Abstract
Maintenance of genomic stability depends on the DNA damage response, an extensive signaling network that is activated by DNA lesions such as double-strand breaks (DSBs). The primary activator of the mammalian DSB response is the nuclear protein kinase ataxia–telangiectasia, mutated (ATM), which phosphorylates key players in various arms of this network. The activation and stabilization of the p53 protein play a major role in the DNA damage response and are mediated by ATM-dependent posttranslational modifications of p53 and Mdm2, a ubiquitin ligase of p53. p53's response to DNA damage also depends on Mdm2-dependent proteolysis of Mdmx, a homologue of Mdm2 that represses p53's transactivation function. Here we show that efficient damage-induced degradation of human Hdmx depends on functional ATM and at least three sites on the Hdmx that are phosphorylated in response to DSBs. One of these sites, S403, is a direct ATM target. Accordingly, each of these sites is important for Hdm2-mediated ubiquitination of Hdmx after DSB induction. These results demonstrate a sophisticated mechanism whereby ATM fine-tunes the optimal activation of p53 by simultaneously modifying each player in the process.
Keywords: ataxia–telangiectasia, DNA damage response, p53, protein degradation
DNA damage initiates a multibranched signaling network that includes DNA repair pathways, cell cycle checkpoints, and modulation of many other processes (1–3). Genetic defects that disturb these mechanisms almost invariably cause severe syndromes that are characterized by the degeneration of specific tissues, sensitivity to specific DNA-damaging agents, chromosomal instability, and a striking predisposition to cancer (2). The prototype DNA damage response is the one mobilized by the highly cytotoxic double-strand break (DSB) (2–4). DSBs are induced by ionizing radiations and radiomimetic chemicals but can also accompany normal genomic transactions, such as meiotic recombination and the maturation of the immune system genes through V(D)J recombination (5). The DSB response includes repair by direct end joining or by homologous recombination of sister DNA molecules and activation of numerous signaling pathways, most notably, the cell-cycle checkpoints (5, 6).
The primary mobilizer of the DSB response network is the nuclear protein kinase ataxia–telangiectasia, mutated (ATM) (2, 7). After DSB induction, ATM is autophosphorylated and activated (8), a portion of it binds to the DSB sites (9), and it phosphorylates numerous substrates playing different roles in various damage response pathways. ATM is missing or inactivated in patients with the human genetic disorder ataxia–telangiectasia (A-T), characterized by cerebellar degeneration, immunodeficiency, genomic instability, predisposition to lymphoreticular malignancies, and extreme sensitivity to ionizing radiation and DSB-inducing agents (10).
The p53 protein plays a major role in cellular stress responses (11) because of its control of two important response pathways: temporary growth arrest through the damage-induced cell-cycle checkpoints (6) and apoptosis (12). The pivotal role of p53 in the DNA damage response requires timely and accurate regulation of its activation. The half-life and activity of the p53 protein are governed largely by two structurally related RING-finger proteins, Mdm2 and Mdmx. These proteins act as essential, nonredundant negative regulators of p53 during embryonic development (13). Mdm2 interacts with p53, inhibits its activity as transcription factor, and serves as one of the E3 ubiquitin ligases in p53's proteasome-mediated degradation (14). Transcription of the gene-encoding Mdm2 is activated by p53, thus creating an autoregulatory negative feedback loop with an important role in the dynamics of p53 levels after stress (14, 15). Mdmx, which lacks detectable ubiquitin ligase activity, interacts directly with p53 and inhibits its transactivation activity (16, 17). Mdmx also interacts with Mdm2 through their respective RING finger domains, and this interaction stabilizes Mdm2 (18). It has been suggested that in this way, Mdmx assists Mdm2 in inhibiting p53 (19). On the other hand, Mdm2 functions as an E3 ubiquitin ligase in proteasome-mediated degradation of Mdmx (20–22). Indeed, Mdmx undergoes degradation in response to DNA damage, and this degradation is Mdm2-dependent (22). Thus, the physical and functional interactions between the three proteins create a sensitive regulatory apparatus around p53 that can serve as an efficient and rapid means to modulate its amount and activity upon stress.
ATM orchestrates the activation and stabilization of p53 in response to DSBs by controlling the induction of numerous posttranslational modifications along the p53 molecule, of which specific subsets enhance its transactivation activity or lead to inhibition of its proteasome-mediated degradation (7, 23). In human cells, ATM phosphorylates p53 directly on S15 and concomitantly activates other kinases that phosphorylate p53 on additional sites, e.g., the checkpoint kinase Chk2. Notably, human ATM also phosphorylates Hdm2 (the human ortholog of Mdm2) on S395, and this phosphorylation inhibits Hdm2-mediated degradation of p53 (24, 25). Here we add another route by which human ATM modulates p53's activation after DNA damage: by removing its inhibitor, Hdmx (human Mdmx), from the scene.
Materials and Methods
Cell Lines. The lymphoblastoid cell lines C3ABR (wild-type, obtained from Martin Lavin, Queensland Institute of Medical Research, Herston, Australia), L3 and AT59RM (derived from A-T patients, the latter from Luciana Chessa, Università La sapienza, Rome) were grown in RPMI medium 1640 supplemented with 10% FBS. HEK293, U2-OS, and MCF-7 cells were grown in DMEM with 10% FBS. Normal skin fibroblasts VH25 and VH10 (obtained from J. W. I. M. Simons, State University of Leiden, Leiden, The Netherlands) and skin fibroblast lines from A-T patients, AT2-RO, and AT4-LA (gift from N. G. J. Jaspers, Erasmus University, Rotterdam, The Netherlands) were grown in DMEM with 10% FBS.
DNA Damaging Agents and ATM Inhibitors. Neocarzinostatin was obtained from Kayaku Chemicals (Tokyo), etoposide from Sigma-Aldrich, and wortmannin from Calbiochem. The ATM inhibitor KU-55933 was a kind gift from Graeme Smith and Steve Jackson (KuDOS Pharmaceuticals and Wellcome Trust/Cancer Research UK Gurdon Institute, both of Cambridge, U.K., respectively).
Antibodies. Anti-Hdmx antibodies were the rabbit polyclonal sera p55, p56 (26), 1327, 1328 (P.d.G. and A.G.J., unpublished data), BL1258 (Bethyl Laboratories, Montgomery, TX) and the mouse monoclonal antibodies 6B1A, 11F4D, and 12G11G (27). The anti-pS403-Hdmx antibody (BL785) was generated by Bethyl Laboratories. Anti-Mdm2 antibody was mouse monoclonal antibody 4B2 (28) in combination with mouse monoclonal SMP14 from Santa Cruz Biotechnology. Anti-ATM antibody was mouse monoclonal MAT3-4G10/8 (29). The p53 monoclonal antibody DO-1 and the anti-HA monoclonal antibody F7 were obtained from Santa Cruz Biotechnology. The rabbit polyclonal anti-HA (A190–108A) antibody was produced by Bethyl Laboratories, and the mouse monoclonal anti-LacZ antibody (D19-2F3-2) was obtained from Roche Applied Science (Indianapolis). Anti-pSer15-p53 was obtained from Cell Signaling Technology (Beverly, MA), the monoclonal anti-HA antibody H11 was obtained from Covance Research Products (Berkeley, CA), and the monoclonal anti-α-tubulin was obtained from Sigma-Aldrich. Secondary antibodies were goat anti-mouse-HRP and goat anti-rabbit-HRP from Jackson ImmunoResearch.
Immunoblotting Analysis and Immunoprecipitations. Cell lysates were made in Giordano buffer (50 mM Tris·HCl, pH 7.4/250 mM NaCl/0.1% Triton X-100/5 mM EDTA), supplemented with a mixture of protease and phosphatase inhibitors. Cell lysis, Western blotting analysis, and immunoprecipitations were carried out by using standard methods and as described in ref. 20. Immunoblots were visualized by enhanced chemiluminescence (Super Signal, Pierce) and visualized by autoradiography or with the use of a ChemiGenius XE3 (Syngene, Cambridge, U.K.).
Transfection of Cell Lines. Cells were seeded 6–24 h before transfection in DMEM and transfected by using the calcium phosphate or with FuGENE 6 reagent (Roche Applied Science).
Expression Vectors and in Vitro Mutagenesis. Expression of recombinant glutatione S-transferase (GST)-fused Hdmx in E. coli was previously described (30). GST-Hdmx was purified from crude Escherichia coli lysates with glutathione beads (Amersham Pharmacia). For ectopic expression of Hdmx in human cell lines, the complete ORF of the protein was amplified by using the bacterial expression vector as a template, and the product was cloned in the pcDNA3.1 vector (Invitrogen). In vitro mutagenesis of this construct was carried out by using the QuikChange in Vitro Mutagenesis System (Stratagene). The expression vector for Hdm2 was a kind gift of M. Oren (The Weizmann Institute of Science, Rehovot, Israel).
Ubiquitination Assay in Cells. Purification of His-tagged ubiquitinated conjugates was performed as described in ref. 18. Twenty-four hours after transfection, cells were preincubated with 20 μM MG132 (Calbiochem) for 15–30 min, after which cells were either mock-treated or treated with 500 ng/ml or 200 ng/ml neocarzinostatin (NCS) (Kayaku Chemicals) for 3 or 6 h, all still in the presence of MG132. Cells were then washed twice and scraped in ice-cold PBS. Twenty percent of the cell suspension was lysed in Giordano buffer and analyzed by Western blotting. Lysis of the remainder of the cells and the subsequent isolation of His-tagged (ubiquitinated) proteins was performed as described in ref. 18. Eluates were analyzed by Western blotting.
Supporting Information. See Supporting Methods, which is published as supporting information on the PNAS web site, for details of in vitro kinase assay and mass spectrometric identification of phosphorylation sites.
Results
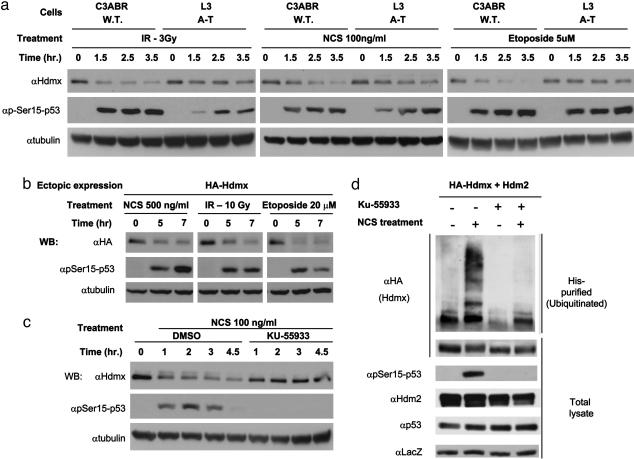
Treatment of cultured human cells with three DSB-inducing agents, ionizing radiation, the radiomimetic drug NCS, and the topoisomerase II inhibitor etoposide, led to a marked decrease both of endogenously expressed and ectopically expressed Hdmx (Fig. 1 a and b). This decrease reflected proteasome-mediated degradation after polyubiquitination rather than reduced production of Hdmx (refs. 21 and 22 and Fig. 6, which is published as supporting information on the PNAS web site). Importantly, ATM presence was required for efficient Hdmx degradation: A-T cells devoid of ATM exhibited a marked retardation of DSB-induced degradation of endogenous Hdmx (Fig. 1a). Furthermore, pretreatment of cells with a specific ATM inhibitor, KU-55933 (31), inhibited both the decrease in Hdmx levels and its polyubiquitination after NCS treatment (Fig. 1 c and d).
Fig. 1.
ATM-dependent decrease in the cellular amount of Hdmx and its ubiquitination in response to treatment with DSB-inducing agents. (a) Wild-type and A-T (ATM-deficient) lymphoblastoid cells were treated with IR (3 Gy), NCS (100 ng/ml), or etoposide (5 μM). Cells were harvested at the indicated time points, and cellular lysates were subjected to Western blotting analysis. Phosphorylation of p53 on Ser-15, an ATM target site (7, 23), served to monitor the DNA damage response. (b) Ectopic Hdmx responds to DSBs similarly to the endogenous protein. MCF-7 cells were transfected in 6-cm dishes with 200 ng HA-Hdmx. Twenty-four hours after transfection, the cells were treated with etoposide (20 μM), NCS (500 ng/ml), or IR (10 Gy), harvested 5 and 7 h later, and cellular lysates underwent Western blotting analysis. (c) Effect of the ATM-specific inhibitor KU-55933 on DNA damage-induced decrease in cellular amount of Hdmx. MCF-7 cells were pretreated with KU-55933 (10 μM) or DMSO (mock treatment) for 1 h and subsequently with NCS (100 ng/ml) and harvested at the indicated time points. Note the appearance of a slower migrating Hdmx band (band shift) after NCS treatment. Both band shift and Hdmx degradation are eliminated by pretreatment with KU-55933. (d) ATM-dependent ubiquitination of Hdmx in response to DSBs. MCF-7 cells were transiently transfected with HA-Hdmx (500 ng) and Hdm2 (100 ng) expression vectors in combination with CMV-LacZ and CMV-His-6-ubiquitin. MG132 (20 μM) and KU-55933 (10 μM) were added 24 h after transfection as indicated. The proteasome inhibitor MG132 was used to prevent rapid degradation of ubiquitinated Hdmx, thus allowing accumulation of ubiquitinated Hdmx. Thirty minutes later, NCS (200 ng/ml) was added for 3 h, after which the cells were harvested. Lysates were analyzed for ubiquitinated proteins as described in Materials and Methods.(Upper) His-6-purified proteins analyzed for ubiquitinated Hdmx by using anti-HA antibody. Total lysates (Lower) were analyzed for p53 phosphorylation on Ser-15 (DNA damage control), total p53, LacZ (transfection control), HA-Hdmx proteins, and Hdm2 expression. Note the marked suppression of damage-induced Hdmx ubiquitination by the ATM inhibitor.
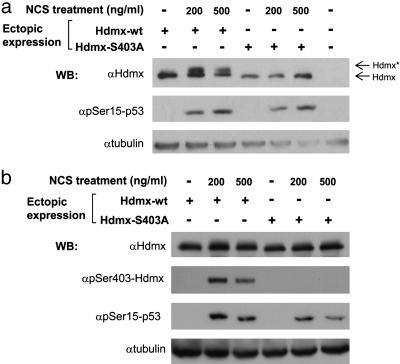
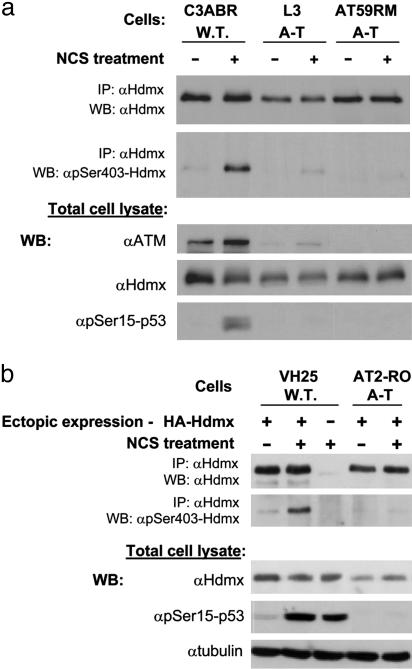
We investigated whether Hdmx degradation could be mediated by ATM-dependent posttranslational modifications of Hdmx. Indeed, when DSB-induced degradation of endogenous Hdmx was blocked by a proteasome inhibitor, accumulated Hdmx showed slower electrophoretic migration (“band shift”) (Fig. 6b). In the absence of the proteasome inhibitor, a slower migrating form of endogenous Hdmx after NCS treatment is transiently observed (data not shown). The most common ATM-mediated modification is phosphorylation, carried out directly by ATM, or by ATM-driven protein kinases (2, 7). ATM, ATM-Rad3-related (ATR), and DNA-PK phosphorylate their substrates on serines or threonines, followed by glutamine residues (“SQ/TQ motif”) (2, 7, 32). Two SQ motifs are present in Hdmx, at S132 and S403. When the ability of ATM to phosphorylate Hdmx in vitro was examined, a S403A, but not a S132A mutation, abolished this phosphorylation, suggesting that S403 of Hdmx was a potential ATM target (Fig. 7, which is published as supporting information on the PNAS web site). Importantly, substitution of S403 for alanine in ectopically expressed Hdmx abolished the appearance of the slower migrating form of this protein (Fig. 2a), suggesting that S403 phosphorylation was responsible for this band shift. A phospho-specific antibody was raised against an Hdmx-derived peptide containing phosphorylated S403. After DNA damage, this antibody reacted with ectopic wild-type Hdmx but not with a S403A mutant protein, demonstrating specificity for this phosphorylation (Fig. 2b). Further characterization of this antibody is shown in Fig. 8, which is published as supporting information on the PNAS web site. S403 phosphorylation in cells responding to DSBs was largely ATM-dependent: again, it was markedly reduced on an ATM-deficient background (Fig. 3) and in cells treated with two ATM inhibitors, wortmannin and KU-55933 (Fig. 9. which is published as supporting information on the PNAS web site). The low-level phosphorylation of S403 in A-T cells may be attributed to ATM's close ally, ATR, which shares many substrates with ATM but phosphorylates them at higher damage levels and slower kinetics (2, 32). We concluded that S403 of Hdmx is an ATM target in response to DNA damage.
Fig. 2.
Phosphorylation of Hdmx on S403 in response to DNA damage. (a) The DNA damage-induced band shift of ectopic Hdmx is abolished by a S403A mutation, suggesting that this band shift had been caused by a modification that occurred at that site. U2-OS cells in 6-cm dishes were transfected with 200 ng of wild-type or S403 mutant Hdmx expression vectors and 24 h later were treated with the indicated doses of NCS for 1 h. Hdmx*, modified Hdmx. (b) Ectopic Hdmx is phosphorylated on S403 in response to DNA damage. U2-OS cells in 6-cm dishes were transfected with 1.5 μg of wild-type or S403A mutant Hdmx vectors and treated 24 h later with the indicated doses of NCS for 1 h. The S403 phospho-specific antibody (Fig. 8) detects the phosphorylation of this site on ectopic wild-type Hdmx but not on S403A mutant protein. Because of the large excess of ectopic Hdmx and the different running conditions, the band shift observed in a is not seen in this experiment.
Fig. 3.
ATM dependence of Hdmx phosphorylation on S403. (a) Endogenous Hdmx was immunoprecipitated with anti-Hdmx antibodies (a mixture of the mouse monoclonal antibodies 6B1A, 11F4D, and 12G11G) from lymphoblastoid cells with the indicated genotypes after treatment with 100 ng/ml NCS for 15 min. Western blotting analysis was carried out by using the anti-phospho-Ser-403 antibody. (b) Primary fibroblasts from a healthy donor and an A-T patient were transfected by electroporation with an Hdmx expression vector and treated 48 h later with 200 ng/ml NCS for 1 h. Hdmx was immunoprecipitated with anti-Hdmx antibodies (6B1A, 11F4D, and 12G11G). Immune complexes and cellular lysates were analyzed by Western blotting.
In view of the multiple damage-induced phosphorylations of p53 and the increasing evidence that Hdm2 may also be phosphorylated on several sites in response to DNA damage (33, 34), we attempted to identify phosphorylations on Hdmx in addition to S403. Ectopically expressed Hdmx, obtained from cells that had been untreated or treated with NCS, was subjected to Q-TOF nanoESI tandem mass spectrometric analysis. The results (Fig. 10 a–c, which is published as supporting information on the PNAS web site) indicated enhanced phosphorylation after DNA damage on a peptide spanning positions 361–374 in Hdmx amino acid sequence, with the phosphorylated residues being T365 and S367. An additional phosphorylation within a peptide spanning residues 333–344 was identified as S342. Of note, because of a low signal corresponding to the peptide expected to span S403, this phosphorylation was not detected in this analysis. The S342, T365, and S367 sites are conserved from Xenopus to humans, whereas the ATM target site, S403 is conserved among fish, mice, rats, and humans (Fig. 10d).
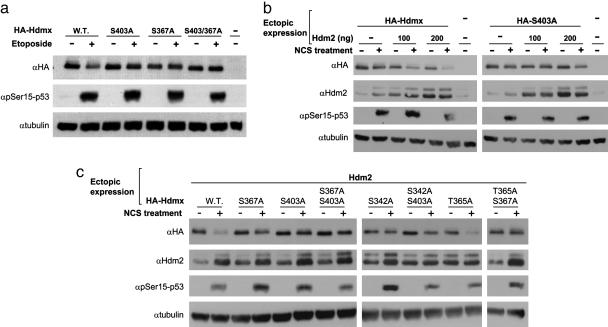
These phosphorylations could play a role in Hdmx degradation after DNA damage. We assessed the contribution of each phosphorylation site to this process by transfecting wild-type and nonphosphorylatable mutants of Hdmx into MCF-7 cells and after their damage-induced degradation (Fig. 4). Alanine substitutions at S342, S367, and S403 diminished damage-induced degradation of Hdmx to various extents, whereas a T365A mutation had no significant effect. Double-mutant combinations virtually abolished Hdmx degradation. Coexpression of Hdm2 enhanced Hdmx degradation and concomitantly highlighted the effects of the mutations on this process in agreement with Hdm2's role as an E3 ubiquitin ligase in this process (see Fig. 6c and refs. 21 and 22).
Fig. 4.
Effect of mutating phosphorylation sites on the DNA damage-induced degradation of Hdmx, and stimulation of this process by Hdm2. (a) MCF-7 cells (6-cm dishes) were transiently transfected with 200 ng of vectors expressing wild-type or mutant versions of HA-Hdmx. Twenty-four hours later, the cells were treated with etoposide (20 μM) for 5 h, and total cell lysates were analyzed by Western blotting. Note the moderate reduction in cellular amount of ectopic Hdmx, which is abolished in the mutant proteins. (b) Enhancement of DNA damage-induced reduction in ectopic Hdmx levels by coexpression of Hdm2. MCF-7 cells (6-cm dishes) were transiently transfected with 100 ng of vector expressing Hdm2 and 500 ng of vectors expressing wild-type or S403A HA-Hdmx. Twenty-four hours later, the cells were treated with NCS (500 ng/ml) for 5 h, and total cell lysates were analyzed by Western blotting. Note the stimulation of damage-induced Hdmx degradation by increasing amounts of ectopic Hdm2. Under these conditions, the inhibitory effect of the S403 mutation is evident. (c) Effect of various combinations of mutations on DNA damage-induced reduction of ectopic Hdmx levels. MCF-7 cells (6-cm dishes) were transiently transfected with 500 ng Hdmx expression vectors together with 100 ng of Hdm2 vector. Twenty-four hours later, the cells were treated with NCS (500 ng/ml) for 5 h, and total cell lysates were processed for Western blotting analysis. Of the four phosphorylation sites identified in this study, T365 does not seem to be needed for DNA damage-induced degradation of Hdmx.
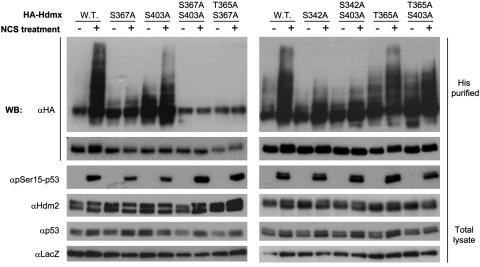
Because Hdm2 stimulates the degradation of Hdmx by its ubiquitin ligase activity, we investigated the effect of mutating the Hdmx phosphorylation sites on DSB-induced, Hdm2-mediated ubiquitination of Hdmx. In accordance with the degradation data, alanine substitutions of S342, S367, and S403 inhibited Hdm2-mediated ubiquitination to varying extents: Hdmx-S403A ubiquitination was reduced, whereas the S342A and S367A mutations virtually abrogated the ubiquitination (Fig. 5). On the other hand, mutating Hdmx at T365 barely affected its DSB-induced ubiquitination.
Fig. 5.
Effect of mutating phosphorylation sites of Hdmx on its DSB-induced ubiquitination. The same experimental system as in Fig. 1d was used. The MCF-7 cells (6-cm dishes) were transfected with 500 ng of wild-type or mutant HA-Hdmx as indicated and 100 ng of Hdm2 expression vectors, again in the presence of CMV-LacZ and CMV-His-6-Ubiquitin expression vectors. Twenty-four hours after transfection, MG132 (20 μM) was added, and, 15 min later, cells were incubated with NCS (500 ng/ml) for 6 h. The rest of the experiment was carried out as in Fig. 1d. The mutant versions of Hdmx in which one or two phosphorylation sites are mutated demonstrate different degrees of requirement of these sites for Hdmx ubiquitination.
A potential explanation for the effects of these mutations on Hdmx ubiquitination could be a reduction in Hdm2-Hdmx binding. To study this possibility, we first immunoprecipitated endogenous Hdmx from lysates of MCF-7 cells, either mock-treated or treated with NCS. We noticed that Hdm2 coprecipitated with Hdmx irrespective of NCS treatment (Fig 11a, which is published as supporting information on the PNAS web site). Then, ectopic Hdmx in various versions and Hdm2 were coexpressed under similar conditions as in the ubiquitination assay. Hdmx was immunoprecipitated, and coimmunoprecipitation of Hdm2 was examined. All Hdmx mutants tested in this experiment coprecipitated Hdm2 as efficiently as the wild-type protein (Fig. 11b). We concluded from these results that the loss of ubiquitination of Hdmx mutants could not be explained by reduction in Hdm2 binding.
Discussion
The list of known ATM-mediated pathways is far from complete, but the emerging picture is disclosing a remarkably broad ATM-dependent response to DSBs. An important aspect of the ATM-mediated signaling is the ability of this transducer to approach the same endpoint from several different directions, e.g., the cell-cycle checkpoints, each of which is governed by several ATM-mediated pathways (2, 6). ATM precisely controls its downstream pathways, often by approaching the same effector from several different directions. The prime example for that is ATM's control of p53 stabilization and activation. The series of ATM-dependent modifications, which activate and stabilize p53, illustrate the elaborate way in which ATM handles a single effector and indicates that ATM might regulate several effectors within the same pathway.
The addition of Hdmx to the list of ATM targets in the p53-mediated damage response pathway highlights the p53-Hdm2-Hdmx trio as a sensitive center of responses to environmental stimuli. When it comes to responding to double-strand breaks in the DNA, ATM is required for optimal response of these proteins. The phosphorylation of Hdmx on several sites apparently stimulates its degradation through the ubiquitin-proteasome pathway in an Hdm2-dependent manner. It has been reported that DNA damage leads to nuclear translocation of ectopically expressed Hdmx (35), but it is unclear yet whether this change in subcellular localization is essential for Hdmx degradation. Conceivably, the nuclear translocation should be essential for ATM-mediated phosphorylation, in view of the nuclear localization of ATM. The importance of ATM in orchestrating the p53 response is also illustrated by the observation that degradation of Hdmx after DNA damage is essential for a proper p53 response, as determined by p53 stabilization, p21 induction, and activation of cell-cycle checkpoints (22).
Given the p53 and Hdm2 examples, further Hdmx modifications in response to DNA damage or other stresses cannot be ruled out, and the modification repertoire of these proteins will likely expand. Different modifications may play roles in different mechanistic aspects of the processes they control, collectively achieving the desired consequence. These modifications sometimes depend on each other, e.g., the interdependence of phosphorylations at the N terminus of p53 (23, 36). It is tempting to assume that direct phosphorylation by ATM serves to prime the substrate for further modifications, which in turn are carried out by enzymes that themselves are activated by ATM. In this way, ATM may maintain elaborate firm control of downstream effectors. ATM's control over several proteins in the same pathway is a remarkable demonstration of a sophisticated mechanism by which this transducer of the DNA damage signal mobilizes complex cellular systems.
Supplementary Material
Acknowledgments
We thank Ruth Frenk for expert technical assistance, Graeme Smith and Steve Jackson for a gift of KU-55933, Martin Lavin, Luciana Chessa, J. W. I. M. Simons and N. G. J. Jaspers for cell lines, and Gilad Mass for expert help with art work. This work was carried out in partial fulfillment of the requirements for the Ph.D. degree of Y.P. This work was supported by research grants from The A-T Medical Research Foundation, The A-T Children's Project, the National Institutes of Health (NS31763), the A-T Medical Research Trust (to Y.S.), the Association for International Cancer Research (to P.d.G.), the Dutch Cancer Society (to E.M.), and the Minerva Foundation (to M.E-A.). Y.P. is a Joseph Sassoon Fellow.
Author contributions: Y.P., D.S., P.d.G., W.D.L., A.G.J., and Y.S. designed research; Y.P., D.S., P.d.G., E.M., M.E.-A., M.S., S.B., and A.F.A.S.T. performed research; Y.P., D.S., P.d.G., E.M., M.E.-A., M.S., S.B., A.F.A.S.T., W.D.L., A.G.J., and Y.S. analyzed data; and A.G.J. and Y.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: A-T, ataxia–telangiectasia; ATM, ataxia–telangiectasia, mutated; DSB, double-strand break; NCS, neocarzinostatin.
References
- 1.Norbury, C. J. & Hickson, I. D. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 367–401. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155–168. [DOI] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J. & Kastan, M. B. (2004) Cell 118, 9–17. [DOI] [PubMed] [Google Scholar]
- 4.Jackson, S. P. (2002) Carcinogenesis 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 5.Bassing, C. H. & Alt, F. W. (2004) DNA Repair (Amsterdam) 3, 781–796. [DOI] [PubMed] [Google Scholar]
- 6.Lukas, J., Lukas, C. & Bartek, J. (2004) DNA Repair (Amsterdam) 3, 997–1007. [DOI] [PubMed] [Google Scholar]
- 7.Kurz, E. U. & Lees-Miller, S. P. (2004) DNA Repair (Amsterdam) 3, 889–900. [DOI] [PubMed] [Google Scholar]
- 8.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499–506. [DOI] [PubMed] [Google Scholar]
- 9.Andegeko, Y., Moyal, L., Mittelman, L., Tsarfaty, I., Shiloh, Y. & Rotman, G. (2001) J. Biol. Chem. 276, 38224–38230. [DOI] [PubMed] [Google Scholar]
- 10.Chun, H. H. & Gatti, R. A. (2004) DNA Repair (Amsterdam) 3, 1187–1196. [DOI] [PubMed] [Google Scholar]
- 11.Oren, M. (2003) Cell Death Differ. 10, 431–442. [DOI] [PubMed] [Google Scholar]
- 12.Slee, E. A., O'Connor, D. J. & Lu, X. (2004) Oncogene 23, 2809–2818. [DOI] [PubMed] [Google Scholar]
- 13.Marine, J. C. & Jochemsen, A. G. (2004) Cell Cycle 3, 900–904. [PubMed] [Google Scholar]
- 14.Moll, U. M. & Petrenko, O. (2003) Mol. Cancer Res. 1, 1001–1008. [PubMed] [Google Scholar]
- 15.Michael, D. & Oren, M. (2002) Curr. Opin. Genet. Dev. 12, 53–59. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, M. W. & Berberich, S. J. (2000) Mol. Cell. Biol. 20, 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabbatini, P. & McCormick, F. (2002) DNA Cell Biol. 21, 519–525. [DOI] [PubMed] [Google Scholar]
- 18.Stad, R., Little, N. A., Xirodimas, D. P., Frenk, R., van der Eb, A. J., Lane, D. P., Saville, M. K. & Jochemsen, A. G. (2001) EMBO Rep. 2, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, J., Kawai, H., Nie, L., Kitao, H., Wiederschain, D., Jochemsen, A. G., Parant, J., Lozano, G. & Yuan, Z. M. (2002) J. Biol. Chem. 277, 19251–19254. [DOI] [PubMed] [Google Scholar]
- 20.de Graaf, P., Little, N. A., Ramos, Y. F., Meulmeester, E., Letteboer, S. J. & Jochemsen, A. G. (2003) J. Biol. Chem. 278, 38315–38324. [DOI] [PubMed] [Google Scholar]
- 21.Pan, Y. & Chen, J. (2003) Mol. Cell. Biol. 23, 5113–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai, H., Wiederschain, D., Kitao, H., Stuart, J., Tsai, K. K. & Yuan, Z. M. (2003) J. Biol. Chem. 278, 45946–45953. [DOI] [PubMed] [Google Scholar]
- 23.Meek, D. W. (2004) DNA Repair (Amsterdam) 3, 1049–1056. [DOI] [PubMed] [Google Scholar]
- 24.Khosravi, R., Maya, R., Gottlieb, T., Oren, M., Shiloh, Y. & Shkedy, D. (1999) Proc. Natl. Acad. Sci. USA 96, 14973–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maya, R., Balass, M., Kim, S. T., Shkedy, D., Leal, J. F., Shifman, O., Moas, M., Buschmann, T., Ronai, Z., Shiloh, Y., et al. (2001) Genes Dev. 15, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos, Y. F., Stad, R., Attema, J., Peltenburg, L. T., van der Eb, A. J. & Jochemsen, A. G. (2001) Cancer Res. 61, 1839–1842. [PubMed] [Google Scholar]
- 27.Stad, R., Ramos, Y. F., Little, N., Grivell, S., Attema, J., van Der Eb, A. J. & Jochemsen, A. G. (2000) J. Biol. Chem. 275, 28039–28044. [DOI] [PubMed] [Google Scholar]
- 28.Chen, J., Marechal, V. & Levine, A. J. (1993) Mol. Cell. Biol. 13, 4107–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banin, S., Moyal, L., Shieh, S., Taya, Y., Anderson, C. W., Chessa, L., Smorodinsky, N. I., Prives, C., Reiss, Y., Shiloh, Y. & Ziv, Y. (1998) Science 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- 30.Bottger, V., Bottger, A., Garcia-Echeverria, C., Ramos, Y. F., van der Eb, A. J., Jochemsen, A. G. & Lane, D. P. (1999) Oncogene 18, 189–199. [DOI] [PubMed] [Google Scholar]
- 31.Hickson, I., Zhao, Y., Richardson, C. J., Green, S. J., Martin, N. M., Orr, A. I., Reaper, P. M., Jackson, S. P., Curtin, N. J. & Smith, G. C. (2004) Cancer Res. 64, 9152–9159. [DOI] [PubMed] [Google Scholar]
- 32.Abraham, R. T. (2004) DNA Repair (Amsterdam) 3, 883–887. [DOI] [PubMed] [Google Scholar]
- 33.Meek, D. W. & Knippschild, U. (2003) Mol. Cancer Res. 1, 1017–1026. [PubMed] [Google Scholar]
- 34.Shinozaki, T., Nota, A., Taya, Y. & Okamoto, K. (2003) Oncogene 22, 8870–8880. [DOI] [PubMed] [Google Scholar]
- 35.Li, C., Chen, L. & Chen, J. (2002) Mol. Cell. Biol. 22, 7562–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito, S., Yamaguchi, H., Higashimoto, Y., Chao, C., Xu, Y., Fornace, A. J., Jr., Appella, E. & Anderson, C. W. (2003) J. Biol. Chem. 278, 37536–375344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.