Abstract
The diagnosis and treatment of lung cancer have evolved into the era of precision medicine. Liquid biopsy, a minimally invasive approach, has emerged as a promising practice in genetic profiling and monitoring of lung cancer. Translating liquid biopsy from bench to bedside has encountered various challenges, including technique selection, protocol standardisation, data analysis and cost management. Regarding these challenges, the 2016 Chinese Lung Cancer Summit expert panel organised a trilateral forum involving oncologists, clinicians, clinical researchers, and industrial expertise on the 13th Chinese Lung Cancer Summit to formally discuss these controversies. Six consensuses were reached to guide the use of liquid biopsy and perform precision medicine in both clinic and research.
Keywords: Liquid biopsy, next-generation sequencing, lung cancer
Introduction
The 13th Chinese Lung Cancer Summit was held on 4–5 March 2016, by the Chinese Society of Clinical Oncology and the Chinese Society of Lung Cancer. The theme of the summit was ‘Precision Cancer Medicine: The potential role of liquid biopsy and next-generation sequencing in clinical practice’.
The diagnosis and treatment of lung cancer have moved into the era of precision medicine. As a minimally invasive approach, liquid biopsy can easily be repeated, and it may facilitate comprehensive genetic profiling of lung cancer. Although an increasing number of molecular testing techniques are employed with liquid biopsy, there is a lack of established consensus to guide its utilisation. Emerging challenges include how to choose among various techniques, how to normalise the applications of testing in clinical practice and research, how to improve the cost-effectiveness of genetic profiling in clinical practice and how to maximise the benefit for patients. Forming a consensus on liquid biopsy in lung cancer is thus warranted. During the summit, a trilateral forum was organised involving clinicians, clinical researchers and industrial expertise. Six consensus decisions were achieved regarding current challenges and potential directions for translating liquid biopsy from bench to bedside. The levels of consensus are defined as follows:
Level 1A: Consensus based on high-level evidence (rigorous meta-analyses/randomised controlled trials (RCTs)), with the expert panel in unanimous agreement
Level 1B: Consensus based on high-level evidence (rigorous meta-analyses/RCT), with minor controversies among the experts
Level 2A: Consensus based on low-level evidence, with the expert panel in unanimous agreement
Level 2B: Consensus based on low-level evidence, with minor controversies among the experts
Level 3: Major controversies among the expert panel
Consensus 1: Precision medicine is a systematic approach
Consensus level: 1B
‘What is precision medicine?’ is the first question all clinicians and researchers have to answer. Multiple international organisations and top-notch academic publications have attempted to define the real-world significance of ‘Precision Medicine’. The US National Cancer Institute suggests that precision medicine is a phrase that is often used to describe how genetic information about a person’s disease is being used for diagnosis and treatment. The New England Journal of Medicine defines precision medicine as tailoring personalised treatment to individuals with similar clinical symptoms on the basis of differences in genetics, biomarkers and phenotypic or psychosocial characteristics, while minimising unnecessary side effects.1
An outstanding example that demonstrates the practical value of precision medicine is the development of the anaplastic lymphoma kinase (ALK) inhibitor crizotinib. Soda et al 2 discovered an EML4-ALK fusion gene in non-small cell lung cancer (NSCLC) in 2007. In 2008, results from a phase I study suggested that advanced ALK-positive NSCLCs might be sensitive to crizotinib.3 A phase I expansion cohort involving 149 patients with advanced ALK-positive NSCLC demonstrated that the objective response rate (ORR) was 60.8% and crizotinib could be well tolerated.4 A phase II clinical trial (PROFILE 1005) showed that the median progression-free survival (PFS) was 8.5 months and the ORR was 53%, providing further evidence for the effectiveness of crizotinib in this subpopulation.5 The clinical efficacy of crizotinib in first-line and second-line treatments for advanced ALK-positive NSCLC was later confirmed in multiple phase III RCTs.6–8 To date, US Food and Drug Administration (FDA), China FDA and the European Medicines Agency have all approved crizotinib for the treatment of advanced ALK-positive NSCLC. Based on these lines of evidence, the expert panel reached a consensus that precision medicine is a systematic approach, consisting of four contiguous steps: (1) finding actionable genetic abnormalities; (2) finding agents that specifically target the genetic abnormalities; (3) validating the efficacy and safety of the targeted agents in clinical trials; (4) further confirming the repeatable activity of the targeted agents in clinical practice. All four steps are mandatory.
Consensus 2: Three sources of genetic material can be used in liquid biopsy: circulating tumour cells (CTCs), cell-free DNA (cfDNA) and exosomes.
Consensus level: 2A
Liquid biopsy plays an important role in discovering actionable genetic abnormalities, the initial step in precision medicine. Currently, liquid biopsy can be applied to three types of materials: CTCs, cfDNA and exosomes. Masuda et al 9 summarised a comprehensive landscape of circulating biomarkers in tumour metastasis, including CTCs without metastatic potential, CTCs with metastatic potential, CTCs demonstrating an epithelial–mesenchymal transition, circulating tumour stem cells, CTC clusters, cfDNA and exosomes. Each biomarker has its own value in clinical applications. Alix-Panabieres et al 10 reported the potential utility of CTCs in scientific research and clinical translation. CTCs could be enriched from blood samples of patients with solid tumours and subsequently expanded in special culture medium or in immunodeficient mice to establish CTC cell lines or xenografts, respectively. The quantity of viable CTCs might provide prognostic information. In vitro culture of CTCs and in-depth characterisation of established CTC lines might identify metastasis-initiator cells, which might be crucial for drug development. CTCs could be expanded in vitro for the evaluation of therapies and the exploration of drug-resistance mechanisms, with the potential to eradicate metastatic disease. Heitzer et al 11summarised the use of circulating tumour DNA (ctDNA) as a liquid biopsy, which is part of the total cfDNA, and may be released during apoptosis or necrosis, or by secretion from tumour cells. Due to its low abundance in plasma, ctDNA can be detected by highly sensitive tests such as amplification refractory mutation system (ARMS), droplet digital PCR (ddPCR), beads, emulsions, amplification and magnetics (BEAMing) and next-generation sequencing (NGS). The diagnostic, predictive and prognostic value of ctDNA have been demonstrated in clinical applications. Specifically, ctDNA has been used in early detection, monitoring minimal residual disease, assessing disease molecular heterogeneity, monitoring tumour dynamics, and identifying patients with a high risk of recurrence, etc. Snyder et al 12 further discovered that nucleosome positioning in plasma cfDNA could provide information on their tissues of origin, which might contribute to distinguish among single primary, multiple primary and secondary tumours. Taverna et al 13 discussed the role of exosomes in NSCLC. Tumour-derived exosomes are nanosized vesicles containing proteins, mRNA, miRNAs and DNA. Exosomes interact with other cells in the tumour microenvironment to modulate tumour progression, angiogenic switch, metastasis and immune escape. Despite these results suggest the great promise of liquid biopsy in clinical applications, clinicians must be cautioned that these novel techniques are still in their infancy. More evidence is needed for clinical translation. However, as a novel technique, liquid biopsy is playing an increasingly important role in precision medicine and thus has great potential for further development.
Consensus 3: For single, known, clinically actionable genetic alteration, ARMS is recommended for ctDNA detection; for multi-parallel clinically actionable genetic alternations, NGS is recommended for ctDNA detection
Consensus level: 2A
At the summit, a survey of the preferred materials assayed in liquid biopsy demonstrated that 95% of participating experts chose to test for blood cfDNA or blood ctDNA, 49% tested for CTCs and 5% tested for exosomes, suggesting that cfDNA/ctDNA is the most commonly targeted material in liquid biopsies. Testing methods for blood ctDNA include ARMS, NGS and ddPCR. Whether these tests are ready to use in clinical practice is still undetermined. For NGS, National Comprehensive Cancer Network guidelines recommend that epidermal growth factor receptor (EGFR)/ALK testing should be conducted as part of multiplex sequencing/NGS. They strongly recommend methodologies with the capability of detecting multiple molecular alterations simultaneously; this may hold great promise in clinical testing.14 American Society of Clinical Oncology, European Society for Medical Oncology and multiple other guidelines also emphasise the advantages of NGS in multiplex parallel sequencing.
Comparing currently available ctDNA testing methods, each has its merits and drawbacks. As an approved ctDNA detection method, ARMS is commonly used in clinical practice, and it exhibits superiority in EGFR testing. However, ARMS is limited to detecting known molecular alterations, with detection rate of about 70%. Although NGS has advantages in detecting multiplex parallel and unknown molecular alterations, technical and cost-effectiveness issues together with a lack of regulatory guidelines have hindered the development of NGS-based testing in clinic. Consequently, there is still a gap to be bridged for its clinical translation. Both ddPCR and SuperARMS are highly sensitive tests developed to detect known mutations in recent years. Although it could be used in the absolute quantification of specific mutations, the clinical utility of ddPCR-based testing has been constrained by the underdevelopment of testing kits and a lack of regulatory guidelines. Notably, 91% of the experts at the summit meeting suggested that NGS could not, at present, replace single-gene-targeted testing of EGFR, ALK or ROS1 (figure 1).
Figure 1.
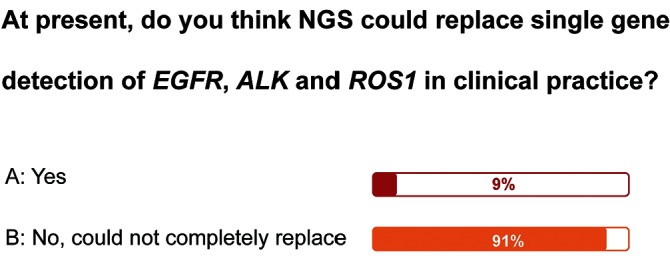
Survey on whether NGS could replace the role of single gene detection in clinical practice. NGS, next-generation sequencing.
Therefore, the expert panel agreed that for single, known, clinically actionable genetic alteration, ARMS is the recommended test for ctDNA detection; for multiparallel clinically actionable genetic alterations, NGS is the recommended test for ctDNA detection.
Consensus 4: The use of NGS for ctDNA detection is recommended to discover novel molecular alterations, to monitor response and prognosis, and to identify resistance mechanisms to targeted agents
Consensus level: 2A
With the discovery of EGFR activating mutations and EGFR-tyrosine kinase inhibitors (TKIs), the median overall survival (OS) of advanced EGFR-positive NSCLC has been extended to approximately 36 months with EGFR-TKIs . Among patients with EGFR-negative lung cancer in China, several tens of genetic abnormalities have been identified by NGS method. Despite a lack of consensus regarding whether these genetic alterations are targetable, a point that deserves further evaluation, it is clear that NGS has demonstrated excellent performance in discovering novel genetic abnormalities. Dynamic monitoring of blood EGFR mutations in the FASTACT-2 study showed that EGFR status at cycle 3 might be a treatment-response predictor in patients with EGFR-positive NSCLC receiving first-line EGFR-TKI treatment. Patients who were EGFR mutation(-) at cycle 3 demonstrated significantly longer median PFS and OS as compared with those who were EGFR mutation(+) (median PFS: 12.0 months vs 7.2 months, p<0.0001; median OS: 31.9 months vs 18.2 months, p=0.0066).15 The use of NGS in monitoring resistance to targeted therapies has also been widely reported. Thress et al 16 reported data from a phase I study of AZD9291, demonstrating that 40% (6/15) of baseline T790M-positive patients developed a tertiary C797S mutation as they acquired resistance to AZD9291. Oxnard et al 17 identified multiple resistance mechanisms to AZD9291 with the combined use of NGS and ddPCR in 40 baseline T790M-positive patients enrolled in the AURA (AZD9291 First Time In Patients Ascending Dose Study) study. The study demonstrated that all C797S mutations developed in patients with detectable T790M during disease progression, and only in those with exon 19 deletions. Additionally, HER2 amplification, BRAFV600E and other bypass resistance mechanisms were found in cases with T790M loss during disease progression. Shaw et al 18 reported a case that successfully overcame the cycle of resistances to ALK inhibition. The patient developed an ALK C1156Y mutation after progression from first-line crizotinib treatment, which is sensitive to lorlatinib. As a result, lorlatinib was administrated, although the patient eventually relapsed. NGS genetic testing performed on the resistant biopsy identified a secondary L1198F mutation, in addition to C1156Y. L1198F probably offset the increased kinase activity due to C1156Y, leading to crizotinib resensitisation. The patient restarted crizotinib and had a clinically significant radiological response that lasted almost 6 months. A survey of at what circumstances NGS-based genetic testing can be used indicated that 93% of experts favoured its use in patients who develop acquired resistance to targeted agents, and 80% of experts preferred to use it inpatients failing to respond to current treatments, without available effective therapies(figure 2).
Figure 2.
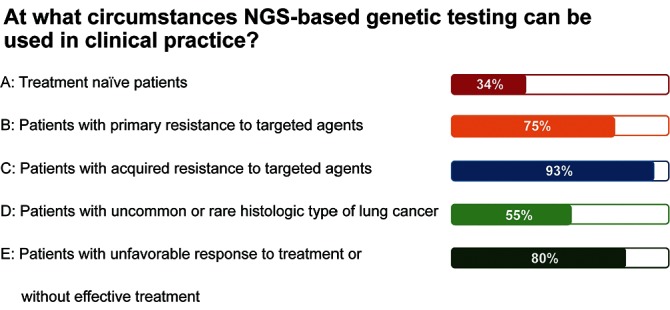
Survey on when NGS-based genetic testing can be used in clinical practice. NGS, next-generation sequencing.
Therefore, the expert panel recommended that NGS-based ctDNA detection could be used to discover novel molecular alterations, to monitor response and prognosis, and to identify resistance mechanisms to targeted agents. However, it must be cautioned that these potential applications of NGS require further extensive validation. The above-mentioned results from case reports and phase I clinical studies with small cohort sizes cannot be directly translated into clinical practice.
Consensus 5: Although liquid biopsy, including cfDNA or CTCs, might be used in early diagnosis and in monitoring NSCLC recurrence, these applications should be limited to scientific research at present
Consensus level: 2A
As the concentration of ctDNA in plasma is <5% in most patients with lung cancer, the testing sensitivity of liquid biopsy is a key factor for consideration. In recent years, technical advances in improving sensitivity have made early-stage diagnosis and monitoring of recurrence possible. A comparison among current available high-sensitivity ctDNA testing methods has also been made.19 20 BEAMing, ddPCR and tagged-amplicon deep sequencing could detect ctDNA at a concentration of 0.01% or lower. This number is 0.1% for ARMS, 0.02% for NGS and >10% for Sanger sequencing.19 20
Bayarri-Lara et al 21 reported the results of a phase I–IIIA study evaluating the prognostic value of CTCs in patients undergoing radical resection for NSCLC. The CTC detection rate dropped significantly, from 51.8% at baseline to 32.1% 1 month after surgery, and the corresponding number of CTCs dropped from an average of 3.16 to 0.66. The presence of CTCs after surgery was significantly associated with early recurrence (p=0.018) and shorter disease-free survival (DFS) (p=0.008). A multivariate analysis demonstrated that CTC presence after surgery was an independent prognostic factor for DFS (HR: 5.750, p=0.010). Based on the above-mentioned preliminary evidence, the expert panel suggested that liquid biopsy including cfDNA or CTCs might be used in the early diagnosis and recurrence monitoring of NSCLC, but these applications should be limited to scientific research at present.
Consensus 6: When applying NGS-based liquid biopsy in clinical practice, a trade-off between patient benefits, ethical requirements and scientific discovery has to be weighed; clinical benefit to patients should be the foremost essence
Consensus level: 2A
NGS technology has presented us with many novel scientific discoveries, but controversies remain over how to objectively and comprehensively evaluate the actual benefits that NGS can offer. In 2016, the biotechnology company Grail announced the launch of a project that aims to enable early cancer detection in asymptomatic individuals through a blood screening, and the company planned to promote the screening in clinical settings within 3 years. A related comment published in Lancet Oncology entitled ‘Liquid cancer biopsy: The future of cancer detection?’ raised many questions and concerns regarding both technical and ethical aspects.22 Technical issues including increased detection sensitivity can lead to an increased risk of false positives, and no universal biomarker for all cancer types has been identified. Ethical questions, such as addressing the risks of overtreatment and the trade-offs between early detection and psychological distress, have also been proposed.
Along with the opportunities offered by new techniques come novel challenges. NGS faces challenges in China, despite its rapid growth. At the summit meeting, 87% of experts suggested that the main obstacles hindering NGS from large-scale clinical implementation are high cost, lack of market standardisation and guarantees of test quality. As cost-efficiency is a key factor in clinical practice, the expert panel agreed that when applying NGS-based liquid biopsy in clinical practice, a balance among patient benefit, ethical requirements and scientific discovery is warranted, and bringing benefit to patients is of the essence.
Footnotes
Contributors: All the authors designed and wrote the manuscript.
Funding: Supported by the National Key Research and Development Program of China (Grant No 2016YFC1303800) , Guangdong Provincial KeyLaboratory of Lung Cancer Translational Medicine (Grant No. 2012A061400006) and Special Fund for Researchin the Public Interest from National Health and Family Planning Commission ofPRC (Grant No.201402031)
Competing interests: S-KC is from Burning Rock Biotech. YS is from Geneseeq Technology Inc. WH is from Geno Biotech. G-SZ is from Amoy Diagnostics. LX is from 3D Medicines.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1. Jameson JL, Longo DL. Precision medicine-personalized, problematic, and promising. N Engl J Med 2015;372:2229–34. 10.1056/NEJMsb1503104 [DOI] [PubMed] [Google Scholar]
- 2. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 3. Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. J Clin Oncol 2009;27:3509. [Google Scholar]
- 4. Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011–9. 10.1016/S1470-2045(12)70344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim D-W, Ahn M-J, Shi Y, et al. Results of a global phase II study with crizotinib in advanced ALK-positive non-small cell lung Cancer [abstract]. J Clin Oncol 2012;30(Suppl 15; Abstr 7533). [Google Scholar]
- 6. Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–94. 10.1056/NEJMoa1214886 [DOI] [PubMed] [Google Scholar]
- 7. Solomon BJ, Mok T, Kim DW, et al. ; PROFILE 1014 Investigators. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167–77. 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 8. Lu S, Mok T, Lu Y, et al. Phase 3 study of first-line crizotinib vs pemetrexed- Cisplatin/Carboplatin (PCC) in east asian patients (pts) with ALK+ advanced non-squamous nonsmall cell lung Cancer (NSCLC) [abstract]. J Clin Oncol 2016;34(Suppl 15; abstr 9058). [Google Scholar]
- 9. Masuda T, Hayashi N, Iguchi T, et al. Clinical and biological significance of circulating tumor cells in cancer. Mol Oncol 2016;10:408–17. 10.1016/j.molonc.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alix-Panabières C, Bartkowiak K, Pantel K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol Oncol 2016;10:443–9. 10.1016/j.molonc.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61:112–23. 10.1373/clinchem.2014.222679 [DOI] [PubMed] [Google Scholar]
- 12. Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget 2016;7:28748–60. 10.18632/oncotarget.7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw 2014;12:1738–61. [DOI] [PubMed] [Google Scholar]
- 15. Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with First-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015;21:3196–203. 10.1158/1078-0432.CCR-14-2594 [DOI] [PubMed] [Google Scholar]
- 16. Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560–2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oxnard GR, Thress K, Paweletz C, et al. Mechanisms of acquired resistance to AZD9291 in EGFR T790M positive lung cancer [abstract]. J Thorac Oncol 2015;10(Suppl 2):S207. [Google Scholar]
- 18. Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med 2016;374:54–61. 10.1056/NEJMoa1508887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bayarri-Lara C, Ortega FG, Cueto Ladrón de Guevara A, et al. Circulating tumor cells identify early recurrence in patients with Non-Small cell lung Cancer undergoing radical resection. PLoS One 2016;11:e0148659 10.1371/journal.pone.0148659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lancet O. Liquid cancer biopsy: the future of cancer detection? Lancet Oncol 2016;17:123. 10.1016/S1470-2045(16)00016-4 [DOI] [PubMed] [Google Scholar]


