variation in neuO tandem heptanucleotide repeat length. Samples from single colonies of the indicated strains were inoculated into LB and halo-tested, or into water and boiled to release DNA. PCR was carried out on samples of boiled extracts by using FF and FB primers and 10% polyacrylamide gel electrophoresis followed by staining with ethidium bromide. The sizes (in bp) of the repeat regions were calculated by reference to Promega ΦX174 HinfI and HaeIII size markers at far left and inner left, respectively, after subtracting 5′ and 3′ nucleotides contributed by FF and FB ± the standard deviation (n = 3). Band size was analyzed by using a Kodak ImageStation 440CF and 1D IMAGE ANALYSIS software. The estimated lengths of the respective poly(Ψ) domains are given under the strain designations in parenthesis and ranged from ≈14 to 39, as indicated by the bracket at right. The NeuO activity given in parentheses is the activity of an acetylation-on variant of strain RS408, with an estimated repeat length of 33.
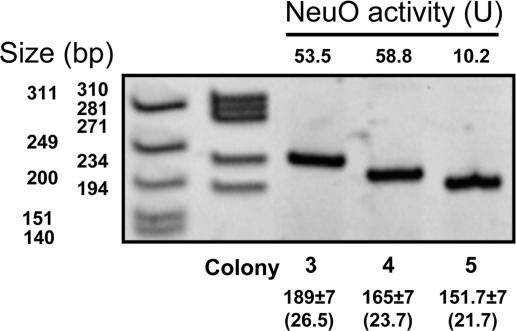
Surprisingly, the 10811 colony tested in Fig. 2, which was in the on-form based on activity, produced a halo, suggesting that in addition to being either on or off, acetylase activity varies between different on-forms. To determine whether there was a correlation between poly(Ψ) motif length and enzyme activity, we isolated two on-form variants (colonies 3 and 4) of strain EV291 by halo test (Fig. 9, which is published as supporting information on the PNAS web site). Acetylase in colony 5 displaying the off, or halo-positive, phenotype had only ≈20% the activity of colonies 3 or 4, with 27 or 24 heptanucleotide repeats, respectively (Fig. 3). The activities of colonies 3 and 4 were comparable to the 10811 (Table 2) and RS408 acetylation-on variants (Fig. 2), indicating that a poly(Ψ) domain of >21 tandem repeats is necessary for a NeuO phenotype conferring >20 units and an unambiguous halo test. When taken together, the results in Figs. 2 and 3 indicate that widespread variation in neuO repeat length exists in a natural E. coli K1 population, and that variation in activity among the on-forms may account for the previously observed range in the degree of acetylation among natural K1 isolates (3). By comparison with the H. influenzae lic contingency locus (36), our results imply that neuO variation has biological relevance for at least altering mutation rates. We predict that the ability of E. coli K1 to generate multiple neuO alleles is of selective advantage during an as yet unknown phase of the host–pathogen interaction, perhaps including a mechanism where modulating the degree of acetylation by varying the size of the poly(Ψ) domain affects the host–pathogen interaction.
Fig. 3.
Correlation between the poly(Ψ) domain and acetylase activity. Strain EV291 was streaked nonselectively on LB agar and 53 colonies were halo-tested; two variants (colonies 3 and 4) were in the on-form. Colonies 3 and 4 and one off-form variant chosen at random (colony 5) were analyzed as described in Fig. 2. Estimated sizes of the poly(Ψ) domains in these colonies were 27, 24, and 21, respectively.
Conclusions and Future Directions
Contingency loci is the designation given to genes controlling reversible switching of surface antigens in pathogenic microorganisms by a mechanism involving slip-strand DNA mispairing of iterative nucleotide repeats and subsequent frame shifting in structural genes or rearrangements of transcriptional control regions (11, 12). These loci are common in obligate symbionts such as H. influenzae, but have not been previously described in E. coli (36, 37). Our characterization of neuO and the mechanism of capsule form variation represents the first description of a contingency, or functional microsatellite (simple repeat sequence) locus in an invasive E. coli pathogen, involving one of the longest perfect repeats detected in any bacterial species. The suggestion (36) that the mechanism causing or repairing mutations in microsatellites differs between E. coli and phase-variable bacteria no longer seems tenable.
In contrast to contingency loci, which by definition affect only a subset of genes, global mutators represent ≈1% of natural E. coli isolates and increase the mutation rates of entire genomes (38). Field et al. (39) have discussed the permutations of local (contingency) and global mutators to speculate on the evolution of host–pathogen interactions. Although known examples of the possibly synergistic effect of combining contingency loci with global mutators have not been described, this combination could account for the evolution of new virulent strains, emerging pathogens, or even new species (39). Our results provide a potential mechanism for generating transient association of a contingency locus upon CUS-3 lysogenization of a hypothetical natural mutator followed by disseminating expanded neuO alleles after phage induction and reinfection of susceptible hosts. Although polysialic acid acetylation does not appear to be required for human disease (40) and form variation was not observed in the septic neonatal mouse model without antibody selection (41), acetylated K1 strains were associated with increased severity of sepsis in adult patients (40). Now that neuO has been identified and defined mutants isolated, it should be possible to determine the precise role of acetylation in disease. The ability to control capsule acetylation under in vitro and in vivo conditions also may have implications for altering polysialic acid structure, which in addition to changing epitopes could influence interactions between cell membranes and metabolism of polysialic acid (42).
All members of the NodL-LacA-CysE family and the broader superfamily of hexapeptide acyltransferases appear to be homotrimers (28). Therefore, it is not unreasonable to assume that three NeuO poly(Ψ) domains might form polar coiled-coils (43), with the arginine residues free to interact with polyanionic substrate (Fig. 10, which is published as supporting information on the PNAS web site). Although this structure is purely speculative, it is unlikely that the poly(Ψ) domain would exist in a disorganized state. Despite there being little evidence for a functional role of repetitive motifs (other than as a mechanism for hypermutability at the genetic level) in the products of most contingency loci, the (LA)n = 2–4 motif in the ATP lid of E. coli MutL may determine the activity of this enzyme's ATP-binding pocket (44). Therefore, iterations of the mutL 5′-CTGGCG-3′ tandem hexanucleotide repeat provide a way to generate reversible hypervariation in the activity of a global mutator as part of another potential mechanism for transiently combining local and global generators of diversity. Given the evidence for direct interaction between the acetylase and oligomeric or polysialic acid (4) and the previously observed variation in the degree of acetylation between E. coli K1 strains (3), our results suggest that the poly(Ψ) motif may affect enzyme stability or substrate recognition. Understanding how NeuO binds its polyanionic substrate may suggest new ways of targeting therapeutic reagents, especially to highly metastatic tumors expressing large amounts of polysialic acid (45).
Supplementary Material
Acknowledgments
This work is dedicated to Dr. Richard P. Silver on the occasion of his retirement. Dr. Silver and his colleagues were the first to clone a multigenic cluster for expression of a polysaccharide virulence factor (the K1 antigen) from any species. He was among the first to document the ubiquity of diverse ATP-hydrolase-coupled export systems in bacteria. He is a trusted colleague who freely shared reagents, strains, and information with all who cared to ask. In large measure, research from the Laboratory of Sialobiolgy has been informed by Dr. Silver`s enthusiasm and support; we thank him and wish him well. We also thank Dr. Willie F. Vann (Center for Biologics Evaluation and Research/Food and Drug Administration, Bethesda) for bacterial strains, reagents, and helpful discussions, and Kerry Helms and Robert Myers for expert graphic and photographic assistance. This work was supported by National Institutes of Health Grant 2R01 AI42015-06.
Author contributions: E.L.D. and E.R.V. designed research; E.L.D., D.I.F., S.M.S., and E.R.V. performed research; E.L.D., S.M.S., and E.R.V. analyzed data; and E.L.D., D.I.F., and E.R.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: O-AcTase, O-acetyltransferase; FF, flanking front; FB, flanking back; HRR, hexapeptide repeat region; endo-N, endo-N-acetylneuraminidase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY779018 and AY779019).
Note Added in Proof. Subcloning of a pSX785-derived neuO deletion lacking 213 base pairs from the 5′ end, predicted to produce NeuO lacking the first 71 amino acid residues (Fig. 1A), and expression of this deletion from a T7 promoter resulted in detectable acetylase activity with exogenous polysialic acid acetylase acceptors, indicating that although the poly(Ψ) domain may modulate O-AcTase function, the motif is not obligatory for enzymatic activity.
Footnotes
Linde-Zwirble, W. T., Angus, D. C., Carcillo, J., Lidicker, J., Clermont, G. & Pinsky, M. R. (1999) Crit. Care Med. 27, Suppl. 1, A33 (abstr.).
References
- 1.Vimr, E. R., Kalivoda, K. A., Deszo, E. L. & Steenbergen, S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vimr, E. & Lichtensteiger, C. (2002) Trends Microbiol. 10, 254–257. [DOI] [PubMed] [Google Scholar]
- 3.Ørskov, F., Ørskov, I., Sutton, A., Schneerson, R., Lin, W., Egan, W., Hoff, G. E. & Robbins, J. B. (1979) J. Exp. Med. 149, 669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higa, H. H. & Varki, A. (1988) J. Biol. Chem. 263, 8872–8878. [PubMed] [Google Scholar]
- 5.Klein, A. & Roussel, P. (1998) Biochimie 80, 49–57. [DOI] [PubMed] [Google Scholar]
- 6.Lewis, A. L., Nizet, V. & Varki, A. (2004) Proc. Natl. Acad. Sci. USA 101, 11123–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claus, H., Borrow, R., Achtman, M., Morelli, G., Kantelberg, C., Longworth, E., Frosch, M. & Vogel, U. (2004) Mol. Microbiol. 51, 227–239. [DOI] [PubMed] [Google Scholar]
- 8.Moxon, E. R., Rainey, P. B., Nowak, M. A. & Lenski, R. E. (1994) Curr. Biol. 4, 24–33. [DOI] [PubMed] [Google Scholar]
- 9.Wren, B. W. (2000) Nat. Rev. Genet. 1, 30–39. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, L. R., Gay, L. S. & Donelson, J. E. (1991) Mol. Biochem. Parasitol. 48, 11–16. [DOI] [PubMed] [Google Scholar]
- 11.Moxon, E. R. & Thaler, D. S. (1997) Nature 387, 659, 661–652. [DOI] [PubMed] [Google Scholar]
- 12.Bridges, B. A. (2001) Philos. Trans. R. Soc. London B 356, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertani, G. (2004) J. Bacteriol. 186, 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, M. D., Lichtensteiger, C. A. & Vimr, E. R. (2001) FEMS Microbiol. Lett. 198, 125–128. [DOI] [PubMed] [Google Scholar]
- 15.Lichtensteiger, C. A. & Vimr, E. R. (2003) Microb. Pathog. 34, 149–154. [DOI] [PubMed] [Google Scholar]
- 16.Steenbergen, S. M., Wrona, T. J. & Vimr, E. R. (1992) J. Bacteriol. 174, 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ringenberg, M. A., Steenbergen, S. M. & Vimr, E. R. (2003) Mol. Microbiol. 50, 961–975. [DOI] [PubMed] [Google Scholar]
- 19.Vimr, E. R., Aaronson, W. & Silver, R. P. (1989) J. Bacteriol. 171, 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petter, J. G. & Vimr, E. R. (1993) J. Bacteriol. 175, 4354–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vimr, E. R., McCoy, R. D., Vollger, H. F., Wilkison, N. C. & Troy, F. A. (1984) Proc. Natl. Acad. Sci. USA 81, 1971–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hallenbeck, P. C., Vimr, E. R., Yu, F., Bassler, B. & Troy, F. A. (1987) J. Biol. Chem. 262, 3553–3561. [PubMed] [Google Scholar]
- 23.Clark, A. J., Inwood, W., Cloutier, T. & Dhillon, T. S. (2001) J. Mol. Biol. 311, 657–679. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez, M. D., Lichtensteiger, C. A., Caughlan, R. & Vimr, E. R. (2002) J. Bacteriol. 184, 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achtman, M., Mercer, A., Kusecek, B., Pohl, A., Heuzenroeder, M., Aaronson, W., Sutton, A. & Silver, R. P. (1983) Infect. Immun. 39, 315–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allison, G. E. & Verma, N. K. (2000) Trends Microbiol. 8, 17–23. [DOI] [PubMed] [Google Scholar]
- 27.Downie, J. A. (1989) Mol. Microbiol. 3, 1649–1651. [DOI] [PubMed] [Google Scholar]
- 28.Vaara, M. (1992) FEMS Microbiol. Lett. 76, 249–254. [DOI] [PubMed] [Google Scholar]
- 29.Bairoch, A. (1993) Nucleic Acids Res. 21, 3097–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vimr, E. R. & Troy, F. A. (1985) J. Bacteriol. 164, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silver, R. P., Finn, C. W., Vann, W. F., Aaronson, W., Schneerson, R., Kretschmer, P. J. & Garon, C. F. (1981) Nature 289, 696–698. [DOI] [PubMed] [Google Scholar]
- 32.Daines, D. A. & Silver, R. P. (2000) J. Bacteriol. 182, 5267–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Annunziato, P. W., Wright, L. F., Vann, W. F. & Silver, R. P. (1995) J. Bacteriol. 177, 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cieslewicz, M. & Vimr, E. (1997) Mol. Microbiol. 26, 237–249. [DOI] [PubMed] [Google Scholar]
- 35.Levinson, G. & Gutman, G. A. (1987) Mol. Biol. Evol. 4, 203–221. [DOI] [PubMed] [Google Scholar]
- 36.De Bolle, X., Bayliss, C. D., Field, D., van de Ven, T., Saunders, N. J., Hood, D. W. & Moxon, E. R. (2000) Mol. Microbiol. 35, 211–222. [DOI] [PubMed] [Google Scholar]
- 37.Karlin, S., Mrazek, J. & Campbell, A. M. (1997) J. Bacteriol. 179, 3899–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeClerc, J. E., Li, B., Payne, W. L. & Cebula, T. A. (1996) Science 274, 1208–1211. [DOI] [PubMed] [Google Scholar]
- 39.Field, D., Magnasco, M. O., Moxon, E. R., Metzgar, D., Tanaka, M. M., Wills, C. & Thaler, D. S. (1999) Ann. N.Y. Acad. Sci. 870, 378–382. [DOI] [PubMed] [Google Scholar]
- 40.Frasa, H., Procee, J., Torensma, R., Verbruggen, A., Algra, A., Rozenberg-Arska, M., Kraaijeveld, K. & Verhoef, J. (1993) J. Clin. Microbiol. 31, 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colino, J. & Outschoorn, I. (1999) Microb. Pathog. 27, 187–196. [DOI] [PubMed] [Google Scholar]
- 42.Cheng, M. C., Lin, C. H., Lin, H. J., Yu, Y. P. & Wu, S. H. (2004) Glycobiology 14, 147–155. [DOI] [PubMed] [Google Scholar]
- 43.Gromiha, M. M. & Parry, D. A. (2004) Biophys. Chem. 111, 95–103. [DOI] [PubMed] [Google Scholar]
- 44.Shaver, A. C. & Sniegowski, P. D. (2003) J. Bacteriol. 185, 6076–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troy, F. A., Jr. (1992) Glycobiology 2, 5–23. [DOI] [PubMed] [Google Scholar]
- 46.Egan, W., Liu, T. Y., Dorow, D., Cohen, J. S., Robbins, J. D., Gotschlich, E. C. & Robbins, J. B. (1977) Biochemistry 16, 3687–3692. [DOI] [PubMed] [Google Scholar]
- 47.Jann, B., Shashkov, A. S., Gupta, D. S. & Jann, K. (1992) Eur. J. Biochem. 210, 241–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.