Abstract
Until recently, stem cells were thought to be endowed with unlimited self-renewal capacity and, thus, assumed exempt from aging. But accumulating evidence over the past decade compellingly argues that a measurable and progressive replicative impairment in the hematopoietic, intestinal, and muscle stem cell activity exists from adulthood to old age, resulting in a decline in stem cell function and rendering stem cell aging as the possible link between cellular aging and organismal aging. By using a previously uncharacterized congenic animal model to study genetic regulation of hematopoietic stem cell aging, we have demonstrated definitively that a locus on murine chromosome 2 regulates hematopoietic stem cell aging. In addition to demonstrating that hematopoietic stem cell aging is regulated by a distinct genetic element, experimental evidence links the response of hematopoietic stem cells to DNA double-strand breaks to cellular aging, suggesting DNA integrity influences stem cell aging.
Keywords: congenic, DNA damage
Many, and perhaps most, tissues of the major organ systems are composed of short-lived cells that require continuous replenishment throughout life. The skin, the lining of the intestinal lumen, and the hematopoietic tissues historically have served as the best examples, although recently, an increasingly extensive list has evolved of renewing tissues whose replenishment is ensured by somatic stem cells (1–6). Stem cells have been recently identified in the brain and heart, although their importance in contributing to adult tissue homeostasis is still debated (7–11).
The aging process is probably best defined, at the cellular level, as a diminished replicative ability in proliferating cells and diminished functional activity in postmitotic cells. Stem cells were thought to have unlimited self-renewal capacity and, thus, assumed exempt from aging. However, accumulating evidence during the past decade compellingly argues that a measurable and progressive impairment in the hematopoietic, intestinal, and muscle stem cell replicative activity exists from adulthood to old age, resulting in a decline in stem cell function (12, 13). Because stem cell activity is necessary to replenish lost differentiated cells in a stem cell-driven tissue, it has been hypothesized that the aging of stem cells leads to perturbed tissue homeostasis in aged animals (12–15). This hypothesis is supported by the fact that the function of the innate immune system, which depends on hematopoietic stem and progenitor cell activity, is compromised in aged individuals (16–18) and that aged hematopoietic stem cells (HSCs) exhibit reduced activity toward differentiation into the B cell lineage (19). The regulation of stem cell aging in vivo is still poorly understood. The reason might be, at least in part, because the regulation of the HSC system is a complex quantitative trait influenced by the action of many factors/genes.
Two strains of inbred mice, C57BL/6 (B6) and DBA/2 (D2), show a remarkable difference in the HSC compartment in response to aging and they are therefore suited for quantitative trait locus (QTL) analysis (12, 19–22). The differences between B6 and D2 stem cell aging are quantitative in nature, with D2 stem cells aging faster than B6 stem cells, and the interstrain differences in function of aged stem cells determined by stringent engraftment requirements in vivo corroborate stem cell measures with an equally stringent in vitro assay, the cobblestone area-forming cell assay (CAFC). Performing QTL analysis, we previously described a locus on murine chromosome 2 (chr. 2) with significant linkage (decimal log likelihood of odds ratio score of 4.4) to the variation in the frequency of HSCs between aged B6 and D2 animals (23).
Using a congenic animal model, we demonstrate here that this locus on chr. 2 regulates stem cell aging in vivo, rendering the locus on chr. 2 a verified genetic element controlling stem cell aging. The established animal model system will enable the delineation of molecular pathways leading to stem cell aging. In addition, results are presented linking the response of stem cells to DNA double-strand breaks to stem cell aging, suggesting that DNA repair pathways influence stem cell aging.
Materials and Methods
Animals. Six- to 8-week-old C57BL/6J and BXD-31/Ty animals were purchased from The Jackson Laboratory (Bar Harbor, ME). B6.SJL(BoyJ) mice were purchased from Charles River Laboratories (Frederick, MD). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Kentucky.
Linkage Analysis. Linkage analysis and determination of the 95% confidence interval as well as the determination of the likelihood of odds values for suggestive and significant linkage were performed as described in ref. 23 by using mapmanager qtxb2.9, (55) based on a newly compiled strain distribution pattern for BXD animals by R. W. Williams, J. Gu, S. Qi, and L. Lu at Neurogenetics, University of Tennessee Health Science Center.
Generation of Congenic Animals. Congenic animals were generated in five generations by a marker-assisted backcross strategy as described in refs. 23–25. The introgressed DBA/2J genomic interval was derived from BXD31, one of the BXD recombinant inbred strains used in the quantitative trait locus mapping and which phenotypically best demonstrated the decline in HSCs in old age. The haplotype shift in BXD31 is located between D2Mit164 (125 Mbp) and D2Mit304 (130 Mbp). Because of the paucity of informative markers in this small region, we do not know precisely where between these two markers the crossover occurred. Therefore, we ascribe the conservative interpretation that the D2-haplotype starts at the D2Mit304 marker at 130 Mbp.
CAFC Assay. The CAFC assay is an in vitro limiting dilution-type cell culture assay. The CAFC assay was performed as described in ref. 23.
Competitive Transplantation/Transplantation. Female B6.SJL(BoyJ) mice were exposed to a radiation dose of 9 Gy at least 4 h before transplantation. Mixtures of donor and competitor bone marrow (BM) cells were transplanted by retroorbital injection in a volume of 150 μl PBS. The competitive repopulation experiments included recipient animals that received 5-fluorouracil (5-FU) at 150 mg/kg i.p. four times between 8 and 23 weeks after the initial competitive transplantation. There was no significant difference in peripheral blood (PB) chimerism in animals with and without 5-FU treatment in experiments with congenic donors. Bone marrow from each primary recipient was transplanted into three secondary recipient animals.
For competitive transplantation in response to radiation, BM from Tri-Con animals was used as the competitor. Tri-Con animals differ for alleles at 3 loci from B6 background mice and from the congenic strain. The Tri-Cons are Ly5.1 positive versus B6 being Ly5.2 positive, they carry the allele that encodes the glucosephosphate isomerase (GPI) isoenzyme GPIa instead of GPIb in B6, and carry the β-globin gene allele that encodes the Hbd hemoglobin variant versus Hbs in B6 mice. Groups of four 2- to 4-month-old B6.D2 chr. 2 animals were exposed to 1.0 or 2.0 Gy of γ irradiation from a 137Cs-source. Fourteen days after irradiation, BM cells were isolated and 1 × 106 cells transplanted along with an equal number of Tri-Con competitor BM cells into lethally irradiated (9 Gy) B6.SJL(BoyJ) recipients. The donor cells (B6.D2 chr. 2) admixed with Tri-Con competitor cells were distinguished from each other and from those of the recipient animals by using a combination of Ly5, GPI, and Hb variants. The GPI isozymes and Hb variants were measured by electrophoresis and densitometry as described in ref. 26.
Flow Cytometry. Erythrocytes were eliminated from tissue samples by hypotonic lysis and leucocytes subsequently stained with antibodies according to standard procedures. Anti-Ly5.2 (clone 104, Becton Dickinson) and anti-Ly5.1 (clone A20, Becton Dickinson) monoclonal antibodies were used to distinguish donor from recipient and competitor cells.
Statistical Analysis. The paired Student t test was used to determine the significance of the difference between means.
Results
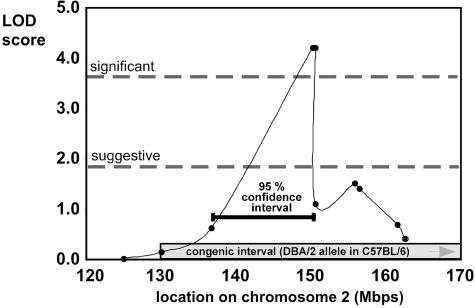
Generation of B6.D2 (chr. 2) Congenic Animals. The variation in the frequency of HSCs and hematopoietic progenitor cells (HPCs) due to aging is genetically determined and might be classified as a complex quantitative trait (19, 20, 23, 24–30). In the C57BL/6 (B6) mouse inbred strain, HSCs increase in a linear fashion with age, whereas in the DBA/2 (D2) strain, the frequency of HSCs follows a bell-shaped distribution throughout life, reaching a maximum at ≈1 year, followed in old age by a decline to ≈80% of the stem cell population size of young animals (25). This difference in the frequency of HSCs between aged B6 and D2 animals was linked significantly by quantitative trait locus analysis to the distal part of murine chr. 2, the 95% confidence interval stretching from 135 to 150 Mbp (Fig. 1) (13, 23). The tightest linkage was to markers D2Mit281 and D2Mit282 with a likelihood of odds score of 4.4, a level statistically significant by guidelines for this type of genetic mapping (Fig. 1) (31, 32).
Fig. 1.
Significant linkage of a locus on murine chr. 2 to HSC aging. Linkage between 120 and 170 Mbp on chr. 2 to the difference in the number of HSCs (CAFC day 35) between aged C57BL/6 and DBA/2 animals is shown. The values for suggestive and significant linkage to the trait along chr. 2 are given, as well as the interval covered in the congenic line (130–180 Mbp) and the 95% confidence interval for linkage (135–151 Mbp).
To unequivocally associate stem cell aging with chr. 2 and generate an animal model that enables the determination of the physiological and functional mechanisms of stem cell aging, mice congenic for the distal part of chr. 2 were generated by introgressing the D2 alleles onto a B6 background strain by a speed congenic approach (33, 34). The introgressed congenic interval spanned 50 Mbp and ranged from 130 Mbp to the end of chr. 2 (≈180 Mbp), including the entire 95% confidence interval (Fig. 1). The frequency of residual D2 alleles in the genome other than the one covering the locus was estimated to be <1%, because all of the analyzed 98 simple sequence repeat length polymorphic markers not localized to the congenic interval were typed as B6.
The Congenic Interval Regulates Stem Cell Aging. A D2 haplotype at the locus on chr. 2 was associated with accelerated stem cell aging (23). By introgressing the D2 alleles onto a B6 background, we predicted that congenic animals would show a significant reduction in HSC frequency with age compared with B6, resembling at least partially the D2 phenotype. To test this hypothesis, homozygous congenic animals were aged, and the frequency of primitive hematopoietic cells in the BM was determined with the CAFC assay. In this assay, the frequency of colonies derived from HSCs that develop beneath a stromal cell layer after 28 and 35 days of in vitro culture is measured as a surrogate for the frequency of HSCs in BM (35). The CAFC assay is a highly reproducible surrogate stem cell assay that allows longitudinal comparison of the frequency of primitive hematopoietic cells (25).
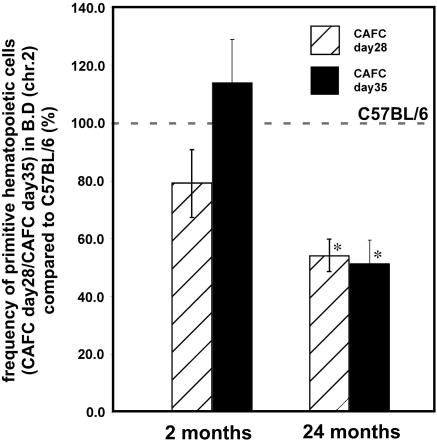
At 2 months of age, as expected, the frequency of HSCs in congenic BM (CAFC day 28 and CAFC day 35) did not differ significantly from the frequency in BM derived from B6 animals (Fig. 2). In contrast, at 24 months of age, the frequency of HSCs in congenic BM was a statistically significant 2-fold less than in age-matched B6 mice (Fig. 2). In addition, we also analyzed 15-month-old congenic mice and 21-month-old B6 animals. Although they are not age-matched, the 15-month-old congenics showed a statistically significant 4-fold lower frequency of HSCs compared with the 21-month-old B6 animals. Thus, in summary, 15- to 24-month old congenic animals showed a 2- to 4-fold lower HSC frequency (Table 1). Old animals are somewhat larger than young adults, and the BM cell number showed only a commensurately modest 20% increase in the BM cellularity (Table 3, which is published as supporting information on the PNAS web site). Therefore, the decrease in frequency of HSCs reported in Table 1 translates into a decrease in the absolute number of HSCs in BM of aged congenic animals.
Fig. 2.
Decreased frequency of primitive hematopoietic cells (CAFC day 28 and 35) in BM of 24-month-old B6.D2 chr. 2 animals compared with the frequency in C57BL/6 BM. CAFC assays with BM cells from 2- and 24-month-old C57BL/6 and B6.D2 (chr. 2) animals were performed as described in Materials and Methods. *, P < 0.05. BM from two aged animals for each genotype was analyzed in six individual CAFC assays per animal.
Table 1. No. of CAFCs per 100,000 BM cells.
| Age of animals, months
|
Days of culture
|
||||
|---|---|---|---|---|---|
| Strain | 14 | 21 | 28 | 35 | |
| C57BL/6 | 2 | 51.6 ± 5.3 | 21.4 ± 2.8 | 4.7 ± 0.7 | 1.8 ± 0.2 |
| 21 | 37.6 ± 2.2 | 25.4 ± 7.9 | 19.6 ± 3.9 | 9.6 ± 1.0 | |
| 24 | 51.5 ± 4.5 | 35.3 ± 6.2 | 19.7 ± 3.8 | 10.7 ± 1.1 | |
| B6.D2 chr. 2, 130-180 Mbp | 2 | 34.5 ± 4.0 | 8.0 ± 1.4 | 3.7 ± 0.4 | 2.0 ± 0.3 |
| 15 | 23.5 ± 5.9 | 13.4 ± 3.1 | 5.1 ± 1.6 | 2.7 ± 0.6 | |
| 24 | 23.5 ± 2.0 | 19.8 ± 2.5 | 10.6 ± 1.1 | 5.5 ± 0.9 | |
C57BL/6 (2 months), n = 18 animals, 9 individual measurements; B6.D2 (chr. 2) (2 months), n = 11 animals, 9 individual measurements; C57BL/6 (21 months), n = 11 animals, 3 individual measurements; B6.D2 (chr. 2) (21 months), n = 2, 2 individual measurements; C57BL/6 (24 months), n = 2, 6 individual measurements; and B6.D2 (chr. 2) (24 months), n = 2, 6 individual measurements, mean ± 1 SEM.
We also determined the frequency of HPCs in young and aged congenic mice with the CAFC assay. Cells that form cobblestone-like colonies after in vitro culture for 14 to 21 days (CAFC day 14 and day 21) are defined as HPCs (35). We observed an age-independent 2- to 3-fold decrease in the frequency of HPCs in BM of congenic mice compared with BM from B6 animals (Table 1). The decrease of the progenitor cell frequency in BM of young congenic animals was not expected because the frequency of HPCs in young BM was not linked to chr. 2 (23).
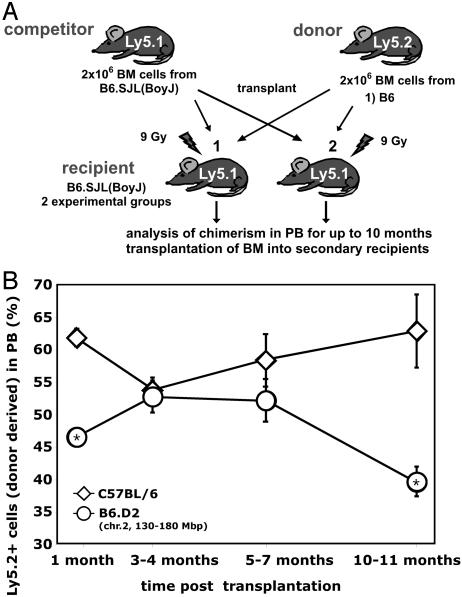
Competitive Repopulation Disadvantage of Congenic Stem Cells on Aging. Stem cells are defined by their ability to self-renew and support multilineage differentiation. Although the determination of stem cell frequency by the CAFC assay is a valid surrogate measurement (23, 35), stringent testing of stem cell activity and frequency is best accomplished by transplanting putative HSCs into lethally irradiated recipients. High-dose irradiation (9 Gy) causes nearly complete ablation of stem cells in the recipient animal and, after stem cell transplantation, leads to an almost complete replacement of the recipient's hematopoietic system by transplanted stem cells. To determine the frequency and the long-term self-renewal capacity of congenic HSCs compared with B6 HSCs, competitive transplantation assays were performed (Fig. 3A) (36–38).
Fig. 3.
Competitive disadvantage of congenic hematopoietic stem cells in aged animals. (A) Schematic diagram of the competitive repopulation assay. (B) Equal numbers (1 × 106 each) of B6.SJL (BoyJ) competitor and either C57BL/6 (group 1) or B6.D2 (chr.2) (group 2) BM cells were transplanted into lethally irradiated B6.SJL recipients, and the animals subsequently aged up to 10 months. Chimerism in peripheral blood was determined by flow cytometry at 1 month, 3–4 months, 5–7 months, and 10 months after transplantation. *, P < 0.05. C57BL/6 donor: n = 18 at 1 month, n = 30 at 3–4 months, n = 13 at 5–7 months, and n = 5 at 10–11 months. B6.D2 (chr. 2) donor: n = 6 for 1 month, n = 6 for 3–4 months, n = 5 for 5–7 months, and n = 5 for 10 months.
Hematopoietic cells from B6.SJL(BoyJ) mice express the Ly5.1 allele, whereas hematopoietic cells from B6 animals express the Ly5.2 allele. Thus, B6.SJL hematopoietic cells can be distinguished from B6 cells by flow cytometry with antibodies specific for Ly5 isotypes. Lethally irradiated B6.SJL females were split into two groups and transplanted with either 2 × 106 male B6 BM cells (group 1) or 2 × 106 male B6.D2 (chr. 2) BM cells (group 2), admixed with 2 × 106 female B6.SJL competitor BM cells. Chimerism in PB was subsequently monitored by flow cytometry during the next 10–11 months. The ratio of chimerism in PB is an indicator of the frequency of the competing primitive hematopoietic cells in the BM.
One month after transplantation, PB cell chimerism revealed a significant increase in the contribution from B6 BM compared with congenic BM (Fig. 3B). At this time after transplantation, hematopoiesis in the recipient animal is primarily sustained by transplanted donor-derived HPCs (39, 40). As reported in Table 1, the frequency of hematopoietic progenitor cells in young congenic animals is decreased, which might explain the difference between B6 and congenic BM with regard to the short-term repopulating ability. At the 3- to 4-month interval, when hematopoiesis in the periphery is mostly sustained by transplanted stem cells, we did not detect a significant difference in the activity of the transplanted HSCs. BM cells from B6 and congenic animals were in the range of 52–54% to PB, which is close to the expected range of 50%. This result reflects the almost identical frequency of HSCs found in both young B6 and congenic animals (Fig. 2 and Table 1). Chimerism in PB at 5–7 months after transplantation revealed no significant difference between the contribution from congenic and B6 BM or from the relative chimerism found 3 months after transplantation. In contrast to this finding, the contribution of the B6 cells to PB at 10–11 months after transplantation was 54%, whereas the contribution of congenic cells decreased significantly to <40%, indicative of congenic stem cell exhaustion due to stem cell aging.
Undisturbed Multilineage Differentiation of Aged Congenic Stem Cells. Competitively transplanted animals were killed at 10 to 11 months after transplantation, because morbidity of recipient animals due to the initial lethal radiation precluded further aging of the recipient animals. To determine the contribution of congenic stem cells to individual hematopoietic tissues and cell lineages and to exclude lineage differentiation preferences as the reason for the decreased contribution of congenic stem cells to PB, chimerism in BM, spleen, and thymus was measured by flow cytometry (Table 2). Chimerism in BM was 60% for B6 and slightly increased compared with the chimerism seen in PB at 10 months, whereas the congenic contribution in the BM reflected the 40% chimerism found in PB of these animals and was significantly reduced compared with B6. Contribution of congenic cells to spleen (31% vs. 44%) and thymus (35% vs. 44%) in aged animals was also diminished compared with B6. Further analysis of the chimerism in distinct hematopoietic blood lineages (monocytes/granulocytes, B cells, and T cells) in PB and BM in the competitively aged animals revealed no significant difference in the relative cell lineage-specific chimerism between animals transplanted with either congenic or B6 BM (Table 4, which is published as supporting information on the PNAS web site). This result suggests that the decline in B6.D2 (chr. 2) hematopoietic contribution was caused by aging of a pluripotent stem cell.
Table 2. Chimerism in primary and secondary recipient animals.
| Ly5.2-positive cells in primary recipients 10-11 months after transplantation, %
|
|||||
|---|---|---|---|---|---|
| Donor strain | PB | BM | SP | Thy | Ly5.2-positive cells in PB of secondary recipients,* % |
| C57BL/6 | 54.4 ± 5.4 | 60.8 ± 2.7 | 44.3 ± 5.7 | 44.2 ± 8.4 | 70.7 ± 5.7 |
| B6.D2 (chr.2, 130-180 Mbps) | 39.6 ± 3.4 | 39.4 ± 8.0 | 30.7 ± 4.1 | 34.7 ± 6.8 | 31.1 ± 4.5 |
SP, spleen; Thy, thymus. *, chimerism in secondary recipients determined 4.5 months after transplantation. Shown are mean values ± 1 SEM.
Competitive Decline of Congenic HSC Frequency in Aged Animals. To determine whether the lower contribution found in animals competitively transplanted with congenic BM was due to a decrease of the frequency of congenic stem cells with age, we performed secondary transplants with BM derived from the aged (10 months) primary transplant recipients. Lethally irradiated secondary recipients were transplanted with 1 × 107 BM cells derived from the primary recipients and chimerism in the PB was analyzed up to 4 months after transplantation (Table 3). Flow cytometry revealed that the contribution of congenic cells to PB decreased even further in the secondary recipients from 40% chimerism in the BM of the primary recipients to 30%, whereas the contribution of B6 cells in the PB of the recipients increased from 60% to 70%. These results confirm that in the BM of the primary recipients, the frequency of congenic stem cells was reduced ≈2-fold compared with the B6 control.
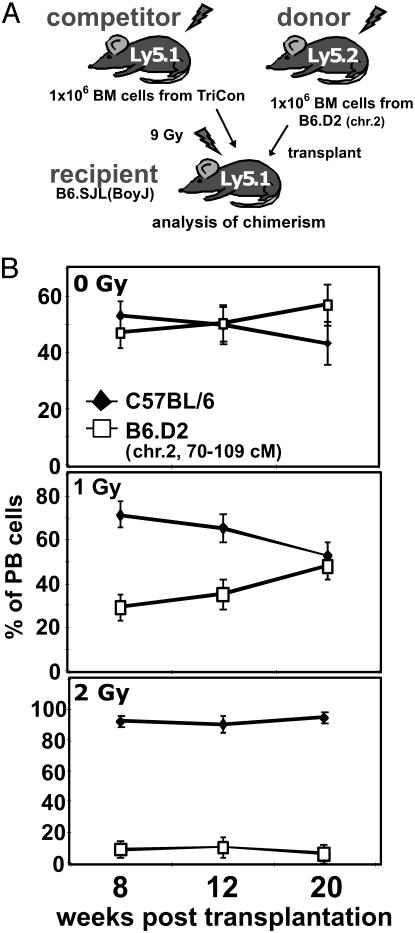
Radiation Susceptibility of Congenic Stem Cells as a Surrogate Assay for Stem Cell Aging. To test our previously formulated hypothesis that DNA integrity influences stem cell aging (13), either 1 or 2 Gy of radiation was administered to B6.D2 chr. 2 congenic animals, which caused DNA damage in stem cells and partial depletion of the population. After a resting period of 2 weeks, BM cells were harvested and subsequently transplanted into lethally irradiated recipients along with an equal number of competitive, similarly treated TriCon BM cells (see Materials and Methods for a description of Tri-Cons) (Fig. 4A). Fig. 4B depicts the chimerism detected in PB in competitively transplanted animals up to 20 weeks after transplantation. Because the transplanted congenic and competitor cells engrafted the same microenvironment, any deviation from the expected chimerism of 50% for each cell population may be assigned to intrinsic differences of the transplanted cells. In accordance with data presented in Fig. 3B, between 8 and 20 weeks, we detected no significant difference in the contribution to PB in animals competitively transplanted with BM cells from nonirradiated (control) animals. In contrast, competitively transplanted BM cells receiving 1.0 Gy, (resulting in 20–30 chromosomal breaks per genome; ref. 41) contributed to a PB chimerism of only 30% from week 8 to week 12 after transplantation. By 20 weeks after transplantation, when hematopoiesis in the periphery was predominately supported by transplanted stem cells, contribution from congenic BM cells surprisingly rose to the expected 50%. In stark contrast, BM from congenic animals receiving 2 Gy only contributed 10% of PB cells at 8 weeks. This low contribution remained unchanged for the duration of the experiment, a finding consistent with stem cell exhaustion due to radiation. Chimerism of GPI isoforms supported the results obtained with flow cytometry (data not shown). Analysis of chimerism in BM of transplanted animals at 20 weeks after transplantation showed that congenic cells comprised 40.1 ± 5.8% of BM in animals receiving no radiation, 27.8 ± 4.1% in animals receiving 1 Gy, and only 1.4 ± 0.2% in animals receiving 2 Gy, further reflecting the progressive nature of the loss of stem cell activity due to radiation.
Fig. 4.
Radiation susceptibility of congenic stem cells as a surrogate assay for stem cell aging. (A) Schematic diagram of the competitive repopulation assay used for these analyses. (B) Tri-Con and B6.D2 (chr. 2) congenic animals were irradiated by 0, 1, or 2 Gy 2 weeks before BM harvest. Equal numbers of Tri-Con competitor and B6.D2 (chr. 2) donor BM cells (1 × 106 each) were transplanted into lethally irradiated B6.SJL (BoyJ) recipients and the animals subsequently aged up to 20 weeks. Chimerism in peripheral blood was determined at 8, 12, and 20 weeks after transplantation by flow cytometry based on the Ly5.1/Ly5.2 mismatch of the transplanted BM cells. n = 4 for each dose.
Because these findings could be explained either by replicative exhaustion of nonapoptotic stem cells or by direct DNA damage to stem cells, experiments similar to those analyzing the radiation response were performed by multiple injections of 5-FU into congenic and control animals. 5-FU leads to apoptosis in HPCs and results in a profound increase in replicative activity of HSCs to replace lost progenitor cells (42, 43). In contrast to the results obtained in response to low-dose radiation, no exhaustion of congenic stem cell activity could be detected after multiple injections of 5-FU (data not shown).
In summary, these results establish the response to low-dose radiation as a short-term surrogate assay to analyze stem cell aging and further validate our congenic animal model system of HSC aging. In addition, they strongly support the hypothesis that stem cell aging is caused by the loss of DNA integrity/accumulation of DNA damage.
Candidate Genes Located in the 95% Confidence Interval. The 95% confidence interval on murine chr. 2 is syntenic to human chromosome 20p. There are no reports in the literature associating 20p with cellular senescence. Table 5, which is published as supporting information on the PNAS web site, lists all genes identified to date with a known function in the 95% confidence interval reaching from 135 Mbp to 151 Mbp. These genes are, owing to their physical location, candidate genes for regulating HSCs in BM with age. Surprisingly, the interval contains multiple genes for which an influence on the cellular response to DNA damage has been reported or might be inferred. For example, RBB9, a retinoblastoma binding protein, might interfere with checkpoint responses in stem cells due to radiation (44). DHM1 has some homology to yeast and human SEP1, a protein involved in homologous recombination and cellular aging (45). Sorting nexin 5, which is also located in the interval, is reported to interact with the Fanconi anemia complementation group protein A (FANC A) (46, 47). Mutations in human FANC A cause severe anemia and genomic instability, thus rendering sorting nexin 5 a prime candidate for the locus.
Discussion
The studies presented here were aimed to verify the biological linkage of a locus on chr. 2 to stem cell aging and to generate an animal model system to allow identification of molecular mechanisms of stem cell aging (23). We generated animals congenic for the interval on chr. 2 by introgressing the DBA/2 (D2) alleles onto a C57BL/6 (B6) background. Subsequently, HSC number and activity in aged congenic animals were analyzed and compared with aged B6 animals. Using the CAFC assay, a surrogate in vitro stem cell assay, a significant decrease in the frequency of HSCs was observed in the BM of 15- to 24-month-old congenic animals compared with B6 (Table 1). To verify the reduction of stem cell activity in aged congenic BM, competitive repopulation experiments were performed and the recipients were analyzed for up to 10 months after transplantation. In accordance with the data from stem cell frequency measured by the CAFC assay, animals reconstituted with B6 or congenic HSCs contributed equally to PB from 3 to 7 months after transplantation. Because engraftment detected 3 to 4 months after transplantation is widely acknowledged to be stem cell derived, this result indicates that in young animals, there was no variation in the functional activity of HSCs between B6 and congenic animals. In contrast, at 10 months after transplantation, which corresponds to an age of 12 months for the transplanted stem cells, a significantly lower contribution of congenic cells to the PB was detected (Fig. 3B). Analysis of the chimerism in BM of the aged animals and in secondary BM transplants revealed that, indeed, the frequency of HSCs had been reduced in the aged primary recipients (Table 2).
Stem cell frequency in old congenic animals thus confirmed the linkage analysis and experimentally assigned the locus on chr. 2 to stem cell aging and, at the same time, established an animal model system to further study cellular and molecular mechanisms of stem cell aging. The influence exerted by the D2 allele at this locus caused a 2-fold reduction of the stem cell frequency with age (Tables 1 and 2). This locus is unique because it does not regulate the variation in HSCs in young animals and, thus, can be viewed as a true stem cell aging locus (23). The results presented here demonstrate a cell-intrinsic action of the locus, because stem cell aging was detectable in transplantable stem cells in the competitive aging experiments and in individual cells in the CAFC assay. A cell-intrinsic aging mechanism for stem cells is further supported by refs. 19, 20, and 48.
We previously described age-related reversible quiescence of HSCs as another aging mechanism for stem cells (13, 49). In these former experiments, D2 HSC senescence in aged B6 and D2 allophenic chimeras was temporarily reversed on transplantation of aged allophenic BM into secondary recipients. We did not detect reversible stem cell senescence in our competitively in vivo aged congenic stem cells, as evidenced by no elevation in the contribution of congenic stem cells to hematopoiesis in secondary recipients. Thus, the locus on chr. 2 confers irreversible stem cell aging, whereas additional genetic components may be responsible for reversible stem cell quiescence, perhaps through epigenetic mechanisms.
Unexpectedly, we detected a significantly lower frequency of HPCs in young congenic animals (Table 1 and Fig. 3), although the locus was not linked to the number of HPCs in young animals (23). Distinct expression profiles of cell surface markers have been used to identify cell populations that are enriched for HPC and HSC activity (50–52). Hematopoietic stem cells are highly enriched in a cell population that is depleted of cells expressing lineage-specific cell surface antigens and enriched for cells that express Sca-1 and c-Kit (LIN-, Sca-1+, c-Kit+, or L-S+K+ cells), whereas HPCs are enriched in the LIN-, Sca-1-, c-Kit+, or L-S-K+ population (51). We previously reported a significantly reduced frequency of L-S+K+ cells in BM of young B6.D2 (chr. 2) animals compared with B6 (53). In these studies we also reported linkage of the in vitro proliferative activity of sorted L-S+ cells from young animals to a locus on chr. 2 contained in the congenic interval. Because we detected similar chimerism in competitively transplanted animals 3 to 7 months after transplantation (Fig. 3), our results question a stringent correlation between the frequency of L-S+K+ cells and the frequency of long-term repopulating stem cells among inbred strains, especially because the frequency of L-S+K+ cells in aged BM was recently linked to a locus on chromosome 12 and not to chr. 2 (30). These observations imply that the locus has a dual function: regulating progenitor cell number in young animals and stem cell number in old animals.
Experiments in which stem cells were irradiated and their potency subsequently analyzed in a competitive transplantation experiment strongly support the hypothesis that DNA damage in stem cells may lead to stem cell aging. At the dose of 1 Gy, we detected a decrease of competitiveness of congenic cells only in the initial months after transplantation. We interpret this finding as a reduction in progenitor cell activity and possibly an exacerbation of the progenitor cell phenotype already seen in the CAFC and long-term transplantation assays (Table 1 and Fig. 4B). Congenic cells receiving 2 Gy were significantly reduced in their competitiveness, even at 5 months after transplantation, which suggests that this increase in dosage disrupted both the stem and progenitor cell compartment. With this assay, we also established a surrogate assay system to study stem cell aging in middle-aged animals.
Active stem cells are necessary to replenish lost differentiated cells in a stem cell-driven tissue, and stem cell aging may lead to perturbed tissue homeostasis in aged animals (12–15, 54). Because of the life-sustaining nature of many of these tissues, including BM, viability of the stem cell compartment may influence organismal longevity (12, 30). As we and others have described, a locus on chr. 2 that is linked to longevity maps in close vicinity to the locus linked to stem cell aging, further supporting this hypothesis (13, 30). Future studies are needed to verify whether the underlying gene(s) regulate(s) both traits.
In summary, the locus on chr. 2 regulates stem cell aging and thus the congenic animals are a resource to analyze HSC aging regulated by a distinct genetic element. This model system will be extremely useful for the delineation of genes and molecular pathways underlying the aging process of stem cells by further generating subcongenic lines in combination with genome-wide gene-expression analyses. By establishing DNA damage due to radiation as a short-tem surrogate assay to study stem cell aging in this system, we provide further evidence that one underlying cause for stem cell aging may be the loss of DNA integrity in stem cells because of DNA damage.
Supplementary Material
Acknowledgments
We thank Barry Grimes for invaluable help with flow cytometry, Yi Zheng and John Bissler for critical comments on the manuscript, and Paula Thomason for editorial support. This work was supported by National Institutes of Health Grants AG16653 and AG20917 (to G.V.Z.) and a fellowship of the Deutsche Akademie der Naturforscher Leopoldina funded by the Bundesministerium für Bildung und Forschung (to H.G.).
Author contributions: H.G., G.R., and G.v.Z. designed research; H.G. and G.R. performed research; H.G., G.R., and G.v.Z. analyzed data; and H.G. and G.v.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BM, bone marrow; CAFC, cobblestone area-forming cell; chr. 2, chromosome 2; 5-FU, 5-fluorouracil; GPI, glucosephosphate isomerase; HPC, hematopoietic progenitor cell; HSC, hematopoietic stem cell; PB, peripheral blood.
References
- 1.Morrison, S. J., Uchida, N. & Weissman, I. L. (1995) Annu Rev. Cell Dev. Biol. 11, 35–71. [DOI] [PubMed] [Google Scholar]
- 2.Tani, H., Morris, R. J. & Kaur, P. (2000) Proc. Natl. Acad. Sci. USA 97, 10960–10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs, E. & Segre, J. A. (2000) Cell 100, 143–155. [DOI] [PubMed] [Google Scholar]
- 4.Potten, C. S. & Morris, R. J. (1988) J. Cell Sci. Suppl. 10, 45–62. [DOI] [PubMed] [Google Scholar]
- 5.Stappenbeck, T. S., Mills, J. C. & Gordon, J. I. (2003) Proc. Natl. Acad. Sci. USA 100, 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Kooy, D. & Weiss, S. (2000) Science 287, 1439–1441. [DOI] [PubMed] [Google Scholar]
- 7.McKay, R. (1997) Science 276, 66–71. [DOI] [PubMed] [Google Scholar]
- 8.Doetsch, F., Caille, I., Lim, D. A., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. (1999) Cell 97, 703–716. [DOI] [PubMed] [Google Scholar]
- 9.Gage, F. H. (2000) Science 287, 1433–1438. [DOI] [PubMed] [Google Scholar]
- 10.Oh, H., Bradfute, S. B., Gallardo, T. D., Nakamura, T., Gaussin, V., Mishina, Y., Pocius, J., Michael, L. H., Behringer, R. R., Garry, D. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrami, A. P., Barlucchi, L., Torella, D., Baker, M., Limana, F., Chimenti, S., Kasahara, H., Rota, M., Musso, E., Urbanek, K., et al. (2003) Cell 114, 763–776. [DOI] [PubMed] [Google Scholar]
- 12.Van Zant, G. & Liang, Y. (2003) Exp. Hematol. 31, 659–672. [DOI] [PubMed] [Google Scholar]
- 13.Geiger, H. & Van Zant, G. (2002) Nat. Immunol. 3, 329–333. [DOI] [PubMed] [Google Scholar]
- 14.Sharpless, N. E. & DePinho, R. A. (2004) J. Clin. Invest. 113, 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torella, D., Rota, M., Nurzynska, D., Musso, E., Monsen, A., Shiraishi, I., Zias, E., Walsh, K., Rosenzweig, A., Sussman, M. A., et al. (2004) Circ. Res. 94, 514–524. [DOI] [PubMed] [Google Scholar]
- 16.Butcher, S. K., Chahal, H., Nayak, L., Sinclair, A., Henriquez, N. V., Sapey, E., O'Mahony, D. & Lord, J. M. (2001) J. Leukoc. Biol. 70, 881–886. [PubMed] [Google Scholar]
- 17.Lord, J. M., Butcher, S., Killampali, V., Lascelles, D. & Salmon, M. (2001) Mech. Ageing Dev. 122, 1521–1535. [DOI] [PubMed] [Google Scholar]
- 18.Ginaldi, L., De Martinis, M., D'Ostilio, A., Marini, L., Loreto, M. F. & Quaglino, D. (1999) Immuol. Res. 20, 117–126. [DOI] [PubMed] [Google Scholar]
- 19.Kim, M., Moon, H. B. & Spangrude, G. J. (2003) Ann. N.Y. Acad. Sci. 996, 195–208. [DOI] [PubMed] [Google Scholar]
- 20.Chen, J., Astle, C. M. & Harrison, D. E. (2000) Exp. Hematol. 28, 442–450. [DOI] [PubMed] [Google Scholar]
- 21.Morrison, S. J., Wandycz, A. M., Akashi, K., Globerson, A. & Weissman, I. L. (1996) Nat. Med. 2, 1011–1016. [DOI] [PubMed] [Google Scholar]
- 22.Zijlmans, J. M. J. M., Martens, U. M., Poon, S. S. S., Raap, A. K., Tanke, H. J., Ward, R. K. & Lansdorp, P. M. (1997) Proc. Natl. Acad. Sci. USA 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger, H., True, J. M., de Haan, G. & Van Zant, G. (2001) Blood 98, 2966–2972. [DOI] [PubMed] [Google Scholar]
- 24.Van Zant, G., Holland, B. P., Eldridge, P. W. & Chen, J. J. (1990) J. Exp. Med. 171, 1547–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Haan, G. & Van Zant, G. (1999) Blood 93, 3294–3301. [PubMed] [Google Scholar]
- 26.Muller-Sieburg, C. E. & Riblet, R. (1996) J. Exp. Med. 183, 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Haan, G., Nijhof, W. & Van Zant, G. (1997) Blood 89, 1543–1550. [PubMed] [Google Scholar]
- 28.de Haan, G. & Van Zant, G. (1997) J. Exper. Med. 186, 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller-Sieburg, C. E., Cho, R. H., Sieburg, H. B., Kupriyanov, S. & Riblet, R. (2000) Blood 95, 2446–2448. [PubMed] [Google Scholar]
- 30.Henckaerts, E., Langer, J. C. & Snoeck, H. W. (2004) Blood 104, 374–379. [DOI] [PubMed] [Google Scholar]
- 31.Churchill, G. A. & Doerge, R. W. (1994) Genetics 138, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darvasi, A. (1998) Nat. Genet. 18, 19–24. [DOI] [PubMed] [Google Scholar]
- 33.Markel, P., Shu, P., Ebeling, C., Carlson, G. A., Nagle, D. L., Smutko, J. S. & Moore, K. J. (1997) Nat. Genet. 17, 280–284. [DOI] [PubMed] [Google Scholar]
- 34.Wakeland, E., Morel, L., Achey, K., Yui, M. & Longmate, J. (1997) Immunol. Today 18, 472–477. [DOI] [PubMed] [Google Scholar]
- 35.Ploemacher, R. E., van der Sluijs, J. P., Voerman, J. S. & Brons, N. H. (1989) Blood 74, 2755–2763. [PubMed] [Google Scholar]
- 36.Jordan, C. T., Astle, C. M., Zawadzki, J., Mackarehtschian, K., Lemischka, I. R. & Harrison, D. E. (1995) Exp. Hematol. (Charlottesville, VA) 23, 1011–1015. [PubMed] [Google Scholar]
- 37.Harrison, D. E., Jordan, C. T., Zhong, R. K. & Astle, C. M. (1993) Exp. Hematol. (Charlottesville, VA) 21, 206–219. [PubMed] [Google Scholar]
- 38.Geiger, H., Szilvassy, S. J., Ragland, P. & Van Zant, G. (2004) Exp. Hematol. (Charlottesville, VA) 32, 60–67. [DOI] [PubMed] [Google Scholar]
- 39.Jordan, C. T. & Lemischka, I. R. (1990) Genes Dev. 4, 220–232. [DOI] [PubMed] [Google Scholar]
- 40.Harrison, D. E. & Astle, C. M. (1997) Blood 90, 174–181. [PubMed] [Google Scholar]
- 41.Silver, L. M. (1995) Mouse Genetics Concepts and Applications (Oxford Univ. Press, Oxford).
- 42.Longley, D. B., Harkin, D. P. & Johnston, P. G. (2003) Nat. Rev. Cancer 3, 330–338. [DOI] [PubMed] [Google Scholar]
- 43.Randall, T. D. & Weissman, I. L. (1997) Blood 89, 3596–3606. [PubMed] [Google Scholar]
- 44.Woitach, J. T., Zhang, M., Niu, C. H. & Thorgeirsson, S. S. (1998) Nat. Genet. 19, 371–374. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, K., Dion, N., Fuchs, B., Damron, T., Gitelis, S., Irwin, R., O'Connor, M., Schwartz, H., Scully, S. P., Rock, M. G., et al. (2002) Gene 298, 121–127. [DOI] [PubMed] [Google Scholar]
- 46.Otsuki, T., Kajigaya, S., Ozawa, K. & Liu, J. M. (1999) Biochem. Biophys. Res. Commun. 265, 630–635. [DOI] [PubMed] [Google Scholar]
- 47.Reuter, T. Y., Medhurst, A. L., Waisfisz, Q., Zhi, Y., Herterich, S., Hoehn, H., Gross, H. J., Joenje, H., Hoatlin, M. E., Mathew, C. G. & Huber, P. A. (2003) Exp. Cell Res. 289, 211–221. [DOI] [PubMed] [Google Scholar]
- 48.Chen, J., Astle, C. M. & Harrison, D. E. (1999) Exp. Hematol. (Charlottesville, VA) 27, 928–935. [DOI] [PubMed] [Google Scholar]
- 49.Van Zant, G., Scott-Micus, K., Thompson, B. P., Fleischman, R. A. & Perkins, S. (1992) Exp. Hematol. (Charlottesville, VA) 20, 470–475. [PubMed] [Google Scholar]
- 50.Spangrude, G. J., Heimfeld, S. & Weissman, I. L. (1988) Science 241, 58–62. [DOI] [PubMed] [Google Scholar]
- 51.Okada, S., Nakauchi, H., Nagayoshi, K., Nishikawa, S., Nishikawa, S., Miura, Y. & Suda, T. (1991) Blood 78, 1706–1712. [PubMed] [Google Scholar]
- 52.Osawa, M., Hanada, K., Hamada, H. & Nakauchi, H. (1996) Science 273, 242–245. [DOI] [PubMed] [Google Scholar]
- 53.Henckaerts, E., Geiger, H., Langer, J. C., Rebollo, P., Van Zant, G. & Snoeck, H. W. (2002) Blood 99, 3947–3954. [DOI] [PubMed] [Google Scholar]
- 54.Bell, D. R. & Van Zant, G. (2004) Oncogene 23, 7290–7296. [DOI] [PubMed] [Google Scholar]
- 55.Manly, K. F. Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930–932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.