ABSTRACT
Epigenetic regulation is based on a network of complexes that modulate the chromatin character and structure of the genome to impact gene expression, cell fate, and development. Thus, epigenetic modulators represent novel therapeutic targets used to treat a range of diseases, including malignancies. Infectious pathogens such as herpesviruses are also regulated by cellular epigenetic machinery, and epigenetic therapeutics represent a novel approach used to control infection, persistence, and the resulting recurrent disease. The histone H3K27 methyltransferases EZH2 and EZH1 (EZH2/1) are epigenetic repressors that suppress gene transcription via propagation of repressive H3K27me3-enriched chromatin domains. However, while EZH2/1 are implicated in the repression of herpesviral gene expression, inhibitors of these enzymes suppressed primary herpes simplex virus (HSV) infection in vitro and in vivo. Furthermore, these compounds blocked lytic viral replication following induction of HSV reactivation in latently infected sensory ganglia. Suppression correlated with the induction of multiple inflammatory, stress, and antipathogen pathways, as well as enhanced recruitment of immune cells to in vivo infection sites. Importantly, EZH2/1 inhibitors induced a cellular antiviral state that also suppressed infection with DNA (human cytomegalovirus, adenovirus) and RNA (Zika virus) viruses. Thus, EZH2/1 inhibitors have considerable potential as general antivirals through the activation of cellular antiviral and immune responses.
KEYWORDS: antiviral, chromatin, epigenetics, herpesvirus, immune mechanisms, innate immunity, Zika virus
IMPORTANCE
A significant proportion of the world’s population is infected with herpes simplex virus. Primary infection and subsequent recurrent reactivation can result in diseases ranging from mild lesions to severe ocular or neurological damage. Herpesviruses are subject to epigenetic regulation that modulates viral gene expression, lytic replication, and latency-reactivation cycles. Thus, epigenetic pharmaceuticals have the potential to alter the course of infection and disease. Here, while the histone methyltransferases EZH2/1 are implicated in the suppression of herpesviruses, inhibitors of these repressors unexpectedly suppress viral infection in vitro and in vivo by induction of key components of cellular innate defense pathways. These inhibitors suppress infection by multiple viral pathogens, indicating their potential as broad-spectrum antivirals.
INTRODUCTION
Following primary infection with herpes simplex virus (HSV), the virus establishes lifelong latency in sensory neurons. However, multiple biological or stress stimuli can induce reactivation of latent genomes and recurrent disease. Ocular HSV infections remain the leading virus-mediated cause of stromal keratitis and corneal scarring, while neonatal infections can result in death, developmental delays, or persisting neurological issues (1, 2). In addition to these pathologies, HSV-2 infection is linked to enhanced acquisition and transmission of HIV (3–5).
The most widely utilized antiherpetic pharmaceuticals target the viral DNA polymerase to interfere with late-stage viral replication. However, drug-resistant strains emerge (6, 7), particularly in immunocompromised individuals (8). Furthermore, these compounds do not adequately control subclinical infectious viral shedding, which is the most prevalent means of transmission.
Many DNA viruses, including herpesviruses, are subject to epigenetic regulation where productive infection, persistence, and quiescence/latency are determined, in part, by the modulation of chromatin associated with the viral genomes (9–14). Epigenetic regulation is mediated by families of histone modification enzymes, adaptor recognition proteins, and chromatin remodelers. Thus, components of the cellular epigenetic machinery represent a plethora of novel therapeutic targets that can be used to modulate the expression of specific gene sets and alter the course of disease. Inhibitors of LSD1 and members of the JMJD2 family of histone H3K9 demethylases have been shown to suppress HSV infection and reactivation from latency (15–18). In a contrasting approach, histone deacetylase inhibitors have been components of some strategies to induce HIV reactivation and deplete latent viral reservoirs (19–21).
More recently, inhibitors of the histone H3K27 methyltransferases EZH2 and EZH1 (EZH2/1) have been developed as potential therapeutics for treating cancers with EZH2 gain-of-function mutations (22–30). As components of Polycomb repressive complex 2 (PRC2), EZH2/1 mediate gene repression primarily through propagation of H3K27me3, which results in domains of nucleosomal compaction (31–33). However, these enzymes also modulate genes whose promoters exhibit bivalent histone markers (H3K27me3 and H3K4me3) that enable the transition to either an active or a repressed state during environmental signaling and differentiation (34, 35).
EZH2/1 PRC2 complexes have also been implicated in the regulation of the lytic-latency cycles of multiple members of the herpesvirus family, including HSV (9, 12, 13, 36–43). Upon infection of permissive cells, the HSV genome is assembled into chromatin structures that initially exhibit both H3K9me3 and H3K27me3 repressive histone methylation signatures (10, 18, 40, 44, 45). Initiation of lytic infection requires the recruitment of a cellular transcriptional coactivator complex (HCF-1) that contains histone H3K9 demethylases (LSD1, JMJD2s) and histone H3K4 methyltransferases (SETD1A, MLLs), which limits the accumulation of H3K9me3 and increases the levels of active H3K4me3 to promote the transcription of viral immediate-early (IE) genes (17, 18, 46). Similarly, the levels of H3K27me3 associated with the viral genome decrease over the course of productive infection in a manner dependent on IE protein ICP0 and viral DNA replication (40). Importantly, both the H3K9me3 and H3K27me3 markers have been associated with the establishment and maintenance of HSV latency in sensory neurons (37, 39, 47, 48) Inhibition of either the HCF-1-associated H3K9 demethylases or the UTX/JMJD3 H3K27 demethylases prevents productive viral reactivation (15–18, 49, 50).
Here, in contrast to the anticipated suppressive role of EZH2/1, inhibition of these histone methyltransferases results in reduced HSV gene expression and lytic infection in vitro and in vivo. Additionally, inhibitors did not induce reactivation but rather suppressed the spread of viral reactivation in a ganglion explant model. Investigation of the mechanisms involved in the antiviral activities of these inhibitors revealed that treatment induces multiple components of antipathogen pathways that result in an enhanced cellular antiviral state. Importantly, the antiviral effects are not limited to HSV but also extends to other representative nuclear DNA viruses (human cytomegalovirus [hCMV], adenovirus 5 [Ad5]), as well as an unrelated RNA virus (Zika virus [ZIKV]).
RESULTS
EZH2/1 inhibitors suppress HSV-1 IE gene expression and productive infection.
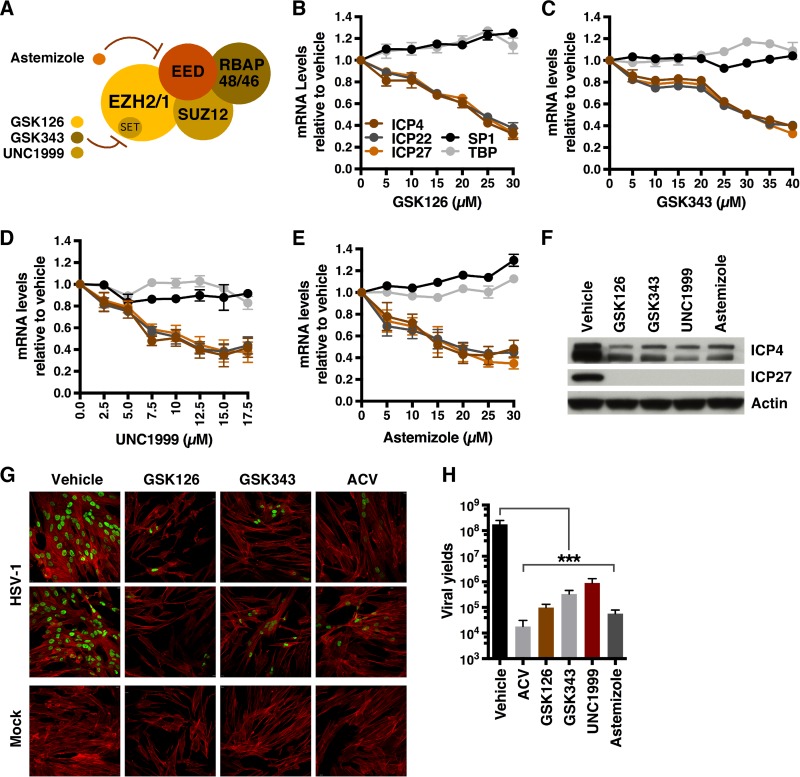
Upon infection, nucleosomes exhibiting heterochromatic methylation signatures (H3K9me3, H3K27me3) are assembled on the viral genome. To investigate the contribution of EZH2/1-catalyzed H3K27 methylation to suppression of viral gene expression during lytic infection, primary human foreskin fibroblast (HFF) cells were pretreated with the EZH2/1 catalytic inhibitors GSK126, GSK343, and UNC1999 (25, 26, 29) for 5 h and infected with HSV-1 (Fig. 1A to D and F). Strikingly, in contrast to the anticipated enhancement of viral gene expression, each compound suppressed viral IE gene transcription in a dose-dependent manner. Similarly, treatment with astemizole, a compound that interferes with the association of EZH2/1 with the chromatin binding subunit of the PRC2 complex (EED, embryonic ectoderm development) (24), also suppressed viral IE gene expression (Fig. 1A and E to F). Thus, suppression of HSV gene expression was dependent on either inhibition of the catalytic activity of EZH2/1 or prevention of the assembly of the PRC2 complex. Pretreatment of cells with GSK126 for 1 h was sufficient to reduce viral IE expression, while recovery of suppressed viral IE gene expression could be rescued within 6 to 12 h following washout of the compounds (see Fig. S1A to C in the supplemental material).
FIG 1 .
Inhibitors targeting EZH2/1 suppress HSV-1 IE gene expression. (A) Schematic of the PRC2 complex containing the EZH2 or EZH1 histone methyltransferase. GSK126, GSK343, and UNC1999 inhibit the catalytic SET domain, while astemizole interferes with the interaction of EED and EZH2/1. (B to E) mRNA levels of viral (ICP4, ICP22, ICP27) and control cellular genes (SP1, TATA box binding protein [TBP]) in HFF cells treated with the vehicle or the concentrations of EZH2/1 inhibitors indicated and infected with HSV-1 (MOI, 2.0) for 1.5 h. Data are means ± SEM of at least two independent experiments. (F) Western blot assay of viral IE (ICP4, ICP27) and cellular (actin) proteins from HFF cells treated with GSK126, GSK343, UNC1999, astemizole, or the vehicle and infected with HSV-1 (MOI, 2.0) for 2 h. (G, H) HFF cells were infected with HSV-1 (MOI, 0.01) for 8 h, and then the vehicle, ACV, GSK126, GSK343, UNC1999, or astemizole was added for 12 h. (G) Cells were stained with anti-UL29 (green) and phalloidin-647 (F-actin, red). (H) Viral yields. Data are means ± SEM of four independent experiments. ***, P < 0.001 (ANOVA and Dunnett’s post hoc test).
EZH2/1 inhibitors do not affect nuclear accumulation of viral genomes and suppress HSV-1 IE expression in an MOI-independent manner. (A) mRNA levels of IE and control cellular genes in HFF cells treated with the vehicle or GSK126 for the times indicated and then infected with HSV-1 (MOI, 2.0) for 1.5 h. Data are means ± SEM of three independent experiments. (B, C) Viral IE and control cellular mRNA levels in HFF cells treated with the vehicle, GSK126, or UNC1999 for 5 h and then subjected to drug washout and infection with HSV-1 (MOI, 2.0) at the times indicated. Data are means ± SEM of three independent experiments. (D) HSV-1 DNA levels in total cells and nuclear fractions of HFF cells treated with the vehicle, GSK126, or GSK343 and infected with HSV-1 for 1.5 h. Data are means ± SEM of three independent experiments. (E) Viral IE (ICP0, ICP4, ICP22, ICP27) and control (SP1) mRNA levels in cells treated with the vehicle or GSK126 and infected at the MOI indicated for 1.5 h. Data are means ± SEM of three independent experiments. Download FIG S1, TIF file, 25.2 MB (25.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine if EZH2/1 inhibitors would suppress viral lytic growth and spread, cells were infected with HSV-1 for 8 h to allow one round of replication and subsequently treated with the vehicle, GSK126, GSK343, or the viral DNA replication inhibitor acyclovir (ACV) for 12 h. As shown by immunofluorescent staining for the viral DNA replication protein UL29/ICP8, treatment with each compound potently suppressed productive viral spread from the initial infected cells (Fig. 1G) and decreased viral yields by 2 to 3 logs (Fig. 1H).
EZH2/1 inhibitors reduce the number of transcriptionally active viral genomes.
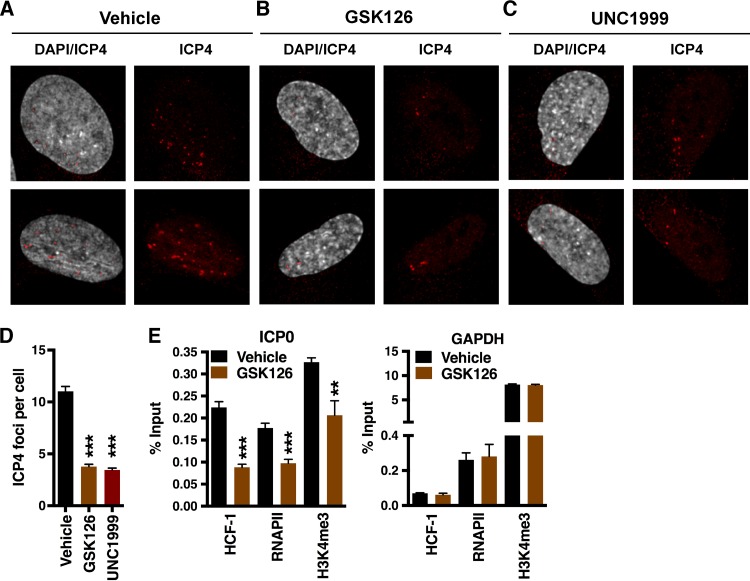
Suppression of HSV-1 IE gene expression was not due to a block in the transport of viral genomes to the nucleus and was relatively independent of the multiplicity of infection (MOI) (Fig. S1D and E). However, the suppression of viral IE gene expression suggested that EZH2/1 inhibitors acted at an early stage of infection. Therefore, cells were treated with the vehicle, GSK126, or UNC1999; infected with HSV-1 for 1.5 h; and stained for a marker of active HSV-1 transcriptional foci (ICP4). As shown in Fig. 2A to D, treatment with either drug reduced the number of transcriptionally active viral genomes per cell. Suppression of IE transcription was further supported by chromatin immunoprecipitation (ChIP) assays, which demonstrated a reduction in the levels of the IE coactivator HCF-1, RNA polymerase II (RNAPII), and the euchromatic marker H3K4me3 associated with the viral IE-ICP0 promoter (Fig. 2E). In contrast, no significant change in occupancy of the control cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter was evident. Therefore, in stark contrast to the anticipated effect, inhibition of EZH2/1 resulted in reduced IE gene expression, decreased numbers of transcriptionally accessible viral genomes, and suppression of lytic infection.
FIG 2 .
EZH2/1 inhibitors reduce the number of transcriptionally active viral genomes. (A to C) Viral transcriptional foci in HFF cells treated with the vehicle, GSK126, or UNC1999 and infected with HSV-1 (MOI, 5.0) for 1.5 h. Cells were stained with anti-ICP4 (viral transcriptional foci, red) and 4',6-diamidino-2-phenylindole (DAPI, gray). (D) The number of transcriptional foci per cell. Data are means ± SEM of ≥110 cells/group. ***, P < 0.001 (ANOVA and Dunnett’s post hoc test). (E) ChIP assays showing HCF-1, RNAPII, and histone H3K4me3 levels associated with the viral IE (ICP0) and control cellular (GAPDH) promoters in HFF cells treated with the vehicle or GSK126 for 5 h and infected with HSV-1 (MOI, 2.0) for 1.5 h. Data are means ± SEM of three independent experiments. **, P < 0.01; ***, P < 0.001 (unpaired two-tailed t test).
EZH2/1 inhibitors induce a cellular antiviral state.
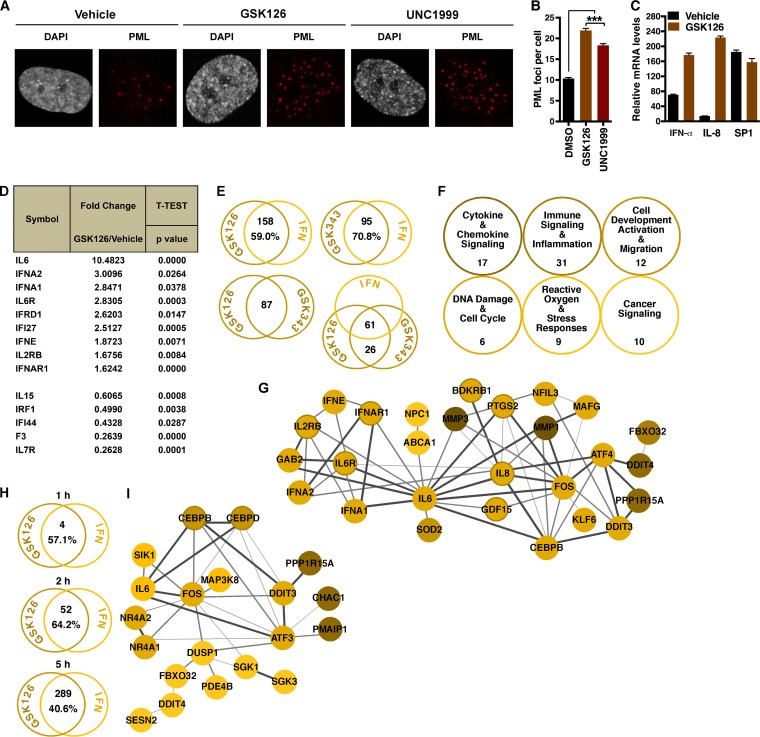
These data suggested that EZH2/1 inhibitors might induce an antiviral state that results in suppression of the viral genome. In support of this, the number of promyelocytic leukemia (PML) foci, a marker of stress- and pathogen-induced cellular responses (51), was higher in GSK126- and UNC1999-treated cells than in vehicle-treated cells (Fig. 3A and B). Furthermore, interferon-alpha (IFN-α) and interleukin-8 (IL-8) mRNAs were significantly upregulated in GSK126-treated cells (Fig. 3C), indicating that EZH2/1 inhibitors might mediate viral repression via IFN/immune signaling-related pathways. To directly address this, the mRNA levels of a selection of IFNs/IFN receptors were measured in GSK126-treated cells with a quantitative PCR (qPCR) Profiler array (Fig. 3D; Table S1A). Compared to vehicle, treatment of cells with GSK126 enhanced the levels of nine genes (>1.5-fold; P < 0.05), including those encoding IL-6, the IL-6 receptor, IFN-α1, and IFN-α2.
FIG 3 .
Inhibition of EZH2/1 leads to induction of innate immunity. (A, B) HFF cells were treated with the vehicle, GSK126, or UNC1999 for 5 h. (A) Cells were stained with anti-PML antibody (red) and DAPI (gray). (B) The number of PML foci per cell. Data are means ± SEM of >391 cells counted per group. ***, P < 0.001 (ANOVA and Dunnett’s post hoc test). (C) IFN-α, IL-8, and control SP1 mRNA levels in cells treated with the vehicle or GSK126 for 5 h. Data are means ± SEM of five independent experiments. (D) Differential expression (≥1.5-fold) of human IFNs and IFN receptors in HFF cells treated with the vehicle or GSK126 for 4 h, as determined with a Profiler PCR array (n = 3). (E to G) HFF cells were treated with the vehicle, GSK126, or GSK343 for 4 h, and differential expression was assessed by microarray analyses (n = 3). (E) The number of ISGs (Interferome database) that are differentially regulated (≥2-fold) by GSK126 or GSK343. Genes differentially expressed (≥2-fold) in both GSK126- and GSK343-treated cells are shown. The number of ISGs that are differentially regulated (≥2-fold) by GSK126 and GSK343 is shown. (F) Supergroups of related overrepresented IPA pathways in GSK126-treated cells. (G) STRING network of genes induced in qPCR arrays (≥1.5-fold) and microarrays (≥2-fold). Confidence levels of inferred functional associations between protein nodes (circles) are indicated by the thickness of the connecting lines (edges). (H, I) HFF cells were treated with the vehicle or GSK126 for 1, 2, or 5 h, and differential expression was assessed by RNA-Seq (n = 3). (H) The number of ISGs (Interferome database) that are differentially regulated (≥2-fold) by GSK126. (I) STRING network of genes induced (≥2-fold) by GSK126 at 2 h.
IFN qPCR array, microarray, and RNA-Seq data sets. Download TABLE S1, XLSX file, 0.9 MB (925.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Global differentials in gene expression were next assessed by microarray analyses of HFF cells treated for 4 h with GSK126 or GSK343 (Table S1B). Among the most significantly induced transcripts in both GSK126- and GSK343-treated cells were those encoding transcription factors, regulatory proteins, and components of immune signaling and cellular stress response (oxidative and unfolded protein response [UPR]) pathways. Strikingly, ISGs (IFN-stimulated genes) were clearly overrepresented in the genes modulated by GSK126 or GSK343 (GSK126, 158 [59.0%] of 268 genes; GSK343, 95 [70.9%] of 134 genes), as indicated by overlap with the Interferome database (52) (Fig. 3E; Table S1B). Furthermore, of the 87 genes differentially regulated by both inhibitors, 61 (71.1%) were ISGs. Ingenuity pathway analyses (IPAs) of the differentially regulated genes (Fig. S2A and B; Table S1B) supports the induction of proinflammatory and stress response pathways in both GSK126- and GSK343-treated cells.
Enrichment of innate signaling pathways with GSK126, GSK343, and EED226. (A, B) IPA top-ranked pathways for genes differentially regulated (≥1.5-fold) in HFF cells treated with GSK126 or GSK343 for 4 h (microarrays, n = 3). (C, D) Karpas422 cells were treated with the vehicle or EED226 (EED inhibitor) for 24, 48, 72, or 144 h. Microarray CEL files were obtained from GEO under accession no. GSE81267. (C) The number of ISGs (Interferome database) differentially regulated (≥2-fold) by EED226. (D) IPA top-ranked pathways for genes differentially regulated (≥2-fold) by EED226. The 72-h IPA results were used as the reference data set with a P value of ≤0.05. (E) IPA top-ranked pathways enriched for genes differentially regulated (≥2-fold) in HFF cells by 1, 2, or 5 h of GSK126 treatment (RNA-Seq, n = 3). Download FIG S2, TIF file, 78.4 MB (80.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
As shown in Fig. 3F and Table S1B, enriched pathways identified by IPA were grouped into six supercategories that illustrate the modulation of immune signaling and cellular antiviral pathways in inhibitor-treated cells. A subset of the genes induced in the qPCR arrays and microarrays could be further organized into a functional network. In this network, IFNs (IFNA1/2, IFNAR1), inflammatory cytokines and mediators (IL-6, IL-8, PTGS2), and transcription factors that induce the expression of inflammatory and endoplasmic reticulum (ER) stress response factors (CEBPB, Fos, ATF4, DDIT3) are nodes with functional relationships (STRING [53]) that illustrate the modulation of a broad spectrum of antipathogen pathways by EZH2/1 inhibitors (Fig. 3G). These results are consistent with previous observations in cancer cell lines that depletion of SUZ12, a subunit of the EZH2/1-PRC2 complex, resulted in the expression of a class of ISGs whose promoters exhibited bivalent (H3K27me3 and H3K4me3) histone methylation markers (54). The results are also complemented by analyses of data sets (GEO data set GSE81267) published by Qi et al. (28) where treatment of Karpas422 cancer cells with a novel inhibitor targeting the PRC2 subunit EED (EED226) induced antipathogen gene cascades that parallel the inflammatory and stress pathways shown here (Fig. S2C and D; Table S1C). Thus, inhibition of EZH2/1 results in the differential regulation of ISGs and components of cellular defense pathways in a manner dependent on EZH2/1 catalytic activity and PRC2 complex integrity.
To define the kinetics, transcriptome sequencing (RNA-Seq) analyses were performed with cells treated with GSK126 for 1, 2, or 5 h (Table S1D). Strikingly, the results support the induction of antipathogen response pathways by 2 h, with significant enhancement of these pathways by 5 h (Fig. 3H; Fig. S2E and Table S1D). Components induced by 2 h formed a functional network enriched in signaling molecules (IL-6, mitogen-activated protein kinase kinase kinase 8 [MAP3K8], SGK1/3) and transcription factors (Fos, CEBPB/CEBPD, DDIT3, NR4A1/2, ATF3) that are regulators of antiviral and stress response pathways (Fig. 3I). Importantly, although EZH2/1 inhibitors induced IFNs and ISGs, GSK126 treatment of cells lacking the type 1 IFN receptor (IFNAR1−/−) still prevented the spread of HSV infection (Fig. S3). Thus, suppression is mediated in a manner that does not require IFNAR1 signaling. The induction of transcription factors and signaling molecules by EZH2/1 inhibitors suggests that these compounds induce critical components of antiviral pathways that are downstream of direct type I IFN receptor signaling.
GSK126 suppression of HSV infection is independent of IFNAR1 signaling. Cultures of fibroblasts from wild-type control (C57BL/6J) and IFNAR1−/− (TAC73) mice were infected with HSV-1 (MOI, 0.01) for 8 h, and then the vehicle, ACV, or GSK126 was added for 12 h. Cells were stained with anti-UL29 antibody (green), phalloidin-647 (F-actin, red), and DAPI (blue). Download FIG S3, TIF file, 9 MB (9.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EZH2/1 inhibition suppresses the infection of multiple viral pathogens.
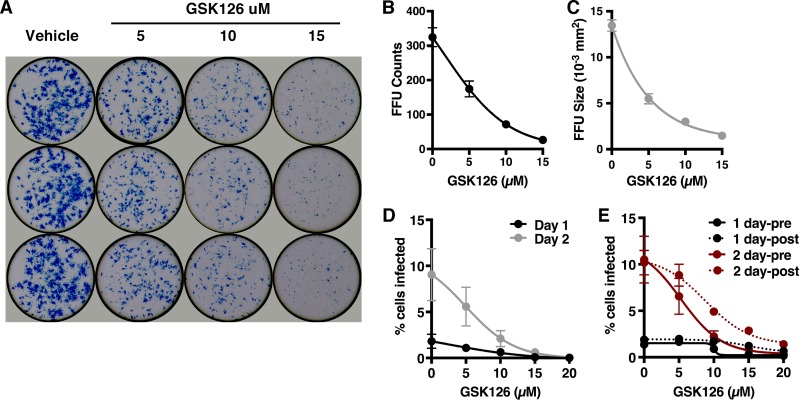
On the basis of the regulation of multiple antiviral components and pathways by EZH2/1 inhibitors, it was hypothesized that these compounds would function as broad-spectrum antiviral inhibitors. Indeed, GSK126 and GSK343 both suppressed the IE gene expression of two other nuclear DNA viruses, Ad5 and hCMV (Fig. S4). To determine if these inhibitors would suppress an RNA virus with a distinct life cycle, HFF cells were treated with GSK126 and infected with ZIKV, a member of the Flavivirus family. As shown in Fig. 4A to C, GSK126 significantly reduced both the number and size of ZIKV focus-forming units (FFU; plaques) in a dose-dependent manner. These results were further supported by a GSK126-meditated reduction in the number of ZIKV-infected cells at 1 and 2 days postinfection (dpi), as measured by intracellular staining for ZIKV antigens (Fig. 4D). Finally, while pretreatment was modestly more efficient at suppression of infection at lower GSK126 concentrations, it was clearly not essential to effect significant suppression (Fig. 4E).
FIG 4 .
EZH2/1 inhibitors suppress hCMV, Ad5, and ZIKV infections. (A to E) HFF cells were treated with the concentrations of GSK126 indicated for 5 h and infected with ZIKV for 40 h (A) Cells were stained with pan-Flavivirus MAb E60 (blue). (B, C) FFU counts and sizes. (D) The percentage of cells infected at days 1 and 2 and treated with the concentrations of GSK126 indicated. (E) Cells were treated either preadsorption (pre) or postadsorption (post), and the percentage of cells infected was determined at days 1 and 2. The data in panels B to E are means ± SEM of at least two independent experiments.
EZH2/1 inhibitors suppress hCMV and Ad5 infections. (A, B) HFF cells were treated with the vehicle, GSK126, or GSK343 and then infected with Ad5 (MOI, 200) for 3 h or with hCMV (MOI, 0.05) for 2 h. Levels of viral IE (Ad5, E1A; hCMV, UL37, UL123, UL44) and cellular control (SP1, TATA box binding protein [TBP]) mRNAs are expressed relative to those in cells treated with the vehicle. Data are means ± SEM of at least two (A) or four (B) independent experiments. Download FIG S4, TIF file, 9.4 MB (9.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EZH2/1 inhibitors suppress primary HSV infection and enhance the recruitment of immune cells to the site of infection in vivo.
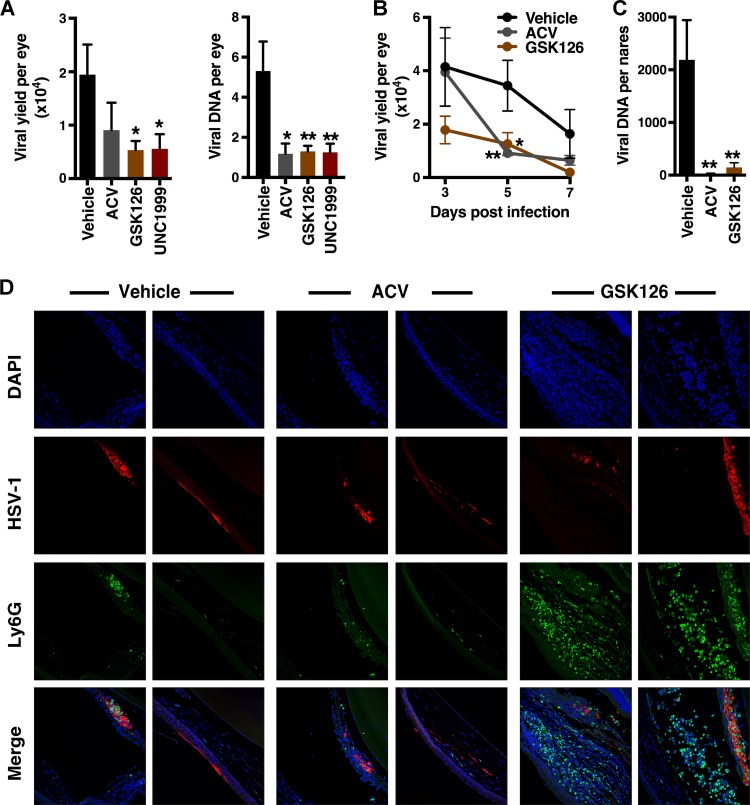
Given the suppression of HSV-1 gene expression and lytic infection in culture, as well as the regulation of antiviral signaling, the impact of EZH2/1 inhibitors on HSV-1 ocular and intranasal infections was assessed in vivo. BALB/c mice were infected via the ocular route and treated topically with the vehicle, the viral DNA replication inhibitor ACV, or EZH2/1 inhibitors for 7 dpi. As shown in Fig. 5A, GSK126 or UNC1999 treatment significantly reduced the progression of infection, as measured by ocular viral yields (infectious virus titers) and viral DNA loads. Suppression of ocular infection by topical GSK126 treatment was evident as early as 3 dpi, with a significant decrease in the infectious virus titer per eye at 5 dpi (Fig. 5B). In mice infected via the intranasal route, intraperitoneal (i.p.) injection of GSK126 also significantly reduced viral loads in nasal tissues (Fig. 5C).
FIG 5 .
Inhibitors of EZH2/1 suppress primary HSV-1 infection and enhance immune cell recruitment in vivo. (A, B) Mice infected with HSV-1 (2 × 105 PFU/eye) were treated topically with the vehicle, ACV, GSK126, or UNC1999 for the times indicated. (A) HSV-1 yields and DNA loads from eyes at 7 dpi (≥12 eyes/group). (B) Viral yields from eyes on the postinfection days indicated (22 eyes/group). (C) Viral DNA yields from nasal tissues of mice infected intranasally with HSV-1 (5 × 105PFU) and treated i.p. with the vehicle, ACV, or GSK126 for 7 days (13 nares/group). Panels A to C: *, P < 0.05; **, P < 0.01 (ANOVA and Dunnett’s post hoc test). (D) Mice were infected with HSV-1 (2 × 105 PFU/eye) and treated topically with the vehicle, ACV, or GSK126 for 7 days. Eye sections were costained with anti-HSV-1 antibody (red), anti-Ly6G (neutrophil) antibody (green), and DAPI (blue).
EZH2/1 inhibitor treatment resulted in the induction of antiviral signaling pathways in vitro and the suppression of primary infection in vivo. To characterize drug-induced changes in cellular immunity, neutrophil recruitment to the site of HSV infection was assessed. Neutrophils are typically the first immune cells to migrate into infected tissues, and neutropenic individuals suffer from recurrent HSV infections. Mice were mock infected or infected with HSV-1 via the ocular route and treated topically with GSK126, ACV, or the vehicle for 7 days. Ocular sections were costained to detect HSV-1-infected cells and neutrophils. Strikingly, while neutrophils were modestly recruited in the vehicle- or ACV-treated mice, treatment with GSK126 resulted in a clear increase in these cells within the cornea, iris, and anterior chamber of the infected eyes (Fig. 5D; Fig. S5). Importantly, enhanced neutrophil recruitment was dependent on viral infection and was not evident in mock-infected mice.
GSK126 enhances immune cell recruitment to sites of viral infection in vivo. (A) Immunofluorescent staining of eyes mock infected or infected with HSV-1 (2 × 105/eye) and treated topically with the vehicle, ACV, or GSK126 for 7 days. Eye sections were costained with anti-HSV-1 (red) and anti-Ly6G (neutrophil) (green) antibodies and DAPI (blue) (11 eyes/group). (B) Eye sections were scored for HSV-1 infection and neutrophil recruitment (Ly6G positive, Gr1, Fig. 5D) at 3, 5, and 7 dpi. Data are means ± SEM of ≥5 eyes/group. Download FIG S5, TIF file, 41.1 MB (42.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reduction in HSV-1 reactivation-spread correlates with stimulation of immune signaling pathways in mouse ganglia.
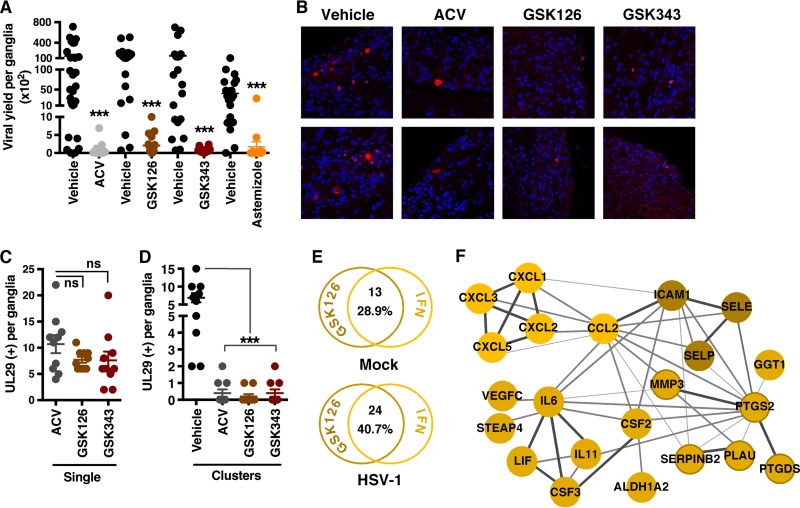
Explantation of latently HSV-1-infected sensory ganglia into culture promotes induction of viral reactivation. To determine if EZH2/1 inhibitors would suppress reactivation, ganglia from latently infected mice were explanted in the presence of the vehicle, ACV, or EZH2/1 inhibitors for 48 h. GSK126, GSK343, and astemizole each reduced viral yields, as measured by infectious titers and viral DNA loads (Fig. 6A; Fig. S6A).
FIG 6 .
EZH2/1 inhibitors suppress HSV-1 during reactivation from latency and stimulate immune signaling pathways in a mouse ganglion explant model. Trigeminal ganglia from latently infected mice were explanted in the presence of the vehicle, ACV, GSK126, GSK343, or astemizole for 48 h. (A) Viral yields per ganglion. Data means ± SEM of ≥19 ganglia/group. ***, P < 0.001 (Wilcoxon matched-pair signed-rank test). (B) Ganglion sections were costained with anti-UL29 antibody (red) and DAPI (blue) and scored for individual UL29+ neurons (single) (C) and clusters of UL29+ cells (D). The data in panels C and D are means ± SEM of 10 ganglia/group. ***, P < 0.001; ns, not significant (ANOVA and Dunnett’s post hoc test). (E to F) RNA-Seq analyses of mock-infected and latently infected ganglia explanted in the presence of the vehicle or GSK126 for 12 h (three pools of five ganglia per group). (E) The number of ISGs (Interferome database) differentially regulated (≥2-fold) by GSK126. (F) STRING network of genes induced (≥2-fold) in HSV-infected ganglia by GSK126. The confidence levels of inferred functional associations between protein nodes (circles) are indicated by the thickness of the connecting lines (edges).
EZH2/1 inhibitors reduce viral yields in a mouse ganglion explant model. (A) Trigeminal ganglia from latently infected mice were bisected, and halves were explanted in the presence of the vehicle (dimethyl sulfoxide [DMSO]) or the compound indicted for 48 h. The yields of viral DNA per ganglion were normalized by the levels of cellular DNA (GAPDH). Data are DNA yields of individual ganglia with means ± SEM of ≥19 ganglia/group. ***, P < 0.001 (Wilcoxon matched-pair signed-rank test). (B) Trigeminal ganglia from mock-infected and latently HSV-1-infected mice were explanted in the presence of the vehicle (DMSO) or GSK126 for 12 h. Levels of cellular mRNAs (murine IL-6 [mIL-6], mCXCL1, and mSP1) are normalized by the level of control TATA box binding protein (TBP) mRNA. Data are means ± SEM of two pools of five ganglia per group. (C) IPA top-ranked pathways enriched for genes differentially regulated (≥2-fold) by GSK126 treatment of explanted ganglia for 12 h (RNA-Seq). Download FIG S6, TIF file, 41.8 MB (42.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reduced viral yields can result from suppression of the initial primary reactivation events and/or suppression of the spread of reactivated virus in the ganglia. To discriminate between these, latently infected ganglia were explanted in the presence of the vehicle, ACV, GSK126, or GSK343 for 48 h and sections were stained for the viral lytic replication protein UL29 (ICP8) (Fig. 6B to D). Ganglia explanted in the presence of the viral DNA replication inhibitor ACV served as comparative controls, as this compound does not suppress the primary reactivation events (single UL29+ cells) but blocks viral replication and spread from the initiating neuron (clusters of UL29+ cells). As shown in Fig. 6C and D, slightly lower numbers of individual/single UL29+ neurons (primary reactivation events) were seen in the presence of GSK126 (7.7/ganglion) and GSK343 (7.6/ganglion) than in the presence of ACV (10.7/ganglion). However, the primary impact of these compounds was clearly in reducing the spread of the infection from the initiating neurons to adjacent cells (clusters) in a manner comparable to that of ACV.
As shown during lytic infection of cultured HFF cells, suppression of viral reactivation-spread in ganglion explants was likely to be due to EZH2/1 inhibitor modulation of antiviral signaling. This was confirmed by RNA-Seq analyses of genes differentially regulated in mock-infected and HSV-1-infected ganglia explanted for 12 h in the presence of GSK126 relative to the vehicle (Table S1E). In mock-infected ganglia, GSK126 treatment induced 13 ISGs, including that for CXCL1, while 24 ISGs, including those for CXCL1-3, CXCL5, CSF2-3, CCL2, LIF, IL-11, and IL-6, were induced in HSV-1-infected ganglia (Fig. S6B; Table S1E). Thus, analogous to the enhanced recruitment of neutrophils by GSK126 in HSV-infected eyes relative to that in mock-infected eyes (Fig. 5D; Fig. S6), the stimulation of these antiviral pathways is also enhanced in HSV-infected ganglia compared to that in mock-infected ganglia.
Pathway, Interferome, and STRING analyses clearly illustrate the induction of proinflammatory and immune cell recruitment pathways by GSK126 (Fig. 6E and F; Fig. S6C and Table S1E) in explanted ganglia. While induction of IL-6, IL-8 (CXCL1/2), IL-11, PTGS2, MMP3, and LIF was common to both the HFF and ganglion RNA-Seq data sets (Table S1D and E), a more extensive set of chemokines (CXCL5, CCL2, CXCL3), adhesion molecules (SELP, ICAMI, SELE), and factors required for hematopoiesis and maturation (CSF2, CSF3) was induced in the ganglion tissue. It is likely that this differential stems from the more diverse cell population, including resident immune cells, in the infected ganglia in vivo relative to the clonal nature of the HFF cultures. Furthermore, the induction of the neutrophil chemotactic factors CXCL1, CXCL2, CXCL3, and CXCL5 is consistent with the robust recruitment of these cells to the sites of ocular HSV-1 infection (Fig. 5D; Fig. S5).
DISCUSSION
Small-molecule compounds targeting components of the epigenetic machinery hold significant promise as novel approaches to control diseases ranging from malignancies and developmental disorders to acute or chronic viral infections. Recently, inhibitors of the histone H3K27 methyltransferases EZH2/1 have been developed on the basis of the potential to treat some diffuse large B-cell, follicular, and non-Hodgkin’s lymphomas (23, 55–57). Importantly, pharmacological inhibition of EZH2/1 has shown promising results in tumor regression in cancer cell lines, mouse tumor xenograft models, and human phase I trials (7, 23, 25–30, 58).
Many viruses, including herpesviruses, are subject to regulation by the cellular epigenetic machinery. EZH2/1 complexes have been linked to the control of lytic and latency stages of multiple herpesviruses, including HSV, Kaposi’s sarcoma-associated herpesvirus, Epstein-Barr virus, and hCMV, where these complexes and the associated H3K27me3 marks repress viral lytic gene expression. For HSV, both the H3K9me3 and H3K27me3 heterochromatic markers are associated with the genome upon lytic infection and likely represent an initial host cell antiviral response. Similarly, in sensory neurons, H3K9me3 and H3K27me3 marks and PRC complex subunits are associated with the latent HSV-1 genome and have been hypothesized to play a role in the establishment and/or maintenance of latency.
Specific epigenetic inhibitors can modulate HSV lytic infection, latency, and reactivation. Inhibitors of the histone H3K9 demethylases (LSD1, JMJD2) block initiation of HSV infection, while inhibitors of the H3K9 and H3K27 demethylases (UTX/JMJD3) block reactivation from latency. Most strikingly, inhibition of LSD1 in vivo enhances epigenetic repression of the latent HSV genomes, which correlates with a reduction of reactivation and viral shedding (15). Here, treatment of primary cells with EZH2/1 inhibitors suppressed HSV gene expression, decreased the spread of the infection to adjacent cells, and blocked the spread of viral reactivation in latently infected sensory ganglia. In vivo, EZH2/1 inhibitors suppressed primary HSV infection and enhanced immune cell infiltration at the site of infection. Thus, despite the fact that the PRC2 complex plays roles in the suppression of HSV lytic gene expression and the promotion of latency, the data presented here support a role for EZH2/1 inhibitors in the induction of an antiviral state that is clearly dominant over the loss of any direct PRC2 suppression of the HSV genome.
Treatment of primary cultured cells with EZH2/1 inhibitors induced the expression of regulators and components of pathways encompassing inflammation, IFN signaling, oxidative stress, and the ER-UPR. These included IFNs and IFN receptors (i.e., IFNA1/2, IFNE, IFNAR1), transcription factors (i.e., Fos, JunD, NF-κB [NF-κB1, NFKBIZ], NR4A1 to -3, CEBPB, CEBPD, ATF3) that modulate the expression of immune and inflammatory components, and kinases/signaling molecules (i.e., MAP3K8, IRAK2, IER3, SGK1/3) that amplify the host cell response to infection. Ultimately, the induction of cytokines/chemokines (i.e., IL-6, IL-8 [CXCL1/2], CSF2, CSF3, IL-11, CXCL3, CXCL5, CCL2) correlates with clearance of infection in vivo.
Collectively, the genes induced by these inhibitors compose a multifaceted antiviral state where 40.6 to 70.9% of the differentially regulated genes are ISGs. Furthermore, in addition to HSV, EZH2/1 inhibitors suppressed infection with other nuclear DNA viruses (hCMV, adenovirus), as well as ZIKV, a member of the Flavivirus family. Thus, EZH2/1 complexes are clearly critical modulators of cellular antipathogen defense pathways. It is, however, important to note that in addition to those factors involved in the stimulation of immune or antipathogen responses, EZH2/1 inhibitor treatment also resulted in the induction of genes that are involved in feedback pathways that limit excessive stress and inflammatory signaling through dampening of receptor signaling (i.e., SOCS2, IL-1RN, DUSP1), transcriptional repression (i.e., ATF3, MAFG), destabilization of AU-containing mRNAs encoding inflammatory proteins (i.e., ZFP36), and processing of reactive oxygen species (i.e., OXR1).
Many viruses have evolved mechanisms to circumvent cellular antiviral strategies and IFN responses. However, the pathogens tested here do not efficiently escape the multiple ISGs and antiviral pathways induced by EZH2/1 inhibitors. Furthermore, while the EZH2/1-PRC2 complex plays roles in the repression of herpesviruses, the impacts of EZH2/1 inhibitors in vivo are more complex and include the induced recruitment of antiviral immune cell populations. Importantly, the impacts of EZH2/1 inhibitors in vivo were clearly enhanced in infected tissues and animals relative to those in uninfected controls. Thus, these inhibitors amplify cellular antiviral responses and contribute to the induction of immune cell responses in a context-dependent manner. The results are analogous to the synergistic/enhanced induction of ISGs in cells treated with both EZH2/1 inhibitors and IFN-γ, lipopolysaccharide, IL-1β, or tumor necrosis factor alpha relative to treatment with each compound alone (54).
Immune surveillance in latently infected sensory ganglia contributes to the suppression of HSV reactivation (59–63). Resident T cells and delivery of IFNs to neurons undergoing viral reactivation repress lytic induction while also preserving neuronal survival. In explanted ganglia, EZH2/1 inhibitors suppressed productive reactivation and spread of HSV from the initiating neurons. Thus, while speculative, it is possible that immune suppression responses could be enhanced by compounds such as EZH2/1 inhibitors, resulting in a decrease in productive clinical reactivation and viral shedding.
Strikingly, although PRC2 components repress herpesvirus gene expression, the induction of antiviral pathways by EZH2/1 inhibitors clearly drives immune-mediated suppression of viral infection. Thus, in addition to their antioncogenic potential, EZH2/1 inhibitors may have a role as general antivirals for short-term enhancement of viral clearance, for suppression of persistent infections and drug-resistant mutants, or as enhancers of immune responses where no specific pharmaceutical antiviral is available. PRC2 epigenetic inhibitors currently in clinical trial for cancer therapy may provide an initial therapeutic resource for new or emerging pathogens (e.g., ZIKV).
MATERIALS AND METHODS
Cells and viral infections.
Telomerase reverse transcriptase-immortalized HFF, MRC-5, and Vero cells were maintained in accordance with standard procedures. Fibroblast cultures were derived from TAC73 (IFNAR1−/−) and control C57BL/6J mice. HSV-1 (F and 17), hCMV (Towne), and Ad5 infections were done in Dulbecco’s modified Eagle’s medium (DMEM) containing 1% fetal bovine serum (FBS) for 1 h at 37°C. ZIKV (H/PF/2013) infections were done in Opti-MEM containing 2% FBS for 4 h at 37°C.
Inhibitor treatment.
Cells were pretreated with GSK126 (30 µM), GSK343 (35 µM), UNC1999 (15 µM), astemizole (30 µM), ACV (100 µM), or the vehicle for 4 to 5 h unless specified otherwise in the appropriate figure legend. Viral adsorption was done in the absence of an inhibitor but included postadsorption for the times indicated. The inhibitors used and their sources are listed in Text S1.
Supplemental materials and methods used in this study. Download TEXT S1, PDF file, 0.1 MB (149.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral yields.
Viral DNA levels in genomic DNA from HFF cells and mouse tissues were determined by qPCR, and samples were normalized on the basis of the levels of the cellular GAPDH gene. The sequences of the primer sets used are listed in Text S1. Viral titers were determined from homogenates of mouse tissues or HFF cells.
Reverse transcription-qPCR and qPCR.
cDNA was synthesized from total RNA (Isolate II RNA minikit; Bioline) with a qScript cDNA synthesis kit (Quanta Biosciences) or a Maxima First Strand cDNA synthesis kit (Thermo Scientific). For ganglia, tissues were first homogenized in TriPure Reagent (Roche catalog no. 11667165001) with Lysing Matrix D (MP Biomedicals catalog no. 6913-100) and a FastPrep-24 Instrument (MP Biomedicals). DNAs or cDNAs were quantitated in triplicate by qPCR with SYBR green master mix (Roche) and a Mastercycler ep realplex4 (Eppendorf; Realplex 2.2v software) or a QuantStudio 3 (Applied Biosystems; QuantStudio 1.4v software). The sequences of the primer sets used are listed in Text S1.
qPCR arrays, microarrays, and RNA-Seq.
Total RNAs from mouse ganglia or HFF cells were isolated with an Isolate II RNA minikit (Bioline) as noted above. RNA quality (an RNA integrity number of >8.5) was verified by Bioanalyzer for all qPCR array, microarray, and RNA-Seq samples. Differentials in the expression of genes for IFNs and IFN receptors were determined with RT2 Profiler PCR Arrays (Qiagen catalog no. PAHS-064ZA) (three independent experiments). Microarrays were completed with Illumina Human HT-12 v4.0 Expression BeadChip as detailed in Text S1. RNA-Seq analyses (HiSeq2000/2500 system; Illumina) of libraries (TruSeq Stranded Total RNA with Ribo-Zero; Illumina) from three independent replicates were done as described in Text S1. Statistical analyses of microarray and RNA-Seq data are described in Text S1.
Western blot assays.
Western blot assays of resolved nuclear protein extracts were done with the antibodies listed in Text S1. Blots were visualized with WesternBright Quantum (Advansta) and quantitated with a G:BOX Chemi XT4 (Syngene; GeneTools 4.03.02.0v software).
ChIP assays.
ChIP assays were done as described in detail in Text S1.
Animals and infections. (i) Trigeminal ganglion explants.
BALB/c mice were infected with 5 × 105 PFU of HSV-1 (strain F) via the ocular route as previously described (64). Latently infected trigeminal ganglia were harvested 30 to 45 days after clearance of the primary infection. Trigeminal ganglia were bisected, and paired halves were explanted in medium with the vehicle and inhibitors for 48 h.
(ii) Primary ocular and intranasal HSV infections.
BALB/c mice were infected with 2 × 105 PFU of HSV-1 (strain F)/eye. The vehicle, ACV (30 to 100 µM), and EZH2/1 inhibitors (GSK126, 30 to 60 µM; UNC1999, 15 µM) were applied topically to the surface of the eye twice daily beginning at 12 h postinfection (hpi). Eyes were harvested on the days postinfection indicated to obtain viral yields and tissue sections. Mice were infected intranasally with 5 × 105 PFU of HSV-1 (strain F) and treated i.p. with the vehicle, ACV (50 mg/kg) or GSK126 (50 mg/kg) beginning 1 day prior to infection. Viral yields from nasal tissue were determined at 7 dpi. Animal care and handling were done in accordance with the National Institutes of Health Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee.
Immunofluorescence microscopy.
Immunofluorescence staining with the antibodies listed in Text S1 was done in accordance with standard protocols. Cells were visualized with a Leica TCS SP5 confocal microscope with LASAF software (version 2.6.0). Deconvolution was completed with Huygens Essential (Scientific Volume Imaging, 15.10.1P6v), and sequential z sections were assembled with Imaris software (Bitplane AG, 8.1.2v).
Ganglion and ocular immunohistochemical staining.
Latently HSV-1-infected trigeminal ganglia were explanted for 48 h into DMEM--10% FBS in the presence or absence of inhibitors. Explanted ganglia or primary infected eyes were fixed in 4% paraformaldehyde (PFA)--phosphate-buffered saline (PBS) and embedded in paraffin. Sections were stained with the antibodies listed in Text S1 as previously described (64). Sequential z-stack images were collected with a Leica TCS SP5 confocal microscope.
ZIKV FFU assay.
Following treatment with the vehicle or EZH2/1 inhibitors, HFF cells were infected with ZIKV (100 FFU/2 × 104 cells) and overlaid with 1% methylcellulose in Opti-MEM containing 2% FBS. Cells were fixed at 40 hpi with 1% PFA--PBS and stained with pan-Flavivirus monoclonal antibody (MAb) E60. Infected foci were quantitated and scored by size with an ImmunoSpot Macroanalyzer (Cellular Technologies, ImmunoCapture v.6.5.7).
ZIKV intracellular staining assay.
Vehicle- or EZH2/1 inhibitor-treated HFF cells were infected with ZIKV (30,000 FFU/2.2 × 105 cells) for 40 h. Cells were fixed in Fix/Perm Solution (BD Biosciences) and stained with anti-ZIKV antibody ZV67. The percentage of infected cells was determined by flow cytometry.
Statistical analyses.
Data are presented as means ± the standard error of the mean (SEM). Where appropriate, statistical analyses (Prism 7.0) included analysis of variance (ANOVA) with Dunnett’s post hoc test for multiple comparisons, two-tailed t tests for individual comparisons, and Wilcoxon matched-pair signed-rank tests for comparisons of paired vehicle- and compound-treated ganglia. Details are presented in Table S2.
Statistical analyses. Download TABLE S2, XLSX file, 0.03 MB (34.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data availability.
Microarray and RNA-Seq data sets are available at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE99841.
ACKNOWLEDGMENTS
We thank J. Vogel, R. Alfonso, and M. O’Donoghue Altman (Laboratory of Viral Diseases, NIAID) for discussions and review of the manuscript and NIAID Bld33 Vivarium staff and A. Reed (Laboratory of Viral Diseases, NIAID) for excellent technical support.
This study was supported by the Intramural Research Program of the NIH, NIAID (T.M.K., T.C.P., J.W.Y.).
The NIAID, NIH, has the following patent applications: preventing or treating viral infections by inhibition of the histone methyltransferase EZH1 or EZH2 (T.M.K. and J.H.A.), U.S. patent application 62/155,704, and international patent application PCT/US2016/030089.
Footnotes
Citation Arbuckle JH, Gardina PJ, Gordon DN, Hickman HD, Yewdell JW, Pierson TC, Myers TG, Kristie TM. 2017. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. mBio 8:e01141-17. https://doi.org/10.1128/mBio.01141-17.
REFERENCES
- 1.Whitley RJ, Kimberlin DW, Prober CG. 2007. Pathogenesis and disease, p 589–601. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, New York, NY. [PubMed] [Google Scholar]
- 2.Farooq AV, Shukla D. 2012. Herpes simplex epithelial and stromal keratitis: an epidemiologic update. Surv Ophthalmol 57:448–462. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston C, Corey L. 2016. Current concepts for genital herpes simplex virus infection: diagnostics and pathogenesis of genital tract shedding. Clin Microbiol Rev 29:149–161. doi: 10.1128/CMR.00043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. 2006. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS 20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Hladik F, Woodward A, Klock A, Peng T, Johnston C, Remington M, Magaret A, Koelle DM, Wald A, Corey L. 2009. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med 15:886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Velzen M, van de Vijver DA, van Loenen FB, Osterhaus AD, Remeijer L, Verjans GM. 2013. Acyclovir prophylaxis predisposes to antiviral-resistant recurrent herpetic keratitis. J Infect Dis 208:1359–1365. doi: 10.1093/infdis/jit350. [DOI] [PubMed] [Google Scholar]
- 7.Piret J, Boivin G. 2011. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 55:459–472. doi: 10.1128/AAC.00615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin MJ, Bacon TH, Leary JJ. 2004. Resistance of herpes simplex virus infections to nucleoside analogues in HIV-infected patients. Clin Infect Dis 39(Suppl 5):S248–S257. doi: 10.1086/422364. [DOI] [PubMed] [Google Scholar]
- 9.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 11.Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ, Kristie TM. 2013. Snapshots: chromatin control of viral infection. Virology 435:141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman PM. 2016. Epigenetics and genetics of viral latency. Cell Host Microbe 19:619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman PM. 2013. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat Rev Microbiol 11:863–875. doi: 10.1038/nrmicro3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves M, Sinclair J. 2013. Regulation of human cytomegalovirus transcription in latency: beyond the major immediate-early promoter. Viruses 5:1395–1413. doi: 10.3390/v5061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4:e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra5. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhary SK, Margolis DM. 2011. Curing HIV: pharmacologic approaches to target HIV-1 latency. Annu Rev Pharmacol Toxicol 51:397–418. doi: 10.1146/annurev-pharmtox-010510-100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. 2016. Latency reversal and viral clearance to cure HIV-1. Science 353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. 2013. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol 21:277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helin K, Dhanak D. 2013. Chromatin proteins and modifications as drug targets. Nature 502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 23.Kim KH, Roberts CW. 2016. Targeting EZH2 in cancer. Nat Med 22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong X, Chen L, Jiao L, Jiang X, Lian F, Lu J, Zhu K, Du D, Liu J, Ding H, Zhang N, Shen J, Zheng M, Chen K, Liu X, Jiang H, Luo C. 2014. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J Med Chem 57:9512–9521. doi: 10.1021/jm501230c. [DOI] [PubMed] [Google Scholar]
- 25.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. 2013. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol 8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 27.Morera L, Lübbert M, Jung M. 2016. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics 8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, Zhang M, Zhang J, Yu Z, Li L, Teng L, Chuai S, Zhang C, Zhao M, Chan H, Chen Z, Fang D, Fei Q, Feng L, Feng L, Gao Y, Ge H, Ge X, Li G, Lingel A, Lin Y, Liu Y, Luo F, Shi M, Wang L, Wang Z, Yu Y, Zeng J, Zeng C, Zhang L, Zhang Q, Zhou S, Oyang C, Atadja P, Li E. 2017. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol 13:381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 29.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, Graves AP, Scherzer DA, Shu A, Thompson C, Ott HM, Aller GS, Machutta CA, Diaz E, Jiang Y, Johnson NW, Knight SD, Kruger RG, McCabe MT, Dhanak D, Tummino PJ, Creasy CL, Miller WH. 2012. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett 3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu B, Konze KD, Jin J, Wang GG. 2015. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp Hematol 43:698–712. doi: 10.1016/j.exphem.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 32.Kim JM, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W. 2015. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci Rep 5:16714. doi: 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 35.Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham CG, Kulesza CA. 2012. Polycomb repressive complex 2 targets murine cytomegalovirus chromatin for modification and associates with viral replication centers. PLoS One 7:e29410. doi: 10.1371/journal.pone.0029410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cliffe AR, Coen DM, Knipe DM. 2013. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Günther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JS, Raja P, Knipe DM. 2016. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7:e02007-15. doi: 10.1128/mBio.02007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata T, Kondo Y, Sugimoto A, Kawashima D, Saito S, Isomura H, Kanda T, Tsurumi T. 2012. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J Virol 86:4752–4761. doi: 10.1128/JVI.06768-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU. 2016. LANA-mediated recruitment of host polycomb repressive complexes onto the KSHV genome during de novo infection. PLoS Pathog 12:e1005878. doi: 10.1371/journal.ppat.1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog 6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristie TM. 2015. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology 479–480:555–561. doi: 10.1016/j.virol.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva L, Cliffe A, Chang L, Knipe DM. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog 4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A 104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cliffe AR, Arbuckle JH, Vogel JL, Geden MJ, Rothbart SB, Cusack CL, Strahl BD, Kristie TM, Deshmukh M. 2015. Neuronal stress pathway mediating a histone methyl/phospho switch is required for herpes simplex virus reactivation. Cell Host Microbe 18:649–658. doi: 10.1016/j.chom.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messer HG, Jacobs D, Dhummakupt A, Bloom DC. 2015. Inhibition of H3K27me3-specific histone demethylases JMJD3 and UTX blocks reactivation of herpes simplex virus 1 in trigeminal ganglion neurons. J Virol 89:3417–3420. doi: 10.1128/JVI.03052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lallemand-Breitenbac V, de Thé H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abou El Hassan M, Huang K, Eswara MB, Zhao M, Song L, Yu T, Liu Y, Liu JC, McCurdy S, Ma A, Wither J, Jin J, Zacksenhaus E, Wrana JL, Bremner R. 2015. Cancer cells hijack PRC2 to modify multiple cytokine pathways. PLoS One 10:e0126466. doi: 10.1371/journal.pone.0126466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, Smith JJ, Moyer MP, Richon VM, Copeland RA, Wigle TJ. 2012. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett 586:3448–3451. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 56.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, Chen SB, Della Pietra A III, Dul E, Hughes AM, Gilbert SA, Thrall SH, Tummino PJ, Kruger RG, Brandt M, Schwartz B, Creasy CL. 2012. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A 109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, Marquez VE, Marra MA, Gascoyne RD, Humphries RK, Arrowsmith CH, Morin GB, Aparicio SA. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comet I, Riising EM, Leblanc B, Helin K. 2016. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 59.Enquist LW, Leib DA. 2017. Intrinsic and innate defenses of neurons: detente with the herpesviruses. J Virol 91:e01200-16. doi: 10.1128/JVI.01200-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knickelbein JE, Khanna KM, Yee MB, Baty CJ, Kinchington PR, Hendricks RL. 2008. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science 322:268–271. doi: 10.1126/science.1164164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosato PC, Leib DA. 2015. Neurons versus herpes simplex virus: the innate immune interactions that contribute to a host-pathogen standoff. Future Virol 10:699–714. doi: 10.2217/fvl.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.St Leger AJ, Hendricks RL. 2011. CD8+ T cells patrol HSV-1-infected trigeminal ganglia and prevent viral reactivation. J Neurovirol 17:528–534. doi: 10.1007/s13365-011-0062-1. [DOI] [PubMed] [Google Scholar]
- 63.van Velzen M, Jing L, Osterhaus AD, Sette A, Koelle DM, Verjans GM. 2013. Local CD4 and CD8 T-cell reactivity to HSV-1 antigens documents broad viral protein expression and immune competence in latently infected human trigeminal ganglia. PLoS Pathog 9:e1003547. doi: 10.1371/journal.ppat.1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arbuckle JH, Turner AM, Kristie TM. 2014. Analysis of HSV viral reactivation in explants of sensory neurons. Curr Protoc Microbiol 35:14E.6.1–14E.6.21. doi: 10.1002/9780471729259.mc14e06s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EZH2/1 inhibitors do not affect nuclear accumulation of viral genomes and suppress HSV-1 IE expression in an MOI-independent manner. (A) mRNA levels of IE and control cellular genes in HFF cells treated with the vehicle or GSK126 for the times indicated and then infected with HSV-1 (MOI, 2.0) for 1.5 h. Data are means ± SEM of three independent experiments. (B, C) Viral IE and control cellular mRNA levels in HFF cells treated with the vehicle, GSK126, or UNC1999 for 5 h and then subjected to drug washout and infection with HSV-1 (MOI, 2.0) at the times indicated. Data are means ± SEM of three independent experiments. (D) HSV-1 DNA levels in total cells and nuclear fractions of HFF cells treated with the vehicle, GSK126, or GSK343 and infected with HSV-1 for 1.5 h. Data are means ± SEM of three independent experiments. (E) Viral IE (ICP0, ICP4, ICP22, ICP27) and control (SP1) mRNA levels in cells treated with the vehicle or GSK126 and infected at the MOI indicated for 1.5 h. Data are means ± SEM of three independent experiments. Download FIG S1, TIF file, 25.2 MB (25.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
IFN qPCR array, microarray, and RNA-Seq data sets. Download TABLE S1, XLSX file, 0.9 MB (925.4KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Enrichment of innate signaling pathways with GSK126, GSK343, and EED226. (A, B) IPA top-ranked pathways for genes differentially regulated (≥1.5-fold) in HFF cells treated with GSK126 or GSK343 for 4 h (microarrays, n = 3). (C, D) Karpas422 cells were treated with the vehicle or EED226 (EED inhibitor) for 24, 48, 72, or 144 h. Microarray CEL files were obtained from GEO under accession no. GSE81267. (C) The number of ISGs (Interferome database) differentially regulated (≥2-fold) by EED226. (D) IPA top-ranked pathways for genes differentially regulated (≥2-fold) by EED226. The 72-h IPA results were used as the reference data set with a P value of ≤0.05. (E) IPA top-ranked pathways enriched for genes differentially regulated (≥2-fold) in HFF cells by 1, 2, or 5 h of GSK126 treatment (RNA-Seq, n = 3). Download FIG S2, TIF file, 78.4 MB (80.3MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
GSK126 suppression of HSV infection is independent of IFNAR1 signaling. Cultures of fibroblasts from wild-type control (C57BL/6J) and IFNAR1−/− (TAC73) mice were infected with HSV-1 (MOI, 0.01) for 8 h, and then the vehicle, ACV, or GSK126 was added for 12 h. Cells were stained with anti-UL29 antibody (green), phalloidin-647 (F-actin, red), and DAPI (blue). Download FIG S3, TIF file, 9 MB (9.2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EZH2/1 inhibitors suppress hCMV and Ad5 infections. (A, B) HFF cells were treated with the vehicle, GSK126, or GSK343 and then infected with Ad5 (MOI, 200) for 3 h or with hCMV (MOI, 0.05) for 2 h. Levels of viral IE (Ad5, E1A; hCMV, UL37, UL123, UL44) and cellular control (SP1, TATA box binding protein [TBP]) mRNAs are expressed relative to those in cells treated with the vehicle. Data are means ± SEM of at least two (A) or four (B) independent experiments. Download FIG S4, TIF file, 9.4 MB (9.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
GSK126 enhances immune cell recruitment to sites of viral infection in vivo. (A) Immunofluorescent staining of eyes mock infected or infected with HSV-1 (2 × 105/eye) and treated topically with the vehicle, ACV, or GSK126 for 7 days. Eye sections were costained with anti-HSV-1 (red) and anti-Ly6G (neutrophil) (green) antibodies and DAPI (blue) (11 eyes/group). (B) Eye sections were scored for HSV-1 infection and neutrophil recruitment (Ly6G positive, Gr1, Fig. 5D) at 3, 5, and 7 dpi. Data are means ± SEM of ≥5 eyes/group. Download FIG S5, TIF file, 41.1 MB (42.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EZH2/1 inhibitors reduce viral yields in a mouse ganglion explant model. (A) Trigeminal ganglia from latently infected mice were bisected, and halves were explanted in the presence of the vehicle (dimethyl sulfoxide [DMSO]) or the compound indicted for 48 h. The yields of viral DNA per ganglion were normalized by the levels of cellular DNA (GAPDH). Data are DNA yields of individual ganglia with means ± SEM of ≥19 ganglia/group. ***, P < 0.001 (Wilcoxon matched-pair signed-rank test). (B) Trigeminal ganglia from mock-infected and latently HSV-1-infected mice were explanted in the presence of the vehicle (DMSO) or GSK126 for 12 h. Levels of cellular mRNAs (murine IL-6 [mIL-6], mCXCL1, and mSP1) are normalized by the level of control TATA box binding protein (TBP) mRNA. Data are means ± SEM of two pools of five ganglia per group. (C) IPA top-ranked pathways enriched for genes differentially regulated (≥2-fold) by GSK126 treatment of explanted ganglia for 12 h (RNA-Seq). Download FIG S6, TIF file, 41.8 MB (42.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplemental materials and methods used in this study. Download TEXT S1, PDF file, 0.1 MB (149.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical analyses. Download TABLE S2, XLSX file, 0.03 MB (34.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Microarray and RNA-Seq data sets are available at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE99841.