Petunia research suggests that the mechanisms controlling the spatial restriction of floral organ identity genes are more diverse than the well-conserved B and C floral organ identity functions.
Abstract
The ABC model is widely used as a genetic framework for understanding floral development and evolution. In this model, the A-function is required for the development of sepals and petals and to antagonize the C-function in the outer floral whorls. In the rosid species Arabidopsis thaliana, the AP2-type AP2 transcription factor represents a major A-function protein, but how the A-function is encoded in other species is not well understood. Here, we show that in the asterid species petunia (Petunia hybrida), AP2B/BLIND ENHANCER (BEN) confines the C-function to the inner petunia floral whorls, in parallel with the microRNA BLIND. BEN belongs to the TOE-type AP2 gene family, members of which control flowering time in Arabidopsis. In turn, we demonstrate that the petunia AP2-type REPRESSOR OF B-FUNCTION (ROB) genes repress the B-function (but not the C-function) in the first floral whorl, together with BEN. We propose a combinatorial model for patterning the B- and C-functions, leading to the homeotic conversion of sepals into petals, carpels, or stamens, depending on the genetic context. Combined with earlier results, our findings suggest that the molecular mechanisms controlling the spatial restriction of the floral organ identity genes are more diverse than the well-conserved B and C floral organ identity functions.
INTRODUCTION
More than two decades ago, the classic ABC model (Bowman et al., 1991; Coen and Meyerowitz, 1991) was formulated to describe how the identity of each of the floral organs (sepals, petals, stamens, and carpels) is acquired by different combinations of three classes of homeotic gene functions, called A, B, and C. This model, based on floral homeotic mutants in Arabidopsis thaliana and snapdragon (Antirrhinum majus), proposes that the expression of A alone specifies sepals, A and B together petals, B and C stamens, and C alone carpels. The ABC model has been widely used as a framework for understanding floral development and evolution. The B- and C-function genes, initially identified in both Arabidopsis and snapdragon (Sommer et al., 1990; Yanofsky et al., 1990; Jack et al., 1992; Tröbner et al., 1992; Bradley et al., 1993; Goto and Meyerowitz, 1994; Causier et al., 2005), have been well characterized at the molecular level in a diverse range of species (Krizek and Fletcher, 2005), including petunia (Petunia hybrida) (Angenent et al., 1993; van der Krol et al., 1993; Kater et al., 1998; Kapoor et al., 2002; Vandenbussche et al., 2004; Rijpkema et al., 2006; Heijmans et al., 2012). In contrast, how the A-function is encoded in species other than Arabidopsis remains poorly understood. Moreover, the exact definition of the A-function itself has been a subject of debate in more recent years (Litt, 2007; Causier et al., 2010). The classical A-function is proposed to play a dual role: to be required for normal sepal and petal development and to antagonize the expression of the C-function in whorls 1 and 2. Accurate patterning of the floral organ identity genes is indeed crucial for floral development since their misexpression leads to homeotic conversion of one floral organ type into another. In Arabidopsis, the AP2 (APETALA2) transcription factor represents one of the major A-function genes: Loss of AP2 function results in the transformation of the sepals into carpelloid structures or leaf-like organs (allele dependent), and petals are either absent or transformed into stamen-like structures due to ectopic C-class activity in the perianth (Kunst et al., 1989; Bowman et al., 1991; Drews et al., 1991; Jofuku et al., 1994). More recently, it has been shown that AP2 is regulated by the microRNA miR172 (Chen, 2004), and the roles of AP2 and miR172 have been further refined and extended (Wollmann et al., 2010; Dinh et al., 2012; Krogan et al., 2012). So far, efforts to associate a similar dual A-function to the AP2-like genes LIPLESS1 (LIP1)/LIP2 from snapdragon (Keck et al., 2003) and AP2A (Maes et al., 2001) from petunia were unsuccessful: No C-patterning defects were observed in petunia ap2a or snapdragon lip1 lip2 mutants. However, like AP2, LIP genes were shown to play a role in sepal, petal, and ovule development, while ap2a mutants displayed a wild-type phenotype. Nevertheless, other mutants with a partial A-function mutant phenotype were found in petunia and snapdragon, called blind (bl) (Vallade et al., 1987) and fistulata (fis) (McSteen et al., 1998), respectively. In both mutants, the 2nd whorl petals are converted to antheroid structures due to expansion of the C-function domain in the outer whorls. We showed that BL and FIS encode homologous miRNAs from the miR169 family, regulating a subset of NF-YA transcription factors, the derepression of which is proposed to lead to an expansion of the C-expression domain into the outer floral whorls (Cartolano et al., 2007). This revealed an unrelated C-function patterning mechanism in petunia and snapdragon compared with Arabidopsis AP2 function. Together, this suggests that the genetic basis of the A-function may have undergone major evolutionary changes between Arabidopsis (a Rosid species) and petunia and snapdragon (Asterid species). Rosids and Asterids represent the two largest higher eudicot clades that separated >100 million years ago (Moore et al., 2010).
Interestingly, even though the C-function genes are ectopically expressed in the first whorl of bl and fis mutants (Tsuchimoto et al., 1993; Kater et al., 1998; Rijpkema et al., 2006; Cartolano et al., 2007), this does not lead to a homeotic conversion of sepals into carpels, in contrast to what would be predicted by the ABC model. This indicates that one or more additional factors exist that prevent carpel development in the first whorl, acting either alone or redundantly with BL/FIS. Here, we report the characterization and cloning of such a factor in petunia, AP2B (renamed BLIND ENHANCER [BEN]), which we identified as a spontaneous recessive mutation in the bl background. Interestingly, ben bl mutants display a full homeotic conversion of sepals into carpels similar to strong Arabidopsis ap2 alleles, demonstrating that AP2B/BEN represses the C-function together with BL. We show that BEN is a member of the euAP2 lineage, a subgroup of the large AP2/ERF transcription factor family that is characterized by the presence of two AP2 domains and a miR172 target site (Kim et al., 2006). The euAP2 lineage can be further divided into two different classes called the TOE and AP2 types (Wang et al., 2016). The Arabidopsis A-class gene AP2 belongs to the AP2 type, together with TOE3. Although BEN appears to encode the functional equivalent of Arabidopsis AP2, BEN belongs to the TOE type of the euAP2 lineage, which in Arabidopsis are represented by TARGET OF EAT1 (TOE1), TOE2, SCHLAFMÜTZE (SMZ), and SCHNARCHZAPFEN (SNZ). Together with TOE3, these genes redundantly act as floral repressors (Aukerman and Sakai, 2003; Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010), but in contrast to ben, loss-of-function mutants for these genes do not display defects in floral organ development. Interestingly, in the absence of the bl mutation, single ben mutants show a defect in patterning of the B-class genes, a phenotype that in normal forward genetic screens would never have identified BEN as a C-patterning factor. Furthermore, by mining the petunia genome sequences (Bombarely et al., 2016), we found that in addition to the previously described AP2A gene (Maes et al., 2001), petunia contains two more members of the AP2-type class. It could therefore not be excluded that the absence of an Arabidopsis ap2-like phenotype in the previously described single ap2a mutants (Maes et al., 2001) is simply due to redundancy and, thus, that petunia AP2-type genes potentially might function in a similar manner as Arabidopsis AP2. However, in contrast to BEN, we did not find any evidence that these three genes are involved in C-function repression in the perianth, although we show that they do play a role in sepal, petal, and ovary development. Surprisingly, we found instead that they function as major B-function repressors in the first floral whorl, redundantly with BEN. Combined with earlier results, our findings provide further support for the idea that the molecular mechanisms controlling the spatial restriction of the floral organ identity genes are more diverse compared with the well-conserved B and C floral organ identity functions. Our results extend the genetic framework for studying floral diversity and evolution and offer deeper insight in the evolutionary history of the AP2 family.
RESULTS
ap2b/ben Completes the Partial A-Function Phenotype of blind Mutants
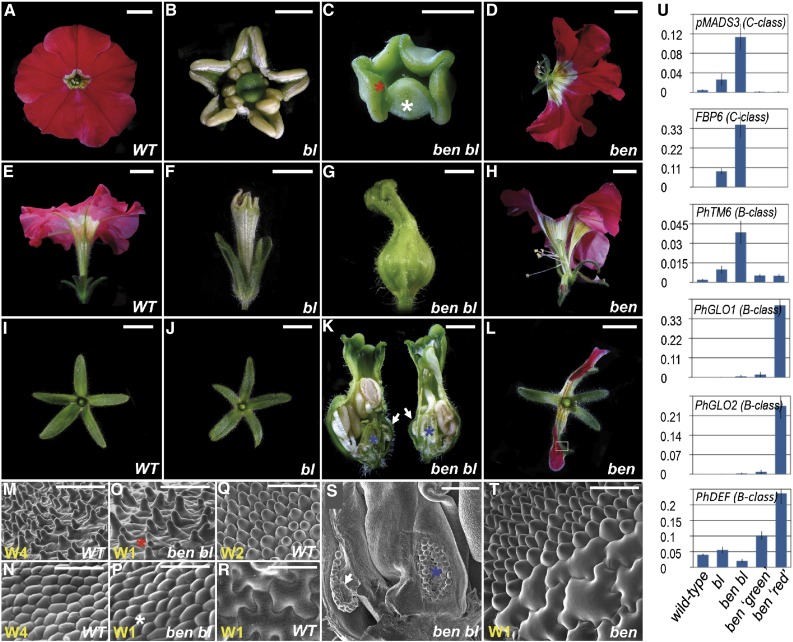
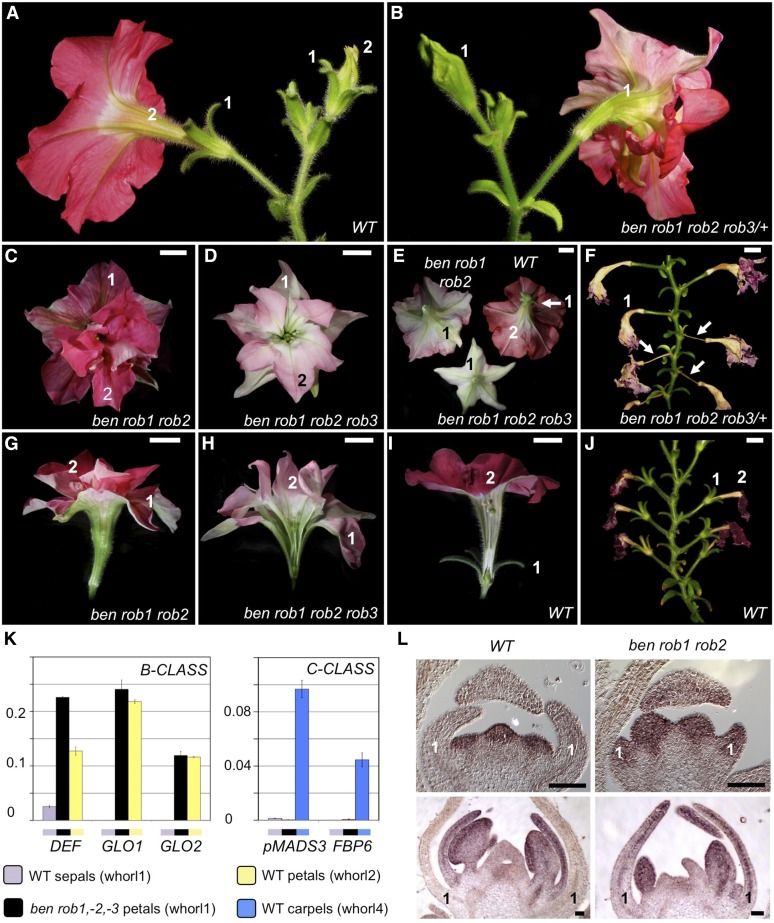
In flowers of the bl mutant (Figure 1), petals are homeotically converted into antheroid structures due to the combined expression of the B- and C-function genes in the 2nd whorl. Although the two C-function MADs box genes pMADS3 and Floral Binding Protein gene 6 (FBP6) are also ectopically expressed in the first whorl of bl mutant flowers (Tsuchimoto et al., 1993; Kater et al., 1998; Rijpkema et al., 2006; Cartolano et al., 2007) (Figure 1U), these organs largely retain sepal identity (Figures 1I and 1J), while normally carpels would be expected based on the ABC model. This suggested that one or more additional factors exist that represses the C-function together with BL. To isolate such redundant regulators, we introgressed the bl-1 allele in the W138 dTph1 transposon high copy number line (Gerats et al., 1990) to perform a secondary mutagenesis screen. We identified a single recessive mutation, named ben, which in combination with bl leads to the homeotic conversion of all sepals into carpels. These first whorl carpels are fully fused enclosing the inner organs, terminate with a stigma, and internally develop ovaries, bearing wild-type-looking ovules (Figures 1C, 1G, 1K, and 1S). At the cellular level, ben bl first whorl organs display the typical stigma and carpel wall epidermal cell types found in wild-type pistils (Figures 1M to 1P).
Figure 1.
Floral and Molecular Phenotypes of Wild-Type, bl, ben bl, and ben Mutants
(A) to (D) Top view of entire flowers.
(E) to (H) Side view of entire flowers.
(I), (J), and (L) Top view of dissected first whorl organs.
(K) Flower longitudinally sectioned. White arrows mark an ovary carrying ovules in the first whorl carpels.
(M) to (T) Scanning electron microscopy images of epidermal tissues of various floral organs in wild-type and mutants. Whorl numbers (W#) are marked in yellow. (M) and (N) correspond to wild-type stigma and ovary surfaces, respectively. Asterisks of the same color in different panels mark the correspondence of regions analyzed by scanning electron microscopy. The white rectangle in (L) marks the region analyzed by scanning electron microscopy in (T), showing wild-type sepal epidermal cells in the green parts and conical petal cells in the colored parts. Bars = 1 cm, except 0.25 cm in (B), (C), (G), and (K), 50 µm in (M) to (R) and (T), and 500 µm in (S). Single and double mutants shown here are from alleles bl-1 and ben-488.
(U) Relative expression levels of C- and B-class homeotic genes in stage 3 first whorl organs. ben ‘green’: green sepals; ben ‘red’: petaloid sepals. Data represent means ± sd of nine data points obtained from three biological replicates that each were analyzed in triplicate for qRT-PCR analysis. Relative expression (R.E.) values were normalized to the geometrical average of three reference genes. Biological replicates were obtained by pooling stage 3 first whorl organs from each time three different flowers.
In all flowers examined, we found that development of the 2nd floral whorl was largely suppressed (Figure 1K), while one or more stamens were strongly underdeveloped or completely missing in the 3rd floral whorl. Together, the phenotype of ben bl flowers closely resembles the strongest ap2 alleles in Arabidopsis (Bowman et al., 1991). Consistent with the observed phenotype, we found an ∼4-fold further increase in C-class gene expression levels in ben bl first whorl carpels compared with bl sepals (Figure 1U). This demonstrates that BL and BEN together are involved in the repression of C-class gene expression in the first floral whorl and that only in ben bl double mutants sufficiently high C-expression levels are reached to provoke homeotic conversion of sepals into carpels. To assess the phenotype of ben independently from bl, we outcrossed ben bl individuals to obtain families in which both ben and bl separately segregated. Remarkably, ben single mutants did not reveal any phenotypic indication that patterning of the C-function was affected, as would normally be expected based on the strong enhancement of the bl phenotype by ben. In line with that, no upregulation of the C-class genes was detected in the sepals of ben single mutants, demonstrating that BL fully masks the C-patterning function of BEN.
Surprisingly, all ben single mutant individuals instead displayed a clear although subtle phenotypic defect, suggesting that BEN in a wild-type background is involved in the patterning of the B-function: In the large majority of the flowers, two opposing sepals oriented along the vertical flower axis were partially petaloid (Figure 1L), mimicking the color pattern of the wild-type petals, with the distal parts brightly colored as the corolla and a yellow/white pigmentation at their basis corresponding to the inner petal tube. At the cellular level, the pigmented regions displayed the typical conical cells found in the petal epidermis (Figures 1Q and 1T). The strength of the phenotype showed some variability, resulting in flowers on the same plant with sepals displaying petal sectors with variations in size, while a small minority of the flowers had either one or three sepals affected instead of two. This variability was observed to a comparable degree in all mutant individuals.
Finally, the majority of ben flowers displayed a corolla that is split open in one or two places, facing the homeotically converted sepals (Figures 1D and 1H). To further investigate the B-function patterning phenotype in these mutants, we monitored the expression of the three petunia B-class genes DEFICIENS (DEF), GLOBOSA1 (GLO1), and GLO2, which in wild-type development specify petal identity in the 2nd whorl (Angenent et al., 1993; van der Krol et al., 1993; Vandenbussche et al., 2004). Not surprisingly, the expression levels of all three genes were dramatically increased in ben petaloid sepals compared with both wild-type sepals (Figure 1U) and ben sepals that did not display ectopic petal tissue. Compared with wild-type sepals, DEF expression also showed an increase in ben sepals that did not display petaloid sectors, but less dramatically. Remarkably, we observed no increase in DEF, GLO1, or GLO2 expression in the ben bl mutants, indicating that the loss of BL function somehow antagonizes ectopic B-expression in the first whorl of ben mutants. We also analyzed the expression of petunia TM6 (Rijpkema et al., 2006), an ancestral B-class gene that is present in many species but has been lost in Arabidopsis. In petunia, TM6 acts a B-class gene to specify stamen development redundantly with DEF but is not involved in petal identity. Compared with wild-type sepals, TM6 expression was enhanced in bl sepals, as reported earlier (Vandenbussche et al., 2004; Rijpkema et al., 2006), and increased further in ben bl sepals to a similar degree to the C-class genes. In contrast, only a very moderate increase was found in ben sepals. This is consistent with the finding that TM6 expression largely depends on C-function activity (Heijmans et al., 2012), unlike classical B-class genes. Note that the ectopic TM6 expression in bl and ben bl sepals does not lead to B-function activity in their sepals because TM6 requires PhGLO2 as an interaction partner to exert the B-function (Vandenbussche et al., 2004; Rijpkema et al., 2006), and PhGLO2 is not ectopically expressed in bl and ben bl sepals (Figure 1U).
BEN Encodes a Member of the AP2/ERF Transcription Factor Family but Is Not Orthologous to the Arabidopsis A-Class AP2 Gene
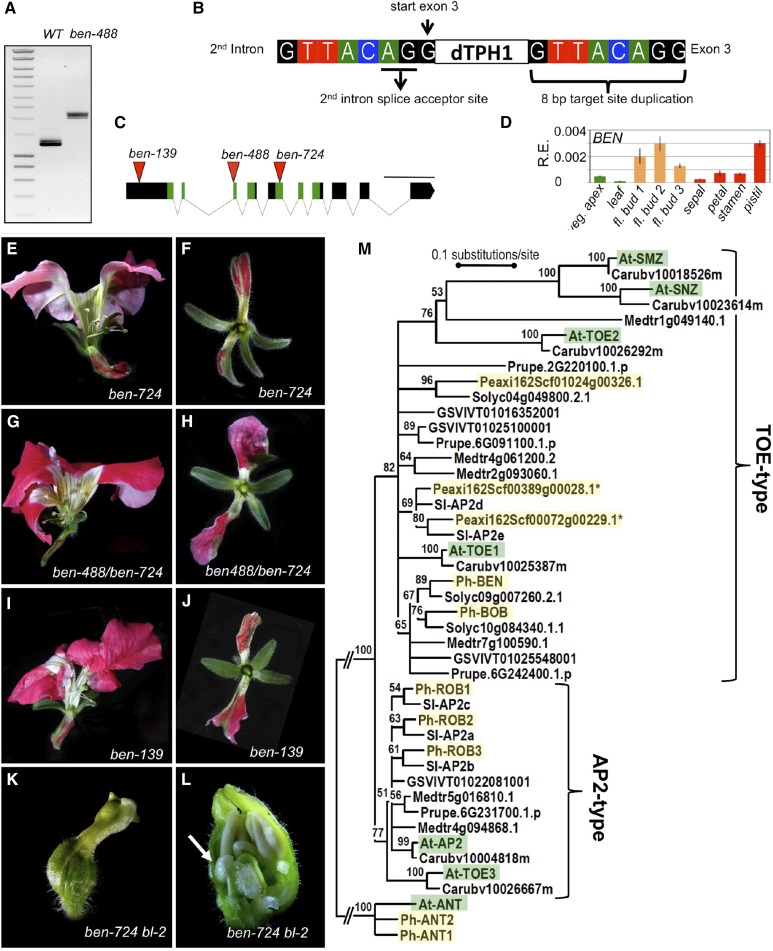
In a transposon display experiment (Van den Broeck et al., 1998; Vandenbussche et al., 2013) performed on selected progeny of the original family in which ben was segregating, we identified a single mutation induced by the transposon dTph1 that fully cosegregated with the ben phenotype and found that BEN encodes a member of the AP2/ERF transcription factor family corresponding to the functionally uncharacterized AP2B gene (Maes et al., 2001) (Figure 2). Within the large AP2/ERF family, AP2B belongs to the euAP2 lineage because of the combined presence of two AP2 domains and a miR172 target site (Kim et al., 2006). The AP2B gene consists of 10 exons (Figure 2C) with the ben allele containing a dTph1 insertion positioned immediately after the first nucleotide of the third exon, 488 bp downstream of the ATG in the coding sequence (ben-488 allele), disrupting the first DNA binding domain. Using ben-488 primers flanking the putative insertion site (Supplemental Table 1), amplification and sequencing of ben-488 transcripts revealed fragments considerably larger than the wild type (Figure 2A) containing the dTph1 element but also the 2nd and 3rd introns, demonstrating that the incorporation of the dTph1 element in the transcript affects splicing. This may be caused by the dTph1-induced 8-bp target site duplication, which includes the splice acceptor site of the 2nd intron and is duplicated immediately downstream of the dTph1 element (Figure 2B).
Figure 2.
Molecular Characterization of ben.
(A) PCR amplification of BEN transcripts with primers ben-488 (Supplemental Table 1) using flower bud cDNA templates from wild-type and ben-488 plants, showing that ben-488 transcripts are considerably larger than the wild type. ben-488 corresponds to the dTph1 insertion allele identified in the transposon display experiment (see Methods).
(B) Sequencing of ben-488 transcripts revealed a dTph1 transposon cotranscribed in the transcript and inserted directly after the first nucleotide of exon 3, followed by an 8-bp target site duplication, which duplicates the 2nd intron splice acceptor site.
(C) BEN genomic structure (from start to stop codon) showing the positions of additional independent dTph1 transposon insertions (red triangles). Rectangles and lines represent exons and introns, respectively. Green regions indicate the two AP2 domains. Numbers in allele names correspond to insert positions (expressed in base pairs) downstream of the ATG in the coding sequence. Bar = 500 bp.
(D) Expression levels of BEN transcripts in different wild-type tissues. Data represent means ± sd of nine data points obtained from three biological replicates that each were analyzed in triplicate for qRT-PCR analysis. Relative expression (R.E.) values were normalized to the geometrical average of three reference genes. Green bars, vegetative tissues; orange bars, complete floral buds from stage 1, diameter 1.5 mm; stage 2, diameter 2.5 mm; and stage 3, diameter 5 mm. Red bars indicate individual organs dissected from stage 3 (see Supplemental Figure 1 for floral stage definition and Methods for sample description).
(E) to (L) Phenotypes of additional ben alleles and crosses. Flowers of ben-724, ben-138, and ben-488/ben-724 mutants display phenotypes very similar to ben-488. In addition, ben-724 bl-2 double mutants exhibit a phenotype identical to ben-488 bl-1 double mutants (compare with Figure 1).
(E), (G), (I), and (K) Side views of undissected flowers.
(F), (H), and (J) Top view of first whorl organs.
(L) Longitudinal flower section showing inner organization. The white arrow indicates a placenta with ovules in the first whorl.
(M) Neighbor-joining tree of euAP2 proteins from petunia (Ph; Peaxi, shaded in yellow), tomato (Sl; Sol), Arabidopsis (At, shaded in green), C. rubella (Carub), M. trunculata (Medtr), grape (GSVIV), and peach (Prupe), rooted with AINTEGUMENTA (ANT) proteins. Bootstrap values (%) based on 2000 replicates are indicated near the branching points; branches below 50% have been collapsed.
To provide further independent genetic evidence that the ben phenotype is caused by the disruption of AP2B gene function, we screened our sequence-indexed dTph1 transposon insertion collection (Vandenbussche et al., 2008) and identified two additional insertions in the AP2B coding sequence (Figure 2C), disrupting either the 2nd DNA binding domain (ben-724) or the first exon (ben-139). We found that flowers of ben-724 and ben-138 homozygous mutants display a very similar phenotype to that of ben-488 as well as the offspring of an allelism test obtained by crossing ben-488 and ben-724 mutants (Figures 2E to 2J). In addition, we found that ben-724 bl-2 double mutants exhibit a phenotype identical to that of the original ben-488 bl-1 double mutants (Figures 2K and 2L). Given the insert positions of the three alleles in the coding sequence, and the finding that dTph1 encodes stop codons in all six possible reading frames, we conclude that the phenotype observed in homozygous ben alleles is due to disruption of the AP2B gene and is most likely the result of a complete loss of function. To acknowledge its function, we renamed this gene BEN. The expression of AP2B/BEN was previously characterized by in situ hybridization, indicating a broad expression pattern, with transcripts detected in the bracts extending under the inflorescence meristems, in sepals, petals, stamens, and carpels, as well as in the endosperm of mature seeds (Maes et al., 2001). The results of qRT-PCR expression analysis (Figure 2D) are in agreement with these observations, and they show that BEN is already expressed in vegetative apices, albeit at lower levels compared with young flower buds, where expression peaks in stage 2 flowers (see Supplemental Figure 1 for stage definition). During later development (in stage 3 flowers, which have sufficiently developed to allow manual dissection of individual floral organs), BEN transcripts were detected in all floral whorls, but with highest expression in the carpels.
A sequence comparison with all six Arabidopsis euAP2 genes (Supplemental Table 2) indicates that RAP2.7 (RELATED TO AP2.7)/TOE1 is the Arabidopsis gene most similar to BEN. TOE1 was shown to act as a repressor of flowering (Aukerman and Sakai, 2003), but unlike ben, toe1 mutants do not display defects in floral organ development. Conversely, we did not observe a flowering time phenotype in ben mutants. Because the function of BEN is more similar to that of the Arabidopsis AP2 gene, this further suggests molecular divergence in the mechanisms that pattern the C-function between Arabidopsis and petunia.
Genome-Wide Phylogenetic Analysis of Petunia euAP2 Genes
To better understand the relationship between BEN and the six different Arabidopsis euAP2 genes, we took advantage of the recently released Petunia axillaris and Petunia inflata genome sequences (Bombarely et al., 2016) to compare all petunia and Arabidopsis euAP2 genes in a phylogenetic analysis. P. axillaris and P. inflata species represent the main parents of modern P. hybrida varieties, including W138, the laboratory line used in this study. We identified eight predicted euAP2 proteins in P. axillaris, while nine were detected in P. inflata. Comparison between the two species showed that the extra euAP2 gene copy in P. inflata is due to a recent duplication of one of the eight genes (Supplemental Table 3 and Supplemental Figure 2A). Representatives of five out of the eight P. axillaris/P. inflata gene pairs were experimentally confirmed in W138 and further analyzed in this study (Supplemental Table 3), including the previously described P. hybrida AP2B (BEN) and AP2A genes. We retained the five W138 euAP2 genes and three remaining P. axillaris representatives to be included in the phylogenetic analysis. Furthermore, we also included the euAP2 genes (Supplemental Table 3) from Capsella rubella (Slotte et al., 2013), a Brassicaceae species closely related to Arabidopsis, the asterid species tomato (Solanum lycopersicum; Tomato Genome Consortium, 2012) as a close relative of petunia, and the species Medicago truncatula (Young et al., 2011), Vitis vinifera (grape) (Jaillon et al., 2007), and Prunus persica (peach) (Verde et al., 2013), all rosid species distant from Arabidopsis and from each other. In agreement with a recent phylogenetic analysis (Wang et al., 2016), we found that all euAP2 sequences cluster in either of two different groups with well-supported bootstrap values, corresponding to TOE and AP2 types (Figure 2M). Furthermore, orthologous relationships are well supported between euAP2 genes from closely related species, showing six pairs of Arabidopsis/C. rubella genes and eight pairs of petunia/tomato genes. In line with the sequence comparison (Supplemental Table 2 and Supplemental Figure 2B), the phylogenetic analysis indicates that BEN is more closely related to TOE1 than to AP2. Within the TOE-type group, however, the aligned region on which the tree is based does not provide enough resolution to completely resolve relationships between members from distantly related species, but the divergent TOE2 and SMZ/SNZ clades (Wang et al., 2016) can be clearly distinguished. Petunia possesses in total five euAP2 genes that belong to the TOE-type, with one gene similar to BEN (see next paragraph) and three other genes that are more distantly related. Finally, petunia harbors three euAP2 genes that belong to the AP2-type class (see further).
BOB and BEN Are Redundantly Required for Proper Development of Second and Third Whorls
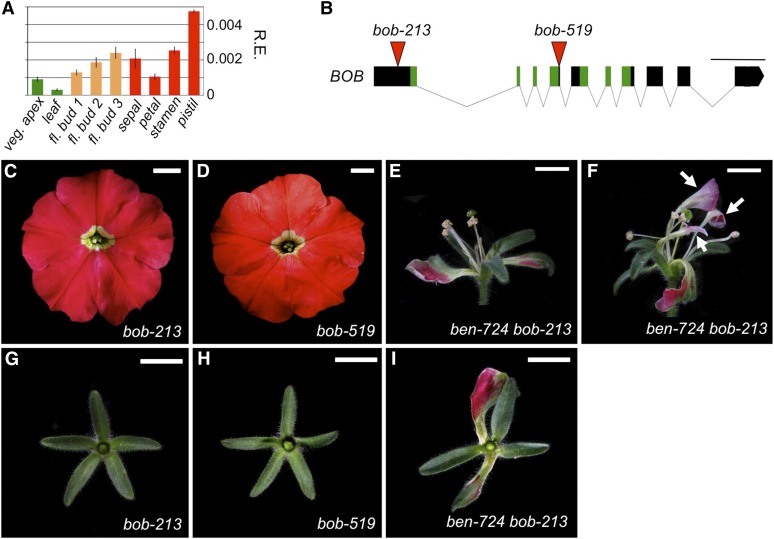
We hypothesized that the subtle B-function patterning defects found in ben single mutants might represent the first signs of an unknown major mechanism that prevents B-class expression in the petunia sepals, encoded by BEN in a largely redundant fashion with one or more unknown factors. We found that petunia contains a gene closely related to BEN (Figure 2M; Supplemental Figures 2A and 2B), which we named BOB (for BROTHER OF BEN). Because BOB expression overlaps with BEN in all tissues tested (Figure 3A), we selected BOB as a plausible candidate to encode a B-repression function in sepals, redundantly with BEN.
Figure 3.
Characterization of Petunia BOB and Genetic Interactions with BEN.
(A) Expression levels of BOB transcripts in different wild-type tissues. BOB expression overlaps with BEN expression in all tissues tested, showing the highest expression levels in carpels of stage 3 flowers. Legend and cDNA templates as in Figure 2D.
(B) BOB genomic structure and position of dTph1 transposon insertions. Legend as in Figure 2C.
(C) to (I) Floral phenotypes of bob and ben bob mutants showing complete flowers in (C) to (F) and dissected first whorls in (G) to (I). (C), (D), (G), and (H) are homozygous bob mutants display a wild-type floral architecture, suggesting redundancy. Bars = 1 cm.
(E) and (F) ben bob flowers display severe 2nd whorl growth defects.
(E) Flower completely lacking petals.
(F) Flower with strongly reduced petals (white arrows). In addition, one to two stamens are often missing in the third floral whorl.
(I) ben bob first whorl organs are similar to those of ben.
We applied a reverse genetics approach to test this and identified two putative null mutations, bob-213 and bob-519 (Figure 3B), but homozygous mutants were not different from the wild type (Figures 3C, 3D, 3G, and 3H), suggesting full redundancy. We therefore tested a putative functional overlap with BEN by analyzing ben bob double mutants. We found that in all ben bob flowers (Figures 3E and 3F), 2nd whorl petal development was strongly reduced. This phenotype ranged from flowers in which all five petals appeared to be absent (Figure 3E) to flowers in which one or more petals developed but were always strongly reduced in size (Figure 3F). No signs of homeotic conversions were observed in these 2nd whorl petals. In the third whorl, most flowers contained fewer than five stamens, some of which occasionally fused entirely with the remnants of the 2nd whorl petals. The central pistil appeared normal. These observations indicate that BEN and BOB are redundantly required for proper 2nd and 3rd whorl development, a phenotype reminiscent of strong Arabidopsis ap2 alleles, which also display similar growth defects in these whorls. Despite this strong phenotype, the first whorl sepals of the double mutants were undistinguishable from those of ben single mutants (Figure 3I), indicating that BOB, in contrast to BEN, most likely is not involved in patterning the B-function.
Petunia ROB1, ROB2, and ROB3 AP2-Type Genes Are Required for Proper Perianth and Pistil Development, but Do Not Antagonize the C-Function in the Outer Whorls
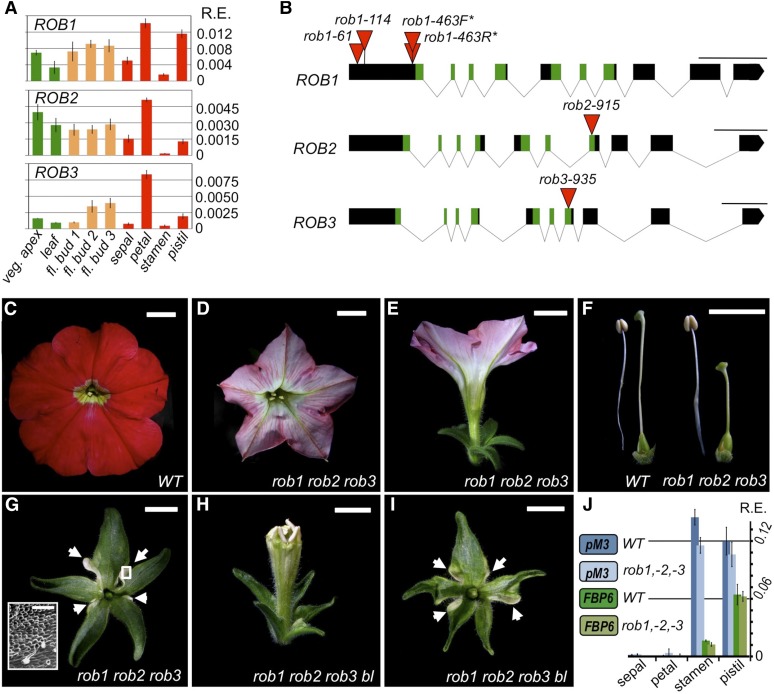
Because the petunia equivalent of the Arabidopsis AP2-mediated C-patterning function is encoded by BEN, which belongs to the TOE-type class, the function of the petunia AP2-type members remained to be determined. Exploring the petunia genomes (Bombarely et al., 2016), we found that in addition to the previously reported AP2A gene (Maes et al., 2001), petunia contains two additional AP2-type genes (Figure 2M; Supplemental Figures 2A and 2B). We named these additional genes ROB2 and ROB3 (for REPRESSOR OF B-FUNCTION, for reasons explained later), while petunia AP2A was renamed ROB1 (Figure 2M). While the phylogenetic analysis within the AP2-type group only provides moderate support (bootstrap value of 51%) for grouping the petunia ROB genes with Arabidopsis AP2, a global sequence comparison with all six Arabidopsis euAP2 genes indicates that AP2 is the Arabidopsis gene most similar to ROB1, -2, and -3 (Supplemental Table 2 and Supplemental Figure 2B). Expression of ROB1 (AP2A) during floral development was previously characterized by in situ hybridization, indicating comparable expression levels in all four floral whorls during early floral stages, while later on, its expression appeared to be stronger in petals and the ovary compared with the other floral organs (Maes et al., 2001). In addition, transcripts were also detected in seeds and in leaf primordia of young seedlings.
By qRT-PCR analysis (Figure 4A), we found that the three ROB genes display broad and quite comparable expression patterns, showing the highest expression levels in petals and carpels in stage 3 flowers, while also being well expressed in vegetative tissues. Given these overlapping expression patterns, the absence of an Arabidopsis ap2-like phenotype in the reported petunia ap2a mutants might also be simply explained by a putative redundancy with ROB2 and ROB3. Therefore, it could not yet be excluded that petunia AP2-type genes function in a similar manner to Arabidopsis AP2. To test this hypothesis, we analyzed the function of the three ROB genes. For all three genes, we identified and confirmed candidate dTph1 insertions in the coding sequence (Figure 4B), positioned either in the first exon (in the case of ROB1) or disrupting the highly conserved 2nd AP2 domain (for ROB2 and ROB3). Homozygous rob1 mutant flowers were not different from the wild type, confirming previous results (Maes et al., 2001), nor were the flowers of rob2 and rob3 mutants (Supplemental Figure 3), suggesting redundancy for all three ROB genes. To test this, we analyzed genetic interactions between the different rob mutants. Our results indicate that all three ROB genes function in a largely redundant fashion, since double mutants only displayed subtle phenotypes if any at all (Supplemental Figure 3), while a clearly recognizable and uniform phenotype different from the wild type was observed in all flowers of triple rob1 rob2 rob3 mutants. We found that rob1 rob2 rob3 petal lobes were less developed, resulting in a more star-like shaped corolla compared with the round petal lobes found in wild-type flowers (Figures 4C and 4E). In addition, triple mutant petals displayed an altered pigmentation, colored pink rather than the bright red color found in the wild type. These pale-pink flowers were exclusively found in all triple homozygous mutants, but colors intermediate between the wild type and the triple mutant were found in some double mutant combinations (Supplemental Figures 3F and 3G), indicating that petunia ROB genes apparently contribute to petal pigmentation in a redundant but additive fashion. Furthermore, we found that the length of the pistil was always reduced in triple mutants compared with the wild type, mainly due to a reduction in style length, while stamen development was not affected (Figure 4F). In addition, the sepals of early arising rob1 rob2 rob3 flowers (Figure 4G) were larger compared with the wild type (Figure 1I) and lower order mutants (Supplemental Figure 3I), but this difference became much less pronounced in flowers developing later (Supplemental Figure 3J).
Figure 4.
ROB Genes Are Redundantly Required for Normal Sepal, Petal, and Carpel Development but Do Not Antagonize the C-Function in the Perianth.
(A) Expression levels of ROB genes in different wild-type tissues. The petunia ROB genes are widely expressed and show very similar expression patterns. Legend and cDNA templates as in Figure 2D.
(B) Genomic structures and positions of dTph1 transposon insertions identified in the petunia ROB genes. Note that rob1-463F and rob1-463R are two independent insertions at the same position, but with opposite orientations of the dTph1 transposon. Legend as in Figure 2C.
(C) to (E) and (H) Undissected flowers.
(F) Comparison of stamens and pistils.
(G) and (I) First whorl organs. White arrows indicate petaloid regions. Inset in (G): scanning electron microscopy analysis showing conical petal cells in the region indicated by the rectangle in (G).
(J) qRT-PCR analysis of C-class MADS-box gene expression in different floral organs (stage 3), showing a similar expression pattern in wild-type and rob1 rob2 rob3 mutants. Data represent means ± sd of nine data points obtained from three biological replicates that each were analyzed in triplicate for qRT-PCR analysis. Relative expression (R.E.) values were normalized to the geometrical average of three reference genes. Biological replicates were obtained by pooling organs from each time three different flowers. Mutants shown here were made using rob1-61, rob2-915, rob3-935, and bl-2 insertion alleles. Bars = 1 cm, except for inset in (G) where bar = 100 µm.
In summary, no morphological indications of C-patterning defects in the perianth organs were found, in sharp contrast to ap2 mutants in Arabidopsis. This was also confirmed at the molecular level, showing that the expression of the two petunia C-class genes pMADS3 and FBP6 remain restricted to the stamens and carpels, as in the wild type (Figure 4J). This demonstrates that ROB genes are not required to repress the C-function in sepals and petals. Yet, it was still possible that they would function as C-repressors in the perianth but that this function is masked by BL, similar to what was found earlier for BEN, whose C-repressor role was only revealed in a bl mutant background. To answer this question, we created and analyzed bl rob1 rob2 rob3 quadruple mutants. In contrast to bl ben, flowers of these quadruple mutants were not markedly different from those of bl mutants (compared with Figures 4H, 1F, and 1G), except for the sepals of early arising flowers (Figure 4I), which were larger, similar to those observed in early arising rob1 rob2 rob3 flowers. Based on these results and the phenotypes of rob lower order mutant combinations, we conclude that the ROB genes are redundantly required for normal development of sepals, petals, and carpels, but are unlikely to antagonize the C-function in the perianth.
BEN, ROB1, ROB2, and ROB3 Genes Redundantly Repress the B-Function in Sepals
Intriguingly, we noticed that the first two to three flowers that appeared on the inflorescence of triple rob and quadruple rob bl mutants displayed sepals that were not only larger, but in addition contained petaloid regions found primarily at the base of the sepal margins (Figures 4G and 4I). This phenotype was clearly observed in around half of these early flowers. This indicated that patterning of the B-function genes is partially impaired, as also observed in ben single mutants. This phenotype was not seen in lower order mutants and disappeared completely in later arising flowers in rob1 rob2 rob3 (bl) mutants (Supplemental Figures 3I to 3K). Earlier, we hypothesized the existence of a novel major mechanism in petunia that prevents B-class expression in the sepals, encoded by BEN in a largely redundant fashion with one or more other unknown factors. Although being the most obvious candidate, we failed to show such a common B-repressor function for BOB, the closest relative of BEN. Interestingly, although the ROB genes are more distantly related to BEN than BOB, the subtle B-function patterning defect observed in the triple rob mutant flowers identified the ROB genes as other possible candidates to repress the B-function together with BEN. To test this, we analyzed genetic interactions of ben with the rob mutations.
To our delight, we found that all flowers of ben rob1 rob2, ben rob1 rob2 rob3/+, and ben rob1 rob2 rob3 mutant individuals displayed a quasi-complete homeotic transformation of their sepals into petals, which as in 2nd whorl wild-type flowers were fully fused, forming a tube and corolla with a size close to the wild type (Figures 5A to 5J). No obvious morphological differences were found between ben rob1 rob2 and ben rob1 rob2 rob3/+ flowers, while quadruple mutant flowers could be clearly distinguished based on the pink color of their petals and the more star-like shape of the corolla, as found earlier in the 2nd whorl of triple rob mutants.
Figure 5.
BEN and ROB Genes Repress the B-Function in the First Whorl.
(A) and (B) Inflorescences showing flowers at various stages of development. “1” and “2” indicate whorl numbers in (A) to (L).
(C) and (D) Top view of ben rob flowers.
(E) Bottom view.
(G) to (I) Side views; flowers in (H) and (I) have been sectioned to reveal inner organization.
(F) and (J) Inflorescences shown from first wilted flower downwards. Mutants shown here were made using rob1-61, rob2-915, rob3-935, ben-724, and bl-2 insertion alleles.
(K) qRT-PCR analysis of B- and C-function genes in individual floral organs (stage 3). Data represent means ± sd of nine data points obtained from three biological replicates that each were analyzed in triplicate for qRT-PCR analysis. Relative expression (R.E.) values were normalized to the geometrical average of three reference genes. Biological replicates were obtained by pooling organs from each time three different flowers. The B-class genes DEF, GLO1, and GLO2 are strongly upregulated in ben rob1 rob2 rob3 first whorl petals compared with first whorl wild-type sepals, reaching expression levels comparable to those observed in wild-type 2nd whorl petals. The C-class genes pMADS3 and FBP6 are expressed at very low levels in sepals compared with carpels in the wild type and remain expressed at very low levels in ben rob1 rob2 rob3 first whorl petals.
(L) In situ hybridization of PhGLO2 in wild-type and ben rob1 rob2 mutants at early stages of development, showing ectopic PhGLO2 expression in first whorl primordia of ben rob1 rob2 flowers. First floral whorls (1) are indicated. Bars = 1 cm, except in (L) where bar = 50 µm.
Outgrowth of the 2nd whorl petals in all flowers of ben rob1 rob2[rob3(/+)] mutants was affected, but to a variable degree, with phenotypes ranging from a 2nd whorl being (almost) completely absent (Supplemental Figures 3L, 3M, and 3O) to flowers with five well-developed petals, but always with fusion defects between one or more petals (Figures 5C and 5D). Finally, the completeness of sepal to petal conversion varied slightly between flowers on the same plant, ranging from first whorl petals that retained some partial sepal-like characteristics, as indicated by a green hue, mainly around the central petal veins, to flowers in which conversion appeared complete in all five first whorl petals (compared with Supplemental Figures 3N and 3O). These variable aspects of the phenotype were observed to a comparable degree in all ben rob1 rob2 [rob3(/+)] mutant individuals. While petunia wild-type sepals do not senesce after fertilization, ben rob1 rob2 [rob3(/+)] first whorl petals wilt in a similar manner to wild-type petals (Figures 5F and 5J). Consistent with the quasi-complete conversion of sepals into petals in these mutants, we found that the petunia B-function genes reach expression levels in the first whorl comparable to those normally observed in wild-type 2nd whorl petals (Figure 5K). In situ hybridization (Figure 5L) further revealed that ectopic B-expression in the petaloid first whorl organs already occurs at the very early stages of floral development, when first whorl primordia start to arise from the floral meristem. Similar to triple rob1 rob2 rob3 mutants, we did not find upregulation of the C-genes in the first whorl of ben rob1 rob2 rob3 mutants compared with wild-type sepals (Figure 5K). Together, we conclude that the ROB genes pattern the B-function genes (but not the C-function genes) by acting as repressors of the B-function in the first floral whorl, redundantly with BEN. For this reason, we have (re)baptized AP2A and the two other AP2-type genes with the names ROB1, ROB2, and ROB3, for REPRESSOR OF B-FUNCTION.
A Combinatorial Model for Patterning the Floral Homeotic B- and C-Functions in Petunia Flowers
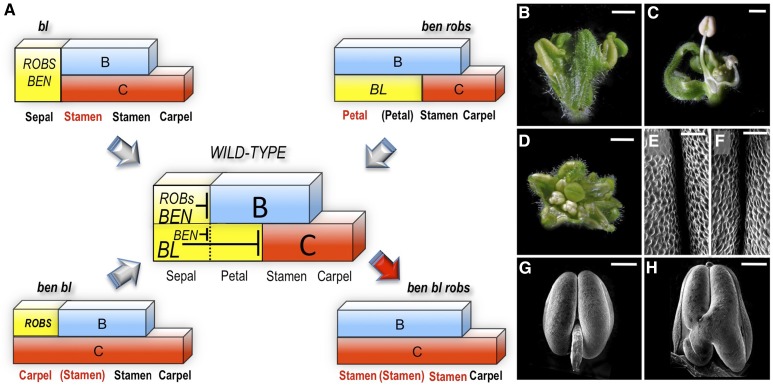
We have summarized our findings thus far in a combinatorial model describing the patterning of the floral homeotic B- and C-functions during petunia flower development (Figure 6A). Note that for clarity, the presented model has been simplified at different levels: (1) The expression domains of the genes regulating the B- and C-functions are not necessarily restricted to the first two whorls. What is shown in the model is rather the final outcome of their action on the patterning of the B- and C-genes in the perianth. For example, BL, like its ortholog FIS in snapdragon, is expressed in all floral whorls and is proposed to pattern the C-genes by dampening the expression of the C-genes within their genuine expression domain in the center of the flower, thereby preventing outward spreading of the C-function into the perianth (Cartolano et al., 2007). (2) The petunia B- and C-functions, although fully characterized (Vandenbussche et al., 2004; Rijpkema et al., 2006; Heijmans et al., 2012), are not shown in detail. The petunia B-function in particular is more complex compared with Arabidopsis and snapdragon, mainly due to the presence and atypical features of TM6 (Rijpkema et al., 2006). (3) Because the B-patterning function is encoded by BEN and the ROB genes in a largely redundant fashion, no individual graphs are shown for single ben or triple rob mutants.
Figure 6.
A Simplified Combinatorial Model Describing the Cadastral (Boundary Setting) Functions That Pattern the Floral B- and C-Functions in Petunia.
(A) The yellow block in the wild type represents combinatorial repression of the petunia B- and C-functions in sepals and the C-function in petals. The model is based on floral phenotypes and genetic interactions of three classes of homeotic mutants (bl, ben bl, and ben rob1-3) described in this study and predicts stamen development in the first whorl in a ben bl robs mutant background (red arrow). See main text for additional information.
(B) and (C) Side view of ben bl rob1 rob2 rob3/+ flowers showing stamens and antheroid structures in the first whorl, respectively.
(D) Top view of a ben bl rob1 rob2 rob3 quintuple mutant showing antheroid organs in the first whorl.
(E) to (H) Scanning electron microscopy images of third whorl anthers ([E] and [G]) and of first whorl antheroids observed in ben bl rob1 rob2 rob3/+ flowers ([F] and [H]). Bars = 2 mm in (B) to (D), 50 µm in (E) and (F), and 200 µm in (G) and (H).
In this model, C-function repression in the perianth is controlled by two parallel pathways, with BL-mediated repression as the dominant mechanism, while BEN fulfills a secondary role by preventing C-function activity in the first whorl in the absence of BL. When BL is functional, BEN acts together with the ROB genes as major B-function repressors in the first whorl. One of the attractive features of the classical ABC model is, besides the beauty of its simplicity, the possibility to predict the outcome of higher order mutant combinations. Interestingly, our model predicts the development of stamens in the first whorl in a bl ben rob mutant background (Figure 6A, red arrow) due to the predicted simultaneous expression of B- and C-functions in the first whorl. However, full stamen identity may require a well-balanced B/C coexpression. To test this, we combined ben, bl, and rob mutations together and found that stamen/antheroid structures indeed developed in the first whorl of ben bl rob1 rob2 rob3(/+) flowers, in line with the model’s prediction. In the majority of the flowers, two to three first whorl organs could be observed that macroscopically resembled wild-type anthers (Figures 6B, 6G, and 6H) as well as at the cellular level (Figure 6E and 6F), while the remaining organs were more carpelloid as in bl ben mutants. Occasionally, we found flowers in which first whorl anthers were supported by a stamen filament as in the wild type (Figure 6C). Much more rarely (in three cases only), we observed flowers in which all first organs appeared antheroid (Figure 6D).
DISCUSSION
Divergent Mechanisms Antagonizing the C-Function in the Perianth of Petunia and Arabidopsis
During the last two decades, the classic ABC model of floral development (Bowman et al., 1991; Coen and Meyerowitz, 1991) has been used with great success as an evolutionary genetic framework to study floral development and diversity. However, extending the molecular basis of the Arabidopsis A-function to other eudicot species has largely failed thus far. Note that a snapdragon A-function was originally postulated based on a semidominant mutation, called ovulata, but was later shown to be the consequence of a gain-of-function mutation in the C class gene PLENA (Bradley et al., 1993). Later on, forward genetic screens in petunia and snapdragon revealed a completely different molecular mechanism encoding the major factor preventing C-expression in the perianth: the miR-BL/FIS-NF-YA module in petunia and snapdragon (Cartolano et al., 2007), compared with AP2-mediated C-patterning in Arabidopsis (Kunst et al., 1989; Bowman et al., 1991; Drews et al., 1991; Jofuku et al., 1994; Dinh et al., 2012). Yet AP2 orthologs exist in petunia and snapdragon, and all components of the miR-BL/FIS (miR169) module are present in Arabidopsis (Mantovani, 1999; Hong et al., 2003; Jones-Rhoades and Bartel, 2004). In theory, it is therefore possible that AP2 and miR-BL/miR169 modules in both species act in parallel to pattern the C-function. However, even if miR169 in Arabidopsis and AP2 orthologs in petunia/snapdragon would play a role in C-patterning, they clearly cannot compensate for the loss of AP2 and BL/FIS function, respectively, indicating that the two species have retained different mechanisms representing the major regulator to repress the C-function in the perianth. Because the bl/fis mutants display only an incomplete C-patterning defect (their first whorl organs retain sepal identity), this suggested that one or more additional factors may exist that repress the C-function together with BL/FIS. Here, we have shown that BEN represents such a factor: The ben mutation strongly upregulates ectopic C-gene expression in the first whorl of bl mutants, leading to the full homeotic conversion of sepals into carpels and resulting in a ben bl floral phenotype closely resembling the strongest ap2 alleles in Arabidopsis. The observation that BEN’s C-repressor function only becomes visible in a bl mutant background further supports the idea that the BL function represents the dominant C-patterning mechanism in petunia. Although BEN appears to encode the functional equivalent of Arabidopsis AP2, BEN is more similar to Arabidopsis TOE1. No floral patterning defects thus far have been described for toe1 mutants: Gain- and loss-of-function analyses showed that TOE1 acts as a floral repressor (Aukerman and Sakai, 2003). In turn, we found that the ROB genes, the petunia euAP2 genes with highest similarity to Arabidopsis AP2, are redundantly required for normal development of sepals, petals, and the ovary but do not antagonize the C-function in the perianth. This is similar to what has been previously described for the snapdragon AP2 homologs LIP1/LIP2 (Keck et al., 2003).
Antagonism of the B-Function in the First Floral Whorl
Inspired by subtle B-function patterning defects commonly found in ben and rob1 rob2 rob3 mutants, we discovered a floral patterning mechanism in petunia flower development, encoded by BEN and ROB genes, that prevents B-function expression in the first floral whorl. Remarkably, the petals developing in the first whorl of ben rob1 rob2 (rob3) flowers closely resemble wild-type 2nd whorl petals, displaying complete fusion and proper tube and corolla development. Compared with this phenotype, the B-patterning defects observed in single ben and triple rob mutants are very subtle, indicating that BEN and the ROB genes repress the B-function in the first floral whorl in a largely redundant way. Nevertheless, these phenotypes indicate that BEN function is more critical in the sepals along the vertical floral axis compared with the lateral sepals, while ROB gene function might be more critical in early arising flowers. A complete homeotic conversion of all sepals into petals and in all flowers produced throughout development as observed in ben rob1 rob2 (rob3) flowers never has been reported in Arabidopsis through loss-of-function mutations. Homeotic patterning defects in strong Arabidopsis ap2 alleles only affect the C-function expression domain (Kunst et al., 1989; Bowman et al., 1991), indicating that the primary patterning function of AP2 is to restrict the C-function to the inner floral whorls. However, there is evidence that AP2, besides its role in C-repression, also prevents B-expression in the sepals: First, some ap2 alleles were reported to contain mosaic organs consisting of carpel and stamen sectors in the medial positions of the first whorl, which is indicative of local simultaneous B- and C-class gene expression (Bowman et al., 1991). More recently, it was found that introducing the ap2-2 mutation in a heterozygous state in the dominant-negative tpl-1 (topless-1) mutant enhanced the frequency of flowers displaying petaloid sepals compared with tpl-1 mutants (Krogan et al., 2012). Although why these B-patterning defects only become apparent in specific ap2 alleles and/or genetic backgrounds is not well understood, this provided clear genetic support for a model in which AP2, in addition to repressing the C-function gene AG, also regulates the expression borders of the B- and E-class genes by recruiting TPL and the histone deacetylase HDA19 (Krogan et al., 2012).
Combined Ectopic Expression of the Homeotic B- and C-Functions in the First Whorl?
Single ben mutants display a partial but clear B-patterning defect in the first whorl, while intriguingly, ben bl mutants only show an enhanced C-function patterning defect in their first whorl organs (Figure 1). Because BL is a negative regulator of NF-YA genes, which are proposed to act as positive regulators of C-gene expression, a lack of ectopic NF-YA activity in ben single mutants may explain why the loss of BEN alone is not sufficient to activate ectopic C-activity in the perianth. Therefore, although BEN patterns both the B-and C-functions, only the B-function expression domain is affected in ben single mutants. On the other hand, it is less obvious why ben bl mutants do not show simultaneous B- and C-function activity in the first whorl, at least in two out of the five first whorl organs, as would normally be expected based on the combination of the single mutant phenotypes (Figure 1). On the other hand, combined B- and C- activity does occur in the first whorl of bl rob1 rob2 rob3(/+) mutants, as indicated by the presence of antheroid organs (Figure 6). Because the ROB genes still partially repress the B-function in the first whorl of ben mutants (and ben bl mutants), this may suggest that ectopic C-function gene expression may have a strong negative impact on the expression level of the B-function genes, in a context in which B-function repression is only partially impaired. In other words, this suggests that the establishment and/or upregulation and further maintenance of the C- and B-function expression domains may not be fully independent. At the moment, we can only speculate how this occurs in petunia, and direct or indirect interactions are equally possible. Interestingly, in Arabidopsis, both B- and C-class MADS-box proteins require complex formation with SEPALLATA (SEP) proteins for their positive autoregulation (Gómez-Mena et al., 2005; Kaufmann et al., 2009). Thus, B- and C-function proteins compete for the same substrate (SEP3) required for further upregulation and maintenance. According to the quartet model (Theissen and Saedler, 2001), carpel identity is determined by a complex consisting of SEP and C-function proteins, while stamen identity is conferred by a complex composed of a B-class heterodimer, a C-function protein, and a SEP protein. Therefore, it is conceivable that a specific initial ratio between B- and C-gene expression levels is needed to result in final high levels of both B- and C-function proteins, which are required to favor the formation of a B/C/SEP stamen identity complex. According to this viewpoint, although B-repression is not fully released in a single ben mutant background, establishment of the B-function program in its sepals would be made possible, since no competition with the C-function genes occurs. By contrast, in a bl ben context, upregulation and maintenance of the B-function in the first whorl could be overruled by the C-function genes due to an unfavorable initial C/B ratio. In turn, the initial C/B ratio might be more balanced in a ben rob bl background to allow simultaneous B- and C-gene upregulation due to full release of B-function repression. Finally, Arabidopsis AP2 also negatively regulates SEP3 expression in the first whorl, in addition to repressing AP3 and AGAMOUS (Krogan et al., 2012). Further experiments will be required to determine to what extent BEN and the ROB genes also regulate the expression domain of the petunia SEP genes. If so, potential differences in SEP protein availability in the first whorl of ben bl versus ben rob bl mutants might further influence the final outcome of B- and C-gene upregulation.
Different Patterns of Functional Divergence in the euAP2 Clade
All of the petunia AP2/ERF family members analyzed here (BEN, BOB, ROB1, ROB2, and ROB3) belong to the euAP2 lineage, a subgroup within the large AP2/ERF family characterized by the presence of two AP2 domains and miR172 target sites (Kim et al., 2006). We showed that the petunia AP2-type genes ROB1, ROB2, and ROB3 only pattern the B-function in the sepals, while Arabidopsis AP2 was shown to regulate both the B- and C- function expression domains (Krogan et al., 2012). In turn, we found that petunia BEN, a TOE-type gene, regulates the B- and C-function expression domains as found for AP2. In addition, we showed that BEN and BOB, two closely related genes, are redundantly required for proper development of second and third floral whorls. In contrast, loss-of-function mutants for the Arabidopsis TOE-type genes (TOE1, TOE2, SMZ, and SNZ) do not display defects in floral organ development. Instead, all of these genes have been identified as floral repressors, together with TOE3 and AP2 (Aukerman and Sakai, 2003; Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010; Huijser and Schmid, 2011; Zhu and Helliwell, 2011). In turn, we did not observe obvious effects on flowering time in any of the single and higher order mutants we analyzed in this study, although we cannot exclude the possibility that we missed subtle phenotypes that might be difficult to observe in a greenhouse. On the other hand, petunia contains three other TOE-type genes (Figure 2M) that we have not yet functionally analyzed. Therefore, it is possible that at least some of the euAP2 genes from petunia might function as floral repressors, possibly in a redundant fashion similar to that in Arabidopsis. Functional analysis of these uncharacterized genes and possibly the creation of higher order mutants will be needed to address this issue.
Although much more functional data from a wider range of species will be required to understand in detail the evolutionary trajectory of individual euAP2 genes, the above data strongly suggest that different patterns of functional divergence in the euAP2 clade have occurred in the lineages leading to Arabidopsis and petunia.
Thus, together with the identification of the BL/FIS functions in petunia and snapdragon, different patterns of functional divergence in the euAP2 clade may help to explain why two decades of comparative flower development have failed to demonstrate molecular conservation of the Arabidopsis A-function in other species. One of the questions that now arises is whether this reflects a general difference between rosids and asterids. Although comparative studies are largely lacking, the available data thus far seem to support this. The distantly related asterid species snapdragon and petunia both employ the same miRNA (FIS/BL) as the major C-patterning factor, while their genes most similar to Arabidopsis AP2 are required for normal floral organ development but are not involved in C-patterning. However, a B-patterning function for the snapdragon LIP genes remains to be shown, possibly still hidden in redundancy with BEN and/or other AP2 homolog(s). Our findings now also offer a logical explanation for the striking phenotypic difference induced by Pro35S:miR172 in Arabidopsis (Chen, 2004) compared with wild tobacco (Nicotiana benthamiana) (Mlotshwa et al., 2006), a close relative of petunia. While Pro35S:miR172 flowers in Arabidopsis closely resemble strong ap2 alleles, N. benthamiana Pro35S:miR172 overexpression lines exhibited phenotypes ranging from the wild type to flowers resembling ben single mutants to ben rob higher order mutants. While the molecular basis of the phenotype was not analyzed, it is likely that miR172 overexpression resulted in the codownregulation of BEN and ROB homologs in N. benthamiana, since they all share a miR172 recognition site in their coding sequence (Supplemental Figure 2B). This suggests that a similar BEN/ROB dependent B-class patterning mechanism may also be active in other asterid species, at least in N. benthamiana.
Finally, it should be noted that in the original ABC model, the B-function expression domain was proposed to be independent from the A- and C-functions. The combinatorial nature and complexity of the identified cadastral (boundary setting) functions in petunia (Figure 6A) are therefore difficult to integrate in the textbook ABC model. However, our findings are perfectly compatible with a more recent view of floral development, the (A)BC model (Causier et al., 2010), in which a more broadly defined (A)-function provides the genetic context in which the B- and C-functions are active and regulates both their spatial and temporal expression domains.
METHODS
Plant Material and Genotyping
Petunia (Petunia hybrida W138) plants were grown under standard greenhouse conditions (16 h day/8 h night; natural light supplemented with Philips Sodium HPS 400W SON-T AGRO light bulbs; 55,000 lumens) that were further influenced by local seasonal changes (45.72°N 4.82°E). The bl-1 and bl-2 alleles have been described earlier (Cartolano et al., 2007). The ben mutation was identified as spontaneous recessive mutation segregating in a bl-1 family introgressed in the W138 background, a highly active dTph1 transposon line that is commonly used for petunia mutagenesis (Vandenbussche et al., 2016). The ben-139, ben-724, bob-213, bob-519, rob1-61, rob1-114, rob1-463F, rob1-463R, rob2-915, and rob3-935 dTph1 insertion alleles were all identified by BLAST searching our dTph1 transposon flanking sequence database (Vandenbussche et al., 2008), which has been considerably enlarged in recent years. Note that the dTph1 transposon is small (284 bp) and therefore does not necessarily affect gene function when inserted in promoter, untranslated region, or intron regions. For that reason, we exclusively selected and analyzed putative dTph1 insertions in the coding sequences of BEN, BOB, ROB1, ROB2, and ROB3. When inserted in the coding sequence, the dTph1 element is cotranscribed and normally leads to disruption of gene function, since dTph1 contains multiple stop codons in all six possible reading frames. Insert positions (expressed in base pairs downstream of the ATG start codon with the coding sequence as a reference) were determined by aligning the dTph1flanking sequences with the genomic and cDNA sequences. All in silico-identified candidate insertions were confirmed by PCR-based genotyping of the progeny from the selected insertion lines, using primers flanking the dTph1 transposon insertions. All segregation primers are listed in Supplemental Table 1. The following thermal profile was used for segregation analysis PCR: 11 cycles (94°C for 15 s, 71°C for 20 s minus 1°C/cycle, 72°C for 30 s), followed by 40 cycles (94°C for 15 s, 60°C for 20 s, and 72°C for 30 s). The different insertion mutants were further systematically genotyped in subsequent crosses and segregation analyses. Homozygous mutants were manually cross-pollinated for double, triple, and higher order mutant analyses. See Supplemental Table 4 for details on the phenotypic analysis. Insertion alleles used in the crosses are indicated in the corresponding figure legends.
Cloning of the ben Mutation by Transposon Display
To generate the required material for a transposon display experiment, all individuals of the ben segregating family that displayed a single mutant bl-1 phenotype were self-pollinated, and progeny (20 individuals) were grown from each of these plants. From this plant material, we selected 12 homozygous ben mutants and 12 plants homozygous wild type for the ben mutation. DNA was harvested from these 24 plants and subjected to transposon display analysis (Van den Broeck et al., 1998; Vandenbussche et al., 2013). Transposon flanking sequence fragments that cosegregate with the ben genotype (absent in all homozygous wild-type plants, but present in all of the homozygous mutants) are good candidates to represent the mutated ben locus. Note that the W138 transposon line is highly active, and transposon excision from a given locus may occur at any moment. In the case of the dTph1 transposon, the major source of mutations in W138, excision usually results in the formation of a disrupting footprint that maintains the mutation. Therefore, transposon flanking sequences that are absent in all homozygous wild-type plants but present in most (but not all) of the homozygous mutants are also plausible candidates to represent the mutated locus. Based on these selection criteria, we retained several candidates potentially corresponding to the ben locus. Because the transposon display analysis does not allow one to clearly discriminate between heterozygous and homozygous presence of the transposon, we performed a segregation analysis using gene-specific flanking primers to more rigorously test linkage of these transposon insertions to the bl ben phenotype. With one exception (see below), this led to the exclusion of all candidates, since in each case, bl ben mutants were found that were not homozygous mutant for the candidate insertion. This demonstrates that these insertions were closely linked to the BEN locus but were not the causative agent of the phenotype. A PCR segregation analysis of the remaining candidate, using ben-488 genotyping primers flanking the insertion site (Supplemental Table 1) showed that all bl ben plants were homozygous mutant for the transposon insertion, except for plant #2, while none of the bl plants showed the insertion. Sequencing of the corresponding transposon flanking fragment showed that it corresponded to an insertion in the functionally uncharacterized PhAP2B gene. Sequencing of the PCR amplification products from bl ben mutants confirmed the position of the insertion in PhAP2B at base pair position 488 downstream of the ATG, and it also revealed that the absence of the transposon in bl ben plant #2 was caused by complete excision of the dTph1 element, which left behind an 8-bp footprint, causing premature stop codons, and thus maintaining the mutation. Finally, segregation analysis of a larger selection of plants further confirmed a full linkage of the phap2b insertion with the ben phenotype in the bl background, as well as in outcrossed ben single mutants.
Imaging and Microscopy
Scanning electron microscopy images were obtained as previously described (Vandenbussche et al., 2009) or using a HIROX SH-1500 bench top environmental scanning electron microscope equipped with a cooled stage. Floral phenotypes were imaged by conventional digital photography using a glass plate as a support to generate a black background. When necessary, backgrounds were further equalized by removing dust particles and light reflections with Photoshop.
Identification of Petunia euAP2 Genes and Phylogenetic Analysis
Petunia euAP2 and ANT genes were identified among the predicted protein data sets from Petunia axillaris and Petunia inflata (Bombarely et al., 2016) by BLASTp using euAP2 family members from Arabidopsis thaliana and tomato (Solanum lycopersicum), and in parallel by tBLASTN search against the P. axillaris and P. inflata genome sequences. Coding sequences of BOB, ROB2/ROB3, ANT1, and ANT2 were experimentally determined by sequencing transcripts amplified from cDNA derived from young floral buds (W138 line) and deposited in GenBank (see Supplemental Table 3 for accession numbers). To evaluate the accuracy of the automatically predicted gene models of the petunia euAP2 genes, we compared the translation of the experimentally obtained coding sequences of BEN, BOB, and ROB1-3 with the gene models (Supplemental Figure 2A). For two out of the five genes (ROB1 and ROB3), predictions were accurate, while for the remaining ones, some exons were absent or their borders not entirely correctly predicted (Supplemental Figure 2A). For the three TOE-type proteins that we did not functionally analyze in this study (Supplemental Table 3), but which were included in the phylogenetic analysis, we improved predictions for two of the P. axillaris copies based on alignments of the genome sequence with RNA-seq data (Supplemental Table 3). The automatically predicted versions as well as our RNA-seq-based improved versions (marked with an asterisk at the end of the sequence name) are shown in the alignment of Supplemental Figure 2A. We retained the five experimentally defined W138 euAP2 sequences and three remaining P. axillaris representatives (two of which were corrected) to be included in the phylogenetic analysis. Whole-genome predicted protein and transcript data sets of Arabidopsis, Capsella rubella, tomato, Medicago truncatula, grape (Vitis vinifera), and peach (Prunus persica) were downloaded from the Phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html). These data sets were subjected to local BLAST analysis using BioEdit (Hall, 1999) for the identification and retrieval of euAP2 protein and nucleotide sequences. The tomato SlAP2 sequences (Supplemental Table 3) were retrieved from GenBank. Alignments were obtained using ClustalW (Thompson et al., 1994) and were further manually refined in BioEdit (see Supplemental File 1). Start and end of the partial sequences of the Arabidopsis and petunia AINTEGUMENTA (ANT) proteins shown in Supplemental Figure 2B mark the conserved region that was used to generate the neighbor-joining tree shown in Figure 2M. The tree was computed with Treecon software (Van de Peer and De Wachter, 1994) using the following settings: (1) distance estimation options: Tajima and Nei distance calculation; insertions and deletion not taken into account; alignment positions: all; bootstrap analysis: yes, 2000 samples. (2) Infer tree topology options: neighbor-joining; bootstrap analysis: yes. (3) Root unrooted trees options: outgroup option: single sequence (forced); bootstrap analysis: yes; select root: At-ANT. The two major lineages of AP2-like genes consist of euAP2 and ANT genes (Kim et al., 2006). We therefore opted to root the euAP2 tree with ANT proteins. Positions of the two conserved AP2 domains (Figures 2C, 3B, and 4B) were determined via the NCBI CCD database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Expression Analysis
For qRT-PCR analysis, tissues were ground in liquid nitrogen, and total RNA was extracted using Trizol reagent (Invitrogen) and treated with Turbo DNA-free DNase I (Ambion). RNA was reverse transcribed using RevertAid M-MuLV reverse transcriptase (Fermentas) according to the manufacturer’s protocol. PCR was performed in an optical 384-well plate in the QuantStudio 6 Flex real-time PCR system (Applied Biosystems) using FastStart Universal SYBR Green Master (Rox) (Roche), in a final volume of 10 µL, according to the manufacturer’s instructions. Primers used were designed using the online Universal ProbeLibrary Assay Design Center (Roche). All primers are listed in Supplemental Table 1. The following standard thermal profile was used for all PCR: 95°C for 10 min, 40 cycles of 95°C for 10 s, and 60°C for 30 s. Data were analyzed using QuantStudio 6 and 7 Flex Real-Time PCR System Software v1.0 (Applied Biosystems). PCR efficiency (E) was estimated from the data obtained from standard curve amplification using the equation 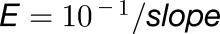 . Relative expression (R.E.) values on the y axes are the average of nine data points resulting from the technical triplicates of three biological replicates ± sd and normalized to the geometrical average of three E−ΔCt, where ΔCt = CtGOI − CtACTIN, GAPDH or RAN. The floral bud series (marked floral buds 1–3 in Figures 2D, 3A, and 4A) are successive developmental stages of complete floral buds harvested from the same inflorescences (Supplemental Figure 1). For each biological replicate, corresponding stages harvested from three inflorescences were pooled. Stage 3 corresponds to flower buds with a diameter of ∼5 mm and from which individual floral organs can be easily dissected by hand. All analyses showing expression in separate floral organ types are from this stage. Biological replicates of the different floral organ types were composed of pooled stage 3 organs harvested from three different flowers each time. For mutants with highly modified floral architecture, developmental stages in relation to wild-type development were deduced based on their position on the inflorescence. Floral buds marked “2” (diameter ∼2.5 mm) and “1” (diameter ∼1.5 mm) are younger stages and were harvested from the next two nodes produced after bud stage 3. In addition to 1.5-mm buds, stage 1 also includes the inflorescence meristem and very young developing floral primordia subtended by bracts, which are attached to the base of the pedicel of the 1.5-mm bud. Vegetative apices (including very small leaf primordia) were harvested from 3-week-old seedlings by manually removing cotyledons, roots, and developed leaves. Young leaf primordia were isolated from the same 3-week-old seedlings. Each biological replicate of the vegetative apices and young leaf primordia consisted of pooled material harvested from each time 10 seedlings. In situ hybridization of PhGLO2 (Figure 5L) was done as described previously (Vandenbussche et al., 2004).
. Relative expression (R.E.) values on the y axes are the average of nine data points resulting from the technical triplicates of three biological replicates ± sd and normalized to the geometrical average of three E−ΔCt, where ΔCt = CtGOI − CtACTIN, GAPDH or RAN. The floral bud series (marked floral buds 1–3 in Figures 2D, 3A, and 4A) are successive developmental stages of complete floral buds harvested from the same inflorescences (Supplemental Figure 1). For each biological replicate, corresponding stages harvested from three inflorescences were pooled. Stage 3 corresponds to flower buds with a diameter of ∼5 mm and from which individual floral organs can be easily dissected by hand. All analyses showing expression in separate floral organ types are from this stage. Biological replicates of the different floral organ types were composed of pooled stage 3 organs harvested from three different flowers each time. For mutants with highly modified floral architecture, developmental stages in relation to wild-type development were deduced based on their position on the inflorescence. Floral buds marked “2” (diameter ∼2.5 mm) and “1” (diameter ∼1.5 mm) are younger stages and were harvested from the next two nodes produced after bud stage 3. In addition to 1.5-mm buds, stage 1 also includes the inflorescence meristem and very young developing floral primordia subtended by bracts, which are attached to the base of the pedicel of the 1.5-mm bud. Vegetative apices (including very small leaf primordia) were harvested from 3-week-old seedlings by manually removing cotyledons, roots, and developed leaves. Young leaf primordia were isolated from the same 3-week-old seedlings. Each biological replicate of the vegetative apices and young leaf primordia consisted of pooled material harvested from each time 10 seedlings. In situ hybridization of PhGLO2 (Figure 5L) was done as described previously (Vandenbussche et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers KU096994, KU096995, KU096996, KU096997, KU096998, KU096994, MF327594, and MF327595 (see also Supplemental Table 3).
Supplemental Data
Supplemental Figure 1. W138 Floral Bud Developmental Stages Sampled for qRT-PCR Analysis.
Supplemental Figure 2. Alignments of euAP2 Protein Sequences from Petunia and Other Selected Species.
Supplemental Figure 3. Additional rob and ben rob Mutant Phenotypes.
Supplemental Table 1. Oligo Sequences Used in This Study
Supplemental Table 2. Sequence comparison of Petunia BEN, BOB, and ROB1-3 against the Six Arabidopsis euAP2 Protein Sequences (TAIR10 Database).
Supplemental Table 3. Gene Names, Synonyms, and Accession Codes for Sequences Shown in Figure 2M and Supplemental Figure 2.
Supplemental Table 4. Overview of Genotypes Analyzed in This Study.
Supplemental File 1. Alignment of euAP2 Protein Sequences from Petunia (Ph; Peaxi), Tomato (Sl; Sol), Arabidopsis (At), Capsella rubella (Carub), Medicago truncatula (Medtr), Grape (GSVIV), and peach (Prupe) in Fasta Format.
Acknowledgments
M.V. was supported by a CNRS ATIP-AVENIR award. K.H. was supported by NWO Grant 818.02.012. We thank A. Lacroix and J. Berger for plant care assistance and V. Bayle for electron microscopy technical support.
AUTHOR CONTRIBUTIONS
M.V., P.M., and K.H. conceived and designed the experiments. P.M., K.H., J.Z., F.R., S.C., S.R.B., A.V.-G., P.C., C.T., and M.V. performed the experiments. P.M., K.H., and M.V. analyzed the data. M.V., K.H., and P.M. wrote the article with feedback from C.T. and A.V.-G.
Footnotes
Articles can be viewed without a subscription.
References
- Angenent G.C., Franken J., Busscher M., Colombo L., van Tunen A.J. (1993). Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 4: 101–112. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20. [DOI] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95. [DOI] [PubMed] [Google Scholar]
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39: 901–905. [DOI] [PubMed] [Google Scholar]
- Causier B., Schwarz-Sommer Z., Davies B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21: 73–79. [DOI] [PubMed] [Google Scholar]
- Causier B., Castillo R., Zhou J., Ingram R., Xue Y., Schwarz-Sommer Z., Davies B. (2005). Evolution in action: following function in duplicated floral homeotic genes. Curr. Biol. 15: 1508–1512. [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Dinh T.T., Girke T., Liu X., Yant L., Schmid M., Chen X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139: 1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002. [DOI] [PubMed] [Google Scholar]
- Gerats A.G., Huits H., Vrijlandt E., Maraña C., Souer E., Beld M. (1990). Molecular characterization of a nonautonomous transposable element (dTph1) of petunia. Plant Cell 2: 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438. [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Heijmans K., Ament K., Rijpkema A.S., Zethof J., Wolters-Arts M., Gerats T., Vandenbussche M. (2012). Redefining C and D in the petunia ABC. Plant Cell 24: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong R.L., Hamaguchi L., Busch M.A., Weigel D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P., Schmid M. (2011). The control of developmental phase transitions in plants. Development 138: 4117–4129. [DOI] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697. [DOI] [PubMed] [Google Scholar]
- Jaillon O., et al.; French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. [DOI] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer B.G., Van Montagu M., Okamuro J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Tsuda S., Tanaka Y., Mayama T., Okuyama Y., Tsuchimoto S., Takatsuji H. (2002). Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. Plant J. 32: 115–127. [DOI] [PubMed] [Google Scholar]
- Kater M.M., Colombo L., Franken J., Busscher M., Masiero S., Van Lookeren Campagne M.M., Angenent G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck E., McSteen P., Carpenter R., Coen E. (2003). Separation of genetic functions controlling organ identity in flowers. EMBO J. 22: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Soltis P.S., Wall K., Soltis D.E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23: 107–120. [DOI] [PubMed] [Google Scholar]
- Krizek B.A., Fletcher J.C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6: 688–698. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Hogan K., Long J.A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Klenz J.E., Martinez-Zapater J., Haughn G.W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A. (2007). An evaluation of A-function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168: 73–91. [Google Scholar]
- Maes T., Van de Steene N., Zethof J., Karimi M., D’Hauw M., Mares G., Van Montagu M., Gerats T. (2001). Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P.C., Vincent C.A., Doyle S., Carpenter R., Coen E.S. (1998). Control of floral homeotic gene expression and organ morphogenesis in Antirrhinum. Development 125: 2359–2369. [DOI] [PubMed] [Google Scholar]
- Mlotshwa S., Yang Z., Kim Y., Chen X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 107: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A.S., Royaert S., Zethof J., van der Weerden G., Gerats T., Vandenbussche M. (2006). Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. [DOI] [PubMed] [Google Scholar]
- Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W.E., Saedler H., Sommer H., Schwarz-Sommer Z. (1992). GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., van der Krol A.R., Chua N.H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallade J., Maizonnier D., Cornu A. (1987). La morphogenèse florale chez le pétunia. I. Analyse d’un mutant à corolle staminée. Can. J. Bot. 65: 761–764. [Google Scholar]
- Van den Broeck D., Maes T., Sauer M., Zethof J., De Keukeleire P., D’hauw M., Van Montagu M., Gerats T. (1998). Transposon display identifies individual transposable elements in high copy number lines. Plant J. 13: 121–129. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Gerats T. (2013). Transposon display: a versatile method for transposon tagging. Methods Mol. Biol. 1057: 239–250. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Chambrier P., Rodrigues Bento S., Morel P. (2016). Petunia, your next supermodel? Front. Plant Sci. 7: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Zethof J., Royaert S., Weterings K., Gerats T. (2004). The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A.S., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 21: 2269–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Janssen A., Zethof J., van Orsouw N., Peters J., van Eijk M.J., Rijpkema A.S., Schneiders H., Santhanam P., de Been M., van Tunen A., Gerats T. (2008). Generation of a 3D indexed petunia insertion database for reverse genetics. Plant J. 54: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., De Wachter R. (1994). TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569–570. [DOI] [PubMed] [Google Scholar]
- van der Krol A.R., Brunelle A., Tsuchimoto S., Chua N.H. (1993). Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7: 1214–1228. [DOI] [PubMed] [Google Scholar]
- Verde I., et al.; International Peach Genome Initiative (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. [DOI] [PubMed] [Google Scholar]
- Wang P., Cheng T., Lu M., Liu G., Li M., Shi J., Lu Y., Laux T., Chen J. (2016). Expansion and functional divergence of AP2 group genes in spermatophytes determined by molecular evolution and Arabidopsis mutant analysis. Front. Plant Sci. 7: 1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H., Mica E., Todesco M., Long J.A., Weigel D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137: 3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39. [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.H., Helliwell C.A. (2011). Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 62: 487–495. [DOI] [PubMed] [Google Scholar]