Abstract
cis-regulatory modules that control developmental gene expression process the regulatory inputs provided by the transcription factors for which they contain specific target sites. A prominent class of cis-regulatory processing functions can be modeled as logic operations. Many of these are combinatorial because they are mediated by multiple sites, although others are unitary. In this work, we illustrate the repertoire of cis-regulatory logic operations, as an approach toward a functional interpretation of the genomic regulatory code.
The differential control of gene expression during development depends primarily on transcription factor interactions with cis-regulatory modules (CRMs) that may be located upstream, downstream, or in the introns of a gene. The potential regulatory functions encoded in the DNA sequence of these modular control units are specified by the combinations of transcription factor target sites they contain, and, typically, a CRM will include sites for four to eight different interactions (1, 2). In the last analysis, it may be said that we will understand the genomic regulatory code only when we can interpret its functional significance by inspection, because it has been possible for decades to recognize protein coding sequence. However, at present, we cannot even recognize many cis-regulatory target sites; nor, perhaps more importantly, can we specify predictively, or, in some cases, even properly name, the elemental functions mediated by the individual sites within a CRM. Here, we take a step toward analysis of the repertoire of elemental cis-regulatory functions.
For a developmentally expressed gene, regulatory control always depends in part on transcription factors presented variably in embryonic time and space. In the following, we use the term “driver” for such factors. These drivers provide spatial and temporal inputs (positive and negative) reflected in the regulatory output of the relevant CRM and in the resulting pattern of gene expression. However, target sites for driver inputs (3) may often account for only a minority of specific CRM target sites. Furthermore, the regulatory outputs of a CRM never exactly equal any of its inputs. This finding can be perceived explicitly when the inputs and outputs are hooked together in a gene regulatory network (2, 4–6). Instead, the CRM processes the driver inputs in a variety of complex ways, depending on its genomic design and, to some extent, on its genomic environs. We find that there is a class of fundamental processing functions mediated by specific CRM target sites and combinations of sites that have the behavior of logic operations, and it is on these sites that we focus herein.
Initial Insights from endo16
Endo16 is a developmentally regulated gene of the sea urchin embryo expressed in endodermal territory. In respect to its genomic regulatory code, endo16 may be the best understood of any developmentally active gene. The functional significance of every detectable target site in the two key CRMs of this gene was determined by mutation, singly, or in combination with other mutations. Their consequences were determined in gene transfer experiments in which the regulatory output was measured spatially and kinetically (3, 7–9). Module A of endo16 controls the initial peak of embryo expression in the endoderm, integrates regulatory transactions requiring upstream interactions, and is the sole source of communications to the basal transcription apparatus (BTA). Module B controls definitive later expression in the midgut. Together, these two CRMs include 13 specific sites, targeted by nine different transcription factors. Every species of interaction proved to have a distinct role.
The endo16 results were an eye opener. First, of the nine factors, no more than two are important drivers, namely, a time varying, although spatially widespread, homeodomain factor (Otx) that provides kinetic input by means of a single site in module A, and a spatially confined POU domain factor (Brn1/2/4) expressed only in midgut that interacts at a single site in module B (refs. 3 and 9 and E. Dorman, E.H.D., and C.-H. Yuh, unpublished data). All of the remaining interactions are mediated by specific DNA-binding proteins, the target sites for which have no regulatory driver activity when associated with a BTA by themselves (in contrast to the sites for Otx and Brn1/2/4). These proteins may be present all of the time, and their functions in the endo16 control system were discovered only by mutation of their target sites in context. Second, a quantitative model that was verified by kinetic measurements of output showed that logic statements represent accurately the functional contributions of those sites where factors other than the drivers bind. Conditional on the occurrence of these interactions, the inputs provided by the drivers are altered in particular ways (including cancellation). In the absence of such interactions, alternative input processing events occur. Thus, from the effects of mutation of individual cis-regulatory sites, reception of driver inputs could be distinguished from other encoded functions. It was demonstrated that the combination of conditional logic functions executed by these sites in combination explicitly represents the input processing capabilities of this whole CRM.
The endo16 analysis, of course, illuminated only functions operating in that control system. Additional such functions are evident in another sea urchin gene that was recently the subject of a similar analysis (the cyIIIa gene; ref. 10 and C. T. Brown and E.H.D., unpublished data). In general, as follows, many diverse cis-regulatory activities, more or less well known, can similarly be treated as operations that determine how driver inputs are used in each given CRM.
Approach to General Treatment of CRM Functions
The events that occur on a cis-regulatory DNA sequence depend, first of all, on the occupancy of its various sites by the transcription factors that recognize and bind to them. Occupancy of given sites, expressed as the continuous probabilities (with values in the interval [0, 1]) of each site being bound, is a function of the intranuclear concentrations of the relevant transcription factors; the equilibrium constants for their interactions with their target sites relative to those for their nonspecific DNA interactions; and the cooperativity constants for their interactions, when bound, with any adjacent proteins with which they exchange energy (refs. 11 and 12; for multiple site occupancies in animal CRMs according to these principles, see ref. 13).
Our object here is to relate regulatory CRM transactions to the regulatory DNA sequence code. Therefore, we have taken the approach of defining the functional significance of each target site in terms of its occupancy, to which is applied an operator function.
For driver sites, occupancies are themselves functions of space and time Oc(s,t), according to changing factor concentrations; when there are multiple sites for the same driver present in limiting amounts, then, assuming that they operate noncooperatively and will function independently, the individual occupancy values can be considered additively. In the following, for other kinds of sites than those that bind time-varying drivers, Oc(s,t) = 1 (the site is always fully occupied because the factor is there always, as are many off the DNA cofactors), except, of course, for the case where the site has been mutated, when Oc(s,t) = 0.
For convenience, we consider operator functions in four categories: (i), D, transcriptional activation operators: quantitative transcriptional activation functions mediated directly by driver target sites, considered to be directly proportional over much of its range to their Oc(s,t) values, (for limiting factor concentrations, as in Eq. 2, see ref. 13); (ii), F, BTA control operators: operator functions mediated by sites that control interaction with the BTA, according to the intrinsic properties of the individual species of DNA-binding proteins (and their cofactors) that interact at these sites; (iii), G, combinatorial logic operators: operator functions that by definition depend on the participation of multiple CRM sites, i.e., on the combinatorial interaction of the proteins interacting at these sites (and their cofactors); and (iv), E, external control operators: permissive or nonpermissive operator functions mediated by sites outside a CRM.
Examples of Combinatorial Logic Functions (G Operators)
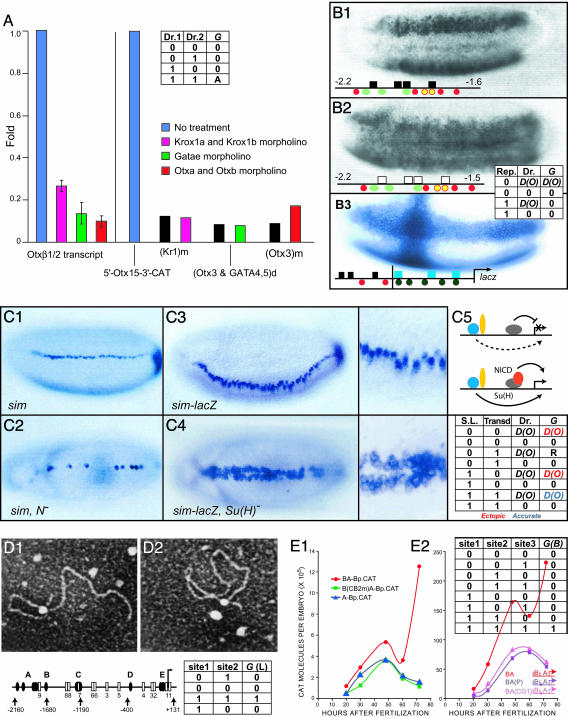
AND Operators. cis-regulatory analyses often reveal requirement for diverse sites to be occupied for significant expression to occur. In development, this device is used to ensure that a gene is activated only in a subdomain, spatial and/or temporal, where two generally noncoincident inputs overlap (reviewed in ref. 2). In the absence of either factor, there is no expression, even where the other factor is present at the normal level, and if either site is destroyed, there is no expression, even if the other remains intact. An experimental example is shown in Fig. 1A (14). In the input/output table (truth table) shown, the output is considered qualitatively as Activation (A), when both factors are present above threshold (th), or if they are not, the output is considered as insignificant activity, as illustrated in the experiment (Fig. 2A) (of course, the drivers performing the activation function could be present at different levels over threshold, so, in more detail, G = D(Oc1(s,t), Oc2(s,t)) = Oc1(s,t)*Oc2(s,t)*Amax, where Amax is the activation impetus when both sites are fully occupied; in other words, for either of the required inputs, when either Oc(s,t)<th, G = 0. AND operators occur frequently and there are diverse biochemical bases for their behavior, including required cooperative factor interactions, synergistic interactions with the BTA, and joint interactions with effector cofactors.
Fig. 1.
Cis-regulatory logic functions. (A) Experimental example of AND logic operation. [Reproduced with permission from ref. 14 (Copyright 2004, Elsevier).] Bars display relative activity of expression constructs, including a cis-regulatory element of the sea urchin otx gene, driving a chloramphenicol acetyltransferase (CAT) reporter. The construct was injected into sea urchin eggs, and the level of CAT transcripts it produced was measured by using quantitative PCR. Results of removal of individual inputs by antisense (morpholino) treatment are shown for wild-type construct as indicated (colored bars), or, when individual respective target sites were mutated (black bars). As required, blocking the inputs and mutating the sites gives the same effect; all three inputs (Kr, Gata, and Otx) are needed, and if any is absent, no significant activity is obtained (14). The truth table shows values of the G operator function for a two input system [drivers 1 and 2 (Dr. 1 and Dr. 2)], where 0 and 1 indicate sub- and above-threshold inputs respectively, and A indicates an activating output (see text). (B) Experimental example of short-range repressor operation. Photographs display lacZ reporter staining in transgenic Drosophila embryos bearing rhomboid (rho) expression constructs. Relevant target sites are indicated below. Activators: Dorsal (red), bHLH (green), Twist (yellow), and Bicoid (black circles). Repressors: Snail (black boxes) and Krüppel (blue boxes). (B1 and B2) Ventral views. Expression of rho-lacZ constructs with and without target sites (open boxes, mutated) for the Snail short-range repressor is shown. These sites are required to prevent expression in the prospective mesoderm. [Images in B1 and B2 are reprinted with permission from ref. 30 (Copyright 1994, Cold Spring Harbor Lab. Press).] (B3) Autonomy of short-range repression, demonstrated by fusion of rho and evenskipped stripe 2 (eve2) CRMs; the latter uses the Krüppel short-range repressor to establish posterior border of expression. The crossed expression pattern shows that each repressor functions independently, and that neither repressor interferes with the activation of the other CRM. [The image in B3 is reprinted with permission fromref. 15 (Copyright 1996, Cold Spring Harbor Lab. Press).] The truth table shows that the operator function G has the activating regulatory value produced by the level of driver occupancy [D(Oc(s, t))], here for a one-driver element}, whereas if the repressor (Rep) is present, or the driver is absent, there is no output. (C) Transcriptional toggle switch, Notch (N) signal transduction system effects on the single-minded (sim) gene of Drosophila (data are from ref. 31). (C1) Wild-type endogenous expression of sim in prospective midline neuroblasts. (C2) Expression of sim in N mutant embryos; N signaling is required positively for normal expression.(C3) Expression of sim-lacZ construct in wild-type embryos; an enlargement is on the on right. (C4) A view of C3, in embryos lacking the transcription factor [Su(H)] that transduces the N signal. In the absence of Su(H), ectopic expression occurs. (C5) CRM diagrams: Dorsal and Twist, activators (blue and yellow, respectively), Su(H), a repressor (gray), except when bound by intracellular N fragment (red), as a result of N signaling. [Images in C1–C5 are reprinted with permission from ref. 31 (Copyright 2000, Cold Spring Harbor Lab. Press).] The truth table shows that there are several possible values of the operator function G: expression, ectopic, or normal (according to the level of driver occupancy, here treating the two molecules portrayed as a single driver), or repression of output that would otherwise be produced according to D(O) (R, only in those cells where the values 0, 1, [D(O) obtain], or just no output (0). The values are combinatorially conditional, depending on whether the signal ligand (S.L.) is presented, and whether or not the transducer [here, as Su(H)], and the activating driver (e.g., here, Twist or Dorsal) are present. (D) Loop formation mediated by multiple CRM sites (17). (D1) Map of cis-regulatory system of the sea urchin cyIIIa gene, with specific sites for SpGCF1, a multimerizing, DNA-binding protein is highlighted; sites for other transcription factors are shown as as open boxes. (D2 and D3) Electron micrographs of loops formed by purified SpGCF1 protein mixed with cyIIIa cis-regulatory DNA in vitro.(D2) A–C site loop. (D3) A–E site loop. [Images in D1–D3 are reprinted with permission from ref. 17 (Copyright 1995, National Academy of Sciences).] The truth table illustrates the point that loop formation, an all-or-nothing operation, requires both sites occupied for any given loop to form. (E) Combinatorial functional linkage of two CRMs, data are from the sea urchin endo16 gene (3, 9). Kinetics for output of CAT reporter enzyme when module A (proximal) and the adjacent module B are included in the construct, are shown in red over developmental time, in two experiments. (E1) Kinetics for module A alone (blue), are identical with kinetic output of whole BA construct bearing mutation of site for a CRM B-CRM A DNA-binding linker protein (CB2). [The image in E1 is reprinted with permission from ref. 9 (Copyright 1998, AAAS, www.sciencemag.org).] (E2) Mutations of sites for two other linker proteins (P and CG1) in the otherwise complete BA construct yield kinetics that are also the same as for CRM A alone. [The image in E2 is reprinted with permission from ref. 3 (Copyright 2001, Company of Biologists Limited).] The truth table shows that all three sites are required for normal kinetic input of module B [here, abbreviated as G(B)] to register, and only in this condition are the red curves generated.
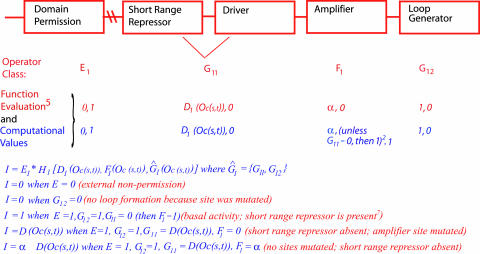
Fig. 2.
Computation of conditional regulatory output (I) for an arbitrary model CRM. Function evaluation, in red, indicates the possible values the operators may assume, and computational values, in blue, are those values that arise from application of the indicated rules to these values. Blue statements below are summaries of points discussed in text, and their biological meaning is shown in parentheses in red.
Short-Range Repressor Binding Within a CRM. Here, the CRM contains target sites for a transcriptional repressor (i.e., a DNA-binding factor that interacts with a cofactor that executes the repression), and these site(s) must be within, or loosely adjacent to, a CRM that also contains sites for an activating driver. As in the example in Fig. 1B (ref. 15; see truth table), the repressor is dominant, so that the activators function only in its absence, or else the output is nil. However, the effect of these short-range repressors is limited to the cancellation of the activation functions of that CRM with which they are associated.
Signal-Mediated Toggle Switch. As reviewed by Barolo and Posakony (16), many developmentally active intercellular signaling systems used in processes of fate specification operate in a janus fashion: when the signal ligand is presented and the DNA-binding transcription factor that mediates signal transduction is also present in the CRM, a coactivating driver is permitted to stimulate transcriptional expression; but if the ligand is absent, the same transducing factor acts as a dominant repressor. An example is shown in Fig. 1C: here, the signal is presented locally, and expression of the system outside of the confined region where the signal is received is prevented by the transducer in its role as repressor, whereas expression within the domain of signal reception is permitted (in this case, the transducer has little activating function of its own; cf. ref. 16). Thus, as in the example illustrated, if the transducer is absent from the CRM, or its site is destroyed, ectopic expression results.
Essential DNA Looping. Some CRMs (perhaps all that are located distantly from the BTA) contain sites for DNA-binding looping proteins. One class of such proteins multimerizes after being bound to distant sites (refs. 17–19; see the example in Fig. 1D), thus causing specific loop formation. This formation is evidently used to bring the CRM into proximity other CRMs (and thereby, to the BTA). Loop formation is Boolean in behavior: either the proteins and sites for these proteins are sufficiently present, and the loop forms, or else, it does not.
Module Linker Function. The endo16 analysis revealed another function, linkage of the A and B CRMs of this gene, requiring three different DNA-binding proteins. Mutation of the sites for any one of these proteins results in functional detachment of module B, even though it remains physically associated with module A and the BTA. This result is revealed by return to the kinetic output of module A alone, because opposed to the distinct output of the linked combination (Fig. 1E). This function also behaves in a Boolean manner: the linkage is extant or it is broken.
Examples of CRM Logic Functions Controlling Direct Interactions with the BTA (F Operators)
CRM Silencers. Some CRM target sites bind repressors that silence the BTA, so that their effect is not limited to the CRM that includes these sites, but rather, extend to any CRM using that BTA for transcriptional expression (e.g., see refs. 20 and 21). Silencers behave as unit dominant-negative regulators. Silencers do not require interactions with nearby target sites where activators bind, but (by means of dedicated cofactors) they directly affect the BTA. In our models, if a silencer is present in a CRM and is occupied, then F(Oc(s,t)) = R(repression), and by the rules below, the result is to set the output of the regulatory system to 0.
Communicators. The site-function analyses carried out on the endo16 and cyIIIa genes (20, 21) have both revealed CRM target sites that are required in order for a function that is mediated by other sites elsewhere to have an effect at the level of BTA execution. In endo16, the spatial domain of the early phase of expression is confined to the future endoderm by repressors that bind at known target sites in upstream modules ≈1 kb away from module A, one of which is a cAMP-response element-binding protein factor. However, all of the repressor interactions are inutile, and ectopic expression occurs the same as if the repressor sites are absent, if a certain target site of module A is mutated (refs. 7 and 9 and C.-H. Yuh and E.H.D., unpublished data). Similarly, in CyIIIa, a site for a POU domain factor in the proximal module of that regulatory system, is required for the spatially controlled activating function of a more distal module to have any effect on expression (C. T. Brown and E.H.D., unpublished data). In neither case is the mechanism known. However, whether directly or indirectly, the proteins binding at these target sites cause regulatory inputs from outside the CRM (negative and positive, respectively) to have an effect, which means interacting with the BTA, i.e., transferring or communicating the inputs to the BTA. Interaction with the BTA may indeed be a general function of proximally located CRMs. In the examples cited, if the communicator sites are absent the regulatory values of the respective distal modules is 0, irrespective of whatever interactions occur there; that is, they behave in a Boolean manner.
Amplifiers. Another function discovered in the endo16 analysis was linear amplification of a positive regulatory input from module B by sites in module A. In the quantitative kinetic study of Yuh et al. (3), the expression of a construct including modules A and B plus the BTA, is equal at all points in time to approximately four times the output of a construct including only module B plus the BTA. Directly or indirectly, this function of module A requires communication of the module B output and an effect on the level of BTA activity (but not de novo generation of a positive regulatory output in module A, because, although the function is located in module A, the output that is amplified comes from module B, as shown by many experiments). This amplification function is not exclusive with respect to other functions, and for example, part of the 4× amplification is mediated by means of the linker target sites of Fig. 1E, and part of the 4× amplification is mediated by sites for proteins that may be directly engaged in communication with the BTA (3, 9). In either case, this is another type of specific CRM function, and in our models, the amplification operation is expressed by multiplying a driver output by a constant >1, F = α. If an amplifier site is mutated so its occupancy is 0, F = 1. If there is no driver input D(Oc(s,t)) = 0, α is set at 1 also.
External Functions (E Operators). Functions that are located outside the CRM, but that control its activity, cannot be enumerated here; for one thing, it remains unclear exactly how major classes of external mechanism work, although it is completely clear that they do work. These conclusion are what could be summed together as domain-choice operators. The phrase denotes (nonexclusively): alternative looping that brings into action a given CRM under given circumstances and other CRMs under other circumstances; insulators that prevent more proximal CRMs from interacting with a BTA while permitting more distal ones to so interact (e.g., refs. 22–24); insulators that, in certain developmental circumstances, transfer a given CRM to an inaccessible or sequestered chromatin domain (e.g., see refs. 25 and 26); and distant locus activators that, if bound by the factors for which they contain target sites permit more proximal CRMs to be active, probably by means of loop formation with them (27–29). From the standpoint of a given CRM, each of these functions is externally mediated, and each can be considered permissive in a Boolean sense. For these cases, either the CRM is allowed to operate (E = 1), or it is not (E = 0). In addition, there is a range of permissiveness because of chromatin states that are a function of the previous history of the cell, that can best be modeled by use of continuous E values between 0 and 1.
Another class of external operators probably exists as well, known as is external silencers, that are not located within CRMs. Their target sites occur in the DNA flanking the CRM proper, not within the conserved sequence patches or complex clusters of diverse target sites that can be used to identify CRMs, or in any case, they are located in much smaller, much less diverse site clusters. Like other silencers, external silencers function in a dominantly negative, Boolean manner.
Assembling the Output
We may consider the integrated regulatory output of a CRM, I, as the combinatorial result of all of its site occupancies, and the operations applied to them. Here, we must take into account the two most important aspects of CRM performance: its conditionality and its regulatory code, which determines its capabilities according to the identity and arrangement of its DNA sites. The conditionality of CRM performance in our models follows from the site occupancy values that vary according to experimental and developmental case. To represent the in-built design specifics for any given CRM, we introduce the concept H, a higher-order operator of the D, F, and G operator functions pertaining to that CRM. In other words, the regulatory output for any given time, spatial domain, and for a specific CRM, i, can be considered to depend on a conditional, and on a hard-wired component: thus, where E represents the external permissiveness value (if one applies, 0 or 1, as above), I(s,t,i) = Ei*Hi[D̂i(Oc(s,t)), F̂i(Oc(s,t)), Ĝi(Oc(s,t))] where D̂i indicates all D functions in module i; and F̂i and Ĝi represent the set of all F and all G functions for module i. In the absence of any information about E function, the default value of E is 1.
To compute a real I(s,t,i) value, we can consider the combinatorial function Hi as a product; i.e., H(s,t,i) = Hi[D̂i(Oc(s,t)), F̂i (Oc(s,t)), Ĝi (Oc(s,t))] = ∏ D̂i (s,t)*∏F̂i (s,t)*∏ Ĝi (Oc(s,t)), given some computational rules (example in Fig. 2), where ∏ D̂i is the product of all D functions in D̂i, similarly for ∏ F̂i and ∏ Ĝ i.
Rules
Evaluate D(Oc(s,t)) = Oc(s,t)*Amax for activating drivers (see above); that is, express the regulatory output in terms of the maximum output of the fully occupied driver (13). If there are two or more such functions and they operate independently in CRM(i), then their outputs are added. For the special case of AND operations on two drivers, see above.
If, according to truth tables (Fig. 1C), the value of G(Oc(s,t)) is a signal-mediated toggle switch, or F[Oc(s,t)] is a silencer, and either has the value R (i.e., repression is in effect), then the computational value is 0, and I(s,t,i) = 0, because repression is dominant.
For any operator function F(Oc(s,t)) or G(Oc(s,t)) that may have a value expressed in terms of a driver function D(Oc(s,t)), when the value of that function is 0 in a truth table, then the computational value is set to 1, that is, this operator does not in that particular condition affect the product Hi. This grouping includes AND operators, short-range repressors, and amplifiers.
For any Boolean F(Oc(s,t)) or G(Oc(s,t)), as defined above, the truth table values of 0 or 1 are applied directly as such to the computation. If the overall value of I(s,t,i) is 1, rather than some function of Amax (>1), then the expression is at a basal level independent of circumstances that affect the CRM.
Discussion
Our main object has been to attempt a framework for interpretation of the cis-regulatory sequence code by linking target sites directly to a defined set of elemental functions, the integrated combination of which is the output of the modular control element. Six basic properties, or principles of CRM function, that we claim apply to all CRMs, are taken into account in this approach:
The functional repertoire of each CRM is a constant, sequence-based, feature of the species.
The specific design of any CRM can be expressed in terms of its elemental functions.
CRMs process continuously varying driver inputs.
Many CRM processing functions can be modeled as logic operations.
Occupancy is causal: CRM outputs are intrinsically conditional on site occupancies.
In all cases where there is qualitatively unique factor–site interaction, the consequences of site mutation and of absence of occupancy due to absence of the unique DNA-binding factor are equal (0cis = 0trans). We note that at the level described the example of F and G functions appear irreducible, in the sense that they cannot be described equivalently by simpler functions. That is, the unit of meaning is not the sequence of a given site, but the function it generates in its cis-regulatory context. This rule is explicit for G functions that are combinatorial. However, it is often true that a given site sequence present in different cis-regulatory modules proves to execute different functions, depending on cis-regulatory design, i.e., exactly where the site is with respect to which other sites are nearby. It is the cis-regulatory design (our H class of operator functions) that is the hard-wired feature of the genomic control code.
The small set of F and G functions that we have included are anything but complete. Those functions we discuss serve as canonical examples, but there are many more to be similarly treated. For example, there are several different modes of behavior of transcriptional signal transduction systems (16) related to, but not identical to, the toggle switch in our model; and there are additional functions already evident in the endo16 analysis besides those we have included here (for example, the conditional intermodule repressor; ref. 3). However, it is too early to attempt completeness, because the number of cis-regulatory modules that have, so far, been examined experimentally at the level required to perceive other than prominent driver inputs is still so small. Yet, ultimately, the objective of reading the genomic code on inspection will require a more or less complete repertoire of such functions; this means that we will need to have for reference many more detailed cis-regulatory examinations in which the significance of each specific binding site has been determined.
However demanding are such experimental analyses, they can be performed in many systems, with present technology. Furthermore, the repertoire of what we have termed F and G functions is going to be finite, and we predict the results will not be significant. Calculating apparatus for the probabilistic relations between driver inputs, site occupancies, and transcriptional BTA function can already be dealt with (ref. 13, our D functions). Comparatively speaking, the diversity or complexity of the genomic regulatory code is going to be greatly less than the diversity of the biochemical operations that execute each type of function (that is why it is properly referred to as a code). This conclusion is obvious from the fact that there are many different factors that in our terms carry out the same functions of repression, of activation, probably of looping, of signal transduction, and so forth, and even more different sets of factors that execute G class combinatorial functions such as AND logic. Tackling the genomic regulatory code head on is liable to be a more direct avenue to learning what it says than by dissection of the particular biochemistry operative in every different CRM. To reverse the argument, the mechanistic biochemical exploration of cis-regulatory function will indeed be much facilitated if it can be couched in terms of an elemental functional CRM repertoire.
Acknowledgments
This work was supported by a grant from Applied Biosystems/Celera Genomics (to S.I.) and by U.S. Department of Energy Grant DE-FG02-03ER63584 (to E H.D.).
Abbreviations: CRM, cis-regulatory module; BTA, basal transcription apparatus.
References
- 1.Arnone, M. & Davidson, E. H. (1997) Development (Cambridge, U.K.) 124, 1851–1864. [DOI] [PubMed] [Google Scholar]
- 2.Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
- 3.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (2001) Development (Cambridge, U.K.) 128, 617–628. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, E. H., Rast, J. P., Oliveri, P, Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Science 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, E. H., McClay, D. R. & Hood, L. (2003) Proc. Natl. Acad. Sci. USA 100, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveri, P. & Davidson, E. H. (2004) Curr. Opin. Genet. Dev. 14, 351–360. [DOI] [PubMed] [Google Scholar]
- 7.Yuh, C.-H. & Davidson, E. H. (1996) Development (Cambridge, U.K.) 122, 1069–1082. [DOI] [PubMed] [Google Scholar]
- 8.Yuh, C.-H., Moore, J. G. & Davidson, E. H. (1996) Development (Cambridge, U.K.) 122, 4045–4056. [DOI] [PubMed] [Google Scholar]
- 9.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (1998) Science 279, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhamer, C. V., Yuh, C.-H. & Davidson, E. H. (1996) Proc. Natl. Acad. Sci. USA 93, 9322–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackers, G. K., Johnson, A. D. & Shea, M. A. (1982) Proc. Natl. Acad. Sci. USA 79, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson, B. M., Lewis, C. D. & Felsenfeld, G. (1985) Cell 41, 21–30. [DOI] [PubMed] [Google Scholar]
- 13.Bolouri, H. & Davidson, E. H. (2003) Proc. Natl. Acad. Sci. USA 100, 9371–9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuh, C.-H., Dorman, E. R., Howard, M. L. & Davidson, E. H. (2004) Dev. Biol. 269, 536–551. [DOI] [PubMed] [Google Scholar]
- 15.Gray, S. & Levine, M. (1996) Genes Dev. 10, 700–710. [DOI] [PubMed] [Google Scholar]
- 16.Barolo, S. & Posakony, J. W. (2002) Genes Dev. 16, 1167–1181. [DOI] [PubMed] [Google Scholar]
- 17.Zeller, R. W., Griffith, J. D., Moore, J. G., Kirchhamer, C. V., Britten, R. J. & Davidson, E. H. (1995) Proc. Natl. Acad. Sci. USA 92, 2989–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su, W., Jackson, S., Tjian, R. & Echols, H. (1991) Genes Dev. 5, 820–826. [DOI] [PubMed] [Google Scholar]
- 19.Mastrangelo, I. A., Courey, A. J., Wall, J. S., Jackson, S. P. & Hough, P. V. C. (1991) Proc. Natl. Acad. Sci. USA 88, 5670–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai, H. N., Arnosti, D. N. & Levine, M. (1996) Proc. Natl. Acad. Sci. USA 93, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, H. & Levine, M. (1999) Proc. Natl. Acad. Sci. USA 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West, A. G., Gaszner, M. & Felsenfeld, G. (2002) Genes Dev. 16, 271–288. [DOI] [PubMed] [Google Scholar]
- 23.Hagstrom, K., Muller, M. & Schedl, P. (1996) Genes Dev. 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, J., Barolo, S., Szymanski, P. & Levine, M. (1996) Genes Dev. 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- 25.Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. (2004) Mol. Cell 13, 291–298. [DOI] [PubMed] [Google Scholar]
- 26.Bell, A. C., West, A. G. & Felsenfeld, G. (2001) Science 291, 447–450. [DOI] [PubMed] [Google Scholar]
- 27.Wijgerde, M., Grosveld, F. & Fraser, P. (1995) Nature 377, 209–213. [DOI] [PubMed] [Google Scholar]
- 28.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. (2002) Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 29.Spitz, F., Gonzalez, F. & Duboule, D. (2003) Cell 113, 405–417. [DOI] [PubMed] [Google Scholar]
- 30.Gray, S., Szymanski, P. & Levine, M. (1994) Genes Dev. 8, 1829–1838. [DOI] [PubMed] [Google Scholar]
- 31.Morel, V. & Schweisguth, F. (2000) Genes Dev. 14, 377–388. [PMC free article] [PubMed] [Google Scholar]