Abstract
The vulval development of Caenorhabditis elegans provides an opportunity to investigate genetic networks that control gene expression during organogenesis. During the fourth larval stage (L4), seven vulval cell types are produced, each of which executes a distinct gene expression program. We analyze how the expression of cell-type-specific genes is regulated. Ras and Wnt signaling pathways play major roles in generating the spatial pattern of cell types and regulate gene expression through a network of transcription factors. One transcription factor (lin-29) primarily controls the temporal expression pattern. Other transcription factors (lin-11, cog-1, and egl-38) act in combination to control cell-type-specific gene expression. The complexity of the network arises in part because of the dynamic nature of gene expression, in part because of the presence of seven cell types, and also because there are multiple regulatory paths for gene expression within each cell type.
Keywords: organogenesis, signaling pathways, transcription
Developmental events are driven by spatially and temporally regulated gene expression. Understanding how complex patterns of expression are produced is therefore a critical part of deciphering mechanisms of development. In general, intercellular signaling mechanisms interact with a network of transcription factors to generate cell-type-specific patterns of gene expression. The late stage of Caenorhabditis elegans vulval development offers a useful model in which to study this process. During this period of vulval development, seven distinct cell types are produced that express unique combinations of genes. Over the last several years, a number of genes were discovered that are expressed in cell-type and stage-specific patterns in the vulva, and several transcription factors were found to regulate these genes. In this paper, we synthesize and extend our current knowledge of this genetic network.
The C. elegans vulva connects the uterine lumen to the outside, allowing for passage of sperm and fertilized eggs (1). Vulval cells are generated postembryonically from precursor cells P3.p P4.p, P5.p, P6.p, P7.p, and P8.p [also called vulval precursor cells (VPC)]. During the mid-third larval (L3) stage, EGF and Notch signaling induces the middle three VPCs (P5.p, P6.p, and P7.p) to adopt vulval fates, whereas P3.p, P4.p, and P8.p fuse with the hypodermal syncytium, hyp7 (2–6).
During the late-L3 to the early-L4 stage, P5.p, P6.p, and P7.p undergo two or three rounds of cell division to produce 22 nuclei (7) (Fig. 1A). These nuclei are in cells of seven types (vulA, vulB1, vulB2, vulC, vulD, vulE, and vulF), as evidenced by subsequent morphogenetic movements and by the pattern of gene expression (8, 9) (Fig. 1B). The seven cell types that are present in the adult vulva represent specializations within the general epithelial cell class. These cells exhibit cell-type general features; for example, each expresses ajm-1, a component of the apical junction that connects neighboring cells in epithelial tissues (8). However, in addition, each cell type exhibits functional specializations: vulF cells, which form the innermost section of the vulva, connect directly with cells of the uterus. vulE cells form structural attachments to lateral hypodermal (seam) cells. vulC and vulD cells attach to vulval muscles that open the vulva for the passage of eggs. vulA cells form attachment to the hyp7 syncytium. It is expected that gene expression differences underlie these specializations.
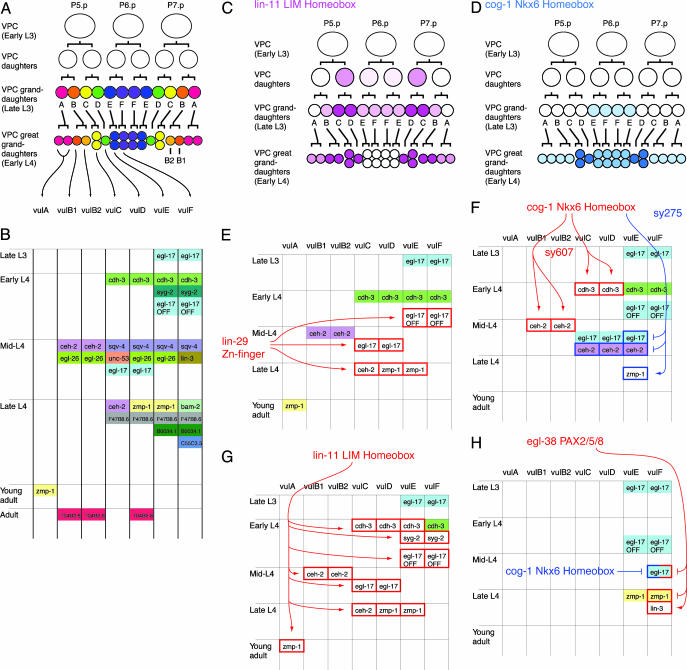
Fig. 1.
The pattern of gene expression during late stages of vulval development. (A) An overview of vulval development. Lineal origins of 22 vulval nuclei are indicated. “ABCDEFFEDCBA” refer to vulval cell types vulA, vulB1, vulB2, vulC, vulD, vulE, and vulF. vulB is the only case in which a single VPC granddaughter gives rise to two cell types (8). Vulval cell nuclei at each stage are positioned as indicated (left, anterior; right, posterior). (B) A summary of cell-type specificity and timing of expression in the wild type (Materials and Methods) (9, 11, 13–21, 24–26). Boxes indicate stages at which gene expression is activated. The vertical order of events within each time block is arbitrary. For egl-17, vulE/vulF expression begins in P6.p (early L3) and persists in their descendants (vulE and vulF) until turned off in the early L4 stage. This inactivation, which is regulated by lin-29 and lin-11, is indicated by the box marked “egl-17 OFF.” ceh-2 is expressed at a higher level in vulB1 compared with vulB2. (C) Expression pattern of lin-11. The diagrammed pattern is based on the lin-11::gfp transgene syIs80 (26). (D) Expression pattern of cog-1. The pattern is based on the cog-1::gfp transgene syIs63 (13). (E) The altered pattern of gene expression in lin-29 mutants (9, 23) (Tables 1 and 2). White boxes with the red outlines indicate loss of expression and loss of egl-17 down-regulation in the lin-29 mutant. lin-29 appears to regulate events that occur during the mid-L4 to the late L4 stage. (F) The altered pattern of gene expression in cog-1 mutants (Table 3 and Fig. 2). Arrows are drawn with the assumption that both sy607 and sy275 phenotypes are caused by different reduction of function of the cog-1 gene. Filled boxes with red or blue outline indicate ectopic expression. (G) Altered pattern of gene expression in lin-11 mutants (26). (H) Altered pattern of gene expression in the egl-38 mutant (16) (Tables 1 and 2). egl-17 expression in vulF is observed in the cog-1(sy275); egl-38 double mutant, suggesting a redundant repression mechanism.
Here, we are concerned with the execution of cell-type-specific gene expression programs during the late L3 and L4 stages, mostly after the terminal division of vulval cells. During this period, each cell type exhibits a cell-type-specific pattern of gene expression, and several transcription factors are known that regulate the expression of these cell-type-specific genes. We bring together our current knowledge of this system to produce the framework in which to investigate the gene regulatory network controlling vulval organogenesis.
Materials and Methods
Determination of Gene Expression Patterns. Essentially all gene expression analyses described in this paper (including data from other papers) were carried out by using gfp reporter transgenes. For all results, it is possible that reporter expression does not accurately reflect the expression pattern of the endogenous gene. For simplicity, we refer to the reporter by the corresponding gene name.
The expression pattern of C55C3.5 was determined by using gfp reporter clone pUL#G221N (I. Hope, personal communication). This plasmid was injected into unc-119(ed4) animals by using the plasmid pDP#MM016B [unc-119(+)] as a coinjection marker (10). Of genes listed in Fig. 1B and in the main text, we have not examined the expression pattern of syg-2, bam-2, and sqv-4. Because GFP is likely to be stable for many hours, the time at which expression is turned off is not reliably indicated by decreased GFP expression. For most genes we analyzed, GFP fluorescence persists into the adult stage.
Genotypes. For Tables 1, 2, 3, gfp reporter transgenes used were ayIs4[egl-17::gfp], syIs50[cdh-3::gfp], syIs49[zmp-1::gfp], and syIs54[ceh-2::gfp] (9). The egl-26::gfp transgenic line analyzed was kuIs36 (11). Mutations used are; cog-1(sy275), cog-1(sy607), lin-29(sy292), lin-11(n389), and egl-38(n578). Of two cog-1 transcripts, the longer cog-1A transcript contains a corepressorbinding domain, whereas the shorter cog-1B transcript does not (12). sy275 is a missense mutation predicted to affect both transcripts. sy607 is a deletion that eliminates the cog-1A transcript. The two alleles exhibit complementary defects in vulval development (13). Although both alleles are recessive, it is not known whether the loss of cog-1 function causes observed phenotypes. lin-29(sy292) and lin-11(n389) are strong loss-of-function alleles, and egl-38(n578) is a reduction-of-function allele. Strains were constructed by using standard methods.
Table 1. Expression of zmp-1 in vulE and vulF cells.
Table 2. Expression of egl-17 in vulE and vulF cells (L4).
Table 3. Expression of egl-17, ceh-2, and cdh-3 in cog-1 mutants.
| Reporter | Mutations | vulA | vulB1 and vulB2 | vulC | vulD | vulE | vulF |
|---|---|---|---|---|---|---|---|
| egl-17 | + | 0 | 0 | 100 | 100 | 0 | 0 |
| egl-17 | cog-1 (sy275) | 0 | 0 | 100 | 92 | 92 | 0 |
| egl-17 | cog-1 (sy607) | 0 | 0 | 93 | 100 | 0 | 0 |
| ceh-2 | + | 0 | 100 | 0 | 0 | 0 | 0 |
| ceh-2 | cog-1 (sy275) | 20 | 90 | 80 | 80 | 88 | 0 |
| ceh-2 | cog-1 (sy607) | 0 | 0 | 0 | 0 | 0 | 0 |
| cdh-3 | + | 0 | 0 | 100 | 100 | 100 | 100 |
| cdh-3 | cog-1 (sy275) | 0 | 0 | 100 | 100 | 100 | 100 |
| cdh-3 | cog-1 (sy607) | 0 | 0 | 14 | 14 | 71 | 94 |
Percentages of cells in mid-L4 animals that expressed egl-17::gfp, ceh-2::gfp and cdh-3::gfp. See Table 4, which is published as supporting information on the PNAS web site, for number of cells scored.
Results and Discussion
Vulval Cell-Type-Specific Gene Expression. A number of genes are expressed in specific subsets of vulval cells (Fig. 1B). Previously described genes of this type include cdh-3 (14), egl-17 (15), lin-3 (16), zmp-1 (9, 17), ceh-2 (9), T04B2.6 (9), F47B8.6 (9), B0034.1 (9), unc-53 (18), egl-26 (11), sqv-4 (19), bam-2 (20), and syg-2 (21). egl-26 was previously reported to express in vulE and vulB2 cells (11). We found that a nuclear-localized egl-26::gfp transcriptional fusion expressed in vulB1, vulB2, vulD, and vulE cells (Materials and Methods). The expression was somewhat variable and was observed starting from the mid-L4 stage and continuing into the adult stage. The C55C3.5 gene encoding a novel protein was previously found to express in vulval cells (I. Hope, personal communication). We found that C55C3.5::gfp was expressed in vulF cells, starting from the late-L4 and continuing into the adult stage.
Several conclusions can be drawn from Fig. 1B. First, all seven cell types exhibit distinct programs of gene expression, despite the fact that these cells are related by cell lineage and function. [vulB1 and vulB2 differ in the level of ceh-2 expression but otherwise have similar expression profiles (9)]. Distinct expression profiles likely underlie distinct functions of vulval cell types. For example, lin-3, which encodes an EGF-related signaling protein, is expressed in vulF cells in the mid-L4 stage (16). This signal is required for a vulva-to-uterus signaling that induces a specific fate, uv1, in uterine cells adjacent to vulF.
The pattern of marker expression also reveals a strict temporal regulation of gene expression (Fig. 1B). For example, cdh-3 is expressed in early L4, F47B8.6 is expressed in late L4, and T04B2.6 is expressed ≈1 day after the L4-to-adult molt (9). For egl-17, ceh-2, zmp-1, and sqv-4, the timing of gene expression is different for different vulval cells (9, 15, 19). For example, egl-17 is expressed in vulE and vulF cells in the L3 stage and in vulC and vulD in the L4 stage.
Trans-Regulation of Vulva Gene Expression. The analysis of the regulatory network controlling the pattern of gene expression in the vulva has focused primarily on the effect of transcription factor mutations on gene expression reporter transgenes. In most cases, a direct transcriptional regulation of the target has not been demonstrated. Key results are summarized in Fig. 1 E–H. So far, important regulators are lin-29 (encoding Zn-finger transcription factor; Fig. 1E) (9, 22, 23), cog-1 (Nkx6 homeodomain; Fig. 1 D and F) (13), lin-11 (LIM homeodomain; Fig. 1 C and G) (24–26), and egl-38 (PAX 2/5/8; Fig. 1H) (16, 27).
A Temporal Regulator of Gene Expression. lin-29 is required for the expression of egl-17 in vulC and vulD (23), ceh-2 in vulC (9), and zmp-1 in vulD and vulE (Fig. 1E, Tables 1 and 2, and Fig. 5, which is published as supporting information on the PNAS web site) (9). By contrast, lin-29 in not required for the expression of cdh-3 in vulC, vulD, vulE, vulF (9), ceh-2 in vulB (9), egl-17 in vulE and vulF (23), and zmp-1 in vulA (9). Moreover, the expression of egl-17 in vulE and vulF is observed during the L4 stage (23), suggesting that the mechanism that turns off egl-17 expression in these cells is compromised (Fig. 5). These lin-29 phenotypes are not easily explained by cell fate changes between vulval cell types but suggest a temporal regulatory defect: lin-29 mutations cause loss of events associated with the mid-to-late L4 time points. This interpretation of these data is particularly attractive, because lin-29 mutations are known to cause heterochronic defects in other tissues, specifically in the L4-to-adult transition in the lateral hypodermis (22, 28, 29). lin-29 is expressed in all vulval cells, starting in the mid-L3 stage and continuing through the L4 stage (30).
Cell-Type-Specific Regulators of Gene Expression. We analyzed the effect of two cog-1 (Nkx6.1/6.2 homeodomain) mutations on the expression of vulval-cell-specific gene expression reporters (Fig. 1F, Table 3, and Materials and Methods). cog-1(sy275) is a missense mutation in the homeodomain, and cog-1(sy607) is a small deletion that eliminates one of two cog-1 transcripts (13). We found that in the mid-L4 stage, cog-1(sy275) caused ectopic expression of egl-17 in vulE cells (Fig. 2) and ectopic expression of ceh-2 in vulC, vulD, and vulE cells and loss of zmp-1 expression in vulE cells. In contrast, cog-1(sy607) caused loss of cdh-3 expression in vulC, vulD, and vulE cells and loss of ceh-2 expression in vulB. These results indicate that egl-17, cdh-3, ceh-2, and zmp-1 are regulated by the cog-1 gene. Although some cog-1 expression is observed in all vulval cells, gfp reporters suggest that cog-1 is most strongly expressed in vulC and vulD and weakly in vulE and vulF, implying a cell-type-specific function (13) (Fig. 1D).
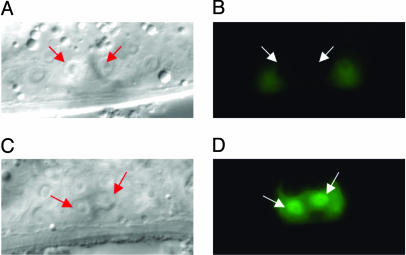
Fig. 2.
Regulation of egl-17 by cog-1. (A and B) Nomarski and epifluorescence images of wild-type mid-L4 animal carrying the egl-17::gfp transgene. Arrows point to vulE nuclei. vulE cells are not fluorescent. (C and D) cog-1(sy275) animals at the same stage carrying the egl-17::gfp transgene. vulE cells are fluorescent.
A somewhat similar situation is presented with lin-11 (LIM-homeodomain) (Fig. 1 C and G). During the L4 stage, lin-11 is expressed strongly in vulB, vulC, and vulD and weakly in other vulval cells, suggesting that lin-11 is involved in the specification of these cell types (24, 26). However, unexpectedly, lin-11 is cell-autonomously required for expression of most vulval genes tested, including in cells where the lin-11 level is low (26).
egl-38 is a PAX2/5/8 transcription factor required for expression of the lin-3 gene in vulF cells (16, 27). We found that egl-38 represses expression of zmp-1 in vulF cells, indicated by ectopic zmp-1 expression in egl-38 mutants (Fig. 1G and Table 2). In addition, in an egl-38; cog-1 double mutant, egl-17 is expressed in both vulE and vulF cells. Thus, egl-38 is also capable of repressing egl-17 expression in vulF cells, although in the wild type, this function is redundant with the cog-1-dependent mechanism that restricts egl-17 expression to vulC and vulD. egl-38 is currently the best example of cell-type-specific factors, promoting expression of some genes (lin-3) and repressing expression of others (zmp-1, egl-17) in a single cell type, vulF.
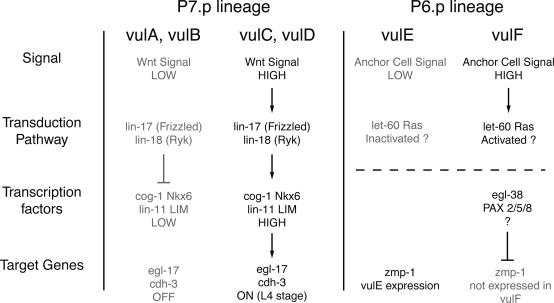
Regulators of the Transcription Factor Network. The transcription factor network that regulates gene expression in individual cell types must be regulated by the cell-fate-patterning mechanism that specifies each cell to a specific fate and does so in a spatially precise pattern. In the vulva, the cell types occur in a specific ABCD-EFFE-DCBA pattern (Fig. 1 A). Although the full mechanism that establishes this pattern is not known, Wnt signals, mediated by lin-17 (Frizzled-type Wnt receptor) and lin-18 (Ryk-type Wnt receptor), control the anterior/posterior order of cell types among P7.p descendants (31, 32) (Fig. 3). Analysis of cog-1 (31) and lin-11 (25) expression in lin-17 and lin-18 mutants indicates that Wnt signaling establishes the correct spatial pattern of transcription factor expression. As described above (Tables 1 and 3 and Fig. 1 F and G) (26), cog-1 and lin-11, in turn, control the expression pattern of egl-17 and cdh-3. Patterns of egl-17 and cdh-3 expression observed in lin-17 and lin-18 mutants are consistent with high levels of cog-1 and lin-11 turning on the expression of these genes (31, 32). Another set of cell-fate-patterning mechanisms controlling gene expression was revealed by the analysis of vulE vs. vulF fate specification using the zmp-1 reporter. A dominant-negative Ras or the ablation of the anchor cell disrupts the pattern of zmp-1 expression in presumptive vulE and vulF cells, indicating that a Ras-mediated signal, probably from the anchor cell, establishes the spatial pattern of cell fates (17).
Fig. 3.
Link between cell fate patterning mechanisms and gene expression. In general, inductive signals regulate transcription factor networks to regulate gene expression. In the P7.p (but not P5.p) lineage, Wnt signals transduced by lin-17 and lin-18 control the pattern of cog-1 and lin-11 expression (25, 31). cog-1 and lin-11 in turn regulate egl-17 and cdh-3 expression (Table 3) (26). It has not been determined whether cog-1 and lin-11 regulate each other. In the P6.p lineage, an anchor cell signal and a let-60 Ras signal transduction pathway are required to establish the correct pattern of zmp-1 expression pattern (17). zmp-1 expression is also repressed in vulF by egl-38 PAX2/5/8 (Table 1). It is not known whether the patterning mechanism acts through egl-38. The expression pattern of egl-38 is also not known.
These results confirm that cell–cell communication is important in patterning cell fates, and that signaling pathways operate through the transcription factor network to control the pattern of gene expression. Expression patterns of various genes (Fig. 1 B–D) suggest that transcription factors are expressed in all vulval cells at different levels, whereas genes regulated by them have relatively simple on/off patterns of expression. This difference suggests that the spatial pattern becomes progressively more refined as the information is passed through the regulatory network. This progressive refinement of pattern is likely a consequence of integration of information from multiple regulatory mechanisms, such as intercellular communication and feedback regulation. Many of these disparate data inputs are likely processed at the level of cis-regulatory modules. Thus, the spatial pattern of transcription factor effects becomes more restricted than the spatial pattern of transcription factor expression. This hypothesis is consistent with the observation that cells affected by lin-11 and cog-1 mutations do not correspond directly to cells that express high levels of lin-11 and cog-1.
cis-Regulation of Vulva Gene Expression. cis-regulatory elements (e.g., enhancers) have been analyzed in detail for several genes expressed in the vulva, most notably egl-17, cdh-3, and zmp-1, using transgenic assays (33, 34). A comparative genomics analysis of the regulatory region of orthologs from C. elegans and Caenorhabditis briggsae has also proved useful.
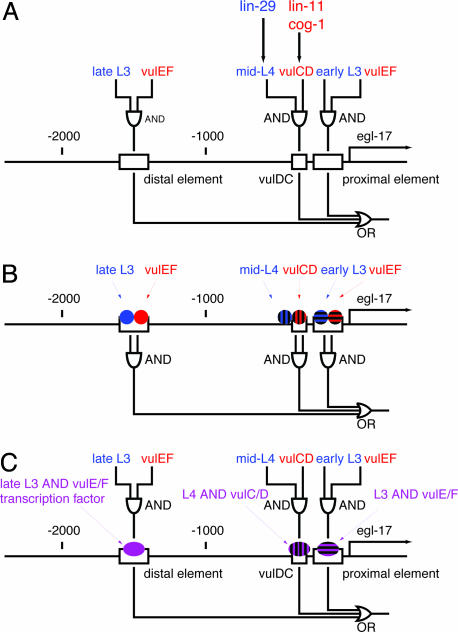
Here, we focus on the analysis of the egl-17 gene. As shown in Fig. 1B, this gene is expressed in vulE and vulF cells during the L3 stage and in vulC and vulD cells during the L4 stage. Dissection of the 5′ regulatory region revealed that there are three separable enhancer elements, two driving expression in vulE and vulF and one driving expression in vulC and vulD (33, 34) (Fig. 4A). Notably, each of these elements drives expression at different times. The distal vulE/vulF element drives expression in the mid-L3 to early-L4 and the proximal vulE/vulF element drives expression in the early to mid-L3 stage (34). The vulDC element drives expression in the mid-L4 stage. Thus, the expression of egl-17 is produced by the composite activity of three discrete enhancers, each of which drives both spatially and temporally restricted pattern of expression. We propose two models for how the information that operates on these enhancers is integrated. In one model (Fig. 4B), temporal (blue) and spatial (red) regulators both bind directly to the egl-17 promoter, and information integration is achieved directly on the cis-regulatory element. Alternatively, transcription factors that bind to each of these promoters may already combine temporal and spatial information (Fig. 4C). Our results indicate that the vulDC element regulating mid-L4 expression is likely regulated by lin-29, lin-11, cog-1, and egl-38. Additional experiments are necessary to determine the molecular mechanism of information integration.
Fig. 4.
cis-regulatory elements of egl-17.(A) A map of the egl-17 5′ regulatory region. Boxes indicate enhancer elements defined by Cui and Han (34) and Kirouac and Sternberg (33). “AND” and “OR” logic gate symbols indicate sites and logic of information integration. Temporal (blue) and spatial (red) information is integrated as indicated by the logic circuit diagram to produce the complete egl-17 expression pattern. In one model (B), spatially and temporally regulated transcription factors each bind directly to the egl-17 cis-regulatory region. The integration of information takes place on enhancer elements. In the alternative model (C), spatial and temporal cues are integrated at the transcription factor level. These transcription factors (purple) with both spatially and temporally restricted activity regulate each enhancer element.
Conclusion
The late vulval development of C. elegans offers an excellent system in which to investigate cell fate determination and regulation of cell-type-specific gene expression. In particular, this system combines single-cell resolution with a high degree of temporal resolution in an easily manipulated model organism. In many respects, vulval development is reminiscent of other systems in that transcription factors are expressed in overlapping domains, and the identity of each domain is established combinatorially by the presence or absence of specific subsets of these transcription factors. One interesting example with possible parallels to the vulva is the fate-specification mechanism in the vertebrate ventral neural tube (35). In this system, Nkx6.1 and Nkx6.2 homeodomain proteins (homologs of cog-1) interact with transcription factors Dbx1 and Dbx2 in a mutually repressive network, and different activities of repressor proteins help establish the spatial pattern of cell fates (36, 37). It is possible that C. elegans cog-1 functions in a similar manner in the vulva.
Analysis of vulval development also highlights several features that are not necessarily evident in other systems. First, analysis of vulval development has revealed a highly complex pattern of temporal regulation, which is undoubtedly a feature of most organogenetic processes (for example, see refs. 38 and 39). The involvement of lin-29, a known regulator of stage-specific development in C. elegans, suggests that the global mechanism of temporal regulation feeds into the development of this particular organ. Additional mechanisms probably exist that control expression at other time points. Whether these other time points are regulated by a global mechanism or in an organ-autonomous manner is not yet clear.
One concept that has been invoked in analyses of cell or organ fate specification is that of ground state and selector genes. For example, in Drosophila appendage development, it has been proposed that a default “ground state” exists and is modified by “selector” genes to produce an antenna or a leg (40). The concept can be applied to the level of individual cell types as well (for example, ref. 41). From this point of view, the cell-type-specific transcription factors cog-1, lin-11, and egl-38 can be thought of as selector genes for subsets of vulval cell types. What is the ground state of vulval cells in the absence of selector genes? A cell in such a state presumably will not express the cell-type-specific genes described in Fig. 1 but will retain the epithelial identity common to all vulval cells. It is unclear whether such a state has been observed in any of the mutants. Vulval cells in lin-11 mutants lack most cell-type-specific expression but retain the ability to undergo some morphogenetic movements characteristic of vulval cells and thus may most closely resemble the ground state.
In other systems, analyses of coregulated genes have successfully identified “gene batteries” (42), sets of genes with common cis-regulatory elements that are coexpressed (for example, ref. 43). However, our understanding of vulval development is still limited, relative to the number of cell types and the number of distinct stages that require different gene expression patterns. Consequently, within the relatively small number of functionally unrelated genes analyzed so far, genes are more likely to be regulated by distinct mechanisms. Thus, although gene batteries with multiple genes probably exist in this system, their analysis requires knowledge of more genes and a detailed understanding of which transcription factors regulate their expression.
Supplementary Material
Acknowledgments
We thank A. Fire (Stanford University, Stanford, CA) for providing GFP vectors, and I. Hope (University of Leeds, Leeds, U.K.) for providing the C55C3.5::gfp construct. We thank Ryan Baugh, Jennifer Sanders, Mihoko Kato, Steven Kuntz, Alok Saldanha, and reviewers for comments on the manuscript. We thank the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis) for C. elegans strains. T.I. was supported by fellowship DRG-1646 from the Damon Runyon Cancer Research Foundation. Research was supported by the Howard Hughes Medical Institute, with which P.W.S. is an investigator.
Author contributions: T.I., M.W., and P.W.S. designed research; T.I., M.W., T.O.R., and J.S.F. performed research; T.I. and M.W. contributed new reagents/analytic tools; T.I., M.W., T.O.R., J.S.F., and P.W.S. analyzed data; and T.I. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VPC, vulval precursor cell; Ln stage, larval n stage.
References
- 1.Wood, W. B. (1988) The Nematode Caenorhabditis elegans (Cold Spring Harbor Lab. Press, Plainview, NY).
- 2.Kimble, J. (1981) Dev. Biol. 87, 286–300. [DOI] [PubMed] [Google Scholar]
- 3.Sternberg, P. W. & Horvitz, H. R. (1986) Cell 44, 761–772. [DOI] [PubMed] [Google Scholar]
- 4.Sternberg, P. W. (1988) Nature 335, 551–554. [DOI] [PubMed] [Google Scholar]
- 5.Yochem, J., Weston, K. & Greenwald, I. (1988) Nature 335, 547–550. [DOI] [PubMed] [Google Scholar]
- 6.Hill, R. & Sternberg, P. (1992) Nature 358, 470–476. [DOI] [PubMed] [Google Scholar]
- 7.Sulston, J. & Horvitz, H. R. (1977) Dev. Biol. 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 8.Sharma-Kishore, R., White, J., Southgate, E. & Podbilewicz, B. (1999) Development (Cambridge, U.K.) 126, 691–699. [DOI] [PubMed] [Google Scholar]
- 9.Inoue, T., Sherwood, D., Aspöck, G., Butler, J., Gupta, B. P., Kirouac, M., Wang, M., Lee, P.-Y., Kramer, J. M., Bürglin, T., et al. (2002) Mech. Dev. 119S, S203–S209. [DOI] [PubMed] [Google Scholar]
- 10.Maduro, M. & Pilgrim, D. (1995) Genetics 141, 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna-Rose, W. & Han, M. (2002) Dev. Biol. 241, 247–258. [DOI] [PubMed] [Google Scholar]
- 12.Chang, S., Johnston, R. J., Jr. & Hobert, O. (2003) Genes Dev. 17, 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer, R. E., Inoue, T., Sherwood, D. R., Jiang, L. I. & Sternberg, P. W. (2002) Dev. Biol. 252, 202–213. [DOI] [PubMed] [Google Scholar]
- 14.Pettitt, J., Wood, W. & Plasterk, R. (1996) Development (Cambridge, U.K.) 122, 4149–4157. [DOI] [PubMed] [Google Scholar]
- 15.Burdine, R. D., Branda, C. S. & Stern, M. J. (1998) Development (Cambridge, U.K.) 125, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 16.Chang, C., Newman, A. P. & Sternberg, P. W. (1999) Curr. Biol. 9, 237–246. [DOI] [PubMed] [Google Scholar]
- 17.Wang, M. & Sternberg, P. W. (2000) Development (Cambridge, U.K.) 127, 5047–5058. [DOI] [PubMed] [Google Scholar]
- 18.Stringham, E., Pujol, N., Vandekerckhove, J. & Bogaert, T. (2002) Development (Cambridge, U.K.) 129, 3367. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, H. Y. & Horvitz, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 14224–14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colavita, A. & Tessier-Lavigne, M. (2003) Science 302, 293–296. [DOI] [PubMed] [Google Scholar]
- 21.Shen, K., Fetter, R. D. & Bargmann, C. I. (2004) Cell 116, 869–881. [DOI] [PubMed] [Google Scholar]
- 22.Rougvie, A. E. & Ambros, V. (1995) Development (Cambridge, U.K.) 121, 2491–2500. [DOI] [PubMed] [Google Scholar]
- 23.Newman, A. P., Inoue, T., Wang, M. & Sternberg, P. W. (2000) Curr. Biol. 10, 1479–1488. [DOI] [PubMed] [Google Scholar]
- 24.Freyd, G., Kim, S. & Horvitz, H. R. (1990) Nature 344, 876–879. [DOI] [PubMed] [Google Scholar]
- 25.Gupta, B. P. & Sternberg, P. W. (2002) Dev. Biol. 247, 102–115. [DOI] [PubMed] [Google Scholar]
- 26.Gupta, B. P., Wang, M. & Sternberg, P. W. (2003) Development (Cambridge, U.K.) 130, 2589–2601. [DOI] [PubMed] [Google Scholar]
- 27.Chamberlin, H. M., Palmer, R. E., Newman, A. P., Sternberg, P. W., Baillie, D. L. & Thomas, J. H. (1997) Development (Cambridge, U.K.) 124, 3919–3928. [DOI] [PubMed] [Google Scholar]
- 28.Ambros, V. & Horvitz, H. R. (1984) Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- 29.Ambros, V. (1989) Cell 57, 49–57. [DOI] [PubMed] [Google Scholar]
- 30.Bettinger, J. C., Euling, S. & Rougvie, A. E. (1997) Development (Cambridge, U.K.) 124, 4333–4342. [DOI] [PubMed] [Google Scholar]
- 31.Inoue, T., Oz, H. S., Wiland, D., Gharib, S., Deshpande, R., Hill, R. J., Katz, W. S. & Sternberg, P. W. (2004) Cell 118, 795–806. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande, R., Inoue, T., Priess, J. R. & Hill, R. J. (2005) Dev. Biol. 278, 118–129. [DOI] [PubMed] [Google Scholar]
- 33.Kirouac, M. & Sternberg, P. W. (2003) Dev. Biol. 257, 85–103. [DOI] [PubMed] [Google Scholar]
- 34.Cui, M. & Han, M. (2003) Dev. Biol. 257, 104–116. [DOI] [PubMed] [Google Scholar]
- 35.Briscoe, J., Pierani, A., Jessell, T. M. & Ericson, J. (2000) Cell 101, 435–445. [DOI] [PubMed] [Google Scholar]
- 36.Muhr, J., Andersson, E., Persson, M., Jessell, T. M. & Ericson, J. (2001) Cell 104, 861–873. [DOI] [PubMed] [Google Scholar]
- 37.Vallstedt, A., Muhr, J., Pattyn, A., Pierani, A., Mendelsohn, M., Sander, M., Jessell, T. M. & Ericson, J. (2001) Neuron 31, 743–755. [DOI] [PubMed] [Google Scholar]
- 38.Aurelio, O., Boulin, T. & Hobert, O. (2003) Development (Cambridge, U.K.) 130, 599–610. [DOI] [PubMed] [Google Scholar]
- 39.Bateman, J. M. & McNeill, H. (2004) Cell 119, 87–96. [DOI] [PubMed] [Google Scholar]
- 40.Casares, F. & Mann, R. S. (2001) Science 293, 1477–1480. [DOI] [PubMed] [Google Scholar]
- 41.Lints, R., Jia, L., Kim, K., Li, C. & Emmons, S. W. (2004) Dev. Biol. 269, 137–151. [DOI] [PubMed] [Google Scholar]
- 42.Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
- 43.Wenick, A. S. & Hobert, O. (2004) Dev. Cell 6, 757–770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.