The medieval philosopher William of Occam admonished that it is best to minimize the postulated entities needed to explain a system. This concept of choosing the simplest explanation of unknown phenomena is now known as the principle of parsimony or Occam's razor. However, in biology, where systems tend to evolve toward states of greater complexity, Occam's razor can be a dangerous tool. Most often, the known components of a biological process increase with study. It is not always safe to assume that a limited number of factors are involved. However, sometimes we do find a simple explanation of a puzzling phenomenon that requires no new unknowns to be added to the model. Such an event is described in this issue of PNAS, where Ibrahim et al. (1) nicely explain a longstanding puzzle in eukaryotic mRNA processing. Specifically, they demonstrate a plausible mechanism for how exons are always linked in order during the pre-mRNA splicing process, without a single exon being skipped.
Eukaryotic mRNAs are transcribed as long precursor molecules containing intervening sequences, or introns separating the coding or otherwise functional portions of the transcript called exons (Fig. 1A). To form a mature mRNA ready for translation, the cell must precisely excise the introns and ligate the exons together. This process of pre-mRNA splicing is carried out in the cell nucleus by a large macromolecular complex called the spliceosome (2). Components of the spliceosome recognize special sequences at the intron ends called splice sites. The 5′ splice site (at the 5′ end of the intron) is initially bound by the U1 small nuclear RNP (snRNP), and the 3′ splice site is bound by the protein U2 auxiliary factor (U2AF) (3, 4). These components interact to bring the two ends of the intron together before going on to assemble the rest of the spliceosome that ultimately catalyzes the cleavage–ligation reactions.
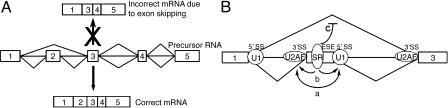
Fig. 1.
The enforcement of exon linking order and the definition of splice sites are both controlled by SR proteins. (A) Exons in a multiexon pre-mRNA are always linked in order. Mechanisms are present that prevent the skipping of exons, such as joining exon 1 to exon 3. (B) Exons and splice sites are defined by interactions across the exon, between the U1snRNP and U2AF (a). SR proteins bound to ESEs also stimulate the assembly of these factors (b). SR proteins also inhibit the splicing of introns that contain an ESE (c). This inhibitory interaction prevents the splicing of nonadjacent exons and forces exon inclusion.
Early on, two questions of how splice sites are chosen for pairing during splicing were seen as particularly mysterious (5–7). First, introns can be very long (>1,000 nt) and have within them many copies of the splice site consensus sequences. It was not understood how the correct sites at the intron ends were recognized while the other “cryptic” sites were avoided. Second, pre-mRNAs can contain many exons that must be joined in precise 5′ to 3′ order to create a proper reading frame for translation. How this correct linkage order is achieved, without skipping or excluding any exons, is unknown.
The conceptual problem of splice site recognition was solved by the demonstration that the splicing apparatus uses more than just the splice site sequences to distinguish between real and cryptic splice sites. A key breakthrough was the discovery that exons can serve as units of recognition (8). Unlike introns, exons tend to be short (50–250 nt). Through a process called exon definition, the splice sites on either side of an exon can stimulate each other's recognition (Fig. 1B). For example, U2AF assembly and splicing at a 3′ splice site are enhanced by the presence of the 5′ splice site downstream, even though this downstream 5′ splice site will not be spliced itself by the same spliceosome. In addition to exon-bridging interactions between splice sites, there are many additional non-splice-site sequences that serve to define exons (9). Most exons contain exonic splicing enhancers (ESEs) that stimulate splicing at the adjacent splice sites. Exons without these elements are generally not recognized and tend to be excluded from the mature mRNA. There are also exonic splicing silencer elements (ESS), as well as intronic splicing enhancers and silencers (ISE and ISS), that can be important in the splicing of constitutive and regulated exons. Recent studies indicate that the number of splicing regulatory elements is very large (10–12). Thus, the unused cryptic sites in introns are not recognized for several reasons: they do not have a proper cognate splice site that defines them as part of an exon, they lack enhancer elements, or they are repressed by silencer elements.
Each of these RNA elements binds a protein that positively or negatively affects spliceosome assembly. The most studied of these splicing factors are the members of the SR protein family (13). These proteins have one or two RNA-binding domains coupled to a serine-arginine rich domain (hence, “SR”). These proteins are implicated in many different aspects of the splicing process. Most significantly, they bind to ESEs and stimulate the association of U2AF and the U1 snRNP with the pre-mRNA (Fig. 1B). This enhancement requires the SR domain, which can engage in both protein–protein and protein–RNA interactions (14). Most exons contain ESEs that bind one or more SR proteins, and these proteins can be essential for defining the exon. This ESE requirement is seen in many human genetic diseases, where mutation of an ESE causes exon skipping and therefore improper production of the encoded protein (15). The discovery of exon definition and of ESEs has improved our understanding of splice site recognition. However, our picture of the process has not gotten simpler. The many new factors and interactions needed to assemble a spliceosome make the reaction dauntingly complex.
Unlike splice site recognition, the question of what enforces the correct order of exon joining has remained obscure. It is this question for which Ibrahim et al. now provide a possible answer. Earlier models for how exon order is enforced were not well supported by experimental data (7). Mechanisms involving the coupling of splicing to transcription, or long-range interactions across introns, may occur in specialized systems but lack support as a general mechanism (16). The enforcement of exon order is an important issue because exon skipping can delete peptide sequence from the protein product or cause truncation of the reading frame. Although such exon exclusion can serve a physiological function when regulated, for most exons it would be highly deleterious, and there needs to be a mechanism to prevent it.
The solution to the exon order problem proposed by Ibrahim et al. is appealing in its simplicity. It requires no new components and fits what we know about exon recognition. Pointing to a new activity of SR proteins, Ibrahim et al. find that SR proteins bound to an internal exon inhibit splicing of the flanking exons to each other (Fig. 1B). Thus, introns cannot assemble a spliceosome if they have an ESE within them. This simple rule will force the splicing of directly adjacent exons and prevent exon skipping. In addition to being enhancers of splice site recognition, exon-bound SR proteins are enforcers of exon order.
The experiments made use of a β-globin splicing substrate that is well characterized for its splicing enhancer sequences. A construct was created carrying globin exon 1 and a duplicated globin exon 2. This construct presents the splicing apparatus with a choice of two 3′ splice sites: The upstream site is proximal to the 5′ splice site, and the downstream site is distal. Under standard conditions, splicing is primarily between the 5′ splice site of exon 1 and the proximal 3′ splice site. Exon 2 contains several known ESEs (17). As these elements are progressively deleted from the upstream exon 2, splicing to the proximal 3′ splice site is lost and switches to the distal 3′ splice site, in part because of the loss of enhancing SR proteins acting on the upstream 3′ splice site. In this study, Ibrahim et al. test constructs with the upstream 3′ splice site mutated. If the ESEs were only enhancing the adjacent proximal splice site, then loss of this site should lead to immediate switching to the distal site; however, this did not happen. When the ESEs for the upstream exon were present, splicing to the distal site was still low. When the upstream ESEs were deleted, splicing again switched to the distal site. Even without the competing proximal 3′ splice site, the distal 3′ splice site was inhibited by the proximal ESEs. The repression of the distal site also occurred when a second upstream 5′ splice site was present. This RNA, with two 5′ and two 3′ sites, undergoes excision of the central intron. However, the splice sites remaining after this first excision fail to be used because the central exonic sequences remain in the intron. The ESEs are apparently inhibitory for spliceosomes that jump over them and thus prevent the definition an intron between a 5′ splice site upstream of the ESE and a 3′ splice site downstream.
ESEs are usually bound by SR proteins that mediate their splicing enhancer activity. To examine whether SR proteins also mediate the repression of distal splicing by ESEs, Ibrahim et al. tested extracts lacking these proteins. To separate the activities of the upstream and downstream ESEs, Ibrahim et al. built a new construct with binding elements for two individual SR proteins 9G8 and SC35. The 9G8 element was placed adjacent to the proximal 3′ splice site, and the SC35 element was placed adjacent to the distal 3′ site. When just SC35 was added to a splicing reaction containing this RNA, both distal and proximal splicing were activated. Significantly, the addition of 9G8 protein to this reaction specifically inhibited the distal site. In another transcript where proximal splicing cannot occur, 9G8 still inhibits distal splicing. This finding shows that SR proteins are required for an ESE to inhibit splicing to a distal site.
The inhibition of splicing to downstream splice sites by ESE-bound SR proteins provides a model for how exon order is maintained and exon skipping is avoided. In a multiple-exon transcript, each exon will contain ESEs and assemble SR proteins, which are generally not found in introns (10). Splicing from one exon will occur only to an adjacent exon, because jumping over an exon would require jumping over its SR/ESE complex. Although it must be confirmed in vivo, this simple mechanism seems likely to hold for most exons. However, it is not yet clear what precise interaction between the SR protein and a spliceosomal component prevents spliceosome assembly. The inhibition of exon skipping by SR proteins has important consequences for understanding alternative exons whose inclusion and skipping are controlled by cellular conditions (9). Similar to normal exons, these exons generally contain ESEs that are needed to activate their splicing. In addition to mechanisms that affect their own splicing, regulated exons must also have a means of allowing splicing of the flanking exons.
Interestingly, in addition to ESEs, most alternative exons carry binding sites for non-SR proteins (9). Notably, the proteins hnRNP A1 and PTB (hnRNP I) are negative regulators of exon inclusion that in some cases antagonize enhancement by SR proteins. The above results imply that, to induce exon skipping, these factors must also block the enforcement of exon order by SR proteins. This blocking could be done by preventing SR protein binding or by other mechanisms. It will be interesting to identify the residues on an SR protein required for its order-enforcement function and test whether these are engaged differently when an alternative exon is repressed.
There are also questions about the pathway of spliceosome assembly brought up by these results. If U2AF and U1 originally interact across an exon during exon definition, they must subsequently interact across an intron to assemble a spliceosome. This transition is perhaps the most poorly defined step in splicing. Could the intronic SR proteins be inhibitory because they interfere with this transition? In addition to mediating ESE activity, another apparent function of SR proteins is to bridge the 5′ and 3′ splice sites during spliceosome assembly (18, 19). Could this be the interaction blocked by the intronic ESEs? The idea that SR proteins bound to ESEs are the key enforcer of exon linking order is a satisfying example of a simple model explaining a complex effect. Nevertheless, this simple picture is likely to develop many complicating details as we examine additional exons.
See companion article on page 5002.
References
- 1.Ibrahim, E. C., Schaal, T. D., Hertel, K. J., Reed, R. & Maniatis, T. (2005) Proc. Natl. Acad. Sci. USA 102, 5002–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge, C. B., Tuschl, T. & Sharp, P. A. (1999) in The RNA World, eds. Gesteland, R. F., Atkins, J. F. & Cech, T. R. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 525–560.
- 3.Brow, D. A. (2002) Annu. Rev. Genet 36, 333–360. [DOI] [PubMed] [Google Scholar]
- 4.Kent, O. A., Ritchie, D. B. & Macmillan, A. M. (2005) Mol. Cell. Biol. 25, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp, P. A. (1981) Cell 23, 643–646. [DOI] [PubMed] [Google Scholar]
- 6.Lewin, B. (1980) Cell 22, 324–326. [DOI] [PubMed] [Google Scholar]
- 7.Black, D. L. (1995) RNA 1, 763–771. [PMC free article] [PubMed] [Google Scholar]
- 8.Berget, S. M. (1995) J. Biol. Chem. 270, 2411–2414. [DOI] [PubMed] [Google Scholar]
- 9.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 10.Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. (2002) Science 297, 1007–1013. [DOI] [PubMed] [Google Scholar]
- 11.Wang, Z., Rolish, M. E., Yeo, G., Tung, V., Mawson, M. & Burge, C. B. (2004) Cell 119, 831–845. [DOI] [PubMed] [Google Scholar]
- 12.Zhang, X. H. & Chasin, L. A. (2004) Genes Dev. 18, 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graveley, B. R. (2000) RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen, H. & Green, M. R. (2004) Mol. Cell. 16, 363–373. [DOI] [PubMed] [Google Scholar]
- 15.Cartegni, L., Chew, S. L. & Krainer, A. R. (2002) Nat. Rev. Genet. 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 16.Kornblihtt, A. R., de la Mata, M., Fededa, J. P., Munoz, M. J. & Nogues, G. (2004) RNA 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaal, T. D. & Maniatis, T. (1999) Mol. Cell. Biol. 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boukis, L. A., Liu, N., Furuyama, S. & Bruzik, J. P. (2004) J. Biol. Chem. 279, 29647–29653. [DOI] [PubMed] [Google Scholar]
- 19.Blencowe, B. J., Issner, R., Nickerson, J. A. & Sharp, P. A. (1998) Genes Dev. 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]