PLIP1, a plastid lipase specific for a unique chloroplast lipid, phosphatidylglycerol tagged with 16:1 trans fatty acid, is involved in acyl exchange and fatty acid export for seed oil biosynthesis.
Abstract
The lipid composition of thylakoid membranes inside chloroplasts is conserved from leaves to developing embryos. A finely tuned lipid assembly machinery is required to build these membranes during Arabidopsis thaliana development. Contrary to thylakoid lipid biosynthetic enzymes, the functions of most predicted chloroplast lipid-degrading enzymes remain to be elucidated. Here, we explore the biochemistry and physiological function of an Arabidopsis thylakoid membrane-associated lipase, PLASTID LIPASE1 (PLIP1). PLIP1 is a phospholipase A1. In vivo, PLIP1 hydrolyzes polyunsaturated acyl groups from a unique chloroplast-specific phosphatidylglycerol that contains 16:1Δ3trans as its second acyl group. Thus far, a specific function of this 16:1Δ3trans-containing phosphatidylglycerol in chloroplasts has remained elusive. The PLIP1 gene is highly expressed in seeds, and plip1 mutant seeds contain less oil and exhibit delayed germination compared with the wild type. Acyl groups released by PLIP1 are exported from the chloroplast, reincorporated into phosphatidylcholine, and ultimately enter seed triacylglycerol. Thus, 16:1Δ3trans uniquely labels a small but biochemically active plastid phosphatidylglycerol pool in developing Arabidopsis embryos, which is subject to PLIP1 activity, thereby contributing a small fraction of the polyunsaturated fatty acids present in seed oil. We propose that acyl exchange involving thylakoid lipids functions in acyl export from plastids and seed oil biosynthesis.
INTRODUCTION
Life on the Earth is driven by photosynthesis, which converts light energy into chemical energy and provides us with food and industrial feedstock. Photosynthesis takes place in chloroplast thylakoids, where protein complexes containing chlorophyll, carotenoids, and other cofactors are embedded into photosynthetic membranes (Kobayashi et al., 2016). Photosynthetic membranes are composed of a specialized glycerolipid matrix with four major polar lipids (in order of abundance): mono- and digalactosyldiacylglycerol (MGDG and DGDG, respectively), phosphatidylglycerol (PG), and the sulfolipid sulfoquinovosyldiacylglycerol (SQDG) (Andersson and Dörmann, 2009; Benning, 2010; Boudière et al., 2014). To date, many of the genes and the respective enzymes needed for photosynthetic membrane lipid biosynthesis have been characterized in Arabidopsis thaliana. However, we are only beginning to explore the dynamic changes in photosynthetic membranes under various developmental and environmental conditions and the underlying mechanisms of plastid lipid turnover and its regulation.
Lipid turnover requires lipases, which are enzymes that hydrolyze ester bonds of glycerolipids (Troncoso-Ponce et al., 2013; Kelly and Feussner, 2016). Lipases are involved in a large number of cell biological processes, from maintaining lipid homeostasis to lipid signaling (Wang, 2004; Scherer et al., 2010; Richmond and Smith, 2011). Phospholipases can be classified into four major types based on their lipid substrate cleavage sites: phospholipase D (PLD), phospholipase C (PLC), phospholipase A1 (PLA1), and phospholipase A2 (PLA2). PLD releases the polar head group and produces phosphatidic acid, while PLC cleaves the phosphodiester bond at the glyceryl sn-3 position and produces the phosphorylated head group and diacylglycerol. PLA1 and PLA2 release acyl groups from the glyceryl moiety at the sn-1 and sn-2 positions, respectively (Wang et al., 2012). The Arabidopsis genome encodes ∼300 proteins that are annotated as lipases, but most of them have not been biochemically verified or have unknown physiological functions (Li-Beisson et al., 2013; Troncoso-Ponce et al., 2013; Kelly and Feussner, 2016). Some of the characterized chloroplast-located lipases have intriguing physiological functions. For example, DEFECTIVE IN ANTHER DEHISCENT1 (DAD1) (Ishiguro et al., 2001) is a chloroplast-located PLA1 that catalyzes the initial step of jasmonic acid (JA) production, which is necessary for proper pollen development and biotic stress resistance. Despite their potentially important functions in membrane maintenance and signaling, the bulk of chloroplast-localized lipases remain uncharacterized.
In land plants, fatty acid (FA) biosynthesis occurs in plastids. In Arabidopsis, two pathways are responsible for glycerolipid biosynthesis (Benning, 2009; Hurlock et al., 2014). De novo synthesized FAs either directly enter the prokaryotic pathway in plastids or are exported to the endoplasmic reticulum (ER) to be assembled into glycerolipids by the eukaryotic pathway. In developing embryos, the bulk of synthesized FAs is incorporated into triacylglycerol (TAG), which serves as the primary energy repository to fuel seed germination. Oleic acid (18:1; carbon:double bonds) is the FA predominantly exported from chloroplasts. Exported FAs are activated to acyl-CoAs and initially incorporated into phosphatidylcholine (PC), which is present in the outer envelope membrane of chloroplasts and in the ER, before reentering the cytosolic acyl-CoA pool by a process referred to as acyl-editing (Bates et al., 2007). Acyl-editing allows 18:1 to be further desaturated into polyunsaturated acyl groups attached to PC before reentering the acyl-CoA pool for incorporation of FAs into other lipids, including TAG. In fact, acyl-editing is one of the reported mechanisms for directing polyunsaturated FAs into TAG during embryogenesis through the action of diacylglycerol acyltransferase (Katavic et al., 1995; Routaboul et al., 1999), in parallel with direct head group exchange between PC and diacylglycerol (DAG) (Bates et al., 2012) or acyl transfer from PC to DAG by phospholipid:diacylglycerol acyl transferase (Dahlqvist et al., 2000). Whether lipids other than PC are subject to acyl exchange remains to be determined.
Here, we report on the function of PLASTID LIPASE1 (PLIP1). This gene encodes a thylakoid-associated PLA1 that specifically releases polyunsaturated sn-1 acyl groups from 16:1Δ3t-PG (16:1Δ3t, 16 carbons with one trans double bond following carbon 3 counting from the carboxyl end, which is always at the sn-2 glyceryl position), a chloroplast-specific lipid. The unusual trans double bond is introduced by FA DESATURASE4 (FAD4), which has diverged during evolution from other desaturases introducing cis double bonds (Gao et al., 2009). In developing seeds, the polyunsaturated acyl groups released by PLIP1 from 16:1Δ3t-PG in the chloroplast are incorporated into PC and subsequently into TAG. PLIP1 initiates a mechanism leading to FA export from the chloroplast and the incorporation of FAs derived from the thylakoid membrane lipid pool into seed TAG. In addition, 16:1Δ3t-PG serves as a specific substrate for PLIP1 in vivo, providing insights into the specialized function of this unusual chloroplast lipid.
RESULTS
PLIP1 Is a Chloroplast Thylakoid-Associated Protein
The Arabidopsis genome encodes ∼300 putative lipases (Li-Beisson et al., 2013; Troncoso-Ponce et al., 2013; Kelly and Feussner, 2016), among which 46 were included in the Chloroplast 2010 Project, which was aimed at assigning functions to nearly all plastid-localized proteins (Lu et al., 2008; Ajjawi et al., 2010). We hypothesized that some of these putative chloroplast lipases may play roles in the maintenance of photosynthetic membranes and perhaps have specialized roles in tissues with high demands on lipid metabolism, such as developing seeds that accumulate TAG. We initially focused on one of the predicted chloroplast lipase genes, At3g61680, which encodes a protein with a conserved Lipase 3 domain and a strongly predicted transit peptide, which we subsequently named PLIP1. With its Lipase 3 domain, which is a signature for triacylglycerol lipases, this Arabidopsis protein shares similarities with a bona fide lipase of Chlamydomonas reinhardtii, PLASTID GALACTOGLYCEROLIPID DEGRADATION1 (PGD1), involved in the turnover of chloroplast MGDG, leading to the export of acyl groups and their incorporation into TAG following nutrient deprivation (Li et al., 2012), although the two proteins do not share sequence similarity outside the Lipase 3 domain and are not orthologs.
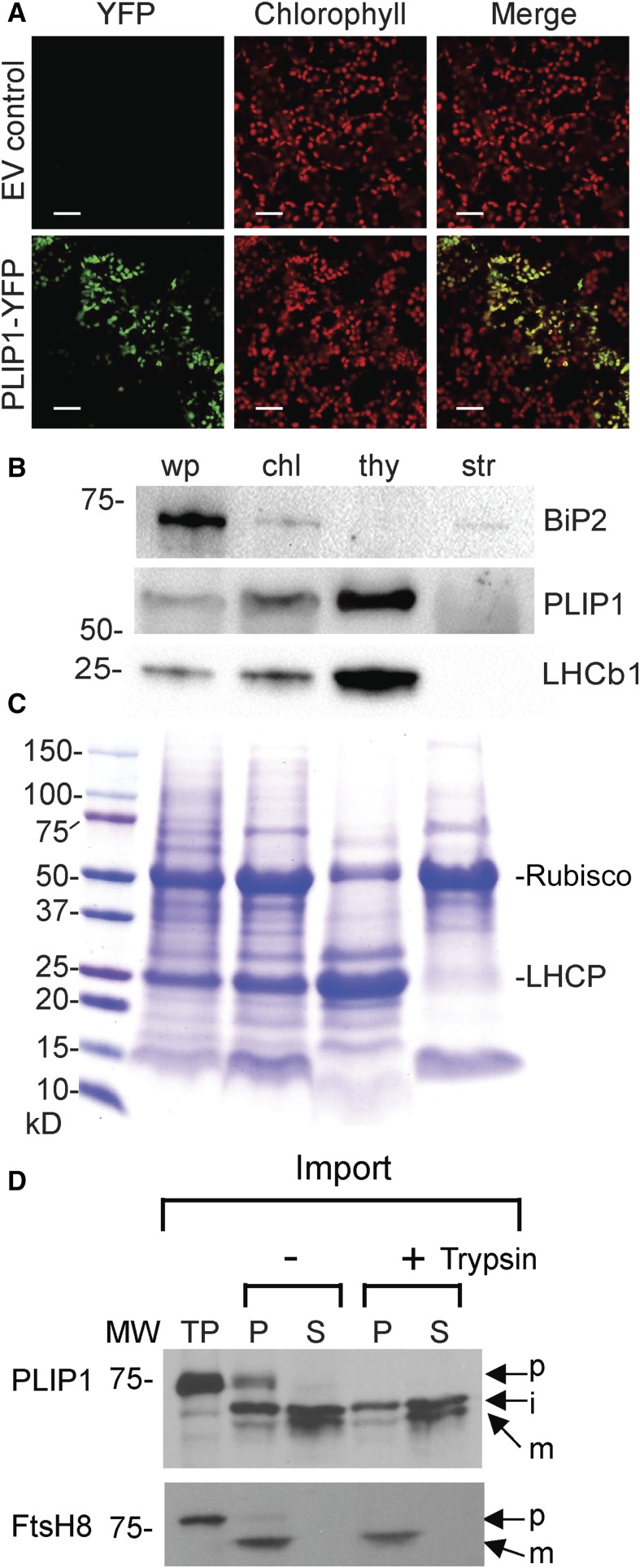
To experimentally verify the subcellular location of PLIP1, the PLIP1 coding sequence derived from an Arabidopsis wild-type (Col-0) cDNA was spliced at its 3′-end (creating a C-terminal fusion) to the open reading frame of YFP. When the PLIP1-YFP construct was stably expressed in the wild type under the control of the CaMV 35S promoter, the YFP and chlorophyll signals overlapped (Figure 1A). Although transgenic lines used in this experiment had constitutive expression of PLIP1-YFP, ubiquitous YFP signals were not detected. Instead, only ∼10 to 15% of mesophyll chloroplasts showed YFP signals, and the cells observed in the PLIP1-YFP transgenic plants were slightly smaller than the wild type.
Figure 1.
Subcellular Localization of PLIP1 in Arabidopsis.
(A) Subcellular localization of PLIP1-YFP in leaf mesophyll cells of 3-week-old Arabidopsis Col-0 transformed with PLIP1-YFP driven by the 35S promoter or EV control using confocal laser scanning microscopy. Chlorophyll autofluorescence is shown in red, and YFP fluorescence is shown in green. Overlay of chlorophyll and YFP is shown as well (Merge). Representative images from one experiment are presented. Bars = 30 µm.
(B) PLIP1 enrichment in chloroplast fractions analyzed by immunoblotting. Intact and subfractionated chloroplasts were prepared using 4-week-old Arabidopsis (Col-0) plants grown on MS medium. Equal amounts of protein of leaf tissues from the whole plant (wp), intact chloroplasts (chl), thylakoid (thy), and stroma (str) were separated by SDS-PAGE or further subjected to immunoblotting analysis using an antibody against PLIP1S422A, a nonfunctional mutant of PLIP1. Immunoblotting was used to detect marker proteins BiP2 (endoplasmic reticulum) and LHCb1 (thylakoid). For protein loading, 12 µg per fraction was loaded for PLIP1; 2 µg per fraction for BiP2 and LHCb1.
(C) SDS-PAGE Coomassie blue staining was used to detect Rubisco large subunit (stroma) and light-harvesting chlorophyll a/b binding protein (LHCP) (thylakoid), which were used as markers. Numbers indicate protein molecular mass in kilodaltons. For protein loading, 12 µg per fraction were loaded.
(D) Chloroplast import experiments with labeled PLIP1 and control protein FtsH8. Chloroplasts were treated with (+) or without (−) trypsin. Total chloroplast membranes (P) or soluble (S) fractions were analyzed by SDS-PAGE followed by fluorography. TP, translation products; p, precursor; i, intermediate; m, mature form; MW, molecular weight markers.
To corroborate the suborganellar location of PLIP1, we isolated intact chloroplasts from 4-week-old wild-type seedlings and fractionated them into thylakoid membranes and stroma. Immunoblot analysis of PLIP1 showed increasing signal intensity from whole plant tissue to intact chloroplasts and thylakoids, consistent with an association of PLIP1 with the thylakoids (Figure 1B). Fractionation quality was controlled for by including marker proteins for each fraction. The thylakoid protein Light Harvesting Complex b1 (LHCb1) showed a similar intensity pattern to that of PLIP1. To exclude the possibility of contamination with ER-associated proteins, the ER-specific marker BiP2 was also included (Figure 1B). The fractionation of the stroma-specific Rubisco large subunit and the thylakoid-specific light-harvesting chlorophyll a/b binding protein (LHCP) were visible on a Coomassie blue-stained SDS-PAGE gel (Figure 1C).
The PLIP1 gene is predicted to encode a 71,735-D protein for which a molecular or biochemical function had not been experimentally determined. Based on the ARAMEMNON database, PLIP1 is not predicted to contain any transmembrane domains (Schwacke et al., 2003). Therefore, it seemed likely that PLIP1 is a peripheral thylakoid membrane protein. To learn more about its suborganellar location and processing en route, we translated PLIP1 cDNA in vitro in the presence of labeled methionine. During the import of the translation product into isolated pea (Pisum sativum) chloroplasts, the PLIP1 precursor was processed into a smaller intermediate protein that was present in both the stroma and chloroplast membrane fractions. In addition, the trypsin resistance of PLIP1 suggested that it is located inside the chloroplast (Figure 1D). Interestingly, the intermediate PLIP1 form was further processed into a smaller mature protein, which was mainly associated with the stroma fraction. We used thylakoid lumen-localized FtsH8 as a control for proper fractionation and import (Rodrigues et al., 2011), which was processed and imported into the thylakoids with a pattern different from that of PLIP1, suggesting that PLIP1 is probably not imported into the thylakoid lumen but instead attaches to the outside leaflet of thylakoid membranes and can be released into the stroma with additional processing. Summing up all localization data, PLIP1 is likely a thylakoid membrane-associated protein.
PLIP1 Is a PLA1 with a Preference for Unsaturated Acyl Groups
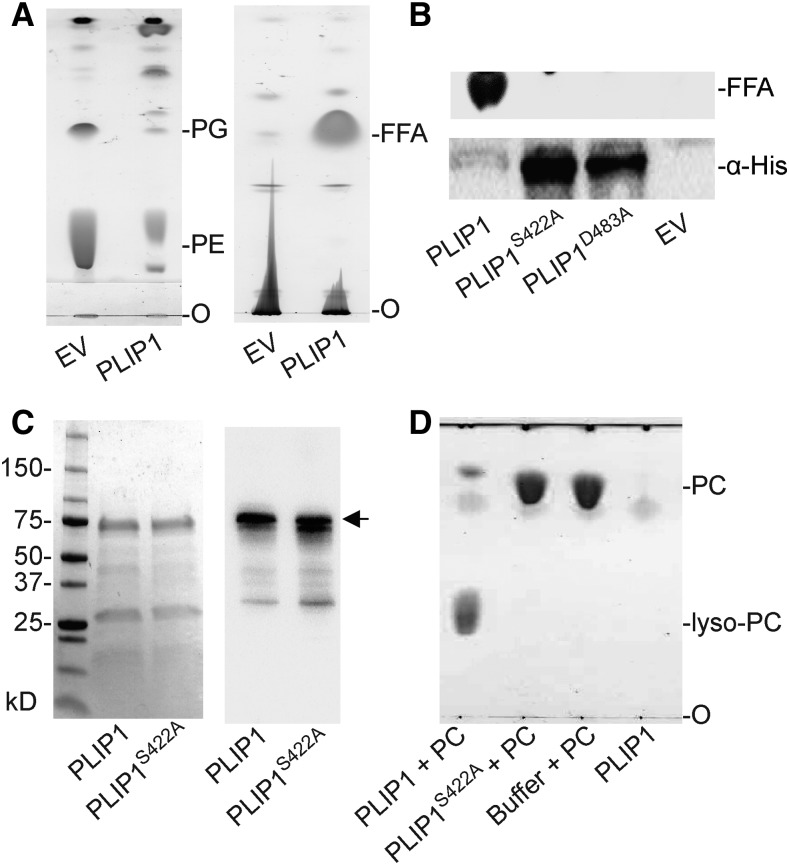
PLIP1 is annotated as a TAG lipase in TAIR (https://www.arabidopsis.org/). To verify its activity and determine its enzymatic properties, we tried to develop an in vitro lipase assay with recombinant PLIP1 purified from E. coli. However, expression of the recombinant 6×His-PLIP1 construct was very low, as detected by immunoblotting against the His tag. Analysis of transgenic E. coli-derived lipid extracts by thin-layer chromatography (TLC) showed that, when PLIP1 was expressed, PG and phosphatidylethanolamine (PE), the two major polar lipids of E. coli, were degraded, leading to the accumulation of free FAs (Figure 2A). This observation suggested that PLIP1 is a lipase that releases acyl groups from PG and PE.
Figure 2.
Recombinant PLIP1 Has Lipase Activity.
(A) Thin-layer chromatographic analysis of polar (left) and neutral (right) lipids in E. coli containing a 6×His-PLIP1 expression construct or EV control at 6 h following induction. FFA, free fatty acid; O, origin of sample loading. TLC plates were stained with iodine vapor.
(B) Mutation of the PLIP1 active site motif. Lipid extracts of E. coli cultures 6 h after induction expressing 6×His-PLIP1 or two point mutation alleles, 6×His -PLIP1S422A or 6×His-PLIP1D483A, or containing an EV control were analyzed by TLC to detect FFA products (top panel). Protein extracts were analyzed for protein production using an antibody against the 6×His tag.
(C) SDS-PAGE analysis of purified PLIP1 and PLIP1S422A proteins. Loading was 5 µg per lane for both samples. SDS-PAGE separated proteins were stained by Coomassie blue (left) or detected by immunoblotting with an antibody raised against PLIP1S422A (right). Numbers indicate protein molecular mass in kilodaltons. 6×His-PLIP1 and 6×His-PLIP1S422A are indicated by the arrow.
(D) Thin-layer chromatogram of products of a representative in vitro lipase reaction using PC with the wild type (PLIP1 + PC) and the mutant enzyme (PLIP1S422A + PC). Substrate without enzyme (Buffer + PC) and enzyme without substrate (PLIP1) were included as controls. O, origin of sample loading. Lipids were visualized by iodine vapor staining.
Lipases belong to a group of serine esterases with a lipase signature motif, an Asp-His-Ser triad, with some exceptions having only a Ser-Asp dyad, and in all cases Ser serves as the active site residue, participating in the reaction mechanism (Brady et al., 1990; Winkler et al., 1990; Richmond and Smith, 2011; Kelly and Feussner, 2016). Alignment of the PLIP1 protein sequence with those of classic lipases using NCBI’s conserved domain database (Marchler-Bauer et al., 2015) identified two potential catalytic residues, Asp-483 and Ser-422, but the conserved His site was not present (Supplemental Figure 1). Replacing these two residues with Ala abolished PLIP1 lipase activity, and the respective mutant proteins were abundantly produced in E. coli, as PG and PE were not degraded (Figure 2B). Taken together, these data suggest that PLIP1 is a lipase with Ser-422 as the catalytic residue. Taking advantage of the enhanced production of nonfunctional PLIP1S422A in E. coli, we purified the mutant protein and raised an antibody in rabbits to specifically detect PLIP1.
To develop an in vitro lipase assay, recombinant PLIP1 and PLIP1S422A were produced in E. coli and affinity purified from the soluble fraction (Figure 2C). We chose Anzergent 3-12 as the solubilizing detergent from a series of other reagents because of its high compatibility with PLIP1 enzyme activity and because it was not cochromatographing with native plant membrane lipids during subsequent TLC analysis. In the final optimized system (Supplemental Figure 2), in vitro lipase activity of PLIP1 was observed on a wide range of substrates. As an example (shown in Figure 2D), PC was provided to PLIP1 and PLIP1S422A. At the end of the reaction, lipids were extracted and separated by TLC. Lipase activity based on the production of lyso-PC was only observed with PLIP1, but not with PLIP1S422A (Figure 2D).
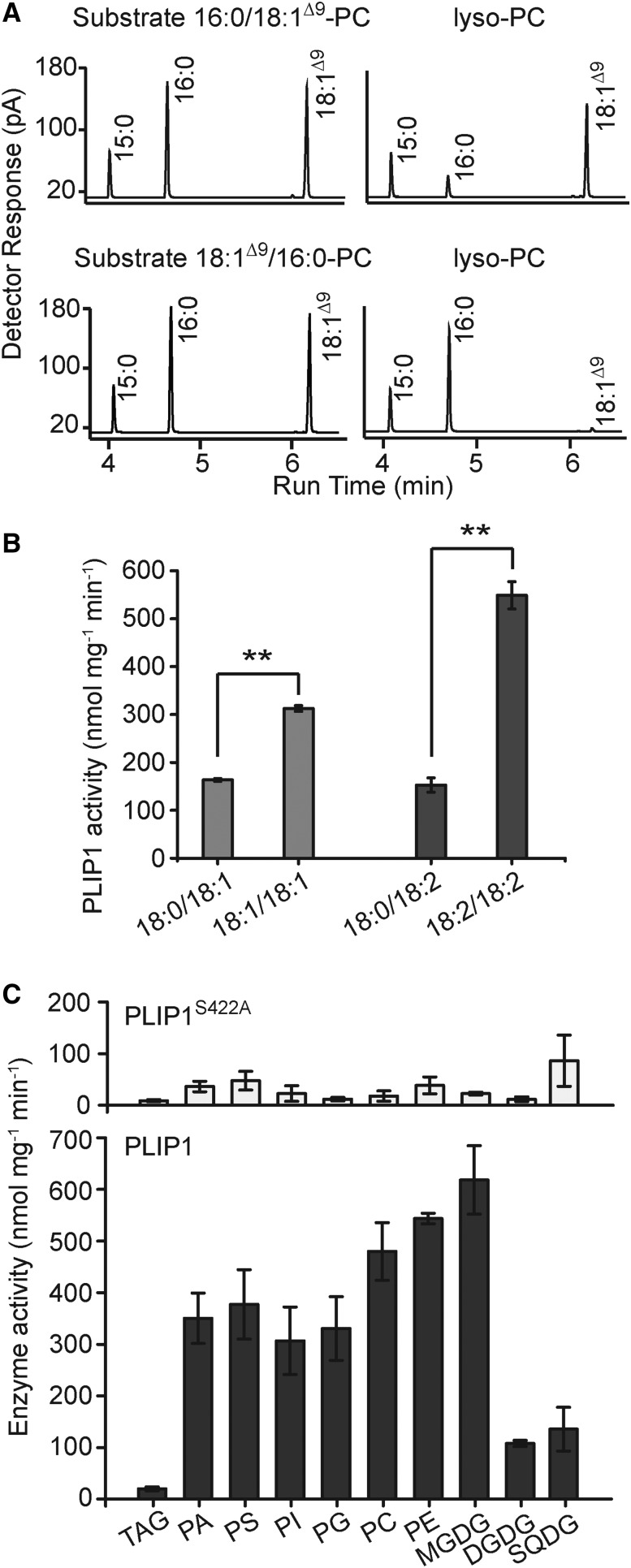
PLIP1 in vitro activity with PC and other glycerolipids as substrates always resulted in lyso-lipid products, implying that PLIP1 can only hydrolyze one of the two acyl-glyceryl ester bonds in glycerolipids. To investigate which glyceryl position PLIP1 prefers, two PCs with reversed acyl compositions were offered to PLIP1. At the end of the reaction, lipids were extracted, lyso-PC was isolated by TLC, and FA methyl esters derived from the lyso-lipid were analyzed by gas-liquid chromatography (Figure 3A). For PC with a composition of 18:1Δ9/16:0 (sn-1/sn-2), 18:1Δ9 was selectively released and 16:0 was retained in the lyso-product. The result was reversed with PC containing 16:0/18:1Δ9 as the lyso-product contained 18:1Δ9. Therefore, PLIP1 is a lipase that prefers the sn-1 glyceryl position of the respective glycerolipid. Furthermore, to determine a possible acyl group preference of PLIP1 at the sn-1 glyceryl position, PLIP1 was offered different combinations of pure PC molecules carrying the same acyl groups at the sn-2 but acyl groups with different degrees of saturation at the sn-1 position (Figure 3B). Comparing 18:0/18:1 with 18:1/18:1, PLIP1 enzyme activity was approximately twice as high for 18:1/18:1-PC, and when comparing 18:0/18:2 with 18:2/18:2, its activity was elevated nearly 4-fold for 18:2/18:2-PC. Therefore, PLIP1 is a phospholipase A1 with a preference for more unsaturated acyl groups.
Figure 3.
Specificity of PLIP1 in Vitro Activity.
(A) Gas-liquid chromatograms of methyl esters derived from commercial PC substrates or lyso-PC fractions from PLIP1 lipase reactions with different PC substrates. 15:0 was used as an internal standard.
(B) PLIP1 lipase activity on commercial PC substrates (carbon number:double bond number; sn-1/sn-2) with different degrees of saturation of the sn-1 acyl groups. PC containing 18:0/18:1 and 18:1/18:1 and PC containing 18:0/18:2 and 18:2/18:2 were compared, respectively. n = 4, ±sd. Student’s t test was applied (**P < 0.01).
(C) PLIP1 activity on different substrates. For each PLIP1 lipase reaction, 60 µg lipids and 0.5 µg protein were used. The reactions were incubated at ambient temperature (∼22°C) for 1.5 h, which was still during the linear portion of the reaction time course for PC (Supplemental Figure 2). PLIP1S422A was included as a negative control and is shown in the top panel. All lipids contained two oleic acids (18:1), except MGDG, DGDG, and SQDG, which were isolated from plants, and PI, which was isolated from bovine liver. n = 3 to 4 for each substrate, ±sd. PA, phosphatidic acid; PS, phosphatidylserine.
To survey PLIP1 substrate preference in vitro, we offered the majority of the plant glycerolipids, including galactoglycerolipids, phospholipids, and TAG (Figure 3C). High enzyme activity was detected for all tested phospholipids and MGDG. Given the plastid location of PLIP1, possible native substrates were limited to PG and MGDG. The low activities detected for SQDG, DGDG, and TAG suggested that these are not likely substrates of PLIP1 (Figure 3C). Based on these results, despite its conserved Lipase 3 domain, PLIP1 is apparently not a TAG lipase.
18:3/16:1Δ3t-PG Is the Preferred Substrate of PLIP1 in Vivo
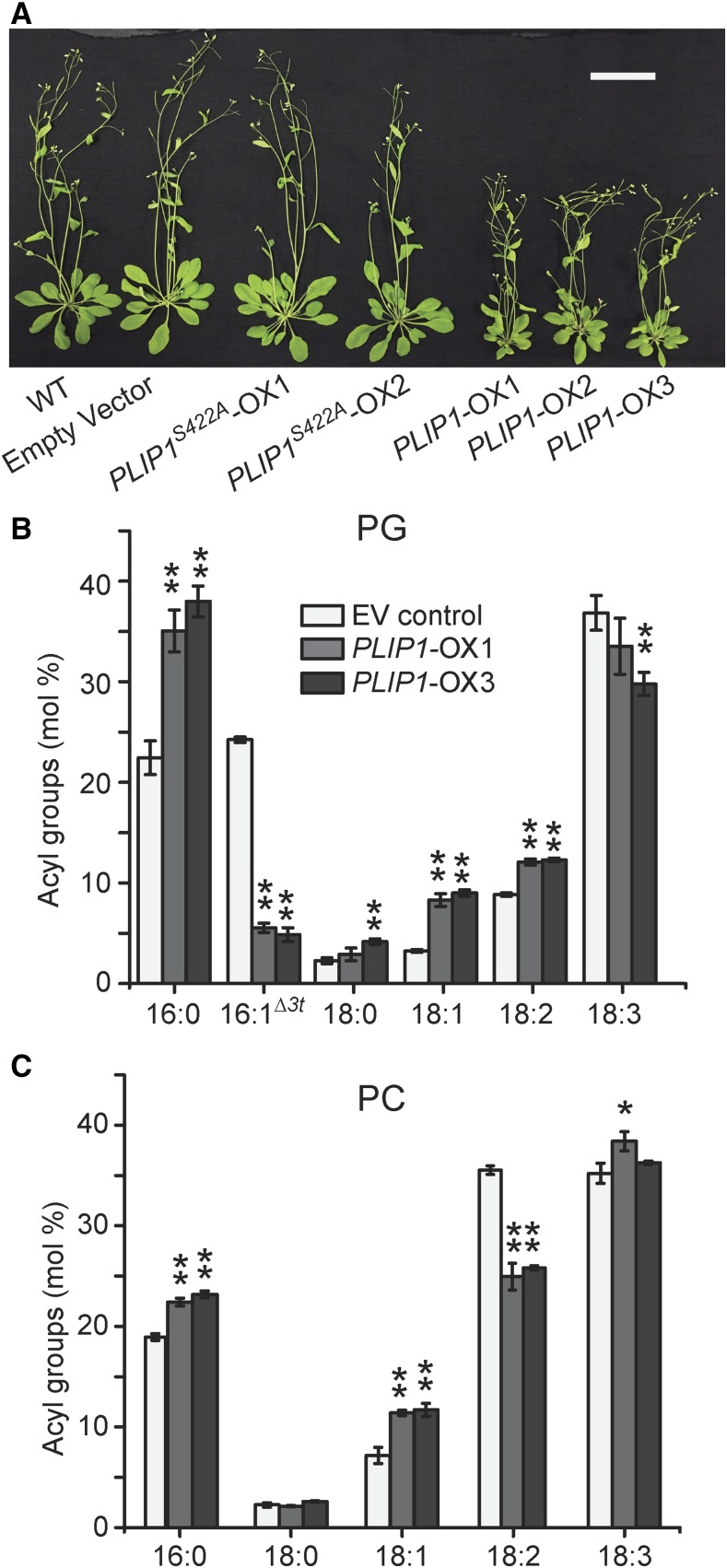
The in vitro assays in combination with its established chloroplast location narrowed down possible native PLIP1 substrates to MGDG and PG (Figure 3C). However, given the complexity of native plant acyl compositions, this limited survey based on an in vitro lipase assay alone could only provide a first approximation of the likely PLIP1-preferred substrate in vivo. To assess PLIP1 activity in its native biological context, we took advantage of the Arabidopsis transgenic lines used for PLIP1 localization described above. In total, 30 independent PLIP1-YFP (PLIP1-OX) and 14 PLIP1S422A -YFP (PLIP1S422A-OX) overexpression lines were generated and three PLIP1-OX lines were selected as representatives. As shown in Figure 4A, compared with the wild type, the PLIP1-OX lines had smaller rosettes and fewer leaves, which were slightly pale yellow, whereas PLIP1S422A-OX plants were indistinguishable from wild-type and empty vector control plants. On a fresh weight basis, the total leaf acyl group content of the smaller PLIP1-OX plants was not reduced (Table 1). A comparison of the relative abundance of polar lipids (Supplemental Figure 3A) and the acyl group composition of individual polar lipids of empty vector (EV) control plants and two PLIP1-OX lines is shown in Figures 4B and 4C and Supplemental Figures 3B to 3E. In PLIP1-OX lines, the levels of lipids associated with chloroplasts (MGDG, PG, and DGDG) decreased, while the levels of lipids mostly associated with the ER (PC, PE, and phosphatidylinositol [PI]) increased (Supplemental Figure 3A), suggesting a reduced ratio of plastid-to-extraplastidic membranes in the PLIP1-OX lines. Acyl group analysis of individual membrane lipids showed the greatest changes for PG (Figure 4B). Specifically, the ratio of 16:0 to 16:1Δ3t was increased in the PLIP1-OX lines. For 18-carbon acyl groups, which are primarily present at the sn-1 position of plastid PG, polyunsaturated 18:3 levels decreased with a concurrent increase in the relative abundance of 18:1 and 18:2. Based on these changes in the molecular composition of PG, we hypothesize that 18:3/16:1Δ3t-PG is a preferred substrate of PLIP1 in its native environment.
Figure 4.
Growth and Lipid Phenotypes of PLIP1 Overexpression Plants.
(A) Growth of 4-week-old soil-grown plants. Arabidopsis wild-type plant, one EV control line, and two PLIP1S422A-OX and three PLIP1-OX overexpression lines are shown. Bar = 5 cm.
(B) and (C) Relative acyl composition of PG and PC in PLIP1-OX and EV control lines. Mature leaf lipids were extracted and analyzed from the plants shown in (A). Leaf samples harvested from one plant were taken as one biological repeat; n = 4, ±sd. Student’s t test was applied to compare the EV control with each PLIP1-OX line (*P < 0.05; **P < 0.01).
Table 1. Leaf Acyl Group Contents in Different Genotypes.
| Genotype | Acyl Groups (µg/mg FW) |
|---|---|
| Col-0 | 3.48 ± 0.067 |
| plip1-1 | 3.51 ± 0.076 |
| plip1-2 | 3.51 ± 0.063 |
| EV control | 3.55 ± 0.055 |
| PLIP1S422A-OX1 | 3.53 ± 0.055 |
| PLIP1S422A-OX2 | 3.59 ± 0.049 |
| PLIP1-OX1 | 3.56 ± 0.062 |
| PLIP1-OX2 | 3.54 ± 0.063 |
| PLIP1-OX3 | 3.56 ± 0.067 |
Acyl group contents were determined by measuring total leaf fatty acid methyl esters. Plants were grown on soil for 4 weeks. Four independent samples were averaged, and the sd is indicated. FW, fresh weight.
MGDG, the most abundant lipid in chloroplasts, also showed a subtly reduced ratio of 16:3 to 18:3 in PLIP1-OX lines (Supplemental Figure 3B). For ER lipids, a decrease in 18:2 and an increase in 18:1 were observed for PC (Figure 4C), as well as for PE and PI (Supplemental Figure 3E). Since PLIP1 is located in the chloroplast and is spatially separated from ER lipids, observation of this ER lipid alteration suggested that turnover of chloroplast lipids can affect the synthesis of ER lipids, assuming that lipid precursors are transported from the chloroplast to the ER. The other two photosynthetic membrane lipids, DGDG and SQDG, showed very minor changes in their molecular compositions in the PLIP1-OX lines (Supplemental Figures 3C and 3D), which is consistent with the low activity of PLIP1 on DGDG and SQDG in vitro (Figure 3C).
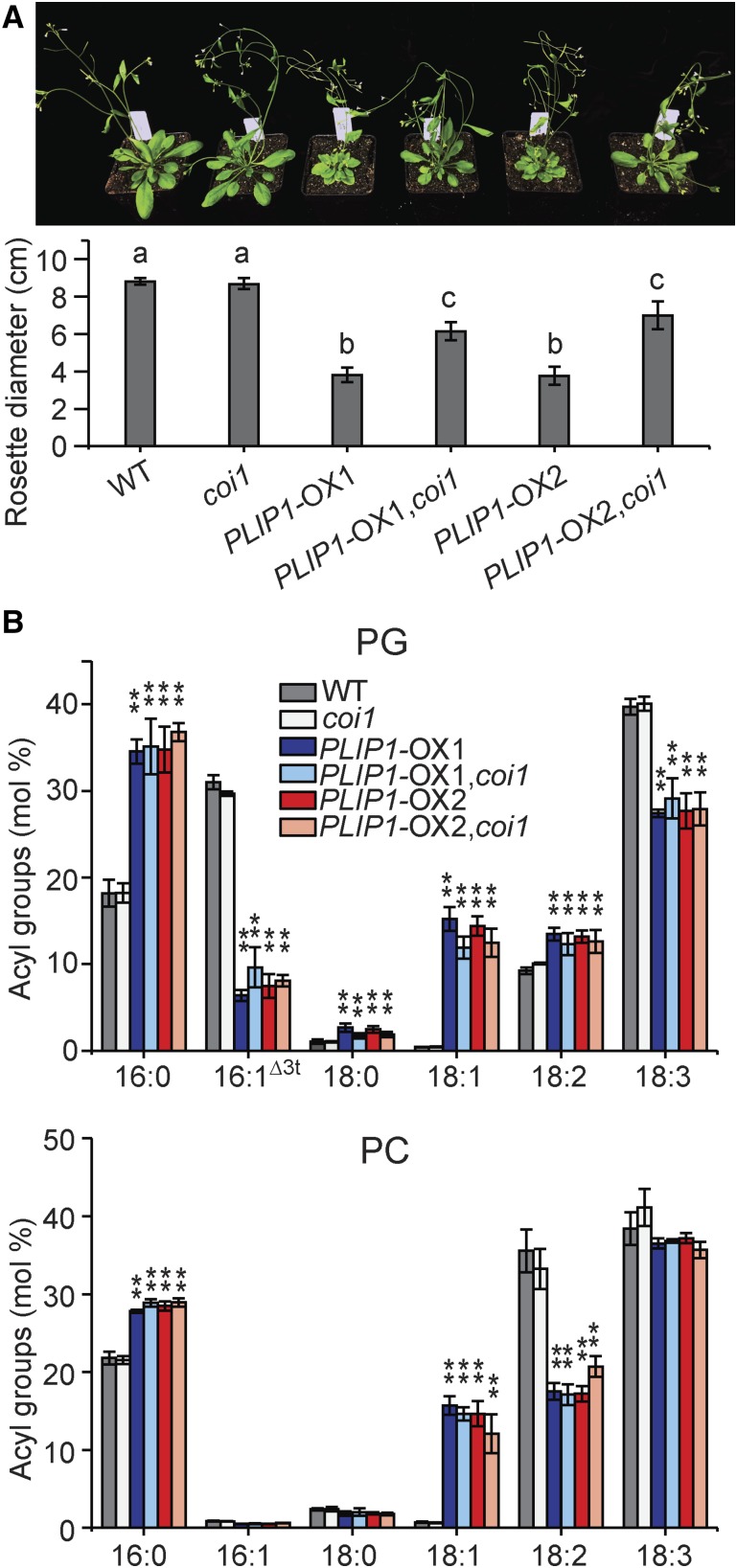
The altered morphology of the PLIP1-OX lines raised a concern that the lipid phenotype might be a secondary phenotype due to the plants’ abnormal development and growth. Therefore, we investigated whether growth and lipid phenotypes could be separated. Besides stunted growth, the three PLIP1-OX lines had shorter leaf petioles than the wild type and accumulated anthocyanin under normal growth conditions (Figure 4A), phenotypes reminiscent of plants with activated JA signaling (Yan et al., 2007; Zhang and Turner, 2008; Campos et al., 2016). Given that PLIP1 releases polyunsaturated FAs including 18:3, a precursor of JA and other oxylipins, we hypothesized that the reduced growth of PLIP1-OX plants is due to the production of oxylipins and the activation of JA signaling. Quantitative PCR analysis showed that the expression of JA-responsive marker genes was constitutively increased in PLIP1-OX lines compared with the wild type (Supplemental Figure 4). Targeted hormone analysis of mesophyll tissues showed that the levels of 12-oxo phytodienoic acid (OPDA) and OPDA-containing glycerolipids called Arabidopsides were strongly elevated in the PLIP1-OX mesophyll tissues (Supplemental Figure 5). JA and to some extent OPDA signaling require the coreceptor CORONATINE INSENSITIVE1 (COI1) (Xie et al., 1998; Ribot et al., 2008). To corroborate that the growth phenotype is secondary and caused by the activation of JA signaling pathways, we crossed PLIP1-OX lines with a coi1 mutant to block COI1-dependent OPDA and JA signaling. As a result, the growth phenotype was largely (but not fully) restored in the offspring of this cross as validated by analysis of variance (Supplemental File 1), while the lipid phenotype remained identical to that of the PLIP1-OX lines (Figure 5). Thus, we were able to at least partially separate the growth and lipid phenotypes, suggesting that the changes in lipid composition observed represent the primary phenotype resulting from PLIP1 overproduction in the PLIP-OX lines and are not the result of abnormal growth.
Figure 5.
Phenotypes of PLIP1-OX and coi1 plants.
(A) Image of the 4-week-old plants grown on soil and the rosette diameter of the indicated plants. n = 6, ±sd, one-way ANOVA with post-hoc Turkey HSD test was applied. Rosette diameters indicated by different letters (a, b, and c) are significantly different (P < 0.01).
(B) Relative acyl group compositions of leaf PG and PC in the indicated plants in (A). Leaf samples harvested from one plant were treated as one biological repeat; n = 3, ±sd. Student’s t test was applied to compare the wild type with each of the remaining genotypes (**P < 0.01).
Overexpression of PLIP1 Accelerates Recycling of Plastid PG Acyl Groups and Their Transfer to PC
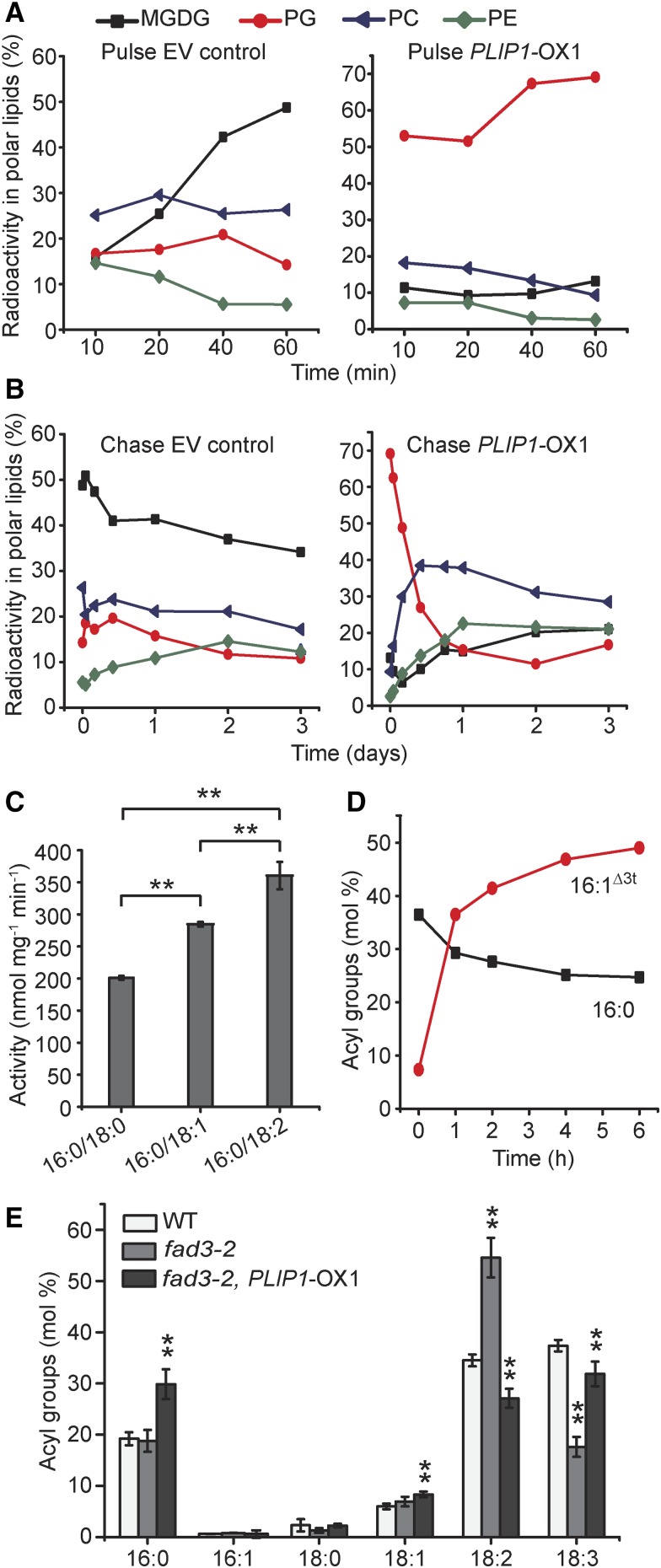
The analysis of the PLIP1-OX lines described above depicts the lipid composition at steady state. However, lipid metabolism is a dynamic process, and pulse-chase labeling is an effective way of probing the dynamics of lipid metabolism and movement of acyl groups through different lipid pools and between organelles (Xu et al., 2008; Li et al., 2012). Therefore, we employed pulse-chase labeling of membrane lipids using [14C]-acetate, which can be readily incorporated into acyl groups in plastids by the FA synthase complex. The pulse phase of the experiment (shown in Figure 6A) indicated that MGDG, PG, and PC contained the majority of the label in EV control leaves, with PG accounting for ∼15% of the label after 1 h. However, in PLIP1-OX1, incorporation of label into PG accounted for nearly 70% of total label at the end of the pulse phase. This result suggests that incorporation of de novo synthesized acyl groups into PG is greatly accelerated in PLIP1-OX lines. During the chase phase (Figure 6B), PG rapidly lost most of the label (within a day), and the label concomitantly increased in PC and to a smaller extent in PE in the PLIP1-OX1 line. The EV control line showed less drastic changes in labeling during the chase phase. The rapid increase and subsequent loss of PG label in PLIP1-OX1 during the pulse and chase phases, respectively, suggested a rapid acyl exchange, preferably on PG, in these lines and supported our hypothesis that PG is the preferred PLIP1 substrate in its native environment.
Figure 6.
PLIP1 Precursor-Product Relationships in PLIP1 Overexpression Plants.
(A) and (B) In vivo pulse-chase acetate labeling of lipids in wild-type and PLIP1-OX1 plants. The length of the [14C]-acetate labeling pulse was 60 min (A), after which medium was replaced with nonlabeled free acetate to initiate the chase with a duration of three days (B). The fractions of label in all polar lipids are given as percentages of total incorporation of label in polar lipids. Experiments were repeated three times with similar results, and one representative result is shown.
(C) Activity of purified recombinant PLIP1 on PC with different sn-2 acyl groups. PC containing 16:0/18:0, 16:0/18:1, and 16:0/18:2 were used as substrates. n = 4, ±sd. Student’s t test was applied (**P < 0.01).
(D) PLIP1 enzyme activity preference for molecular species of phosphatidylglycerol isolated from N. benthamiana leaves. Acyl groups of lyso-phosphatidylglycerol are shown as molar percentages of total acyl groups at any given time point. Experiments were repeated three times with similar results and data from one representative experiment are shown.
(E) Relative acyl composition of PC in wild-type, fad3-2, and fad3-2 PLIP1-OX1 plants. Leaf lipids were extracted and isolated by TLC and fatty acid methyl esters derived from the lipids were analyzed by GC. Mature leaf samples harvested from one plant were taken as one biological repeat; n = 4, ±sd. Student’s t test was applied (**P < 0.01).
The most notable acyl group change observed in the PLIP1-OX lines was the increased 16:0-to-16:1Δ3t ratio in PG (Figure 4B). However, 16-carbon FAs only exist at the sn-2 position of plastid PG. This implies that acyl groups at the glycerol sn-2 position affect PLIP1-catalyzed hydrolysis at the glyceryl sn-1 position of PG. To test this possibility, we provided purified recombinant PLIP1 with a set of commercial PCs with 16:0 at the glyceryl sn-1 position, but 18-carbon acyl groups of different saturation levels at the glyceryl sn-2 position. The highest enzyme activity was observed for PC with 18:2 at the sn-2 position, followed by 18:1, with the lowest activity observed for 18:0 (Figure 6C), suggesting that unsaturated sn-2 acyl groups enhance PLIP1 activity. Therefore, it follows that 16:1Δ3t should be favored over 16:0 at the sn-2 position of PG. To test this hypothesis, plant-derived PG composed of species containing 16:0 or 16:1Δ3t at the glyceryl sn-2 position was extracted from wild tobacco (Nicotiana benthamiana) leaves and offered to PLIP1 in vitro. Total PG was degraded, while lyso-PG was produced over time (Supplemental Figure 6A). The fraction of 16:1Δ3t in lyso-PG increased over time, while 16:0 decreased (Figure 6D), indicating that 16:1Δ3t-PG is preferred by PLIP1 under these conditions when native PG substrate is offered. The opposite pattern between 16:0 and 16:1Δ3t was observed in retained PG (Supplemental Figure 6B). Therefore, we concluded that 18:3/16:1Δ3t-PG is the preferred native substrate of PLIP1 based on data gathered from the above-described combination of in vitro and in vivo experiments.
Another interesting observation during the chase phase of the labeling experiment was the sequential labeling of PG and PC, which points toward a precursor-product relationship between these two lipids. This observation was also consistent with the decreased plastid-to-extraplastidic lipid ratio observed during steady state lipid analysis of the PLIP1-OX lines (Supplemental Figure 3A). Therefore, we hypothesized that 18:3 released from 18:3/16:1Δ3t-PG was exported from the plastid and incorporated into PC. PC is known for its intermediate role in acyl editing involving desaturation of PC-acyl groups (18:1 to 18:2 and 18:3) followed by acyl exchange (Bates et al., 2007). We reasoned that the reduced 18:2 content of PC in the PLIP1-OX lines might be due to the increased competition of incorporation of plastid-derived 18:3 into PC with the activity of ER desaturases FAD2 and FAD3, generating 18:3 from 18:1 and 18:2 bound to PC. To test this hypothesis, we crossed a PLIP1-OX1 plant to a fad3-2 mutant plant (Figure 6E; Supplemental Figure 7), which is deficient in the desaturation of 18:2 to 18:3 on ER lipids (PC, PI, and PE). We expected that PLIP1 overexpression might rescue the fad3-2 defect. As reported (Browse et al., 1993), the fad3-2 mutant had a reduced 18:3 content in ER lipids. The overexpression of PLIP1 in the fad3-2 mutant background partially reversed this phenotype by increasing the 18:3 content in ER lipids PC, PI, and PE (Figure 6E; Supplemental Figure 7F). These 18:3 acyl groups must have been derived from the chloroplast, where the FAD7/8 desaturases (Li-Beisson et al., 2013) catalyze the lipid-linked desaturation of acyl groups from 18:2 to 18:3. This increase in 18:3 in PC is paralleled by a decrease in 18:3/16:1Δ3t-PG (Supplemental Figure 7E). Taken together, these data support our hypothesis that in the PLIP1-OX lines, 18:3 increasingly moves from plastid 18:3/16:1Δ3t-PG to PC, which interferes with the desaturation of acyl groups on PC by ER desaturases FAD2 and 3 and the PC-based acyl editing process.
PLIP1 Contributes a Small Fraction of Precursors for TAG Biosynthesis during Embryogenesis
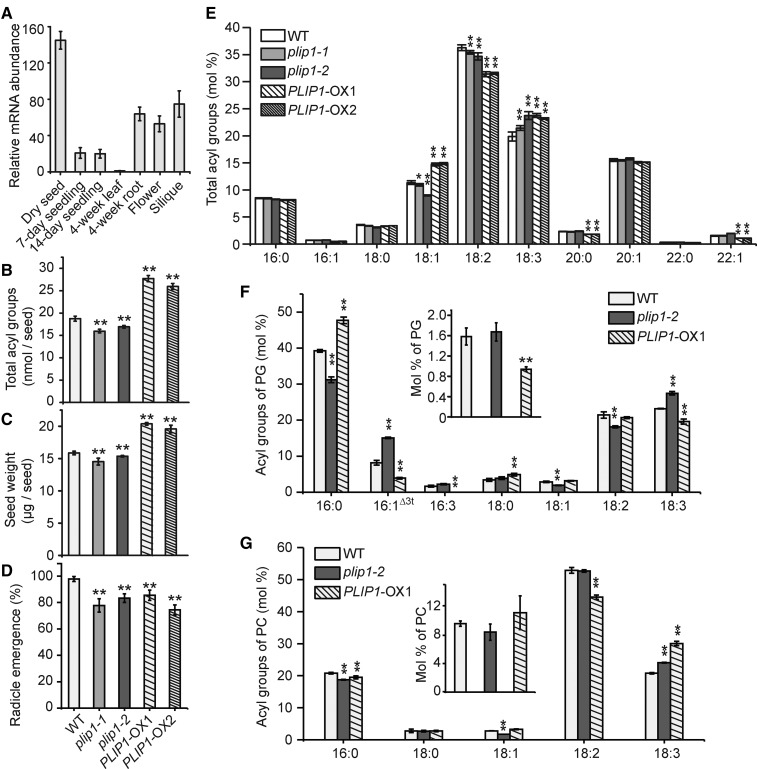
The analysis of PLIP1 thus far has focused on its biochemical function in vitro and in vivo using overexpression lines. However, these experiments can shed only limited light on the physiological functions of PLIP1. Querying the native tissue-specific and developmental expression of PLIP1, the highest gene expression was detected primarily in seeds and reproductive tissues, including flowers and siliques (Figure 7A). Since PLIP1 encodes a lipase and is highly expressed in seeds, we postulated that it might play a role in seed lipid metabolism, which is dominated by the biosynthesis of TAG. In fact, toward the end of seed development, over 90% of total acyl groups are stored in TAG (Li et al., 2006; Li-Beisson et al., 2013). To explore the physiological function of PLIP1 during embryogenesis, two independent T-DNA insertion lines were obtained (Alonso et al., 2003). The T-DNA allele corresponding to SALK_102149 was designated as plip1-1, and the second, corresponding to SALK_147687, was designated as plip1-2. The T-DNA insertions were in the 3′ and 5′ untranslated regions, respectively (Supplemental Figure 8A). Quantitative RT-PCR analysis indicated that both lines carry leaky alleles (Supplemental Figure 8B, numbers on panel). Under normal growth conditions, the two plip1 mutant alleles were physiologically indistinguishable from the wild-type plants (Supplemental Figure 8B). Lipid analysis also showed no changes in vegetative tissues (Supplemental Figures 8C to 8J). However, in dry seeds, where PLIP1 mRNA is highly abundant, the insertion lines showed an approximate 10% reduction in total seed acyl group content indicative of a decrease in TAG, while the overexpression lines had a 40 to 50% increased seed acyl group content (Figure 7B). Seed weight changes were consistent with the changes in seed TAG levels; the plip1 mutants had smaller seeds than the wild type, while the seeds of the overexpression lines were larger (Figure 7C). Compromised germination was also observed for the seeds of the two plip1 mutants (Figure 7D), a typical phenotype related to the reduction in the major seed carbon reserve, TAG. Surprisingly, seeds from the overexpression lines also had slower germination rates than the wild type, despite their elevated TAG content (Figure 7D). To investigate whether this is caused by altered seed development due to altered plant hormone levels, we analyzed the major hormones typically present in dry seeds. As shown in Supplemental Figure 9, the hormone levels of the plip1 mutant seeds were not significantly different from those of the wild-type seeds, whereas the PLIP1-OX1 seeds had elevated OPDA and JA-Ile levels (Supplemental Figure 9), both of which are effective germination inhibitors (Dave et al., 2011; Dave and Graham, 2012). Arabidopsides were not detected in seeds of any genotypes. It must be noted that mature PLIP1-OX lines had shorter and bushier inflorescences than the wild type (Supplemental Figure 10A) and that seed yield was reduced by ∼60% for the PLIP1-OX lines (Supplemental Figure 10B). Thus, overall oil yield was not elevated in the PLIP1-OX plants. Nevertheless, the phenotype of the plip1 reduced function mutant suggested that PLIP1 might play a role in TAG biosynthesis during embryogenesis.
Figure 7.
Effect of PLIP1 on Seed Oil Biosynthesis and Germination.
(A) PLIP1 transcript levels in different tissues or developmental stages determined by quantitative PCR. Expression levels were normalized to those lowest in 4-week-old leaf tissues and shown as relative fold changes. n = 3 for each tissue, ±sd.
(B) Total acyl group content in dry seeds of the wild type, plip1-1, plip1-2, PLIP1-OX1, and PLIP1-OX2. Thirty seeds were analyzed in bulk for each repeat; n = 5, ±sd.
(C) Weight of the seeds shown in (B). Two hundred seeds were analyzed for each repeat; n = 4-7, ±sd.
(D) Germination of wild-type, plip1-1, plip1-2, PLIP1-OX1, and PLIP1-OX2 seeds. The fraction of seeds showing radicle emergence was determined 40 h after stratified seeds were sown on MS medium. One hundred seeds were used for each repeat, n = 3, ±sd.
(E) Relative acyl group composition of dry seeds shown in (B). Acyl groups with a molar percentage <0.5% were omitted. Thirty seeds were analyzed in bulk for each repeat; n = 5, ±sd.
(F) and (G) content and compositional analysis of PG and PC in developing seeds isolated from wild-type, plip1-1, and PLIP1-OX1 siliques harvested 7 d after flowering. Molar percentages of PG and PC in lipids are shown in the inserted figures. Equivalent volume (100 μL) of developing seeds was used for each repeat. n = 3, ±sd. Where appropriate, Student’s t test was applied to compare wild-type values with those of plip1-2 and PLIP1-OX, respectively (*P < 0.05; **P < 0.01).
To gain more information on how the plastid-located PLIP1 contributes to TAG biosynthesis during embryogenesis, we analyzed TAG acyl groups in dry seeds. In the insertional mutants, especially in the slightly stronger plip1-2 allele, 18:3 levels increased relative to 18:1 (Figure 7E). For the two PLIP1-OX lines, 18:2 levels decreased, while 18:3 and 18:1 levels increased, a pattern that resembled the leaf PC acyl group profile in the PLIP1-OX lines (Figure 4C). This suggested that increased TAG may be derived from increased flux of acyl groups through PC in the overexpression lines. Based on the lipid analysis in vegetative tissues of the PLIP1-OX lines (Figure 4), we hypothesized that PLIP1 in developing embryos contributes to TAG biosynthesis by catalyzing the turnover of PG, increasing the flux of acyl groups into PC and ultimately TAG. To corroborate this hypothesis, we analyzed PG and PC content and composition in developing seeds isolated from siliques 7 d after flowering. Total PG content was ∼1.6% of total lipids in both the wild type and plip1-2, while developing seeds of PLIP1-OX1 contained ∼50% less PG (Figure 7F, inset). With regard to acyl composition, relative levels of 16:1Δ3t and 18:3 decreased in the PLIP1-OX1 line, while they increased in plip1-2, supporting a role for PG as the substrate of PLIP1 in developing seeds. PC accounts for ∼10% of the total lipids at this stage of developing seeds (Figure 7G, inset), and the acyl compositions of PC in different genotypes resembled that of TAG in dry seeds (Figure 7G), implying that the altered TAG acyl compositions in PLIP1-OX and plip1 mutant seeds at least partially reflect the metabolism of PC.
It has been proposed that acyl editing of PC, head group exchange, and PDAT activity represent the mechanisms for directing polyunsaturated FAs from PC into TAG during seed development (Dahlqvist et al., 2000; Bates et al., 2012). In the fad3-2 mutant, ∼15% of 18:3 was retained in total seed FAs (Supplemental Figure 11), suggesting that the proposed PLIP1-based mechanism is a relatively small but significant contributor to the incorporation of 18:3 into seed TAG compared with the PC-based FAD2/3 desaturation pathway. In fact, consistent with the observation in leaf tissues (Figure 6E), overexpressing PLIP1 in the fad3-2 background restored the 18:3 level to 36% (Supplemental Figure 11).
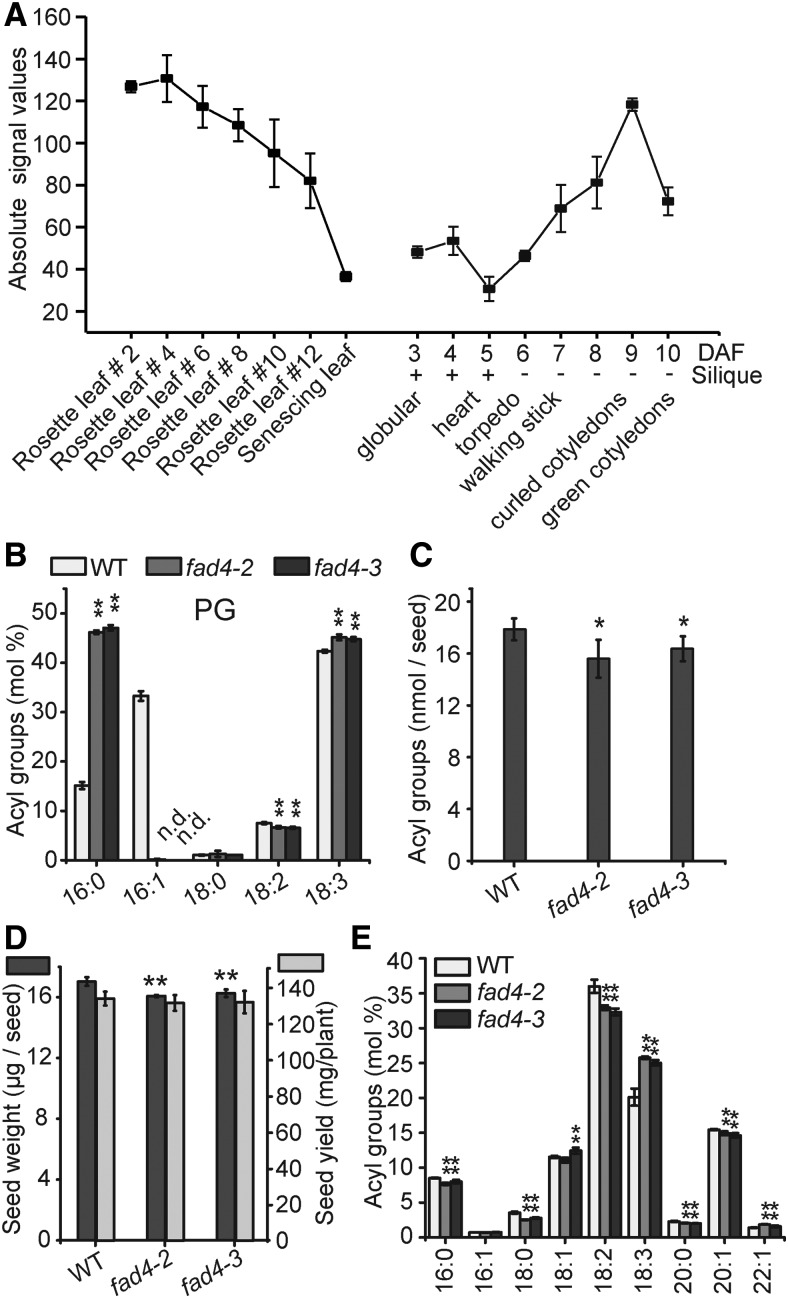
As discussed above, 16:1Δ3t-PG is likely the preferred substrate of PLIP1 and 16:1Δ3t only exists at the sn-2 position of plastid PG. FAD4 is the enzyme in Arabidopsis that specifically introduces trans-double bonds into the 16:0 acyl chain of PG (Gao et al., 2009). Transcript analysis of FAD4 during seed development showed that this gene is upregulated during embryogenesis, in a time course that correlates well with oil biosynthesis during embryogenesis (Figure 8A). It should be noted that the transcript levels of FAD4 are on the same order of magnitude in leaf and seed tissues (Winter et al., 2007) (Figure 8A), but the 16:1Δ3t-PG level varies drastically between these two tissues.
Figure 8.
Analysis of fad4-2 and fad4-3 Plants.
(A) Absolute expression levels of FAD4 in leaves and developing seeds at different stages. Data were extracted from the Arabidopsis eFB browser. Average values from triplicates are shown, and the error bars represent sd.
(B) Leaf PG acyl group composition of 3-week-old MS-medium-grown wild-type, fad4-2, and fad4-3 plants. Leaves harvested from one plant were pooled as one biological repeat; n = 3 to 4, ±sd. n.d., not detected.
(C) Total acyl group content in dry seeds of the wild type, fad4-2, and fad4-3. Thirty seeds were used for each repeat; n = 5, ±sd.
(D) Seed weight and yield measurements of wild-type, fad4-2 and fad4-3 plants. For seed weight measurement, 200 seeds were used for each repeat, n = 3, ±sd. For seed yield measurement, seeds harvested from four plants grown in the same pot were used as one repeat, n = 3, ±sd.
(E) Relative acyl group composition of seed lipid extracts from of the wild type, fad4-2, and fad4-3. 30 seeds were used for each repeat, n = 5, ±sd. Acyl groups with a molar percentage less than 0.5% were omitted. Where appropriate, Student’s t test was used to compare the wild type and each fad4 mutant (*P < 0.05; **P < 0.01).
Our hypothesis that PLIP1 contributes to embryonic TAG biosynthesis predicts that removal of 16:1Δ3t-PG should result in a similar seed phenotype to that observed for the plip1 mutants. To test this hypothesis, we characterized two FAD4 loss-of-function lines, fad4-2 and fad4-3 (Gao et al., 2009). As shown in Figure 8B, 16:1Δ3t was not detected in PG in either fad4-2 or fad4-3 leaf tissues. Similar to the plip1 mutants, fad4-2 and fad4-3 showed a nearly 10% reduction in total seed acyl group content (Figure 8C), reduced seed weight, but no altered seed yield (Figure 8D). The fad4-2 and fad4-3 mutants also had altered seed acyl group profiles, specifically reduced 18:2 and increased 18:3 content (Figure 8E), similar to the changes in plip1-2 seed acyl group composition (Figure 7E). Taken together, these observations led us to conclude that acyl groups in plastid 16:1Δ3t-PG contribute to TAG biosynthesis during embryogenesis and that this requires PLIP1 activity.
Alignment of the predicted PLIP1 sequences found in land plants (Supplemental Table 1 for species and accession numbers and Supplemental File 2 for alignment) followed by phylogenetic analysis shows that PLIP1 proteins are conserved in land plants (Supplemental Figure 12), except for the orchid family, for which genomes of two species are available in NCBI (Phalenopsis equestris and Dendrobium catentum). Interestingly, presumed FAD4 homologs are also ubiquitous among land plants, except for the two representatives of the orchid family (Supplemental File 3 for alignment and Supplemental Figure 12). Calculation of the linear correlation coefficient between the PLIP1 and FAD4 phylogenetic trees after extracting the 18S rRNA background from 26 sequenced land plant species (Pazos et al., 2005) resulted in a score of 0.971, suggesting that these two proteins are coevolving. This supports our hypothesis that PLIP1 and FAD4 function in the same pathway.
Overexpression of PLIP1 Increases PG Recycling and TAG Biosynthesis during Embryogenesis
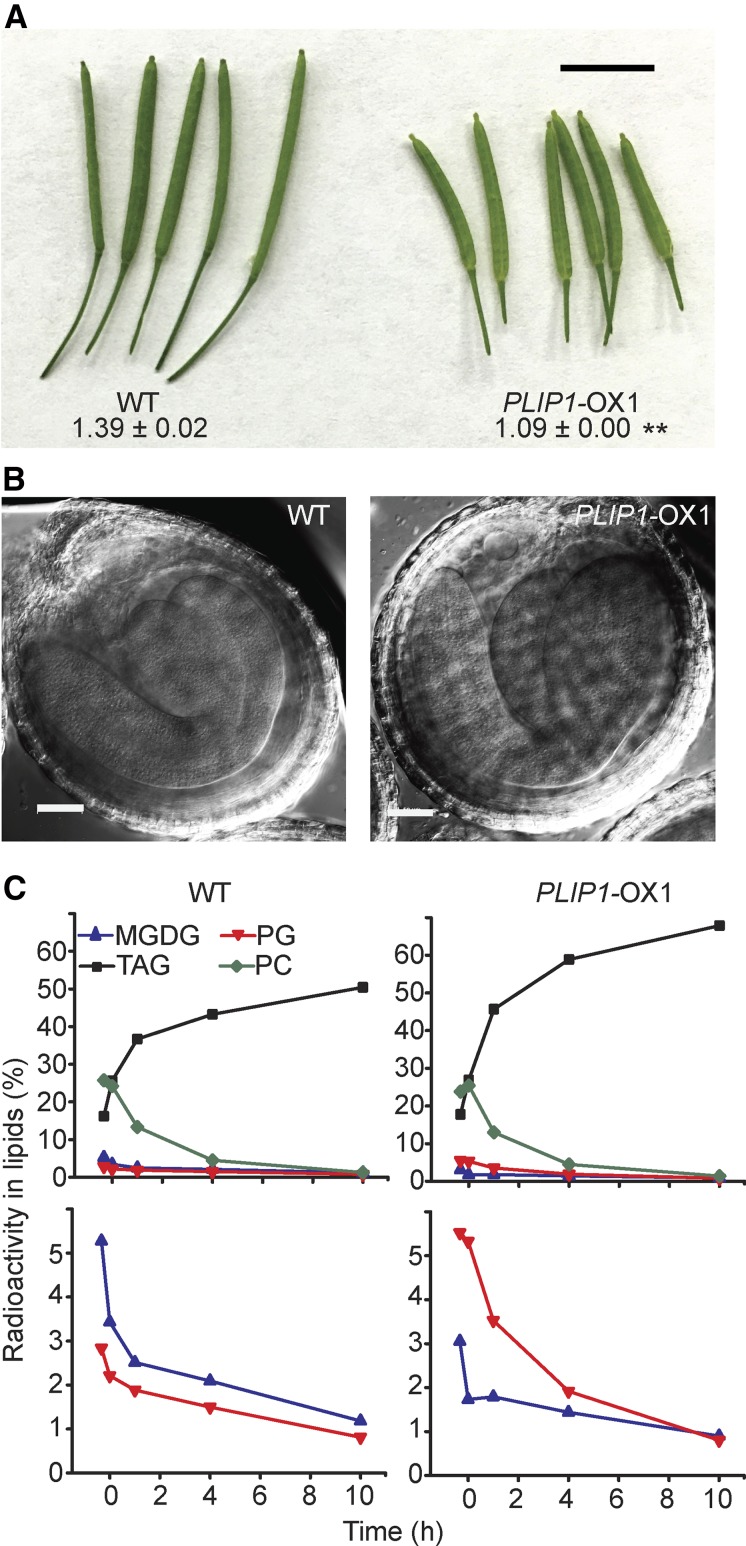
To determine whether increased turnover of plastid PG is responsible for increased TAG biosynthesis during embryogenesis in the PLIP1-OX lines, we harvested siliques at 9 d after flowering from wild-type and PLIP1-OX1 plants and isolated the developing seeds. Seeds at this developmental stage have robust lipid metabolism (Le et al., 2010; Bates et al., 2012). However, siliques of the same age collected from PLIP1-OX1 plants were shorter than those from the wild type (Figure 9A), which raised the concern that embryos from PLIP1-OX1 and the wild type might be at different developmental stages. However, upon closer examination, the wild type and PLIP1-OX1 had nearly mature embryos with fully developed cotyledons and radicals (Figure 9B), indicating they were at a similar developmental stage and likely metabolically comparable. Therefore, we performed [14C]-acetate pulse-chase labeling on the isolated developing seeds. Pulse time points were collected after 20 and 60 min and are shown before time 0 of the chase start on the x axis, followed by three chase time points; a representative example of multiple repeats is shown (Figure 9C). Compared with PC and TAG, plastid lipids PG and MGDG were not highly labeled during embryogenesis, likely due to their small pool size; therefore, an expanded view for PG is shown in the lower graph of Figure 9C. PLIP1-OX1 had higher incorporation of label into PG and increased turnover during the chase phase (Figure 9C), as was observed for the equivalent experiment done on leaves. The altered labeling patterns for PG and MGDG resembled those observed in the leaf labeling assays (Figures 6A and 6B). The most strongly labeled lipids were TAG and PC, reflecting their end-product status (TAG) or large pool size (PC) in developing seeds. However, the much smaller PG pool (mostly in the chloroplast as 16:1Δ3t-PG) seemed to be more metabolically active in PLIP1-OX1 than in the wild type. Incorporation of label into PG during the pulse under the conditions tested was faster than could be captured by the earliest sampling time points. The rate and extent of incorporation into TAG was increased in the PLIP1-OX1 line, consistent with increased total acyl group content in these seeds, while the PC pool was similarly labeled in the wild-type and overexpression lines.
Figure 9.
Phenotypic Analysis of PLIP1-OX1 Embryos.
(A) Morphology of the wild-type and PLIP1-OX1 siliques 9 d after flowering. The numbers indicate the length of siliques. n = 9 to 12, ±sd. Student’s t test was applied (**P < 0.01). Bar = 0.5 cm.
(B) Differential interference contrast images of embryos isolated from siliques of WT and PLIP1-OX1 plants. Bars = 50 µm. Representative images are shown.
(C) Pulse-chase labeling of developing embryos isolated from siliques of wild-type and PLIP1-OX1 plants. Equivalent volume (100 μL) of developing seeds were used for each time point. The first two time points represent the labeling pulse. Embryos were transferred to unlabeled medium after 1 h. Values represent the fraction of label in select individual lipids compared with label in total lipids. The top panels show four lipids, as indicated. The lower panels show PG and MGDG again, but on an expanded scale. Experiments were repeated three times on PLIP1-OX1 seeds and one time on PLIP1-OX2 seeds, with similar results. One representative result is shown.
DISCUSSION
PLIP1 Prefers PG as Natural Substrate
Recombinant lipases are notoriously difficult to produce and to study in vitro. PLIP1 is no exception, as its production in E. coli led to membrane degradation, which confirmed its general lipase activity, but made PLIP1 challenging to purify (Figure 2). Furthermore, while recombinant PLIP1 was specifically acting on the glyceryl sn-1 position, it is active on a range of polar lipids found in plants and bacteria in vitro (Figure 3C). Notably, it did not act on TAG and had very little activity on the two glycolipids found in the chloroplast, DGDG, and sulfolipid, but showed high activity toward PG and MGDG. Because it was impractical to test the full spectrum of all possible combinations of glycerolipid molecular species occurring in chloroplasts in vitro, we explored the in vivo specificity of PLIP1 by overproducing the protein in chloroplasts, which we determined was the location of PLIP1 in plant cells using multiple independent approaches (Figure 1). This allowed us to test PLIP1 lipase activity in a quasi-native environment with the caveat that PLIP1 is normally not abundant in leaf chloroplasts and is more likely active in the chloroplasts of developing embryos based on the gene’s expression profile and the reduced-function phenotype visible in seeds (Figure 7). However, leaf chloroplasts are much more readily accessible for assays than embryo chloroplasts, and we assumed that findings on PLIP1 activity would be transferable between the two tissues, which we ultimately confirmed. Based on the in vivo analysis of PLIP1, 18:3/16:1Δ3t-PG emerged as the most likely in vivo substrate for PLIP1, which was corroborated in vitro using a native, leaf-isolated molecular species mixture of PG (Figure 6D). It should be noted that in the overexpression lines, effects on MGDG and PC were observed in addition to PG (Figures 4B and 4C; Supplemental Figure 3B). This could be directly due to the activity of PLIP1 on MGDG or caused by secondary effects related to acyl exchange and acyl transfer in the case of PC, which is not in the thylakoid membranes and should not be directly accessible to PLIP1.
The Location of PLIP1 Limits the Availability of Likely Substrates in Vivo
To explain the observed substrate preference of PLIP1 in vivo, one might invoke the presence of factors in its native environment that are simply not present in vitro. Another more likely explanation might be the limited accessibility of certain lipid molecular species to PLIP1 due to its specific membrane location, assuming that specific membrane leaflets or lateral subdomains have a specific lipid composition. We have some clarity now on the location of PLIP1, its likely peripheral association with thylakoid membranes based on fractionation, chloroplast import, and protease protection experiments, and its dual processing (Figure 1). Fractionation showed that PLIP1 is associated with thylakoid membranes, while import and protease protection assays were consistent with three possible suborganellar locations for PLIP1: stroma, thylakoid, or the stroma leaflet of the inner envelope membrane (Figure 1). PLIP1 is not predicted to contain transmembrane domains, but it must be a peripheral membrane protein to gain access to its substrate. Most likely, PLIP1 is a peripheral thylakoid protein, but it also has the ability to be transiently free in the stroma or even has access to the inner envelope membrane. Double processing of PLIP1, as observed, can be interpreted as first generating an intermediate during protein import into the stroma, while the second processing possibly releases the majority of the mature protein from the thylakoid fraction into the soluble stroma fraction. A conserved twin-arginine motif is generally required for importing proteins into thylakoids (Robinson and Bolhuis, 2001; Goosens and van Dijl, 2017), but PLIP1 is missing a canonical motif, although it contains two sets of twin-arginine in its transit peptide usually part of such a motif. Therefore, PLIP1 may peripherally attach to the thylakoid membrane but is likely prevented from being further imported into the thylakoid lumen.
For the likely substrate of PLIP1, 18:3/16:1Δ3t-PG, we only know that it is exclusively present in chloroplasts, where the FAD4 desaturase required for its synthesis is located as well (Gao et al., 2009). However, the presence of 18:3/16:1Δ3t-PG in a specific suborganellar membrane, leaflet, or lateral membrane domain is not known. All we can conclude based on our localization of PLIP1 is that 18:3/16:1Δ3t-PG must be present in the stroma leaflet of the thylakoid or envelope membranes to be accessible to PLIP1.
PLIP1 Contributes to TAG Biosynthesis in Developing Embryos
PG is required for proper embryo development. The development of embryos in a pgp1 pgp2 double mutant affected in PG biosynthesis in chloroplasts, mitochondria, and the endoplasmic reticulum is delayed, and maturing seeds shrink during desiccation, resulting subsequently in compromised germination (Tanoue et al., 2014). Originally, it was proposed that the chloroplast-specific molecular lipid species, 16:1Δ3t-PG, is critical for the function of the photosynthetic membrane, but its complete replacement with 16:0-PG in the Arabidopsis fad4 mutant had only mild effects on leaf photosynthesis (Browse et al., 1985; McCourt et al., 1985). What was possibly neglected before was that the expression level of FAD4 is on the same order of magnitude in leaf and developing seeds (Figure 8A), but the level of 16:1Δ3t-PG in developing seeds was much lower than that in leaves. The discovery that PLIP1 preferentially metabolizes 16:1Δ3t-PG during seed development potentially explains the small 16:1Δ3t-PG pool and points toward roles for this lipid in seeds. In fact, decreased PLIP1 expression in T-DNA insertional lines reduced seed TAG content without affecting growth or seed hormone levels (Figure 7B), corroborating a possible contribution of PLIP1 to seed TAG biosynthesis. The plip1 low-TAG seed phenotype is recapitulated in the fad4 mutant (Figure 8C), and further phylogenetic analysis suggests coevolution of PLIP1 and FAD4 in land plants. These findings support a role for PLIP1 in seed TAG biosynthesis and provide a possible function for this unusual lipid molecular species.
It should be noted that PLIP1-OX plants produced seeds with higher TAG content, but 40% fewer seeds, compared with the wild type (Supplemental Figure 10). Overall, seed oil production per plant was reduced in line with the reduced growth of the PLIP1-OX plants, likely due to the activation of oxylipin-induced defense pathways (Figure 5; Supplemental Figure 5). During the labeling experiment with developing seeds, saturating substrate levels were provided (Figure 9C). Therefore, higher carbon incorporation into TAG in PLIP1-OX seeds reflects an increased capacity for TAG biosynthesis in individual embryos, in spite of the reduced plant growth and the decreased overall seed yield of the plants (Figure 4A; Supplemental Figure 10B).
PLIP1 Enables Channeling of Acyl Groups from Plastid 16:1Δ3t-PG to TAG at the ER
How can PLIP1, a lipase, be a component of a mechanism directing FAs synthesized in the plastid into TAG lipid droplets in the cytosol during embryogenesis? A large body of evidence suggests that PC is a critical precursor for TAG biosynthesis in developing seeds. As shown in Figure 10, several pathways, including acyl exchange (Figure 10, reaction 1) and desaturation of acyl groups on PC by FAD2/3 followed by transfer of 18:3 from the acyl-CoA pool to DAG (Figure 10, reaction 4), head group exchange generating DAG with 18:3 acyl groups (Figure 10, reaction 3), and transfer of acyl groups from PC to DAG by PDAT (Figure 10, reaction 5) contribute to the incorporation of polyunsaturated FAs into TAG during seed development (Dahlqvist et al., 2000; Bates et al., 2012; Li-Beisson et al., 2013). However, even in the rod1 lpcat1 lpcat2 triple mutant carrying the strongest known alleles at each locus, which in combination should completely disrupt acyl editing and head group exchange, the capacity of seeds to produce 18:3-containing TAGs is only reduced by half (Bates et al., 2012). Therefore, other compensatory mechanisms likely exist for incorporating polyunsaturated FAs into ER lipids and TAGs that could involve PLIP1 or PDAT. We propose that PLIP1 is part of a chloroplast acyl editing mechanism augmenting the supply of polyunsaturated FAs to the cytosolic acyl-CoA and PC pools, as depicted in Figure 10, reaction 2. This is particularly apparent whenever PLIP1 is highly abundant, as the turnover of plastid PG accelerates (Figures 6 and 9). Acyl group flux is increased from PG to PC in PLIP1 overexpression lines, and these acyl groups eventually end up in TAGs, which is evident from the pulse-chase labeling experiments (Figures 6 and 9). Partial restoration of the low 18:3-PC lipid phenotype of the fad3-2 mutant by overexpression of PLIP1 in mesophyll tissues and seeds (Figure 6E; Supplemental Figure 11) supports a role for PLIP1 in plastid acyl export. The resemblance of acyl compositions of PC and TAG in seeds is indicative of the precursor-product relationship between these two lipids (Figure 7), but the detailed mechanism needs further investigation. In vegetative and seed tissues, overexpression of PLIP1 results in a marked decrease in 18:2 and increase of 18:1 in PC, a phenotype reminiscent of the fad2 mutant. This suggests that the elevated 18:3 flux from plastids outcompetes the incorporation of unsaturated fatty acids by the FAD2/3 pathway. Consistent with this assumption, in the plip1 mutant, which lacks the PLIP1-dependent plastid acyl export pathway, one would expect that the ER-based FAD2/3 mechanism for the incorporation of unsaturated FAs into PC and TAG is more active. In fact, the decreased 18:1 and increased 18:3 contents in PC and total acyl groups (reflecting mostly TAG), as observed in the plip1 mutant seeds (Figures 7E and 7G), corroborate this prediction. The FAD2/3 desaturation pathway counteracts the reduced 18:3 export in the plip1 mutants but does not affect loss of plastid-derived acyl group flux, leading to the observed decrease in seed TAG. The reversibly compensatory activity of the PLIP1-mediated plastid export of 18:3 acyl groups and the ER-based FAD2/3 desaturation pathway explains the seemingly paradoxical observation that 18:3 contents are high in both the plip1 mutants and the PLIP1 overexpression lines.
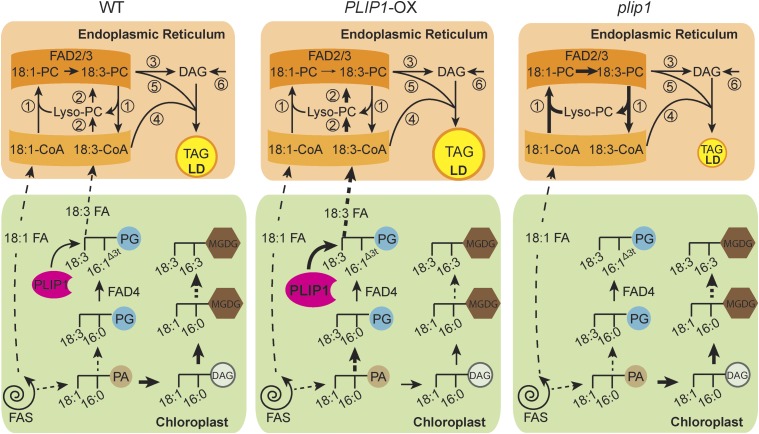
Figure 10.
Hypothesis for the Role of PLIP1 in Triacylglycerol Biosynthesis.
The left panel depicts the wild type, the middle panel the PLIP1 overexpression lines, and the right panel the plip1 mutant. The thickness of the arrows indicates the relative fluxes in the three different lines. Reactions or sets of reactions are numbered as follows: (1) In the wild type (left panel), acyl exchange on PC involving desaturation of acyl groups by FAD2/3 provides one mechanism to introduce polyunsaturated FAs into PC. (2) A parallel mechanism to introduce PUFAs into PC involves PLIP1. In the chloroplast, PLIP1 hydrolyzes 18:3/16:1Δ3t- PG and other 16:1Δ3t-phosphatidylglycerol species at the sn-1 glyceryl position and releases 18:3 (carbon:double bonds) or other acyl groups at the sn-1 position. The released acyl group is exported to the ER and incorporated into the acyl-CoA pool and PC. (3) A head group exchange mechanism leads to diacylglycerol (DAG) formation from PC containing PUFAs. (4) TAG, which accumulates in lipid droplets (LDs), is formed by the action of DAG-acyltransferases, which can introduce additional acyl groups into DAG from the acyl-CoA pool. (5) Phospholipid-DAG acyltransferase provides an additional route for the incorporation of polyunsaturated FAs from PC into TAG. (6) DAG can also be formed by de novo assembly through the Kennedy pathway, which, however, is thought to play a minor role in the synthesis of TAGs in seeds. In the chloroplast, biosynthesis of PG and MGDG shares the precursor phosphatidic acid (PA), with more PA being shuttled to MGDG biosynthesis in the wild type. In PLIP1-OX lines (middle panel), both PG biosynthesis and degradation are accelerated, resulting in increased export of 18:3 and other acyl groups and their direct incorporation into PC (reaction 2). Direct incorporation of 18:3 competes with polyunsaturated FA formation by the acyl-editing pathway of PC involving FAD2/3 (reaction 1), but leads to increased flux of 18:3 into the end product TAG. As a result of increased PG turnover in chloroplasts of PLIP1-OX lines, PA is preferably shuttled into PG biosynthesis, which subsequently reduces its availability for MGDG assembly in the plastid visible in changes in the MGDG acyl composition. In the plip1 mutant (right panel), the PLIP1-dependent pathway is deficient, resulting in decreased TAG biosynthesis. Without the competing effect of PLIP1 on the acyl exchange reactions and FAD2/3, more 18:1 is converted to 18:3, explaining the altered acyl composition of TAG and other extraplastidic lipids.
It should be noted that nearly 85% of the seed TAG 18:3 is dependent on FAD3, and the contribution of PLIP1 to seed 18:3 reflects the finding that the PLIP1-mediated pathway contributes ∼10% of the acyl groups in TAG in seeds, even though overexpression of PLIP1 is able to significantly increase 18:3 content in the fad3-2 seeds (Supplemental Figure 11). This may seem like a minor contribution. However, even the ablation of the presumed key enzyme of TAG biosynthesis, DGAT1, leads to only a 20 to 40% reduction in seed oil content (Katavic et al., 1995; Routaboul et al., 1999). Furthermore, in a PDAT loss-of-function mutant affecting a pathway parallel to DGAT (Figure 10, reactions 5 and 4, respectively), seeds show no changes in oil content nor composition (Mhaske et al., 2005), even though a double mutant deficient in these two pathways exhibits up to 80% reduction in seed oil content and a loss in seed viability. These results indicate that other parallel mechanisms exist to account for the remaining small fraction of TAG. The regulatory network governing the different TAG biosynthesis pathways is complex, allowing for redundancy and compensatory mechanisms. The PLIP1-mediated pathway described here could represent one of the alternative mechanisms for TAG biosynthesis. At this time, the lack of a PLIP1 loss-of-function mutant prevents us from more accurately quantifying the contribution of a PLIP1-dependent pathway of polyunsaturated FA incorporation into TAG under different conditions or in different loss-of-function mutants. Despite repeated attempts, we have not yet succeeded in isolating a homozygous PLIP1 loss-of-function mutant using targeted gene disruption methods. Thus, we cannot rule out that PLIP1 has additional essential functions in embryo metabolism that still need to be discovered.
PLIP1 Takes Part in Acyl Group Export from Chloroplasts
The hypothesis outlined above (Figure 10) also implies that PLIP1 activity leads to acyl export from the plastid. Assuming that PLIP1 acts at the stroma surface of the thylakoids or the inner envelope membrane, additional chloroplast proteins are likely necessary to direct acyl groups from chloroplast PG into TAGs. Recently, chalcone isomerase-like chloroplast proteins were shown to be FATTY ACID BINDING PROTEINS (FAP), which may be associated with chloroplast fatty acid export (Ngaki et al., 2012). The expression pattern of FAPs resembles that of PLIP1, and all FAPs are located in the stroma of chloroplasts. One FAP, FAP1, shows high proclivity for binding 18:3. Therefore, it seems possible that 18:3 released by PLIP1 from 18:3/16:1Δ3t-PG is bound by FAP, thereby sequestering it to avoid cytotoxicity of free FAs or to mediate FA transfer to the chloroplast envelope membrane. Another protein possibly involved is FATTY ACID EXPORT1 (FAX1), a likely acyl group or FA transporter of the inner envelope membrane of plastids (Li et al., 2015). In the fax1 mutant, of the four thylakoid lipids, PG levels are increased the most, especially 18:3/16:1Δ3t-PG, and the levels of PC are reduced. Also, TAG biosynthesis, especially its polyunsaturated FA content, correlates with the presence of FAX1 in reproductive tissues. Hence, it seems possible that PLIP1, FAP, and FAX1 work together to channel a small fraction of plastid synthesized acyl groups through 16:1Δ3t-PG into PC outside the plastid and ultimately into TAG during seed development.
Movement of de novo-synthesized lipid groups through the chloroplast membrane lipid pool has been observed in Chlamydomonas, in which PGD1 is a lipase specific for newly synthesized 18:1/18:1-MGDG, while 18:3/16:4-MGDG is resistant to its activity (Li et al., 2012). In this case, PGD1 expression is induced following N deprivation and participates in the channeling of acyl groups into TAG biosynthesis under these conditions. Although plants synthesize TAG in vegetative tissues under stress (Moellering et al., 2010), they generally produce bulk TAG in developing embryos. PLIP1 is too distantly related to PGD1 to be an ortholog, and Chlamydomonas does not contain PC, while Arabidopsis lacks 18:3/16:4-MGDG. However, both lipases point toward a common theme, the need for channeling of plastid-synthesized acyl groups through the chloroplast lipid pool prior to incorporation into extraplastidic TAGs. The specific substrate selectivity of these two lipases also partially explains the existence of unusual molecular species of chloroplast lipids, 18:3/16:4-MGDG in Chlamydomonas and 18:3/16:1Δ3t-PG in Arabidopsis and most other plants and algae. It seems likely that unusual acyl groups tag specific molecular species for specific purposes. In the case of 18:3/16:4-MGDG in Chlamydomonas, it is tagged as a structural thylakoid membrane lipid resistant to PGD1, while in the case of 18:3/16:1Δ3t-PG in Arabidopsis, it is the preferred substrate for PLIP1, leading to 18:3 acyl export, rather than having a specific function related to photosynthetic light capture and conversion as previously assumed. Chlamydomonas also contains 16:1Δ3t-PG, and genomes of plants and algae encode many more potential plastid-targeted lipases. Therefore, it is likely that acyl hydrolysis catalyzed by specific plastid lipases and their respective native substrates is a common process for maintaining photosynthetic membrane lipid homeostasis while enabling the exchange and export of acyl groups for the synthesis of extraplastidic lipids or as precursors for retrograde signaling molecules, a possibility that still begs to be explored.
METHODS
Plant Material and Growth Conditions
Experiments were performed with Arabidopsis thaliana ecotype Columbia (Col-0). Seeds of T-DNA insertion lines SALK_102149 (plip1-1) and SALK_147687 (plip1-2) were obtained from the Arabidopsis Biological Resource Center, Ohio State University. Lines overexpressing PLIP1 (or PLIP1S422A) were generated by subcloning the coding sequence of PLIP1 or PLIP1S422A (see below for their origin) into pEarleyGate 101 (YFP at the C terminus) (Earley et al., 2006), followed by introducing constructs into Col-0 plants by Agrobacterium tumefaciens-mediated floral dip (Clough and Bent, 1998). Transformed seeds were initially screened for resistance to Basta, followed by confirmation by RT-PCR. Primers used for genotyping of T-DNA insertion lines or for RT-PCR analysis of overexpression lines are given in Supplemental Table 2. Arabidopsis seeds were vernalized at 4°C in the dark for 2 d before being sown on soil and grown under 100 μmole m−2 s−1 (Sylvania FO/741/ECO) in a 16-h-light (22°C) and 8-h-dark (20°C) cycle. Alternatively, sterilized and vernalized seeds were sown onto Phytoagar plates containing 1× Murashige and Skoog (MS) growth medium (Murashige and Skoog, 1962) and 1% sucrose under 100 μmole m−2 s−1 in the same light/dark cycle at 22°C (Wang et al., 2016).
Quantitative RT-PCR
Total RNA was isolated from leaves of 4-week-old Arabidopsis plants grown on soil as previously described (Wang et al., 2016) using an RNeasy Plant Mini kit (Qiagen). Total RNA (600 ng) was used to synthesize cDNA using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed using SYBR Green PCR Core Reagents mix (Life Technologies) based on the manufacturer’s instructions. The 2−ΔΔCt calculation was used to determine the relative mRNA levels. Supplemental Table 2 lists the primers used. Reference primers were as previously described (Robinson and Bolhuis, 2001). For tissue-specific expression analysis, 100 mg dry seeds, five seedlings, roots harvested from five plants, 10 fully opened flowers, or 10 siliques harvested 9 d after flowering were pooled as one biological repeat.
Confocal Laser Scanning Microscopy
Imaging of YFP fusions was performed on leaves of 4-week-old Arabidopsis grown on soil using an Olympus FluoView 1000 confocal laser scanning microscope (Olympus) with excitation at 514 nm and emissions at 600 nm. Chlorophyll autofluorescence was visualized using excitation at 633 nm and emission at 700 nm. Images were merged and pseudocolored using Olympus FluoView 1000 confocal microscope software (Olympus).
Protein Extraction and Immunoblot Analysis
Intact chloroplasts were isolated from 4-week-old Arabidopsis plants grown on MS medium essentially according to Aronsson and Jarvis (2002) and Roston et al. (2011), followed by subfractionation into stroma and thylakoid according to Keegstra and Yousif (1986) and Roston et al. (2012) with minor modifications. In brief, isolated intact chloroplasts were pelleted and ruptured by resuspension in hypertonic solution (0.6 M sucrose in TE buffer), and the suspension was homogenized with a Dounce tissue homogenizer. After incubation on ice for 10 min, bulk thylakoid fractions were harvested by three 1500g 5-min centrifugations at 4°C. Supernatants were subjected to another 100,000g 2-h centrifugation at 4°C to remove envelope membranes, and the final supernatants were harvested as the stroma fraction. Total protein from each fraction was extracted using a Plant Total Protein Extraction Kit (Sigma-Aldrich) according to the manufacturer’s instructions, and protein was quantified using the Bio-Rad Bradford assay. Appropriate amounts of extracted organellar or total cellular protein were separated by SDS-PAGE (4–20% gradient; Bio-Rad), transferred to polyvinylidene fluoride membranes (Bio-Rad), and subjected to immunoblot analysis using primary antisera in 1:1000 to 1:5000 dilutions in TBST buffer (137 mM NaCl, 20 mM Tris base, pH 7.5, and 0.5% Tween-20). Secondary anti-rabbit or anti-chicken IgG antibodies were diluted 1:10,000. Positive immunoreactions were visualized using the horseradish peroxidase reaction with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific), and the chemiluminescent signal was captured using the ChemiDoc MP imaging system (Bio-Rad) according to the manufacturer’s instructions.
Recombinant Protein and Antiserum Production
The PLIP1 sequence was amplified from Arabidopsis wild-type cDNA (see above under RT-PCR procedure) and inserted into the pGEM-T-EASY plasmid (Promega). It was then subcloned into the pET41a plasmid through BamHI and XhoI restriction sites. The PLIP1S422A point mutation construct was generated with a Q5 site-directed mutagenesis kit (New England Biolabs). Constructs were confirmed by sequencing. Final pET41a-PLIP1 and pET41a-PLIP1S422A constructs were transformed into BL21 (DE3) Escherichia coli strains for protein production. Cultures grown in LB medium (containing 0.1% glucose) were inoculated with fresh E. coli colonies and grown to log phase (OD260 0.8) at 37°C. Protein production was then induced by adding IPTG to a final concentration of 0.2 mM, and the culture was transferred to 14°C. Cells were harvested after 1 h of induction. Cultures were harvested and sonicated to lyse cells. Supernatant was collected after centrifugation at 10,000g for 30 min and subjected to another 1-h centrifugation at 100,000g to remove the majority of membrane bound PLIP1. The supernatant was collected and used to extract and purify PLIP1 recombinant proteins using a Ni-NTA column as described (Lu and Benning, 2009), except with a modified washing buffer (50 mM Tris HCl, pH 7.5, 600 mM NaCl, and 40 mM imidazole). Purified protein was concentrated using an Amicon Ultra-15 Centrifugal Filter (Millipore UFC901024) and recovered with 1× PBS buffer. The protein was quantified using the Bio-Rad Bradford assay, aliquoted, and stored at −20°C in 30% glycerol.
Recombinant PLIP1S422A was produced in E. coli and purified with a Ni-NTA column as described above. Purified protein was separated by SDS-PAGE and the corresponding band of PLIP1S422A was isolated. Protein was recovered by immersing the gel bands into 1× PBS buffer at 4°C overnight with gentle agitation. Recovered proteins were concentrated with an Amicon Ultra-15 centrifugal filter to a final purity above 98%. Antisera were raised in rabbits by Cocalico Biologicals using their standard protocol.
Chloroplast Import Assay
The N-terminal 6×His tag and TEV cleavage site of pET41a-PLIP1 were removed using a Q5 site-directed mutagenesis kit (New England Biolabs) and the construct was confirmed by sequencing prior to use for import assays. The FtsH8 gene was used as a control. Isolation of pea chloroplasts, import assays and postimport trypsin treatment were done as previously described (Xu et al., 2005).
PLIP1 Lipase Assay
Commercial lipid substrates were purchased from Avanti Polar Lipids. For each PLIP1 lipase reaction, 60 µg lipids were used. The organic solvent was removed under an N2 stream, and the lipids were resuspended in 300 μL reaction buffer (0.1 M PBS, pH 7.4, and 4.2 mM Anzergent 3-12 [Anatrace]) and dispersed by sonication for 3 × 10 s on ice (Misonix; Sonicator 3000 with microprobe; power setting 1.5). Then, 0.5 µg protein in 20 μL 1× PBS with 30% glycerol was added to each reaction. The mixture was sonicated again for 10 s with the same parameters mentioned above and incubated at ambient temperature (∼22°C) for 1.5 h or as indicated for time courses. The linear range of the reaction was determined by plotting the substrate degradation percentage along a time course as indicated in Supplemental Figure 2. The reaction was stopped by lipid extraction, followed by lipid analysis with TLC and gas chromatography as described below.
To prepare Nicotiana benthamiana PG substrates, total lipids were isolated from 4-week-old plant leaves and resolved by polar TLC. The PG bands were isolated and lipids were recovered from silica powder by extraction with chloroform-methanol (1:1 by volume).
Lipid Analysis
Lipid extraction, TLC of polar and neutral lipids, transesterification, and gas chromatography were done as described (Wang and Benning, 2011). Polar lipids were analyzed on activated ammonium sulfate-impregnated silica gel TLC plates (TLC Silica gel 60; EMD Chemical) using a solvent system consisting of acetone, toluene, and water (91:30:7–7.5 by volume). The amount of water was adjusted according to ambient humidity (in general, 7 for summer and 7.5 for winter). This solvent system was also used for separation of lyso-lipids derived from MGDG and PG during in vitro lipase assays. For TAG quantification, lipids were resolved by TLC on DC-Fertigplatten SIL G-2 (Macherey-Nagel) using a solvent system consisting of petroleum ether, ether, and acetic acid (80:20:1 by volume). For total FA analysis of dry seeds, 3-h transesterification was conducted directly on a number of seeds as specified. Lipids were visualized on TLC plates by brief exposure to iodine vapor. To separate lyso-lipids from PC, PE, or PI, a solvent system consisting of chloroform, methanol, glacial acetic acid, and water (65:35:8:5 by volume) was used. To separate lyso-PS from PS, the running solvent consisted of chloroform, methanol, and ammonium hydroxide (28-30% NH3 in water) (65:25:5 by volume). To separate lyso-lipids from DGDG and SQDG, the running solvent contained chloroform, methanol, glacial acetic acid, and water (85:20:10:4 by volume).
Pulse-Chase Labeling
For the leaf labeling experiments, detached leaves from 4-week-old soil-grown plants were incubated in nonradioactive medium (25 mM MES-KOH, pH 5.7, and 0.01% Triton X-100) in the light (25-W incandescent bulb, ∼40 μmole m−2 s−1) at ambient temperature for 1 h. Radiolabeling was initiated by adding sodium [14C]-acetate (specific activity 100 mCi/mmol in ethanol; American Radiolabeled Chemicals) to the medium to provide 1 µCi/mL, followed by a 1-h incubation with gentle agitation. The leaves were then washed twice in nonradioactive medium prior to incubation in nonradioactive medium for another 48 h. At various time points after application of the label, samples were harvested and the metabolism was halted by immediate lipid extraction. Lipids were extracted and separated by TLC as described above, and radioactivity in each lipid fraction was analyzed using a scintillation counter (MicroBeta Trilux; Perkin-Elmer) with 3 mL of scintillator solution (4a20; Research Products International) for 1 min per sample, or using phosphorimaging (FBCS 810; Fisher Biotech) with quantification by Quantity One (V 4.6.6).
Embryo labeling experiments were done as described (Bates et al., 2012). Briefly, the newly opened flowers of 4-week-old soil-grown plants were tagged, and 9 d later, siliques were harvested for embryo isolation. For each time point, a 100 μL volume of embryos was collected from ∼50 siliques and preincubated in nonradioactive buffer (5 mM MES, pH 5.8, 0.5% sucrose, and 0.5× MS) in the light (∼40 μmole m−2 s−1) for 20 min with gentle agitation at room temperature. Labeling was initiated by removing the old medium and replacing it with the same medium containing 5 µCi sodium [14C] acetate. Pulse labeling lasted for 1 h, followed by washing and replacing with nonradioactive medium to start the chase. Samples were collected at the indicated time points, and the reaction was quenched by immediate lipid extraction as described above.
Observation of Embryo Morphology
Siliques were harvested nine days after flowering and subsequently cleared with a clearing solution (chloral hydrate:glycerol:water = 8:2:1) according to Herr (1993). Developing embryos were dissected from siliques after clearance and observed under a Nikon C2 microscope.
Hormone Extraction and Arabidopside Measurement
Hormone extraction and quantification was done according to Zeng et al. (2011) with minor modifications. Briefly, hormones were extracted from 100 mg (fresh weight) leaf tissue or 50 mg dry seeds with an extraction buffer composed of methanol:water (80:20 v/v) containing 0.1% formic acid, 0.1 g/L butylated hydroxytoluene, and 100 nM abscisic acid (ABA)-d6 as an internal standard. Hormones were analyzed by liquid chromatography/tandem mass spectrometry as described (Zeng et al., 2011). Standard curve samples were prepared by serial dilution of stock ABA, IAA, SA, JA, JA-Ile, and OPDA to reach final concentrations from 1 to 1000 nM of each hormone with a fixed concentration of ABA-d6 of 100 nM. Peak integration and calibration curve processing was performed using Masslynx software (version 4.1).
Arabidopsides were extracted following the hormone extraction procedure without adding internal standards. Samples were analyzed using a Waters Xevo G2-XS Q-TOF mass spectrometer interfaced with a Waters Acquity binary solvent manager and Waters 2777c autosampler. Ten-microliter samples were injected onto an Acquity UPLC CSH C18 column (100 × 2.1 mm; Waters) held at 40°C. A gradient of solvent A (acetonitrile/water [60:40] containing 10 mM ammonium formate and 0.1% formic acid) and solvent B (isoproponal/acetonitrile [90:10] containing 10 mM ammonium formate and 0.1% formic acid) was applied in a 10-min program with a mobile phase flow rate of 0.3 mL/minute. The elution was performed as follows: hold for 1 min after injection at 95% A/5% B, followed by a 7-min linear ramp to 0% A/100% B, hold for 1 min at 100% B, then return to 95% A/5% B and equilibrate for 1 min. LC-separated analytes were ionized by negative ion mode electrospray ionization and mass spectra were acquired using an MSe method in continuum mode over m/z 50 to 1500 to provide data under nonfragmenting and fragmenting conditions (collision energy ramp from 15–60 V). Peak areas from extracted ion chromatograms for OPDA/OPDA-MGDG ([M-H]-, m/z 847.5), OPDA/OPDA-DGDG ([M-H]-, m/z 1009.57), and OPDA/dnOPDA-MGDG ([M-H]-, m/z 819.5) were obtained using Masslynx (version 4.1) software. Identification was based on accurate mass and OPDA or dnOPDA fragment ions (m/z values are 291.19 and 263.16, respectively). The peaks of the Arabidopsides were normalized to that of 18:3/18:3-DGDG ([M-H]-, m/z 981.61).
Phylogenetic Analysis
The presumed orthologous land plant sequences of Arabidopsis PLIP1 and FAD4 were obtained by BLAST analysis against the NCBI database with default parameters. Only one sequence per plant with the highest alignment score and over 40% sequence similarity with the Arabidopsis sequence was considered a presumed ortholog. The amino acid alignment was created using MUSCLE with default settings in MEGA (version 7.0.21) software. The maximum likelihood phylogenetic trees were built using MEGA (version 7.0.21), and the bootstrap percentages were based on 1000 replicates.
The correlation coefficiency score was obtained essentially according to Pazos et al. (2005). The 18S rRNA sequences were obtained from the SILVA rRNA database (Quast et al., 2013). The tree built with 18S rRNA sequences was subtracted as background from both the FAD4 and PLIP1 trees before calculating the Pearson’s correlation coefficient. The accession numbers of the PLIP1, FAD4 orthologous sequences, and the 18S rRNA sequences are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org/) under the following accession numbers: At3g61680 for PLIP1, At2g29980 for FAD3, At4g27030 for FAD4, At5g42020 for BIP2, and At1g06430 for FTSH8.
Supplemental Data
Supplemental Figure 1. Alignment of PLIP1 with class 3 lipase protein sequences in the NCBI conserved domain database.
Supplemental Figure 2. Time course of PLIP1 lipase activity with phosphatidylcholine as substrate.
Supplemental Figure 3. Analysis of polar lipids in the leaves of 4-week-old, soil-grown PLIP1-OX, and empty vector control plants.
Supplemental Figure 4. Expression of jasmonic acid-responsive genes.
Supplemental Figure 5. Quantification of plant hormones and Arabidopsides in leaves of the wild type and PLIP-OX1.
Supplemental Figure 6. Effect of sn-2 acyl 16:1Δ3t on PLIP1 function.
Supplemental Figure 7. Lipid analysis of offspring from a cross between fad3-2 and PLIP1-OX1.
Supplemental Figure 8. Genotyping, phenotypic, and lipid analysis of the plip1 mutants.
Supplemental Figure 9. Hormone quantification in plip1 and PLIP1-OX dry seeds.
Supplemental Figure 10. Growth phenotypes of mature plip1 mutant and PLIP1-OX plants.
Supplemental Figure 11. Relative composition of total acyl groups in the wild type, fad3-2, and fad3-2 PLIP1-OX1 dry seeds.
Supplemental Figure 12. Phylogenetic trees of PLIP1 and FAD4 in land plants.
Supplemental Table 1. Sequences used in the PLIP1 and FAD4 phylogenetic analyses.
Supplemental Table 2. Primer sequences used in this study.
Supplemental File 1. ANOVA tables.
Supplemental File 2. The alignment results of PLIP1 sequences.
Supplemental File 3. The alignment results of FAD4 sequences.
Acknowledgments
We thank Danny Schnell, Michigan State University, for providing antibodies against LHCb1, Gregg Howe for providing the coi1-30 seeds, Dan Jones for providing the ABA-d6 internal standard, and Anthony Schilmiller for assisting with plant hormone and Arabidopside measurement. We also thank Rebecca Roston, University of Nebraska, Lincoln, and Barb Sears, Michigan State University, for their advice on conducting the experiments or critically reading the manuscript. This work was supported by DOE Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences Grants DE-FG02-98ER20305 and DE-FG02-91ER20021, by the DOE Great Lakes Bioenergy Research Center Cooperative Agreement DE-FC02-07ER64494, and by MSU AgBioResearch.
AUTHOR CONTRIBUTIONS
K.W. conceived the research plan, designed experiments, performed most of the experiments, analyzed data, and wrote the first draft of the manuscript. J.E.F. conducted the chloroplast import assay. A.Z. conducted confocal microscopy. H.L.H. conducted experiments and assisted K.W. C.B. conceived the research plan, analyzed data, and edited the manuscript with contributions by all authors.
Glossary
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- PG
phosphatidylglycerol
- SQDG
sulfoquinovosyldiacylglycerol
- JA
jasmonic acid
- FA
fatty acid
- ER
endoplasmic reticulum
- TAG
triacylglycerol
- PC
phosphatidylcholine
- TLC
thin-layer chromatography
- PE
phosphatidylethanolamine
- OPDA
12-oxo phytodienoic acid
- EV
empty vector
- MS
Murashige and Skoog
- PI
phosphatidylinositol
Footnotes
Current address: Department of Plant Biochemistry, Albrecht-von-Haller-Institute for Plant Sciences, Georg-August-University, Göttingen 37073, Germany.
References
- Ajjawi I., Lu Y., Savage L.J., Bell S.M., Last R.L. (2010). Large-scale reverse genetics in Arabidopsis: case studies from the Chloroplast 2010 Project. Plant Physiol. 152: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Andersson M.X., Dörmann P. (2009). Chloroplast membrane lipid biosynthesis and transport. In The Chloroplast, Sandelius A.S., Aronsson H., eds (Berlin, Heidelberg: Springer Berlin Heidelberg; ), pp. 125–158. [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220. [DOI] [PubMed] [Google Scholar]
- Bates P.D., Ohlrogge J.B., Pollard M. (2007). Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J. Biol. Chem. 282: 31206–31216. [DOI] [PubMed] [Google Scholar]
- Bates P.D., Fatihi A., Snapp A.R., Carlsson A.S., Browse J., Lu C. (2012). Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 160: 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25: 71–91. [DOI] [PubMed] [Google Scholar]
- Benning C. (2010). The anionic chloroplast membrane lipids: phosphatidylglycerol and sulfoquinovosyldiacylglycerol. In The Chloroplast: Biochemistry, Molecular Biology and Bioengineering, Rebeiz C.A., Benning C., Bohnert H., Daniell H., Green B., Hoober K., Lichtenthaler H., Portis A., Tripathy B., eds (Dordrecht, The Netherlands: Springer), pp. 171–184. [Google Scholar]
- Boudière L., Michaud M., Petroutsos D., Rébeillé F., Falconet D., Bastien O., Roy S., Finazzi G., Rolland N., Jouhet J., Block M.A., Maréchal E. (2014). Glycerolipids in photosynthesis: Composition, synthesis and trafficking. Biochim Biophys Acta. 1837: 470–480. [DOI] [PubMed] [Google Scholar]
- Brady L., Brzozowski A.M., Derewenda Z.S., Dodson E., Dodson G., Tolley S., Turkenburg J.P., Christiansen L., Huge-Jensen B., Norskov L., Thim L., Menge U. (1990). A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343: 767–770. [DOI] [PubMed] [Google Scholar]
- Browse J., McCourt P., Somerville C.R. (1985). A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science 227: 763–765. [DOI] [PubMed] [Google Scholar]
- Browse J., McConn M., James D. Jr., Miquel M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268: 16345–16351. [PubMed] [Google Scholar]
- Campos M.L., Yoshida Y., Major I.T., de Oliveira Ferreira D., Weraduwage S.M., Froehlich J.E., Johnson B.F., Kramer D.M., Jander G., Sharkey T.D., Howe G.A. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97: 6487–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Graham I.A. (2012). Oxylipin signaling: a distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 3: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Hernández M.L., He Z., Andriotis V.M., Vaistij F.E., Larson T.R., Graham I.A. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23: 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Gao J., Ajjawi I., Manoli A., Sawin A., Xu C., Froehlich J.E., Last R.L., Benning C. (2009). FATTY ACID DESATURASE4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 60: 832–839. [DOI] [PubMed] [Google Scholar]
- Goosens V.J., van Dijl J.M. (2017). Twin-arginine protein translocation. Curr. Top. Microbiol. Immunol. 404: 69–94. [DOI] [PubMed] [Google Scholar]
- Herr, J.M.J. (1993). Clearing techniques for the study of vascular plant tissues in whole structures and thick sections. In Tested Studies for Laboratory Teaching, C.A. Goldman, P.L. Hauta, M.A. O'Donnell, S.E. Andrews, and R. van der Heiden, eds (Toronto, Canada: Association for Biology Laboratory Education), pp. 63–84.
- Hurlock A.K., Roston R.L., Wang K., Benning C. (2014). Lipid trafficking in plant cells. Traffic 15: 915–932. [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V., Reed D.W., Taylor D.C., Giblin E.M., Barton D.L., Zou J., Mackenzie S.L., Covello P.S., Kunst L. (1995). Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K., Yousif A.E. (1986). Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 118: 316–325. [Google Scholar]
- Kelly A.A., Feussner I. (2016). Oil is on the agenda: Lipid turnover in higher plants. Biochim. Biophys. Acta 1861: 1253–1268. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Endo K., Wada H. (2016). Roles of lipids in photosynthesis. Subcell. Biochem. 86: 21–49. [DOI] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Gügel I.L., Giavalisco P., Zeisler V., Schreiber L., Soll J., Philippar K. (2015). FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 13: e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Moellering E.R., Liu B., Johnny C., Fedewa M., Sears B.B., Kuo M.H., Benning C. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Benning C. (2009). A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284: 17420–17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Savage L.J., Ajjawi I., Imre K.M., Yoder D.W., Benning C., Dellapenna D., Ohlrogge J.B., Osteryoung K.W., Weber A.P., Wilkerson C.G., Last R.L. (2008). New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol. 146: 1482–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P., Browse J., Watson J., Arntzen C.J., Somerville C.R. (1985). Analysis of photosynthetic antenna function in a mutant of Arabidopsis thaliana (L.) lacking trans-hexadecenoic acid. Plant Physiol. 78: 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaske V., Beldjilali K., Ohlrogge J., Pollard M. (2005). Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol. Biochem. 43: 413–417. [DOI] [PubMed] [Google Scholar]
- Moellering E.R., Muthan B., Benning C. (2010). Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330: 226–228. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Ngaki M.N., Louie G.V., Philippe R.N., Manning G., Pojer F., Bowman M.E., Li L., Larsen E., Wurtele E.S., Noel J.P. (2012). Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485: 530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F., Ranea J.A., Juan D., Sternberg M.J. (2005). Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J. Mol. Biol. 352: 1002–1015. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C., Zimmerli C., Farmer E.E., Reymond P., Poirier Y. (2008). Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 147: 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond G.S., Smith T.K. (2011). Phospholipases A1. Int. J. Mol. Sci. 12: 588–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Bolhuis A. (2001). Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2: 350–356. [DOI] [PubMed] [Google Scholar]
- Rodrigues R.A., Silva-Filho M.C., Cline K. (2011). FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J. 65: 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roston R., Gao J., Xu C., Benning C. (2011). Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J. 66: 759–769. [DOI] [PubMed] [Google Scholar]
- Roston R.L., Gao J., Murcha M.W., Whelan J., Benning C. (2012). TGD1, -2, and -3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J. Biol. Chem. 287: 21406–21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul J.M., Benning C., Bechtold N., Caboche M., Lepiniec L. (1999). The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol. Biochem. 37: 831–840. [DOI] [PubMed] [Google Scholar]
- Scherer G.F., Ryu S.B., Wang X., Matos A.R., Heitz T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15: 693–700. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue R., Kobayashi M., Katayama K., Nagata N., Wada H. (2014). Phosphatidylglycerol biosynthesis is required for the development of embryos and normal membrane structures of chloroplasts and mitochondria in Arabidopsis. FEBS Lett. 588: 1680–1685. [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce M.A., Cao X., Yang Z., Ohlrogge J.B. (2013). Lipid turnover during senescence. Plant Sci. 205-206: 13–19. [DOI] [PubMed] [Google Scholar]
- Wang G., Ryu S., Wang X. (2012). Plant phospholipases: an overview. Methods Mol. Biol. 861: 123–137. [DOI] [PubMed] [Google Scholar]
- Wang K., Hersh H.L., Benning C. (2016). SENSITIVE TO FREEZING2 aides in resilience to salt and drought in freezing-sensitive tomato. Plant Physiol. 172: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7: 329–336. [DOI] [PubMed] [Google Scholar]
- Wang Z., Benning C. (2011). Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 49: 2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F.K., D’Arcy A., Hunziker W. (1990). Structure of human pancreatic lipase. Nature 343: 771–774. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu C., Fan J., Cornish A.J., Benning C. (2008). Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20: 2190–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Froehlich J.E., Awai K., Benning C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Brutus A., Kremer J.M., Withers J.C., Gao X., Jones A.D., He S.Y. (2011). A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]