Two plastidial coiled coil-containing proteins play a critical role in delivering suitable maltooligosaccharide primers to STARCH SYNTHASE4 during starch granule initiation.
Abstract
The molecular mechanism that initiates the synthesis of starch granules is poorly understood. Here, we discovered two plastidial proteins involved in granule initiation in Arabidopsis thaliana leaves. Both contain coiled coils and a family-48 carbohydrate binding module (CBM48) and are homologs of the PROTEIN TARGETING TO STARCH (PTST) protein; thus, we named them PTST2 and PTST3. Chloroplasts in mesophyll cells typically contain five to seven granules, but remarkably, most chloroplasts in ptst2 mutants contained zero or one large granule. Chloroplasts in ptst3 had a slight reduction in granule number compared with the wild type, while those of the ptst2 ptst3 double mutant contained even fewer granules than ptst2. The ptst2 granules were larger but similar in morphology to wild-type granules, but those of the double mutant had an aberrant morphology. Immunoprecipitation showed that PTST2 interacts with STARCH SYNTHASE4 (SS4), which influences granule initiation and morphology. Overexpression of PTST2 resulted in chloroplasts containing many small granules, an effect that was dependent on the presence of SS4. Furthermore, isothermal titration calorimetry revealed that the CBM48 domain of PTST2, which is essential for its function, interacts with long maltooligosaccharides. We propose that PTST2 and PTST3 are critical during granule initiation, as they bind and deliver suitable maltooligosaccharide primers to SS4.
INTRODUCTION
Starch, one of the most abundant biopolymers on Earth, is produced by most plants in leaves and storage organs. Starch is composed of two glucose polymers, amylopectin and amylose, which are arranged in insoluble, semicrystalline granules. Starch synthesis in green plants is confined to the plastids, and in Arabidopsis thaliana leaves, a typical chloroplast contains five to seven starch granules (Crumpton-Taylor et al., 2012). These granules are synthesized during the day from photoassimilates and are almost fully degraded during the subsequent night to provide energy required for metabolism and growth (Stitt and Zeeman, 2012; Smith, 2012). Despite this critical role of starch in plant growth and productivity, and our reliance on starch as a major component of our diet, little is known about the mechanisms that initiate starch granule synthesis, or the factors that control granule number, shape, and size within plastids. An understanding of these mechanisms is crucial for ultimately controlling these properties, which is a goal that has significant industrial interest (Lindeboom et al., 2004).
Previous research on starch granule initiation in Arabidopsis has focused on the role of STARCH SYNTHASE4 (SS4), one of five starch synthase (SS) isoforms. These glucosyltransferases elongate glucan chains using ADP-glucose as a glucosyl donor. The isoforms SS1, SS2, and SS3 are involved in amylopectin synthesis, while GRANULE BOUND STARCH SYNTHASE (GBSS) is involved in amylose synthesis (Streb and Zeeman, 2012; Pfister and Zeeman, 2016). However, SS4 is proposed to play a unique role in generating glucan primers required to initiate starch granule synthesis. This notion is supported by three lines of evidence: First, while mutants lacking the other SS isoforms have altered amylopectin structure (Delvallé et al., 2005; Zhang et al., 2005, 2008; Pfister et al., 2014), ss4 mutants have only minor alterations (Roldán et al., 2007). However, chloroplasts of the ss4 mutant contain either zero, one, or rarely two large starch granules (Roldán et al., 2007; Szydlowski et al., 2009; Crumpton-Taylor et al., 2013). Such a severe reduction in granule number has not been reported for mutants lacking the other SS isoforms (Szydlowski et al., 2009; Seung et al., 2015). Furthermore, the ss4 mutant leaves contain less starch than the wild type at the end of the day, but more at the end of the night. Second, ss4 mutants accumulate large amounts of ADP-glucose (Crumpton-Taylor et al., 2013; Ragel et al., 2013). This accumulation of the SS substrate is consistent with the idea that the other SS isoforms have limited activity in the absence of SS4. SS4 may therefore function upstream of the other SS in the initiation process. The amount of adenylates sequestered into ADP-glucose in ss4 exceeds the total adenylate pool in wild-type plants. This may limit the availability of adenylates for photophosphorylation, resulting in the pale dwarfed phenotype reported for ss4 (Ragel et al., 2013). Finally, the ss4 phenotype can be partly rescued by abolishing the activity of the glucan hydrolase, α-AMYLASE3 (AMY3) (Seung et al., 2016). Although the amy3 mutant has no obvious difference in granule number per chloroplast compared with the wild type, chloroplasts in the amy3 ss4 double mutant have many more granules than ss4 and accumulate less ADP-glucose. This suggests that glucan primers can be generated in the absence of SS4, but SS4 is necessary to prevent their premature degradation. All of these studies are consistent with the notion that SS4 plays an important role in the initial stages of granule formation. However, it is not known exactly which features of SS4 make it particularly suited to fulfill this role, or whether other proteins are required in the process.
Recently, we reported that the PROTEIN TARGETING TO STARCH (PTST or PTST1) protein plays an essential role in amylose synthesis, as it is important for the localization of GBSS on the starch granule (Seung et al., 2015). The PTST1 protein contains coiled coils, typically implicated in protein-protein interactions (Mason and Arndt, 2004), adjacent to a family-48 carbohydrate binding module (CBM48) at the C terminus (Lohmeier-Vogel et al., 2008; Seung et al., 2015). CBM48s are conserved glucan binding domains found in proteins involved in starch metabolism in plants, such as the phosphoglucan phosphatases, branching enzymes, and debranching enzymes (Janeček et al., 2011; Meekins et al., 2014; Møller et al., 2016), as well as the mammalian AMP kinase glycogen binding β-subunit (McBride et al., 2009).
In this study, we identified two homologs of PTST1 in the Arabidopsis genome, which we named PTST2 (At1g27070) and PTST3 (At5g03420). Both proteins have predicted coiled coils adjacent to a CBM48 domain. We show that both homologs are required for proper granule initiation in Arabidopsis chloroplasts. Interestingly, PTST2 is the Arabidopsis ortholog of FLO6, a CBM48-containing protein in the rice (Oryza sativa) endosperm that contains coiled coils (Seung et al., 2015) and influences grain starch content, granule morphology, and the physicochemical properties of starch (Peng et al., 2014). The FLO6 protein was proposed to interact with the rice debranching enzyme ISOAMYLASE1 (ISA1), but the exact mechanism by which it participates in starch metabolism is unknown. We propose here that in Arabidopsis leaves, PTST2 and PTST3 interact with SS4 rather than with ISA1 and play a critical role in granule initiation by delivering suitable glucan primers to SS4.
RESULTS
PTST2 and PTST3 Are Conserved Coiled Coil and CBM48-Containing Proteins
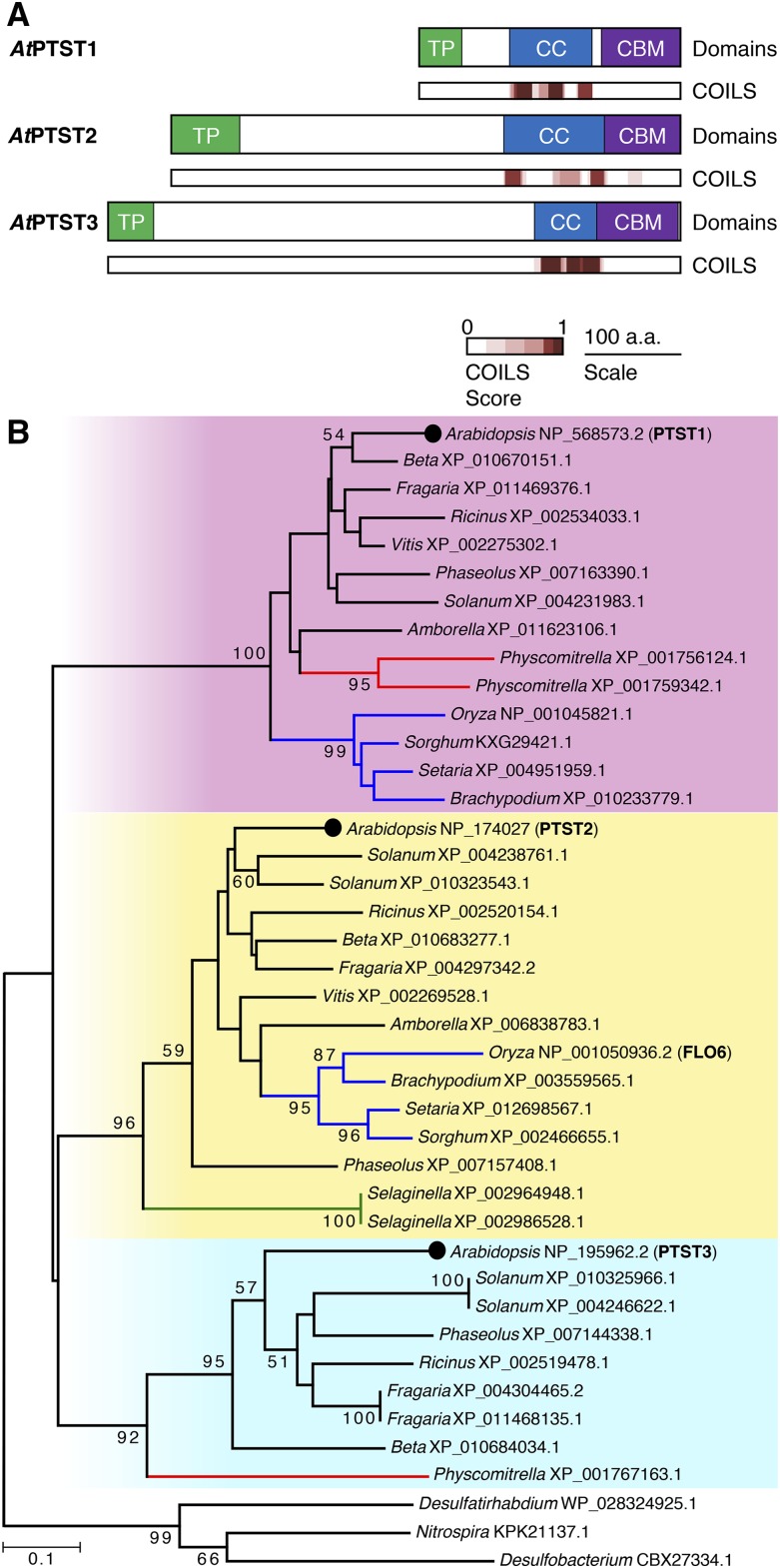
We previously demonstrated that the CBM48-containing PTST1 protein plays an important role in localizing the amylose-synthesizing enzyme, GBSS, to the starch granule (Seung et al., 2015). To find other proteins with a similar function, we used BLASTp against the Arabidopsis proteome to find proteins with sequence similarity to PTST1. The top two hits were encoded by At1g27070 and At5g03420, with E values of 3 × 10−13 and 4 × 10−12, respectively, and both proteins had not been previously characterized in Arabidopsis. We then analyzed the sequence for known features and domains. Both proteins were predicted to have a chloroplast transit peptide (cTP) using the TargetP program (Emanuelsson et al., 2007). The COILS/PCOILS server (Lupas, 1996) also predicted that both proteins contain coiled coils, similar in length to those found in PTST1. Both proteins also contained CBM48 domains at the C-terminal end, directly adjacent to the predicted coiled coils. The two proteins therefore had similar domain arrangements to PTST1, so we named them PTST2 and PTST3 (Figure 1). PTST2 is encoded at locus At1g27070 and is the Arabidopsis ortholog of the rice FLO6 protein, as shown by the phylogenetic analyses of Peng et al. (2014). Most (83%) of the Arabidopsis PTST2 sequence could be aligned to the rice FLO6 sequence using BLASTp, over which the proteins shared 38% identity. PTST3 is encoded at locus At5g03420. No alternative gene models are predicted for either the PTST2 or PTST3 locus from full-length cDNAs or RNA-seq data. The amino acid sequences of both PTST2 and PTST3 differ from that of PTST1 in that they have long N-terminal extensions with no predicted domains or motifs (Figure 1A). The N-terminal sequences from PTST2 and PTST3 show weak similarity with each other, with 25% identity.
Figure 1.
PTST2 and PTST3 Are Chloroplast-Localized CBM48-Containing Proteins with Coiled Coils.
(A) Schematic illustration of domains in the PTST proteins. The chloroplast transit peptides (TP) and the CBM48 domains are shown in green and violet, respectively. Coiled coil (CC)-containing regions are shown in blue. The locations of coiled coils predicted using COILS analysis with a 14-amino acid prediction window are shown with an indication of probability score (from 0 to 1, where 1 is the highest probability).
(B) Phylogenetic tree of PTST proteins. The tree was constructed using the neighbor-joining method, and clades corresponding to PTST1-, PTST2-, and PTST3-like sequences are highlighted in pink, yellow, and cyan, respectively. Branches in red, green, and blue represent sequences from Bryophytes, Pteridophytes, and monocots, respectively. Bootstrap values greater than 50 are shown above or underneath the branches. The amino acid alignment used to generate this tree is provided in Supplemental Figure 1.
To assess the evolutionary relationship between the PTST proteins, we conducted a phylogenetic analysis, extending on the previous analyses for PTST1 (Seung et al., 2015) and FLO6/PTST2 (Peng et al., 2014). Amino acid sequences of homologs for each of the three Arabidopsis PTST proteins were retrieved using BLASTp against NCBI RefSeq genomes. The C-terminal region of the sequences containing the coiled coils and the CBM48 domain were aligned (Supplemental Figure 1), and a phylogenetic tree was constructed using the neighbor-joining method (Figure 1B). The N-terminal sequences were not included in this analysis as they aligned poorly between the three isoforms. The tree was rooted using an outgroup containing the closest prokaryotic sequence to each PTST, identified using BLASTp. The plant sequences grouped into three clades with strong bootstrap support, each representing PTST1-, PTST2-, or PTST3-like sequences. To further substantiate this grouping, we analyzed the position of introns for each sequence. We observed a perfect retention of intron positions within each clade and mostly differences between the three clades (Supplemental Figure 1). However, a conserved intron was observed between the PTST2 and PTST3 clades in the highly conserved CBM48 domain. Another intron outside the CBM48 domain coincided between the PTST1 and PTST2 clades, but the sequences in this region were poorly conserved. Thus, it is unlikely that these introns have a common origin. These observations, together with the finding that the PTST2 and PTST3 clades group closer together in the tree than with the PTST1 clade (Figure 1B), suggest that PTST2 and PTST3 share a more recent common ancestor.
We then assessed the conservation of PTST2 and PTST3 sequences among plant species. No sequence from Physcomitrella was observed in the PTST2 clade. Also, no sequences from Amborella or grasses such as Brachypodium, Sorghum, and Setaria were found in the PTST3 clade. This suggests that the PTST3 gene was lost multiple times during plant evolution. However, none of the land plant genomes assessed had lost both PTST2 and PTST3, suggesting the necessity of at least one of the two proteins.
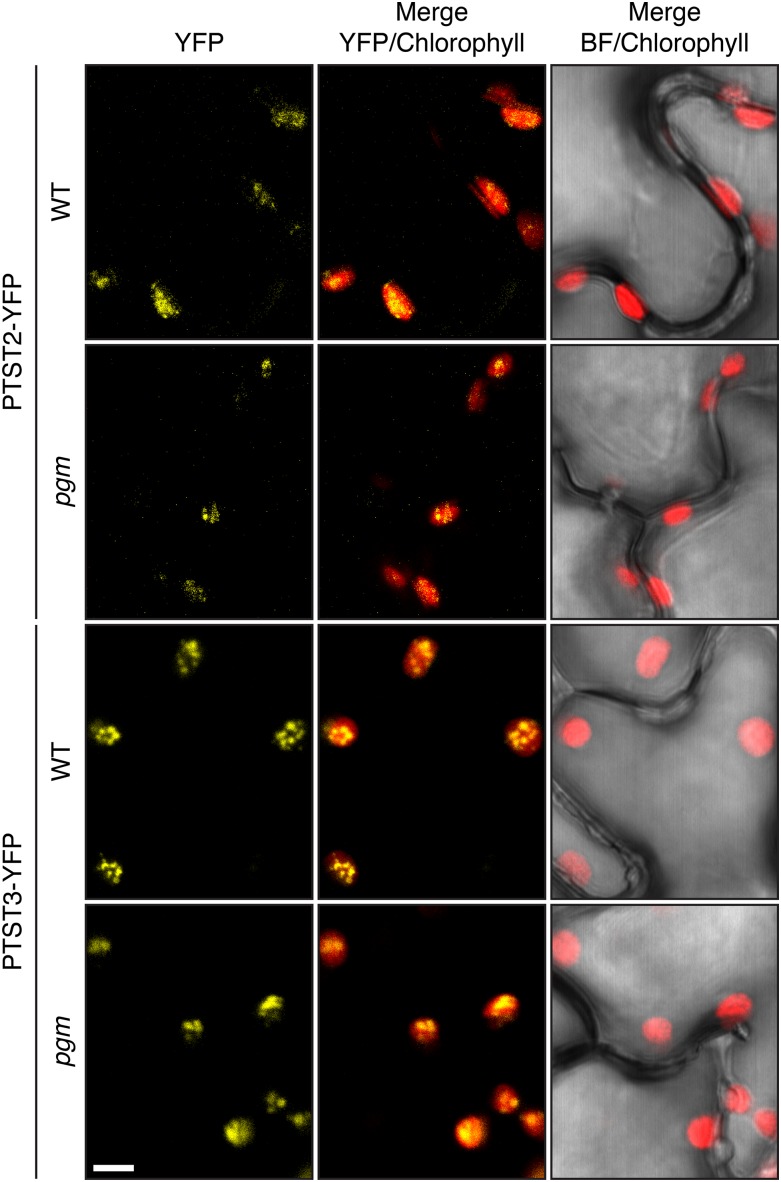
To verify experimentally that both PTST2 and PTST3 localize to the plastid, we cloned the full-length coding sequences into a plant expression vector yielding fusion proteins with a C-terminal YFP tag. The fusion proteins were transiently expressed in Nicotiana sylvestris leaves by infiltrating Agrobacterium tumefaciens cultures carrying the expression vectors, and the resulting YFP signal was imaged using confocal laser scanning microscopy. Both PTST2-YFP and PTST3-YFP localized to the chloroplast of transformed epidermal cells (Figure 2). Interestingly, the YFP signal was not homogenously distributed in the chloroplast for either protein, suggesting that the proteins localize to distinct regions. Although the pattern did not resemble a starch-granule localization, which typically appears as discrete flattened bodies within the chloroplast (Feike et al., 2016; Seung et al., 2015), the observation that PTST2 and PTST3 proteins have CBM48s prompted us to investigate whether this subchloroplastic localization was associated with the presence of starch granules. We therefore also expressed the proteins in the starchless phosphoglucomutase (pgm) mutant of N. sylvestris. However, similar localization patterns were observed (Figure 2), suggesting that the subchloroplastic localization is not due to the presence of starch.
Figure 2.
Localization of Arabidopsis PTST2-YFP and PTST3-YFP.
The fusion proteins were transiently expressed in either wild-type N. sylvestris or its starchless pgm mutant. YFP and chlorophyll fluorescence were imaged using confocal microscopy. All panels are shown at the same scale. Bar = 5 µm.
Mutants Lacking PTST2 Initiate Fewer Starch Granules per Chloroplast
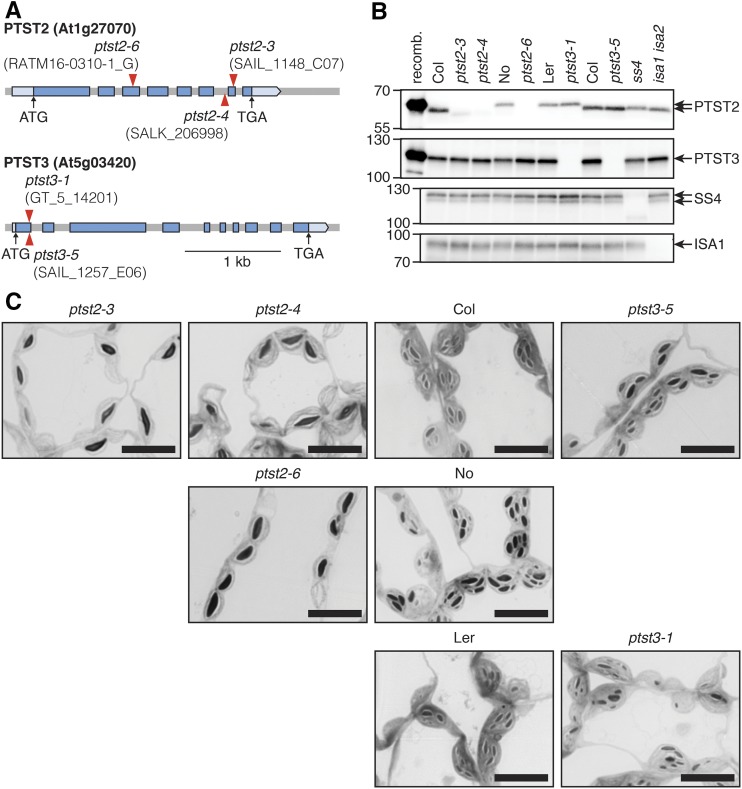
To investigate the role of PTST2 and PTST3 in starch metabolism, we obtained Arabidopsis mutants harboring T-DNA insertions in either of the two loci (Figure 3). Multiple T-DNA insertion lines were available (Supplemental Table 1). For further analysis, we selected three independent lines for PTST2 and two for PTST3. For PTST2, we chose ptst2-3 with an insertion in exon 7, ptst2-4 with an insertion in the intron between exons 6 and 7, and ptst2-6 with an insertion in exon 3 (Figure 3A). Both ptst2-3 and ptst2-4 were generated in the Columbia (Col) genetic background, while ptst2-6 was in the Nossen background. For PTST3, we selected ptst3-1 (Landsberg erecta [Ler] background) and ptst3-5 (Col background), both with insertions in exon 1. Lines homozygous for the insertion alleles were isolated using PCR-based genotyping (see Supplemental Table 2 for primers).
Figure 3.
Starch Granules in Chloroplasts of Arabidopsis Mutants Carrying T-DNA Insertions in PTST2 or PTST3.
(A) Schematic illustration of the exon-intron structure of PTST2 and PTST3 genes. Exons are represented with blue boxes, while the pale-blue boxes represent the 5′ and 3′ untranslated regions. The position of the translation start codon (ATG) and the stop codon (TGA) are shown with black arrows. Red arrowheads represent the position of T-DNA insertions.
(B) Immunoblot detection of PTST2 and PTST3 proteins in total protein extracts from Arabidopsis leaves. Panels from top to bottom show immunoblots with antisera raised against Arabidopsis PTST2, PTST3, SS4, and ISA1, respectively. An equal amount of total protein (30 µg) was loaded in each lane. The leftmost lanes on the PTST2 and PTST3 immunoblot panels were loaded with the respective recombinant (recomb.) protein (0.01 µg), expressed in E. coli, and purified. The wild-type controls for the ptst2-6 and ptst3-1 mutants are Nossen (No) and Ler, respectively. All other mutants are in the Col background. The migration of molecular mass markers (in kD) is indicated on the left of each panel.
(C) Observation of starch granules in chloroplasts of mutants using light microscopy. Leaf segments were excised from young leaves (as shown in Figure 4B) at the end of the day and embedded in resin for sectioning. Thin sections were stained with toluidine blue. Similar starch granule phenotypes were observed in at least three independent experiments performed on different batches of plants, and representative images are shown. Bars = 10 µm.
To observe the effect of the insertions on PTST2 and PTST3 expression, we detected the proteins in total protein extracts of young leaves by immunoblotting. We used antisera raised in rabbits against either Arabidopsis PTST2 or PTST3 recombinant proteins, which were expressed in and purified from E. coli. The expected molecular masses of PTST2 and PTST3, after cleavage of the predicted transit peptides, are 51.4 and 61.0 kD, respectively. The PTST2 antiserum recognized a band of the expected size in extracts from leaves of wild-type plants, but not in extracts from mutants carrying homozygous ptst2 alleles (Figure 3B). However, a faint band of lower molecular mass was observed in ptst2-3 and ptst2-4. It should be noted that the location of the T-DNA insertion in these alleles is toward the 3′-end of the gene, and the lower band may result from the translation of a C-terminally truncated version of the protein, missing most of the CBM48 domain. Such a band was not observed in the ptst2-6 mutant in the Nossen background, suggesting that it completely lacks the PTST2 protein. The PTST3 antiserum recognized a band in extracts of the wild type that was absent in both ptst3-1 and ptst3-5 mutant extracts. Interestingly, this band consistently migrated higher than expected (at ∼110 kD). The reason for this anomalous migration is unknown, but since the recombinant protein migrated similarly, we suggest that the protein has inherent features (e.g., hydrophobic regions that interfere with SDS binding) that retard migration on SDS-PAGE gels.
We hypothesized that PTST2 and PTST3 could play a similar role to PTST1, by targeting SS isoforms to their substrates. We therefore investigated whether the mutants have alterations in starch content and/or structure. We quantified the starch content of plants harvested at the end of the day and end of the night and found only minor differences between the mutants and wild types; all ptst2 and ptst3 mutants had slight increases in starch content, relative to the respective wild types, at the end of the night. The ptst2-6 mutant had slightly less starch than the Nossen wild type at the end of the day (Table 1). There was no altered phenotype visible in the ptst2 and ptst3 mutants under our conditions. We then profiled the amylopectin chain length distribution in the ptst2 and ptst3 mutants. Alterations in amylopectin structure might be expected if the activities of SS1 and SS2 were affected by the mutations (Delvallé et al., 2005; Zhang et al., 2008). However, no major changes in chain length distributions were observed in either mutant compared with the Col-0 wild type (Supplemental Figure 2A). To investigate whether GBSS activity was affected, we quantified the amylose content of starch in the mutants. Small increases in amylose content relative to the wild type were observed in both the ptst2 and ptst3 mutants (Supplemental Figure 2B). However, as these mutants do not fully degrade their starch during the night (Table 1), this may allow more time for amylose synthesis to occur within the granule. Higher amylose has previously been observed in other mutants that do not fully degrade their starch (Zeeman et al., 2002; Seung et al., 2015). PTST2 and PTST3 therefore play a different role in starch synthesis compared with PTST1, since ptst1 mutants produce starch that is almost completely amylose-free.
Table 1. Starch Content of ptst2 and ptst3 Mutants.
| Starch Content (mg/g FW) |
|||
|---|---|---|---|
| Ecotype | Genotype | ED | EN |
| ptst2 mutants | |||
| Col-0 | Wild type | 9.5 ± 0.2 | 0.5 ± 0.1 |
| ptst2-3 | 10.1 ± 0.7 | 1.4 ± 0.1 * | |
| ptst2-4 | 9.9 ± 0.3 | 1.5 ± 0.1 * | |
| No-0 | Wild type | 9.9 ± 0.4 | 0.5 ± 0.0 |
| ptst2-6 | 7.9 ± 0.4 * | 1.0 ± 0.1 * | |
| ptst3 mutants | |||
| Ler | Wild type | 10.2 ± 0.2 | 0.8 ± 0.1 |
| ptst3-1 | 10.7 ± 0.3 | 1.5 ± 0.1 * | |
| Col-0 | ptst3-5 | 10.3 ± 0.6 | 1.0 ± 0.1 * |
Values for starch content at end of day (ED) and end of night (EN) time points are the mean ± se from n = 6 individual plants. Values marked with an asterisk are significantly different from the corresponding wild type under a two-tailed t test at P < 0.05. FW, fresh weight.
We then investigated the number of starch granules per chloroplast, which might be altered if the mutations affected SS4 activity. Segments of young leaves were harvested at the end of day, fixed in glutaraldehyde, and embedded in resin. Thin sections were briefly stained in toluidine blue to allow visualization of starch granules via light microscopy. Chloroplasts of the ptst2 mutants contained far fewer starch granules than the respective wild-type chloroplasts, with most chloroplast sections containing only one large starch granule (Figure 3C). This strong reduction in granule number was observed in all ptst2 mutants, suggesting that the residual truncated PTST2 protein potentially present in ptst2-3 and ptst2-4 has little or no functional capacity. A similar qualitative observation of chloroplasts in ptst3 mutants did not reveal an obvious alteration in granule number. Taken together, these data suggest that PTST2 strongly influences the number of granules in each chloroplast but has only minor effects on starch content, amylopectin structure, and starch composition.
We investigated whether the loss of PTST2 might affect debranching enzyme (ISA1) function during starch synthesis, since the orthologous protein in rice, FLO6, was reported to interact with ISA1 (Peng et al., 2014). Arabidopsis isa1 mutants accumulate a soluble glucan (phytoglycogen) in the chloroplast in place of much of their starch (Delatte et al., 2005). However, soluble glucans did not accumulate in either the ptst2 or ptst3 mutants (Supplemental Table 3). It therefore appears unlikely that PTST2 affects ISA1 function in a significant way during starch synthesis in Arabidopsis leaves.
The Function of PTST3 in Granule Initiation Is Partially Redundant with That of PTST2
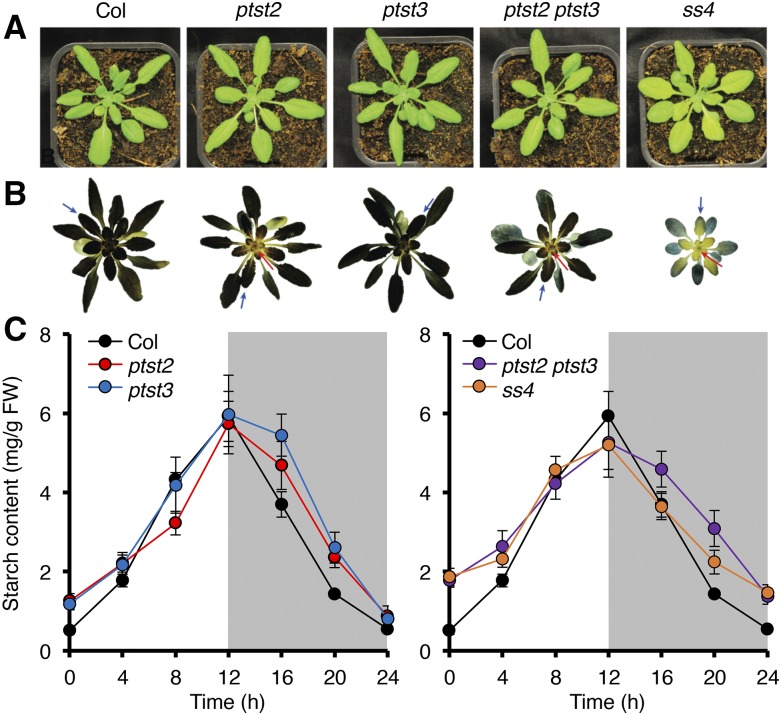
The reduced granule number per chloroplast observed in all ptst2 mutants suggests that PTST2 is involved in granule initiation. Given that PTST2 and PTST3 are related, and that some species appear to have lost one or the other isoform (Figure 1B), we investigated whether there was any redundancy between the two proteins. The ptst2-3 and ptst3-5 mutants were crossed to produce a ptst2 ptst3 double mutant (Figure 4). The double mutant, like the single mutants, did not have a visible growth defect (Figure 4A). Upon staining the rosettes with an iodine solution, we noted that the ptst2 and ptst2 ptst3 mutants had alterations in the distribution of starch within the rosette, where the center of the rosette stained visibly lighter than the wild type (Figure 4B). This was reminiscent of the ss4 mutant phenotype described previously (Crumpton-Taylor et al., 2013) and reproduced here (Figure 4B).
Figure 4.
Phenotype of the ptst2 ptst3 Double Mutant.
The ptst2-3 and ptst3-5 alleles were used for all experiments in this figure.
(A) Photographs showing the appearance of ptst2, ptst3 and double mutant rosettes, compared with ss4 rosettes.
(B) Iodine staining of starch in rosettes harvested at the end of day. The lighter-staining areas at the center of ptst2, ptst2 ptst3, and ss4 rosettes are indicated with red arrows. The blue arrows indicate examples of young leaves taken for light microscopy analysis.
(C) Starch turnover over a 12-h-day/12-h-night cycle. Entire rosettes were harvested at the indicated time points and starch content was determined. Each value represents the mean ±se of measurements made on n = 4 to 6 rosettes. All data were collected within a single experiment. For clarity, data from the single mutants are presented on the left panel, and data from the double mutant and ss4 are presented on the right panel. The same data from the wild type are shown in both panels.
Starch quantification again showed that both the ptst2 and ptst3 single mutants retained more starch at the end of the night than the wild type (Tables 1 and 2). However, this effect was exacerbated in the ptst2 ptst3 double mutant (Table 2), which had an end-of-night starch content that was significantly higher than that of ptst2 and comparable to that of the ss4 mutant. Given this similarity to ss4, we measured starch over a 24-h day-night cycle (Figure 4C). The pattern of starch turnover in the ptst2 ptst3 double mutant closely resembled that of ss4, with a reduced starch synthesis rate during the day and a reduced degradation rate during the night. This was also observed, although to a lesser extent, in the ptst2 and ptst3 single mutants.
Table 2. Starch Content of the ptst2 ptst3 Double Mutant.
| Starch Content (mg/g FW) |
|||
|---|---|---|---|
| Ecotype | Genotype | ED | EN |
| Col-0 | Wild type | 8.3 ± 0.3 | 0.5 ± 0.1 |
| ptst2 | 9.0 ± 0.3 | 1.2 ± 0.2 * | |
| ptst3 | 9.1 ± 0.2 * | 1.0 ± 0.0 * | |
| ptst2 ptst3 | 8.3 ± 0.1 | 1.6 ± 0.1 * | |
| ss4 | 6.5 ± 0.3 * | 1.6 ± 0.1 * | |
| isa1 isa2 | 4.2 ± 0.1 * | 0.0 ± 0.0 * | |
Values for starch content at end of day (ED) and end of night (EN) time points are the mean ± se from n = 6 individual plants. Values marked with an asterisk are significantly different from the wild type under a two-tailed t test at P < 0.05. Mutants carrying the ptst2-3 and/or ptst3-5 alleles were used in this experiment. FW, fresh weight.
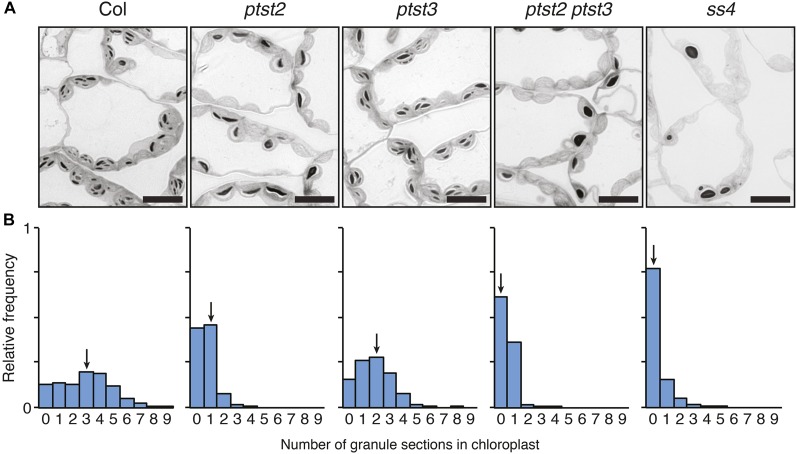
To visualize the starch granules in chloroplasts, we harvested a young leaf (examples shown in Figure 4B) from the single and double mutants and prepared them for observation via light microscopy. The double mutant appeared to contain even fewer starch granules than the ptst2 single mutant (Figure 5A). To quantify the number of granules in the chloroplasts of these mutants, we counted the number of granule sections in a total of 540 chloroplasts from three individual plants for each genotype. The number of granule sections is indicative of the actual number of granules, since the presence of more granules increases the likelihood of sectioning through one. However, it should be noted that it is not the actual number of granules, as such information cannot be obtained from two-dimensional sections. Wild-type chloroplasts had a median number of three granule sections per chloroplast and a broad frequency distribution (Figure 5B). Interestingly, although the ptst3 mutant had multiple granules per chloroplast (Figure 3C), the frequency distribution was shifted toward lower numbers compared with that of the wild type, with a median of two granule sections per chloroplast. This suggests that the ptst3 mutant does in fact have a reduction in granule number that was not immediately obvious from qualitative assessments. Most chloroplasts of the ptst2 mutant contained zero or one granule section. However, in ptst2 ptst3, the frequency of chloroplasts containing no granule sections was much higher than in the ptst2 single mutant. The ss4 mutant contained the fewest granules overall, with no granule sections observed in more than 75% of the chloroplasts assessed.
Figure 5.
Number of Starch Granules per Chloroplast in the ptst2 and ptst3 Single and Double Mutants.
The ptst2-3 and ptst3-5 alleles were used for all experiments in this figure.
(A) Starch granules in chloroplasts observed with light microscopy in leaf sections stained with toluidine blue. Leaves were harvested at the end of the day. Similar phenotypes were observed in three independent experiments performed on different batches of plants. Representative images are shown. Bars = 10 µm.
(B) Quantification of granule sections within chloroplasts. Light micrographs were acquired as in (A), and the number of starch granules observed within each chloroplast section was counted. Histograms show the frequency of chloroplasts containing a given number of granule sections relative to the total number of chloroplasts analyzed (n = 540 for each genotype). Equal numbers of chloroplasts were analyzed in leaves from three different plants for each genotype. Arrows indicate the bin containing the median value.
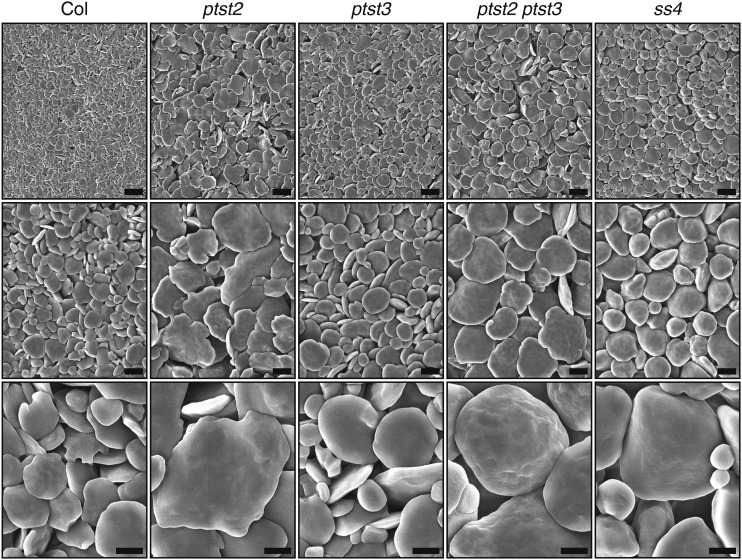
In the light micrographs, the shape of the starch granules in the double mutant appeared more rounded than those in the wild type and single mutants, which had the typical flattened morphology of leaf starch (Figure 5A). We therefore purified starch granules from the mutants and assessed their morphology in detail using scanning electron microscopy. As expected, the granules were much larger in the ptst2 mutant than in the wild type, but they had the same flattened shape (Figure 6). The ptst3 mutant granules also had a similar morphology to wild-type granules but were slightly larger. Granules in the ptst2 ptst3 double mutant were similar in size to ptst2 granules but appeared more rounded. However, the ptst2 ptst3 granules were not as rounded as ss4 granules. Thus, the ptst2 ptst3 double mutant has granule numbers and morphology more like the ss4 mutant than the wild type.
Figure 6.
Starch Granule Morphology in the ptst2 and ptst3 Single and Double Mutants.
Granules were purified from entire 4-week-old rosettes harvested at the end of the day (∼60 rosettes per genotype) and observed using scanning electron microscopy. The same sample prepared from each genotype was imaged at three different magnifications, 2000× (upper panels, bars = 5 µm), 5000× (middle panels, bars = 2 µm), and 15,000× (lower panels, bars = 1 µm).
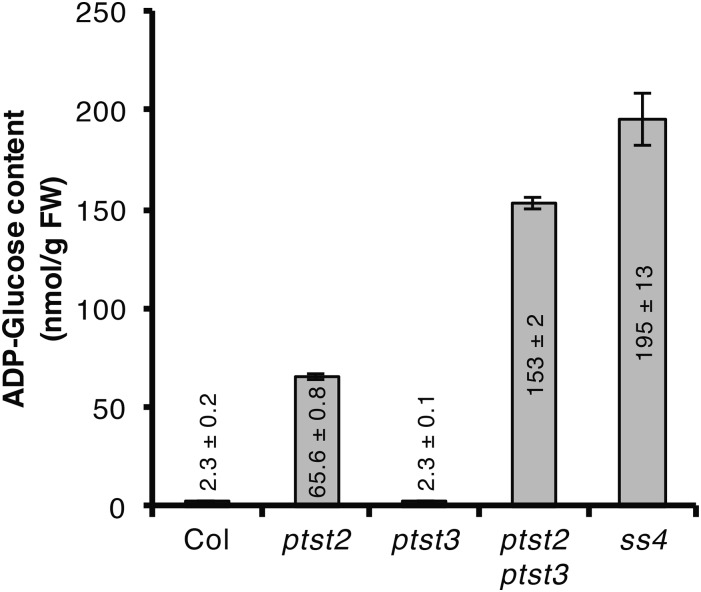
The reduction in granule number in SS4 was accompanied by an accumulation of ADP-glucose (Crumpton-Taylor et al., 2013; Ragel et al., 2013). We therefore quantified ADP-glucose in our ptst2 and ptst3 mutants (Figure 7). We detected wild-type levels of ADP-glucose in the ptst3 mutant, which was expected, as granule numbers were only slightly altered in ptst3. However, ADP-glucose level in the ptst2 mutant was almost 30-fold higher than in the wild type. The ptst2 ptst3 mutant accumulated even more ADP-glucose (twice that of ptst2), which is consistent with the fewer granules detected in the double mutant (Figure 5B). In ptst2 and ptst2 ptst3, the ADP-glucose level was 34 and 78%, respectively, of that measured in the ss4 mutant.
Figure 7.
ADP-Glucose Accumulation in ptst2 and ptst2 ptst3 Mutants.
Entire rosettes of 4-week-old plants were harvested at the end of the day and metabolites were extracted with the chloroform/methanol procedure. ADP-glucose was quantified using UHPLC-MS/MS. Values represent mean ±se of n = 4 rosettes.
We verified that these changes in granule number, granule morphology, and ADP-glucose content in the ptst2 ptst3 mutant were not due to alterations in SS4 abundance. Immunoblots showed no difference in SS4 levels between the double mutant and the wild type (Supplemental Figure 3). Taken together, our data provide strong evidence that PTST2 plays an important role in granule initiation. PTST3 appears to be less important than PTST2, as the ptst3 single mutant had only a subtle reduction in granule number. However, PTST3 may partially compensate for the loss of PTST2, as the double mutant is much more impaired in its ability to initiate starch granule formation than the ptst2 single mutant.
These phenotypic analyses further suggest that PTST2 or PTST3 are unlikely to affect ISA1 function in Arabidopsis leaves, contrasting to what was reported for the FLO6 protein in the rice endosperm. Furthermore, no change in ISA1 abundance was detected in either of our single mutants or the double mutant (Supplemental Figure 3), and no soluble glucans were detected (Supplemental Table 3).
PTST2 Interacts with SS4
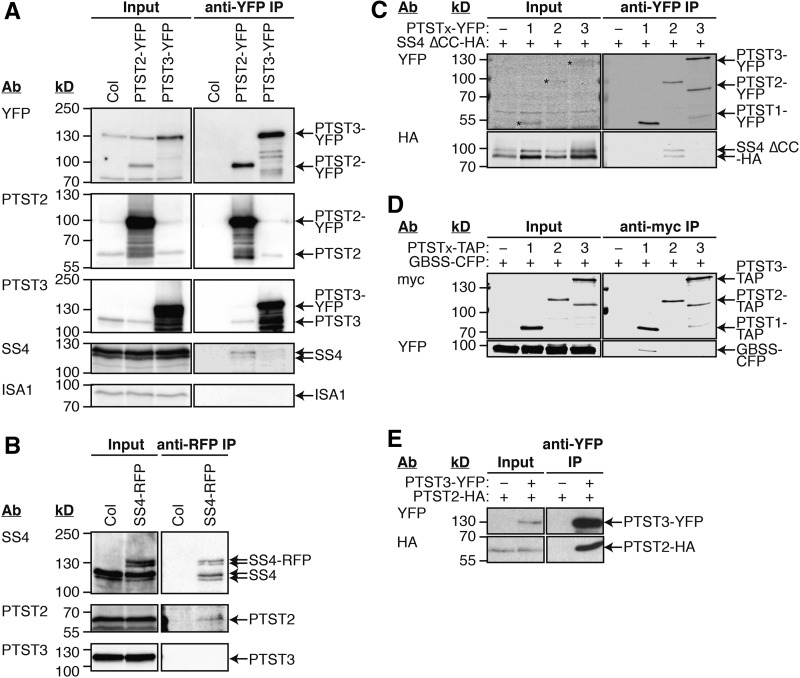
Given the importance of PTST2 in granule initiation, we sought to understand the mechanism by which it acts and explain the observed similarities between the ptst2 and ss4 phenotypes. Given that PTST2 has both coiled coils and a CBM48 domain, we hypothesized that PTST2 may interact with SS4 and help it come into contact with suitable glucan substrates, thereby facilitating granule initiation. We therefore designed a series of experiments to test for interaction between PTST2 and SS4 (Figure 8). First, we created transgenic Arabidopsis lines overexpressing either PTST2 or PTST3 via the constitutive CaMV 35S promoter, with both proteins tagged with YFP at the C terminus. Proteins were extracted from leaves of the transgenic lines harvested during the middle of the day, and the fusion proteins were purified by immunoprecipitation using beads that specifically bind the YFP tag. Immunoblotting the immunoprecipitate (IP) fraction with antiserum specific for YFP showed that both fusion proteins could be effectively recovered (Figure 8A). Using the SS4 antiserum, we could detect SS4 in the IP with PTST2-YFP. This suggests that PTST2 and SS4 interact in Arabidopsis leaves. A smaller amount of the SS4 protein copurified with PTST3-YFP. However, we did not detect any ISA1 protein in the IP with PTST2-YFP or PTST3-YFP. Interestingly, immunoblotting with PTST2 and PTST3 antibodies showed a substantial amount of endogenous untagged PTST3 in the IP with PTST2-YFP and endogenous untagged PTST2 in the IP with PTST3-YFP. This suggests that PTST2 and PTST3 can also interact with each other.
Figure 8.
Immunoprecipitation Experiments for Interaction Partners of PTST2 and PTST3.
The antibodies (Ab) used for detection and the migration of molecular mass markers (in kD) are indicated on the left of all immunoblot panels.
(A) Immunoprecipitation of PTST2-YFP and PTST3-YFP expressed in Arabidopsis. Proteins extracted from leaf tissue (Input) were incubated with beads conjugated to YFP antibodies. The bound proteins were eluted (anti-YFP IP), and proteins in the input and IP fractions were detected by immunoblotting.
(B) Immunoprecipitation of SS4-RFP expressed in Arabidopsis. The experiment was conducted as for (A), except that anti-RFP beads were used. SS4, PTST2, and PTST3 proteins in the input and IP fractions were detected by immunoblotting.
(C) Immunoprecipitation of YFP-tagged PTST proteins coexpressed in N. benthamiana leaves with HA-tagged SS4 protein. A truncated version of SS4 lacking the N-terminal coiled coils (SS4 ∆CC) was used. Immunoprecipitation was conducted as for (A). Epitope-tagged proteins in the Input and IP fractions were detected using immunoblotting.
(D) Immunoprecipitation of PTST proteins tagged with TAP-tags, coexpressed in N. benthamiana leaves with CFP-tagged GBSS protein. The experiment was conducted as described for (C), except using anti-myc beads, which bind the c-myc epitopes in the TAP tag.
(E) Immunoprecipitation of YFP-tagged PTST3 coexpressed in N. benthamiana leaves with HA-tagged PTST2 protein. The experiment was conducted as described for (A). Epitope-tagged proteins in the input and IP fractions were detected using anti-YFP (upper panels) and anti-HA (lower panels).
We then generated stable Arabidopsis lines expressing RFP-tagged SS4 under the control of the constitutive UBIQUITIN10 promoter (Grefen et al., 2010) and conducted the reciprocal immunoprecipitation with beads that specifically bind RFP. PTST2 was detected in the IP fraction with the SS4-RFP protein, confirming the interaction between the two proteins (Figure 8B). No PTST3 was detected in the IP fraction. The amount of SS4-RFP fusion protein in these lines was lower than the level of endogenous SS4 protein. Interestingly, however, a substantial amount of endogenous, untagged SS4 protein copurified with the SS4-RFP protein in the IP. This is consistent with the recently published evidence that SS4 forms dimers (Raynaud et al., 2016).
To further test these interactions, we used an immunoprecipitation-based assay using epitope-tagged proteins expressed in Nicotiana benthamiana leaves. For unknown reasons, neither the full-length SS4-HA protein nor the SS4-RFP protein could be reliably expressed in N. benthamiana leaves. This is not unexpected considering that we achieved only modest expression of the full-length SS4-RFP protein in Arabidopsis. However, a truncated version of SS4 lacking the coiled coils at the N terminus (SS4 ∆CC-HA) showed robust expression in N. benthamiana leaves. This protein was then coexpressed with either PTST1-YFP, PTST2-YFP, or PTST3-YFP. Immunoprecipitations were then performed with beads that specifically bind the YFP tag. This effectively recovered the tagged PTST proteins (Figure 8C). The SS4 ∆CC-HA protein only copurified with PTST2-YFP. We then coexpressed tandem affinity purification (TAP)-tagged PTST proteins with GBSS-CFP and conducted immunoprecipitations using beads conjugated to myc antibodies, which bind the c-myc epitopes in the TAP tag. The GBSS-CFP was only detected in the IP with PTST1-TAP (Figure 8D). These data suggest that both PTST1 and PTST2 specifically interact with their respective SS partners. Interestingly, it also appears that the N-terminal coiled coils of SS4 are not required for its interaction with PTST2.
We also tested the interaction between the Arabidopsis PTST2 and PTST3 proteins in N. benthamiana leaves. We coexpressed PTST2-HA with PTST3-YFP and conducted an immunoprecipitation with beads that specifically bind YFP. A substantial amount of PTST2-HA copurified together with PTST3-YFP, confirming that the two proteins interact (Figure 8E).
Overexpression of PTST2 Increases Granule Number per Chloroplast
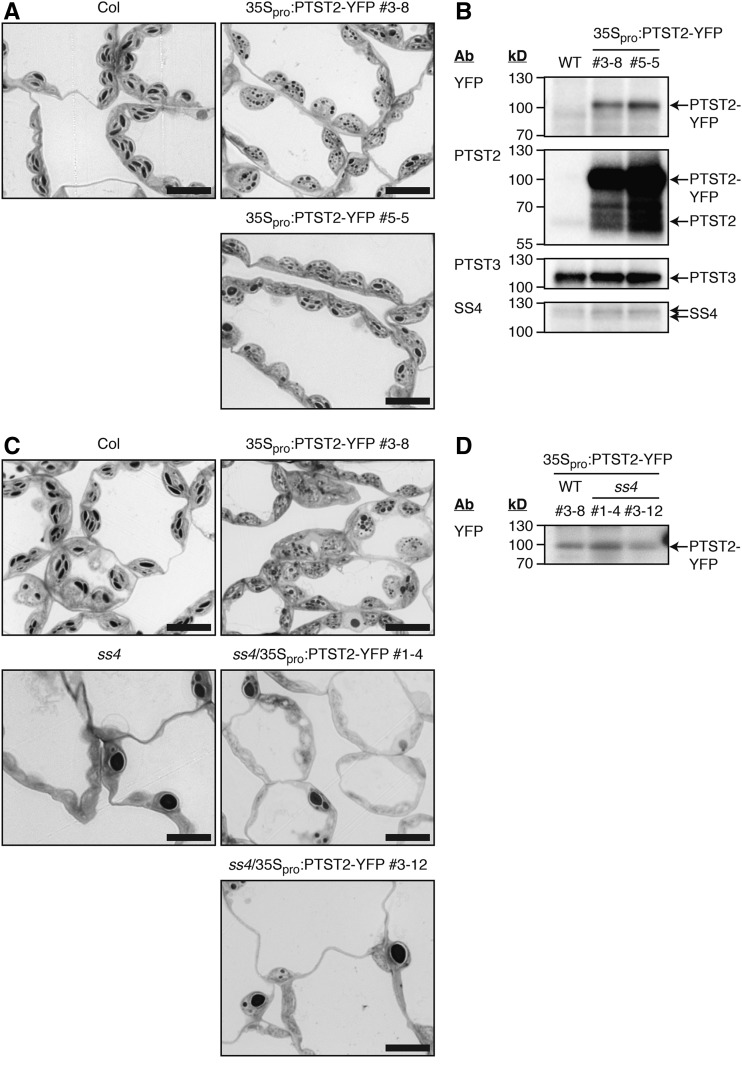
Since PTST2 could interact with SS4, and the absence of either protein strongly reduced the frequency of granule initiations, we further investigated the effect of PTST2 overexpression on starch metabolism using the stably transformed 35Spro:PTST2-YFP Arabidopsis lines (Figure 9). The starch content of the overexpression lines was not different from that of the wild type at the end of the day, but they contained slightly more starch at the end of night (Table 3). However, chloroplasts of the overexpression lines contained many more granules than wild-type chloroplasts, and these granules were small and rounded (Figure 9A). Immunoblotting showed that the PTST2-YFP protein was expressed to much greater levels than the endogenous, untagged PTST2 protein (Figure 9B). Neither PTST3 nor SS4 protein abundance was affected by PTST2 overexpression (Figure 9B).
Figure 9.
Effect of PTST2 Overexpression on Starch Granule Numbers.
(A) Starch granules in chloroplasts of transgenic Arabidopsis lines overexpressing PTST2-YFP under the control of the 35S promoter. Leaves harvested at the end of the day were sectioned and stained with toluidine blue prior to observation by light microscopy. Similar starch granule phenotypes were observed in at least three independent experiments performed on different batches of plants (either on plants from the T2 or T3 generation). Representative images from T3 homozygous plants are shown. Bars = 10 µm.
(B) Immunoblots showing the level of transgene expression. The antibodies (Ab) used for detection and the migration positions of molecular mass markers (in kD) are indicated on the left of each panel. An equal amount of total protein was loaded in each lane: 10 µg for anti-YFP and anti-PTST2 blots; 20 µg for anti-PTST3 and anti-SS4 blots.
(C) Starch granules in chloroplasts of transgenic Arabidopsis lines expressing PTST2-YFP under the control of the 35S promoter, as in (A), but in the ss4 mutant background. Similar starch granule phenotypes were observed in a second, independent experiment performed on a different batch of plants. Representative images are shown. The line in the wild-type background is from the T3 generation and homozygous for the insertion. The lines in the ss4 background are from the T2 generation (either homo- or heterozygous for the insertion). Bars = 10 µm.
(D) Immunoblot with anti-YFP showing the levels of transgene expression in ss4/35Spro:PTST2-YFP lines. Protein extracts were produced from the same plants analyzed in (C). Equal fresh weight (equivalent to 1.8 mg) was loaded per lane.
Table 3. Starch Content of PTST2 Overexpression Lines.
| Starch Content (mg/g FW) | |||
|---|---|---|---|
| Construct | Line | ED | EN |
| – | Wild type | 5.76 ± 0.26 | 0.31 ± 0.05 |
| 35Spro:PTST2-YFP | #3-8 | 6.07 ± 0.13 | 0.48 ± 0.03 * |
| #5-5 | 6.06 ± 0.34 | 0.59 ± 0.05 * | |
Values for starch content at end of day (ED) and end of night (EN) time points are the mean ± se from n = 5 individual plants. Analyses were conducted on plants homozygous for the transgene from the T3 generation. Values marked with an asterisk are significantly different from the corresponding wild type under a two-tailed t test at P < 0.05. FW, fresh weight.
To investigate the role of SS4 in initiating the large number of granules observed in the PTST2-YFP overexpression lines, we also overexpressed this fusion protein to a comparable level in the ss4 mutant background (Figure 9D). Chloroplasts in these lines did not contain many small granules and were most similar in appearance to chloroplasts of the untransformed ss4 mutant (Figure 9C). Nevertheless, there was a measurable impact of PTST2 overexpression in the ss4 background. Chloroplasts sections containing no granules were more frequent in the ss4/35Spro:PTST2-YFP lines (89 to 95% of total chloroplasts observed) than in the ss4 mutant (66% of total chloroplasts observed) (Supplemental Figure 4). PTST2 overexpression therefore repressed granule initiation in the ss4 mutant background rather than stimulating it. Thus, we conclude that PTST2 stimulates the formation of granules in wild-type chloroplasts in a manner that depends on the presence of SS4.
PTST2 Interacts with Glucans Able to Adopt a Helical Secondary Structure
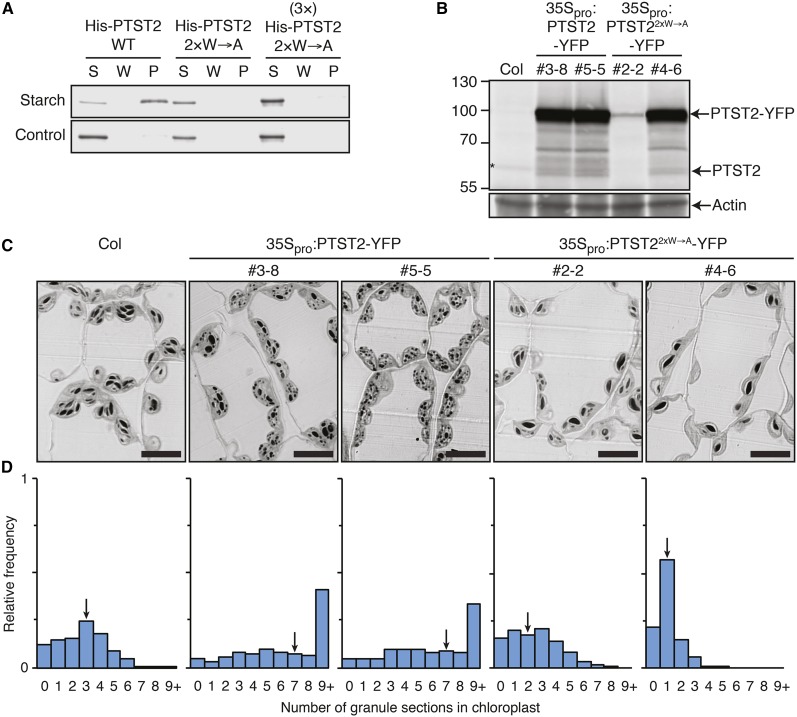
Since PTST1 can bind to starch through its CBM48 domain (Lohmeier-Vogel et al., 2008; Seung et al., 2015), we investigated the importance of the CBM48 domain on PTST2 function (Figure 10). First, we tested whether PTST2 could interact with starch. Recombinant proteins were coincubated with maize (Zea mays) starch granules, and the starch was subsequently pelleted. After washing the pellet, the bound proteins were eluted from the starch using a medium containing SDS. In this assay, PTST2 bound strongly to starch and was mostly detected in the pellet fraction (Figure 10A). The protein remained almost completely soluble in the absence of starch. We then tested whether the starch binding was mediated by the CBM48 domain. We found that Trp-475 and Trp-510 residues of PTST2 aligned to the known glucan binding tryptophan residues in the CBM48 of the mammalian AMP-kinase β-subunit (Trp-100 and Trp-133) (McBride et al., 2009) (Supplemental Figure 5). We therefore substituted these tryptophans with alanine residues, creating the 2×W→A variant of the protein. These mutations completely disrupted the binding of PTST2 to starch, suggesting that the CBM48 domain is essential for its interaction with glucans.
Figure 10.
The Role of the Glucan Binding CBM48 Domain in PTST2.
(A) Glucan binding assay using recombinant His-PTST2 against intact waxy maize starch granules. Unbound proteins are in the soluble fraction (S), while bound proteins are in the starch pellet (P). The final wash fraction was also loaded (W). The 2×W→A variant of PTST2 carries two Trp-to-Ala substitutions in both conserved glucan binding Trp residues within the CBM48 (Trp-475 and Trp-510). Similar results were obtained in two other independent experiments.
(B) Levels of PTST2-YFP and the 2×W→A variant expressed under the control of the 35S promoter in transgenic Arabidopsis lines. An equal amount of protein (30 µg) was loaded per lane, and immunoblots were probed with the PTST2 antibody (upper panel) and the actin antibody as a loading control (lower panel).
(C) Starch granules in chloroplasts overexpressing either the wild type or 2×W→A variant of PTST2. Leaves harvested at the end of the day were sectioned and stained with toluidine blue prior to observation under light microscopy. The lines are from the T3 generation and homozygous for the transgene. Bars = 10 µm.
(D) Quantification of granule sections within chloroplasts. Light micrographs were acquired as in (C), and the number of starch granules observed within each chloroplast section was counted. Histograms show the frequency of chloroplasts containing a given number of granule sections relative to the total number of chloroplasts analyzed (n = 705 for each line). Equal numbers of chloroplasts were analyzed in leaves from three different plants for each genotype. Arrows indicate the bin containing the median value.
We then investigated whether a functional CBM48 domain is important for PTST2 function in planta. We overexpressed the non-glucan-binding 2×W→A variant of PTST2 in Arabidopsis plants to assess whether it could stimulate granule initiation, as observed for the wild-type PTST2. One of the 35Spro:PTST22×W→A-YFP lines (#2-2) had lower levels of protein expression compared with the 35Spro:PTST2-YFP overexpression lines analyzed above, while the other line (#4-6) had comparable or slightly higher expression (Figure 10B). The overexpression of the 2×W→A variant did not result in the production of more granules (Figure 10C). In fact, the lines overexpressing the mutated form had fewer granules than untransformed wild-type plants. Quantification of the granules in the light micrographs showed that both 35Spro:PTST22×W→A-YFP lines had a frequency distribution of chloroplasts that was shifted toward lower granule numbers compared with the wild type. This effect was most pronounced in line #4-6, which strongly overexpressed the mutated form of the protein (Figure 10D). These data suggest that the mutated form of PTST2 has a dominant-negative effect, whereby the non-glucan-binding form of PTST2 competes with the endogenous wild-type form, presumably for SS4 binding. Indeed, we confirmed that the 2×W→A variant could still interact with SS4 in vivo (Supplemental Figure 6). The CBM48 domain is therefore necessary for PTST2 function.
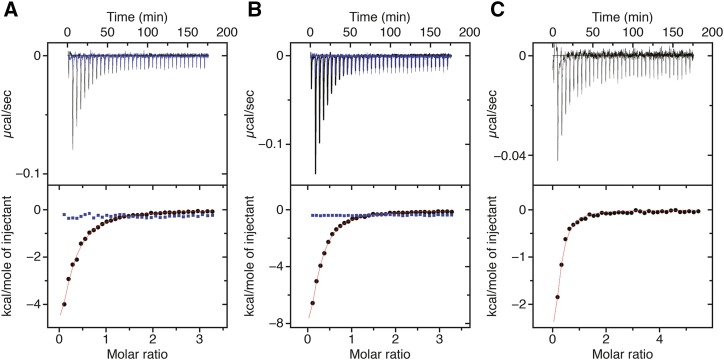
Given our evidence that PTST2 is involved in granule initiation, we reasoned that mature starch granules may not be its in vivo substrate, as its primary function would presumably precede the formation of a starch granule. Additionally, the proteins did not localize to the starch granules (Figure 2). Therefore, we investigated whether PTST2 could also bind to soluble maltooligosaccharides (MOS), as these could act as primers for starch granule synthesis. For this analysis, we used isothermal titration calorimetry (ITC), which measures heat exchanged as the titrated ligand (in this case, the sugar) interacts with the receptor (here, the PTST2 protein) (Figure 11). First, we titrated the candidate ligand maltoheptaose (Glc7) into the recombinant PTST2 protein solution. However, the heat variations collected after each injection were not consistent with an interaction (Figure 11A). We tested different [ligand]/[receptor] ratios with different protein concentrations (4 and 7.5 µM) and maltoheptaose concentrations (120 and 200 µM), but in no case was binding detected. We then tested β-cyclodextrin (Glc7), which also has seven glucose units, but in a cyclic configuration. While the cyclodextrin does not occur naturally in Arabidopsis plants, it can mimic the helical conformation of longer MOS and of the starch polymers, amylose and amylopectin. Injections of β-cyclodextrin produced significant integrated heat variations that were consistent with its binding to PTST2. We confirmed that these heat variations were a result of sugar-protein interaction by measuring the heat values of β-cyclodextrin injection without PTST2 protein present, which showed no significant heat variations (Supplemental Figure 7A). The Kd for the binding of β-cyclodextrin to PTST2 ranged from 1 to 3.3 µM.
Figure 11.
ITC Analysis of Interactions between PTST2 and Different Soluble Glucans.
(A) ITC profile of maltoheptaose (blue) and β-cyclodextrin (black) injected into a solution of the full-length His-PTST2 protein. The upper panels show the raw calorimetric data. The bottom plots are integrated heats as a function of the glucan/PTST2 molar ratio. The best fit for β-cyclodextrin was obtained from a nonlinear least squares method using a one-site binding model for two significantly distant N-values. Heats of dilution without PTST2 protein have been obtained from independent titration experiments (Supplemental Figure 7A). Reactions are exothermic.
(B) As for (A), but showing maltoheptaose (blue) and β-cyclodextrin (black) injected into the isolated CBM48 domain.
(C) As for (A), but showing maltodecaose injected into the full-length His-PTST2 protein.
To investigate whether the specificity for β-cyclodextrin was determined by the CBM48 domain of PTST2, we repeated the analysis above with a truncated form of the PTST2 protein containing the CBM48 domain alone. Like the full-length protein, no binding was detected upon maltoheptaose injections (Figure 11B). However, the CBM48 domain interacted with β-cyclodextrin with a Kd range between 1.7 and 4 µM, similar to what was measured for the full-length PTST2 protein. Furthermore, we performed a complementary experiment in which we injected β-cyclodextrin into an equimolar mix of PTST2 and maltoheptaose (Supplemental Figure 7B). Similar heat variations were detected with or without maltoheptaose in the sample cell. This confirms that maltoheptaose does not occupy the binding sites, as it does not interfere with the interaction between the CBM48 and the cyclodextrin.
The interaction with β-cyclodextrin suggests that PTST2 selectively recognizes the helical conformation of MOS. Maltoheptaose is too short to stably form such conformations by itself, as its length would only allow a single helical turn with limited stabilization by intramolecular interactions parallel to the helical axis (Goldsmith et al., 1982). We therefore tested the longest commercially available maltooligosaccharide, maltodecaose, which has 10 glucose units and is long enough to form helical structures by itself (Gidley and Bulpin, 1987). Unlike for maltoheptaose, we observed significant integrated heat variations with each injection of maltodecaose, consistent with binding to PTST2 (Figure 11C). The Kd (between 1.5 and 4.7 µM) was in a similar range to that determined for β-cyclodextrin. No significant variations were observed in the control, showing the heats of maltodecaose dilution without the PTST2 protein present (Supplemental Figure 7A). Together, these data suggest that the CBM48 domain of PTST2 can bind to long MOS, probably by recognizing their helical secondary structure.
DISCUSSION
PTST2 and PTST3 Are Components of the Granule Initiation Mechanism in Chloroplasts
Our work has identified PTST2 and PTST3 as two components of the starch granule initiation mechanism in Arabidopsis chloroplasts. Both are CBM48-containing proteins that are homologs of the recently characterized PTST1 protein (Figure 1). The PTST family of proteins therefore play diverse roles during starch synthesis, as PTST1 facilitates amylose synthesis rather than granule initiation (Seung et al. 2015).
PTST2 appears to be indispensable for normal granule initiation, as most chloroplasts in the ptst2 mutants contain zero or only one starch granule. In our proposed model (Figure 12), we suggest that PTST2 facilitates granule initiation by delivering suitable substrates to SS4, an enzyme known to play a critical role in granule initiation (Roldán et al., 2007; Szydlowski et al., 2009; Crumpton-Taylor et al., 2013). PTST2 interacts with SS4 (Figure 8), and, through its CBM48 domain, with soluble MOS primers that have a propensity to adopt a helical secondary structure (Figure 11). Since SS4 itself does not possess a specialized CBM, the interaction with PTST2 provides access to a CBM that can bind suitable MOS substrates. The MOS can then be further elongated by SS4 and ultimately develop (with the action of other starch biosynthetic enzymes) into a starch granule. The efficiency at which SS4 elongates these MOS may be enhanced by the ability of SS4 to dimerize (Figure 8; Raynaud et al., 2016). In our model, both the enzymatic activity of SS4 and the carbohydrate binding activity of PTST2 are necessary for granule initiation. This is consistent with our experimental observations, as both proteins are necessary for normal granule number in chloroplasts (Figures 3 and 5). We have also demonstrated that the CBM48 domain is essential for PTST2 function in planta (Figure 10).
Figure 12.
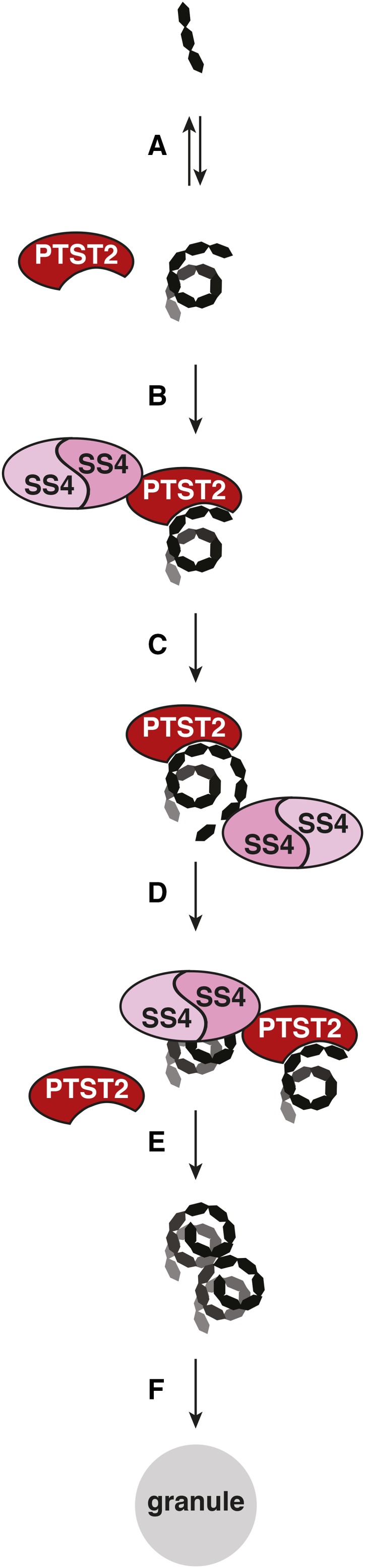
A Model for PTST2 Function in Granule Initiation.
In an initial step (A), short MOS (such as maltose and maltotriose) is elongated into longer MOS. This is likely to be a stochastic process, and the amount of longer MOS available will be influenced by the rate of MOS elongation by the SS isoforms and phosphorylase, as well as the rate of MOS degradation by starch-degrading enzymes. Once PTST2 recognizes a long MOS through its CBM48 domain, it can recruit SS4 to the substrate (B), allowing SS4 to further elongate the MOS (C). PTST2 could then dissociate from the MOS, becoming available to interact with other SS4 and MOS molecules (D). Since SS4 can also dimerize, several MOS molecules may be recruited by PTST2 for elongation within close proximity of each other, which could increase the efficiency at which crystalline glucans are formed (E). The other starch biosynthesis enzymes (SS isoforms, branching enzymes, and isoamylase) then further elaborate the structure of these substrates until a starch granule is formed (F).
The ptst3 mutant also had fewer granules per chloroplast compared with the wild type, but this was a minor change compared with what was observed in ptst2 (Figure 5). We also found no evidence that PTST3 could interact directly with SS4, but it interacted with PTST2 (Figure 8). Thus, PTST3 appears to work together with PTST2 in granule initiation, but it appears less important than PTST2 under our growth conditions. Several plant species appear to have lost the gene encoding PTST3 altogether, including the grasses (Figure 1B). Nevertheless, in Arabidopsis, PTST3 has some ability to function in granule initiation in the absence of PTST2, since the ptst2 ptst3 double mutant contained fewer granules than ptst2 and accumulated more ADP-glucose (Figures 5 and 7). The pattern of starch turnover in the ptst2 ptst3 double mutant more closely resembled that of ss4 than either the ptst2 or ptst3 single mutants (Figure 4C). We do not yet fully understand the mechanism by which PTST3 acts in granule initiation in the absence of PTST2, but clearly, this process is not efficient, as demonstrated by the strong phenotype of the ptst2 mutant.
However, it should be noted that the ptst2 phenotype was milder than that of ss4 in several ways. First, most chloroplasts in ss4 leaves did not contain any starch granules, while in ptst2 leaves, most chloroplasts contained at least one (Figure 5). Second, the ptst2 mutant accumulated only one-third the amount of ADP-glucose measured in ss4 plants and did not have an obvious defect in its growth phenotype (Figures 4 and 7). Since starch content is normal in ptst2, it may be counterintuitive that the mutant accumulates ADP-glucose at all. However, it should be noted that we did not observe starch granules in almost 50% of the chloroplasts in our sections of ptst2 leaves (Figure 5). It is possible that ADP-glucose only accumulates in these starchless chloroplasts.
The ss4 mutant was also more impaired in granule initiation than the ptst2 ptst3 double mutant. Compared with ss4, the double mutant still had more granules (Figure 5), lower ADP-glucose levels (Figure 7), and no visible pale phenotype. We therefore conclude that SS4 function is greatly compromised in the absence of both PTST2 and PTST3, but its activity is not completely abolished. This is similar to what has been observed for PTST1 and GBSS. While starch in the ptst1 mutant is almost completely amylose-free, there is some residual GBSS bound to the granule that can synthesize small amounts of amylose (Seung et al., 2015). Thus, both GBSS and SS4 can act in the absence of their partner PTST isoforms, but at greatly reduced efficiency.
In addition to the reduction in granule number, the morphology of granules in the ptst2 ptst3 double mutant was aberrant, appearing larger and slightly rounder than those of the wild type. Granules in both single mutants were also larger than wild-type granules, but they retained the flat, discoid morphology of the latter. Therefore, PTST2 and PTST3 appear to have some control over establishing correct granule morphology. The exact mechanism of how they exert this control is not known, but it is likely they do so by altering SS4 activity, as ss4 mutants have granules that are round (Roldán et al., 2007) (Figures 5 and 6). However, the function of SS4 in establishing granule morphology appears compromised but not completely abolished in the absence of both PTST2 and PTST3: The granules of the ss4 mutant were generally more rounded than those in the double mutant (Figure 6).
Overall, our discovery of components involved in granule initiation demonstrates that SS4 is necessary, but not sufficient for establishing the correct number of granules in chloroplasts. SS4 has been reported to interact with the plastidial fibrillins through its N-terminal coiled coils (Gámez-Arjona et al., 2014; Raynaud et al., 2016), but the importance of fibrillins in determining granule number has not yet been established. Also, a later study of chloroplast complexes could not detect the interaction between fibrillins and SS4 (Lundquist et al., 2017). Interestingly, we noted that the N-terminal coiled coils of SS4 are not required for its interaction with PTST2, although further experiments are required to determine whether the coiled coils can influence the interaction in vivo, perhaps by enhancing or regulating the interaction. However, since PTST1 interacts with GBSS on the C-terminal GT1 domain (Seung et al., 2015), the primary interaction site of PTST2 on SS4 may also reside in the GT1 domain. This might allow the SS4 N-terminal end to remain available for dimerization (Raynaud et al., 2016) or for interacting with other proteins, possibly including the fibrillins or as yet unidentified components.
PTST2 May Determine the Glucan Substrates Available to SS4
Previous studies have examined the substrate specificity of SS4 in vitro to understand its role in granule initiation (Szydlowski et al., 2009; Cuesta-Seijo et al., 2016). However, our study raises the possibility that the activity of SS4 in vivo may not only depend on the substrate specificity of SS4 itself, but also on the binding specificity of the CBM48 domain of PTST2. Our ITC experiments suggest that the CBM48 domain of PTST2 is specific for longer MOS, as it could bind to maltodecaose but not maltoheptaose (Figure 11). Maltodecaose, having 10 glucose units, is the shortest MOS able to form stable helical structures in solution (Gidley and Bulpin, 1987). Maltoheptaose has been observed to form helices, but only when bound to proteins that specifically stabilize it into that conformation (Goldsmith et al., 1982). PTST2 appears unable to do this, as it cannot initiate binding to the molecule at all. However, PTST2 could interact with β-cyclodextrin, which mimics the helical conformation of glucans. PTST2 therefore appears to recognize the helical secondary structure adopted by longer glucan chains, rather than the length per se. Such recognition of helical glucan conformation has been reported for a bacterial glucan binding protein (Koropatkin et al., 2008). The CBM48 domain of the β-subunit of AMP kinase also appears to have higher affinity for cyclodextrin over maltoheptaose in some NMR studies (Koay et al., 2010), although not in others (Koay et al., 2007).
The formation of secondary structure is a prerequisite for glucans to crystallize. We speculate that PTST2 may direct SS4 activity toward glucans already having secondary structure because these have a greater propensity to form a crystalline product (Gidley and Bulpin, 1987). We previously suggested that SS4 may increase the rate at which stable, crystalline glucans form during granule initiation (Seung et al., 2016). The ability to interact with PTST2 may be one of the features of SS4 that facilitates this in vivo. However, in a yeast model system expressing different combinations of starch biosynthetic enzymes, the presence of SS4 had a tendency to promote the formation of insoluble glucans, and this effect was seen in the absence of PTST2 (Pfister et al., 2016). Thus, SS4 is likely to have other inherent features that promote the formation of crystalline insoluble glucans. Such additional features may include the ability of SS4 to dimerize (Raynaud et al., 2016). We also detected an interaction between SS4 monomers in Arabidopsis leaves, supporting the idea that SS4 can form oligomers with itself (Figure 8).
The Generation of Longer MOS in the Chloroplast
The generation and elongation of MOS is proposed to be important during the granule initiation process in different plant species (Nakamura, 2015). In our model (Figure 12), we suggest that the availability of longer MOS for PTST2 binding is important for granule initiation in Arabidopsis leaves and may depend on the rate of MOS generation against the rate of MOS degradation in the chloroplast. MOS may be generated in the chloroplast using several stromal enzymes. The α-glucan phosphorylase (PHS1) has been implicated in granule initiation in rice endosperm (Satoh et al., 2008), and this enzyme could generate longer MOS from maltohexaose using glucose-1-phosphate (Glc-1-P) in vitro at physiologically relevant concentrations of Pi and Glc-1-P (Hwang et al., 2010). In the rice endosperm, PHS1 interacts with disproportionating enzyme, which allows it to synthesize longer MOS efficiently (Hwang et al., 2016). PHS1 also interacts directly with branching enzyme in the cereal endosperm (Tetlow et al., 2004), and the two enzymes act synergistically in producing branched MOS in vitro (Nakamura et al., 2012). We speculate that PTST2 could also bind branched MOS, if the linear chains are long enough to produce helical secondary structure. PHS1 is present in the chloroplast in Arabidopsis leaves, although its role in starch synthesis or degradation remains unclear (Zeeman et al., 2004). However, its role in generating MOS may be redundant, as all Arabidopsis SS enzymes could elongate maltose into longer MOS (including maltodecaose) in vitro after an extended incubation period (Brust et al., 2013). It is difficult to estimate the frequency of such occurrence in the stroma, as longer MOS are often below the level of detection.
The presence of glucan-degrading enzymes in the chloroplast could mean that longer MOS are available only transiently. Thus, PTST2 may ensure that these substrates are efficiently detected and elongated, so that they form crystalline structures that can evade degradation. Our previous work has shown that the starch-degrading enzyme AMY3 could interfere with the granule initiation process in the ss4 mutant, but this interference is negligible in the wild type (Seung et al., 2016). SS4 can therefore prevent the premature hydrolysis of glucan primers in vivo, and we expect that efficient substrate targeting by PTST2 helps SS4 fulfill this role. However, glucan-degrading enzymes can also promote granule initiation by generating MOS primers, especially once large glucan polymers have already been synthesized (Seung et al., 2016). An example of a glucan-degrading enzyme that can both promote and suppress granule initiation is ISA. On one hand, ISA can liberate linear MOS from larger polymers as part of the normal amylopectin biosynthetic process, providing PTST2 and SS4 with suitable substrates. However, potato (Solanum tuberosum) tubers and barley (Hordeum vulgare) endosperms deficient in ISA activity accumulate many small granules (Burton et al., 2002; Bustos et al., 2004), suggesting that ISA normally suppresses granule formation, possibly by degrading away small soluble glucans that might otherwise go on to nucleate new starch granules.
The importance of MOS availability is also demonstrated by our PTST2 overexpression lines. The overexpression of PTST2 stimulated granule formation in the wild-type background, but not in the ss4 background (Figure 9; Supplemental Figure 4). In fact, a greater majority of chloroplasts were starchless in the overexpression lines in the ss4 background than in the ss4 mutant itself, suggesting that PTST2 overexpression repressed granule initiation in the absence of SS4. This is probably because the excess PTST2 can bind MOS, but SS4 is not present to elongate them. Thus, the PTST2-bound MOS may be less accessible for elongation by other enzymes, such as SS3, which normally initiates the formation of the few granules found in the ss4 mutant (Szydlowski et al., 2009).
The observation that PTST2 could stimulate granule formation in the wild-type background suggests that PTST2 abundance at least partially limits the number of granule initiations. If the frequency of encounters between suitable MOS and PTST2 limits the number of initiations, we expect that raising the abundance of either component may increase the number of granules. Alternatively, it is possible that primer MOS are always present in excess and that PTST2 is a major node where granule number is controlled. In this case, further experiments should focus on how the binding between PTST2 and MOS, as well as between PTST2 and SS4, may be regulated in the chloroplast to exert such control. Possible factors include posttranslational modifications (e.g., phosphorylation) or other protein interaction partners. We also do not know whether PTST2 is the only protein limiting the number of granules initiated. The overexpression of SS4 did not alter granule number per chloroplast (Supplemental Figure 8). However, it should be noted that the levels of SS4 overexpression in these lines were not as high as those achieved for PTST2. We do not know why we could not achieve substantial overexpression of the full-length SS4 protein, either in Arabidopsis or N. benthamiana.
PTST2/FLO6 Is a Potential Target for the Biotechnological Modification of Starch
PTST2 is a promising target gene for the biotechnological modification of starch, particularly for modifying granule size. Despite their differences in granule number, the ptst2 mutant and the overexpression lines still synthesized the same total amount of starch as the wild type (Tables 1 and 3). The ability to alter granule number without changing starch content allows the modification of granule size. Indeed, the ptst2 mutant had much larger starch granules than the wild type, while much smaller granules were observed in the overexpression lines (Figures 5, 6, and 9). There is commercial interest in genetic approaches to alter granule size, as it influences the physicochemical properties of starch (such as gelatinization temperature and pasting properties) as well as milling efficiency (Lindeboom et al., 2004).
PTST2 is especially promising as a target gene, since the knockout of its ortholog in rice, FLO6, has been shown to affect starch synthesis in the endosperm (Peng et al., 2014). The mechanism of starch granule initiation in developing rice grains may differ in some ways from that in Arabidopsis leaves. In amyloplasts of rice endosperm, granules form from multiple initiation centers that are held together into larger structures, so-called compound granules (Matsushima et al., 2013; Toyosawa et al., 2016). Rice flo6 mutants have reduced starch content in the grain, altered physicochemical properties of starch, and minor changes in the chain length distribution of amylopectin. Some amyloplasts in the mutant did not contain any starch granules, while others contained many small granules or starch with aberrant compound granule morphology. The flo6 phenotype could therefore be interpreted as defects in granule initiation, and, in light of our work in Arabidopsis, it would be interesting to investigate whether there is any interaction of PTST2/FLO6 with SS4 in the endosperm.
One difference between Arabidopsis PTST2 and rice FLO6 is that the latter interacted with ISA1 in bimolecular fluorescence complementation experiments, in vitro pull-down assays, as well as yeast two-hybrid experiments. In Arabidopsis, we found no evidence that PTST2 interacts with ISA1 in vivo. Furthermore, loss of ISA1 activity in Arabidopsis results in a reduced amount of insoluble starch, the accumulation of soluble phytoglycogen, and altered chain length distribution of amylopectin (Delatte et al., 2005)—traits that were not observed in ptst2 mutants. In contrast, the physical and functional interaction of PTST2 with SS4 is strongly supported by phenotypic impact on granule numbers. However, ISA1-containing isoamylase is known to differ in the endosperms of rice and other cereals compared with Arabidopsis leaves and other dicots like potato. In rice endosperms, ISA1 is present primarily as a homodimer, whereas in Arabidopsis, it is exclusively present as an ISA1/ISA2 complex (Delatte et al., 2005; Utsumi and Nakamura, 2006; Streb and Zeeman, 2014).
It is also important to note that a rice mutant deficient in the major isoform of SS4 in the endosperm (SS4b) has relatively minor alterations in starch content and granule morphology (Toyosawa et al., 2016). Therefore, it would not be easy to explain how altered SS4 activity would cause the flo6 phenotype. Rice mutants that are deficient in ISA1 have defects in granule initiation in the endosperm and have some starchless amyloplasts, both of which were observed to a smaller extent in flo6 (Kawagoe et al., 2005; Peng et al., 2014). However, the isa1 mutants of rice also accumulate phytoglycogen, which was not reported for flo6 (Kawagoe et al., 2005; Peng et al., 2014). In conclusion, it appears that PTST2 is a conserved protein important for granule initiation in both leaves and cereal endosperm and possibly other important starch-storing organs. Further investigation will be necessary to determine if the mechanism by which it participates in this process differs between plant tissues.
METHODS
Plant Growth
For all experiments, Arabidopsis thaliana plants were grown on soil in a controlled environment chamber: either a Percival AR-95L (CLF Plant Climatics) fitted with fluorescent lamps and supplemented with red LED panels or a Kälte 3000 chamber fitted with fluorescent lamps. The chambers were set to provide a 12-h-light/12-h-dark cycle, with light intensity of 150 μmol photons m−2 s−1, temperature at 20°C, and relative humidity of 65%.
The T-DNA insertion mutants used in this study and their background accessions are as follows: ptst2-3 (SAIL_1148_C07; Col), ptst2-4 (SALK_206998; Col), ptst2-6 (RATM16-0310-1_G; Nossen), ptst3-1 (GT_5_14201; Ler), and ptst3-5 (SALK_33414; Col). The ptst2 ptst3 double mutant was generated by crossing the ptst2-3 mutant with ptst3-5. The ss4 mutant used in this study is ss4-1 (GABI_290D11), characterized as a knockout mutant by Roldán et al. (2007). The isa1 isa2 mutant, which was described by Delatte et al., (2005), carries the isa1-1 T-DNA allele (SALK_042704), and the isa2-1 allele has a single base pair deletion in the ISA2 coding sequence. Both ss4 and isa1 isa2 are in the Col background.
Iodine Staining and Quantification of Starch and ADP-Glucose Content
For iodine staining of plant material, rosettes were harvested at the end of day and chlorophyll was removed with 80% (v/v) ethanol. Excess ethanol was rinsed away with water before staining in Lugol solution (KI/I2 solution; Sigma-Aldrich). Excess iodine was washed away with water before imaging.
The method used for starch quantification was based on Smith and Zeeman (2006). Entire rosettes of 4-week-old plants were homogenized in 0.7 M perchloric acid. The insoluble material was pelleted by centrifugation, washed three times in 80% (v/v) ethanol, and resuspended in water. The starch in the insoluble fraction was gelatinized at 95°C for 15 min and digested to glucose at 37°C using α-amylase and amyloglucosidase (Roche). Starch content (in glucose equivalents) was determined by quantifying the glucose with a hexokinase/glucose-6-phosphate dehydrogenase-based spectrophotometric assay. Soluble glucans were also digested to glucose by treating the supernatant with α-amylase and amyloglucosidase, as for the insoluble fraction. The glucose was then quantified as described for starch. Soluble glucan content (in glucose equivalents) was calculated by subtracting the amount of glucose after digestion with the amount of glucose in the undigested sample.
ADP-glucose was extracted from entire rosettes harvested at the end of the day using chloroform/methanol and quantified by UHPLC-MS/MS, as described in detail by Seung et al. (2016).
Visualization of Starch Granules in Chloroplasts
Leaf samples were harvested from 3-week-old plants at the end of the day, fixed, and embedded in Epon resin. Young leaves of equivalent age were selected for all mutants (examples of leaves taken for this analysis are indicated with a blue arrow in Figure 4B), and samples were taken approximately two-thirds along the length of the leaf, toward the distal part. Thin sections produced from the embedded leaves were stained with toluidine blue and imaged by light microscopy using an AxioImager Z2 microscope fitted with a 100× oil immersion lens (1.4 numerical aperture) and an AxioCam monochrome camera (Carl Zeiss). A detailed protocol can be found in Seung et al. (2015).
Protein Extraction and Immunoblotting
For the extraction of total proteins from leaves, young leaves were homogenized in protein extraction medium (40 mM Tris-HCl, pH 6.8, 5 mM MgCl2, 2% [w/v] SDS, and Complete Protease Inhibitor [Roche]). Insoluble material was pelleted at 20,000g, and the proteins were collected in the supernatant. Protein content was determined using the BCA protein assay kit (Thermo Scientific), and equal amounts of protein per sample were loaded onto SDS-PAGE gels.
For immunoblotting, proteins were transferred onto a PVDF membrane following SDS-PAGE. PTST2 was detected using polyclonal antibodies raised against the recombinant Arabidopsis PTST2 protein. Antibodies specific for PTST2 were affinity purified from the antisera using NHS-activated Sepharose beads conjugated to the recombinant PTST2 protein. PTST3 antibodies were prepared in the same manner. No PTST2 protein could be detected in plant extracts with the PTST3 antibody, and vice versa, suggesting the antibodies had low cross-reactivity. SS4 was detected using the antisera described by Roldán et al. (2007), while ISA1 was detected using antisera described by Delatte et al. (2005). For detection of epitope tagged proteins, we used anti-GFP/YFP (ab290; Abcam), anti-HA (ab9110; Abcam), and anti-myc (M4439; Sigma-Aldrich). Proteins were detected with either chemiluminescence, using horseradish peroxidase-coupled secondary antibodies (Bio-Rad), or with IR fluorescence. For IR fluorescence, IR800-conjugated secondary antibodies and an Odyssey CLx detection system (Li-Cor) were used. Primary antibody dilutions are as follows: anti-PTST2, 1:200; anti-PTST3, 1:200; anti-SS4, 1:5000; anti-ISA1, 1:1000; anti-GFP/YFP, 1:10,000; anti-HA, 1:7000; and anti-myc, 1:10,000.
Phylogenetic and Bioinformatics Analyses
To build the phylogenetic tree, sequences of PTST1, -2, and -3 homologs were retrieved from NCBI using BLASTp, searching in genomes for which Refseqs were available. Species were chosen so that the major evolutionary branches of land plants were represented. Sequences selected as the outgroup were identified by finding the closest prokaryotic sequence to each PTST sequence using BLASTp. Sequences were aligned using Clustal Omega (Sievers et al., 2011), and then a tree was constructed using the neighbor-joining method in MEGA v6 using a Poisson substitution model, uniform substitution rates among sites, complete deletion of gaps, and 1000 bootstrap replicates (Tamura et al., 2013).
The prediction of coiled coils in amino acid sequences were performed on the COILS/PCOILS server (Lupas, 1996). Results obtained with a 14-amino acid prediction window are shown.
Recombinant Protein Expression in Escherichia coli
All primers used for cloning the following constructs are shown in Supplemental Table 2. For the IPTG-inducible expression of His-PTST2 in E. coli, a PTST2-cTP:pProExHTb expression vector was cloned as follows: Arabidopsis cDNA was prepared using the RevertAid reverse transcription kit (Thermo) on total RNA, which was prepared using the RNeasy Plant total RNA extraction kit (Qiagen). The full-length cDNA sequence of Arabidopsis PTST2 was amplified from the cDNA preparation and cloned into the pCR8 vector using the pCR8/GW/TOPO TA cloning kit (Thermo Scientific). The region encoding the PTST2 protein, lacking the predicted 71-amino acid chloroplast transit peptide at the N terminus, was amplified with BamHI and XhoI restriction sites and cloned into the pProExHTb vector (Invitrogen). The point mutations in the CBM48 were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions.
For the IPTG-inducible expression of PTST3-His in E. coli, a PTST3-cTP:pET21a+ expression vector was cloned as follows: The full-length coding sequence of PTST3 was amplified from the Arabidopsis cDNA preparation, but was unstable in E. coli when amplified in a high-copy-number plasmid. To circumvent this, the full coding sequence was created by gene synthesis and cloned into a low-copy-number pCC1 vector by Genscript. From this template, we amplified the region encoding the PTST3 protein, lacking the predicted 47-amino acid chloroplast transit peptide at the N terminus, together with NheI and XhoI restriction sites. This was then cloned into the pET21a+ vector (Novagen). This vector was stable in E. coli, either due to the low copy number of pET21a+, or due to the absence of the transit peptide sequence.
For the IPTG-inducible expression of His-PTST2 CBM48 in E. coli, a PTST2 CBM48:pProExHTb expression vector was cloned as follows: The sequence encoding the CBM48 of the PTST2 protein (amino acids 453–532) was amplified with NcoI and XhoI restriction sites and cloned into the pProExHTb vector (Invitrogen).
For protein expression, the appropriate expression vector was transformed into E. coli BL21 (DE3) CodonPlus cells (Agilent Technologies). The cells were cultured in LB medium at 37°C until OD600 of 0.5 to 0.6. Protein expression was then induced by adding 1 mM IPTG to the culture, followed by incubation overnight at 18.5°C. The lysis of cells and subsequent purification of His-tagged protein was performed as described (Seung et al., 2013).
Starch Binding Assay
The indicated amount of recombinant PTST2 protein was incubated in binding medium (50 mM HEPES-NaOH, pH 7.5, 2 mM MgCl2, 1 mM DTT, 0.05% [w/v] BSA, and 0.005% [v/v] Triton X-100) with 30 mg of waxy maize starch (Sigma-Aldrich) for 30 min at 20°C in a total volume of 250 µL. For the control reaction, starch was substituted with Sepharose G-10 resin. The reactions were mixed on a spinning wheel during the incubation. The starch/resin was pelleted at 5000g for 30 s, and unbound proteins were collected in the supernatant. The pellet was then washed three times in equivalent volumes of binding medium, and bound proteins were eluted in an equivalent volume of elution medium (50 mM HEPES-NaOH, pH 7.5, 2 mM MgCl2, 1 mM DTT, and 2% [w/v] SDS). Proteins in the supernatant, final wash, and pellet fractions were detected by SDS-PAGE followed by immunoblotting (see above).
Isothermal Titration Calorimetry
Unless otherwise specified, 35 injections of ligand solution at the indicated concentration, each with a volume of 8 µL, were monitored with the VP-ITC instrument. An injection spacing of 300 s was selected, and duration of each injection was automatically calculated. An initial injection of 2 μL was excluded from data analysis. Default sample cell volume was 1.4644 mL. Stirring speed and reference power were set to 307 rpm and 10 µCal⋅s−1, respectively. Before measurement, the samples were homogenized and degassed for 10 min at 22°C. The proteins were desalted into interaction medium (20 mM Tris-HCl, pH 7.5, and 10 mM NaCl) using a NAP-5 desalting column (GE Healthcare) prior to the experiment. The ligands tested were β-cyclodextrin (C4805; Sigma-Aldrich), maltoheptaose (M7753; Sigma-Aldrich), and maltodecaose (Glu320; Elicityl) and were dissolved directly into the interaction medium. All data have been fitted in the Origin software version 7 (Origin Lab) with the one-site binding model. Since the saturation of binding sites occurred early, we opted for an optimized protocol to determine a reliable Kd range (Milev, 2015). Data fitting was performed with two assumed extreme N-values of n = 0.2 and n = 0.0002 (the ITC N-value corresponds to binding ratios of active reactants involved in the interaction).
Cloning of Expression Vectors for Plant Expression and Transformation
For expression of PTST2-YFP under the control of the 35S promoter, the PTST2:pB7YWG2 vector was cloned. The Gateway-compatible PTST2:pCR8 vector, cloned as described above, was recombined directly into the pB7YWG2 vector (Karimi et al., 2002), yielding PTST2 in-frame with the C-terminal YFP tag.
For expression of PTST3-YFP under the control of the 35S promoter, the PTST3:pB7YWG2 vector was cloned. Since the full-length PTST3 sequence was unstable in E. coli (as described above), the coding sequence of PTST3 was codon optimized for E. coli using JCat (Grote et al., 2005), and this sequence was synthesized by Biomatik (sequence provided as Supplemental File 1). This was done to change the codons, which would alter the formation of any secondary structure that may have contributed to the instability of the wild-type sequence. The optimized coding sequence was then amplified with attB-flanked primers and recombined into the Gateway-compatible donor vector pDONR 221. The insert was then recombined into the pB7YWG2 vector as described above.
For expression of SS4-RFP under the control of the UBIQUITIN10 promoter, the SS4 coding sequence was recombined into the pUBC-RFP vector (Grefen et al., 2010), in frame with the C-terminal RFP tag. For expression of SS4 ∆CC-YFP under the control of the 35S promoter in Nicotiana benthamiana, the region encoding the coiled coils of SS4 (amino acids 184–465) was deleted out of the SS4:pDONR 221 vector using the QuikChange site-directed mutagenesis kit (Agilent). The coding sequence was then recombined into the pB7YWG2 vector as described above. For expression of GBSS-CFP, the GBSS:pEarleyGate102 vector was used, as described by Seung et al. (2015).
To generate stably transformed Arabidopsis lines, the appropriate constructs were transformed into Agrobacterium tumefaciens (strain GV3101), and wild-type Columbia plants were transformed using the floral dip method as described previously (Zhang et al., 2006). Many transformants (20–40 seedlings) were isolated in the T1 generation and screened for transgene expression using immunoblotting. For plants expressing the transgene, insertion number was determined by segregation analysis of the BASTA selection marker in the T2 generation. Two independent transformants with single insertions, originating from different T0 parents, were selected for detailed analysis, either in the T2 generation (resistant plants that are heterozygous or homozygous for the transgene) or T3 generation (resistant plants that are homozygous for the transgene), as specified. Transient transformation of N. benthamiana and Nicotiana sylvestris leaves was performed as previously described (Seung et al., 2015; Feike et al., 2016).
Immunoprecipitation
For immunoprecipitations, proteins were extracted from leaf tissue harvested from Arabidopsis rosettes, or N. benthamiana leaves expressing the epitope-tagged proteins, by homogenizing in immunoprecipitation medium (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% [v/v] Triton X-100, 1 mM DTT, and Complete Protease Inhibitor cocktail). Insoluble material was removed by centrifugation at 20,000g. The supernatant was incubated for 1 h at 4°C with µMACS magnetic beads conjugated to anti-YFP or anti-myc (Miltenyi Biotec) or RFP-Trap magnetic particles conjugated to anti-RFP (Chromotek). After incubation, the beads were recovered using a µColumn (Miltenyi Biotec) on a magnetic stand. The beads were washed five times with immunoprecipitation medium before eluting the bound proteins with SDS-PAGE loading medium (50 mM Tris-HCl, pH 6.8, 2% [w/v] SDS, 100 mM DTT, 3% [v/v] glycerol, and 0.005% [w/v] bromophenol blue). For immunoprecipitation assays using N. benthamiana leaves, the beads were washed with wash medium (50 mM Tris-HCl, pH 8, 300 mM NaCl, 1% [v/v] Nonidet P-40, 1 mM DTT, and Complete Protease Inhibitor cocktail) instead of immunoprecipitation medium.
Confocal Laser Scanning Microscopy
Localization of YFP signal in leaf tissue was imaged using confocal laser scanning microscopy as described by Seung et al. (2015).
Analysis of Amylose Content and Amylopectin Structure
The amylose content of the purified starch granules was determined using an iodine colorimetry-based method, as previously described by Hostettler et al. (2011). For amylopectin chain length distribution, starch in the insoluble fractions from perchloric acid-extracted Arabidopsis rosettes was debranched enzymatically, and the liberated linear chains were analyzed by high-performance anion exchange chromatography with pulsed amperometric detection as described previously (Streb and Zeeman, 2014).
Accession Numbers
Sequence data from this article can be found in TAIR (www.arabidopsis.org) under the following accession numbers: PTST2 (At1g27070), PTST3 (At5g03420), PTST1 (At5g39790), SS4 (At4G18240), ISA1 (At2g39930), and GBSS (At1g32900).
Supplemental Data
Supplemental Figure 1. Alignment of the PTST1, -2, and -3 C-terminal amino acid sequences used to generate the phylogenetic tree shown in Figure 1B.
Supplemental Figure 2. Analysis of amylopectin structure and amylose content in ptst2 and ptst3 mutants.
Supplemental Figure 3. Immunoblot detection of SS4 and ISA1 in the ptst2 ptst3 double mutant.
Supplemental Figure 4. Quantification of granule sections in chloroplasts of PTST2 overexpression lines in the ss4 mutant background.
Supplemental Figure 5. Alignment of CBM48 sequences from Arabidopsis proteins and the mammalian AMP kinase β-subunit.
Supplemental Figure 6. Glucan binding residues in the CBM48 of PTST2 are not required for the interaction with SS4.
Supplemental Figure 7. ITC experiments measuring heats of substrate dilution and competition between substrates.
Supplemental Figure 8. Starch granule number in Arabidopsis plants overexpressing SS4.
Supplemental Table 1. Arabidopsis mutants used in this study.
Supplemental Table 2. Oligonucleotide primers used in this study.
Supplemental Table 3. Quantification of soluble glucans in ptst2 and ptst3 mutants.
Supplemental File 1. Codon-optimized coding sequence of PTST3
Acknowledgments
This work was funded by the Swiss-South African Joint Research Program (Grant IZLSZ3_148857/1 to S.C.Z.) and by a ETH Zurich Foundation Heinz-Imhof Fellowship (to D.S.). We thank Simona Rodighiero from the ScopeM microscopy facility of ETH Zurich for producing the scanning electron microscope images, and we thank ScopeM for providing access to microscopes. We thank Ángel Mérida (University of Sevilla) for providing the SS4 antibody. We also thank Alison M. Smith (John Innes Centre) and Frédéric Allain (ETH Zurich) for helpful discussions, as well as the reviewers for their useful suggestions in improving this manuscript.
AUTHOR CONTRIBUTIONS
D.S. and S.C.Z. conceived and directed the research. D.S., J.B., J.M., and S.C.Z. designed the experiments. D.S., J.B., J.M., T.B.S., L.C.D., M.A., K.-J.L., and M.Z. performed research and analyzed data. D.S. and S.C.Z. wrote the article with input from all authors.
Glossary
- SS
starch synthase
- IP
immunoprecipitate
- MOS
maltooligosaccharide
- ITC
isothermal titration calorimetry
Footnotes
Current address: John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK.
Articles can be viewed without a subscription.
References
- Brust H., Orzechowski S., Fettke J., Steup M. (2013). Starch synthesizing reactions and paths: in vitro and in vivo studies. J. Appl. Glycosci. 60: 3–20. [Google Scholar]
- Burton R.A., Jenner H., Carrangis L., Fahy B., Fincher G.B., Hylton C., Laurie D.A., Parker M., Waite D., van Wegen S., Verhoeven T., Denyer K. (2002). Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 31: 97–112. [DOI] [PubMed] [Google Scholar]
- Bustos R., Fahy B., Hylton C.M., Seale R., Nebane N.M., Edwards A., Martin C., Smith A.M. (2004). Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc. Natl. Acad. Sci. USA 101: 2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpton-Taylor M., Grandison S., Png K.M.Y., Bushby A.J., Smith A.M. (2012). Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol. 158: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpton-Taylor M., Pike M., Lu K.J., Hylton C.M., Feil R., Eicke S., Lunn J.E., Zeeman S.C., Smith A.M. (2013). Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 200: 1064–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Seijo J.A., Nielsen M.M., Ruzanski C., Krucewicz K., Beeren S.R., Rydhal M.G., Yoshimura Y., Striebeck A., Motawia M.S., Willats W.G.T., Palcic M.M. (2016). In vitro biochemical characterization of all barley endosperm starch synthases. Front. Plant Sci. 6: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T., Trevisan M., Parker M.L., Zeeman S.C. (2005). Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 41: 815–830. [DOI] [PubMed] [Google Scholar]
- Delvallé D., Dumez S., Wattebled F., Roldán I., Planchot V., Berbezy P., Colonna P., Vyas D., Chatterjee M., Ball S., Mérida A., D’Hulst C. (2005). Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 43: 398–412. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Feike D., et al. (2016). The starch granule-associated protein EARLY STARVATION1 (ESV1) is required for the control of starch degradation in Arabidopsis thaliana leaves. Plant Cell 28: 1472–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez-Arjona F.M., Raynaud S., Ragel P., Mérida A. (2014). Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 80: 305–316. [DOI] [PubMed] [Google Scholar]
- Gidley M.J., Bulpin P.V. (1987). Crystallisation of malto-oligosaccharides as models of the crystalline forms of starch: minimum chain-length requirement for the formation of double helices. Carbohydr. Res. 161: 291–300. [Google Scholar]
- Goldsmith E., Sprang S., Fletterick R. (1982). Structure of maltoheptaose by difference Fourier methods and a model for glycogen. J. Mol. Biol. 156: 411–427. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. (2005). JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33: W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettler C., Kölling K., Santelia D., Streb S., Kötting O., Zeeman S.C. (2011). Analysis of starch metabolism in chloroplasts. Methods Mol. Biol. 775: 387–410. [DOI] [PubMed] [Google Scholar]
- Hwang S.-K., Koper K., Satoh H., Okita T.W. (2016). Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 291: 19994–20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.-K., Nishi A., Satoh H., Okita T.W. (2010). Rice endosperm-specific plastidial alpha-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 495: 82–92. [DOI] [PubMed] [Google Scholar]
- Janeček Š., Svensson B., MacGregor E.A. (2011). Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enzyme Microb. Technol. 49: 429–440. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kawagoe Y., Kubo A., Satoh H., Takaiwa F., Nakamura Y. (2005). Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 42: 164–174. [DOI] [PubMed] [Google Scholar]
- Koay A., et al. (2010). AMPK beta subunits display isoform specific affinities for carbohydrates. FEBS Lett. 584: 3499–3503. [DOI] [PubMed] [Google Scholar]
- Koay A., Rimmer K.A., Mertens H.D.T., Gooley P.R., Stapleton D. (2007). Oligosaccharide recognition and binding to the carbohydrate binding module of AMP-activated protein kinase. FEBS Lett. 581: 5055–5059. [DOI] [PubMed] [Google Scholar]
- Koropatkin N.M., Martens E.C., Gordon J.I., Smith T.J. (2008). Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16: 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom N., Chang P.R., Tyler R.T. (2004). Analytical, biochemical and physicochemical aspects of starch granule size, with emphasis on small granule starches: a review. Stärke 56: 89–99. [Google Scholar]
- Lohmeier-Vogel E.M., Kerk D., Nimick M., Wrobel S., Vickerman L., Muench D.G., Moorhead G.B.G. (2008). Arabidopsis At5g39790 encodes a chloroplast-localized, carbohydrate-binding, coiled-coil domain-containing putative scaffold protein. BMC Plant Biol. 8: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Mantegazza O., Stefanski A., Stühler K., Weber A.P.M. (2017). Surveying the oligomeric state of Arabidopsis thaliana chloroplasts. Mol. Plant 10: 197–211. [DOI] [PubMed] [Google Scholar]
- Lupas A. (1996). Prediction and analysis of coiled-coil structures. Methods Enzymol. 266: 513–525. [DOI] [PubMed] [Google Scholar]
- Mason J.M., Arndt K.M. (2004). Coiled coil domains: stability, specificity, and biological implications. ChemBioChem 5: 170–176. [DOI] [PubMed] [Google Scholar]
- Matsushima R., Yamashita J., Kariyama S., Enomoto T., Sakamoto W. (2013). A phylogenetic re-evaluation of morphological variations of starch grains among Poaceae species. J. Appl. Glycosci. 60: 131–135. [Google Scholar]
- McBride A., Ghilagaber S., Nikolaev A., Hardie D.G. (2009). The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab. 9: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Raththagala M., Husodo S., White C.J., Guo H.F., Kötting O., Vander Kooi C.W., Gentry M.S. (2014). Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc. Natl. Acad. Sci. USA 111: 7272–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev S. (2015). N-value ITC data. Malvern Materials Talks. http://www.materials-talks.com/blog/2015/04/23/.
- Møller M.S., Henriksen A., Svensson B. (2016). Structure and function of α-glucan debranching enzymes. Cell. Mol. Life Sci. 73: 2619–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. (2015). Initiation process of starch biosynthesis. In Starch: Metabolism and Structure, Y. Nakamura, ed (Tokyo: Springer), pp. 315–332. [Google Scholar]
- Nakamura Y., Ono M., Utsumi C., Steup M. (2012). Functional interaction between plastidial starch phosphorylase and starch branching enzymes from rice during the synthesis of branched maltodextrins. Plant Cell Physiol. 53: 869–878. [DOI] [PubMed] [Google Scholar]
- Peng C., et al. (2014). FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77: 917–930. [DOI] [PubMed] [Google Scholar]
- Pfister B., Lu K.-J., Eicke S., Feil R., Lunn J.E., Streb S., Zeeman S.C. (2014). Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol. 165: 1457–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister B., Sánchez-Ferrer A., Diaz A., Lu K., Otto C., Holler M., Shaik F.R., Meier F., Mezzenga R., Zeeman S.C. (2016). Recreating the synthesis of starch granules in yeast. eLife 5: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister B., Zeeman S.C. (2016). Formation of starch in plant cells. Cell. Mol. Life Sci. 73: 2781–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P., Streb S., Feil R., Sahrawy M., Annunziata M.G., Lunn J.E., Zeeman S., Mérida Á. (2013). Loss of starch granule initiation has a deleterious effect on the growth of arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 163: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud S., Ragel P., Rojas T., Mérida Á. (2016). The N-terminal part of Arabidopsis thaliana Starch Synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 291: 10759–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I., Wattebled F., Mercedes Lucas M., Delvallé D., Planchot V., Jiménez S., Pérez R., Ball S., D’Hulst C., Mérida A. (2007). The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 49: 492–504. [DOI] [PubMed] [Google Scholar]
- Satoh H., et al. (2008). Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Lu K.-J., Stettler M., Streb S., Zeeman S.C. (2016). Degradation of glucan primers in the absence of Starch Synthase 4 disrupts starch granule initiation in Arabidopsis. J. Biol. Chem. 291: 20718–20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Soyk S., Coiro M., Maier B.A., Eicke S., Zeeman S.C. (2015). PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D., Thalmann M., Sparla F., Abou Hachem M., Lee S.K., Issakidis-Bourguet E., Svensson B., Zeeman S.C., Santelia D. (2013). Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic α-amylase. J. Biol. Chem. 288: 33620–33633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. (2012). Starch in the Arabidopsis plant. Stärke 64: 421–434. [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345. [DOI] [PubMed] [Google Scholar]
- Stitt M., Zeeman S.C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15: 282–292. [DOI] [PubMed] [Google Scholar]
- Streb S., Zeeman S.C. (2014). Replacement of the endogenous starch debranching enzymes ISA1 and ISA2 of Arabidopsis with the rice orthologs reveals a degree of functional conservation during starch synthesis. PLoS One 9: e92174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S., Zeeman S.C. (2012). Starch metabolism in Arabidopsis. The Arabidopsis Book 10: e0160, doi/10.1199/tab.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N., et al. (2009). Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21: 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I.J., Wait R., Lu Z., Akkasaeng R., Bowsher C.G., Esposito S., Kosar-Hashemi B., Morell M.K., Emes M.J. (2004). Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyosawa Y., et al. (2016). Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 170: 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi Y., Nakamura Y. (2006). Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 225: 75–87. [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Thorneycroft D., Schupp N., Chapple A., Weck M., Dunstan H., Haldimann P., Bechtold N., Smith A.M., Smith S.M. (2004). Plastidial alpha-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 135: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman S.C., Tiessen A., Pilling E., Kato K.L., Donald A.M., Smith A.M. (2002). Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol. 129: 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang X., Myers A.M., James M.G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Szydlowski N., Delvallé D., D’Hulst C., James M.G., Myers A.M. (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol. 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]