Abstract
BACKGROUND
The DiGeorge syndrome, the most common of the microdeletion syndromes, affects multiple organs, including the heart, the nervous system, and the kidney. It is caused by deletions on chromosome 22q11.2; the genetic driver of the kidney defects is unknown.
METHODS
We conducted a genomewide search for structural variants in two cohorts: 2080 patients with congenital kidney and urinary tract anomalies and 22,094 controls. We performed exome and targeted resequencing in samples obtained from 586 additional patients with congenital kidney anomalies. We also carried out functional studies using zebrafish and mice.
RESULTS
We identified heterozygous deletions of 22q11.2 in 1.1% of the patients with congenital kidney anomalies and in 0.01% of population controls (odds ratio, 81.5; P=4.5×10−14). We localized the main drivers of renal disease in the DiGeorge syndrome to a 370-kb region containing nine genes. In zebrafish embryos, an induced loss of function in snap29, aifm3, and crkl resulted in renal defects; the loss of crkl alone was sufficient to induce defects. Five of 586 patients with congenital urinary anomalies had newly identified, heterozygous protein-altering variants, including a premature termination codon, in CRKL. The inactivation of Crkl in the mouse model induced developmental defects similar to those observed in patients with congenital urinary anomalies.
CONCLUSIONS
We identified a recurrent 370-kb deletion at the 22q11.2 locus as a driver of kidney defects in the DiGeorge syndrome and in sporadic congenital kidney and urinary tract anomalies. Of the nine genes at this locus, SNAP29, AIFM3, and CRKL appear to be critical to the phenotype, with haploinsufficiency of CRKL emerging as the main genetic driver. (Funded by the National Institutes of Health and others.)
Deletions on chromosome 22Q11.2 are the most common cause of the DiGeorge syndrome (Online Mendelian Inheritance in Man [OMIM] number, 188400) and the velocardiofacial syndrome (OMIM number, 192430) and constitute the most common micro-deletion disorder in humans, with an estimated prevalence of 1 in 2000 to 4000 live births.1–3 The DiGeorge syndrome is a debilitating, multisystemic condition that features (with variable expressivity) cardiac malformations, velopharyngeal insufficiency, hypoparathyroidism with hypocalcemia, and thymic aplasia with immune deficiency. Additional phenotypes include neurodevelopmental defects and urogenital malformations.4–7 The long arm of chromosome 22 contains multiple segmental duplications (low-copy repeats) that confer a predisposition to genomic rearrangements.8–10 Most frequently, the DiGeorge syndrome is caused by a de novo heterozygous deletion of approximately 2.5 mb in length on chromosome 22q11.2 between low-copy repeats (LCR22) A and D. Less frequently, the syndrome is the result of deletions between LCR22 A and B, between B and D, or between C and D.5,8,11
Congenital kidney and urinary tract anomalies are present in approximately 30% of the patients with the DiGeorge syndrome.4,6,12,13 Although some of the hallmarks of this syndrome (e.g., heart defects) can be attributed in part to haploinsufficiency of TBX1,14–18 the identity of the genes that are responsible for such congenital kidney and urinary tract anomalies remains unknown.
METHODS
STUDY SAMPLES
We studied samples obtained from 2666 patients affected by congenital kidney and urinary tract anomalies at 26 international centers, along with additional samples provided by the Chronic Kidney Disease in Children Study (see the Methods section and Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). We performed genomewide genotyping for analysis of copy-number variations in 2080 of these samples. Among an additional 586 patients with congenital kidney and urinary tract anomalies, we performed either whole-exome sequencing (in 60 samples) or targeted next-generation sequencing and Sanger validation (in 526 samples). All the patients provided written informed consent. The study was approved by the institutional review board at each site. (Descriptions of the patients, analyses of convolution defects in zebrafish, analysis of tissue localization in the patients and zebrafish, and the generation and analysis of a mouse model are provided in the Methods section in the Supplementary Appendix.)
GENETIC ANALYSES
Using samples obtained from 2080 patients with congenital kidney and urinary tract anomalies and 22,094 controls, we performed genomewide genotyping for analysis of copy-number variation by means of high-density single-nucleotide polymorphism (SNP) microarrays manufactured by Illumina (1820 samples) or Affymetrix (260 samples), as described previously.19–21 We also performed whole-exome sequencing on samples obtained from 60 patients through the Yale Center for Mendelian Genomics, as described previously.22–24 We performed high-throughput next-generation sequencing for eight genes in the 370-kb minimal region of overlap for the DiGeorge syndrome in samples obtained from an additional 526 patients using microfluidic polymerase-chain-reaction capture (Fluidigm) coupled with next-generation sequencing on an Illumina 2500 HiSeq system, as described previously.25,26 We subjected CRKL coding exons to next-generation resequencing in samples obtained from 576 unaffected controls and from 1152 patients affected by IgA nephropathy but with normal results on renal ultrasonography. These additional 1728 controls were matched with the patients according to their ancestral origin and recruitment site.
RESULTS
PATIENTS WITH 22Q11.2 DELETIONS
In a genomewide search for rare copy-number variations in a discovery cohort of 1752 patients with congenital kidney and urinary tract anomalies, we identified deletions at the chromosome 22q11.2 locus in 11 patients (0.6%) and in 3 of 22,094 population controls (0.01%; odds ratio for patients versus controls, 46.4; P = 9.7×10−11). An analysis of breakpoints indicated that all deletions in the 11 patients overlapped with the common deletion between LCR22 A and D: 2 patients had the classic deletion of DNA between A and D, 1 patient had a smaller deletion (bounded by B and D), and 8 patients had the smallest deletion, between C and D (Table 1 and Fig. 1, and Table S2 in the Supplementary Appendix).
Table 1.
Characteristics of 14 Patients with Microdeletions at the 22q11.2 Locus Associated with the DiGeorge Syndrome.*
| Patient No. | Deletion Type† |
Kidney Phenotype |
Kidney Location |
Urinary Tract Phenotype |
Extrarenal Phenotype |
Sex | Nationality | Age at Diagnosis |
Outcome |
|---|---|---|---|---|---|---|---|---|---|
| Discovery cohort | |||||||||
| P1 | A–D | Renal hypodysplasia (right), cortical cyst (left) | Bilateral | None | None | M | French | 14 days | Normal renal function |
| P2 | A–D | Renal agenesis | Right | None | Developmental delay | M | Italian | 10 yr | Normal renal function |
| P3 | B–D | Normal renal parenchyma | Left | Left ureterovescical junction obstruction, hypospadia | Arched palate, mild antimongoloid slants | M | Macedonian | 3 mo | Mild renal insufficiency |
| P4 | C–E | Renal agenesis | Left | Vesicoureteral reflux, megaureter | Ventricular septal defect | M | French | 3 mo | Mild renal insufficiency, microalbuminuria |
| P5 | C–D | Renal hypodysplasia, cortical cysts | Left | Left megaureter | Undescended testis, epicanthus, chest hemangioma | M | Czech | 1 mo | Normal renal function |
| P6 | C–D | Renal agenesis | Bilateral | None | None | M | French | Prenatal | Termination of pregnancy |
| P7 | C–D | Reflux nephropathy | None | Vesicoureteral reflux | None | F | European American | 16 yr | Mild renal insufficiency |
| P8 | C–D | Renal hypodysplasia | Left | None | None | F | Polish | 9 yr | Normal renal function |
| P9 | C–D | Renal hypodysplasia and scarring | Bilateral | Ureteropelvic junction obstruction, vesicoureteral reflux, bladder diverticuli | Phimosis | M | Italian | Birth | Chronic kidney disease |
| P10 | C–D | Renal agenesis | Left | Vesicoureteral reflux | Undescended testis | M | Swiss | Birth | Normal renal function |
| P11 | C–D | Renal agenesis | Left | None | None | F | Italian | 21 yr | Normal renal function |
| Replication cohort | |||||||||
| RP1 | A–D | Renal agenesis | Right | None | None | M | Polish | 8 yr | Normal renal function |
| RP2 | B–D | Renal agenesis | Left | Vesicoureteral reflux | None | F | Polish | 1 yr | Normal renal function |
| RP3 | B–D | Renal hypodysplasia | Left | Vesicoureteral reflux | None | M | Polish | 4 mo | Normal renal function |
A total of 1752 patients were included in the discovery cohort and 328 patients in the replication cohort. F denotes female, M male, and RP patients in the replication cohort.
According to the megabase coordinates from the Human Genome 19 release, the boundaries for the chromosome 22q11.2 deletions are from 18.9 to 21.5 mb for the region between LCR22 A and D, 20.7 to 21.5 mb for the region between B and D, 21.1 to 21.5 mb for the region between C and D, and 21.0 to 22.1 mb for the region between C and E.
Figure 1. Genomic Organization of Chromosome 22q11.2 and the Deletions Associated with Kidney and Urinary Tract Malformations Identified in This Study.
In approximately 90% of the patients with the DiGeorge syndrome, the congenital disorder is caused by a classic de novo heterozygous deletion of approximately 2.5 mb in length spanning chromosome 22q11.2 low-copy repeats (LCR22) A and D, as shown in blue. Less than 10% of the patients with this syndrome carry the critical 1.5-mb deletion between LCR22 A and B. Shown in red are deletions that were identified in 14 patients who were affected by congenital anomalies of the kidney and urinary tract among the 2080 patients who were tested. According to the megabase coordinates for the Human Genome 19 release, the proximal and distal breakpoints for the chromosome 22q11.2 deletions that were identified in the patients are as follows: P1, 18.88 to 21.47 mb; P2, 18.89 to 21.47 mb; P3, 20.73 to 21.46 mb; P4, 21.02 to 22.47 mb; P5, 21.05 to 21.47 mb; P6, 21.06 to 21.47 mb; P7, 21.06 to 21.46 mb; P8, 21.06 to 21.46 mb; P9, 21.07 to 21.46 mb; P10, 21.08 to 21.47 mb; P11, 21.09 to 21.47 mb; Patient 1 from the replication cohort (RP1), 18.88 to 21.46 mb; RP2, 20.74 to 21.46 mb; and RP3, 20.74 to 21.46 mb. The deletion between LCR22 C and D defines the smallest region of overlap for congenital kidney disease among patients with 22q11.2 deletions.
Of the 11 patients, 9 had renal agenesis or hypodysplasia, and 2 had an isolated ureteric phenotype, findings indicating that the 22q11.2 locus between LCR22 C and D is critical for human nephrogenesis and is possibly specific for renal agenesis or hypodysplasia (in 9 of 765 patients [1.2%]). In a replication study involving an additional 328 patients with renal agenesis or hypodysplasia, we identified 3 (0.9%) with 22q11.2 deletions, for a total of 14 patients with these deletions (Table 1 and Fig. 1). Taken together, we identified deletions at this locus in 12 of 1093 patients (1.1%) with renal agenesis or hypodysplasia, as compared with 3 of 22,094 controls (odds ratio, 81.5; P=4.5×10−14), which implicates deletions at the locus associated with the DiGeorge syndrome as the second most common genomic disorder of the kidney and urinary tract after the 17q12 microdeletion associated with the renal cysts and diabetes syndrome (Table S2 in the Supplementary Appendix).19,27
Of the 14 patients with the 22q11.2 deletion, Patients P1, P2, and replication Patient 1 (RP1) carried the most frequent deletion between LCR22 A and D; in Patient P2, the deletion was inherited from the mother, in whom a clinical diagnosis of the DiGeorge syndrome had not been made. In all the patients, the molecular genetic diagnosis preceded a clinical diagnosis of the DiGeorge syndrome (in which some but not all features of the syndrome were observed) and had a direct effect on the patient’s treatment. In patients with deletions between LCR22 B and D and C and D, additional urinary tract defects consisted of vesicoureteral reflux in 6 patients and hypospadia in 1 patient. Extrarenal defects were rare and mild in patients with deletions between LCR22 B and D and C and D. The deletion between LCR22 C and D that was identified in Patient P10 was also observed in a sibling who was affected by left renal agenesis and an undescended testis.
The analysis of the breakpoints in copy-number variation that was based on SNP array data localized the critical region for the phenotype associated with congenital kidney and urinary tract anomalies to a locus of approximately 370 kb, which contains nine genes (Fig. 1, and Table S3 in the Supplementary Appendix). This region excluded the gene encoding T-box 1 (TBX1), a protein that is not expressed in the murine embryonic kidney,28 so Tbx1-null mice have normal early nephrogenesis (Fig. S2 in the Supplementary Appendix). Interrogation of the “22q and You” database from the Children’s Hospital of Philadelphia identified kidney malformations in 2 of 10 patients with the 22q11.2 deletion between LCR22 C and D (Table S4 in the Supplementary Appendix). Finally, we reexamined the three controls with 22q11.2 deletions; one carried the typical deletion between LCR22 A and D, one the deletion between B and D, and one the deletion between C and D. We obtained clinical records for Control C1, who had Parkinson disease, congenital hypoparathyroidism, and advanced chronic kidney disease (Table S5 in the Supplementary Appendix). Thus, we found a patient with undiagnosed DiGeorge syndrome with renal involvement among our 22,000 population controls, which provided further support for the pathogenicity of the 22q11.2 deletion in patients with congenital kidney and urinary tract anomalies. After removal of this patient from the control data set, the strength of association between 22q11.2 deletions and renal agenesis or hypodysplasia increased further (12 of 1093 patients vs. 2 of 22,093 controls, P=8.5×10−15; odds ratio, 123.7).
FUNCTIONAL MODELING IN ZEBRAFISH
The genetic data suggested that dosage perturbation of one or more of the nine genes in the micro-deletion on 22q11.2 is a driver of congenital kidney and urinary tract anomalies. We had previously found that systematic in vivo suppression of experimentally tractable genes within a deletion copy-number variant, coupled with quantitative phenotyping, can determine the contribution of specific transcripts to disease associated with copy-number variation in humans.29–31
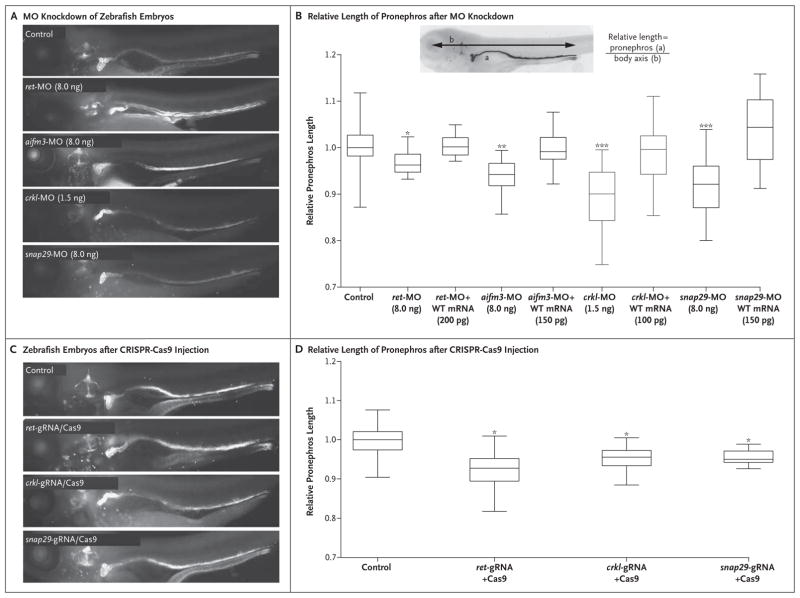
We first sought to establish a phenotypic surrogate for congenital kidney and urinary tract anomalies in zebrafish embryos. Previous studies in mice and humans have shown the critical role of the gene encoding ret proto-oncogene (RET) for kidney development and branching morphogenesis.32–35 We therefore injected an established morpholino oligonucleotide (MO) against RET 36 into zebrafish that were engineered to enable visualization of the developing nephron and then examined the convolution of the pronephros (the earliest developmental stage in the zebrafish) at 4.5 days after fertilization.37 The injection of 8.0 ng of a splice-blocking MO, which suppressed approximately 80% of wild-type message and induced the inclusion of intron 2, followed by staining of embryos with an antibody against sodium–potassium ATPase, induced convolution defects of the proximal pronephros and an overall reduction in the length of the tubules (Fig. S3 in the Supplementary Appendix). We captured this phenotype by measuring the length of the tubule corrected for the overall length of the embryonic body axis, thus controlling for possible developmental delay due to the mechanical manipulation of embryos (P<0.05 for all comparisons between MO knockdown and wild type) (Fig. 2A and 2B). This phenotype was specific; not only were we able to rescue this anomaly by coinjection of 200 pg of human capped RET messenger RNA (mRNA) (Fig. 2B), but deletions at this locus that were mediated by CRISPR–Cas9 also reproduced this anomaly in a manner indistinguishable from the MO, both qualitatively and quantitatively (Fig. 2C and 2D). We therefore proceeded to deploy this assay across all testable genes within the region of copy-number variation.
Figure 2. Functional Modeling of the DiGeorge Syndrome Terminal Deletion Genes Associated with Kidney and Urinary Tract Malformations.
Panel A shows zebrafish larvae 4.5 days after fertilization, in which the proximal tubule is folded into a hairpin structure, displaying proper anterior convolution in noninjected control embryos (staining with antibody against sodium–potassium ATPase). Knockdown of ret, aifm3, crkl, and snap29 by the injection of 8.0 ng of a splice-blocking morpholino oligonucleotide (MO) against RET resulted in major convolution defects, which are apparent by the failure of the anterior portion of the pronephros (the earliest developmental stage in the zebrafish) to progress, along with an overall reduction in the length of the tubules. Panel B shows the relative length of the pronephros, which was defined as the ratio of the length of the pronephros (a) to the length of the body axis (b), in individual larvae (inset). The number of replicate measurements were as follows: control or sham-injected control, 177 in Panel A and 68 in Panel B; ret-MO, 50; ret-MO+mRNA, 42; aifm3-MO, 38; aifm3-MO+mRNA, 42; crkl-MO, 43; crkl-MO+mRNA, 58; snap29-MO, 48; snap29-MO+mRNA, 39; ret-gRNA+Cas9, 44; crkl-gRNA+Cas9, 31; and snap29-gRNA+Cas9, 41). Morphant phenotypes could be rescued by the coinjection of each respective human messenger RNA (mRNA). In each box-and-whisker plot, the horizontal line represents the median, the top and bottom of the boxes the interquartile range, and the I bars the minimum and maximum values. Panel C shows embryos that have been injected with CRISPR–Cas9 and that are reproducing the convolution defects observed in the morphant embryos. Guide RNA (gRNA) that targeted each respective gene was coinjected with purified Cas9 protein, and the relative length of the pronephros was measured in founders, as shown in Panel D. In Panels B and D, a single asterisk indicates P<0.05, two asterisks P<0.01, and three asterisks P<0.001. WT denotes wild type.
First, we used the Basic Local Alignment Search Tool (BLAST) algorithm for sequence searching, in which we detected orthologues for seven of nine genes. RNA sequencing data indicated that all seven genes were expressed in the early embryo, between 2 and 4 days after fertilization.38 We therefore designed MOs to knock down the expression of these genes and injected them into zebrafish reporter lines in parallel with the ret–MO as a control. For four of the transcripts (lztr1, pi4ka, serpind1, and slc7a4) we observed no differences in convolution complexity or length of the pronephros between the knockdown zebra-fish and controls in 26 to 34 embryos, with each analysis repeated twice with blinded scoring (Fig. S4 in the Supplementary Appendix). In contrast, the suppression of crkl expression or interruption of splicing of aifm3 and snap29 phenocopied the pathologic features of RET (Fig. 2A and 2B). These phenotypes could be rescued for each of the three genes by coinjection with human mRNA (Fig. 2B). In addition, deletions of snap29 and crkl mediated by CRISPR–Cas9 on the day of fertilization induced insertions or deletions in 60 to 80% of cells within each mutant embryo (Fig. S5 in the Supplementary Appendix). (The gene aifm3 was intractable to this method.) Subsequently, the mutant fish fully reproduced the renal disease (Fig. 2C and 2D). We observed no renal phenotypes when each human mRNA was injected alone, nor did we find any other gross morphologic defects in embryos subjected to either MO knockdown or overexpression at the studied developmental time points that might indicate nonspecific toxicity. Because kidney morphogenesis could be affected by extrarenal defects (e.g., loss of cardiac output and collective cell migration of the nephron induced by loss of flow), we analyzed heart function in both ret and crkl mutants and found no effect on the morphologic features or rate of the heart. We also found no evidence of kidney cysts, which would be expected if cilia-dependent flow were to be impaired. Analysis of body length as an indication of global-developmental delay showed no significant difference between “knocked down” zebrafish and control zebrafish (Fig. S6 in the Supplementary Appendix). Thus, we concluded that the defects we observed were not due to the known indirect causes of failed nephron convolution in zebrafish and support our use of this assay as a screening technique for intrinsic kidney defects.
Previous functional dissections of copy-number variation have revealed a complex genetic architecture, in which a single driver may account for the induction of disease either alone or in cis epistasis with other genes within the copy-number variation.29–31 We tested this possibility in vivo by asking whether the three transcripts in zebrafish embryos that induce congenital kidney and urinary tract anomalies could interact genetically. For this purpose, we injected embryos with subeffective doses of each transcript, with the requirement that each dose by itself should induce modest or no disease; we then tested all possible pairwise combinations. We observed no genetic interaction between crkl and either aifm3 or snap29. In contrast, cosuppression of aifm3 with snap29 phenocopied the convolution defect of strong morphants and CRISPR mutants, which suggested a contributory role to the copy-number variation pathology. This interaction was specific and not due to toxicity caused by the presence of multiple MOs, since it could be rescued by coinjection of SNAP29 mRNA (Figs. S7 and S8 in the Supplementary Appendix).
WHOLE-EXOME AND TARGETED SEQUENCING OF CRKL
We asked whether sporadic patients with congenital kidney and urinary tract anomalies might have loss-of-function lesions in any of the nine genes included in the minimal region of overlap for the kidney defects of the DiGeorge syndrome. We first queried exome-sequencing data from 60 patients with renal agenesis or hypodysplasia. None of the genes showed excess burden of rare truncating mutations as compared with controls (Table S6 in the Supplementary Appendix). LZTR1, P2RX6, and SLC7A4 have a high frequency of loss-of-function mutations (defined as premature termination, splicing, and frameshift mutations), a prevalence that approaches or exceeds that of such anomalies in the general population. Conversely, SERPIND1, SNAP29, CRKL, and THAP7 carry loss-of-function mutations in no more than 2 of 10,000 persons. Among more than 60,500 publicly available population controls from the Exome Aggregation Consortium (ExAC) database (exac.broadinstitute.org), only 1 had a high-quality loss-of-function variant in CRKL, which ranks in the top second percentile in the genome for haploinsufficiency — in other words, there is a high probability of a detrimental effect on phenotype when only one copy of the gene is deleted. This finding suggests that loss-of-function variations in CRKL have deleterious effects on genetic fitness (Table S3 in the Supplementary Appendix).39
We also performed targeted next-generation resequencing of all 107 coding exons of PI4KA, SERPIND1, SNAP29, CRKL, AIFM3, THAP7, P2RX6, and SLC7A4 in 526 patients with renal agenesis or hypodysplasia. We identified six loss-of-function variants in 11 patients: two in SERPIND1, one in CRKL, one in AIFM3, and two in P2RX6 (in 7 patients) (Table S7 in the Supplementary Appendix). Loss-of-function mutations in SERPIND1 have been associated with a mendelian clotting disease (heparin cofactor II deficiency) that has no known associations with kidney and urinary tract development.40 In contrast, the CRKL truncating mutation, p.Q31*, was found in a patient (P13) with isolated unilateral renal agenesis and was predicted to result in haploinsufficiency. We also identified four additional missense variants that were absent from the ExAC database, that were conserved across vertebrates, and that were predicted to affect protein structure and function (Table S8 and Figs. S9 and S10 in the Supplementary Appendix).
Whole-exome sequencing of DNA obtained from P13 did not show pathogenic mutations in genes that had previously been implicated in congenital kidney and urinary tract anomalies or loss-of-function variants in newly plausible candidates (Table S9 in the Supplementary Appendix). Finally, because of the formal possibility that the discovered CRKL variants were population-specific polymorphisms, we performed targeted resequencing on samples obtained from 576 additional controls and from 1152 patients with IgA nephropathy and normal results on renal ultrasonography who were matched with our patients according to ethnic background and recruitment site. All CRKL variants were absent in the more than 60,500 population controls from the ExAC database and in the 1728 controls. Aggregating our sequencing data and performing burden tests between our 586 patients with congenital kidney and urinary tract anomalies and 33,352 European controls from ExAC or 1728 ethnically and geographically matched controls showed significant excess of rare functional CRKL variants in our patients (P = 3.7×10−3 by Fisher’s exact test for the comparison with ExAC controls; odds ratio, 5.2; and P= 4.9×10−3 for the comparison with matched controls; odds ratio, 14.8) (Table S10 in the Supplementary Appendix).
EXPRESSION AND FUNCTIONAL STUDIES OF CRKL
We performed mRNA and protein expression studies in relevant tissues and examined a mouse model with a Crkl mutation. In humans, CRKL protein showed mild, diffuse cytoplasmic expression in both ureteric bud and metanephric mesenchyme derivatives during the sixth week of fetal development (Fig. S11A in the Supplementary Appendix). At week 21, CRKL was detected only in proximal tubules and collecting tubules at the apical side of epithelial cells (Fig. S11B in the Supplementary Appendix). In the kidney of a 1.5-year-old boy, we observed abundant CRKL expression in the proximal and collecting tubules at the apical side, along with diffuse cytoplasmic signaling in glomerular endothelial cells, podocytes, Bowman’s capsule, and distal tubules (Fig. S11C in the Supplementary Appendix). The expression of SNAP29 and AIFM3, although present at very low levels in zebrafish pronephros (not shown), was seen in the urinary tract in fetuses and children (Figs. S12 and S13 in the Supplementary Appendix).
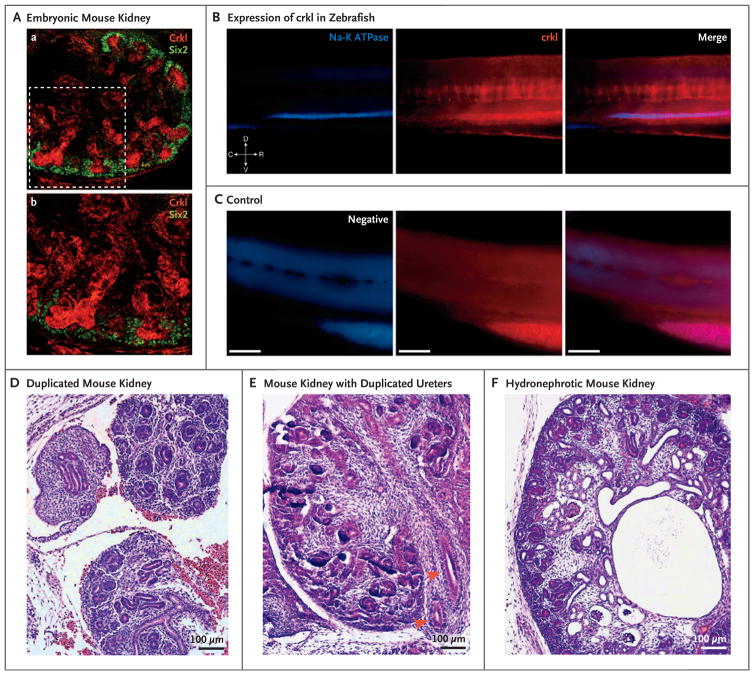
In the mouse kidney on embryonic day E15.5, Crkl showed specific expression in structures derived from the ureteric bud and, occasionally, in S-shaped bodies and developing proximal tubules (Fig. 3A, and Fig. S14 in the Supplementary Appendix). In zebrafish, crkl was specifically expressed in the pronephros (Fig. 3B and 3C). RNA studies that were performed with the use of flow cytometry and cell sorting, along with in situ hybridization, confirmed that crkl was expressed in the pronephric convoluted tubule and pronephric duct (Figs. S15 and S16 in the Supplementary Appendix).
Figure 3. Localization of Crkl in Developing Urinary Tracts in Mice and Zebrafish and Phenotypes of Crkl Knockout Mice.
Panel A shows immunostaining for Crkl in kidney obtained from a transgenic mouse on embryonic day E15.5, in which Six2 has been tagged with enhanced green fluorescent protein (GFP), with specific Crkl staining of the ureteric bud (in red) surrounded by Six2-positive cap mesenchyme cells (in green) (subpanel a). A magnified field shows ureteric-bud branching within condensing metanephric mesenchyme (subpanel b). Panel B shows specific pronephros expression of crkl in zebrafish, as shown by colocalization after staining with antibody against sodium–potassium ATPase. In the orientation symbol, D denotes dorsal, V ventral, C caudal, and R rostral. Panel C shows images of negative controls (i.e., fish treated with fluorophore-conjugated secondary antibodies only). In Panels B and C, the scale bars represent 100 μm. In a mouse model that targets Crkl exon 2, three crosses with transgenic Cre-recombinase mice were created to effect the deletion of exon 2 in specific compartments: E2a-Cre for global knockout, Six2-Cre in the cap mesenchyme, and Hoxb7 in the structures derived from ureteric buds. Panel D shows tissue from a Six2-Cre mouse in which duplication of the right kidney is accompanied by an irregular, dysplastic pattern or ureteric-bud branching on embryonic day E15.5. Panel E shows tissue from an E2a-Cre mouse in which a single kidney with duplicated ureters (arrowheads) is accompanied by failure of medullary and renal papillary development on day E14.5. Panel F shows tissue from a Six2-Cre mouse, in which the kidney is hydronephrotic with dilated pelvis, absence of medullary architecture, and several microcystic glomeruli and tubules on day E15.5.
Finally, we engineered a mouse model that targets Crkl exon 2. We generated three different crosses with transgenic Cre-recombinase mice to effect the deletion of exon 2 in specific compartments: E2a-Cre for global knockout, Six2-Cre in the cap mesenchyme, and Hoxb7 in the ureteric bud–derived structures. We analyzed four litters (one E2a, one Hoxb7, and two Six2) at embryonic days E14.5 through E15.5. We observed developmental anomalies in the kidney and urinary tract, including bilaterally duplicated kidneys, duplicated ureters, ureteric bud–branching defects, dysplastic features, hydronephrosis, microcystic tubules and glomeruli, and tubular and glomerular capsule dilatation, in eight mice (Fig. 3D, 3E, and 3F, and Fig. S17 in the Supplementary Appendix). We observed phenotypes related to congenital kidney and urinary tract anomalies in embryos that were heterozygous and those that were homozygous for the targeted deletion.
DISCUSSION
We determined that deletions in the telomeric 22q11.2 classic region are associated with sporadic congenital kidney and urinary tract anomalies and renal disease in the DiGeorge syndrome. Correlations between genotype and phenotype suggest that these variants are specific for kidney parenchyma defects (i.e., renal agenesis or hypodysplasia), rather than ureteric and lower urinary tract disease. However, the presence of these variants may be an indication of kidney disease in persons with apparently isolated ureteric defects, since the two patients with obstructive uropathy and vesicoureteral reflux whom we identified in this study showed renal insufficiency. We observed that the 22q11.2 deletions were present in 1.1% of our sample of 1093 patients with renal agenesis or hypodysplasia, which suggests that such deletions constitute the second most common structural variant associated with congenital kidney and urinary tract anomalies after the 17q12 deletion that causes the renal cysts and diabetes syndrome, which we identified in 2.2% of patients with renal agenesis or hypodysplasia from the same cohort. Our data also support the hypothesis that 22q11.2 microdeletions are medically actionable variants that confer a predisposition to renal hypodysplasia and kidney disease.
A review of the literature indicates the presence of kidney and urinary tract defects in about one third of the patients with chromosome 22q11.2 deletions spanning LCR22 B and D or C and D,5,41 a prevalence that is nearly identical to that of kidney and urinary tract defects among patients with the DiGeorge syndrome caused by the typical 22q11.2 deletions spanning LCR22 A and D.4,6,42 These observations, together with our data, strongly suggest that the kidney disease associated with the DiGeorge syndrome is attributable largely to haploinsufficiency of one or more genes located between LCR22 C and D.
Genetic interaction studies using zebrafish suggested a complex genetic architecture, in which haploinsufficiency of crkl had a potent detrimental effect on renal development, whereas knockdown of its flanking genes, aifm3 and snap29, generated the phenotype only with cosuppression. Consistent with these data, we found deleterious CRKL variants, including a premature truncating allele, in approximately 1% of the patients with sporadic congenital renal agenesis or hypodysplasia. We obtained other molecular data in humans, mice, and zebrafish that supported the role of CRKL in urinary tract development.
CRKL encodes an adapter protein that regulates intracellular signaling transduction from multiple growth factors, including the fibroblast growth factors,43 which are key regulators of kidney and urinary tract development.44,45 Inactivation of Crkl in mice recapitulates some of the phenotypes of the DiGeorge syndrome, in particular cardiac malformations,46,47 but the kidney phenotype in the mutant embryos had not hitherto been studied. We observed that genetic inactivation of Crkl in the mouse model results in developmental phenotypes of the kidney and urinary tract that resemble congenital anomalies in the human urinary tract.
We suggest that CRKL mutations sensitize the genetic background and contribute to the penetrance of congenital kidney and urinary tract anomalies in patients with the DiGeorge syndrome. It is possible that other genes within or outside the locus of the DiGeorge syndrome and 22q11.2 deletions might also be involved. Two of the genes in the minimal region were refractory to our studies, and it is possible that the deletion copy-number variant affects the expression of genes across the chromosome or elsewhere in the genome, as has been shown for other copy-number variants.48
In conclusion, our approach provides support for the causal role of CRKL in the pathogenesis of kidney developmental defects. Such defects occur specifically in the context of the DiGeorge syndrome and 22q11.2 deletions and, more broadly, in sporadic congenital kidney and urinary tract anomalies.
Supplementary Material
Acknowledgments
Supported by grants (1R01DK103184, 1R21DK098531, and UL1 TR000040, to Dr. Sanna-Cherchi; P50DK096415 and P30DK096493, to Dr. Katsanis; 2R01DK080099, to Dr. Gharavi; 3U54DK104309, to Drs. Gharavi and Barasch; P01HD070454, to Ms. McDonald-McGinn and Dr. Morrow; 4R01GM030518, to Dr. Honig; R37HD033082, to Dr. Papaioannou; and 1R01DK105124, to Dr. Kiryluk) from the National Institutes of Health (NIH); a grant-in-aid (13GRNT14680075, to Dr. Sanna-Cherchi) from the American Heart Association; a grant (RF-2010-2307403, to Drs. Sanna-Cherchi and Ghiggeri) from the Joint Italian Ministry of Health and NIH Young Investigators Finalized Research; a grant (HG006504, to Dr. Lifton) from the National Human Genome Research Institute Centers for Mendelian Genomics; a grant (to Dr. Ghiggeri) from the Fondazione Malattie Renali nel Bambino; a grant (AAE07007KSA, to Drs. Salomon and Jeanpierre) from the GIS-Institut des Maladies Rares; and a grant (AOM07129, to Drs. Salomon and Jeanpierre) from the Programme Hospitalier de la Recherche Clinique Assistance Publique; by the Polish Ministry of Health (to Drs. Materna-Kiryluk and Latos-Bielenska); by the Polish Kidney Genetics Network (POLYGENES), the Polish Registry of Congenital Malformations (PRCM), and the NZOZ Center for Medical Genetics (GENESIS); by grants (to the Chronic Kidney Disease in Children Study) from the National Institute of Diabetes and Digestive and Kidney Diseases and the Eunice Kennedy Shriver National Institute of Child Health and Human Development; by grants (U01DK66143, U01DK66174, U01DK082194, U01DK66116, and RO1DK082394) from the National Heart, Lung, and Blood Institute; and by the Paul Marks Scholar Award (to Dr. Sanna-Cher-chi); and a Kolff Postdoc Fellowship Abroad grant (15OKK95, to Dr. Westland) from the Dutch Kidney Foundation.
We thank the patients and their families for participating in the study; Katarzyna Zachwieja (Dialysis Unit, Jagiellonian University Medical College, Krakow, Poland), Daria Tomczyk (Department of Pediatrics, Immunology and Nephrology Polish Mother’s Memorial Hospital Research Institute, Lodz, Poland), Tomasz Jarmolinski (Miedzyrzecz Regional Hospital, Department of Pediatrics, Miedzyrzecz, Poland), Robert Pawluch and Maria Katarzyna Boroszewska-Kornacka (Neonatal and Intensive Care Department, Medical University of Warsaw, Poland), Piotr Adamczyk (Department of Pediatrics, School of Medicine with the Division of Dentistry in Zabrze, Medical University of Silesia in Katowice, Poland), and Klaudia Korecka (Department of Pediatric Surgery and Urology, Medical University of Silesia, Upper Silesian Child’s Health Center Katowice, Poland) for recruting patients for this study; and Cyrus Zabetian (University of Washington, Seattle) and Haydeh Payami (University of Alabama, Birmingham) for sharing clinical and genetic data from the control population.
APPENDIX
The authors’ full names and academic degrees are as follows: Esther Lopez-Rivera, Ph.D., Yangfan P. Liu, Ph.D., Miguel Verbitsky, Ph.D., Blair R. Anderson, Ph.D., Valentina P. Capone, M.D., Edgar A. Otto, Ph.D., Zhonghai Yan, Ph.D., Adele Mitrotti, M.D., Jeremiah Martino, Ph.D., Nicholas J. Steers, Ph.D., David A. Fasel, B.S., Katarina Vukojevic, M.D., Ph.D., Rong Deng, B.S., Silvia E. Racedo, Ph.D., Qingxue Liu, M.S., Max Werth, Ph.D., Rik Westland, M.D., Ph.D., Asaf Vivante, M.D., Gabriel S. Makar, B.S., Monica Bodria, M.D., Matthew G. Sampson, M.D., Christopher E. Gillies, Ph.D., Virginia Vega-Warner, Ph.D., Mariarosa Maiorana, M.D., Donald S. Petrey, Ph.D., Barry Honig, Ph.D., Vladimir J. Lozanovski, M.D., Ph.D., Rémi Salomon, Ph.D., Laurence Heidet, M.D., Wassila Carpentier, Ph.D., Dominique Gaillard, M.S., Alba Carrea, Ph.D., Loreto Gesualdo, M.D., Daniele Cusi, M.D., Claudia Izzi, M.D., Francesco Scolari, M.D., Joanna A.E. van Wijk, M.D., Ph.D., Adela Arapovic, M.D., Mirna Saraga-Babic, Ph.D., Marijan Saraga, M.D., Nenad Kunac, Ph.D., Ali Samii, M.D., Donna M. McDonald-McGinn, M.S., Terrence B. Crowley, Ph.D., Elaine H. Zackai, M.D., Dorota Drozdz, M.D., Monika Miklaszewska, M.D., Marcin Tkaczyk, M.D., Przemyslaw Sikora, M.D., Maria Szczepanska, M.D., Malgorzata Mizerska-Wasiak, M.D., Grazyna Krzemien, M.D., Agnieszka Szmigielska, M.D., Marcin Zaniew, M.D., John M. Darlow, M.D., Ph.D., Prem Puri, M.D., David Barton, Ph.D., Emilio Casolari, M.D., Susan L. Furth, M.D., Ph.D., Bradley A. Warady, M.D., Zoran Gucev, M.D., Ph.D., Hakon Hakonarson, Ph.D., Hana Flogelova, M.D., Velibor Tasic, M.D., Ph.D., Anna Latos-Bielenska, M.D., Anna Materna-Kiryluk, M.D., Landino Allegri, M.D., Craig S. Wong, M.D., M.P.H., Iain A. Drummond, Ph.D., Vivette D’Agati, M.D., Akira Imamoto, Ph.D., Jonathan M. Barasch, M.D., Ph.D., Friedhelm Hildebrandt, M.D., Krzysztof Kiryluk, M.D., Richard P. Lifton, M.D., Ph.D., Bernice E. Morrow, Ph.D., Cecile Jeanpierre, Ph.D., Virginia E. Papaioannou, Ph.D., Gian Marco Ghiggeri, M.D., Ph.D., Ali G. Gharavi, M.D., Nicholas Katsanis, Ph.D., and Simone Sanna-Cherchi, M.D.
The authors’ affiliations are as follows: the Division of Nephrology (E.L.-R., M.V., V.P.C., Z.Y., A.M., J.M., N.J.S., D.A.F., R.D., M.W., G.S.M., M.B., J.M.B., K.K., A.G.G., S.S.-C.) and the Division of Nephrology in Medicine and Zuckerman Mind Brain Behavior Institute (B.H.), the Departments of Systems Biology (D.S.P., B.H.), Biochemistry and Molecular Biophysics (B.H.), and Pathology (V.D.), and the Howard Hughes Medical Institute (D.S.P., B.H.), Columbia University, and the Department of Genetics and Development, Columbia University Medical Center (Q.L., V.E.P.), New York, and the Department of Genetics, Albert Einstein College of Medicine, Bronx (S.E.R., B.E.M.) — all in New York; the Center for Human Disease Modeling, Duke University, Durham, NC (Y.P.L., B.R.A., N. Katsanis); the Departments of Internal Medicine–Nephrology (E.A.O.) and Pediatrics–Nephrology (M.G.S., C.E.G., V.V.-W.), University of Michigan School of Medicine, Ann Arbor; the Department of Anatomy, Histology, and Embryology, School of Medicine, University of Split (K.V., M.S.-B.), and the Departments of Pediatrics (A.A., M. Saraga) and Pathology (N. Kunac), University Hospital of Split, Split, Croatia; the Department of Pediatric Nephrology, VU University Medical Center, Amsterdam (R.W., J.A.E.W.); the Department of Medicine, Boston Children’s Hospital (A.V., F.H.), and Harvard Medical School, Boston (A.V., F.H., I.A.D.), and the Nephrology Division, Massachusetts General Hospital, Charlestown (I.A.D.) — all in Massachusetts; the Division of Nephrology, Dialysis, Transplantation, and Laboratory on Pathophysiology of Uremia, Istituto G. Gaslini, Genoa (M.B., A.C., G.M.G.), the Department of Clinical and Experimental Medicine, University of Parma (M.B., M. Maiorana, L.A.), and the Pediatric Surgery Unit, University Hospital of Parma (E.C.), Parma, the Section of Nephrology, Department of Emergency and Organ Transplantation, University of Bari, Bari (L.G.), the Department of Medical Sciences, University of Milano, and Institute of Biomedical Technologies, Italian National Institute of Research ITB-CNR, Milan (D.C.), and Dipartimento Ostetrico-Ginecologico e Seconda Divisione di Nefrologia ASST Spedali Civili e Presidio di Montichiari (C.I.) and Cattedra di Nefrologia, Università di Brescia, Seconda Divisione di Nefrologia Azienda Ospedaliera Spedali Civili di Brescia Presidio di Montichiari (F.S.), Brescia — all in Italy; the Department of General and Transplant Surgery, University Hospital of Heidelberg, Germany (V.J.L.); the Department of Pediatric Nephrology, Centre de Référence des Maladies Rénales Héréditaires de l’Enfant et de l’Adulte (R.S., L.H., C.J.), INSERM UMR 1163, Laboratory of Hereditary Kidney Diseases (R.S.), Necker–Enfants Malades Hospital, Paris Descartes–Sorbonne Paris Cite University, Imagine Institute (R.S.), Sorbonne Universités, UPMC 06, Plateforme Post-génomique de la Pitié–Salpêtrière, UMS 2 Omique, Inserm US029 (W.C.), Paris, and the Department of Genetics, Centre Hospitalier Universitaire de Reims, Unité de Formation et de Recherche de Médecine, Reims (D.G.) — both in France; the Department of Neurology, University of Washington School of Medicine, and Northwest VA Parkinson’s Disease Research, Education and Clinical Centers, Seattle (A. Samii); the Division of Human Genetics, Department of Pediatrics, 22q and You Center, Children’s Hospital of Philadelphia and Perelman School of Medicine at the University of Pennsylvania (D.M.M.-M., T.B.C., E.H.Z., S.L.F.), Division of Nephrology, Children’s Hospital of Philadelphia (S.L.F.), and the Department of Genetics, University of Pennsylvania (H.H.), Philadelphia; the Dialysis Unit, Jagiellonian University Medical College (D.D.), and the Department of Pediatric Nephrology, Jagiellonian University Medical College (M. Miklaszewska), Krakow, the Department of Pediatrics, Immunology and Nephrology, Polish Mother’s Memorial Hospital Research Institute, Lodz (M.T.), the Department of Pediatric Nephrology Medical University of Lublin, Lublin (P.S.), the Department of Pediatrics, School of Medicine with the Division of Dentistry in Zabrze, Medical University of Silesia, Katowice (M. Szczepanska), the Department of Pediatrics and Nephrology, Medical University of Warsaw, Warsaw (M.M.-W., G.K., A. Szmigielska), and Krysiewicza Children’s Hospital (M.Z.) and the Department of Medical Genetics, Poznan University of Medical Sciences, and Center for Medical Genetics GENESIS (A.L.-B., A.M.-K.), Poznan — all in Poland; the Department of Clinical Genetics (J.M.D., D.B.), National Children’s Research Centre (J.M.D., P.P.), and University College Dublin School of Medicine (D.B.), Our Lady’s Children’s Hospital Crumlin, and the National Children’s Hospital Tallaght (P.P.), Dublin, Ireland; the Division of Pediatric Nephrology, Children’s Mercy Hospital, Kansas City, MO (B.A.W.); University Children’s Hospital, Medical Faculty of Skopje, Skopje, Macedonia (Z.G., V.T.); Faculty of Medicine, Palacky University, Olomouc, Czech Republic (H.F.); the Division of Pediatric Nephrology, University of New Mexico Children’s Hospital, Albuquerque (C.S.W.); Ben May Department for Cancer Research, University of Chicago, Chicago (A.I.); and the Department of Genetics, Howard Hughes Medical Institute, and Yale Center for Mendelian Genomics, Yale University, New Haven, CT (R.P.L.).
Footnotes
The authors’ full names, academic degrees, and affiliations are listed in the Appendix.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2:858–68. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 2.Devriendt K, Fryns JP, Mortier G, van Thienen MN, Keymolen K. The annual incidence of DiGeorge/velocardiofacial syndrome. J Med Genet. 1998;35:789–90. doi: 10.1136/jmg.35.9.789-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11. 2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 4.Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11. 2 deletion syndromes. Lancet. 2007;370:1443–52. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- 5.Burnside RD. 22q11. 21 Deletion syndromes: a review of proximal, central, and distal deletions and their associated features. Cytogenet Genome Res. 2015;146:89–99. doi: 10.1159/000438708. [DOI] [PubMed] [Google Scholar]
- 6.Noël AC, Pelluard F, Delezoide AL, et al. Fetal phenotype associated with the 22q11 deletion. Am J Med Genet A. 2014;164A:2724–31. doi: 10.1002/ajmg.a.36720. [DOI] [PubMed] [Google Scholar]
- 7.Bassett AS, Chow EW, Husted J, et al. Clinical features of 78 adults with 22q11 Deletion Syndrome. Am J Med Genet A. 2005;138:307–13. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelmann L, Pandita RK, Spiteri E, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8:1157–67. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 9.Saitta SC, Harris SE, Gaeth AP, et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11. 2 deletion. Hum Mol Genet. 2004;13:417–28. doi: 10.1093/hmg/ddh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaikh TH, O’Connor RJ, Pierpont ME, et al. Low copy repeats mediate distal chromosome 22q11. 2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–91. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaikh TH, Kurahashi H, Saitta SC, et al. Chromosome 22-specific low copy repeats and the 22q11. 2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 12.Kujat A, Schulz MD, Strenge S, Froster UG. Renal malformations in deletion 22q11. 2 patients. Am J Med Genet A. 2006;140:1601–2. doi: 10.1002/ajmg.a.31289. [DOI] [PubMed] [Google Scholar]
- 13.Wu HY, Rusnack SL, Bellah RD, et al. Genitourinary malformations in chromosome 22q11. 2 deletion. J Urol. 2002;168:2564–5. doi: 10.1016/S0022-5347(05)64215-2. [DOI] [PubMed] [Google Scholar]
- 14.Yagi H, Furutani Y, Hamada H, et al. Role of TBX1 in human del22q11. 2 syndrome. Lancet. 2003;362:1366–73. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 15.Paylor R, Glaser B, Mupo A, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci U S A. 2006;103:7729–34. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 17.Merscher S, Funke B, Epstein JA, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–29. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay EA, Vitelli F, Su H, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 19.Sanna-Cherchi S, Kiryluk K, Burgess KE, et al. Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet. 2012;91:987–97. doi: 10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verbitsky M, Sanna-Cherchi S, Fasel DA, et al. Genomic imbalances in pediatric patients with chronic kidney disease. J Clin Invest. 2015;125:2171–8. doi: 10.1172/JCI80877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westland R, Verbitsky M, Vukojevic K, et al. Copy number variation analysis identifies novel CAKUT candidate genes in children with a solitary functioning kidney. Kidney Int. 2015;88:1402–10. doi: 10.1038/ki.2015.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanna-Cherchi S, Sampogna RV, Papeta N, et al. Mutations in DSTYK and dominant urinary tract malformations. N Engl J Med. 2013;369:621–9. doi: 10.1056/NEJMoa1214479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westland R, Bodria M, Carrea A, et al. Phenotypic expansion of DGKE-associated diseases. J Am Soc Nephrol. 2014;25:1408–14. doi: 10.1681/ASN.2013080886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halbritter J, Diaz K, Chaki M, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49:756–67. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 26.Halbritter J, Porath JD, Diaz KA, et al. Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet. 2013;132:865–84. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mefford HC, Clauin S, Sharp AJ, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman DL, Garvey N, Hancock S, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–90. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 29.Golzio C, Willer J, Talkowski ME, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11. 2 copy number variant. Nature. 2012;485:363–7. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho CM, Vasanth S, Shinawi M, et al. Dosage changes of a segment at 17p13. 1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am J Hum Genet. 2014;95:565–78. doi: 10.1016/j.ajhg.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dauber A, Golzio C, Guenot C, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24. 3 copy-number variant. Am J Hum Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuchardt A, D’Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–3. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee R, Ramos E, Hoffman M, et al. Traditional and targeted exome sequencing reveals common, rare and novel functional deleterious variants in RET-signaling complex in a cohort of living US patients with urinary tract malformations. Hum Genet. 2012;131:1725–38. doi: 10.1007/s00439-012-1181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang DY, Dworschak GC, Kohl S, et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014;85:1429–33. doi: 10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Pontual L, Zaghloul NA, Thomas S, et al. Epistasis between RET and BBS mutations modulates enteric innervation and causes syndromic Hirschsprung disease. Proc Natl Acad Sci U S A. 2009;106:13921–6. doi: 10.1073/pnas.0901219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasilyev A, Liu Y, Mudumana S, et al. Collective cell migration drives morphogenesis of the kidney nephron. PLoS Biol. 2009;7(1):e9. doi: 10.1371/journal.pbio.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borck G, Hög F, Dentici ML, et al. BRF1 mutations alter RNA polymerase III-dependent transcription and cause neuro-developmental anomalies. Genome Res. 2015;25:155–66. doi: 10.1101/gr.176925.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6(10):e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo S, Tokunaga F, Kario K, Matsuo T, Koide T. Molecular and cellular basis for type I heparin cofactor II deficiency (heparin cofactor II Awaji) Blood. 1996;87:1006–12. [PubMed] [Google Scholar]
- 41.Rump P, de Leeuw N, van Essen AJ, et al. Central 22q11. 2 deletions. Am J Med Genet A. 2014;164A:2707–23. doi: 10.1002/ajmg.a.36711. [DOI] [PubMed] [Google Scholar]
- 42.Besseau-Ayasse J, Violle-Poirsier C, Bazin A, et al. A French collaborative survey of 272 fetuses with 22q11. 2 deletion: ultrasound findings, fetal autopsies and pregnancy outcomes. Prenat Diagn. 2014;34:424–30. doi: 10.1002/pd.4321. [DOI] [PubMed] [Google Scholar]
- 43.Moon AM, Guris DL, Seo JH, et al. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10:71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Am J Physiol Renal Physiol. 2011;301:F245–F251. doi: 10.1152/ajprenal.00186.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 46.Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11. 2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–8. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 47.Racedo SE, McDonald-McGinn DM, Chung JH, et al. Mouse and human CRKL is dosage sensitive for cardiac outflow tract formation. Am J Hum Genet. 2015;96:235–44. doi: 10.1016/j.ajhg.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Migliavacca E, Golzio C, Männik K, et al. A potential contributory role for ciliary dysfunction in the 16p11. 2 600 kb BP4-BP5 pathology. Am J Hum Genet. 2015;96:784–96. doi: 10.1016/j.ajhg.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.