Abstract
To make more practical the total chemical synthesis of proteins by the ligation of unprotected peptide building blocks, we have developed a method to facilitate the isolation and handling of intermediate products. The synthetic technique makes use of a His6 tag at the C terminus of the target polypeptide chain, introduced during the synthesis of the C-terminal peptide segment building block. The presence of a His6 tag enables the isolation of peptide or protein products directly from ligation reaction mixtures by Ni-NTA affinity column purification. This simple approach enables facile buffer exchange to alternate reaction conditions and is compatible with direct analytical control by protein MS of the multiple ligation steps involved in protein synthesis. We used syntheses of crambin and a modular tetratricopeptide repeat protein of 17 kDa as models to examine the utility of this affinity purification approach. The results show that His6 tag-assisted chemical protein synthesis is a useful method that substantially reduces handling losses and provides for rapid chemical protein syntheses.
Keywords: affinity purification, native chemical ligation
Chemical ligation (1) enables the synthesis of large polypeptide chains by the chemoselective reaction of unprotected peptide segments. A variety of ligation chemistries has been developed for this purpose (1–15). Application of the chemical ligation principle (1) has led to practical chemical syntheses of a wide variety of different classes of proteins (16). Synthetic access to protein molecules has been used to elucidate the molecular basis of protein folding and stability (17), to elucidate the molecular basis of protein function (18), to design and build proteins of novel structure (19), and to determine the molecular structure of proteins by both NMR (20) and x-ray crystallography (21). Chemical protein synthesis has also been used to develop candidate protein therapeutic molecules with improved properties (21, 22).
Thus far, most research on chemically synthesized protein molecules (16) has been focused on relatively small proteins in the size range of 50 to ≈150 aa (i.e., made from two or three peptide segments). This size limitation is the result of two phenomena: (i) practical constraints on the size of the unprotected peptide segment building blocks (23) and (ii) technical challenges in the chemical ligation of more than two or three peptide segments. Even highly optimized stepwise solid-phase peptide synthetic procedures are limited to ≈50 amino acid residues for the practical preparation of high-purity unprotected peptides (24). Thus, the chemical synthesis of a protein of typical size (≈300 aa) (25) would require the use of at least six synthetic peptide segments as building blocks.
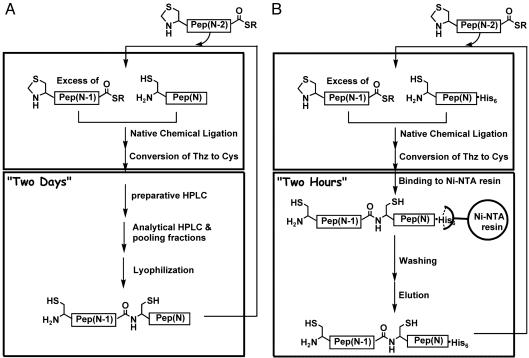
The most effective way of covalently joining unprotected peptide segments to form a protein molecule is native chemical ligation (3, 26). Native chemical ligation involves the reaction of a peptide-αthioester with a Cys-peptide; reversible thioesterthiol exchange with the N-terminal Cys residue gives a thioester-linked intermediate that undergoes an irreversible intramolecular rearrangement to give a near-quantitative yield of a single product linked by a native peptide bond at the ligation site. The native chemical ligation reaction is both practical and highly effective.¶ Each ligation product can be purified by reverse-phase HPLC and characterized with great precision by electrospray MS. In principle, the ability to purify and characterize intermediate products at each successive stage of construction of a protein molecule is one of the major advantages of the chemical ligation approach to total protein synthesis; it ensures accurate construction of high-purity protein molecules. However, in practice, such purification and characterization are arduous: the consecutive chemical ligation of several peptide segments involves multiple laborious purifications carried out by reverse-phase HPLC (see Fig. 1A). The repetitive HPLC purifications result in significant handling losses. Moreover, each of these purification steps entails time-consuming lyophilization of the product to enable solvent exchange for subsequent reactions.
Fig. 1.
Synthetic strategies (A, conventional; B, His6 tag-assisted) for the consecutive chemical ligation of several peptide segments to give a target protein/polypeptide chain. His6 tag-assisted chemical protein synthesis dramatically speeds up and facilitates the isolation and handling of intermediate ligation products (B).
In an attempt to overcome these problems, we and others have introduced methods to facilitate the handling of the intermediate products formed in the course of chemical protein synthesis. These methods include solid-phase chemical ligation (28, 29) and “one-pot” methods for the synthesis of proteins (30). In the solid-phase approach (28, 29), unprotected peptide building blocks are consecutively ligated onto a water-compatible polymer support. After each ligation reaction, excess soluble reactants and coproducts are removed by filtration and washing. When assembly of the target sequence is complete, the product polypeptide is liberated from the polymer for analysis and purification. However, significant additional chemistry is required for the preparation of the cleavable peptide–polymer linker for each new target protein (28, 29). Also, analytical control of solid-phase chemistry is not straightforward, and significant amounts of unreacted peptide byproducts accumulated after as few as three ligation steps, even with the use of a large excess of soluble peptide–thioester reactants (28). In the one-pot synthesis approach (30), a three-segment ligation synthesis and subsequent folding for the preparation of a protein molecule were carried out in one pot without purification of intermediate products; only a single final HPLC purification step was needed to produce high yields of synthetic protein of exceptional purity. The one-pot approach significantly reduced the number of manipulations required for total synthesis of a protein molecule. However, one-pot synthesis requires a near-quantitative yield for each ligation reaction and at some as-yet-undefined point, amounts of unreacted peptide segments (and byproducts derived from these segments) will accumulate sufficiently to interfere with the effective purification of the final product.
Here we report an alternative, more effective approach to the facilitation of chemical protein synthesis: the use of a C-terminal His6 “tag” to facilitate the isolation and handling of intermediate products formed in the course of chemical protein synthesis. The principles of His6 tag-assisted chemical protein synthesis are shown in Fig. 1B. Synthetic case studies reported include the total synthesis of the model protein crambin and the total synthesis of a 17-kDa modular repeat protein, tetratrico peptide repeat (TPR).
Methods
Peptide Synthesis. Peptides and peptide-αthioesters were made manually by use of stepwise Boc chemistry in situ neutralization solid-phase peptide synthesis (24) on –OCH2-Pam-resins (for free αcarboxyl peptides), on 4-methylbenzhydrylamine resin (for αcarboxamide peptides), or on HSCH2CH2CO-Leu-OCH2-Pam-resin (31) (for αthioester peptides). Products were purified by reverse-phase HPLC and were characterized by electrospray MS.
His6 Tag-Assisted Chemical Protein Synthesis. The His6 tag was attached to the C terminus of the first (i.e., C-terminal) peptide segment. The peptide-His6 construct was prepared either by stepwise assembly of the peptide on the His6 resin, followed by cleavage/deprotection, or by native chemical ligation of a peptide-αthioester with Cys-His6 to give peptide-Cys-His6 (see Supporting Text, which is published as supporting information on the PNAS web site, for more details on the chemical synthesis of peptide-His6). Subsequent ligation reactions were carried out under standard conditions (see Supporting Text) on the His6-tagged peptide (Fig. 1B). After each ligation reaction, the {product peptide}-His6 was recovered from excess reactant and nontagged coproducts by adsorption onto a Ni-NTA agarose column, followed by thorough washing. Where appropriate, the {product peptide}-His6 was eluted in ligation buffer containing 200 mM imidazole for use in the next ligation reaction. Before the next Ni-column purification, we diluted the imidazole concentration of 200 mM to 20 mM. Approximately 1 ml of Ni-NTA agarose∥ resin was used per ≈5mgof ligated peptide (see Supporting Text for details of experimental procedures). At any desired stage of the synthesis, aliquots can be removed for analytical control by HPLC-MS.
Protein Folding/Disulfide Formation. While adsorbed to the Ni-agarose support, the final full-length reduced crambin polypeptide was folded with a concomitant formation of disulfides. After adsorption, the support was washed with folding buffer (pH 8 Tris buffer containing 2 M guanidinium chloride, 8 mM cysteine, and 1 mM cystine) to remove excess reactants and nontagged coproducts. The {crambin(SH)6}-His6-Ni-agarose equilibrated in folding buffer was allowed to stand for 30 min at room temperature, after which the folded protein {crambin-His6} was eluted with the same buffer containing 200 mM imidazole.
CD Spectroscopy and Thermal Denaturation. CD spectra were recorded in the far-UV wavelength region (250–190 nm) at 5°C. The sample was 10 μM protein/10 mM phosphate (pH 8.0) in a 1-mm path-length cuvette. For thermal denaturation experiments, the ellipticity was monitored at 222 nm, and the temperature was increased from 5°C to 100°C.
Results
Model 1: Crambin. We have previously developed synthetic routes to the protein crambin that involve the ligation of three unprotected peptide segments. Chemical ligation of the peptide segments and the folding of the full-length polypeptide have been highly optimized (30, 32). Thus, crambin was chosen as a suitable target with which to examine His6 tag-assisted chemical protein synthesis. The synthetic scheme is shown in Fig. 2A.
Fig. 2.
His6 tag-assisted total chemical synthesis of crambin-His6. i–viii from synthetic strategy (A) correspond to the same numbers in the chromatographic data (Bi–viii). Reactions were monitored by HPLC. The chromatographic separations were performed by use of a linear gradient (10–50% of buffer B in buffer A over 20 min); buffer A = 0.1% trifluoroacetic acid (TFA) in water; buffer B = 0.08% TFA in acetonitrile. See text for a detailed description of B.
Three unprotected peptide segments, (Cram[1-V15A]αthioester, [Thz16-31]Cramαthioester, and [Cys32–46]Cram-His6), were prepared as described (32), except that [Cys32–46]Cram-His6 was made on 4-methylbenzhydrylamine resin. The first ligation reaction between 6.0 μmol of Cram[Thz16-31]αthioester and 4.7 μmol of [Cys32–46]Cram-His6 was carried out in 4 ml of pH 7.3 100 mM phosphate buffer containing 6 M guanidinium chloride and 1% thiophenol [analytical data shown in Fig. 2B(i)]. After completion of the ligation reaction, a very small amount of Cram[Cys32–46]-His6 remained (Fig. 2Bii; elution time, 8 min), together with a larger amount of unreacted [Thz16-31]Cramαthioester in the form of an internal thiolactone (elution time, 9.5 min). The pH of the solution was reduced to ≈4 by addition of 0.2 M methoxyamine·hydrochloride to effect the conversion of Thz-peptide (mass 3,976.2 ± 0.3 Da) to Cys-peptide (mass 3,964.4 ± 0.3 Da) (Fig. 2Biii).
For affinity purification on a Ni-agarose column, the pH of the solution was adjusted to ≈6.1–6.5.** After the binding of His6-tagged peptides to the Ni-NTA agarose column, unreacted peptide-αthioester and nontagged coproducts were washed away with buffer. The column effluent was analyzed by HPLC to make sure that no ligated product was present (data not shown). Subsequent elution of the {product peptide}-His6 was carried out by use of the ligation buffer containing 200 mM imidazole. The eluted {product peptide}-His6 solution (Fig. 2Biv) has all of the necessary components for the next ligation step.
The second ligation reaction was initiated by the addition of 5.8 μmol of Cram[1-V15A]αthioester to the mixture eluted from the Ni-column (Fig. 2Bv). Reaction for 20 h resulted in formation of the second ligation product (Fig. 2Bvi; elution time 14 min). The Cram[1–46]-His6 was recovered by passage over a fresh Ni-agarose column. After adsorption to the Ni-agarose and washing, folding/disulfide formation was carried out on the column by buffer exchange to a folding buffer (pH 8, 100 mM Tris containing 2 M guanidinium chloride, 8 mM cysteine, and 1 mM cystine). After 30 min standing in folding buffer, elution was performed with the same buffer containing 200 mM imidazole (Fig. 2Bvii; note the purification effected by adsorption and washing). The folded disulfide-containing protein crambin-His6 eluted at 14.5 min on reverse-phase HPLC, later than the reduced polypeptide crambin(SH)6-His6 at 14 min, reflecting the more hydrophobic nature of the folded protein molecule (30). Crambin-His6 had a measured mass 6.1 ± 0.2 Da less than that of the reduced polypeptide, reflecting the formation of the expected three disulfide bridges in the folded protein. The synthetic crambin-His6 protein was purified by reverse-phase HPLC (Fig. 2Bviii), and 0.75 μmol of the His6 tagged crambin was obtained. Overall yield was 16%, based on the limiting C-terminal peptide.
Previously, we reported a 40% yield for the synthesis of crambin by a three-segment ligation one-pot strategy (30). The relatively low yield for the synthesis of crambin by His6 tag-assisted ligation is largely due to the less efficient folding steps†† (Fig. 2Bvii).
Model 2: Modular Repeat Protein TPR. To examine the utility of His6 tag-assisted chemical protein synthesis for the chemical ligation of more than three peptide segments, we chose a repeat protein as a synthetic target. Repeat proteins are formed from multiple copies of a motif of between ≈20 and 40 aa (34).‡‡ After examination of several repeat proteins, we chose to make a repeat protein based on the 34-amino-acid-residue peptide repeat (TPR).§§ The synthetic strategy is shown in Fig. 3A.
Fig. 3.
His6 tag-assisted total chemical synthesis of the modular repeat protein TPR. i–x in the synthetic strategy (A) correspond to the same numbers in the chromatographic data (B i–x). Reactions were monitored by HPLC. The chromatographic separations were performed by use of the same gradient as in the crambin synthesis. CD spectra (Bxi) were recorded in the far-UV wavelength region (250–190 nm) at 5°C. See Supporting Text for a detailed description of B.
Peptide Design and Synthesis. Our initial TPR motif target sequence was the 34-amino-acid-residue peptide Thz-AWYNLGNAYYKQGDYDEA IEYYQKA LELDPNNA-αthioester. An N-terminal Thz (30) was incorporated to enable the use of native chemical ligation at cysteine, with the intent that the resulting Cys residues at the ligation sites would be carboxamidomethylated to give pseudoGln residues (22) after construction of the TPR. Boc chemistry SPPS of this initial peptide sequence gave a product with a series of –17-Da byproducts. Subsequent analysis identified dehydration of the Asn side chain as the culprit. Thus, several Asn residues were replaced by the next most common amino acid in the global propensity data for that amino acid position of the TPR motif.§§ The sequence was redesigned for synthetic convenience and was Thz-AWYNLGHAYYKLGDYDEAIEYYQKALELDPDNA-αthioester (Thz-TPR[W]-αthioester). By use of this sequence, we were able to obtain a much-improved synthetic product. We also prepared a second TPR motif peptide Thz-AYYNLGHAYYKLGDYDEA IEYYQKALELDPDNA-αthioester (Thz-TPR[Y]-αthioester).¶¶ A third peptide containing the necessary C-terminal His6 tag CAWYRLGHAYYKLGDYDEA IEYYQKA LELDPDDHHHHHH (Cys-TPR[W]-His6) was also synthesized. All peptides were purified by reverse-phase HPLC and characterized by electrospray MS.
Ligations and Ni-NTA Agarose Column Purifications (Fig. 3). Data from the ligation reactions and the Ni-column purifications are shown in Fig. 3. The peptide segment Thz-TPR[Y]-αthioester was reacted with Cys-TPR[W]-His6 under standard ligation conditions as described above for crambin (Fig. 3Bi and ii) (see Supporting Text for more details about reaction scale and conditions). After the reaction was complete, 0.2 M methoxyamine·hydrochloride was added to effect the conversion of Thz- to Cys-peptide at pH 4. After readjustment to pH 6.3, the His6-tagged peptides were adsorbed onto the Ni-agarose column, and unreacted peptide-thioester and nontagged coproducts were washed away with buffer. Elution of the Cys-{product peptide}-His6 was carried out by use of the ligation buffer containing 200 mM imidazole (Fig. 3Biii).
The second and third ligation reactions were performed in a similar fashion by the addition of Thz-TPR[W]-αthioester and Thz-TPR[Y]-αthioester peptides to the Cys-peptide-His6 products eluted from Ni-agarose column (Fig. 3B iv and v for the second ligation and vii for third ligation). It was necessary to dilute the reaction mixture 10-fold to reduce the imidazole concentration to 20 mM before Ni-agarose column purification, as described above. After completion of the third ligation (Fig. 3Bviii), the full-length 142-residue polypeptide was carboxamidomethylated [i.e., converting Cys to pseudoGln (ψQ)] by adding 1 vol of pH 7.3 phosphate buffer containing 0.1 M 2-bromoacetamide to the crude ligation mixture. The desired carboxamidomethylated repeat protein Thz-TPR[Y]-ψQ-TPR[W]-ψQ-TPR[Y]-ψQ-TPR[W]-His6 was recovered by adsorption to a Ni-agarose column and subsequent elution (Fig. 3Bix). The product was further purified by preparative HPLC (Fig. 3Bx) and lyophilized.
The overall yield of purified repeat protein was 8%, based on the limiting C-terminal peptide Cys-TPR[W]-His6. The TPR protein had the expected molecular mass (observed mass, 16,941 ± 2 Da; calculated mass, 16,942.2 Da). CD spectroscopy of the synthetic protein exhibited a typical α-helical signature in the far-UV region (Fig. 3Bxi). Thermal denaturation data are given in Fig. 4, which is published as supporting information on the PNAS web site.
Discussion
The synthesis of crambin-His6 illustrates the straightforward nature of the manipulations required to ligate peptide segments by use of His6 tag-assisted protocols. However, the crambin synthesis made use of only three peptide segments (two ligations). Thus, we examined the broader potential of this technique for the modular assembly of proteins by synthesis of a repeat protein TPR from four unprotected peptide segments.
The synthesis of TPR illustrates several important features of our His6 tag-assisted synthesis methodology. First, the modular assembly of an ≈17-kDa protein could be performed with simple manipulations in a matter of days. The main time savings is in the rapid affinity purification (2 h) compared with HPLC/lyophilization (2 days) for each ligation or other reaction in the synthesis (see Fig. 1). The Ni-NTA agarose column purification was significantly less laborious than preparative HPLC and also did not require a time-consuming lyophilization step to effect solvent exchange for the next reaction. Second, the use of simple affinity column purification after each ligation allowed us to drive the reactions to completion by use of a modest (1.2- to 1.5-fold) excess of peptide-αthioester reagent. Overall recoveries of synthetic proteins were good. The synthesis of TPR included: three native chemical ligation reactions, two conversion reactions of Thz- to Cys-peptide, one alkylation reaction of three Cys residues, three Ni-agarose column purification steps, and one preparative HPLC purification, for a total of 10 steps. The overall recovery of final product corresponds to ≈80% yield for each reaction/handling step. The final yield of desired product (≈8%) was particularly impressive, because it was obtained from a synthesis carried out on an unusually small scale (only ≈5 mg of limiting peptide starting material). Conventional chemical protein synthesis on such a scale would quickly result in complete loss of product because of low recovery at each step. The third advantage of His6 tag-assisted synthesis is that analytical control by HPLC on each reaction step was readily performed, because the desired products in the each step were present in the solution phase [in contrast to the situation in solid-phase chemical ligation (28, 29)].
Conclusion
The His6 tag-assisted chemical protein synthesis method described here works and is useful, as illustrated by the total syntheses of crambin and the repeat protein TPR. The combination of native chemical ligation and simple purification on Ni-agarose columns enables facile buffer exchange to new reaction conditions and is compatible with direct analytical control by modern protein MS methods. Also, the reduced handling losses from affinity purifications give significantly improved overall yields.
Consecutive assembly of unprotected peptide building blocks by means of His6 tag-assisted chemical ligation has enabled the straightforward preparation of an ≈17-kDa protein from four peptide segments. We expect that the assembly of proteins from up to six peptide segments will be straightforward with His6 tag-assisted consecutive chemical ligations, giving high-purity products with an estimated overall yield of 3% to ≈4%. If needed, a single intermediate HPLC purification can be used at an intermediate point in the synthesis. The synthesis of molecules in this size range will provide general access to the world of protein domains, the modular building blocks of function in biology, and will enable the synthesis of protein molecules of the typical size (≈300 amino acid residues) found in nature.
The work described here is not the final word in chemical protein synthesis. Even with His6 tag-assisted methods, there are limits to the size of polypeptide chain that can be prepared by consecutive assembly of peptide segments. For a truly practical total synthesis of proteins of typical size (≈35 kDa) from six to eight peptide building blocks, a convergent synthetic approach (36) will be needed to avoid unacceptably low yields and consequent ambiguity of product structures and identities.
Supplementary Material
Acknowledgments
We thank Dr. Wendy Gordon for helpful discussions. We acknowledge support from the National Science Foundation Materials Research Science and Engineering Centers Program (Grant DMR-0213745) at the University of Chicago and from the Department of Energy Genomes to Life Genomics Program (Grant DE-FG02-04ER63786).
Author contributions: D.B. and S.K. designed research; D.B. performed research; D.B. and S.K. analyzed data; and D.B. and S.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: TPR, tetratrico peptide repeat.
Footnotes
Note that the native chemical ligation reaction itself has been shown to have undiminished effectiveness for the reaction of even very large protein molecules (27).
The binding capacity of Ni-NTA agarose (Qiagen, Chatsworth, CA) is 5–10 mg (0.3–0.5 μmol for ≈20-kDa protein) per milliliter of swollen support.
At the low pH used in the Thz to Cys conversion reaction, the peptide-His6 does not adsorb to the Ni-agarose column.
The folding of the crambin[1–46]-His6 polypeptide is significantly less efficient than the folding of crambin[1–46]αCOO– (30), presumably because of the absence of the salt bridge between crambinαcarboxylate and the side chain guanidinium of Arg10, which is believed to be important for crambin folding and stability (33). With the αcarboxylate, crambin folds quantitatively (30). We have previously observed poor folding yields (≈50%) arising from the perturbation of the salt bridge by constructing a crambinαcarboxamide polypeptide (unpublished data). We note the low folding yield is not coming from the folding on Ni-agarose resin. To show that the low folding yield is not due to on-resin folding, we prepared reduced polypeptide crambin(SH)6-His6, as described above. The reaction mixture was divided into two equal portions. We folded/disulfide-formed and purified the half of the polypeptide product mixture in the Ni-agarose resin, as described above. The other half of the polypeptide product mixture was folded/disulfide-formed in solution, as described (30). Folding efficiencies were poor for the both conditions and were essentially the same.
There are ≈20 classes of repeat proteins that are found throughout nature. Usually, the number of repeats ranges from 3 to ≈20 (34). Thus, repeat proteins are good model systems in which to examine His tag-assisted modular chemical protein synthesis, because the synthesis of a repeat motif can be performed from a single peptide-thioester. Also, any desired number of ligation reactions can be performed.
The Regan group has successfully constructed one, two, and three TPR repeats by recombinant means (35). In their work, the amino acid sequence of the repeat motif was selected by statistical analysis of 1,837 different TPR motifs from 107 proteins. The global amino acid propensity for each position in the TPR motif was assigned. We used these propensity data to design the peptide sequences used in the current work.
Thz-TPR[Y]-αthioester was prepared because a mass difference of TPR[Y] and TPR[W] enabled us to determine by MS that each desired ligation reaction had been carried out.
References
- 1.Schnolzer, M. & Kent, S. B. (1992) Science 256, 221–225. [DOI] [PubMed] [Google Scholar]
- 2.Dawson, P. E. & Kent, S. B. (1993) J. Am. Chem. Soc. 115, 7263–7266. [Google Scholar]
- 3.Dawson, P. E., Muir, T. W., Clark-Lewis, I. & Kent, S. B. (1994) Science 266, 776–779. [DOI] [PubMed] [Google Scholar]
- 4.Rose, K. (1994) J. Am. Chem. Soc. 116, 30–33. [Google Scholar]
- 5.Liu, C.-F. & Tam, J. P. (1994) J. Am. Chem. Soc. 116, 4149–4153. [Google Scholar]
- 6.Englebretsen, D. R., Garnham, B. & Alewood, P. F. (2002) J. Org. Chem. 67, 5883–5890. [DOI] [PubMed] [Google Scholar]
- 7.Melnyk, O., Bossus, M., David, D., Rommens, C. & Gras-Masse, H. (1998) J. Pept. Res. 52, 180–184. [DOI] [PubMed] [Google Scholar]
- 8.Muir, T. W., Sondhi, D. & Cole, P. A. (1998) Proc. Natl. Acad. Sci. USA 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxon, E., Armstrong, J. I. & Bertozzi, C. R. (2000) Org. Lett. 2, 2141–2143. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson, B. L., Kiessling, L. L. & Raines, R. T. (2000) Org. Lett. 2, 1939–1941. [DOI] [PubMed] [Google Scholar]
- 11.Yan, L. Z. & Dawson, P. E. (2001) J. Am. Chem. Soc. 123, 526–533. [DOI] [PubMed] [Google Scholar]
- 12.Low, D. W., Hill, M. G., Carrasco, M. R., Kent, S. B. & Botti, P. (2001) Proc. Natl. Acad. Sci. USA 98, 6554–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gieselman, M. D., Xie, L. & van Der Donk, W. A. (2001) Org. Lett. 3, 1331–1334. [DOI] [PubMed] [Google Scholar]
- 14.Quaderer, R., Sewing, A. & Hilvert, D. (2001) Helv. Chim. Acta 84, 1197–1206. [Google Scholar]
- 15.Rostovtsev, V. V., Green, L. G., Fokin, V. V. & Sharpless, K. B. (2002) Angew. Chem. Int. Ed. 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, P. E. & Kent, S. B. (2000) Annu. Rev. Biochem. 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 17.Beligere, G. S. & Dawson, P. E. (2000) J. Am. Chem. Soc. 122, 12079–12082. [Google Scholar]
- 18.Baca, M. & Kent, S. B. (2000) Tetrahedron 56, 9503–9513. [Google Scholar]
- 19.Blankenship, J. W. & Dawson, P. E. (2003) J. Mol. Biol. 327, 537–548. [DOI] [PubMed] [Google Scholar]
- 20.Lu, W. Y., Starovasnik, M. A., Dwyer, J. J., Kossiakoff, A. A., Kent, S. B. & Lu, W. (2000) Biochemistry 39, 3575–3584. [DOI] [PubMed] [Google Scholar]
- 21.Wilken, J., Hoover, D., Thompson, D. A., Barlow, P. N., McSparron, H., Picard, L., Wlodawer, A., Lubkowski, J. & Kent, S. B. (1999) Chem. Biol. 6, 43–51. [DOI] [PubMed] [Google Scholar]
- 22.Kochendoerfer, G. G., Chen, S. Y., Mao, F., Cressman, S., Traviglia, S., Shao, H., Hunter, C. L., Low, D. W., Cagle, E. N., Carnevali, M., et al. (2003) Science 299, 884–887. [DOI] [PubMed] [Google Scholar]
- 23.Kent, S. B. (1988) Annu. Rev. Biochem. 57, 957–989. [DOI] [PubMed] [Google Scholar]
- 24.Schnolzer, M., Alewood, P., Jones, A., Alewood, D. & Kent, S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193. [DOI] [PubMed] [Google Scholar]
- 25.Berman, A. L., Kolker, E. & Trifonov, E. N. (1994) Proc. Natl. Acad. Sci. USA 91, 4044–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson, P. E., Churchill, M. J., Ghadiri, M. R. & Kent, S. B. H. (1997) J. Am. Chem. Soc. 119, 4325–4329. [Google Scholar]
- 27.Muir, T. W. (2003) Annu. Rev. Biochem. 72, 249–289. [DOI] [PubMed] [Google Scholar]
- 28.Canne, L. E., Botti, P., Simon, R. J., Chen, Y., Dennis, E. A. & Kent, S. B. (1999) J. Am. Chem. Soc. 121, 8720–8727. [Google Scholar]
- 29.Brik, A., Keinan, E. & Dawson, P. E. (2000) J. Org. Chem. 65, 3829–3835. [DOI] [PubMed] [Google Scholar]
- 30.Bang, D. & Kent, S. B. (2004) Angew. Chem. Int. Ed. 43, 2534–2538. [DOI] [PubMed] [Google Scholar]
- 31.Hackeng, T. M., Griffin, J. H. & Dawson, P. E. (1999) Proc. Natl. Acad. Sci. USA 96, 10068–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bang, D., Neeraj, C. & Kent, S. B. (2004) J. Am. Chem. Soc. 126, 1377–1383. [DOI] [PubMed] [Google Scholar]
- 33.Arnold, F. H. (1988) Protein Eng. 2, 21–25. [DOI] [PubMed] [Google Scholar]
- 34.Main, E. R., Jackson, S. E. & Regan, L. (2003) Curr. Opin. Struct. Biol. 13, 482–489. [DOI] [PubMed] [Google Scholar]
- 35.Main, E. R., Xiong, Y., Cocco, M. J., D'Andrea, L. & Regan, L. (2003) Structure (London) 11, 497–508. [DOI] [PubMed] [Google Scholar]
- 36.Warren, J. D., Miller, J. S., Keding, S. J. & Danishefsky, S. J. (2004) J. Am. Chem. Soc. 126, 6576–6578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.