Abstract
To understand how genes control floral asymmetry, we have isolated and analyzed the role of the RADIALIS (RAD) gene in Antirrhinum. We show that the RAD gene encodes a small MYB-like protein that is specifically expressed in the dorsal region of developing flowers. RAD has a single MYB-like domain that is closely related to one of the two MYB-like domains of DIV, a protein that has an antagonistic effect to RAD on floral development. Interactions between RAD and other genes indicate that floral asymmetry depends on the interplay between two pairs of transcription factors. First, a pair of TCP proteins is expressed in dorsal regions of the floral meristem, leading to the activation of RAD in the dorsal domain. The RAD MYB-like protein then antagonizes the related DIV MYB-like protein, preventing DIV activity in dorsal regions. In addition to its role in dorsal regions, RAD acts nonautonomously on lateral regions either directly, through RAD protein movement, or indirectly, through a signaling molecule.
Keywords: flower development, dorsal, ventral, adaxial, petal shape
Floral asymmetry is thought to have evolved many times independently as a specialized mechanism for pollinator interaction (1–3). In a few cases, most notably in Antirrhinum majus, the molecular genetic basis of floral asymmetry has begun to be understood. Four key genes have been shown to control dorsoventral asymmetry in Antirrhinum: CYCLOIDEA (CYC), DICHOTOMA (DICH), RADIALIS (RAD), and DIVARICATA (DIV) (4–8). Two of these genes, CYC and DICH, promote dorsal identity and encode proteins belonging to the TCP family of transcription factors. DIV promotes ventral identity and encodes a protein belonging to the MYB family of transcription factors, carrying two MYB-like domains. However, the mechanism by which CYC, DICH, and DIV interact remains unclear. To address this question, we have isolated and characterized RAD, the fourth member of this group of genes, and explored how it acts in combination with the other genes to establish floral asymmetry.
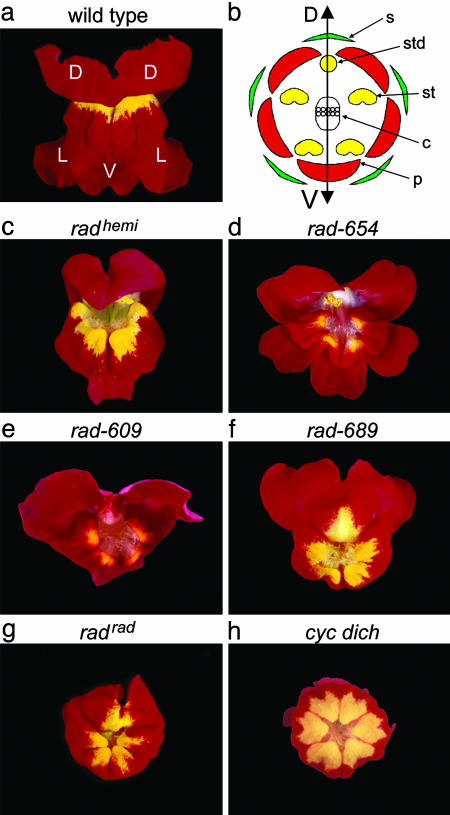
Flowers of wild-type Antirrhinum are zygomorphic, having a single plane of symmetry (bilateral symmetry), in contrast to actinomorphic flowers, which have multiple planes of symmetry (radial symmetry) (3). The zygomorphy of Antirrhinum flowers reflects morphological distinctions between the upper (dorsal) and lower (lateral and ventral) organs of whorls two and three. In whorl two, each flower has two dorsal petals, two lateral petals, and one ventral petal, whereas whorl three comprises a single arrested dorsal stamen (staminode), two lateral stamens, and two ventral stamens (Fig. 1 a and b).
Fig. 1.
Phenotype of rad mutants compared with wild type and a peloric mutant. (a) Frontal view of a wild-type Antirrhinum flower. D, dorsal petal; L, lateral petal; V, ventral petal. (b) Diagram showing the four whorls of the wild-type Antirrhinum flower. Whorl one, sepals (s); whorl two, petals (p); whorl three, stamens (st) and staminode (std); whorl four, carpels (c). The double-ended arrow marks the dorsal–ventral (DV) axis of the flower. (c–g) Frontal views of the five rad mutant lines used in this study. Note the similarity between the radrad flower (g) and the peloric cyc dich double mutant (h).
The CYC and DICH genes are required for dorsoventral asymmetry in Antirrhinum and are expressed from an early stage in the dorsal domain of the floral meristem (5, 7, 9). At later stages, CYC expression persists throughout most of the dorsal domain, whereas DICH becomes restricted to the most dorsal half of the dorsal domain. Inactivation of both CYC and DICH results in peloric (radially symmetrical) flowers, in which all petals have ventral identity (Fig. 1h). In cyc or dich single mutants, flowers are only partially ventralized, indicating that CYC and DICH act in a partially redundant manner (5, 7). Although CYC is expressed specifically in dorsal regions, it also acts nonautonomously on organs in lateral positions, because these organs are ventralized in cyc mutants.
A major mechanism by which CYC and DICH act is by repression of DIV, a gene that promotes ventral identity (6, 8). In the absence of CYC, DICH, and DIV (i.e., in cyc dich div triple mutants), all petals develop with lateral identity, whereas in the absence of just CYC and DICH (cyc dich double mutants), all petals have ventral identity. This observation indicates that, in a cyc dich background, DIV promotes ventral identity at all positions around the flower. In wild type, expression of CYC and DICH in dorsal regions restricts DIV function to the ventral and adjacent lateral domains (i.e., only these domains are affected in div single mutants). The effect of CYC and DICH on DIV appears to be posttranscriptional, because DIV is expressed throughout the wild-type flower at early stages in development in a pattern that is unaffected by cyc or dich mutations. At later stages, DIV is still expressed in all petals, although expression is enhanced in some ventral regions in a manner that depends on DIV itself (8).
RAD, like CYC and DICH, promotes dorsal identity, with recessive rad alleles giving very similar phenotypes to cyc alleles. This similarity, together with the linkage between RAD and CYC, led to an original assignment of rad and cyc mutations to the same locus (10). Subsequent analysis showed that the mutations fall into two complementation groups (11).
Here, we describe the isolation and characterization of RAD. We show that RAD is activated by CYC and DICH in the dorsal region of the developing flower, where it plays a major role in establishing floral asymmetry. RAD encodes a 93-aa protein containing a single MYB-like domain. The RAD protein is closely related to the N-terminal MYB-like domain of DIV, suggesting that RAD may have been derived from a DIV-like ancestral protein through C-terminal deletion. The molecular similarity between RAD and DIV suggests that their antagonistic phenotypic effects arise through competition for common DNA or protein targets. The RAD protein also mediates the nonautonomous effects of CYC, acting as a direct or indirect signal between dorsal and lateral domains.
Materials and Methods
Plant Material. The radrad and radhemi lines, originally named cycrad and cychemi, were obtained from the Gatersleben seed bank (lines 80 and 78) (12). The other alleles arose from transposon mutagenesis carried out at the John Innes Centre. The rad-609 and rad-654 alleles arose from the wild-type line JI-stock 75 (11), and the rad-689 allele arose from a centroradialis (cen) mutant line (JI-stock 663) (13). The cyc-608 and div-35 alleles used to create double mutants are described in refs. 8 and 11.
DNA Methods. Genomic DNA was extracted from A. majus leaf tissue, as described in ref. 14. Southern analyses were carried out by using Hybond-N+ nylon membranes according to the manufacturer's protocol (Amersham Pharmacia Biotech). Probes were radiolabeled with [α-32P]CTP by using a kit from Amersham Pharmacia Biotech that was based on the random hexamer labeling method.
The inverse PCR method used was based on the approach described in ref. 15. Genomic DNA (≈2 μg) was digested with the appropriate restriction enzyme in 50-μl reactions. The digested DNA was extracted from the reaction by using the Wizard DNA Clean-Up System kit (Promega). The restricted fragments (≈500 ng) were then incubated overnight with T4 DNA ligase in 100-μl reactions at 12°C. The resulting circularized DNA fragments were used as templates in a PCR-based reaction on the Expand Long Template PCR System (Boehringer Mannheim) protocol. Between 10 and 20 μl of the ligation reaction was used in a 50-μl PCR mix. The PCR products were fractionated on an agarose gel, allowing the correct product to be identified, gel extracted, and ligated into the pGEM-T vector (Promega).
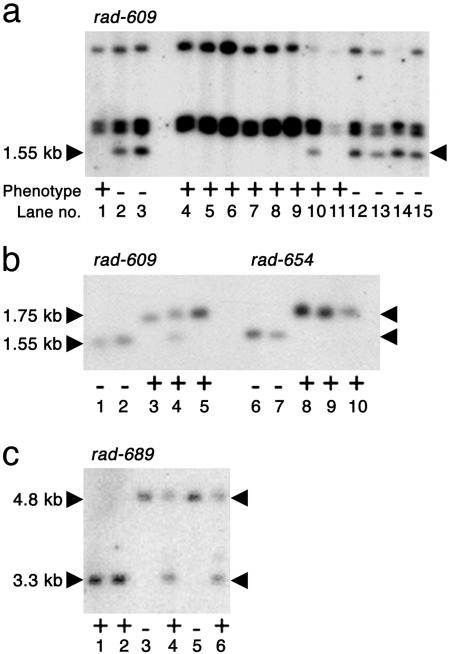
The segregating 1.55-kb EcoRI fragment seen in Fig. 2a was isolated by inverse PCR with primers designed against Ram1: oligo 4325 (5′-TAAGGAAGCTTCGGGTCCGG-3′) and oligo 4327 (5′-TAGGACCATGATCCCCTAGCC-3′). The resulting clone was named pJAM2255.
Fig. 2.
Retrotransposon tagging the RAD locus. (a) Genomic DNA from rad-609 mutants and wild-type siblings digested with EcoRI and probed with the 3′ end of Ram1. The segregating 1.55-kb band is marked. The wild-type plant used in lane 10 was later shown to be a heterozygote (b, lane 4). Lanes 1, 10, and 11 contain DNA from wild-type siblings of the original mutant plant, whereas lane 2 and lanes 12–15 contain DNA from mutant siblings. Lane 3 contains DNA from a family of rad-609 mutants, and lanes 4–9 contain DNA from siblings of the wild-type progenitor of rad-609.(b) Genomic DNA from rad-609 and rad-654 mutants and their wild-type relatives digested with EcoRI and probed with the 215-bp sequence flanking the Ram1 insertion. The 1.75-kb wild-type bands and the 1.55-kb mutant bands are indicated. DNA in lanes 1–5 corresponds to that in lanes 3, 2, 1, 10, and 5 of a. Lanes 6 and 7 contain DNA from rad-654 mutants. Lanes 8–10 contain DNA from homozygous revertants of rad-654. (c) Genomic DNA from rad-689 mutants and their wild-type relatives digested with HindIII and probed with the 215-bp sequence flanking Ram1. The wild-type band is 3.3 kb, and the mutant band is 4.8 kb. Lanes 1 and 2 contain DNA from wild-type siblings of the original mutant. Lanes 3 and 5 contain DNA from rad-609 mutants. DNA from heterozygous revertant siblings of these mutants was used in lanes 4 and 6. Plant phenotypes are + for wild type and – for mutant.
The 3.3-kb HindIII wild-type genomic fragment seen in Fig. 2c was also isolated by inverse PCR. The primers used were designed against the 215-bp fragment of the RAD locus that formed part of pJAM2255: oligo 4468 (5′-GTACATCATTAATATACCAC-3′) and oligo 4470 (5′-CAGT T TGTACAAACGGTTCG-3′). The resulting clone was labeled pJAM2247.
The pJAM2247 insert was used to screen a cDNA library made from young inflorescences of wild-type A. majus plants (16). Approximately 5 × 105 plaque-forming units were transferred to nitrocellulose filters (Sartorius) and probed. Six positives were obtained, all representing the same gene. The longest insert was 696 bp, and this insert was subcloned into the BlueScript KS+ vector (Stratagene) to give pJAM2246. A poly(A) tail was present in the same position in all six cDNA clones. A putative polyadenylation signal (i.e., AATAAA) was also identified, ≈20 bp upstream of the poly(A) region.
Sequence analysis was carried out by using the program wisconsin package 9.0 (Genetics Computer Group, Madison, WI) in UNIX.
RNA Methods. RNA in situ hybridizations were carried out as described in ref. 17. Riboprobes against RAD, CYC, and DICH were synthesized by using plasmids pJAM2251, pJAM193, and pJAM2143, respectively. Plasmid pJAM2251 consisted of the pGEM-T vector (Promega), containing ≈370 bp of RAD cDNA as insert. This insert was obtained by PCR with oligos 5045 (5′-ATGGCTTCGACTCGTGGTTCTG-3′) and 5041 (5′-GAACGATTACAAACGGGTAGG-3′), by using pJAM2246 as template. Antisense probes were obtained by linearizing pJAM2251 with PstI, followed by transcription from the T7 promoter. Sense probes were obtained in the same way but from another plasmid, pJAM2248. This plasmid contained the same insert as pJAM2251 but in the opposite orientation. Plasmids pJAM193 and pJAM2143 are described in refs. 5 and 7.
Results
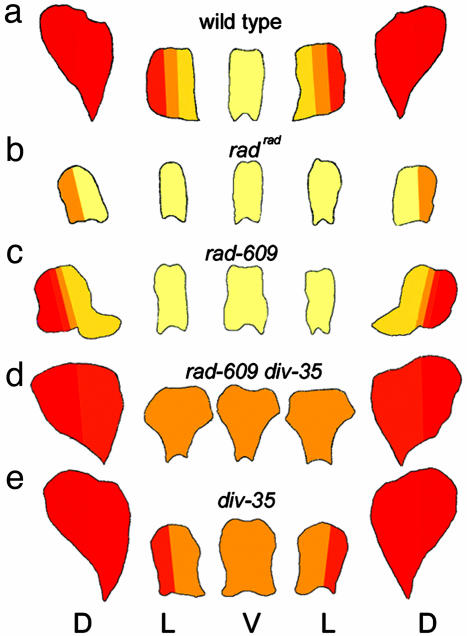
RAD Mutant Phenotypes and Interactions. Five recessive rad-mutant alleles were analyzed: radhemiradialis (radhemi), rad-654, rad-609, rad-689, and radradialis (radrad) (Fig. 1 c–g). Two of these alleles (radrad and radhemi) were originally isolated in the early 20th century by Erwin Baur (18), whereas the other alleles arose more recently from transposon mutagenesis experiments (11). Based on their phenotypes, the alleles could be arranged in a series, going from weakest to strongest in the order radhemi < rad-609 ∼ rad-654 < rad-689 < radrad. The strongest allele, radrad, gave almost peloric (i.e., radially symmetric) flowers in which all petals resembled the ventral petal of wild type, with only the dorsal half of each dorsal petal retaining some dorsal/lateral features (petal lobes are shown schematically in Fig. 3b). Weaker alleles showed more partial ventralization, as illustrated schematically for rad-609 (Fig. 3c). Unlike cyc mutants, which often have six instead of five organs per whorl, the organ number was not affected in rad mutants. The dorsal stamen (staminode) of rad mutants was typically longer than that of wild type but did not develop into the functional stamen found in cyc mutants (data not shown).
Fig. 3.
Shape of petal lobes. Flowers of wild type (a), radrad (b), rad-609 (c), rad-609 div-35 double mutant (d), and div-35 (e) are shown. For each flower, the petal lobes have been laid out from left to right in positional order, starting from the right dorsal petal lobe (as shown in Fig. 1a) and working clockwise around the flower. The colors represent the identities of the petals based on cell types and appearance. Pale yellow represents ventral identity, and each color change from yellow to red represents a shift toward more dorsal identity (dark red). D, dorsal; L, lateral; and V, ventral.
To investigate how RAD interacts with other genes in controlling floral asymmetry, several double mutants were constructed. The rad cyc double mutant had a fully ventralized phenotype (data not shown), even though both the rad and cyc alleles used (rad-609 and cyc-608) were of intermediate strength. This result indicates that the residual asymmetry seen in rad single mutants depends on CYC activity. The double mutant between rad-609 and a putative null div allele, div-35 [div-35 carries a frame-shift mutation (8)], had features of both the rad-609 and div-35 single mutants (Fig. 3 c–e). The petals in lateral and ventral positions were bilaterally symmetrical (as in rad-609) but resembled the ventral petal of div-35 mutants, although the petals had a slightly different shape. The petals in dorsal positions had some dorsal identity but were clearly distinct from the dorsal petals of both single mutants.
Molecular Cloning of RAD. The rad-609 allele arose from a transposon mutagenesis experiment, but, unlike many other transposon-induced mutations, the rad-609 allele appeared to be genetically stable (11). One possibility was that rad-609 was caused by insertion of a DNA sequence, such as a retrotransposon, that did not normally excise. A stable mutation caused by insertion of a retrotransposon, Ram1, had previously been described for the incolorata locus of Antirrhinum [inc-713 (19)]. To test whether Ram1 might also be responsible for rad-609, DNA from rad-609 mutants and wild-type siblings was digested with EcoRI, blotted, and probed with the 3′ end of Ram1, revealing a 1.55-kb EcoRI fragment that segregated with the rad-609 mutant allele (Fig. 2a).
To determine whether the 1.55-kb fragment was derived from the RAD locus, the fragment was amplified by inverse PCR and sequenced. In addition to Ram1, the 1.55-kb fragment contained 215 bp of flanking sequence. This 215-bp sequence was amplified and used to probe DNA from three independent rad mutants (rad-609, rad-654, and rad-689) and various wild-type lines, including some revertants derived from rad-654 and rad-689 (Fig. 2 b and c). A distinct band was observed in each of the rad mutants when compared with their wild-type relatives and revertants (some of the wild types were heterozygous, as expected from the recessive nature of rad alleles). This observation indicated that the 215-bp sequence flanking Ram1 was derived from the RAD locus.
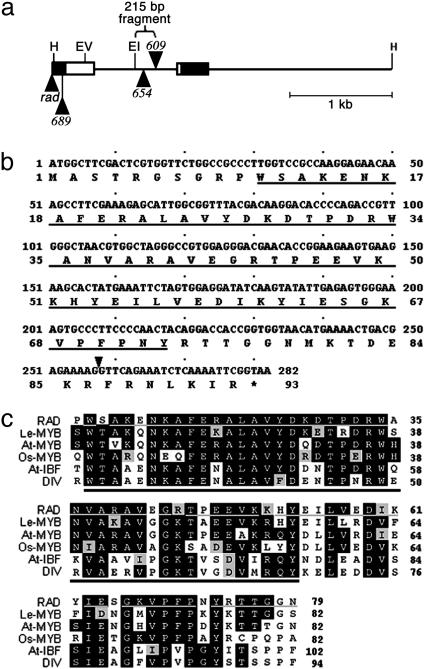
Oligonucleotides were designed, based on the 215-bp sequence, to obtain more of the RAD locus by inverse PCR, leading to the amplification of a 3.3-kb HindIII genomic fragment from wild-type DNA. The fragment was sequenced and used to probe a cDNA library made from A. majus inflorescence RNA. Six cDNA clones were isolated, representing transcripts from a single gene. Comparison of the cDNA with the genomic sequence showed that the cDNAs were derived from a gene with an intron of 829 bp (Fig. 4a). There was a single large ORF, mostly in the first exon, encoding a protein of 93 aa (Fig. 4b).
Fig. 4.
The RAD locus, coding sequence, and predicted protein sequence. (a) Diagram of the 3.3-kb wild-type HindIII fragment, containing part of the RAD locus. The two exons are shown as boxes; white sections are coding regions, and black sections are 5′ and 3′ untranslated regions. The insertions in radrad, rad-689, rad-654, and rad-609 are shown as black triangles. Also marked is the 215-bp fragment used to generate probes for Southern analysis. H, HindIII; EV, EcoRV; EI, EcoRI. (b) The complete coding sequence of the RAD gene with the predicted RAD protein sequence. The underlined amino acid sequence is similar to the MYB I domain of DIV defined in ref. 8. The black arrowhead marks the position of the intron. (c) Alignment of RAD, DIV, and RAD-like protein sequences. Conserved residues are shown as white letters on a black background. Residues that show similarity to the consensus are highlighted in varying shades of gray according to how similar they are. The black bar marks the conserved MYB I domain, and the gray bar indicates those amino acids in RAD that are found in at least one other sequence in the group. RAD, A. majus RADIALIS protein; Le-MYB, Lycopersicon esculentum MYB-related protein (CAB91874); At-MYB, Arabidopsis thaliana MYB-related protein (At2g21650; AAP40504); Os-MYB, Oryza sativa MYB-related protein (BAB90739); At-IBF, A. thaliana I box-binding-factor-like protein (At5g58900; BAB09635); DIV, A. majus MYB-like transcription factor DIVARICATA protein (AAL78741).
A combination of DNA blotting and PCR was used to characterize the rad mutants (Fig. 4a). rad-609 had a Ram1-like insertion of ≈1.45 kb within the intron. rad-654 had an insertion of a ≈4.3-kb transposon belonging to the CACTA family (20, 21) within the intron. The similarity of the rad-654 and rad-609 insertion sites may account for their similar phenotypes. rad-689 and radrad had insertions within or near the 5′ untranslated region.
RAD Encodes a Putative MYB-Like Protein. Database searches revealed that the protein encoded by RAD is related to the MYB family of transcription factors (Fig. 4c). The MYB family is one of the largest transcription factor families in plants, comprising >125 members in Arabidopsis (22). The diagnostic feature of this family is an ≈50-aa sequence, the MYB domain, with three tryptophan residues spaced 18–20 residues apart. In plants, one or more of the tryptophan residues may be replaced by another aromatic residue or by a similarly sized aliphatic residue. The MYB domain may be present once, twice, or three times in the protein.
RAD had a single MYB-like domain and was most closely related to a subfamily of single-repeat MYB-like proteins. Other members of this subfamily can be found in several dicots, as well as in monocots, although the function of these subfamily members is not known (e.g., Fig. 4c). The nearest relatives to RAD outside the subfamily are a group of proteins with two MYB-like domains that includes the Antirrhinum DIV protein. Of the two domains in DIV, the 63-aa MYB I region is most similar to RAD (≈52% identical; Fig. 4c). Because RAD is only 93 aa long, this MYB I-like domain comprises about two-thirds of the RAD protein. No other known functional domains or motifs were identified in the RAD protein sequence.
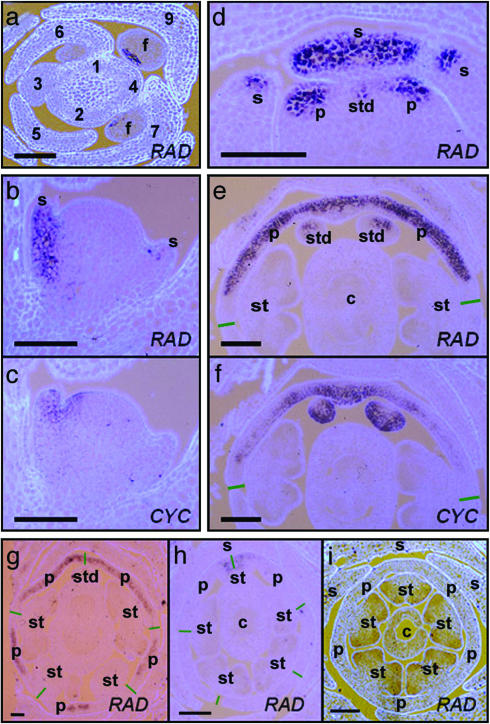
Expression of RAD in Wild-Type Inflorescences. To determine the expression pattern of RAD, RNA in situ hybridizations were carried out on sections from wild-type inflorescences. Floral meristems of Antirrhinum are generated at regular time intervals, termed plastochrons, on the periphery of the inflorescence apex. Flower development has been divided into 15 stages, each stage lasting ≈4 plastochrons (≈40 h), when plants are grown at 25°C (23, 24). The earliest clear sign of RAD expression was at early stage two (plastochrons 7 and 8), when the floral meristem comprised a loaf-shaped bulge of cells (Fig. 5a). Expression was in the dorsal region of the floral meristem. This pattern is similar to the pattern of CYC and DICH expression; although, unlike RAD, expression of these genes can also be detected earlier, in stage-one meristems (5).
Fig. 5.
RNA in situ hybridizations by using DIG-labeled CYC and RAD riboprobes on sections from wild-type and mutant inflorescences. (a) Transverse section through the tip of a wild-type inflorescence, hybridized with antisense RAD. Approximate plastochron numbers are marked. Early RAD expression can be seen in floral (f) meristems at plastochrons 7 and 9. The main stem is in the center of the picture. (b and c) Adjacent longitudinal sections through a stage-four wild-type floral meristem. The section in b was hybridized with antisense RAD, and the section in c was hybridized with antisense CYC. The main stem is on the left-hand side of the photos, and the subtending bract is on the right. (d) Transverse section through an early stage-five wild-type floral meristem hybridized with antisense RAD. Only the dorsal part of the flower (where RAD is expressed) is shown. (e and f) Adjacent transverse sections through a stage-seven to -eight wild-type floral meristem. The section in e was hybridized with antisense RAD, and the section in f was hybridized with antisense CYC. Only the dorsal part of the flower is shown. Green lines mark the approximate site of the junction between dorsal and lateral petals. (g) Transverse section from a stage-seven to -eight backpetals floral meristem. Green lines mark the approximate site of the junctions between the five petals. (h) Transverse section from a stage-seven cyc-608 floral meristem. Green lines mark the approximate site of the junctions between the five petals. (i) Transverse section from a stage-seven cyc dich double-mutant floral meristem. The sections were photographed under UV light. Light blue/mauve color is calcofluor-stained tissue; dark purple/black color corresponds to DIG-labeled probe. (Scale bars, 100 μm.) s, sepal; p, petal; st, stamen; std, staminode; c, carpel.
At stage four, when sepal primordia had emerged around the meristem dome, RAD expression could be seen in the dorsal sepal primordium (Fig. 5b). Probing an adjacent section with CYC revealed that CYC was expressed in a domain that was similar to RAD but also extended into dorsal regions of whorls two and three (Fig. 5c). By stage five, RAD expression could be detected in the dorsal sepal and in the dorsal part of the lateral sepals, similar to the expression pattern of CYC at this stage (5). Also by stage five, RAD expression could be detected in emerging dorsal primordia of whorls two and three, suggesting that RAD is activated shortly after CYC in internal whorls (Fig. 5d).
In older flower buds (stages seven and eight), RAD expression was found mainly in the dorsal petals and staminode (Fig. 5e). The base of the dorsal sepal also showed weak expression. Transverse sections through these flower buds revealed that the CYC and RAD expression patterns were very similar. Except for a region at their lateral edges, both genes were expressed throughout the dorsal petals (Fig. 5 e and f). Also, both genes were expressed in the staminode; although, within the staminode, the CYC and RAD expression domains seemed almost complementary: CYC was expressed throughout the staminode, except for a small patch on the abaxial side, whereas RAD expression appeared to be strongest on the abaxial side (Fig. 5 e and f) (25).
Expression of RAD in Floral Symmetry Mutants. The dorsal-specific expression pattern observed for both RAD and CYC raised the question of whether these genes might influence each other's expression. To address this question, RNA in situ hybridizations were carried out on cyc and rad mutant inflorescences by using RAD and CYC probes, respectively. The expression pattern of CYC was unaffected in rad mutants (data not shown). However, expression of RAD was reduced in cyc mutants, being restricted to the most dorsal part of the dorsal-petal primordia (Fig. 5h), in contrast to wild type, where RAD was expressed throughout most of the dorsal-petal primordia (Fig. 5e). In addition, RAD expression was delayed in cyc mutants: The earliest signal was observed at about plastochrons 11 and 12 (early stage three), compared with plastochrons 7 and 8 (early stage two) in wild type. Expression of RAD in the dorsal stamen appeared to be unaffected in cyc mutants.
These results show that the loss of CYC activity leads to a reduction in RAD expression in the dorsal domains of the flower. To test whether gain of CYC might lead to a corresponding increase of RAD expression, RNA in situ experiments were carried out on the backpetals mutant cyc-705. This allele confers a dorsalized phenotype and carries a transposon insertion in the CYC promoter that results in ectopic expression of CYC in the lateral and ventral petals after about stage six (7, 25). RAD was also ectopically expressed in lateral and ventral petals of the backpetals mutant, mirroring the ectopic pattern of CYC expression (Fig. 5g).
Although RAD expression was reduced in cyc mutants, residual expression was still observed in dorsal-petal primordia, suggesting that a gene other than CYC could also be involved in activating RAD in a dorsal-specific manner. A good candidate was DICH, because this gene is expressed in the domain where residual RAD expression is observed [i.e., in the most dorsal region of the dorsal-petal primordia (7)]. To test whether DICH might be involved, RNA in situ hybridizations were carried out on cyc dich double mutants by using the RAD probe. No RAD transcripts could be detected at any stage of floral development of double mutants, indicating that DICH acted partially redundantly with CYC to regulate RAD expression (Fig. 5i). Probing dich single mutants with RAD revealed no obvious reduction in expression compared with that in wild type, showing that DICH had less impact than CYC on RAD expression, consistent with the weaker phenotypic effects of dich compared with cyc mutations.
RNA in situ hybridization was also carried out on div-mutant floral meristems by using the RAD probe. Mutations in DIV appeared to have no effect on RAD expression (data not shown).
Discussion
RAD Encodes a Small MYB Protein. As with the other three genes controlling dorsoventral asymmetry in Antirrhinum, RAD encodes a protein belonging to a transcription-factor family. However, unlike the dorsal promoting genes CYC and DICH, the protein encoded by RAD is not a member of the TCP family. Rather, this protein belongs to the MYB superfamily of transcription factors, being closely related to DIV, which has the opposite function of RAD in promoting ventral identity. These results raise the question of how two pairs of proteins, two TCP proteins and two MYB-like proteins, interact to control floral asymmetry.
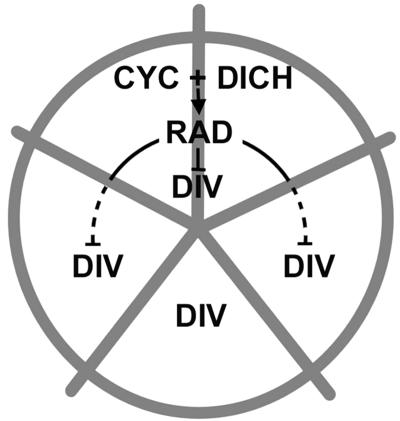
CYC and DICH Activate RAD. Several pieces of evidence suggest that a major mechanism by which CYC and DICH influence petal development is through activation of RAD (Fig. 6). (i) RAD is activated in a similar domain to that of CYC and DICH, in dorsal regions of the developing flower bud. (ii) Activation of RAD occurs shortly after that of CYC and DICH. (iii) Activation of RAD depends on both CYC and DICH function: Expression of RAD is reduced in floral meristems of the cyc single mutant and disappears altogether in the cyc dich double mutant. (iv) Ectopic expression of CYC is sufficient to activate RAD in lateral and ventral petals, resulting in a dorsalized phenotype. (v) The petal phenotype of strong rad mutants resembles that of the cyc dich double mutant. Because both CYC and DICH encode proteins belonging to the TCP family of transcription factors, it is possible that CYC and DICH activate RAD directly by binding to its regulatory regions or indirectly through activation of an intermediary gene. Once activated, RAD may act on downstream target genes independently of CYC/DICH, or its action may require the continuing presence of CYC and DICH.
Fig. 6.
Model for interactions between genes controlling dorsoventral asymmetry. Expression of CYC and DICH in the dorsal domain (upper two sections) leads to activation of RAD (solid arrow), which, in turn, inhibits DIV activity in both dorsal regions (solid line) and in lateral regions (dotted line).
If all the effects of CYC and DICH are mediated through RAD, then the loss-of-function rad mutant flowers should be identical to cyc dich double mutant flowers. The strongest rad mutant phenotypes identified so far, although very similar to the cyc dich phenotype, differ in several respects. Unlike cyc dich mutants, strong rad mutant flowers have slightly asymmetric dorsal petals, do not have fully functional stamens in dorsal positions, and have five organs per whorl rather than six. It is possible that the strong rad alleles obtained so far do not result in complete loss of function and that the distinctions from cyc dich are simply due to residual RAD activity. Alternatively, some of the effects of CYC/DICH may not be mediated by RAD. Some support for such a RAD-independent mechanism comes from the observation that cyc-608 rad-609 double mutants have fully ventralized flowers, showing that the residual asymmetry of rad mutants is lost if cyc is also mutated. CYC could therefore be controlling floral asymmetry independently of RAD as well as through RAD.
RAD May Act, in Part, by Inhibiting DIV. At a morphological level, RAD and DIV have apparently antagonistic functions: RAD promotes dorsal identity, whereas DIV promotes ventral identity. Given the sequence similarity between the RAD and DIV proteins, this antagonism most likely reflects molecular competition. DIV is a 307-aa protein with two MYB-like domains (I and II) resembling other functional MYB transcription activators (26), whereas RAD is a 93-aa protein with a single MYB I-like domain. RAD may therefore have been derived from a DIV-like ancestor through loss of the C-terminal MYB II domain, reducing the protein's functionality. Although not fully functional as a transcriptional activator, RAD could nevertheless inhibit DIV by competitively binding to DIV's target sequences or to proteins that interact with DIV. Expression of RAD in dorsal petals would therefore prevent DIV protein from functioning in these regions. This mechanism would account for the effects of DIV being restricted to lateral and ventral petals, even though DIV is expressed in all petals (8).
Although antagonism of DIV may be a key mechanism for RAD influencing floral asymmetry, this antagonism does not account for all aspects of RAD function, because null div mutants still have asymmetric flowers with distinctive dorsal petals (8). Moreover, rad div double mutants have a phenotype distinct from that of div single mutants. RAD must therefore be able to influence development in ways other than by antagonizing DIV, perhaps by interfering with the action of other members of the MYB family of transcription factors.
Nonautonomous Effects of CYC and RAD. The precise alignment of CYC and RAD expression in dorsal petals indicates that CYC can activate RAD in an autonomous manner. The nonautonomous effect of CYC on lateral petals is therefore likely to be downstream of RAD activation. One possibility is that activation of RAD in dorsal regions influences the production of a downstream signaling molecule that, in turn, influences development in lateral positions. Alternatively, the RAD protein may move from cells in the dorsal domain to those in the lateral domain. Nonautonomous gene action mediated by cell-to-cell protein movement has been described for plants (e.g., ref. 27). A case of particular relevance involves the Arabidopsis CAPRICE (CPC) protein, a single-repeat MYB-like protein of a size similar to RAD (94 aa, compared with 93 aa for RAD). The CPC gene is expressed in only nonhair cells in wild-type roots, but the CPC protein can also be detected in adjacent hair cells, where it represses the action of a two-repeat MYB gene called WEREWOLF (28, 29). In these cases, protein movement involves traversing one or a few cells. If RAD movement is to account for the nonautonomous effects of RAD, the effects would either have to occur very early in development (when petals are a few cells wide) or over a longer range than in previously described cases of protein movement.
Acknowledgments
We thank Minsung Kim, Manuela Costa, and Catherine Baxter for helpful comments on the manuscript and Amelia Green for help with Fig. 3.
Author contributions: E.C. designed research; S.B.C., R.C., and L.C. performed research; R.C. and L.C. contributed new reagents/analytic tools; S.B.C. and E.C. analyzed data; and S.B.C. and E.C. wrote the paper.
Abbreviations: CYC, CYCLOIDEA; DICH, DICHOTOMA; DIV, DIVARICATA; RAD, RADIALIS.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY954971).
References
- 1.Stebbins, G. L. (1974) Flowering Plants: Evolution Above the Species Level (Harvard Univ. Press, Cambridge, MA).
- 2.Donoghue, M. J., Ree, R. & Baum, D. A. (1998) Trends Plant Sci. 3, 311–317. [Google Scholar]
- 3.Endress, P. (1999) Int. J. Plant Sci. 160, S3–S23. [DOI] [PubMed] [Google Scholar]
- 4.Coen, E. S. (1996) EMBO J. 15, 6777–6788. [PMC free article] [PubMed] [Google Scholar]
- 5.Luo, D., Carpenter, R., Vincent, C., Copsey, L. & Coen, E. (1996) Nature 383, 794–799. [DOI] [PubMed] [Google Scholar]
- 6.Almeida, J., Rocheta, M. & Galego, L. (1997) Development (Cambridge, U.K.) 124, 1387–1392. [DOI] [PubMed] [Google Scholar]
- 7.Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J. & Coen, E. (1999) Cell 99, 1–12. [DOI] [PubMed] [Google Scholar]
- 8.Galego, L. & Almeida, J. (2002) Genes Dev. 16, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cubas, P., Lauter, N., Doebley, J. & Coen, E. (1999) Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- 10.Stubbe, H. (1966) Genetik und Zytologie von Antirrhinum L. sect Antirrhinum (Fischer, Jena, Germany).
- 11.Carpenter, R. & Coen, E. (1990) Genes Dev. 4, 1483–1493. [DOI] [PubMed] [Google Scholar]
- 12.Hammer, K., Knüpffer, S. & Knüpffer, H. (1990) Kulturpflanze 38, 91–117. [Google Scholar]
- 13.Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S. & Coen, E. (1996) Nature 379, 719–797. [DOI] [PubMed] [Google Scholar]
- 14.Coen, E., Carpenter, R. & Martin, C. (1986) Cell 47, 285–296. [DOI] [PubMed] [Google Scholar]
- 15.Ochman, H., Gerber, A. S. & Hartl, D. L. (1988) Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon, R., Carpenter, R., Doyle, S. & Coen, E. (1994) Cell 78, 99–107. [DOI] [PubMed] [Google Scholar]
- 17.Coen, E., Romero, J. M., Doyle, S., Elliott, R., Murphy, G. & Carpenter, R. (1990) Cell 63, 1311–1322. [DOI] [PubMed] [Google Scholar]
- 18.Baur, E. (1924) Bibl. Genet. 4, 1–170. [Google Scholar]
- 19.Doyle, S. (1996) Master's thesis (Univ. of East Anglia, Norwich, U.K.).
- 20.Sommer, H., Hehl, R., Krebbers, E., Piotrowiak, R., Lönnig. W.-E. & Saedler, H. (1988) in Plant Transposable Elements, ed. Nelson, O. (Plenum, New York), pp. 227–235.
- 21.Luo, D., Coen, E., Doyle, S. & Carpenter, R. (1991) Plant J. 1, 59–69. [PubMed] [Google Scholar]
- 22.Stracke, R., Werber, M. & Weisshaar, B. (2001) Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- 23.Vincent, C. A. & Coen, E. S. (2004) Can. J. Bot. 82, 681–690. [Google Scholar]
- 24.Carpenter, R., Copsey, L., Vincent, C., Doyle, S., Magrath, R. & Coen, E. (1995) Plant Cell 7, 2001–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark, J. I. & Coen, E. S. (2001) Plant J. 30, 639–648. [DOI] [PubMed] [Google Scholar]
- 26.Rose, A., Meier, I. & Wienand, U. (1999) Plant J. 20, 641–652. [DOI] [PubMed] [Google Scholar]
- 27.Nakajima, K., Giovanni, S., Nawy, T. & Benfey, P. N. (2001) Nature 413, 307–311. [DOI] [PubMed] [Google Scholar]
- 28.Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M. D., Shimura, Y. & Okada, K. (2002) Development (Cambridge, U.K.) 129, 5409–5419. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M. M. & Schiefelbein, J. (2002) Plant Cell 14, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]