A plastidial cis-prenyltransferase synthesizes a family of polyprenols that accumulate in thylakoid membranes and play a central role in the efficiency of photosynthetic performance.
Abstract
Plants accumulate a family of hydrophobic polymers known as polyprenols, yet how they are synthesized, where they reside in the cell, and what role they serve is largely unknown. Using Arabidopsis thaliana as a model, we present evidence for the involvement of a plastidial cis-prenyltransferase (AtCPT7) in polyprenol synthesis. Gene inactivation and RNAi-mediated knockdown of AtCPT7 eliminated leaf polyprenols, while its overexpression increased their content. Complementation tests in the polyprenol-deficient yeast ∆rer2 mutant and enzyme assays with recombinant AtCPT7 confirmed that the enzyme synthesizes polyprenols of ∼55 carbons in length using geranylgeranyl diphosphate (GGPP) and isopentenyl diphosphate as substrates. Immunodetection and in vivo localization of AtCPT7 fluorescent protein fusions showed that AtCPT7 resides in the stroma of mesophyll chloroplasts. The enzymatic products of AtCPT7 accumulate in thylakoid membranes, and in their absence, thylakoids adopt an increasingly “fluid membrane” state. Chlorophyll fluorescence measurements from the leaves of polyprenol-deficient plants revealed impaired photosystem II operating efficiency, and their thylakoids exhibited a decreased rate of electron transport. These results establish that (1) plastidial AtCPT7 extends the length of GGPP to ∼55 carbons, which then accumulate in thylakoid membranes; and (2) these polyprenols influence photosynthetic performance through their modulation of thylakoid membrane dynamics.
INTRODUCTION
Isoprenoids are the most abundant and structurally diverse group of compounds that occur in nature. Comprising more than 50,000 different members, these natural products participate in a broad spectrum of biological processes ranging from respiration and energy capture to hormonal regulation and signal transduction (Kirby and Keasling, 2009). All isoprenoids are derived from the universal five-carbon (C5) building blocks, isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). In animals, archaea, and some bacteria, these C5 building blocks are synthesized via the mevalonic acid (MVA) pathway, while in most microbes and protozoa, the methylerythritol phosphate (MEP) pathway provides IPP and DMAPP for isoprenoid biosynthesis. Plants, by contrast, are unique in this context and have evolved to maintain both pathways, with the MVA pathway present in the cytosol and the MEP pathway operating in plastids (Rohmer, 1999; Rodríguez-Concepción and Boronat, 2002; Lange and Ghassemian, 2003; Hemmerlin et al., 2012). This “division of labor” allows for the compartment-specific synthesis of brassinosteroids, sesquiterpenes, sterols, and ubiquinone from the cytosolic MVA pathway, while the MEP pathway provides the precursors for the synthesis of abscisic acid, carotenoids, chlorophyll, gibberellins, plastoquinone, and various mono- and diterpenes in the plastid (Lange et al., 2000; Vranová et al., 2013). In spite of this strict compartmentalization of the two pathways, the exchange of isoprenoid precursors between the plastids and cytosol is well recognized in some plant species and leads to the synthesis of “mosaic compounds” that are composed of isoprenoid intermediates derived from both the MVA and MEP pathways (Hemmerlin et al., 2003; Laule et al., 2003; Dudareva et al., 2005; Bartram et al., 2006; Skorupinska-Tudek et al., 2008; May et al., 2013; Opitz et al., 2014).
Polyisoprenoid alcohols are a class of isoprenoids that were originally discovered in plants and later shown to reside in all other kingdoms of life (Lindgren, 1965; Swiezewska et al., 1994; Skorupinska-Tudek et al., 2008). These compounds are linear hydrophobic polymers composed of IPP and DMAPP units and broadly fall into two categories, the dolichols and polyprenols. The length of these polymers is defined by the number of C5 units that they contain. Dolichols are saturated at the terminal isoprene unit (α-saturated) and typically comprise 14-19 C5 units (Dol-14 to Dol-19; corresponding to C70–C95). This class of polyisoprenoid alcohols is most noted for its role in eukaryotic protein N-glycosylation, which occurs in the secretory pathway. Polyprenols, in comparison, are α-unsaturated and exhibit a much broader length, ranging from C25 to more than C500, with medium-chain compounds composed of 9 to 11 C5 units (Pren-9 to Pren-11; C45–C55) predominating in plants and microbes. Polyprenols serve as lipid carriers in bacterial peptidoglycan cell wall biosynthesis. In plants, these compounds have historically been considered dispensable and therefore classified as “secondary metabolites” (Surmacz and Swiezewska, 2011). However, their widespread occurrence throughout the plant kingdom suggests otherwise and points toward a more primary role for polyprenols in plant cellular metabolism (Bajda et al., 2009).
Polyprenol biosynthesis occurs in two steps beginning with the assembly of a trans-prenyl diphosphate precursor. Enzymes known as trans-prenyldiphosphate synthases generate these precursors via the condensation of DMAPP with up to three IPP units, which results in the synthesis of geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), or geranylgeranyl diphosphate (GGPP; C20). These trans-prenyl diphosphates are spatially separated within the cell, with FPP derived from the cytosolic MVA pathway and GPP and GGPP residing in the plastid as products of the MEP pathway (Tholl and Lee, 2011). The C10-C20 trans-prenyl precursors serve as substrates for the synthesis of all isoprenoids and can further be extended with additional IPP units, in a cis-orientation, to generate polyisoprenoids. This step is catalyzed by the class of enzymes known as cis-prenyltranferases (CPTs). CPTs are not homologous to trans-prenyldiphosphate synthases, and comparatively little is known about these enzymes, particularly in plants. Bioinformatic analysis indicates that animal and prokaryotic genomes typically encode a single CPT, while plants maintain small CPT gene families, containing anywhere between three to nine members (Surmacz and Swiezewska, 2011; Akhtar et al., 2013). Bacterial CPTs synthesize the Pren-11 (C55) polyprenol known as undecaprenyl diphosphate, which as mentioned above, functions in peptidoglycan biosynthesis, while animal CPTs are involved in the synthesis of longer chain dolichols that serve an indispensable role in protein N-glycosylation. Bacterial and animal CPTs are phylogenetically distinct from one another, and while CPTs from microbes operate autonomously, their animal counterparts strictly require a distantly related partner protein for stability and enzymatic activity (Harrison et al., 2011; Park et al., 2014; Grabińska et al., 2016). Given that plants accumulate both classes of polyisoprenoids, it is not surprising that their CPT families can be phylogenetically divided into two groups: those predicted to act alone and synthesize polyprenols and those requiring a partner protein for dolichol biosynthesis.
The recent identification of a heteromeric dolichol synthase enzyme complex, composed of a CPT and its stabilizing partner protein (CPTBP/CPTL), established how this class of plant polyisoprenoids is synthesized and functions (Brasher et al., 2015; Qu et al., 2015). However, open questions regarding the biosynthesis, compartmentation, and biological function of plant polyprenols still remain, in spite of nearly five decades of research that has clearly demonstrated the occurrence of medium-chain polyprenols (Pren-9 to Pren-11) in numerous plant species (Swiezewska and Danikiewicz, 2005). This study addresses this gap in our understanding of plant polyisoprenoid biology. We provide evidence that medium-chain polyprenols in Arabidopsis thaliana are synthesized by a plastidial CPT and that these compounds accumulate in thylakoid membranes. Their absence appears to alter thylakoid membrane dynamics and results in a reduced photosynthetic capacity, thereby providing insight into the function of this ubiquitous class of plant secondary metabolites.
RESULTS
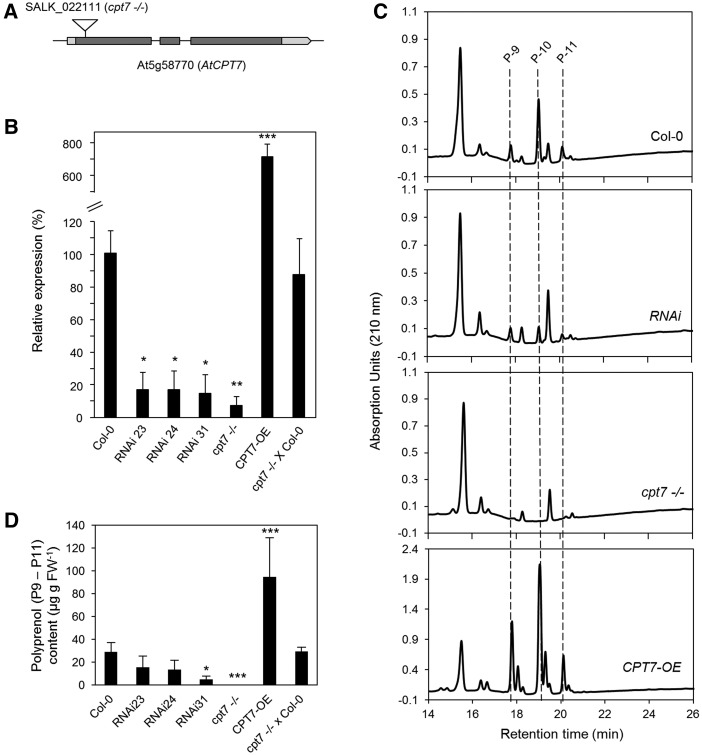
Inactivation and RNAi-Mediated Knockdown of AtCPT7 Result in Medium-Chain Polyprenol Deficiency in Arabidopsis Leaves
The widespread occurrence of medium-chain polyprenols in the green tissues of virtually all plants prompted us to identify which CPT was responsible for their synthesis in the model plant Arabidopsis. We surveyed the Arabidopsis nine-member CPT family (Supplemental Table 1) based on the criteria that the CPT must be (1) expressed in photosynthetic tissues and (2) be phylogenetically distinct from the CPTs that are involved in dolichol biosynthesis (Surmacz and Swiezewska, 2011; Brasher et al., 2015). This search yielded two candidates: AtCPT2 (At2g23400) and AtCPT7 (At5g58770). While both CPTs are expressed in leaves, AtCPT2 is absent in stem tissue, which is known to contain polyprenols (Surmacz and Swiezewska, 2011; Jozwiak et al., 2015); hence, AtCPT7 emerged as the most promising candidate. To explore the involvement of AtCPT7 in polyprenol synthesis, we first isolated a single homozygous T-DNA line for the gene locus At5g58770 (SALK_022111; Figure 1A) and then generated three independent RNAi lines targeting AtCPT7 for mRNA knockdown (RNAi-23, -24, and -31) as well as transgenic lines overexpressing AtCPT7 (CPT7-OE). Quantitative RT-PCR analysis of AtCPT7 expression in the leaf tissue from these lines indicated that AtCPT7 transcripts were virtually undetectable in the T-DNA knockout (cpt7−/−), were significantly reduced by ∼85% in the three RNAi lines, and significantly (∼7-fold) elevated in the CPT7-OE lines relative to wild-type levels (Figure 1B; Supplemental Figure 1A). Additionally, F1 heterozygous plants derived from a cpt7−/− × Col-0 (wild type) backcross showed almost complete restoration of AtCPT7 expression (Figure 1B). AtCPT7 deficiency or overexpression did not affect the expression of the remaining members of the AtCPT gene family (Supplemental Figure 2) nor did it result in any obvious phenotype in the aerial portions of these plants (Supplemental Figure 3). Analysis of total polyisoprenoids from these plants revealed that wild-type leaves accumulated a family of polyisoprenoids that were composed of polyprenols containing 9-11 isoprenoid units (Pren-9 to Pren-11), which correspond to C45-C55 in length (Figure 1C). A comparison of saponified versus nonsaponified polyisoprenoid extracts revealed that these polyprenols exist predominantly as free alcohols, with less than ∼7% of the total being esterified to propionic acid (Supplemental Figure 4). These polyprenol alcohols were virtually absent in cpt7−/−, significantly decreased in all of the RNAi lines (0.5% and as low as 11% of the wild type, respectively), and significantly increased in CPT7-OE plants, reaching ∼185% of the wild type. The F1 heterozygous plants derived from the cpt7−/− × Col-0 backcross showed almost complete restoration of Pren-9 to Pren-11 content compared with the wild type (Figure 1D; Supplemental Figure 1B). Taken together, these results imply that cpt7−/− carries an AtCPT7 loss-of-function allele and that AtCPT7 plays a role in the synthesis of medium-chain polyprenols in Arabidopsis leaves.
Figure 1.
The Effect of AtCPT7 Inactivation or Overexpression on Polyprenol Accumulation in Arabidopsis Leaves.
(A) A gene model of AtCPT7 (At5g58770) illustrating the arrangement of introns (solid lines), exons (gray boxes), and 5′- and 3′-untranslated regions (light-gray boxes). A single T-DNA mutant line for AtCPT7 (cpt7−/−) was identified from the SALK collection (SALK_022111), and the position of the T-DNA insertion is shown.
(B) Relative AtCPT7 gene expression in Arabidopsis leaf tissue from the wild type (Col-0), three independent RNAi lines targeting AtCPT7 (RNAi-23, -24, and -31), a homozygous T-DNA loss-of-function AtCPT7 mutant (cpt7−/−), an AtCPT7 overexpression line (CPT7-OE), and a F1 heterozygous mutant generated from a backcross to the wild type (cpt7−/− × Col-0). AtCPT7 mRNA abundance was quantified by real-time PCR and is presented as a percentage of the wild type (100%).
(C) Analysis of total polyisoprenoids extracted from the leaf tissue of wild-type, RNAi, cpt7−/−, CPT7-OE, and cpt7−/− × Col-0 lines. Representative HPLC/UV chromatograms are presented and the main polyprenols (P9, P10, and P11) are indicated.
(D) Quantification of total polyprenols from the samples above. Data are the means ± sd from at least three independent experiments. Statistically significant differences compared with wild-type plants are indicated; *P < 0.05, **P < 0.01, and ***P < 0.001 in Student’s t test.
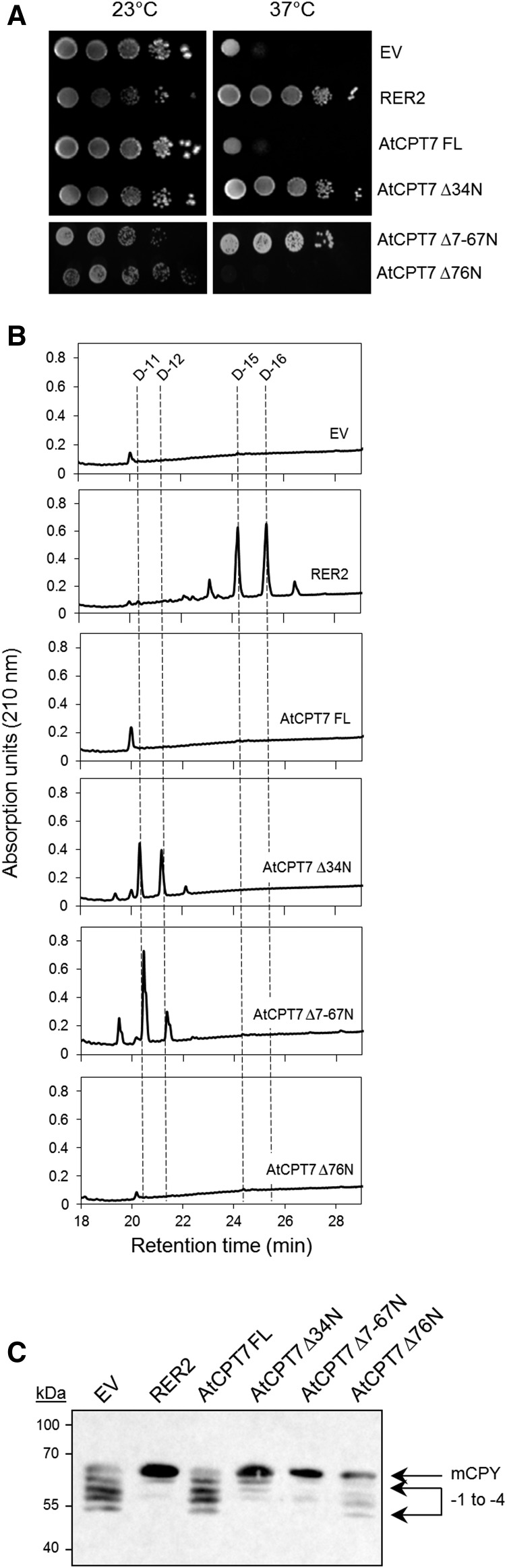
Functional Complementation of the Yeast Δrer2 Mutant by AtCPT7
The yeast Δrer2 mutant exhibits a distinct growth defect and aberrant N-glycosylation at elevated temperatures due to the ablation of the CPT that is involved in dolichol biosynthesis (Sato et al., 1999). Strikingly, medium-chain polyisoprenoids (Pren-11) that are synthesized by bacterial CPTs can functionally substitute for dolichols in this mutant (Rush et al., 2010), thereby offering a tractable platform to test for the in vivo enzyme activity of various plant CPTs in complementation assays. To determine whether AtCPT7 is a functional enzyme with cis-prenyltransferase activity, we introduced AtCPT7 or the native RER2 gene (as a positive control) into the Δrer2 mutant and subsequently monitored growth, polyisoprenoid accumulation, and N-glycosylation status of the prototypical glycoprotein, carboxypeptidase Y (CPY). Introduction of the native RER2 protein rescued the growth defects of the Δrer2 mutant but the full-length AtCPT7 protein (AtCPT7 FL) and empty vector failed to restore growth at the nonpermissive temperature (Figure 2A). However, upon removal of the predicted N-terminal targeting peptide from AtCPT7 (AtCPT7Δ34N), growth of the Δrer2 mutant was restored (Figure 2A). When introduced as C-terminal GFP fusion proteins into the Δrer2 mutant, both the RER2 and AtCPT7Δ34N proteins exhibited a diffuse pattern of GFP fluorescence throughout the yeast cells (Supplemental Figure 5). However, the AtCPT7-FL fusion protein appeared as discrete punctate structures (Supplemental Figure 5) that are characteristic of aggregated or misfolded proteins (Kaganovich et al., 2008; Pampeno et al., 2014). A further truncated version of AtCPT7 that closely resembles the CPT from Escherichia coli (AtCPT7Δ7-67N, containing the first seven N-terminal amino acids and lacking the next 60 residues which are absent in the E. coli enzyme; Supplemental Figures 6 and 7) also restored the growth of the Δrer2 mutant at the nonpermissive temperature, while a truncated version of AtCPT7 missing the first N-terminal 76 residues (AtCPT7∆76N) failed to restore the growth (Figure 2A). Various other truncated versions of AtCPT7, up to the first 67 amino acids, also complemented the Δrer2 mutant (Supplemental Figures 6 and 8).
Figure 2.
Functional Complementation of the Yeast Dolichol (rer2Δ) Mutant by AtCPT7.
(A) The full-length (AtCPT7 FL) or three truncated (AtCPT7 Δ34N, -Δ7-67N, and -Δ76N) open reading frames of AtCPT7 were introduced into the yeast rer2Δ mutant, and serially diluted transformed cells were grown at the indicated temperatures. As positive and negative controls, the mutant was transformed with the native RER2 gene or the expression vector (EV) alone, respectively. Note that growth of the rer2Δ mutant is fully restored only when two of the truncated versions of AtCPT7 (-Δ34N and -Δ7-67N) or the native RER2 gene are expressed. The rer2Δ mutant strains containing AtCPT7 ∆7-67N and -∆76N were plated separately and are therefore presented as an individual group.
(B) Total polyisoprenoid content of the yeast strains indicated above was determined and a representative HPLC/UV chromatogram of this analysis is presented. Note the accumulation of dolichols (D-11 and D-12) in the rer2Δ mutant cells containing two of the truncated forms of AtCPT7 and dolichols composed of mainly D-15 and D-16 in cells containing the native RER2 gene.
(C) Restoration of N-glycosylation in the rer2Δ mutant by AtCPT7. Microsomal proteins from the various rer2Δ mutant strains above were resolved by SDS-PAGE and analyzed by immunoblotting using antibodies specific for CPY. The positions of mature CPY (mCPY) and its hypoglycosylated forms lacking between one and four (−1 to −4) N-linked oligosaccharide chains are indicated.
Figure 8.
A Model for Polyisoprenoid Biosynthesis in the Context of Leaf Isoprenoid Metabolism.
IPP and DMAPP are synthesized by both the cytosolic MVA and plastidial MEP pathways. The formation of trans-prenyl diphosphates from IPP and DMAPP in these respective cellular compartments results in a spatially distinct pool of plant polyisoprenoids: Cytosolic FPP is utilized for dolichol biosynthesis and the plastidial pool of GGPP is the precursor for polyprenol biosynthesis. Polyprenols, as well as other GGPP-derived isoprenoids, are incorporated into the thylakoid membranes and also into the envelope membranes of plastids. Intermediates: farnesyl diphosphate, FPP; glyceraldehyde-3-phosphate, G3P; geranyl diphosphate, GPP; geranylgeranyl diphosphate, GGPP. Dotted lines indicate multiple steps in the biosynthetic pathway. ω and α represent the terminal isoprene units in dolichols and polyprenols.
Next, total polyisoprenoid content was measured in these various Δrer2 mutant strains to determine whether the restoration of mutant growth by AtCPT7∆34N and AtCPT7∆7-67N correlated with the accumulation of dolichols. In Δrer2 mutant cells expressing these two truncated versions of AtCPT7, accumulation of predominantly medium-chain dolichols (Dol-11 and Dol-12) was observed (Figure 2B). These dolichols were not found in the Δrer2 mutant cells expressing the empty vector, full-length AtCPT7, or truncated AtCPT7∆76N lacking the first 76 residues (Figure 2B). As expected, expression of the native RER2 gene restored the synthesis of Dol-15 and Dol-16 in the Δrer2 mutant (Figure 2B). To explore whether the rescue of Δrer2 growth by the various truncated forms of AtCPT7 was due to the restoration of proper N-glycosylation, the glycosylation status of lysosomal CPY was assessed. In Δrer2 mutant cells expressing the native RER2, AtCPT7Δ34N, or AtCPT7Δ7-67N proteins, the fully glycosylated form of CPY containing four N-glycan chains predominated, whereas in cells expressing the empty vector, full-length AtCPT7, or AtCPT7∆76N, the mature and hypoglycosylated (mono-, di-, and tri-) forms of CPY were detected (Figure 2C). Taken together, these results indicate that the in vivo synthesis of medium-chain polyisoprenoids by AtCPT7, lacking its predicted organellar targeting peptide, can functionally replace yeast dolichols in the N-glycosylation pathway.
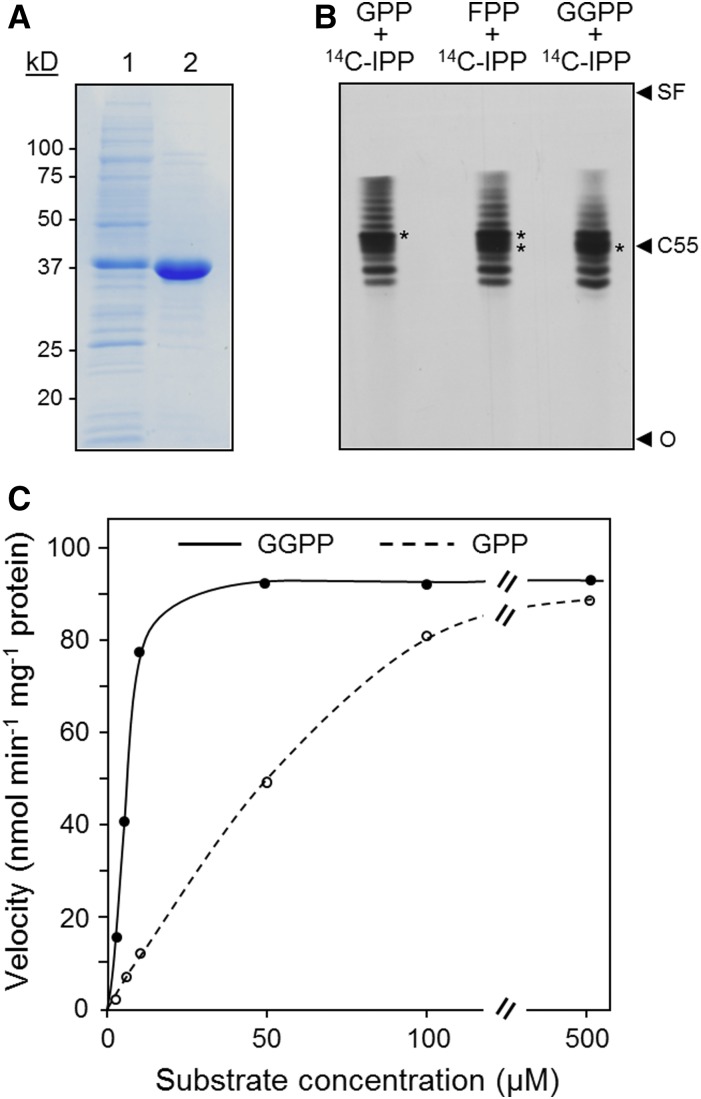
Recombinant AtCPT7 Enzyme Activity and Substrate Specificity
The involvement of AtCPT7 in the synthesis of medium-chain polyprenols in both Arabidopsis leaves and in yeast cells led us to explore which trans-prenyldiphosphate serves as its preferred substrate. AtCPT7, minus the predicted N-terminal targeting sequence (AtCPT7Δ34N), was expressed in E. coli as a C-terminal fusion protein with a hexahistidine tag and purified to near-homogeneity by Ni2+-affinity chromatography (Figure 3A). The recombinant protein was then assayed for CPT activity using 14C-IPP together with one of three trans-prenyldiphosphates (GPP, FPP, or GGPP) that are known to serve as initiator substrates in polyisoprenoid synthesis. The enzymatic products from these in vitro assays were resolved by thin-layer chromatography, and this analysis revealed that AtCPT7 could extend the length of either trans-prenyldiphosphate initiator to ∼50 to 55 carbons in length (Figure 3B). GPP, FPP, as well as GGPP all proved to serve as substrates for AtCPT7 and exhibited Michaelian kinetics at saturating concentrations of 14C-IPP (Figure 3C). However, steady state kinetic analysis revealed that the enzyme had a 10-fold preference for GGPP and FPP compared with GPP (Table 1), with kinetic constants that fell within the range reported for other plant and bacterial CPTs (Kharel et al., 2001; Schulbach et al., 2001; Guo et al., 2005; Schilmiller et al., 2009; Kera et al., 2012; Demissie et al., 2013). This point was further clarified by structural analysis (1H NMR) of Pren-10 (the dominating polyprenol in leaf tissue; Figure 1C) isolated from the leaves of wild-type Arabidopsis (Supplemental Figure 9). There was a signal from the purified Pren-10 compound corresponding to a chemical shift of δ 1.62 ppm, clearly confirming that the molecule contains three internal trans methyl groups (Ciepichal et al., 2007). Moreover, the ratio of methyl groups in cis versus trans configuration was ∼2.4, which is in agreement with a ω-t3-c6 structure. Although these results suggest that AtCPT7 prefers GGPP over FPP as a substrate both in vitro and in vivo, the spatial separation of GGPP (plastidial) and FPP (cytosolic) raised the question of where AtCPT7 resides in the cell and thereby has access to its bona fide substrate.
Figure 3.
Recombinant AtCPT7 Enzyme Activity and Substrate Specificity.
(A) Purification of His6-tagged AtCPT7 by Ni2+ affinity chromatography. Proteins were separated by SDS-PAGE and stained with Coomassie blue. For each protein, lane 1 was loaded with E. coli extract containing 10 µg of total protein and lane 2 with 2 µg of purified protein.
(B) Analysis of enzymatic reaction products. Recombinant AtCPT7 was incubated with 14C-IPP together with either FPP or GGPP and the dephosphorylated reaction products were resolved on reverse-phase silica gel 60-Å plates using an acetone/water (39:1) solvent system and developed by autoradiography. The position of C55 was determined based on the migration of authentic polyprenol standards of known size (C10–C120); the solvent front (SF) and origin (O) are also indicated.
(C) Recombinant AtCPT7 was assayed with various concentrations of either GPP or GGPP, and enzyme activity was determined by scintillation counting as described in Methods. Results are representative from experiments performed with three independent preparations of recombinant protein.
Table 1. Kinetic Parameters of AtCPT7 with GPP, FPP, or GGPP as Substrate.
| Substrate | Km (µM) | Kcat (s−1) | Kcat/Km (s−1 M−1) |
|---|---|---|---|
| GPP | 43.91 ± 11.35 | 0.106 ± 0.008 | 2.41 × 103 |
| FPP | 3.83 ± 0.72 | 0.104 ± 0.004 | 2.70 × 104 |
| GGPP | 5.87 ± 0.91 | 0.109 ± 0.004 | 1.85 × 104 |
Measurements were made at room temperature in 50 mM HEPES buffer, pH 8.0, containing, 5 mM DTT, 7.5 mM MgCl2, 100 mM KCl, 10% (v/v) glycerol, and 0.1% (v/v) Triton X-100. Reactions were started by adding 14C-IPP substrate. Data are the means of three independent determinations ± sd.
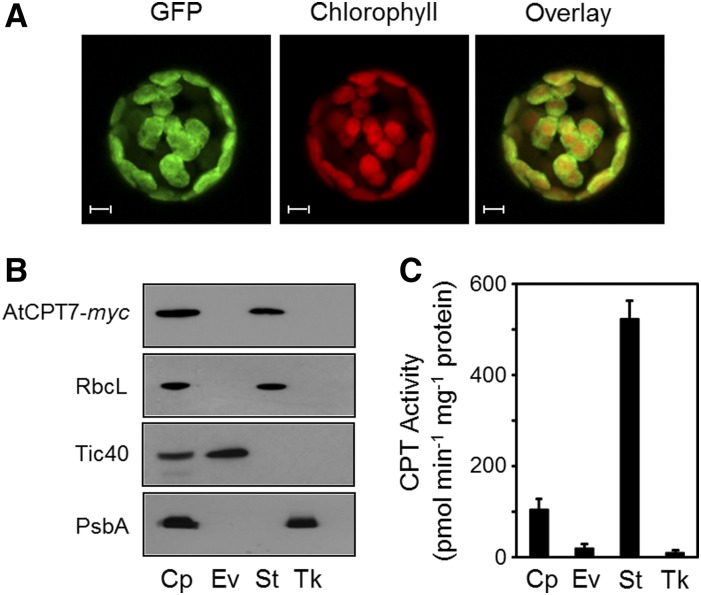
Subcellular Localization of AtCPT7
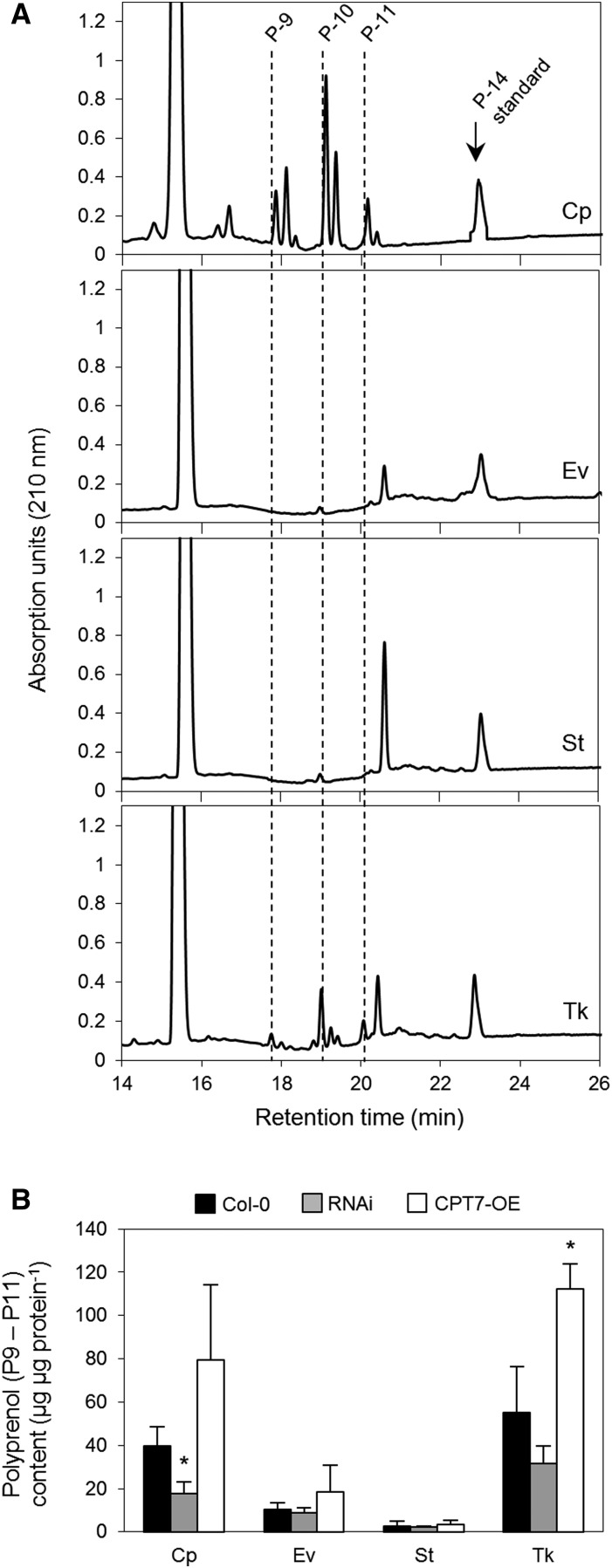
Many plant CPTs, including AtCPT7, have N-terminal extensions (albeit poorly conserved) relative to their mammalian, yeast, and prokaryotic homologs. Various algorithms predict these N-terminal extensions to be mitochondrial, endoplasmic reticulum, and/or plastidial targeting sequences (Supplemental Figure 7 and Supplemental Table 1). We used these in silico algorithms to predict the subcellular location of AtCPT7; however, no consensus was reached regarding its organellar targeting. We therefore generated Arabidopsis plants expressing AtCPT7 as a C-terminal fusion to GFP and tracked its localization via confocal microscopy. Fluorescence attributed to the GFP signal in both intact leaves and protoplasts that were prepared from these leaves exclusively overlapped with the chlorophyll autofluorescence, therefore suggesting a plastidial localization (Figure 4A; Supplemental Figure 10). Next, we isolated intact chloroplasts from Arabidopsis plants that stably expressed a C-terminal AtCPT7-myc fusion protein and immunologically monitored its presence in envelope, stroma, and thylakoid subcompartments to determine its precise location within the chloroplast. Marker proteins were used to establish the purity of each fraction, and this analysis indicated that AtCPT7 resides solely in the stroma (Figure 4B). Finally, chloroplasts that were isolated from wild-type Arabidopsis leaves and fractioned into the envelope, stroma, and thylakoid subcompartments were assayed for medium-chain CPT enzyme activity, using 14C-IPP and the plastid-localized GGPP as cosubstrates. These assays indicated that the majority of CPT enzyme activity that results in the synthesis of medium-chain polyprenols also resides in the stroma, coincident with the localization of AtCPT7 (Figure 4C).
Figure 4.
Subcellular Localization of AtCPT7.
(A) Protoplasts were prepared from Arabidopsis plants stably expressing GFP fused to the C terminus of AtCPT7. Fluorescence attributable to GFP, chlorophyll, and their merged signals were observed by confocal microscopy. Bars = 5 µm.
(B) Fractionation of chloroplasts from Arabidopsis plants stably expressing AtCPT7-myc. Intact chloroplasts (Cp) were fractioned into envelope (Ev), stroma (St), and thylakoid (Tk) compartments by sucrose gradient centrifugation. Protein samples from each fraction were resolved by SDS-PAGE and analyzed by immunoblotting using antibodies specific for the myc tag of AtCPT7, the Rubisco large subunit (RbcL), the Tic40 component of the inner envelope translocon, or the D1 reaction center protein (PsbA) of PSII.
(C) CPT enzyme activity in chloroplasts. Intact chloroplasts from wild-type Arabidopsis plants were fractioned as in (B), and each compartment was assayed for CPT enzyme activity using GGPP and 14C-IPP as substrates. Enzymatic products were analyzed and quantified as described in Methods, and the data represent the means ± sd from three independent experiments.
Plastidial Compartmentation of Medium-Chain Polyprenols
The plastidial localization of AtCPT7 together with the in vitro and in vivo characterization of the enzyme implies that the chloroplast is the site of medium-chain polyprenol biosynthesis, and possibly accumulation, in the leaves of Arabidopsis. However, considering the extreme hydrophobicity of polyprenols, it is difficult to reason that these compounds accumulate in the soluble milieu of the chloroplast stroma, where AtCPT7 resides. To further investigate this apparent inconsistency, we isolated intact chloroplasts from wild-type Arabidopsis leaves and compared their polyprenol content to those found in their envelope, stroma, and thylakoid subcompartments. The intact chloroplasts contained the same medium-chain polyprenols (Pren-9 to Pren-11) that were found in Arabidopsis leaves, and these compounds were mainly present in thylakoid membranes (Figure 5A). When polyprenol abundance in the various chloroplast subcompartments was normalized to protein content, a fraction of these compounds was also found in the envelope membranes, albeit to a lesser extent (Figure 5B). To establish that the accumulation of Pren-9 to Pren-11 in plastidial membranes was due to the activity of AtCPT7, we next measured polyprenol abundance in chloroplasts and in their various subcompartments from both the RNAi and CPT7-OE lines. The cpt7−/− knockout lines did not accumulate any medium-chain polyprenols (Figure 1D) and were therefore omitted from this analysis. Compared with the wild type, intact chloroplasts that were isolated from the three independent RNAi lines accumulated on average ∼66% less polyprenols, while a 2-fold increase in polyprenol abundance was observed in intact chloroplasts from the CPT7-OE lines (Figure 5B). Strikingly, these changes in plastidial polyprenol abundance were proportionately reflected in the thylakoid membranes isolated from the RNAi and CPT7-OE lines; an ∼60% decrease in thylakoid polyprenols was observed in the RNAi lines, while a ∼2-fold increase was observed in thylakoids from the CPT7-OE lines (Figure 5B). Moreover, the accumulation of these polyprenols mirrored total CPT enzyme activity in the stromal subcompartment from these lines (Supplemental Table 2). While the polyprenol content in the envelope membranes from the RNAi and CPT7-OE also corresponded with stromal CPT activity, their abundance was typically much lower compared with those found in thylakoids (Figure 5B; Supplemental Table 2).
Figure 5.
Plastidial Compartmentation of Polyprenols.
(A) Analysis of total polyisoprenoids. Intact chloroplasts (Cp) were isolated from Arabidopsis leaves and fractioned into envelope (Ev), stroma (St), and thylakoid (Tk) compartments by sucrose gradient centrifugation and analyzed for the presence of polyprenols (P-9 to P-11). Representative HPLC/UV chromatograms are presented and the vertical lines indicate the retention time of the main polyprenols (P-9, P-10, and P-11) that are present in the various compartments. Extracts were supplemented with an internal polyprenol (P-14) standard prior to extraction.
(B) Quantification of polyprenols (P-9 to P-11). Polyprenol content of intact chloroplasts (Cp) and envelope (Ev), stroma (St), and thylakoid (Tk) compartments were measured in the wild type (Col-0), three independent RNAi lines (RNAi), and an overexpression line (CPT7-OE). Note that the polyprenols present in intact chloroplasts mainly accumulate in the thylakoid and envelope compartments. Values (±sd) represent the mean of three independent experiments. Statistically significant differences compared with wild-type plants are indicated; *P < 0.05 by Student’s t test.
Polyprenol Deficiency Alters Thylakoid Membrane Dynamics
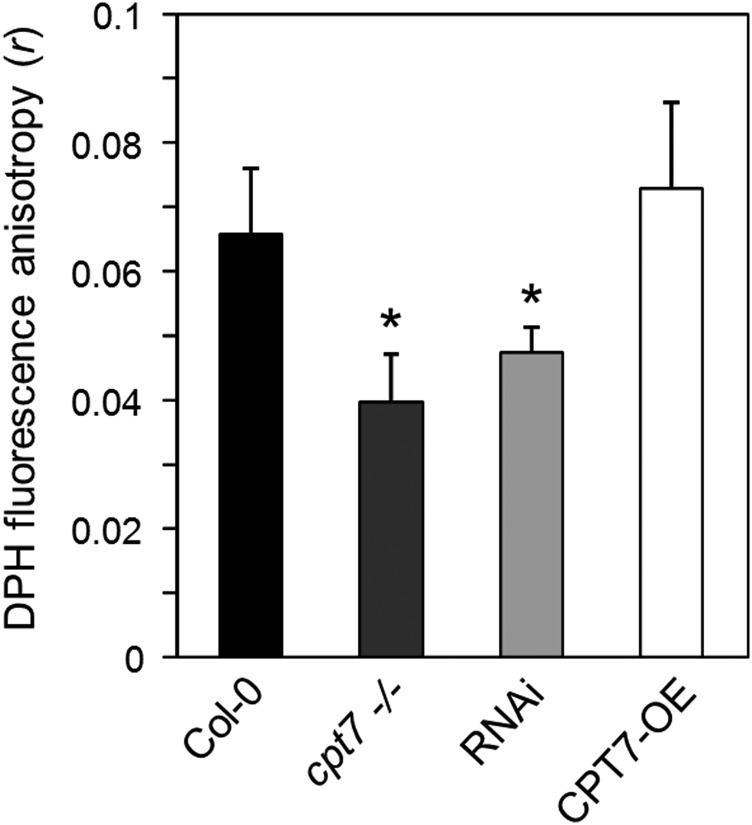
The incorporation of polyprenols into model membrane systems in vitro is well known to alter their membrane physico-chemical properties (Vigo et al., 1984; Valtersson et al., 1985; Schroeder et al., 1987; Janas et al., 1994; Wang et al., 2008; Sévin and Sauer, 2014). However, in vivo evidence to support these observations is lacking. Considering that the majority of polyprenols in Arabidopsis appear to accumulate in thylakoids, we compared the microviscosity of thylakoid membranes isolated from wild-type, polyprenol-deficient (cpt7−/−; RNAi), or polyprenol-hyperaccumulating (CPT7-OE) leaves using the technique of fluorescence anisotropy. Thylakoids from these leaves were incubated with the lipophilic fluorophore 1,6-diphenyl-1,3,5-hexatriene (DPH), which freely partitions into the hydrophobic interior of membrane bilayers (Ford and Barber, 1983; Lentz, 1993), and the extent of DPH fluorescence anisotropy was subsequently measured. In principle, the degree of DPH fluorescence anisotropy (r) from thylakoid membranes that are embedded with the fluoroprobe is determined by the rotational diffusion of DPH, which is ultimately dependent on the fluidity of its surrounding lipid environment (McCourt et al., 1987; Dobrikova et al., 1997; Popova and Hincha, 2007). Anisotropy measurements on polyprenol-deficient thylakoids from the cpt7−/− and RNAi lines revealed a significant decrease in r-values compared with those obtained from wild-type thylakoids, while no change in DPH anisotropy was observed in thylakoids isolated from the CPT7-OE lines (Figure 6). These results indicate that the rotational freedom of DPH is influenced by the amount of polyprenols within thylakoid membranes; thylakoids that are abundant in polyprenols appear to restrict DPH mobility, while in polyprenol-deficient thylakoids, the mobility of DPH is enhanced, thereby suggesting a more “fluid” or disordered state of the thylakoid bilayer. While these results point to a causal relationship between polyprenol abundance and thylakoid membrane fluidity, it is possible that other plastidial isoprenoids that are derived from GGPP and accumulate into thylakoids could influence membrane dynamics in much the same way. We therefore measured the content of selected GGPP-derived metabolites, i.e., tocopherols, phylloquinone, carotenoids, plastoquinone, and chlorophylls, in the leaf tissue and purified thylakoids from the wild type (Col-0), knockout (cpt7−/−), three independent RNAi lines (RNAi), and an overexpression line (CPT7-OE). The leaf content of these various isoprenoids (Table 2) was comparable to those previously reported for Arabidopsis (Pogson et al., 1996; Collakova and DellaPenna, 2003; Oostende et al., 2008; Eugeni Piller et al., 2014) and differed little between the various plant lines described above (Table 2; Supplemental Figures 11 to 13). Purified thylakoids from these lines also exhibited very similar levels of these GGPP-derived isoprenoids (Table 2; Supplemental Figures 11 to 13), suggesting that the observed change in their membrane dynamics was specific to polyprenol abundance.
Figure 6.
The Effect of Altered Polyprenol Levels on Thylakoid Membrane Fluidity.
Thylakoid membranes from the wild type (Col-0), a knockout line (cpt7−/−), three independent RNAi lines (RNAi), and an overexpression line (CPT7-OE) were embedded with the lipophilic fluorophore DPH. The steady state DPH anisotropy (r) values of each sample were determined by fluorescence anisotropy measurements. Data are the means ± sd from at least six independent experiments, and asterisks indicate a significant difference (Student’s t test, P < 0.05) when comparing r values to those obtained from wild-type samples.
Table 2. The Content of Plastidial Isoprenoids in Rosette Leaves and Purified Thylakoids from the Plant Lines Used in This Study.
| Rosette Leaf (µg g FW−1) |
Thylakoids (µg mg Chl−1) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild Type | cpt7−/− | RNAi | CPT7-OE | Wild Type | cpt7−/− | RNAi | CPT7-OE | |
| Tocopherols | 8.21 ± 1.76 | 7.68 ± 0.66 | 9.56 ± 2.87 | 9.71 ± 4.28 | 3.23 ± 0.32 | 3.95 ± 0.88 | 3.87 ± 1.23 | 3.25 ± 0.86 |
| Carotenoids | 203.36 ± 35.26 | 217.07 ± 21.21 | 211.05 ± 23.10 | 215.08 ± 27.71 | 232.29 ± 14.58 | 177.07 ± 20.24 | 221.64 ± 66.77 | 184.38 ± 26.56 |
| Plastoquinone | 35.22 ± 3.61 | 33.82 ± 1.48 | 39.9 ± 9.71 | 32.6 ± 2.03 | 60.97 ± 8.51 | 52.22 ± 20.72 | 74.28 ± 27.77 | 63.87 ± 16.46 |
| Phylloquinone | 1.63 ± 0.29 | 1.49 ± 0.27 | 1.62 ± 0.15 | 1.47 ± 0.48 | 2.38 ± 0.55 | 2.86 ± 0.51 | 2.37 ± 1.36 | 2.77 ± 0.25 |
The quantities of tocopherols, carotenoids, plastoquinone, and phylloquinone in the rosette leaves (per gram fresh weight [FW]) and purified thylakoids (per milligram of chlorophyll [Chl]) were determined from the indicated plant lines. Data are the means of at least three independent determinations ± sd, and bold numbers indicate significant difference (Student’s t test, P < 0.05) when comparing the quantity of various isoprenoids to those found in wild-type samples.
Polyprenol Deficiency Leads to Impaired Photosynthetic Performance
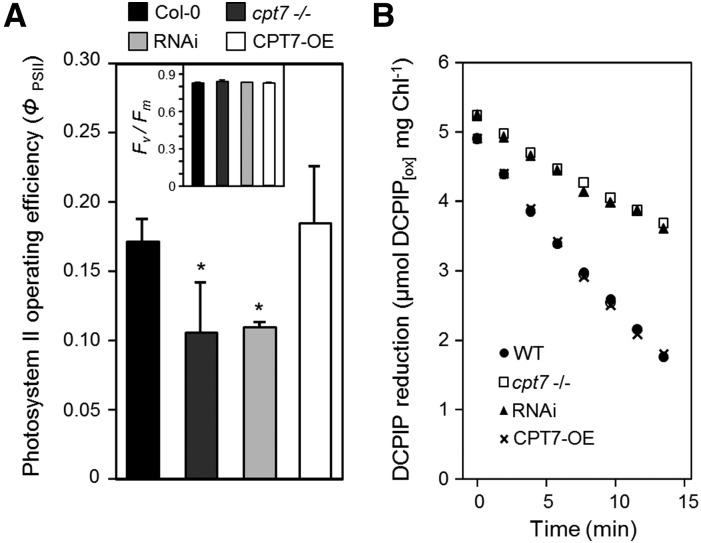
We next measured whether the observed change in thylakoid membrane fluidity, arising from altered polyprenol content, influences the efficiency of photosynthesis. Chlorophyll a fluorescence measurements on attached Arabidopsis leaves revealed that photosystem II (PSII) operating efficiency (ΦPSII) was compromised in the polyprenol-deficient cpt7−/− and RNAi lines, while only a slight increase in ΦPSII was observed in CPT7-OE lines, relative to wild-type plants (Figure 7A). However, the maximum quantum efficiency of PSII (Fv/Fm) did not differ between the wild type and leaves with altered polyprenol content (Figure 7A, inset) suggesting that the abundance of thylakoid polyprenols influences not PSII architecture, but rather photosynthetic electron transport, which is the main determinant of ΦPSII (Baker, 2008). Accordingly, we next measured the efficiency with which electrons are transferred from PSII to the cytochrome b6f complex (via plastoquinone), which is widely considered to be the rate-limiting step in photosynthetic electron transport (Haehnel, 1984; Heber et al., 1988; Hope et al., 1992; Laisk et al., 1992). Thylakoids from wild-type, polyprenol-deficient (cpt7−/−; RNAi), and polyprenol-hyperaccumulating (CPT7-OE) leaves were illuminated in the presence of the artificial electron acceptor 2,6-dichlorophenolindophenol (DCPIP), which “short-circuits” photosynthetic electron transport by accepting electrons from reduced plastoquinone (Govindjee and van Rensen, 1978). Monitoring DCPIP reduction over time therefore provides a proxy for rates of electron transport (Holloway et al., 1983). This analysis revealed an average decrease of ∼52% in the rate of electron transport in polyprenol-deficient thylakoids from the cpt7−/− and RNAi lines, compared with that measured in wild-type thylakoid membranes (Figure 7B). Thylakoids from the CPT7-OE lines exhibited nearly identical rates of electron transport as wild-type thylakoids. In support of these observations, the chlorophyll a fluorescence parameter 1 − qL, which reflects the redox level of the primary electron acceptor of PSII (QA) and is therefore considered to be a reliable measure of plastoquinone redox status (Hendrickson et al., 2004; Kramer et al., 2004; Baker, 2008; Akhtar et al., 2010), indicated a proportionally more reduced plastoquinone pool in the polyprenol-deficient lines compared with wild-type plants (Supplemental Table 3). Taken together, these observations suggest that polyprenol deficiency in thylakoid membranes impedes linear photosynthetic electron transport of the mobile plastoquinone electron acceptor and ultimately the efficiency of photosynthesis.
Figure 7.
The Effect of Altered Polyprenol Levels on Photosynthetic Performance.
(A) PSII operating efficiency (ΦPSII) and maximum quantum efficiency of PSII (Fv/Fm; inset) were derived from steady state chlorophyll a fluorescence measurements performed on dark-adapted rosette leaves from the wild type (Col-0), a knockout line (cpt7−/−), three independent RNAi lines (RNAi), and an overexpression line (CPT7-OE). Note the significant decline in ΦPSII in polyprenol-deficient leaves, while Fv/Fm remains unaltered in all lines. Data are the means ± sd from at least 10 independent experiments, and asterisks indicate a significant difference (Student’s t test; P < 0.05) when comparing ΦPSII values to those obtained from the wild type.
(B) Polyprenol deficiency results in a decline in photosynthetic electron transport. Thylakoid membranes from these plants were illuminated in the presence of the oxidized artificial electron acceptor 2,6-dichlorophenolindophenol (DCPIP[OX]), and its rate of reduction was monitored spectrophotometrically over the indicated time period. The rate of DCPIP reduction by thylakoids from each plant line is the average value from five independent preparations.
DISCUSSION
The accumulation of medium-chain polyprenols (Pren-9 to Pren-11) in the leaves of numerous plant species has long been recognized (Kurisaki et al., 1997; Sakaihara et al., 2000; Swiezewska and Danikiewicz, 2005; Skorupinska-Tudek et al., 2008). Here, we show via enzyme characterization, genetic manipulation, and polyisoprenoid analysis that a single member of the Arabidopsis CPT family, AtCPT7, is responsible for their synthesis. These compounds are produced in the chloroplast stroma and mainly accumulate in thylakoid membranes. Leaves that are deficient in polyprenols exhibit a reduced rate of photosynthetic electron transport together with a higher proportion of reduced plastoquinone, which suggests that the transfer of electrons between PSII and the cytochrome b6f complex is impeded by the mobility of plastoquinone through the lipid matrix of polyprenol-deficient thylakoids. However, the membrane-altering properties imparted by thylakoid polyprenols would be expected to affect not only the diffusivity of plastoquinone, but also the mobility and perhaps organization of the entire thylakoid membrane proteome, which occupies 70 to 80% of the membrane area (Kirchhoff et al., 2002). Indeed, the diffusion coefficient and packing densities of thylakoid protein complexes are intimately related to their lipid environment (Kirchhoff et al., 2008; Kirchhoff, 2014). Moreover, several eukaryotic proteins contain highly specific polyprenol recognition motifs that, upon interaction with their target polyprenols, alter protein conformation and their motional properties (Zhou and Troy, 2005). In bacteria, these compounds also promote protein-protein interactions (Hartley et al., 2013), so it is conceivable that plant polyprenols may interact with specific thylakoid membrane proteins, in addition to altering their lipid microenvironment and thereby influence a wide range of processes. Unfortunately, our data cannot resolve whether the defects observed in photosynthesis as a result of thylakoid polyprenol deficiency are solely due to altered plastoquinone mobility or arise from a combination of this and the efficacy of the target protein(s) with which polyprenols associate and/or indirectly influence.
In this context, plant polyprenols may therefore be viewed as “superlipids” that exert dynamic control over thylakoid membrane physiology. The apparently modest decrease in DPH fluorescence anisotropy (r) observed in polyprenol-deficient thylakoids indicates that polyprenols have an ordering effect on the thylakoid membrane bilayer. Such changes in DPH polarization are typically observed in vitro only when millimolar quantities of membrane-altering agents, such as sterols, SDS, or benzyl alcohol are incorporated into model membrane systems (Yamamoto et al., 1981; Vigo et al., 1984; Popova and Hincha, 2007; Lin et al., 2011). Our observations with polyprenol-deficient thylakoids are therefore striking when considering that on a molar basis, polyprenol abundance in thylakoid membranes is approximately an order of magnitude lower than in the model membranes described above.
The membrane-altering properties of polyprenols are well established in model in vitro systems. These compounds are believed to intercalate into only one leaflet of the bilayer, perpendicular to its plane, and are orientated in such a way that their unusually long poly-cis isoprenoid moiety modulates fatty acyl chain motion via hydrophobic interactions (Zhou and Troy, 2003). In these systems, polyprenols increase membrane ionic permeability and enhance membrane fluidity (Vigo et al., 1984; Valtersson et al., 1985; Janas et al., 1994; Ciepichal et al., 2011). Our results indicate an opposite effect: our anisotropy measurements revealed that thylakoid membranes deficient in polyprenols actually exhibited a more “fluid,” or perhaps disordered, state. The aforementioned studies using in vitro model membranes have largely ignored the effect of proteins on membrane dynamics, which upon incorporation, can in fact reverse the fluidizing effect that polyprenols exert on model membranes toward a more “ordered” state (Zhou and Troy, 2003). Moreover, the specific lipid composition of model membrane systems markedly affects the degree to which polyprenols influence membrane microviscosity (Sévin and Sauer, 2014) and the above studies did not assess how galactolipids, which account for >80% of thylakoid membrane lipids (Douce and Joyard, 1990), influence this behavior. We therefore postulate that in plant biomembranes, polyprenols may serve as a scaffold to organize and tether membrane proteins with their specific surrounding lipid microenvironment and thereby provide a stabilizing or “membrane ordering” effect, as originally proposed by Zhou and Troy (2003). Studies with synaptic mice membranes, thylakoids from barley (Hordeum vulgare) leaves, and various E. coli mutant cells that are deficient in polyisoprenoids lend support to this notion and suggest that polyprenols and structurally related isoprenoids can instead exert a stabilizing effect on biomembranes (Wood et al., 1989; Tardy and Havaux, 1997; Sévin and Sauer, 2014).
However, there appears to be a limit to the membrane-stabilizing properties of polyprenols; in thylakoids from the CPT7-OE lines, which contain nearly twice the amount of polyprenols, membrane fluidity was not significantly affected nor did the abundance of these polyprenols enhance photosynthetic operating efficiency or electron transport. This implies that a basal level of polyprenols is required to maintain or stabilize thylakoid membranes under standard plant growth conditions and that any further increases in polyprenol content offer little benefit. However, it is noteworthy that AtCPT7 is highly upregulated in response to various environmental stresses (Zimmermann et al., 2004) and that polyprenols accumulate in such cases (Bajda et al., 2009; Jozwiak et al., 2013; Milewska-Hendel et al., 2017). Given that plastid membranes experience changes in dynamics in response to stress, it is tempting to speculate that the stress-induced accumulation of polyprenols may serve a protective role for plastids in these environments, either as a ballast to support the integrity of thylakoids and/or envelope membranes or as an antioxidant, as recently postulated (Cavallini et al., 2016).
The physiological role of plastidial polyprenols may also transcend matters related to thylakoid membrane microviscosity and photosynthetic electron transport. Considering that a significant proportion of plastidial polyprenols also reside in the chloroplast envelope, it is tempting to speculate that these compounds also influence envelope dynamics, in much the same way as their thylakoid counterparts. Processes that are critically altered by any change in envelope fluidity, such as plastid division (Osteryoung and Nunnari, 2003), metabolite exchange (Mehrshahi et al., 2013), or the sporadic extension and retraction of stroma filled tubules known as stromules (Schattat et al., 2012; Mathur et al., 2013; Brunkard et al., 2015), would conceivably be affected by the abundance of polyprenols. The latter process is particularly relevant in this context as it has long been speculated that stromule formation, for which the mechanism is unknown, requires modulation of plastid envelope membrane architecture (Thomson and Whatley, 1980; Kwok and Hanson, 2004).
Among the other CPTs in Arabidopsis, only two members of the family have been characterized to date. AtCPT1 and AtCPT6 are expressed exclusively in root tissue, localize to the endomembrane system, and synthesize long-chain (C95-110) and short-chain (C35) polyprenols, respectively (Cunillera et al., 2000; Oh et al., 2000; Kera et al., 2012; Surmacz et al., 2014). The physiological function(s) of these polyprenols are still unclear. Strikingly, both experimental and in silico gene expression analysis indicates that a robust upregulation of AtCPT1 and AtCPT6 transcript levels occurs during various environmental stresses (Zimmermann et al., 2004; Kera et al., 2012; Jozwiak et al., 2013). Given the membrane-altering properties of plastidial polyprenols, it is tempting to speculate that the enzymatic products of AtCPT1 and AtCPT6 may also serve to alter root cell membrane dynamics under specific biotic or abiotic stresses and thereby influence various aspects of root membrane physiology related to morphology, translocation, and/or transport.
Finally, the observation that C55 polyprenols (Pren-11) can functionally substitute for the longer (∼C85) dolichols that are absent in the yeast Δrer2 mutant suggests that these shorter compounds are sufficient to restore proper N-glycosylation, as first observed by Rush et al. (2010). However, there appears to be a restriction on the chain length of polyprenols in this capacity as mediators in the posttranslational modification of eukaryotic proteins. AtCPT6, which produces C35 polyprenols specifically in root tissue, is incapable of rescuing the N-glycosylation defects when introduced into the Δrer2 mutant (Surmacz et al., 2014). This finding raises three critical questions regarding plant polyprenols: (1) What determines the chain length of the products that are synthesized by the various members of plant CPT families, (2) how are these variably sized compounds accommodated within the biomembranes in which they are found, and (3) what role do nonplastidial polyprenols serve?
Since the accidental discovery of polyprenols as contaminants in cellulose pulp extracts almost 50 years ago, their widespread occurrence in all kingdoms of life have been well recognized. Indeed, the co-occurrence of AtCPT7 and polyprenols in the plastids of Arabidopsis leaves fits well with previous reports that have documented the presence of these compounds and CPT enzyme activity in the chloroplasts of various plants (Lütke-Brinkhaus et al., 1985; Swiezewska et al., 1993; Sakaihara et al., 2000). However, these studies provided little insight into the size of the polyprenols that accumulate in plastids or which trans-prenyl diphosphate serves as the bona fide precursor for polyprenol biosynthesis. Our in vitro and in vivo characterization of AtCPT7 together with the structural characterization of the predominant polyprenol species that reside in Arabidopsis chloroplasts (Pren-10) suggests the following model: AtCPT7 and its orthologous plant CPTs extend the length of the C20 trans-prenyldiphosphate, GGPP, to ∼45 to 55 carbons in length and that the resulting polyprenols ultimately accumulate into plastidial membranes (Figure 8). This model predicts that polyprenols represent an additional branch point in plastidial isoprenoid metabolism which divert a portion of the available pool of GGPP away from the synthesis of indispensable isoprenoids, such as chlorophyll, carotenoids, gibberellins, tocopherols, and the electron carriers, plastoquinone and phylloquinone. Of course, isoprenoid precursors derived from the MVA pathway may also contribute to plastidial isoprenoid metabolism, and hence to polyprenol synthesis (Opitz et al., 2014). Additionally, the accumulation of polyprenols in thylakoid membranes appears to play a central role in the efficiency of photosynthetic performance by modulating thylakoid membrane dynamics. Hence, these findings shed light on the historically enigmatic role that plant polyprenols serve and offer insight into how these ubiquitous plant secondary metabolites should be considered in future strategies that are aimed to enhance plant productivity.
METHODS
Chemicals and Reagents
Authentic polyprenol and plastoquinone standards were from the Collection of Polyprenols, Institute of Biochemistry and Biophysics, Polish Academy of Sciences. Standards for phylloquinone, menaquinone, and α-, β-, γ-, and δ-tocopherol were obtained from Sigma-Aldrich. Tocol was from Matreya. All trans-prenyl diphosphates were obtained from CedarLane laboratories and radiolabeled 14C-IPP, 50.6 mCi mmol−1 (1.872 GBq mmol−1; 0.02 mCi mL−1) was from Perkin-Elmer. Polyclonal antibodies against the Rubisco large subunit (anti-RbcL; ASO3037, lot no. 1409), anti-Tic40 (AS10709, lot no. 1005), and the D1 reaction center protein (anti-psbA; AS05084, lot no. 1412) were obtained from Agrisera. Monoclonal anti-CPY (PA1-27244, lot no. PJ1923387) was from Thermo Fisher, and anti-myc-HRP (460709, lot no. 1,514,778) was from Sigma-Aldrich. HRP-conjugated anti-rabbit (AS09602, lot no. 1406) was from Agrisera. Macerozyme R-10 and Cellulose “Onozuka” R-10 enzymes (for Arabidopsis thaliana protoplast isolation) were from Yakult. Restriction enzymes and all other molecular biology reagents were from Thermo Scientific and/or Invitrogen. DCPIP and DPH were obtained from Sigma-Aldrich. Synthetic dropout media lacking amino acids for yeast cultures were obtained from Roth. Thin-layer chromatography plates (RP-18, silica gel 60, 200 μm, 20 cm × 20 cm) were obtained from Mandel Scientific, and high-performance liquid chromatography columns were from Agilent. All other chemicals were obtained from Sigma-Aldrich.
Cloning of AtCPT7 and Quantitative PCR Analysis
Total RNA was isolated from Arabidopsis leaf tissue using the RNeasy Plant Mini Kit (Qiagen), according to the manufacturer’s protocols. The mRNA was reverse-transcribed to cDNA using RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific), and sequences encoding full and truncated forms of AtCPT7 were generated by PCR cloned into the pENTR D-TOPO vector system, according to manufacturer’s protocols (Invitrogen). Quantitative RT-PCR analysis was performed in a Step One-Plus Real-Time PCR System (Applied Biosystems) using SYBR GREEN Master Mix (ABI) and the ABI universal cycling conditions. All reactions were performed in triplicate along with a no-template control, and the relative expression levels were determined according to the 2−ΔΔCt method (Livak and Schmittgen, 2001) normalized against the PP2AA3 gene (At1g13320) or the UBQ10 (At4g05320) gene. All primers that were used in PCR analysis and their sequences are listed in Supplemental Table 4.
Plant Materials
Arabidopsis (Col-0) plants were routinely grown in potting soil or cultured hydroponically (Gibeaut et al., 1997) in growth chambers maintained under a 16-h photoperiod (150 µmol m−2 s−1; mixed cool white and incandescent bulbs) or short-day (8 h light/16 h dark) conditions. The At5g58770 T-DNA insertion mutant SALK_022111 (cpt7−/−) was obtained from the Nottingham Arabidopsis Stock Center and genotyped by PCR. For RNAi-mediated knockdown of the gene encoded by At5g58770, two PCR products corresponding to bases 23 to 778 and 23 to 572 of the open reading frame were ligated in a sense/antisense orientation between the SacI/BamHI sites within the pSAT4A vector to form a hairpin construct under the control of the cauliflower mosaic virus 35S promoter and terminator (Tzfira et al., 2005). The hairpin cassette was transferred into the pPZP200 binary vector (Hajdukiewicz et al., 1994) and mobilized into Agrobacterium tumefaciens strain GV3101, which was then used to transform Arabidopsis (Col-0) according to the floral dip method (Clough and Bent, 1998). Transformation of Arabidopsis to express AtCPT7-myc and AtCPT7-GFP as C-terminal fusion proteins was similarly achieved by ligating the full-length open reading frame of AtCPT7 between the XhoI/BamHI sites of pSAT4A and pSAT6A, respectively. AtCPT7 overexpression lines (AtCPT7-OE) were generated by recombining the AtCPT7 full-length coding sequence from the pENTR clone with the Imp GWB 402 binary vector, via the Gateway LR Clonase system (Invitrogen) and then subsequently introducing it into Arabidopsis, as above. Transgenic plants were selected on Murashige and Skoog agar plates supplemented with kanamycin (50 mg/L) and verified by PCR.
Extraction and Analysis of Polyisoprenoids
Approximately 3 g of fresh Arabidopsis rosette leaves were homogenized using an Ultra-Turrax T25 (IKA Labortechnik), and lipids were extracted with a mixture of chloroform:methanol (1:1, v/v) for 48 h at room temperature. The extract was filtered and the remaining tissue was reextracted with chloroform:methanol (2:1, v/v). The extracts were combined, evaporated under a stream of nitrogen, and dissolved in a mixture containing toluene/7.5% KOH/95% ethanol (20:17:3, v/v). Following hydrolysis for 1 h at 95°C, polyisoprenoids were extracted three times with hexane, applied to a silica gel 60 column, and purified using isocratic elution with 10% diethyl ether in hexane, as described by Surmacz et al. (2014). Polyisoprenoids from intact chloroplasts and plastidial subcompartments (see below) were extracted as above. Fractions containing polyisoprenoids were pooled, evaporated, dissolved in 2-propanol, and analyzed by HPLC as described earlier (Skorupińska-Tudek et al., 2003). Extracts were separated by HPLC (Waters) using a Zorbax XDB-C18 (4.6 × 75 mm, 3.5 μm) reverse-phase column (Agilent) and polyisoprenoids eluted with a linear gradient from 0% to 100% methanol:isopropanol:hexane (2:1:1) in water:methanol (1:9) at a flow rate of 1.5 mL/min. Polyisoprenoids were detected by absorption at 210 nm and quantified relative to authentic standards. To analyze the content of polyprenyl esters, lipids from Arabidopsis chloroplasts were extracted with chloroform/methanol/water (C/M/W) 1:1:0.3 (by volume) for 3 d at RT and the extract was adjusted to a final C/M/W ratio of 3:2:1 (by volume). The lower organic phase was evaporated to dryness. The extracted lipids were dissolved in hexane and purified on a silica gel 60 column using gradient elution with 2 to 10% diethyl ether in hexane. All fractions containing both native and nonsaponifiable lipids were analyzed by HPLC/UV. The fraction containing purified polyprenyl esters was subjected to alkaline hydrolysis and the liberated polyprenols were used for estimation of the respective alcohols. Quantitative determination of polyprenols was performed using Pren-10 as external standard. Yeast polyisoprenoids were extracted from ∼3 g of pelleted yeast cells harvested at log phase (OD600 = 1), with 10 mL of a hydrolytic solution (25% KOH in 65% ethanol) for 1 h at 95°C, and analyzed as described above. All samples were supplemented with an internal polyprenol standard (Pren-14 or Pren-28), prior to extraction, and analysis was performed in triplicate.
Analysis of Plastidial Isoprenoids
All plastidial isoprenoids were extracted from 4-week-old Arabidopsis rosette leaf tissue (∼50 mg) and thylakoids (∼150 µg of chlorophyll) purified from this tissue, as described below. For phylloquinone analysis, samples were ground to a fine powder in liquid nitrogen and prenyllipids were extracted in ethanol:water (0.45 mL, 95:5, v/v), containing 1 μM menaquinone (MK-4) as an internal standard. Clarified extracts (50 µL) were analyzed by HPLC on a 5-μm Discovery C-18 column (250 × 4.6 mm; Supelco) maintained at 30°C, according to the method described by Oostende et al. (2008). Samples were eluted using an isocratic method with a flow rate of 1 mL min−1 with methanol:ethanol (80:20, v/v) containing 1 mM sodium acetate, 2 mM acetic acid, and 2 mM ZnCl2. Naphthoquinone species were detected by fluorescence (238 nm and 426 nm for excitation and emission, respectively) following their online reduction in a post-column guard column (2 mm i.d. × 2 cm; Western Analytical) packed with -100 mesh zinc dust (Sigma-Aldrich), with a detection limit of 500 fmol. Reduced phylloquinol was undetectable in the analysis, whereas detection of MK-4 and phylloquinone was observed at retention times of 7.4 and 10.8 min, respectively. Phylloquinone was quantified according to external calibration standards and data were corrected for recovery of MK-4 added to samples at the beginning of the extraction. For tocopherol analysis, samples were ground to a fine powder in chloroform:methanol (0.3 mL, 1:2, v/v), containing 7.5 μM Tocol as an internal standard, followed by addition of chloroform and water to achieve phase separation at a final chloroform:methanol:water ratio of (v/v/v). Following phase separation, the organic phase containing the prenyllipids was collected and evaporated to dryness under N2 gas. The dried prenyllipids were dissolved in 100 μL of hexane and 50 μL was analyzed by HPLC on a 5-μm LiChrosorb Si60A normal phase column (250 × 4.6 mm; Supelco) maintained at 42°C. Samples were eluted using an isocratic method with a flow rate of 2.1 mL min−1 with hexane:diisopropyl ether (92:8, v/v). Tocopherols were detected by fluorescence (290 and 325 nm for excitation and emission, respectively) with a detection limit of 125 pmol. Retention times of α-tocopherol (α-Toc), γ-tocopherol (γ-Toc), and Tocol were 6.8, 12.2, and 21.8 min, respectively. β-Tocopherol was present in trace amounts whereas δ-tocopherol was undetectable in Arabidopsis rosette leaf tissue. α-Toc and γ-Toc were quantified according to external calibration standards and data were corrected for recovery of Tocol added to the samples at the beginning of the extraction. For plastoquinone analysis, samples (∼0.5 g leaf tissue; ∼150 µg chlorophyll) were homogenized in 6 mL of chloroform:methanol (1:1, v/v) and extracted for 24 h at 25°C, followed by addition of 1.5 mL chloroform and 1 mL water to separate aqueous and organic phases. The chloroform phase was collected and the water phase was reextracted with a fresh volume of chloroform. Both chloroform fractions were pooled and evaporated to dryness under N2 gas. Samples were analyzed by HPLC on a 3.5 µm Zorbax XBD-C18 column (4.6 × 75 mm) and separated with the following solvent system: Solvent A, methanol:water (9:1, v/v); Solvent B, methanol:hexane:propan-2-ol (2:1:1, v/v/v): starting from 100 to 35% A for 3 min, 35 to 0% A for 7 min, and 100% B for 5 min. Plastoquinone was detected using a dual λ Absorbance Detector or Waters Photodiode Array Detector and quantified at λ 210 nm using external standards. For carotenoid analysis, samples (∼0.2 g leaf tissue; ∼150 µg chlorophyll) were homogenized in 3 mL acetone, and carotenoids in clarified extracts were quantified spectrophotometrically at 470 nm, according to the method of Lichtenthaler and Buschmann (2001).
Functional Complementation of the Yeast Δrer2 Mutant
The wild-type yeast strain SS328 (MATα ade2-101 ura3-52 his3Δ 200 lys2-801) was cultured in yeast peptone dextrose (YPD) medium and the rer2Δ mutant strain YG932 (MATα rer2Δ::kanMX4 ade2-101 ura3-52 his3Δ 200 lys-801) (Sato et al. 1999) in YPD supplemented with G418 (200 mg L−1). The full-length and truncated forms of AtCPT7 were recombined from the pENTR vector (see above) with the yeast expression vector pYES-DEST52 using the Gateway LR Clonase system (Invitrogen). The sequence-verified constructs were introduced into the rer2Δ mutant strain, and transformants were selected on 0.67% yeast nitrogen base, 2% galactose, and all necessary auxotrophic requirements except uracil. In vivo complementation assays and analysis of carboxypeptidase Y (CPY) glycosylation status were performed as previously described (Surmacz et al., 2014). For subcellular localization in yeast cells, the full-length and truncated (AtCPT7∆34N) versions of AtCPT7 were amplified by PCR using the pSAT6A-AtCPT7-GFP construct (see above) as a template and the primers listed in Supplemental Table 4. The amplicons were then ligated between the SmaI/BamHI sites of the pRS423 vector which drives expression of the AtCPT7-GFP fusions from the native RER2 promoter (Akhtar et al., 2013; Brasher et al., 2015). The constructs were introduced into the yeast rer2∆ mutant and transformants were selected on 0.67% yeast nitrogen base, 2% glucose, and all necessary auxotrophic requirements except histidine.
Subcellular Localization and Chloroplast Fractionation
Protoplasts were prepared from transgenic Arabidopsis (Col-0) plants expressing AtCPT7 as a C-terminal fusion (see above) via the tape-sandwich method (Wu et al., 2009) and fluorescence attributed to the GFP signal (500–539 nm) and chlorophyll autofluorescence (650–700 nm) were visualized with a Leica SP5 confocal laser scanning microscope equipped with 488-nm argon laser for fluorophore excitation. Chloroplast subcompartments were prepared from intact chloroplasts that were isolated from ∼15 g of 3- to 5-week-old Arabidopsis rosette leaf tissue, according to the method described by Hall et al. (2011) for stroma and thylakoid fractions and by Salvi et al. (2011) for envelope membranes. The purity of each fraction was assessed by immunoblot analysis using antibodies against the stromal Rubisco large subunit (RbcL), the Tic40 component of the envelope translocon, or the thylakoid D1 reaction center protein (psbA) of PSII. A minimum of three independent preparations of each chloroplast subcompartment were obtained for measurements of polyprenol content and CPT enzyme activity from all genotypes used in this study.
Expression and Purification of Recombinant AtCPT7 in Escherichia coli
The open reading frame of AtCPT7 was amplified by PCR beginning at Ser-34 to remove the predicted targeting peptide, transferred into the pEXP-5-CT/TOPO expression vector, which introduces an in-frame C-terminal hexahistidine tag extension, and then introduced into BL21-CodonPlus (DE3)-RIPL E. coli cells. Bacteria containing this construct were grown at 37°C in the presence of the appropriate antibiotics until the optical density at 600 nm reached 0.6, at which point isopropyl-β-d-thiogalactoside was added to a final concentration of 1 mM, and then incubated for an additional 20 h at 16°C. Cell pellets were resuspended in lysis buffer (20 mM sodium phosphate, pH 7.4, and 200 mM KCl), disrupted with a French press, and centrifuged at 5000g for 10 min at 4°C to remove unbroken cells and debris. The clarified cell lysates were applied to a 1 mL HisTrap HP column equilibrated in lysis buffer, and bound protein was eluted with lysis buffer containing 400 mM imidazole and then immediately desalted on PD-10 columns (GE Healthcare) equilibrated with 50 mM HEPES, pH 8.0, 100 mM KCl, 7.5 mM MgCl2, 5 mM DTT, 0.1% (v/v) Triton X-100, and 10% (v/v) glycerol. Protein concentration of the purified protein was determined by the method of Bradford (1976) using BSA as a standard.
CPT Enzyme Assays
Assays for determining AtCPT7 enzyme activity were performed in a 50 μL reaction volume by incubating ∼1 µg of purified recombinant protein in 50 mM HEPES, pH 8.0, 100 mM KCl, 7.5 mM MgCl2, 5 mM DTT, 5% (v/v) glycerol, and 0.1% (v/v) Triton X-100 with 20 µM trans-prenyl diphosphate acceptors (GPP, FPP, or GGPP), and initiated by adding 14C-IPP (50 µCi mmol−1) at a final concentration of 80 µM (200 nCi). After 30 min at room temperature, reactions were terminated by the addition of HCl at a final concentration of 0.6 M and incubated for 30 min at 37°C. The hydrolyzed products were extracted with three volumes of ethyl acetate and quantified by scintillation counting, and the remaining extract was applied to a reverse-phase silica gel 60-Å thin-layer chromatography plate, which was developed in acetone:water (39:1) and visualized by autoradiography. Enzymatic product size was determined with cochromatographed authentic standards and visualized by iodine-vapor staining (Akhtar et al., 2013). For determining kinetic constants, assays were performed as above with varying concentrations of trans-prenyl diphosphate substrates and a fixed concentration of 14C-IPP of 80 µM over a period of 8 min. Apparent Km and Vmax values were determined by nonlinear regression analysis using the Michaelis-Menten kinetics model of the SigmaPlot 12.3 software. CPT enzyme activity in the various plastidial subcompartments was assessed by incubating ∼5 µg of total protein preparation with 20 µM GGPP and initiated by adding 14C-IPP as described above. The radiolabeled enzymatic products were resolved by TLC and zones corresponding to C45 to C60 in length were scraped from the TLC plate and quantified by scintillation counting.
Chlorophyll a Fluorescence and Photosynthetic Electron Transport Measurements
Modulated chlorophyll a fluorescence was measured from dark-adapted attached leaves using a portable PAM fluorometry system (FluorPen FP-100; Quibit Biology). Steady state fluorescence parameters were gathered after an initial saturating pulse of white light (2100 µmol m−2 s−1) followed by saturating pulses every 20 s in the presence of continuous illumination with actinic light (300 µmol m−2 s−1) for a duration of 5 min. The light-adapted minimal fluorescence was determined during a 6-min dark recovery period with saturating pulses every 60 s immediately after the actinic light was turned off. The maximal photochemical efficiency (Fv/Fm), nonphotochemical quenching of PSII fluorescence (NPQ), the PSII operating efficiency (ΦPSII), the coefficient of photochemical quenching (qP), and the fraction of open PSII centers (qL) according to the lake model (Kramer et al., 2004) were determined according to the protocols outlined by Baker (2008). Linear photosynthetic electron transport rates were measured using thylakoid membranes prepared from ∼1.5 g of 4-week-old Arabidopsis rosette leaves according to the method of Chen et al. (2016) in the presence of the artificial electron acceptor DCPIP, as described by Dean and Miskiewicz (2003). Assays contained thylakoids (20 µg chlorophyll) suspended in 25 mM Tricine-NaOH, pH 7.8, 100 mM sorbitol, 5 mM MgCl2, and 10 mM NaCl and were initiated by the addition of oxidized DCPIP at a final concentration of 30 µM. The rate of DCPIP reduction was monitored spectrophotometrically at 600 nm.
Thylakoid Membrane Fluidity Measurements
Estimations of membrane fluidity were determined by fluorescence anisotropy measurements of thylakoid membranes (∼100 µg of chlorophyll) containing the hydrophobic fluorophore DPH. Thylakoid membranes were isolated as above and incubated with 10 µM DPH at room temperature for 40 min in the dark, collected by centrifugation at 2500g for 5 min, and then resuspended in DPH fluorescence buffer (100 mM sorbitol, 25 mM Tricine, pH 7.8, and 10 mM NaCl) to a final concentration of 20 µg mL−1 chlorophyll and 2 µM DPH. Fluorescence anisotropy measurements of the labeled membranes were performed using a Photon Technology International model 814 spectrofluorometer system equipped with polarization filters for excitation and emission wavelengths of 360 and 460 nm, respectively (slit widths 10 nm). The degree of fluorescence anisotropy (r) was calculated according to Ford and Barber (1983).
Polyprenol Purification and Structural Characterization via NMR Analysis
Polyprenols were purified from wild-type Arabidopsis leaf tissue according to the methods previously described by Jozwiak et al. (2013).1H NMR spectra were obtained with a Varian INOVA 400-MHz spectrometer at 25°C in C6D6 (D-99.5%; Cambridge Isotope Laboratories). A total of 32,000 data points were collected and a spectral width of 6 kHz was used. Spectra were calibrated against the residual chemical shift of benzene (7.16 ppm).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: AtCPT1 (At2g23410), AtCPT2 (At2g23400), AtCPT3 (At2g17570, AtCPT4 (At5g60510), AtCPT5 (At5g60500), AtCPT6 (At5g58780), AtCPT7(At5g58770), AtCPT8 (At5g58782), and AtCPT9 (At5g58784).
Supplemental Data
Supplemental Figure 1. AtCPT7 gene expression and polyisoprenoid content in seven AtCPT7 overexpression lines (CPT7-OE).
Supplemental Figure 2. Expression of the Arabidopsis CPT family members in wild type (Col-0), homozygous T-DNA AtCPT7 loss-of-function (cpt7−/−), and AtCPT7 overexpression lines (CPT7-OE).
Supplemental Figure 3. Aerial growth phenotypes of the 5-week-old Arabidopsis plants used in this study.
Supplemental Figure 4. HPLC/UV analysis of polyisoprenoid alcohols and their carboxylic acid esters.
Supplemental Figure 5. Subcellular localization of AtCPT7 in yeast.
Supplemental Figure 6. Protein sequences of the full-length and various truncated forms of AtCPT7 used in functional complementation assays with the yeast ∆rer2 mutant.
Supplemental Figure 7. Multiple sequence alignment of AtCPT7 and its homologs in other organisms.
Supplemental Figure 8. Functional complementation of the yeast dolichol (rer2Δ) mutant by various truncated forms of AtCPT7.
Supplemental Figure 9. 1H NMR spectrum of Pren-10 isolated from the leaves of wild-type Arabidopsis plants.
Supplemental Figure 10. Subcellular localization of AtCPT7 in Arabidopsis leaves.
Supplemental Figure 11. Tocopherol analysis of the various Arabidopsis lines used in this study.
Supplemental Figure 12. Phylloquinone analysis of the various Arabidopsis lines used in this study.
Supplemental Figure 13. Chlorophyll, carotenoid, and plastoquinone analysis of the various Arabidopsis lines used in this study.
Supplemental Table 1. The Arabidopsis cis-prenyltransferase family.
Supplemental Table 2. cis-Prenyltransferase enzyme activity in chloroplasts with altered polyprenol content.
Supplemental Table 3. Chlorophyll a fluorescence parameters in the wild type and in Arabidopsis leaves with altered polyprenol content.
Supplemental Table 4. Synthetic oligonucleotides used in this study.
Supplemental File 1. ANOVA tables.
Acknowledgments
We thank Jeffrey Rush and Charles Waechter (University of Kentucky) for providing the rer2∆ strain, Michael Mucci and Tannis Slimmon (University of Guelph) for help with plant growth maintenance, Michaela Strüder-Kypke (University of Guelph) for technical advice on confocal microscopy, and Steffen Graether and Janet Wood (University of Guelph) for expertise and assistance with fluorescence anisotropy. This work was supported by grants from the National Science Centre of Poland (UMO-2011/03/B/NZ1/00568 and UMO-2016/21/B/NZ1/02793 to L.S. and UMO-2012/07/B/NZ3/02437 to E.S.), from the Ontario Ministry of Agriculture, Food and Rural Affairs (UofG2014-2169), and from the Natural Sciences and Engineering Research Council of Canada (2014-05628 to T.A.A.).
AUTHOR CONTRIBUTIONS
T.A.A. and L.S. designed the research. T.A.A., P.S., H.S., M.K., K.V.G., K.A.R., L.K.A.V., M.V., W.D., D.B., K.G., J.W., and L.S. performed the research. T.A.A., W.D., E.S., and L.S. analyzed the data. T.A.A. and L.S. wrote the article.
Glossary
- DMAPP
dimethylallyl diphosphate
- IPP
isopentenyl diphosphate
- MVA
mevalonic acid
- MEP
methylerythritol phosphate
- GPP
geranyl diphosphate
- FPP
farnesyl diphosphate
- GGPP
geranylgeranyl diphosphate
- CPT
cis-prenyltranferase
- DPH
1,6-diphenyl-1,3,5-hexatriene
- DCPIP
2,6-dichlorophenolindophenol
Footnotes
Articles can be viewed without a subscription.
References
- Akhtar T.A., Lees H.A., Lampi M.A., Enstone D., Brain R.A., Greenberg B.M. (2010). Photosynthetic redox imbalance influences flavonoid biosynthesis in Lemna gibba. Plant Cell Environ. 33: 1205–1219. [DOI] [PubMed] [Google Scholar]
- Akhtar T.A., Matsuba Y., Schauvinhold I., Yu G., Lees H.A., Klein S.E., Pichersky E. (2013). The tomato cis-prenyltransferase gene family. Plant J. 73: 640–652. [DOI] [PubMed] [Google Scholar]
- Bajda A., et al. (2009). Role of polyisoprenoids in tobacco resistance against biotic stresses. Physiol. Plant. 135: 351–364. [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113. [DOI] [PubMed] [Google Scholar]
- Bartram S., Jux A., Gleixner G., Boland W. (2006). Dynamic pathway allocation in early terpenoid biosynthesis of stress-induced lima bean leaves. Phytochemistry 67: 1661–1672. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brasher M.I., Surmacz L., Leong B., Pitcher J., Swiezewska E., Pichersky E., Akhtar T.A. (2015). A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 82: 903–914. [DOI] [PubMed] [Google Scholar]
- Brunkard J.O., Runkel A.M., Zambryski P.C. (2015). Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 112: 10044–10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini G., Sgarbossa A., Parentini I., Bizzarri R., Donati A., Lenci F., Bergamini E. (2016). Dolichol: A component of the cellular antioxidant machinery. Lipids 51: 477–486. [DOI] [PubMed] [Google Scholar]
- Chen Y.E., Yuan S., Schröder W.P. (2016). Comparison of methods for extracting thylakoid membranes of Arabidopsis plants. Physiol. Plant. 156: 3–12. [DOI] [PubMed] [Google Scholar]
- Ciepichal E., Jemiola-Rzeminska M., Hertel J., Swiezewska E., Strzalka K. (2011). Configuration of polyisoprenoids affects the permeability and thermotropic properties of phospholipid/polyisoprenoid model membranes. Chem. Phys. Lipids 164: 300–306. [DOI] [PubMed] [Google Scholar]
- Ciepichal E., Wojcik J., Bienkowski T., Kania M., Swist M., Danikiewicz W., Marczewski A., Hertel J., Matysiak Z., Swiezewska E., Chojnacki T. (2007). Alloprenols: novel α-trans-polyprenols of Allophylus caudatus. Chem. Phys. Lipids 147: 103–112. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collakova E., DellaPenna D. (2003). Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol. 131: 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N., Arró M., Forés O., Manzano D., Ferrer A. (2000). Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 477: 170–174. [DOI] [PubMed] [Google Scholar]
- Dean R.L., Miskiewicz E. (2003). Rates of electron transport in the thylakoid membranes of isolated, illuminated chloroplasts are enhnaced in the presence of ammonium chloride. Biochem. Mol. Biol. Educ. 31: 410–417. [Google Scholar]
- Demissie Z.A., Erland L.A.E., Rheault M.R., Mahmoud S.S. (2013). The biosynthetic origin of irregular monoterpenes in Lavandula: isolation and biochemical characterization of a novel cis-prenyl diphosphate synthase gene, lavandulyl diphosphate synthase. J. Biol. Chem. 288: 6333–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova A.G., Tuparev N.P., Krasteva I., Busheva M.H., Velitchkova M.Y. (1997). Artificial alterations of fluidity of pea thylakoid membranes and its effect on energy distribution between both photosystems. Z. Naturforsch. 52: 475–480. [Google Scholar]
- Douce R., Joyard J. (1990). Biochemistry and function of the plastid envelope. Annu. Rev. Cell Biol. 6: 173–216. [DOI] [PubMed] [Google Scholar]
- Dudareva N., Andersson S., Orlova I., Gatto N., Reichelt M., Rhodes D., Boland W., Gershenzon J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102: 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugeni Piller L., Glauser G., Kessler F., Besagni C. (2014). Role of plastoglobules in metabolite repair in the tocopherol redox cycle. Front. Plant Sci. 5: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford R.C., Barber J. (1983). Incorporation of sterol into chloroplast thylakoid membranes and its effect on fluidity and function. Planta 158: 35–41. [DOI] [PubMed] [Google Scholar]
- Gibeaut D.M., Hulett J., Cramer G.R., Seemann J.R. (1997). Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115: 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindjee J.J.S., van Rensen J.J.S. (1978). Bicarbonate effects on the electron flow in isolated broken chloroplasts. Biochim. Biophys. Acta 505: 183–213. [DOI] [PubMed] [Google Scholar]
- Grabińska K.A., Park E.J., Sessa W.C. (2016). Cis-prenyltransferase: new insights into protein glycosylation, rubber synthesis and human disease. J. Biol. Chem. 291: 18582–18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R.T., Ko T.P., Chen A.P., Kuo C.J., Wang A.H., Liang P.H. (2005). Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 280: 20762–20774. [DOI] [PubMed] [Google Scholar]
- Haehnel W. (1984). On the lateral electron transport between the two light reactions in spinach chloroplasts. In Advances in Photosynthesis Research, Sybesma C., ed (Brussels, Belgium: Springer Netherlands), pp. 545–548. [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hall M., Mishra Y., Schröder W.P. (2011). Preparation of stroma, thylakoid membrane, and lumen fractions from Arabidopsis thaliana chloroplasts for proteomic analysis. Methods Mol. Biol. 775: 207–222. [DOI] [PubMed] [Google Scholar]
- Harrison K.D., Park E.J., Gao N., Kuo A., Rush J.S., Waechter C.J., Lehrman M.A., Sessa W.C. (2011). Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 30: 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley M.D., Schneggenburger P.E., Imperiali B. (2013). Lipid bilayer nanodisc platform for investigating polyprenol-dependent enzyme interactions and activities. Proc. Natl. Acad. Sci. USA 110: 20863–20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber U., Neimanis S., Dietz K.J. (1988). Fractional control of photosynthesis by the QB protein, the cytochrome f/b 6 complex and other components of the photosynthesic apparatus. Planta 173: 267–274. [DOI] [PubMed] [Google Scholar]
- Hemmerlin A., Harwood J.L., Bach T.J. (2012). A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 51: 95–148. [DOI] [PubMed] [Google Scholar]
- Hemmerlin A., Hoeffler J.F., Meyer O., Tritsch D., Kagan I.A., Grosdemange-Billiard C., Rohmer M., Bach T.J. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278: 26666–26676. [DOI] [PubMed] [Google Scholar]
- Hendrickson L., Furbank R.T., Chow W.S. (2004). A simple alternative approach to assessing the fate of absorbed light energy using chlorophyll fluorescence. Photosynth. Res. 82: 73–81. [DOI] [PubMed] [Google Scholar]
- Holloway P.J., Maclean D.J., Scott K.J. (1983). Rate-limiting steps of electron transport in chloroplasts during ontogeny and senescence of barley. Plant Physiol. 72: 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope A.B., Huilgol R.R., Panizza M., Thompson M., Matthews D.B. (1992). The flash-induced turnover of cytochrome b-563, cytochrome f and plastocyanin in chloroplast. Models and estimation of kinetic parameters. Biochim. Biophys. Acta 1100: 15–26. [Google Scholar]
- Janas T., Chojnacki T., Swiezewska E., Janas T. (1994). The effect of undecaprenol on bilayer lipid membranes. Acta Biochim. Pol. 41: 351–358. [PubMed] [Google Scholar]
- Jozwiak A., Gutkowska M., Gawarecka K., Surmacz L., Buczkowska A., Lichocka M., Nowakowska J., Swiezewska E. (2015). Polyprenol reductase2 deficiency is lethal in arabidopsis due to male sterility. Plant Cell 27: 3336–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwiak A., Ples M., Skorupinska-Tudek K., Kania M., Dydak M., Danikiewicz W., Swiezewska E. (2013). Sugar availability modulates polyisoprenoid and phytosterol profiles in Arabidopsis thaliana hairy root culture. Biochim. Biophys. Acta 1831: 438–447. [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. (2008). Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kera K., Takahashi S., Sutoh T., Koyama T., Nakayama T. (2012). Identification and characterization of a cis,trans-mixed heptaprenyl diphosphate synthase from Arabidopsis thaliana. FEBS J. 279: 3813–3827. [DOI] [PubMed] [Google Scholar]
- Kharel Y., Zhang Y.W., Fujihashi M., Miki K., Koyama T. (2001). Identification of significant residues for homoallylic substrate binding of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase. J. Biol. Chem. 276: 28459–28464. [DOI] [PubMed] [Google Scholar]
- Kirby J., Keasling J.D. (2009). Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 60: 335–355. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. (2014). Diffusion of molecules and macromolecules in thylakoid membranes. Biochim. Biophys. Acta 1837: 495–502. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Haferkamp S., Allen J.F., Epstein D.B., Mullineaux C.W. (2008). Protein diffusion and macromolecular crowding in thylakoid membranes. Plant Physiol. 146: 1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Mukherjee U., Galla H.J. (2002). Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882. [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218. [DOI] [PubMed] [Google Scholar]
- Kurisaki A., Sagami H., Ogura K. (1997). Distribution of polyprenols and dolichols in soybean plant. Phytochemistry 44: 45–50. [Google Scholar]
- Kwok E.Y., Hanson M.R. (2004). Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 23: 188–195. [DOI] [PubMed] [Google Scholar]
- Laisk A., Kiirats O., Oja V., Gerst U., Weis E., Heber U. (1992). Analysis of oxygen evolution during photosynthetic induction and in multiple-turnover flashes in sunflower leaves. Planta 186: 434–441. [DOI] [PubMed] [Google Scholar]
- Lange B.M., Ghassemian M. (2003). Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51: 925–948. [DOI] [PubMed] [Google Scholar]
- Lange B.M., Rujan T., Martin W., Croteau R. (2000). Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 97: 13172–13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule O., Fürholz A., Chang H.S., Zhu T., Wang X., Heifetz P.B., Gruissem W., Lange M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6866–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz B.R. (1993). Use of fluorescent probes to monitor molecular order and motions within liposome bilayers. Chem. Phys. Lipids 64: 99–116. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K., Buschmann C. (2001). Chlorophylls and carotenoids: measurement and characterization by UVVIS spectroscopy. Curr. Protoc. Food Analyt. Chem. F4.3: F4.3.1–F4.3.8.
- Lin Z., Liu N., Lin G., Peng C. (2011). Factors altering the membrane fluidity of spinach thylakoid as determined by fluorescence polarization. Acta Physiol. Plant. 33: 1019–1024. [Google Scholar]
- Lindgren B.O. (1965). Homologous aliphatic C30-C45 terpenols in birch wood. Acta Chem. Scand. 19: 1317–1326. [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lütke-Brinkhaus F., Weiss G., Kleinig H. (1985). Prenyl lipid formation in spinach chloroplasts and in a cell-free system of Synechococcus (Cyanobacteria): polyprenols, chlorophylls, and fatty acid prenyl esters. Planta 163: 68–74. [DOI] [PubMed] [Google Scholar]
- Mathur J., Barton K.A., Schattat M.H. (2013). Fluorescent protein flow within stromules. Plant Cell 25: 2771–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B., Lange B.M., Wüst M. (2013). Biosynthesis of sesquiterpenes in grape berry exocarp of Vitis vinifera L.: evidence for a transport of farnesyl diphosphate precursors from plastids to the cytosol. Phytochemistry 95: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P., Kunst L., Browse J., Somerville C.R. (1987). The effects of reduced amounts of lipid unsaturation on chloroplast ultrastructure and photosynthesis in a mutant of Arabidopsis. Plant Physiol. 84: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrshahi P., Stefano G., Andaloro J.M., Brandizzi F., Froehlich J.E., DellaPenna D. (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. USA 110: 12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewska-Hendel A., Baczewska A.H., Sala K., Dmuchowski W., Brągoszewska P., Gozdowski D., Jozwiak A., Chojnacki T., Swiezewska E., Kurczynska E. (2017). Quantitative and qualitative characteristics of cell wall components and prenyl lipids in the leaves of Tilia x euchlora trees growing under salt stress. PLoS One 12: e0172682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., Han K.H., Ryu S.B., Kang H. (2000). Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana. Implications in rubber biosynthesis. J. Biol. Chem. 275: 18482–18488. [DOI] [PubMed] [Google Scholar]
- Oostende Cv., Widhalm J.R., Basset G.J.C. (2008). Detection and quantification of vitamin K(1) quinol in leaf tissues. Phytochemistry 69: 2457–2462. [DOI] [PubMed] [Google Scholar]
- Opitz S., Nes W.D., Gershenzon J. (2014). Both methylerythritol phosphate and mevalonate pathways contribute to biosynthesis of each of the major isoprenoid classes in young cotton seedlings. Phytochemistry 98: 110–119. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Nunnari J. (2003). The division of endosymbiotic organelles. Science 302: 1698–1704. [DOI] [PubMed] [Google Scholar]
- Pampeno C., Derkatch I.L., Meruelo D. (2014). Interaction of human laminin receptor with Sup35, the [PSI+] prion-forming protein from S. cerevisiae: a yeast model for studies of LamR interactions with amyloidogenic proteins. PLoS One 9: e86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.J., et al. (2014). Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 20: 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B., McDonald K.A., Truong M., Britton G., DellaPenna D. (1996). Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A.V., Hincha D.K. (2007). Effects of cholesterol on dry bilayers: interactions between phosphatidylcholine unsaturation and glycolipid or free sugar. Biophys. J. 93: 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Chakrabarty R., Tran H.T., Kwon E.J.G., Kwon M., Nguyen T.D., Ro D.K. (2015). A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 290: 1898–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Boronat A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16: 565–574. [DOI] [PubMed] [Google Scholar]
- Rush J.S., Matveev S., Guan Z., Raetz C.R., Waechter C.J. (2010). Expression of functional bacterial undecaprenyl pyrophosphate synthase in the yeast rer2Delta mutant and CHO cells. Glycobiology 20: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaihara T., Honda A., Tateyama S., Sagami H. (2000). Subcellular fractionation of polyprenyl diphosphate synthase activities responsible for the syntheses of polyprenols and dolichols in spinach leaves. J. Biochem. 128: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Salvi D., Moyet L., Seigneurin-Berny D., Ferro M., Joyard J., Rolland N. (2011). Preparation of envelope membrane fractions from Arabidopsis chloroplasts for proteomic analysis and other studies. Methods Mol. Biol. 775: 189–206. [DOI] [PubMed] [Google Scholar]
- Sato M., Sato K., Nishikawa S., Hirata A., Kato J., Nakano A. (1999). The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M.H., Klösgen R.B., Mathur J. (2012). New insights on stromules: stroma filled tubules extended by independent plastids. Plant Signal. Behav. 7: 1132–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Schauvinhold I., Larson M., Xu R., Charbonneau A.L., Schmidt A., Wilkerson C., Last R.L., Pichersky E. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 106: 10865–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder F., Gorka C., Williamson L.S., Wood W.G. (1987). The influence of dolichols on fluidity of mouse synaptic plasma membranes. Biochim. Biophys. Acta 902: 385–393. [DOI] [PubMed] [Google Scholar]
- Schulbach M.C., Mahapatra S., Macchia M., Barontini S., Papi C., Minutolo F., Bertini S., Brennan P.J., Crick D.C. (2001). Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J. Biol. Chem. 276: 11624–11630. [DOI] [PubMed] [Google Scholar]
- Sévin D.C., Sauer U. (2014). Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 10: 266–272. [DOI] [PubMed] [Google Scholar]
- Skorupińska-Tudek K., Bieńkowski T., Olszowska O., Furmanowa M., Chojnacki T., Danikiewicz W., Swiezewska E. (2003). Divergent pattern of polyisoprenoid alcohols in the tissues of Coluria geoides: a new electrospray ionization MS approach. Lipids 38: 981–990. [DOI] [PubMed] [Google Scholar]
- Skorupinska-Tudek K., et al. (2008). Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of dolichols in plants. J. Biol. Chem. 283: 21024–21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacz L., Plochocka D., Kania M., Danikiewicz W., Swiezewska E. (2014). cis-Prenyltransferase atCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta 1841: 240–250. [DOI] [PubMed] [Google Scholar]
- Surmacz L., Swiezewska E. (2011). Polyisoprenoids: Secondary metabolites or physiologically important superlipids? Biochem. Biophys. Res. Commun. 407: 627–632. [DOI] [PubMed] [Google Scholar]
- Swiezewska E., Danikiewicz W. (2005). Polyisoprenoids: structure, biosynthesis and function. Prog. Lipid Res. 44: 235–258. [DOI] [PubMed] [Google Scholar]
- Swiezewska E., Sasak W., Mańkowski T., Jankowski W., Vogtman T., Krajewska I., Hertel J., Skoczylas E., Chojnacki T. (1994). The search for plant polyprenols. Acta Biochim. Pol. 41: 221–260. [PubMed] [Google Scholar]
- Swiezewska E., Thelin A., Dallner G., Andersson B., Ernster L. (1993). Occurrence of prenylated proteins in plant cells. Biochem. Biophys. Res. Commun. 192: 161–166. [DOI] [PubMed] [Google Scholar]
- Tardy F., Havaux M. (1997). Thylakoid membrane fluidity and thermostability during the operation of the xanthophyll cycle in higher-plant chloroplasts. Biochim. Biophys. Acta 1330: 179–193. [DOI] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson W.W., Whatley J.M. (1980). Development of nongreen plastids. Annu. Rev. Plant Physiol. 31: 375–394. [Google Scholar]
- Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516. [DOI] [PubMed] [Google Scholar]
- Valtersson C., van Duÿn G., Verkleij A.J., Chojnacki T., de Kruijff B., Dallner G. (1985). The influence of dolichol, dolichol esters, and dolichyl phosphate on phospholipid polymorphism and fluidity in model membranes. J. Biol. Chem. 260: 2742–2751. [PubMed] [Google Scholar]
- Vigo C., Grossman S.H., Drost-Hansen W. (1984). Interaction of dolichol and dolichyl phosphate with phospholipid bilayers. Biochim. Biophys. Acta 774: 221–226. [DOI] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64: 665–700. [DOI] [PubMed] [Google Scholar]
- Wang X., Mansourian A.R., Quinn P.J. (2008). The effect of dolichol on the structure and phase behaviour of phospholipid model membranes. Mol. Membr. Biol. 25: 547–556. [DOI] [PubMed] [Google Scholar]
- Wood W.G., Sun G.Y., Schroeder F. (1989). Membrane properties of dolichol in different age groups of mice. Chem. Phys. Lipids 51: 219–226. [DOI] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Ford R.C., Barber J. (1981). Relationship between thylakoid membrane fluidity and the functioning of pea chloroplasts: effects of cholesteryl hemisuccinate. Plant Physiol. 67: 1069–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G.P., Troy F.A. II (2003). Characterization by NMR and molecular modeling of the binding of polyisoprenols and polyisoprenyl recognition sequence peptides: 3D structure of the complexes reveals sites of specific interactions. Glycobiology 13: 51–71. [DOI] [PubMed] [Google Scholar]
- Zhou G.P., Troy F.A. II (2005). NMR studies on how the binding complex of polyisoprenol recognition sequence peptides and polyisoprenols can modulate membrane structure. Curr. Protein Pept. Sci. 6: 399–411. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]